Application of Circulating Nucleic Acids as Biomarkers for Breast Cancer
A breast cancer and nucleic acid technology, applied in the application field of circulating nucleic acid as a breast cancer biomarker, can solve the problems of lack of research, poor repeatability of results, small sample size, etc., and achieve the effect of predicting survival effect and valuable application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 196
[0141] Example 1 Determination of 96 candidate miRNAs
[0142] In order to find circulating miRNAs that can be effective as prognostic markers for breast cancer, this study screened multiple breast cancer-related miRNAs in reported databases or studies, while their circulating forms have not been studied. Using this as a candidate gene, expression profiling was performed in the multi-sample training set. Sources and selection criteria of candidate genes: 1) miRNAs with differential (>2-fold) expression indicated by microarray or functional experiments in breast cancer and adjacent tissues; 2) prognostic indicators such as breast cancer typing and lymph node status Relevant miRNAs; 3) miRNAs that have been proved to be involved in the occurrence and development of breast cancer in cell or animal models; 4) miRNAs that may be effective have been detected in the serum of a small sample of breast cancer populations; 5) are widely present in serum and plasma. miRNAs. Through the ...
Embodiment 2
[0143] Example 2 Screening of internal reference and standardization of miRNA expression
[0144] Twenty-five breast cancer patients and 20 age-matched healthy controls were randomly selected for detection of 96 miRNA trace serum extraction-free multiplex qRT-PCR expression profiles, and the obtained miRNA Ct values were stabilized by NormFinder and geNorm software Sexual assessment.
[0145] NormFinder analysis showed that miR-132 was the most stable miRNA expressed in patients and healthy controls, while geNorm analysis suggested that miR-103a / miR-107 were suitable as internal controls. The stability evaluation of the pairwise combination of the above three miRNAs (miR-132, miR-103a, miR-107) showed that miR-103a / miR-132 had the strongest stability and was suitable as an internal reference.
[0146] After selecting the internal control, normalization was performed on the remaining miRNAs from breast cancer patients and healthy controls. like figure 1 As shown, different m...
Embodiment 3
[0147] Example 3 Acquisition of circulating miRNA expression profile in breast cancer patients
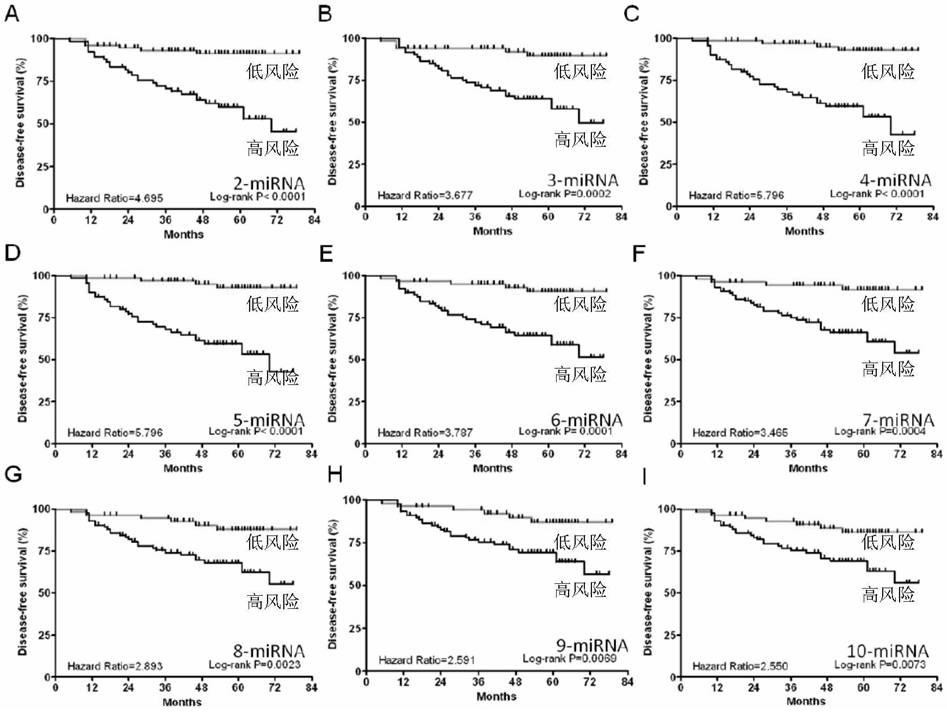
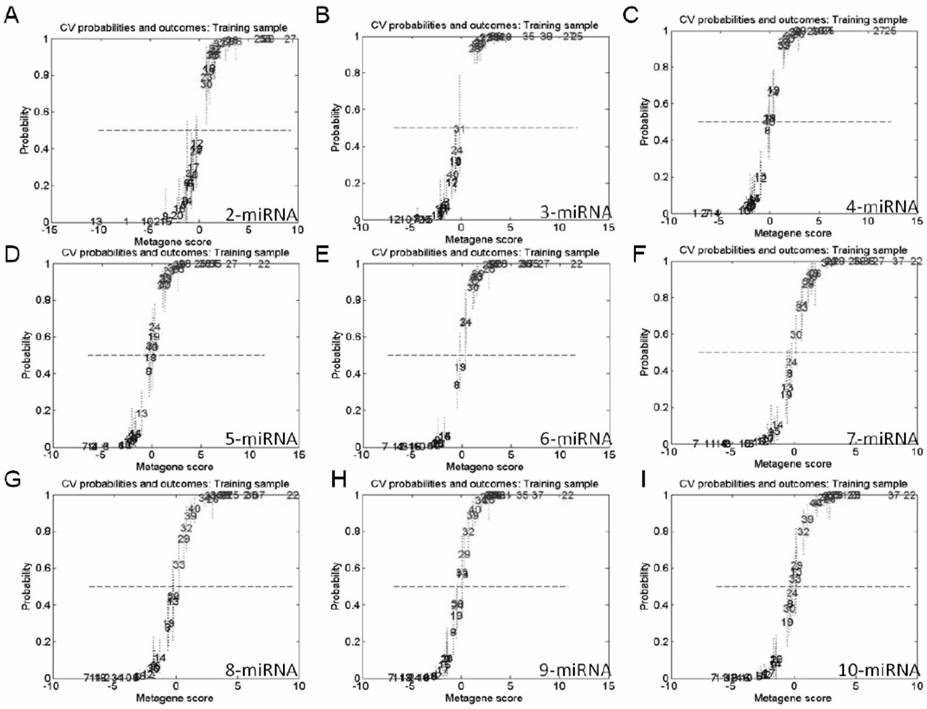
[0148] In order to obtain an effective combination of miRNAs for predicting the outcome of breast cancer, the original data of 96 miRNAs from 189 breast cancer patients were screened according to the following criteria: 1) The Ct value of circulating miRNAs was between 15 and 35; The Ct value of miRNA in the repeated serum experiments was STD<2; 2) the Ct value of miRNA was between the average Ct±3 of 189 breast cancer patients. Finally, the expression profiles of 82 serum miRNAs from 182 breast cancer patients meeting the requirements were obtained. The clinical information of 182 breast cancer patients is shown in Table 1, with a median age of 52 years. Among them, 55 breast cancer patients had adverse clinical outcomes, including recurrence in situ, distant metastasis and death.
[0149] Using miR-103a / miR-132 as an internal reference, the Ct values of the other 80 miRNAs wer...
PUM
| Property | Measurement | Unit |
|---|---|---|
| correlation coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com