Method for improving rolling circle amplification efficiency with integration host factor
A technology of integrating host factors and rolling circle amplification, which is applied in the field of improving the efficiency of rolling circle amplification by integrating host factors, can solve the problems of slow response speed and low sensitivity of RCA technology, and achieve the effect of improving efficiency and enhancing sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] 1. Preparation of IHF-β
[0021] ① Insert the IHF-β gene derived from Escherichia coli into the pET-22b expression vector, and through ligation transformation and verification by colony PCR, enzyme digestion and sequencing, the recombinant expression vector pET-22b-IHF-β can be obtained;
[0022] ②Transform the constructed recombinant vector pET-22b-IHF-β into Escherichia coli BL21, take the positive strain containing the recombinant plasmid and add it to LB liquid medium for overnight cultivation, and then inoculate the bacterial liquid into 50ml of LB culture medium at a ratio of 1%. , placed in a shaker at 37°C until the OD value is 0.6-0.8; add IPTG, put the bacterial solution in a shaker at 37°C to induce expression for 3 hours, and the shaker speed is 160rpm;
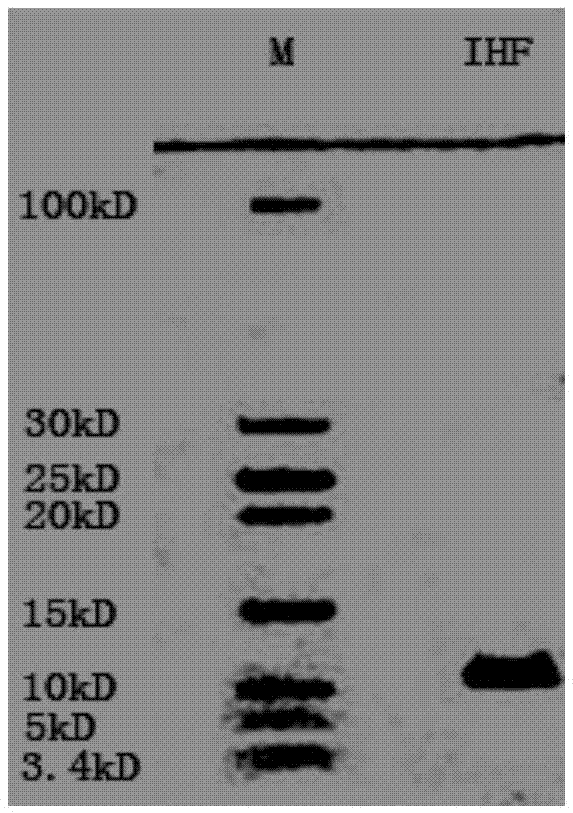
[0023] ③Centrifuge the bacterial liquid after induced expression to obtain the bacterial cells, lyse and release the protein. SDS-PAGE electrophoresis to detect protein expression, electrophoresis results ...
Embodiment 2
[0028] Example 2: Activity detection of IHF-β
[0029] Configure a 10 μl reaction system according to Table 1 below:
[0030] Table 1
[0031]
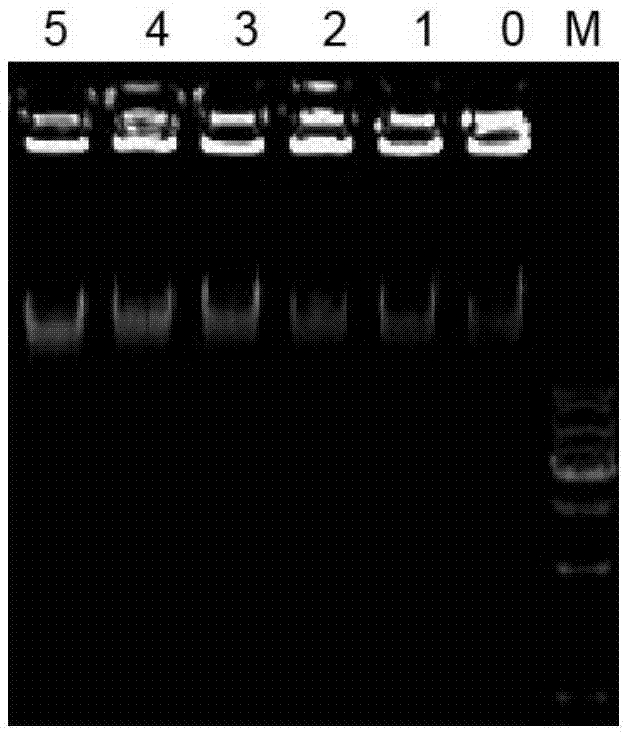
[0032] In the reaction solution, different concentrations of the IHF-β protein obtained in Example 1 were added and allowed to stand at room temperature for 10 minutes. Afterwards, 5 μl of the reaction solution was taken for 1% agarose gel electrophoresis (105V, 25min), then stained in 0.5μg / mL ethidium bromide staining solution for 15min, and observed with a UV transilluminator. For specific experimental results, see figure 2 .
[0033] figure 2 Each lane indicates: M is 1kbDNALadderMarker; 1, 2, 3, 4, 5, 6, 7, 8 indicate that 0μl, 0.5μl, 1μl, 2μl, 4μl, 6μl, 8μl and 9μl of IHF-β protein were added to the reaction solution .
[0034] Depend on figure 2 It can be seen that as the amount of IHF-β protein added increases, the DNA bands gradually move up. It shows that the purified protein IHF-β is active and can combine with...
Embodiment 3
[0035] Example 3: Effect of IHF-β on the efficiency of rolling circle amplification (using the recombinant plasmid pUC19-hupB as a template)
[0036] 1. Construction of recombinant plasmid pUC19-hupB: Insert the hupB gene from Escherichia coli into the pUC19 plasmid, transform by ligation and use colony PCR, enzyme digestion identification and sequencing verification to obtain the recombinant plasmid pUC19-hupB;
[0037] 2. Denaturation: Using the recombinant plasmid pUC19-hupB as a template, add 1 ng of pUC19-hupB plasmid DNA to 5 μl of sample buffer, mix well, heat at 95°C for 3 minutes, and then cool to room temperature or 4°C to obtain a denatured sample solution;
[0038]3. Incubation: Add 0.2 μl Phi29 DNA polymerase to 5 μl reaction buffer, mix well, and place on ice to obtain a premixed enzyme solution; add 5 μl premixed enzyme solution to the denatured sample solution, mix well to obtain an RCA reaction solution; add IHF-β protein 0ng, 2.04ng, 5.1ng, 15.3ng, 25.5ng and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com