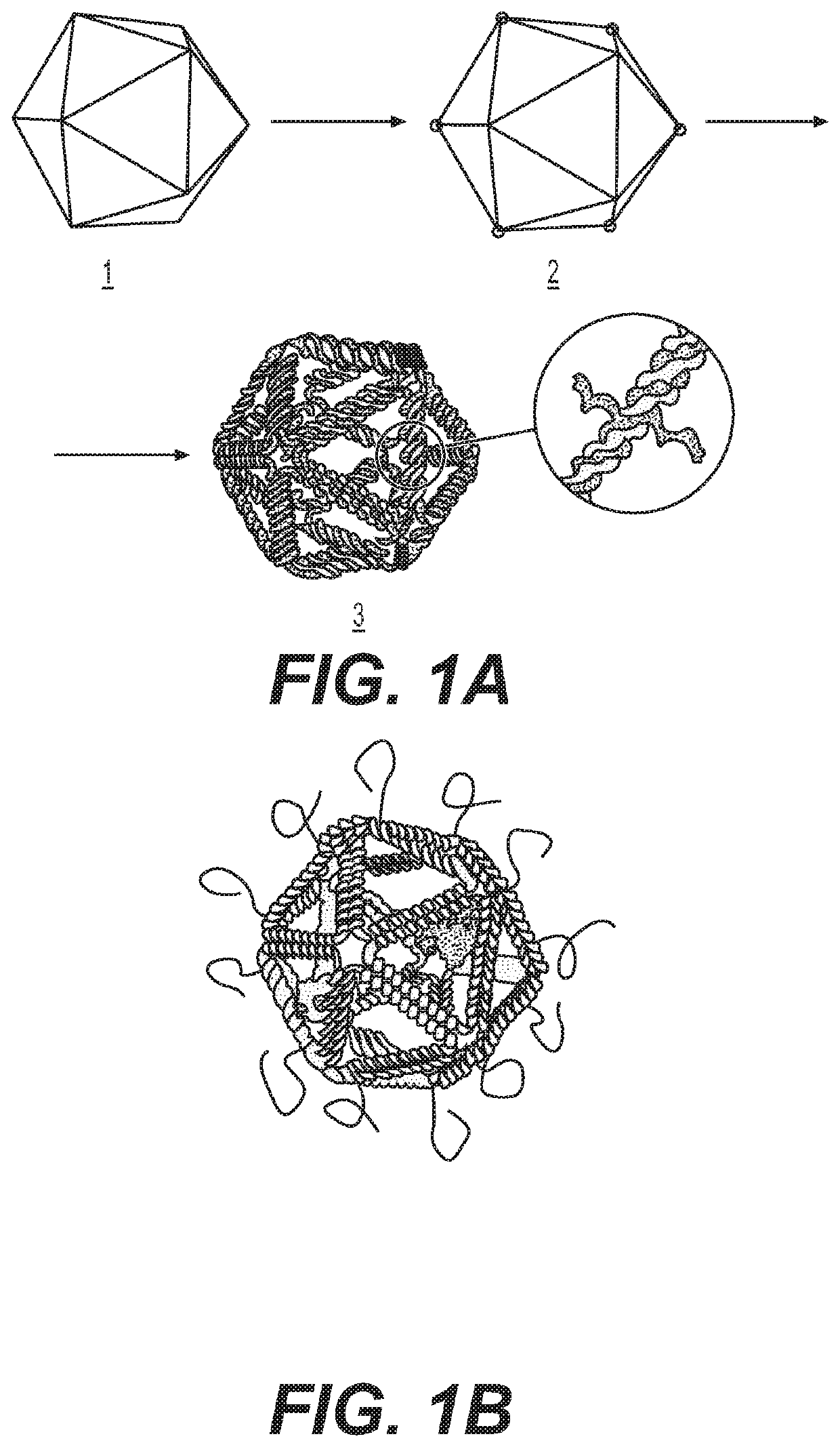
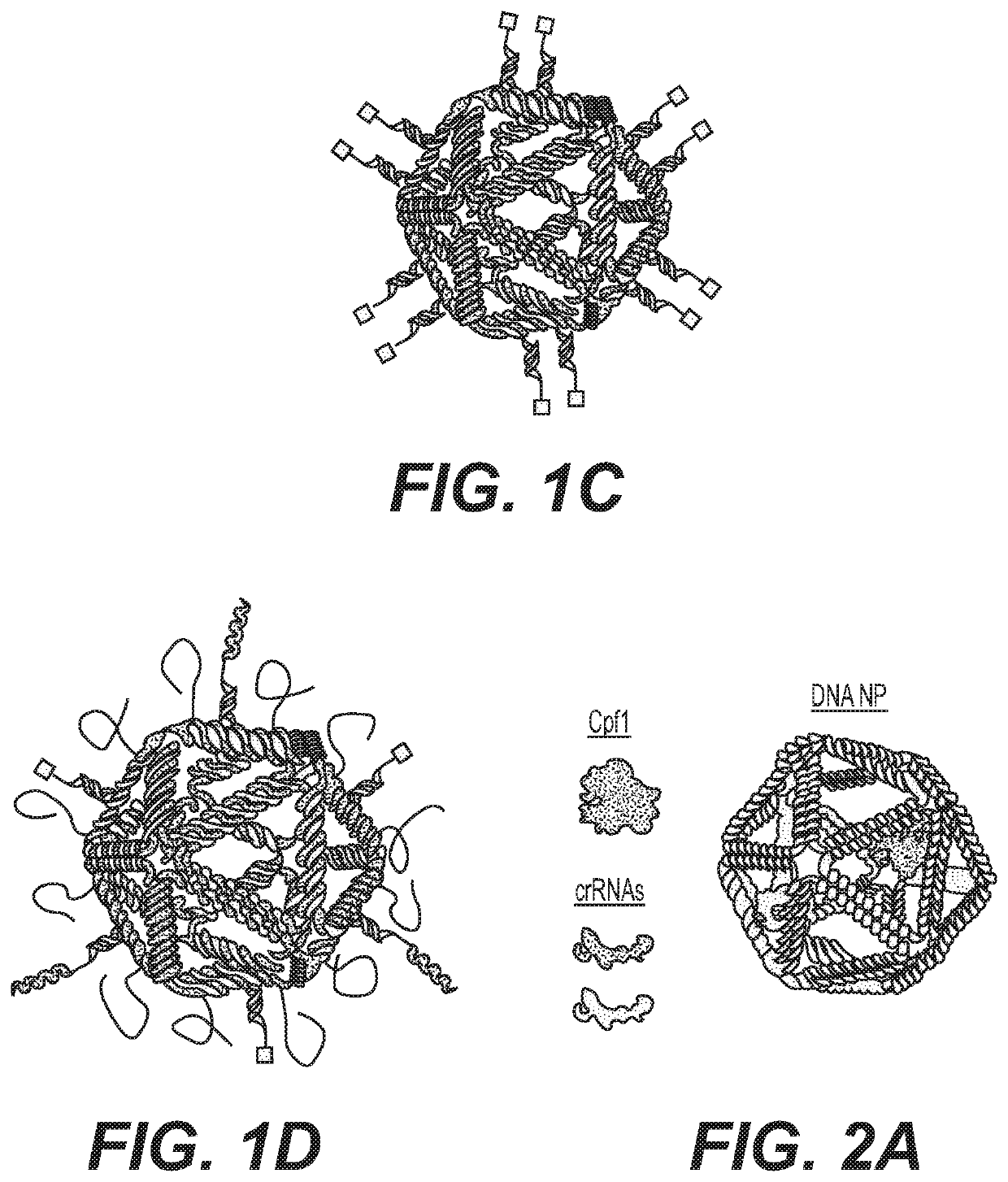
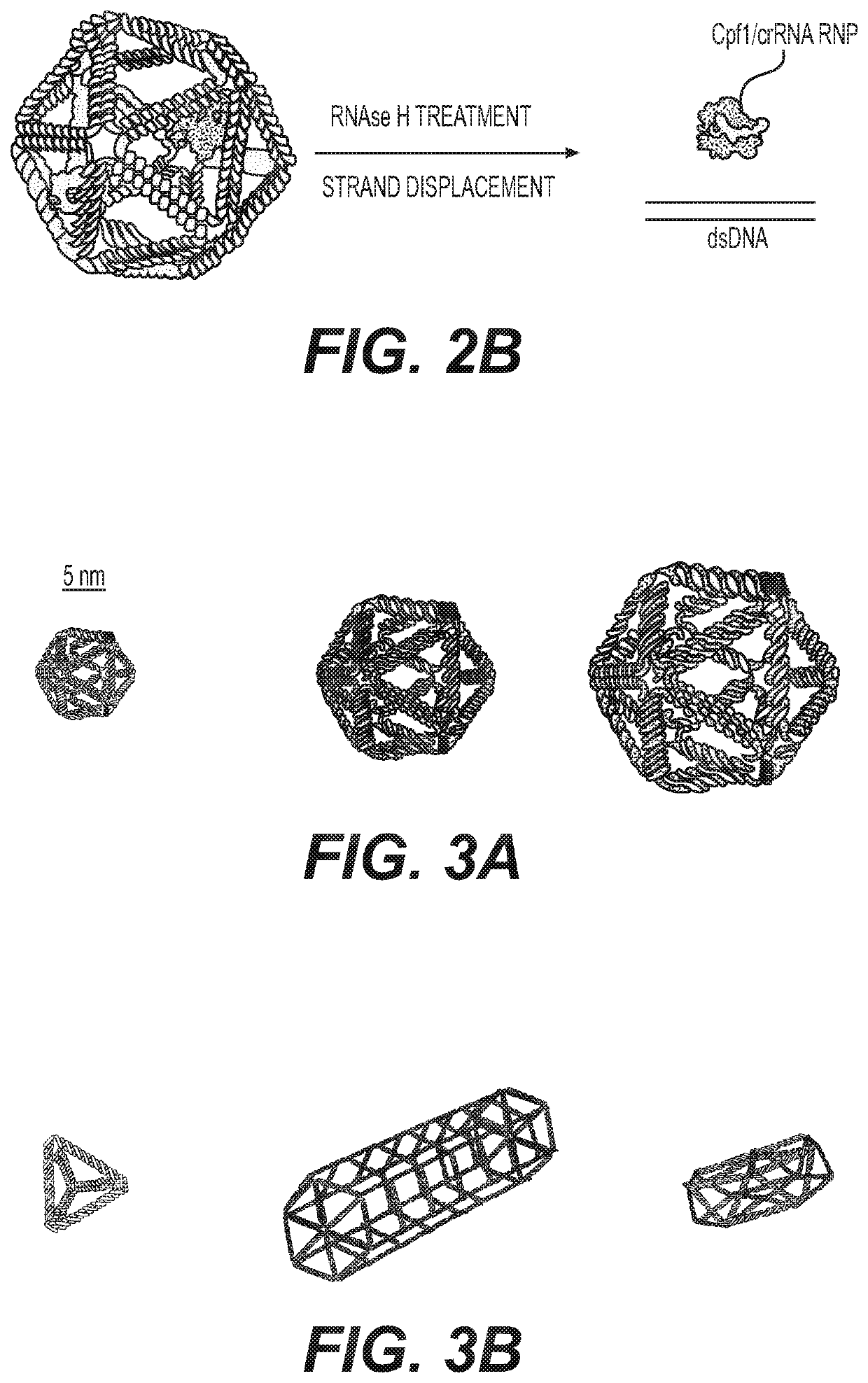
CRISPR Base Editing Potential in Developing New Genetic Polymers
OCT 10, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
CRISPR Base Editing Evolution and Objectives
CRISPR-Cas systems have revolutionized genome editing since their discovery as adaptive immune mechanisms in bacteria and archaea. The journey of CRISPR technology began with the identification of unusual repeat sequences in prokaryotic genomes in the late 1980s, but it wasn't until 2012 that Jennifer Doudna and Emmanuelle Charpentier demonstrated CRISPR-Cas9's potential as a programmable genome editing tool. This breakthrough fundamentally transformed genetic engineering capabilities across scientific disciplines.
Base editing, a more recent advancement in CRISPR technology, emerged around 2016 when David Liu's laboratory developed the first cytosine base editor (CBE). This innovation allowed for precise C-to-T nucleotide substitutions without requiring double-strand breaks. Shortly thereafter, adenine base editors (ABEs) were developed, enabling A-to-G conversions. These developments marked a significant evolution from traditional CRISPR-Cas9 systems, which primarily relied on cellular repair mechanisms following DNA cleavage.
The technical evolution of CRISPR base editing has accelerated dramatically in recent years, with improvements in specificity, efficiency, and versatility. Researchers have engineered various Cas proteins with enhanced properties, expanded targeting capabilities, and reduced off-target effects. The development of prime editing in 2019 further expanded the CRISPR toolkit, allowing for all possible base-to-base conversions and small insertions or deletions without double-strand breaks.
In the context of developing new genetic polymers, CRISPR base editing technology aims to achieve several ambitious objectives. Primarily, it seeks to enable the precise incorporation of non-canonical nucleotides into DNA and RNA, potentially expanding the genetic alphabet beyond the traditional four nucleotides. This capability could revolutionize synthetic biology by creating novel genetic polymers with enhanced functionalities or unique properties not found in nature.
Another critical objective is to develop systems capable of editing and manipulating these expanded genetic alphabets with the same precision and efficiency currently possible with natural DNA. This includes creating specialized base editors that can recognize and modify non-canonical bases, as well as developing delivery systems that can effectively target these novel genetic constructs in various cellular environments.
The long-term vision encompasses creating entirely new genetic systems with expanded information storage capacity, novel enzymatic functions, and potentially resistance to natural nucleases. Such advancements could lead to breakthrough applications in biomanufacturing, medical therapeutics, data storage, and environmental remediation, representing a paradigm shift in our understanding and utilization of genetic information.
Base editing, a more recent advancement in CRISPR technology, emerged around 2016 when David Liu's laboratory developed the first cytosine base editor (CBE). This innovation allowed for precise C-to-T nucleotide substitutions without requiring double-strand breaks. Shortly thereafter, adenine base editors (ABEs) were developed, enabling A-to-G conversions. These developments marked a significant evolution from traditional CRISPR-Cas9 systems, which primarily relied on cellular repair mechanisms following DNA cleavage.
The technical evolution of CRISPR base editing has accelerated dramatically in recent years, with improvements in specificity, efficiency, and versatility. Researchers have engineered various Cas proteins with enhanced properties, expanded targeting capabilities, and reduced off-target effects. The development of prime editing in 2019 further expanded the CRISPR toolkit, allowing for all possible base-to-base conversions and small insertions or deletions without double-strand breaks.
In the context of developing new genetic polymers, CRISPR base editing technology aims to achieve several ambitious objectives. Primarily, it seeks to enable the precise incorporation of non-canonical nucleotides into DNA and RNA, potentially expanding the genetic alphabet beyond the traditional four nucleotides. This capability could revolutionize synthetic biology by creating novel genetic polymers with enhanced functionalities or unique properties not found in nature.
