Recombinant saccharomyces cerevisiae for expressing caveolin and application of recombinant saccharomyces cerevisiae
A technology for recombining Saccharomyces cerevisiae and caveolin, which is applied in the fields of genetic engineering and bioengineering, can solve problems such as no caveolin and ceramide's exact role is not completely clear, and achieve the effect of promoting synthesis and increasing production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
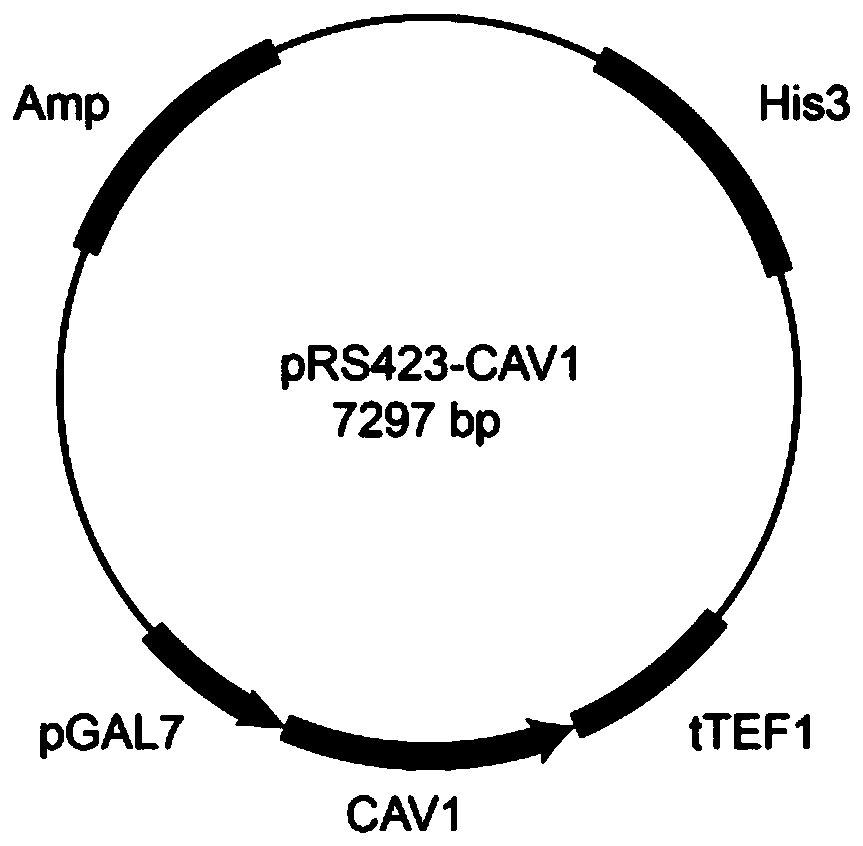
[0051] Example 1: CAV1 Gene Synthesis and Construction of the Caveolin CAV1 Shuttle Plasmid
[0052] The codon was optimized according to the human caveolin CAV1 gene in the NCBI database, and the gene was synthesized by Shanghai Sangon Company. The final nucleotide sequence is shown in SEQ ID NO.1.
[0053] Primer pairs designed to amplify the CAV1 sequence:
[0054] F1: TGA CTC GAG T TA AAT TTC CTT TTG CAA ATT AAT TCT AAC GTT AGA GAAAAT TTT ACC AA, SEQ ID NO.3 (the underlined part is the homology arm sequence, the same below),
[0055] R1: TCC CTC AAA A AT GTC TGG TGG TAA ATA CGT TGA TTC TGA AG, SEQ ID NO. 4.
[0056] Using the synthetic sequence SEQ ID NO.1 as a template, PCR amplification was carried out with primers F1 and R1, and Primer StarMasterMix (Takara Company) high-fidelity pfu enzyme was selected for pre-denaturation at 95°C for 3 minutes; 30 cycles of amplification , according to 95°C, 15s, 55°C, 5s, 72°C, 30s; extend at 72°C, 5min; purify the PCR produ...
Embodiment 2
[0066] Example 2: Construction of recombinant Saccharomyces cerevisiae strains containing caveolin CAV1
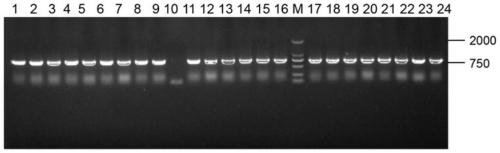
[0067] The correctly sequenced recombinant vector pRS423-CAV1 was transformed into the chassis cell ZLHB04-4 by lithium acetate chemical conversion method, and cultured on a YNB plate without histidine at 30°C for 3 days until a single colony grew; Bacteria were colonized in YNB medium and cultured at 220rpm for 24h; the cultured bacterial solution was verified by PCR, and the correct clone was picked to construct recombinant Saccharomyces cerevisiae ZLHB04-4 / pRS423-CAV1 (see figure 2 ).
[0068] Primers used in colony PCR:
[0069] F4: TTG AAT TTC AAT CAA GAA AGA CTT AAT ACA TGG AAC AAC, SEQ ID NO.9,
[0070] R4: GGG TAA TTT TTC CCC TTT ATT TTG TTC, SEQ ID NO. 10.
Embodiment 3
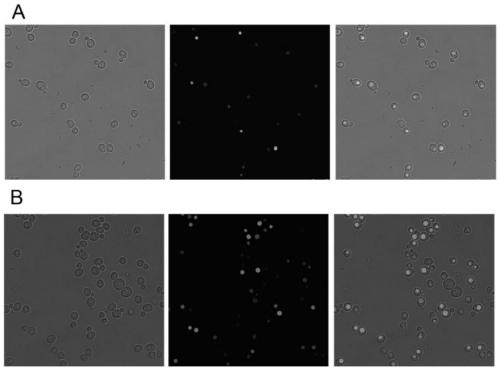
[0071] Example 3: Functional verification of transport of exogenous substances by recombinant Saccharomyces cerevisiae containing caveolin CAV1
[0072] The constructed recombinant Saccharomyces cerevisiae ZLHB04-4 / pRS423-CAV1 (the strain ZLHB04-4 / pRS423 transformed with pRS423 empty load was used as a control) was streaked on a YNB plate without histidine, and cultured at 30°C for 3 days. Pick a single colony and transfer it into 5mL of the corresponding YNB medium, cultivate for 24h to make the OD 600 3, the inoculum size of 1% by volume was transferred to 5 mL of YNB medium without histidine, and 10 mM 5.6-carboxyfluorescein was added after culturing for 16 hours (3.7632 g of 5.6-carboxylate fluorescein was dissolved In 1L of absolute ethanol), after culturing for 6 hours, collect 5mL of the bacterial solution and centrifuge to discard the supernatant, wash the cells with ice bath TBS buffer, and finally resuspend in 1mL of TBS buffer, and observe the cells with a laser con...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap