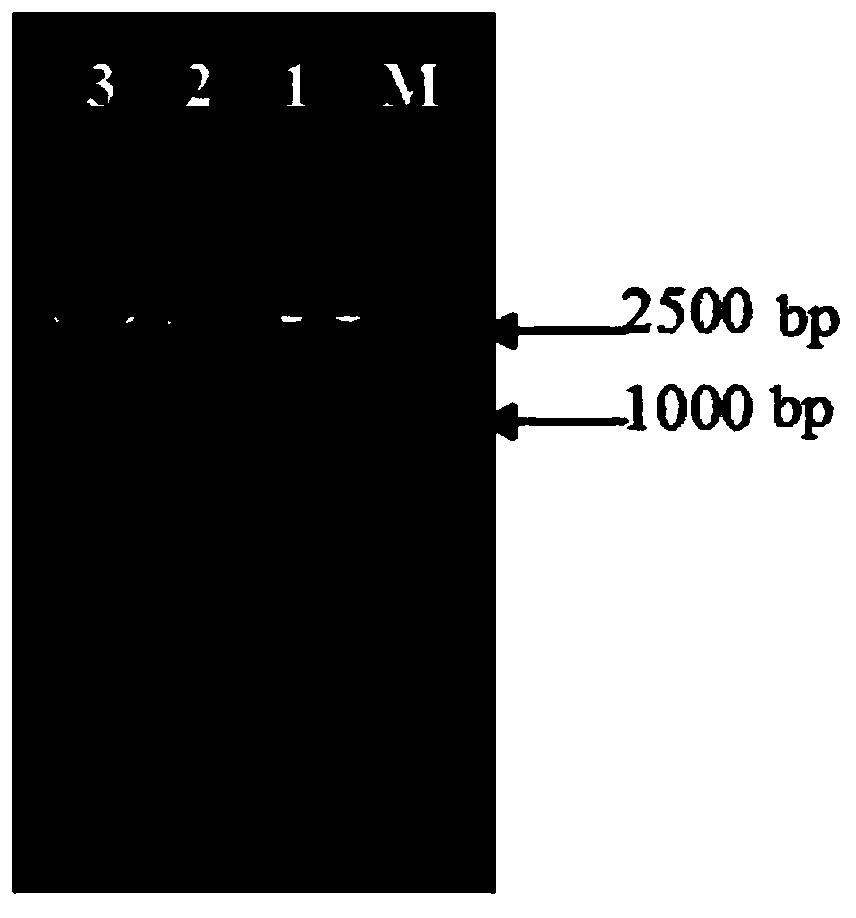
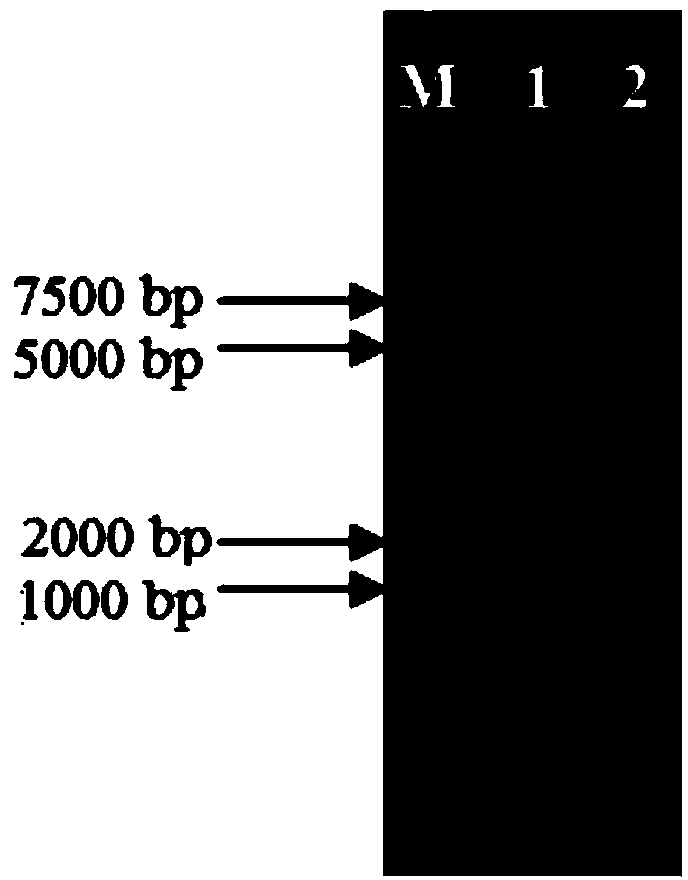
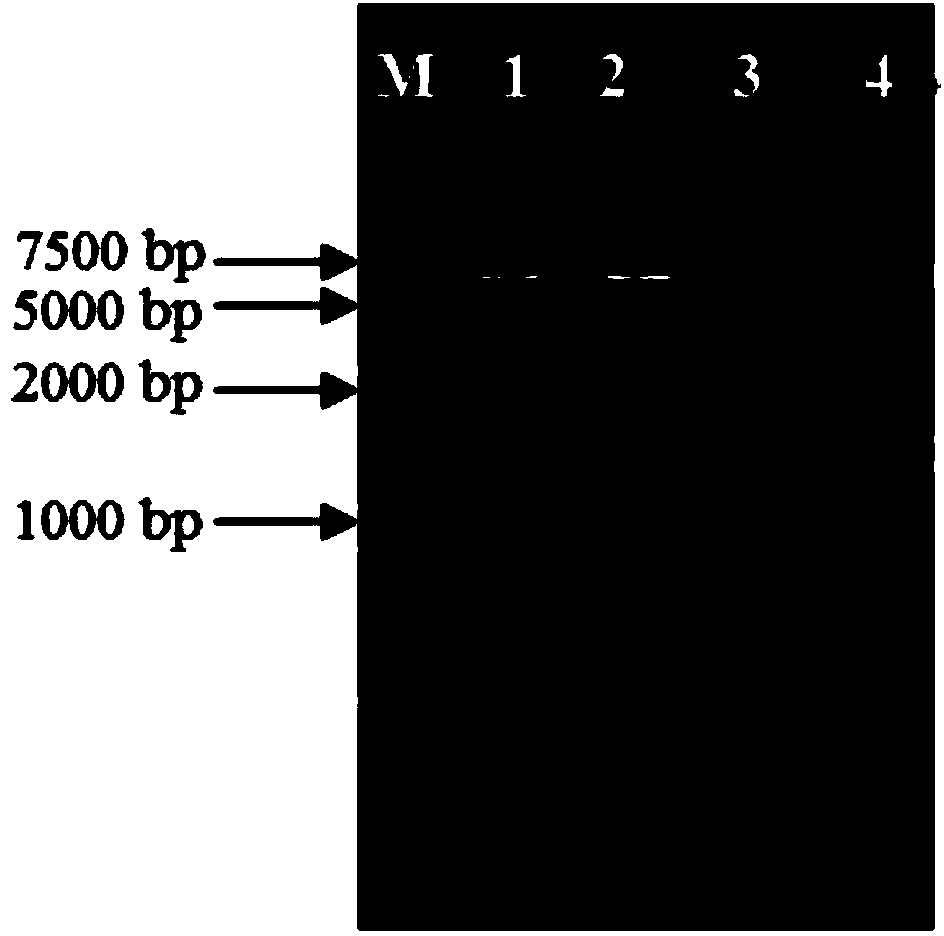
Suicide plasmid pYTRLRRT with prcR gene knockout effect and construction method thereof
A technique of suicide plasmid and construction method, which is applied in the field of suicide plasmid and its construction, can solve the problems such as failure to successfully knock out the PRCR gene of Lactobacillus paracasei, achieve the goal of increasing homologous recombination rate, avoiding wrong gene replacement, and easy screening Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0025] Embodiment 1: The suicide plasmid pYTRLRRT for knocking out the prcR gene in this embodiment is composed of prcRL, the left fragment of the prcR gene, prcRR, the right fragment of the prcR gene, a tetracycline resistance gene, and plasmid pUC18; the nucleic acid sequence of the prcRL fragment is as shown in SEQ ID NO: 1; the nucleic acid sequence of the fragment prcRR is shown in SEQ ID NO:2.
specific Embodiment approach 2
[0026] Specific embodiment two: The suicide plasmid pYTRLRRT knocking out the prcR gene in this embodiment is constructed according to the following steps:
[0027] 1. Use L.paracasei HD1.7 genomic DNA as a template to PCR amplify the left fragment prcRL of the prcR gene, the upstream primer of the amplified fragment prcRL is prcRL-up, and the nucleotide sequence of prcRL-up is 5'-CCG GAGCTC CCTACTCAGCATTCAGAGGTCAACT-3', the downstream primer of the amplified fragment prcRL is prcRL-down, and the nucleotide sequence of prcRL-down is 5'-GTC GGTACC ATCTTCAGCATCGTTTGGTGGTTGG-3', the reaction system of the PCR amplified fragment prcRL is 50 μL by 10 μL containing Mg 2+ 5×PrimeSTAR Buffer, 4μL of dNTP Mixture with a concentration of 2.5mmol / L, 10μL of L.paracasei HD1.7 genomic DNA with a concentration of 25ng / μL, 10μL of prcRL-up with a concentration of 1pmol / μL, and 10μL of a concentration of 1pmol / μL µL of PRCRL-down, 0.5 µL of PrimeSTAR HS DNA polymerase at a concentration of ...
specific Embodiment approach 3
[0038] Specific embodiment three: the difference between this embodiment and specific embodiment two is: in step two, the enzyme digestion reaction system of plasmid pUC18 is 20 μL consisting of 1 μL of Sac I with a concentration of 10 U / μL, 1 μL of Kpn I with a concentration of 10 U / μL, 2 μL 10×Buffer Tango, 7 μL plasmid pUC18 with a concentration of 80 ng / μL and 9 μL sterile ddH 2 O composition; digest at 37°C for 2 h at constant temperature, and use enzyme gel to recover the digested fragments. Other steps and parameters are the same as in the second embodiment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com