Method for determining small nucleic acid sequence set and application thereof
A nucleic acid sequence and small nucleic acid technology, applied in the field of bioinformatics, can solve the problem that the accuracy of results depends on the integrity of the genome, and achieve the effects of improving accuracy and sensitivity, comprehensive results, and reducing dependence on structural features
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0061] In this example, 200 small RNA sequences were randomly selected from a small RNA sequencing data, and some sequence files are shown in Table 1.
[0062] Table 1: small RNA sequences
[0063] name sequence t2_33221_25926 GGTTAAGTGACTAAGCGTACATGGTGGATGCCTTGGC t43_9407_8664 GATGTCTACTAGCCGTTGGG t115983_54_50 TATGGTTAAGTGACTAAGCGTAC t20_12754_11131 TGGACAAAGACTGACGCTCAGGT t24_11745_10234 GCCTTGACATGTCTGGAATCC … …
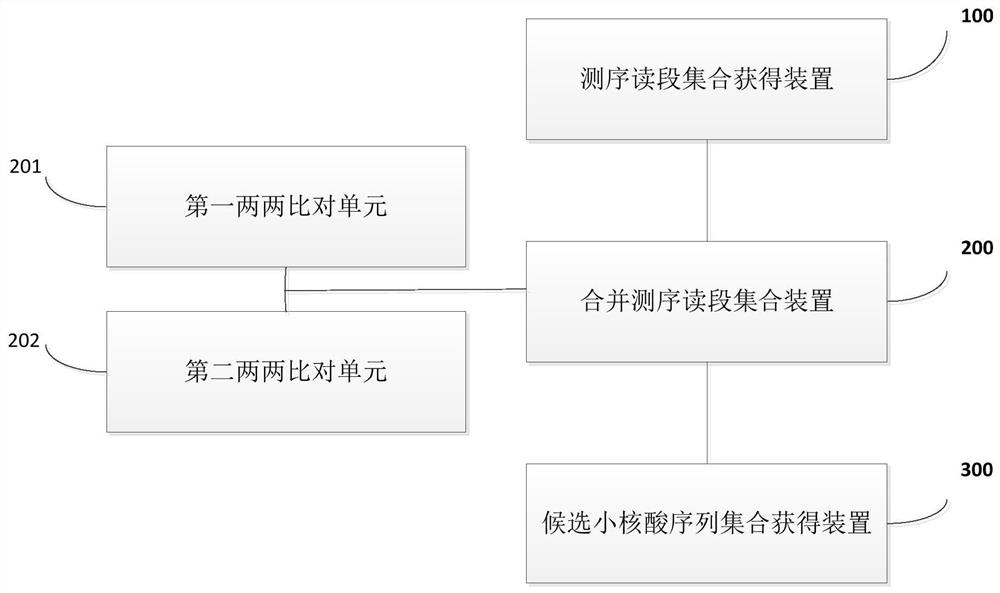
[0064] 1) Input a randomly selected small RNA sequencing data for processing, convert all characters into uppercase and convert non-ATCGN characters into N;
[0065] 2) Divide the file in 1) into files containing at most the set number of files (default 500, generally the value is too large or too small, and the running time will be longer). The file after segmentation is recorded as A 1 , A 2 ,...,A n , merged according to step 3 respectively;
[0066] 3) Suppose A i contains S 1 , S 2 ,…,S n bar sequenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com