Detection method based on Cas protein
A detection method and protein technology, applied in biochemical equipment and methods, microbial measurement/inspection, DNA/RNA fragments, etc., can solve problems such as differences in quantitative detection results, inability to detect Cas12a, lack of PAM sequences, etc., to reduce detection Cost, effect of reducing operation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
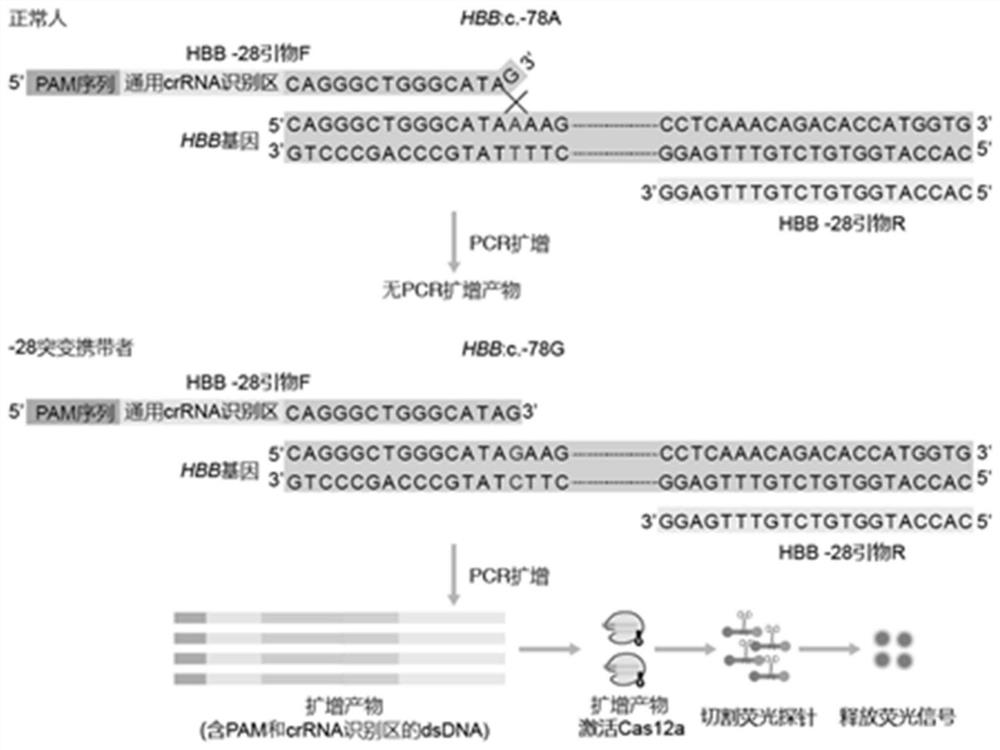
[0096] (1) According to the human HBB gene sequence, select three common mutation sites in the Chinese population: HBB:c.-78A>G(-28), HBB:c.126_129delCTTT(CD41-42) and HBB:c.316-197C >T(IVS-II-654), design and synthesize PCR amplification primers, as shown in Table 1.
[0097] Table 1 Primers, probes and crRNA sequence information
[0098]
[0099]
[0100] (2) Verify the primers and explore the annealing temperature.
[0101] ①Taking HBB-28 primers F and R as an example, select a negative control (water), HBB:c. A PCR amplification system.
[0102] The PCR amplification system is as follows:
[0103]
[0104] ②PCR reaction conditions are roughly as follows Figure 4 As shown, what has changed is that the annealing temperature is set to 63, 64, 65, and 66°C, and the annealing time is 30s, and the rest of the conditions are the same. Each sample was amplified under 4 different reaction conditions, and the amplified products were subjected to agarose gel electropho...
Embodiment 2
[0135] Comparative experimental methods - existing Cas12a detection methods and principle detection
[0136] Use the existing Cas12a detection method and principle to detect three mutation sites, such as HBB:c.-78A>G, HBB:c.126_129delCTTT and HBB:c.316-197C>T. First, it is necessary to search for PAM near the mutation site sequence and design a suitable crRNA. Using the crRNA design software (CRISPR RGEN Tools) to design, it was found that: ① There is no PAM sequence (TTTN) near the HBB:c.-78A>G mutation site, and crRNA cannot be designed ( Figure 8 a); ②HBB: There is a PAM sequence (TTTN) near the c.126_129delCTTT mutation site, but the designed crRNA does not cover the mutation site ( Figure 8 b); ③HBB: There is a PAM sequence (TTTN) near the c.316-197C>T mutation site and two crRNAs can be designed, but the mutation site is in the middle or 3' end of the crRNA recognition region, and the distance from the PAM sequence Far, the specificity of the two crRNAs to recogniz...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com