Molecular breeding method for improving stigma exsertion rate of sterile line of three-line rice
A technology of three-line sterile lines and exposed stigmas, which is applied in the fields of botany equipment and methods, biochemical equipment and methods, and microbial measurement/inspection, etc., and can solve the problems of low yield of hybrid seed production, regulatory analysis, and incomplete research results In order to improve the efficiency and accuracy of improved breeding and speed up the breeding process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
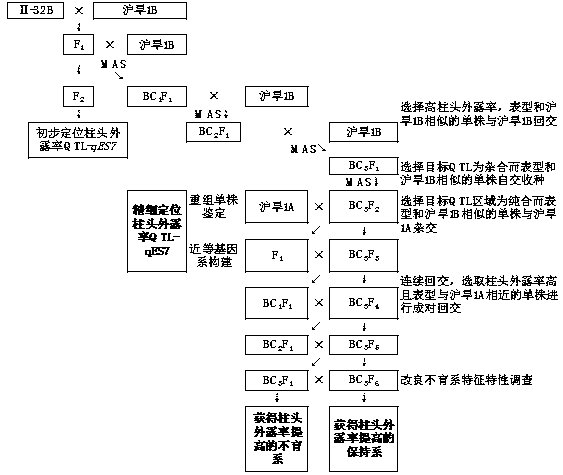
[0024] Examples, see figure 1 and 2 ,
[0025] 1. Fine mapping of rice head exposure rate gene qES7
[0026] (1) Population construction of Shanghai Han 1B / II-32B BC3F2
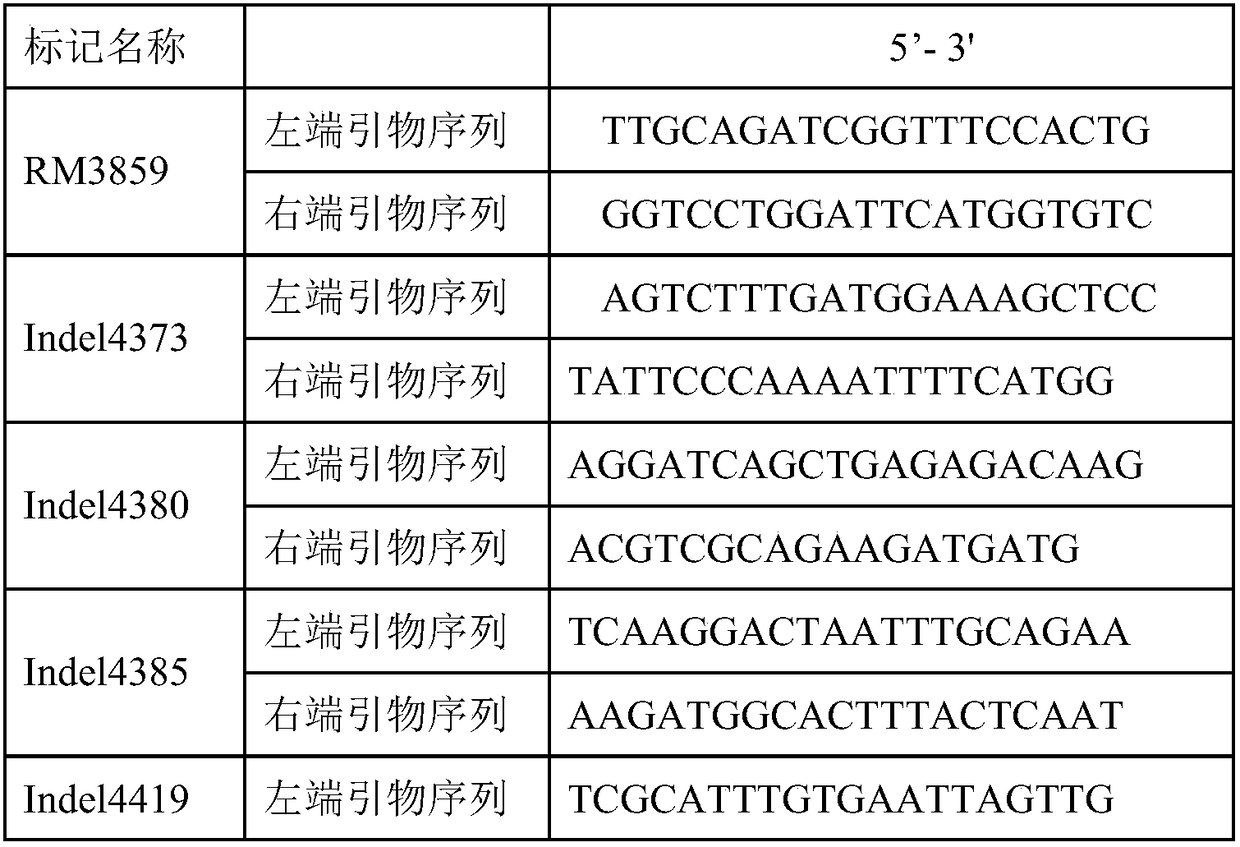
[0027] Yue Gaohong et al. (2009) used the F 2 The population conducted QTL analysis on the stigma exposure rate traits of rice, and initially located a QTL-qES7 that affects both the unilateral stigma exposure rate and the total stigma exposure rate. It is located between the chromosome 7 molecular markers RM3859 and RM5436, and the additive effect It comes from the parent II-32B with a high rate of stigma exposure. Therefore, taking Huhan 1B as the recurrent parent, a single plant containing the qES7 locus in the F2 population was selected for backcrossing with Huhan 1B to obtain BC1F1, and then molecular markers RM3859 and RM5436 were used to detect the backcross offspring, and the markers on both sides were selected to be hybrids. A single plant of the zygotic genotype was continued to be backcrossed ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com