Another critical objective is to develop systems capable of editing and manipulating these expanded genetic alphabets with the same precision and efficiency currently possible with natural DNA. This includes creating specialized base editors that can recognize and modify non-canonical bases, as well as developing delivery systems that can effectively target these novel genetic constructs in various cellular environments.
The long-term vision encompasses creating entirely new genetic systems with expanded information storage capacity, novel enzymatic functions, and potentially resistance to natural nucleases. Such advancements could lead to breakthrough applications in biomanufacturing, medical therapeutics, data storage, and environmental remediation, representing a paradigm shift in our understanding and utilization of genetic information.
Market Analysis for Synthetic Genetic Polymer Applications
The synthetic genetic polymer market is experiencing rapid growth, driven by advancements in CRISPR base editing technologies. Current market valuations indicate the global synthetic biology market reached approximately 9.5 billion USD in 2022, with projections suggesting compound annual growth rates exceeding 25% through 2030. Within this broader market, synthetic genetic polymers represent an emerging segment with particularly strong growth potential.
Healthcare applications dominate the current market landscape, accounting for roughly 60% of synthetic genetic polymer applications. This includes therapeutic development, diagnostic tools, and personalized medicine approaches. The pharmaceutical industry has demonstrated increasing interest in these technologies, with major players like Novartis, Roche, and emerging biotechnology companies making significant investments in CRISPR-based genetic polymer research.
Agricultural applications represent the second-largest market segment, comprising approximately 25% of current applications. This includes crop improvement, pest resistance development, and nutritional enhancement. Companies like Corteva Agriscience and Bayer have established dedicated research divisions focused on synthetic genetic polymer applications in agriculture.
Industrial biotechnology applications, including biofuel production, bioremediation, and specialty chemical manufacturing, constitute approximately 15% of the market. This segment shows promising growth potential as industries seek more sustainable production methods and materials.
Regional analysis reveals North America currently leads the market with approximately 45% share, followed by Europe (30%) and Asia-Pacific (20%). However, the Asia-Pacific region demonstrates the fastest growth rate, driven by increasing research investments in China, Japan, and South Korea.
Market barriers include regulatory uncertainties, with different regions implementing varying approaches to synthetic genetic polymer oversight. Public perception and ethical considerations also impact market development, particularly in consumer-facing applications. Technical challenges in scaling production and ensuring consistency of synthetic genetic polymers remain significant hurdles for commercialization.
Investor interest has grown substantially, with venture capital funding for synthetic biology startups exceeding 3 billion USD in 2021. Strategic partnerships between academic institutions, biotechnology startups, and established pharmaceutical and agricultural companies are increasingly common, accelerating technology development and market entry.
The market demonstrates high fragmentation, with numerous specialized players focusing on specific applications or technological approaches. Industry consolidation through mergers and acquisitions is expected as technologies mature and commercial applications expand.
Healthcare applications dominate the current market landscape, accounting for roughly 60% of synthetic genetic polymer applications. This includes therapeutic development, diagnostic tools, and personalized medicine approaches. The pharmaceutical industry has demonstrated increasing interest in these technologies, with major players like Novartis, Roche, and emerging biotechnology companies making significant investments in CRISPR-based genetic polymer research.
Agricultural applications represent the second-largest market segment, comprising approximately 25% of current applications. This includes crop improvement, pest resistance development, and nutritional enhancement. Companies like Corteva Agriscience and Bayer have established dedicated research divisions focused on synthetic genetic polymer applications in agriculture.
Industrial biotechnology applications, including biofuel production, bioremediation, and specialty chemical manufacturing, constitute approximately 15% of the market. This segment shows promising growth potential as industries seek more sustainable production methods and materials.
Regional analysis reveals North America currently leads the market with approximately 45% share, followed by Europe (30%) and Asia-Pacific (20%). However, the Asia-Pacific region demonstrates the fastest growth rate, driven by increasing research investments in China, Japan, and South Korea.
Market barriers include regulatory uncertainties, with different regions implementing varying approaches to synthetic genetic polymer oversight. Public perception and ethical considerations also impact market development, particularly in consumer-facing applications. Technical challenges in scaling production and ensuring consistency of synthetic genetic polymers remain significant hurdles for commercialization.
Investor interest has grown substantially, with venture capital funding for synthetic biology startups exceeding 3 billion USD in 2021. Strategic partnerships between academic institutions, biotechnology startups, and established pharmaceutical and agricultural companies are increasingly common, accelerating technology development and market entry.
The market demonstrates high fragmentation, with numerous specialized players focusing on specific applications or technological approaches. Industry consolidation through mergers and acquisitions is expected as technologies mature and commercial applications expand.
Current Limitations in Genetic Polymer Engineering
Despite significant advancements in genetic polymer engineering, several critical limitations continue to impede progress in developing novel genetic polymers using CRISPR base editing technologies. The primary challenge remains the inherent specificity of natural enzymes for their canonical substrates. CRISPR-Cas systems and base editors have evolved to recognize and process DNA and RNA with specific chemical structures, creating substantial barriers when attempting to engineer them to work with synthetic genetic polymers.
Off-target effects represent another significant limitation, as current base editing systems frequently introduce unintended modifications at non-target sites. This issue becomes magnified when working with novel genetic polymers where the interaction dynamics between editing machinery and substrate are less predictable and poorly characterized. The fidelity of base editing in non-natural contexts remains substantially lower than with natural nucleic acids.
Delivery mechanisms pose considerable challenges for in vivo applications. While viral vectors and lipid nanoparticles have been optimized for natural nucleic acids, their efficiency drops dramatically when carrying components designed for synthetic genetic polymer manipulation. The size constraints of delivery vehicles further limit the packaging of multiple components often required for complex editing of novel genetic polymers.
The cellular toxicity associated with introducing foreign genetic materials and editing machinery presents another barrier. Novel genetic polymers may trigger unexpected immune responses or disrupt cellular homeostasis through mechanisms distinct from natural nucleic acids. Current systems lack adequate safeguards to mitigate these potential adverse effects in complex biological environments.
Technical limitations in synthesis and scalability also hinder progress. The production of synthetic genetic polymers at scales necessary for comprehensive research remains costly and technically challenging. Additionally, the analytical tools for characterizing these novel polymers and their interactions with editing machinery lack the resolution and throughput available for natural nucleic acids.
Regulatory frameworks present non-technical barriers, as current guidelines were developed primarily for natural genetic materials. Novel genetic polymers exist in a regulatory gray area, creating uncertainty for research advancement and potential applications. The absence of standardized safety assessment protocols specifically designed for synthetic genetic polymers further complicates their development pathway.
Computational tools for predicting editing outcomes with novel genetic polymers remain underdeveloped. While machine learning approaches have improved predictions for conventional CRISPR applications, these models perform poorly when applied to non-canonical genetic systems due to limited training data and fundamental differences in biochemical interactions.
Off-target effects represent another significant limitation, as current base editing systems frequently introduce unintended modifications at non-target sites. This issue becomes magnified when working with novel genetic polymers where the interaction dynamics between editing machinery and substrate are less predictable and poorly characterized. The fidelity of base editing in non-natural contexts remains substantially lower than with natural nucleic acids.
Delivery mechanisms pose considerable challenges for in vivo applications. While viral vectors and lipid nanoparticles have been optimized for natural nucleic acids, their efficiency drops dramatically when carrying components designed for synthetic genetic polymer manipulation. The size constraints of delivery vehicles further limit the packaging of multiple components often required for complex editing of novel genetic polymers.
The cellular toxicity associated with introducing foreign genetic materials and editing machinery presents another barrier. Novel genetic polymers may trigger unexpected immune responses or disrupt cellular homeostasis through mechanisms distinct from natural nucleic acids. Current systems lack adequate safeguards to mitigate these potential adverse effects in complex biological environments.
Technical limitations in synthesis and scalability also hinder progress. The production of synthetic genetic polymers at scales necessary for comprehensive research remains costly and technically challenging. Additionally, the analytical tools for characterizing these novel polymers and their interactions with editing machinery lack the resolution and throughput available for natural nucleic acids.
Regulatory frameworks present non-technical barriers, as current guidelines were developed primarily for natural genetic materials. Novel genetic polymers exist in a regulatory gray area, creating uncertainty for research advancement and potential applications. The absence of standardized safety assessment protocols specifically designed for synthetic genetic polymers further complicates their development pathway.
Computational tools for predicting editing outcomes with novel genetic polymers remain underdeveloped. While machine learning approaches have improved predictions for conventional CRISPR applications, these models perform poorly when applied to non-canonical genetic systems due to limited training data and fundamental differences in biochemical interactions.
Existing CRISPR Base Editing Methodologies
01 CRISPR-Cas base editing systems for targeted genetic modifications
CRISPR-Cas base editing systems enable precise modifications of genetic sequences without requiring double-strand breaks. These systems combine a catalytically impaired Cas protein with a deaminase enzyme to convert one nucleotide to another at specific target sites. This approach allows for the correction of point mutations and single nucleotide polymorphisms (SNPs) with reduced off-target effects compared to traditional CRISPR-Cas9 systems, making it valuable for therapeutic applications targeting genetic disorders.- CRISPR-Cas base editing systems for targeted genetic modifications: CRISPR-Cas base editing systems enable precise modifications of genetic polymers without requiring double-strand breaks. These systems typically combine a catalytically impaired Cas protein with a deaminase enzyme to convert one nucleotide to another at specific target sites. This approach allows for the correction of point mutations and single nucleotide polymorphisms with reduced off-target effects compared to traditional CRISPR systems.
- Base editors for treating genetic disorders: Base editing technologies are being developed to treat genetic disorders by correcting disease-causing mutations. These therapeutic applications focus on modifying specific genetic polymorphisms associated with inherited conditions. The base editors can be delivered using various vectors, including viral vectors and nanoparticles, to reach target tissues and cells. This approach offers potential treatments for previously untreatable genetic diseases by making precise corrections to the underlying genetic causes.
- Novel base editor architectures and improvements: Innovations in base editor design include engineered variants with enhanced specificity, expanded editing windows, and altered PAM requirements. These improved architectures incorporate modified deaminases, optimized linkers between functional domains, and engineered Cas variants. Some designs incorporate multiple enzymatic activities to enable more complex editing outcomes or to reduce unwanted byproducts of the editing process, resulting in more efficient and precise genetic polymer modifications.
- Base editing of non-standard genetic polymers: Base editing technologies have been expanded beyond traditional DNA editing to include modifications of other genetic polymers such as RNA and synthetic nucleic acids. These systems utilize specialized deaminases and modified CRISPR proteins adapted for different substrates. Applications include temporary gene expression modulation through RNA editing and the creation of novel genetic polymers with properties not found in natural systems, opening new possibilities for biotechnology and synthetic biology.
- Computational methods for base editing design and analysis: Computational tools and methods have been developed to design efficient base editing strategies and analyze editing outcomes. These include algorithms for predicting editing efficiency, identifying potential off-target sites, and optimizing guide RNA design for specific genetic polymer targets. Advanced sequencing and bioinformatics approaches enable comprehensive assessment of editing outcomes across the genome, facilitating the development of more precise and effective base editing applications.
02 Base editing of synthetic genetic polymers and xeno-nucleic acids
Base editing technologies have been extended to modify synthetic genetic polymers and xeno-nucleic acids (XNAs) that differ from natural DNA and RNA. These artificial genetic systems can be engineered with novel properties for applications in synthetic biology, diagnostics, and therapeutics. Base editors have been adapted to work with these alternative genetic polymers, enabling the creation of diverse genetic materials with customized functions and enhanced stability compared to natural nucleic acids.Expand Specific Solutions03 Delivery systems for CRISPR base editors
Various delivery systems have been developed to transport CRISPR base editing components into cells and tissues. These include viral vectors (such as AAV, lentivirus), lipid nanoparticles, and cell-penetrating peptides. Efficient delivery methods are crucial for the successful application of base editing technologies in both ex vivo and in vivo settings. Advanced delivery systems aim to improve targeting specificity, reduce immunogenicity, and enhance the efficiency of base editing for therapeutic applications.Expand Specific Solutions04 Base editing for correction of disease-causing genetic polymorphisms
CRISPR base editing technologies are being applied to correct disease-causing genetic polymorphisms in various disorders. By precisely converting specific nucleotides, base editors can repair pathogenic mutations without introducing double-strand breaks. This approach has shown promise in addressing monogenic diseases caused by point mutations, including blood disorders, metabolic conditions, and neurological diseases. The ability to correct genetic polymorphisms with minimal disruption to the genome makes base editing a valuable tool for developing gene therapy strategies.Expand Specific Solutions05 Computational tools and methods for CRISPR base editing design
Computational tools and algorithms have been developed to optimize CRISPR base editing strategies. These tools help predict editing outcomes, identify potential off-target sites, and design guide RNAs with high specificity and efficiency. Machine learning approaches are increasingly being used to improve the accuracy of these predictions and to develop personalized base editing strategies. Advanced computational methods enable researchers to design base editing experiments with greater precision, accelerating the development of therapeutic applications targeting genetic polymorphisms.Expand Specific Solutions
Leading Organizations in CRISPR Base Editing Research
CRISPR base editing technology for developing new genetic polymers is in its early growth stage, with significant research momentum but limited commercial applications. The market is expanding rapidly, projected to reach substantial value as therapeutic and agricultural applications mature. Technologically, the field is advancing from experimental to translational phase, with key players demonstrating varying levels of expertise. Beam Therapeutics and Mammoth Biosciences lead commercial development with proprietary platforms, while academic institutions like Broad Institute and Shanghaitech University contribute foundational research. Established companies including AstraZeneca and Agilent are strategically positioning through partnerships. Flagship Pioneering and other venture firms are actively funding emerging startups, creating a competitive ecosystem balancing scientific innovation with commercial potential.
The Broad Institute, Inc.
Technical Solution: The Broad Institute has pioneered significant advancements in CRISPR base editing for developing novel genetic polymers. Their technology platform centers on precision C-to-T and A-to-G base editing without inducing double-strand breaks, enabling targeted nucleotide substitutions with minimal off-target effects[1]. They've developed cytosine and adenine base editors (CBEs and ABEs) that can introduce point mutations with efficiency rates exceeding 50% in various cell types and organisms. Their recent innovations include engineered Cas variants with expanded PAM compatibility and reduced off-target activity, allowing access to previously unreachable genomic sites[2]. The Broad Institute has also developed RNA-targeting base editors that can modify RNA transcripts without altering the underlying DNA, providing reversible genetic modifications. Their platform includes computational tools for predicting editing outcomes and optimizing guide RNA design, significantly enhancing editing precision for creating novel genetic polymers[3].
Strengths: Industry-leading precision with reduced off-target effects; comprehensive toolkit spanning DNA and RNA editing; strong computational infrastructure for guide design. Weaknesses: Some base editing applications still show variable efficiency across different genomic contexts; delivery systems for in vivo applications remain challenging for certain tissue types.
Beam Therapeutics, Inc.
Technical Solution: Beam Therapeutics has developed a proprietary base editing platform specifically designed for creating and manipulating genetic polymers with unprecedented precision. Their technology utilizes engineered CRISPR-Cas systems that can perform single-base conversions without creating double-strand breaks in DNA[1]. The company's platform includes both cytosine base editors (CBEs) that enable C→T conversions and adenine base editors (ABEs) for A→G transitions. Beam has further refined these systems with their "Engineered Delivery Vehicle" technology that enhances cellular delivery and targeting specificity[2]. Their proprietary REPAIR and RESCUE systems allow for targeted RNA editing, expanding their capabilities beyond DNA modification. Notably, Beam has developed novel genetic polymer applications through their "Multiplex Editing" technology, which can simultaneously introduce multiple precise edits across the genome to create synthetic genetic polymers with novel properties[3]. Their platform includes machine learning algorithms to predict and minimize off-target effects, significantly improving safety profiles compared to traditional CRISPR systems.
Strengths: Highly precise single-base editing without double-strand breaks; versatile platform covering both DNA and RNA editing; proprietary delivery systems enhancing cellular uptake. Weaknesses: Limited to certain types of base conversions (primarily C→T and A→G); potential immunogenicity concerns with some delivery vehicles; higher cost compared to traditional gene editing approaches.
Key Patents in Genetic Polymer Engineering
Crispr-associated base-editing of the complementary strand
PatentWO2022164319A1
Innovation
- Development of a CRISPR-based editing system using a cleavage-deficient Cas nuclease fused with deaminases that allows for A to G and C to T modifications on the complementary strand of double-stranded target DNA, enabling editing of both strands and expanding the editing range by modifying the Cas nuclease to lack certain domains and multimerize upon gRNA binding.
Nucleic acid assemblies for use in targeted delivery
PatentPendingUS20210317479A1
Innovation
- Nucleic acid assemblies that enclose and protect cargo, such as CRISPR-Cas effector proteins and guide molecules, with designed physiochemical properties for targeted delivery, enhanced stability, and reduced immunogenicity, allowing for controlled stoichiometry and intracellular trafficking.
Biosafety and Bioethical Considerations
The development of CRISPR base editing technologies for creating novel genetic polymers raises significant biosafety and bioethical considerations that must be addressed before widespread implementation. The potential for unintended genomic modifications presents a primary concern, as base editors can induce off-target effects that may lead to unpredictable genetic alterations. These off-target modifications could potentially disrupt essential cellular functions or activate oncogenic pathways, posing risks to both experimental systems and, if applied clinically, to human subjects.
Containment strategies represent a critical aspect of biosafety protocols when working with engineered genetic polymers. Physical containment measures, including specialized laboratory facilities with appropriate biosafety levels, must be implemented alongside biological containment approaches such as self-limiting genetic circuits and kill-switch mechanisms. These safeguards help prevent unintended environmental release of engineered genetic materials that could potentially interact with natural ecosystems.
The potential for horizontal gene transfer between engineered genetic polymers and natural organisms demands rigorous risk assessment frameworks. Long-term ecological impact studies are essential to understand how these novel genetic constructs might behave if released into the environment, particularly regarding their stability, transmissibility, and potential to confer selective advantages to recipient organisms.
From a bioethical perspective, the creation of novel genetic polymers raises fundamental questions about the boundaries of human intervention in biological systems. The principle of responsible innovation requires transparent governance frameworks that balance scientific progress with precautionary approaches. International regulatory harmonization becomes increasingly important as these technologies develop, necessitating collaborative efforts to establish consistent standards for risk assessment and management.
Informed consent protocols require special consideration when base editing technologies are applied in human contexts. The novelty of these approaches, combined with potential unknown long-term effects, demands comprehensive disclosure of risks and limitations to research participants. This becomes particularly complex when considering germline applications that could affect future generations who cannot provide consent.
Equitable access to beneficial applications of these technologies must be balanced against potential misuse scenarios. Dual-use concerns are particularly relevant, as techniques developed for therapeutic purposes could potentially be repurposed for enhancement applications or bioweapons development. Establishing robust oversight mechanisms, including ethics review boards with specialized expertise in synthetic biology and genetic engineering, represents an essential component of responsible research governance in this rapidly evolving field.
Containment strategies represent a critical aspect of biosafety protocols when working with engineered genetic polymers. Physical containment measures, including specialized laboratory facilities with appropriate biosafety levels, must be implemented alongside biological containment approaches such as self-limiting genetic circuits and kill-switch mechanisms. These safeguards help prevent unintended environmental release of engineered genetic materials that could potentially interact with natural ecosystems.
The potential for horizontal gene transfer between engineered genetic polymers and natural organisms demands rigorous risk assessment frameworks. Long-term ecological impact studies are essential to understand how these novel genetic constructs might behave if released into the environment, particularly regarding their stability, transmissibility, and potential to confer selective advantages to recipient organisms.
From a bioethical perspective, the creation of novel genetic polymers raises fundamental questions about the boundaries of human intervention in biological systems. The principle of responsible innovation requires transparent governance frameworks that balance scientific progress with precautionary approaches. International regulatory harmonization becomes increasingly important as these technologies develop, necessitating collaborative efforts to establish consistent standards for risk assessment and management.
Informed consent protocols require special consideration when base editing technologies are applied in human contexts. The novelty of these approaches, combined with potential unknown long-term effects, demands comprehensive disclosure of risks and limitations to research participants. This becomes particularly complex when considering germline applications that could affect future generations who cannot provide consent.
Equitable access to beneficial applications of these technologies must be balanced against potential misuse scenarios. Dual-use concerns are particularly relevant, as techniques developed for therapeutic purposes could potentially be repurposed for enhancement applications or bioweapons development. Establishing robust oversight mechanisms, including ethics review boards with specialized expertise in synthetic biology and genetic engineering, represents an essential component of responsible research governance in this rapidly evolving field.
Commercialization Pathways for Novel Genetic Polymers
The commercialization of novel genetic polymers developed through CRISPR base editing technologies presents multiple strategic pathways for market entry and value creation. Initial commercialization efforts will likely focus on research tools and reagents, representing the lowest regulatory barrier and fastest route to market. Companies can develop and sell specialized base editing kits, engineered genetic polymers, and analytical tools to academic and industrial research laboratories, establishing early revenue streams while building market presence.
Diagnostic applications represent a promising mid-term commercialization avenue. Novel genetic polymers with enhanced stability or detection properties could revolutionize nucleic acid-based diagnostics, enabling more sensitive detection of pathogens, genetic disorders, and cancer biomarkers. The regulatory pathway for diagnostics, while substantial, is less onerous than therapeutics, allowing for faster market penetration.
Therapeutic applications, though requiring the longest development timeline and highest investment, offer the greatest potential market value. Novel genetic polymers could address fundamental limitations in current gene therapy approaches, including delivery challenges, immunogenicity, and durability of effect. Strategic partnerships with established pharmaceutical companies will be crucial for navigating the complex regulatory landscape and accessing specialized manufacturing capabilities.
Agricultural and industrial biotechnology applications present alternative commercialization routes with distinct regulatory considerations. Novel genetic polymers could enable improved crop traits or more efficient biocatalysts for industrial processes, potentially facing fewer regulatory hurdles than medical applications in certain jurisdictions.
Intellectual property strategy will be paramount across all commercialization pathways. Companies must develop robust patent portfolios covering both the base editing technologies and the resulting novel genetic polymers, considering composition of matter, method of manufacture, and specific applications. Strategic licensing agreements may optimize value capture across different market segments.
Manufacturing scalability represents a critical consideration for commercialization success. Early investment in process development and quality systems will facilitate smoother transitions from research to commercial production scales, particularly important for therapeutic applications where consistency and purity requirements are stringent.
Phased market entry strategies that begin with research tools while simultaneously developing higher-value applications can optimize cash flow and risk management. This approach allows companies to generate revenue while building the data and capabilities necessary for more complex applications, creating a sustainable commercialization trajectory for these revolutionary genetic technologies.
Diagnostic applications represent a promising mid-term commercialization avenue. Novel genetic polymers with enhanced stability or detection properties could revolutionize nucleic acid-based diagnostics, enabling more sensitive detection of pathogens, genetic disorders, and cancer biomarkers. The regulatory pathway for diagnostics, while substantial, is less onerous than therapeutics, allowing for faster market penetration.
Therapeutic applications, though requiring the longest development timeline and highest investment, offer the greatest potential market value. Novel genetic polymers could address fundamental limitations in current gene therapy approaches, including delivery challenges, immunogenicity, and durability of effect. Strategic partnerships with established pharmaceutical companies will be crucial for navigating the complex regulatory landscape and accessing specialized manufacturing capabilities.
Agricultural and industrial biotechnology applications present alternative commercialization routes with distinct regulatory considerations. Novel genetic polymers could enable improved crop traits or more efficient biocatalysts for industrial processes, potentially facing fewer regulatory hurdles than medical applications in certain jurisdictions.
Intellectual property strategy will be paramount across all commercialization pathways. Companies must develop robust patent portfolios covering both the base editing technologies and the resulting novel genetic polymers, considering composition of matter, method of manufacture, and specific applications. Strategic licensing agreements may optimize value capture across different market segments.
Manufacturing scalability represents a critical consideration for commercialization success. Early investment in process development and quality systems will facilitate smoother transitions from research to commercial production scales, particularly important for therapeutic applications where consistency and purity requirements are stringent.
Phased market entry strategies that begin with research tools while simultaneously developing higher-value applications can optimize cash flow and risk management. This approach allows companies to generate revenue while building the data and capabilities necessary for more complex applications, creating a sustainable commercialization trajectory for these revolutionary genetic technologies.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!