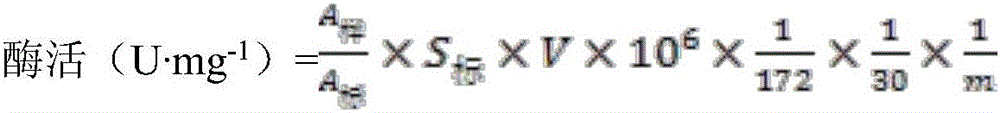Activity remodeling method for changing lipase catalytic activity
A technology of catalytic activity and lipase, applied in the field of enzyme engineering, can solve the problems of lipase losing ester synthesis ability, lipase not showing esterification activity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1: Extraction of Membrane Environment Complex WTS
[0071] TritonX-100, CHAPS, Tween80, deoxycholic acid and other surfactants were used to extract membrane environment complexes and membrane-bound proteins from the cultured Rhizopus sinensis, and then passed through the Bio-BeadsSM-2 adsorbent of American bio-rad company. Adsorb the surfactant in the extract to precipitate the membrane-bound protein, and the supernatant obtained after high-speed centrifugation is the membrane environment complex. The specific method is as follows:
[0072] Rhizopus sinensis CCTCCM201021 was cultured on potato-dextrose agar medium at 30°C for 3 days, and the spores were washed with sterile water. 200μl of spore suspension was inoculated into a shake flask (250mL Erlenmeyer flask) containing 20mL of fermentation medium, at 30°C, 150r min -1 Conditioned for 72h. The concentration of spore suspension is about 10 7 pcs ml -1 . After the bacterial cell culture, the mycelium was...
Embodiment 2
[0074] Example 2: Active Remodeling Method
[0075] Perform in vitro refolding by any of the following methods:
[0076] Protocol A: Adding Membrane Environment Complexes During Refolding
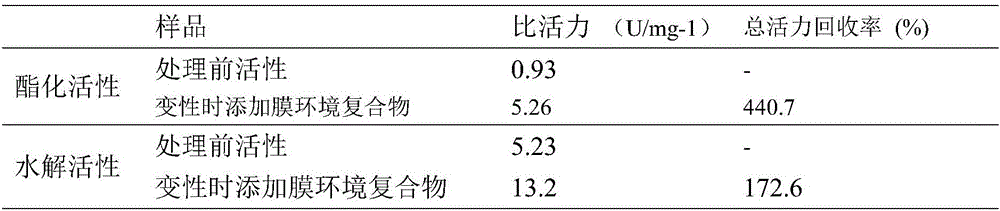
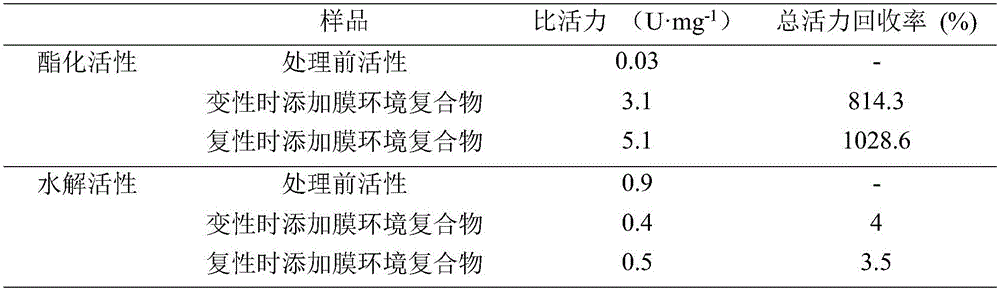
[0077] (1) Protein denaturation: take lipase protein solution or suspension, slowly add 0.5g of ground urea powder per milliliter of solution under low temperature shaking, stir magnetically at 4°C for 2-4h, take out 100μL every half hour for hydrolysis activity Determination, complete loss of activity means that the tertiary structure is completely opened to form a peptide chain, and the protein is completely denatured. Denatured protein 12000r·min -1 Centrifuge for 10 minutes, take the supernatant for concentration determination, and ensure that the protein concentration is not higher than 2.5 mg·mL -1 , to ensure that the protein can be completely refolded. (2) Protein renaturation: Add excess prepared membrane environment complex at a volume ratio of 1:1, stir magnetically for 5-10 ...
Embodiment 3
[0083] Example 3: Escherichia coli heterologous expression of Rhizopus chinensis mature peptide lipase MP 1 Restoration of vitality
[0084]Primers were designed according to the whole genome sequence of Rhizopus sinorifica lipase, and the mature peptide lipase gene A06672 of Rhizopus sinensis was amplified by PCR (http: / / genome.jgi.doe.gov / Rhich1 / Rhich1.home. html) Connection expression vector 6xHis-MBP 3 -TEV-pET15 (constructed by inserting the MBP fusion protein gene fragment (J.Am.Chem.Soc.2012,134,375–385) with a 6xHis tag at the N-terminal of pQEH6MBP and containing a TEV protease cleavage site into pET15), Transformation of E.coliBL21trxB(DE3), IPTG induction to obtain fusion protein MBP (His)8 -MP 1 , HisTrap was purified and digested with TEV protease, and then purified by HisTrap and DEAE column to obtain pure enzyme MP 1 .
[0085] According to the scheme B and scheme A of Example 2, add the membrane environment complex during denaturation and add the membrane ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com