Protein structure ab initio prediction method based on bacterial oraging optimization algorithm
A technology of protein structure and prediction method, which is applied in the field of ab initio prediction of protein structure based on bacterial population optimization algorithm, can solve the problems of weak conformational space sampling ability and insufficient conformation update accuracy, and achieve the effect of reducing calculation cost and improving sampling ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Example Embodiment
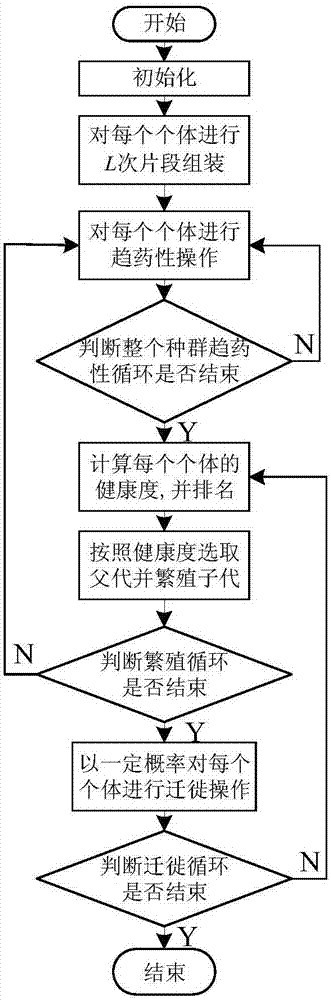
[0036] The present invention will be further described below in conjunction with the drawings.
[0037] Reference figure 1 , A protein structure ab initio prediction method based on flora optimization algorithm, including the following steps:
[0038] 1) Select Rosetta Score3 as the protein force field model, that is, the energy function J(C), C represents the protein conformation;
[0039] 2) Given the input sequence information, obtain the target protein fragment library through the free web server Robetta Server;
[0040] 3) Parameter initialization: the population size of the bacteria NP, NP is an even number, the step size of the i-th bacterium for chemotaxis behavior i ,i∈{1,...,NP}, the number of cycles of bacterial chemotaxis behavior N c , The maximum number of advances in the chemotaxis cycle N s , The number of bacterial reproduction behaviors N re , The number of times the bacteria perform migration behavior N ed , The migration probability P ed ;
[0041] 4) Use dihedral a...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap