Raman Spectroscopy vs MALDI-TOF: Proteomic Analysis Depth
SEP 19, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Raman vs MALDI-TOF Background and Objectives
Raman spectroscopy and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry represent two distinct yet complementary analytical techniques that have evolved significantly over the past several decades. Raman spectroscopy, discovered by C.V. Raman in 1928, has progressed from a purely academic tool to a versatile analytical method with applications spanning from material science to biomedical diagnostics. Similarly, MALDI-TOF, developed in the late 1980s by Karas, Hillenkamp, and Tanaka, has revolutionized protein analysis by enabling the ionization of large biomolecules without fragmentation.
The technological evolution of both techniques has been marked by significant improvements in instrumentation, data processing capabilities, and application methodologies. Raman spectroscopy has benefited from advances in laser technology, detector sensitivity, and computational algorithms, leading to enhanced spectral resolution and reduced acquisition times. MALDI-TOF has similarly evolved with improvements in matrix formulations, ion detection systems, and mass analyzers, resulting in higher mass accuracy and resolution.
In the context of proteomic analysis, these technologies serve different yet complementary roles. Raman spectroscopy provides detailed information about molecular vibrations and chemical bonds, offering insights into protein secondary structure and conformational changes. MALDI-TOF, conversely, excels in determining the molecular weight of proteins and peptides with high precision, enabling protein identification through peptide mass fingerprinting.
The primary objective of this comparative analysis is to evaluate the relative strengths and limitations of Raman spectroscopy and MALDI-TOF for in-depth proteomic analysis. Specifically, we aim to assess their capabilities in terms of sensitivity, specificity, sample preparation requirements, throughput, and data interpretation complexity. Additionally, we seek to identify potential synergies between these techniques that could enhance proteomic analysis depth when used in combination.
Furthermore, this analysis intends to explore emerging trends in both technologies, such as surface-enhanced Raman spectroscopy (SERS) and MALDI imaging mass spectrometry, which are expanding the analytical capabilities of these techniques. By understanding the current technological landscape and future development trajectories, we can better position these tools within the broader context of proteomic research and clinical applications.
Ultimately, this comparative assessment aims to provide a comprehensive framework for researchers and industry professionals to make informed decisions regarding technology selection for specific proteomic applications, while also highlighting opportunities for technological innovation and integration that could address current analytical limitations.
The technological evolution of both techniques has been marked by significant improvements in instrumentation, data processing capabilities, and application methodologies. Raman spectroscopy has benefited from advances in laser technology, detector sensitivity, and computational algorithms, leading to enhanced spectral resolution and reduced acquisition times. MALDI-TOF has similarly evolved with improvements in matrix formulations, ion detection systems, and mass analyzers, resulting in higher mass accuracy and resolution.
In the context of proteomic analysis, these technologies serve different yet complementary roles. Raman spectroscopy provides detailed information about molecular vibrations and chemical bonds, offering insights into protein secondary structure and conformational changes. MALDI-TOF, conversely, excels in determining the molecular weight of proteins and peptides with high precision, enabling protein identification through peptide mass fingerprinting.
The primary objective of this comparative analysis is to evaluate the relative strengths and limitations of Raman spectroscopy and MALDI-TOF for in-depth proteomic analysis. Specifically, we aim to assess their capabilities in terms of sensitivity, specificity, sample preparation requirements, throughput, and data interpretation complexity. Additionally, we seek to identify potential synergies between these techniques that could enhance proteomic analysis depth when used in combination.
Furthermore, this analysis intends to explore emerging trends in both technologies, such as surface-enhanced Raman spectroscopy (SERS) and MALDI imaging mass spectrometry, which are expanding the analytical capabilities of these techniques. By understanding the current technological landscape and future development trajectories, we can better position these tools within the broader context of proteomic research and clinical applications.
Ultimately, this comparative assessment aims to provide a comprehensive framework for researchers and industry professionals to make informed decisions regarding technology selection for specific proteomic applications, while also highlighting opportunities for technological innovation and integration that could address current analytical limitations.
Market Demand for Advanced Proteomic Analysis Technologies
The global proteomics market has experienced significant growth, valued at approximately $25 billion in 2022 and projected to reach $68 billion by 2030, with a compound annual growth rate (CAGR) of 13.5%. This robust expansion reflects the increasing demand for advanced proteomic analysis technologies across various sectors, particularly in pharmaceutical research, clinical diagnostics, and academic research institutions.
Healthcare and pharmaceutical industries represent the largest market segments, driven by the critical need for more precise protein characterization in drug discovery and development processes. The ability to identify biomarkers for disease diagnosis, progression monitoring, and therapeutic response evaluation has become essential in modern medicine, creating sustained demand for high-resolution proteomic analysis tools.
Both Raman Spectroscopy and MALDI-TOF mass spectrometry have seen increasing adoption rates in clinical settings. Hospital laboratories and diagnostic centers are increasingly incorporating these technologies into their workflow, with MALDI-TOF currently holding a larger market share due to its established protocols in microbial identification. However, Raman Spectroscopy is gaining traction for its non-destructive analysis capabilities and potential for point-of-care applications.
The academic research sector demonstrates growing interest in comprehensive proteomic analysis platforms, with universities and research institutions allocating substantial portions of their instrumentation budgets to acquire these technologies. This trend is particularly evident in regions with strong life sciences research infrastructure, such as North America, Western Europe, and increasingly in Asia-Pacific countries, especially China and Japan.
Market surveys indicate that end-users prioritize analysis depth, throughput capacity, and operational costs when selecting proteomic analysis platforms. MALDI-TOF systems are preferred for high-throughput screening applications, while Raman Spectroscopy is increasingly valued for its ability to provide structural information without sample preparation complexity.
Emerging applications in personalized medicine represent a significant growth driver, with clinicians seeking technologies that can rapidly analyze protein biomarkers from minimal sample volumes. This trend aligns with the broader shift toward precision medicine approaches, creating market opportunities for technologies offering enhanced sensitivity and specificity in protein detection and characterization.
The COVID-19 pandemic has further accelerated market growth by highlighting the importance of rapid protein analysis in pathogen identification and vaccine development. This has resulted in increased funding for proteomics research and technology development from both public and private sources, creating favorable market conditions for advanced analytical platforms.
Healthcare and pharmaceutical industries represent the largest market segments, driven by the critical need for more precise protein characterization in drug discovery and development processes. The ability to identify biomarkers for disease diagnosis, progression monitoring, and therapeutic response evaluation has become essential in modern medicine, creating sustained demand for high-resolution proteomic analysis tools.
Both Raman Spectroscopy and MALDI-TOF mass spectrometry have seen increasing adoption rates in clinical settings. Hospital laboratories and diagnostic centers are increasingly incorporating these technologies into their workflow, with MALDI-TOF currently holding a larger market share due to its established protocols in microbial identification. However, Raman Spectroscopy is gaining traction for its non-destructive analysis capabilities and potential for point-of-care applications.
The academic research sector demonstrates growing interest in comprehensive proteomic analysis platforms, with universities and research institutions allocating substantial portions of their instrumentation budgets to acquire these technologies. This trend is particularly evident in regions with strong life sciences research infrastructure, such as North America, Western Europe, and increasingly in Asia-Pacific countries, especially China and Japan.
Market surveys indicate that end-users prioritize analysis depth, throughput capacity, and operational costs when selecting proteomic analysis platforms. MALDI-TOF systems are preferred for high-throughput screening applications, while Raman Spectroscopy is increasingly valued for its ability to provide structural information without sample preparation complexity.
Emerging applications in personalized medicine represent a significant growth driver, with clinicians seeking technologies that can rapidly analyze protein biomarkers from minimal sample volumes. This trend aligns with the broader shift toward precision medicine approaches, creating market opportunities for technologies offering enhanced sensitivity and specificity in protein detection and characterization.
The COVID-19 pandemic has further accelerated market growth by highlighting the importance of rapid protein analysis in pathogen identification and vaccine development. This has resulted in increased funding for proteomics research and technology development from both public and private sources, creating favorable market conditions for advanced analytical platforms.
Current Technical Limitations in Proteomic Analysis Methods
Despite significant advancements in proteomic analysis technologies, both Raman Spectroscopy and MALDI-TOF face substantial technical limitations that impact their effectiveness in comprehensive proteomic studies. Raman Spectroscopy, while offering non-destructive analysis capabilities, struggles with sensitivity issues when detecting low-abundance proteins. The inherent weak Raman scattering effect necessitates longer acquisition times, which can be problematic for time-sensitive analyses and high-throughput applications.
The spatial resolution of conventional Raman systems typically ranges from 0.5-1 μm, which may be insufficient for subcellular protein localization studies. Additionally, fluorescence interference remains a persistent challenge, often overwhelming the weaker Raman signals and necessitating complex background correction algorithms that can introduce artifacts into the data.
MALDI-TOF, despite its widespread adoption in proteomics, exhibits significant limitations in dynamic range, typically spanning only 3-4 orders of magnitude. This restricts its ability to simultaneously detect both high and low abundance proteins without sample fractionation. The technique also demonstrates bias toward certain protein classes, particularly those with intermediate hydrophobicity and molecular weight, potentially leading to incomplete proteome coverage.
Sample preparation for MALDI-TOF introduces variability that affects reproducibility, with matrix crystallization heterogeneity causing "hot spots" that result in signal intensity fluctuations. This variability complicates quantitative analyses and requires multiple measurements to achieve reliable results. Furthermore, MALDI-TOF struggles with the analysis of proteins exceeding 100 kDa without specialized modifications to standard protocols.
Both technologies face challenges in data processing and interpretation. Raman spectral data requires sophisticated chemometric approaches to extract meaningful biological information, while MALDI-TOF mass spectra demand complex peak assignment algorithms to identify proteins accurately. The lack of standardized data processing workflows impedes cross-laboratory comparisons and meta-analyses.
Integration with liquid chromatography improves the analytical capabilities of both techniques but introduces additional complexity and potential sources of error. LC-MALDI-TOF systems face challenges in maintaining consistent spotting and crystallization across the chromatographic run, while hyphenated Raman systems must contend with solvent interference and reduced sensitivity in flow-through cells.
These technical limitations highlight the need for complementary analytical approaches and continued technological innovation to achieve comprehensive proteomic analysis depth. Recent developments in surface-enhanced Raman spectroscopy (SERS) and high-resolution MALDI-TOF instruments show promise in addressing some of these challenges, but significant hurdles remain before either technology can provide truly comprehensive proteome coverage independently.
The spatial resolution of conventional Raman systems typically ranges from 0.5-1 μm, which may be insufficient for subcellular protein localization studies. Additionally, fluorescence interference remains a persistent challenge, often overwhelming the weaker Raman signals and necessitating complex background correction algorithms that can introduce artifacts into the data.
MALDI-TOF, despite its widespread adoption in proteomics, exhibits significant limitations in dynamic range, typically spanning only 3-4 orders of magnitude. This restricts its ability to simultaneously detect both high and low abundance proteins without sample fractionation. The technique also demonstrates bias toward certain protein classes, particularly those with intermediate hydrophobicity and molecular weight, potentially leading to incomplete proteome coverage.
Sample preparation for MALDI-TOF introduces variability that affects reproducibility, with matrix crystallization heterogeneity causing "hot spots" that result in signal intensity fluctuations. This variability complicates quantitative analyses and requires multiple measurements to achieve reliable results. Furthermore, MALDI-TOF struggles with the analysis of proteins exceeding 100 kDa without specialized modifications to standard protocols.
Both technologies face challenges in data processing and interpretation. Raman spectral data requires sophisticated chemometric approaches to extract meaningful biological information, while MALDI-TOF mass spectra demand complex peak assignment algorithms to identify proteins accurately. The lack of standardized data processing workflows impedes cross-laboratory comparisons and meta-analyses.
Integration with liquid chromatography improves the analytical capabilities of both techniques but introduces additional complexity and potential sources of error. LC-MALDI-TOF systems face challenges in maintaining consistent spotting and crystallization across the chromatographic run, while hyphenated Raman systems must contend with solvent interference and reduced sensitivity in flow-through cells.
These technical limitations highlight the need for complementary analytical approaches and continued technological innovation to achieve comprehensive proteomic analysis depth. Recent developments in surface-enhanced Raman spectroscopy (SERS) and high-resolution MALDI-TOF instruments show promise in addressing some of these challenges, but significant hurdles remain before either technology can provide truly comprehensive proteome coverage independently.
Comparative Technical Solutions in Proteomic Analysis
01 Depth profiling using Raman spectroscopy
Raman spectroscopy can be used for depth profiling of various materials and biological samples. This technique allows for non-destructive analysis of different layers within a sample by adjusting the focal point of the laser. The depth resolution can be enhanced through confocal arrangements, allowing for three-dimensional chemical mapping. This approach is particularly useful for analyzing layered structures, coatings, and subsurface features without physical sectioning of the sample.- Depth profiling techniques using Raman spectroscopy: Raman spectroscopy can be used for depth profiling analysis of various materials and biological samples. This technique allows for non-destructive analysis of sample composition at different depths by adjusting the focal point of the laser. Advanced implementations include confocal Raman microscopy which provides high spatial resolution in three dimensions, enabling precise depth measurements of layered structures, thin films, and subsurface features without physical sectioning of the sample.
- MALDI-TOF mass spectrometry for depth analysis: Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry can be used for depth analysis by controlling laser penetration or through layer-by-layer analysis. This technique is particularly useful for analyzing biological samples, polymers, and thin films. By adjusting laser parameters or using sample preparation techniques that expose successive layers, MALDI-TOF can provide compositional information at different depths, enabling three-dimensional molecular characterization of complex samples.
- Combined Raman and MALDI-TOF analysis systems: Integrated systems combining Raman spectroscopy and MALDI-TOF mass spectrometry provide complementary analytical capabilities for comprehensive sample characterization. These hybrid systems allow for correlative analysis where Raman provides structural and chemical bonding information while MALDI-TOF delivers precise molecular mass data. The combination enables more accurate depth profiling by leveraging the strengths of both techniques, particularly useful for complex biological samples, pharmaceutical formulations, and material science applications.
- Sample preparation methods for depth analysis: Specialized sample preparation techniques enhance the depth analysis capabilities of both Raman spectroscopy and MALDI-TOF mass spectrometry. These methods include cross-sectioning, layer-by-layer removal techniques, depth markers, and specialized matrices for MALDI that facilitate controlled laser penetration. Advanced preparation protocols enable precise depth resolution and improved signal quality, allowing for more accurate characterization of subsurface features and compositional gradients in complex samples.
- Data processing algorithms for depth profile interpretation: Sophisticated data processing algorithms are essential for interpreting depth profile data from Raman spectroscopy and MALDI-TOF analysis. These algorithms include multivariate statistical methods, machine learning approaches, and specialized software tools that can extract meaningful information from complex spectral datasets. Advanced processing techniques enable deconvolution of overlapping signals, background correction, and correlation of data from different depths, ultimately providing more accurate three-dimensional chemical mapping of analyzed samples.
02 MALDI-TOF depth analysis techniques
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry can be used for depth analysis by controlling the laser penetration or through layer-by-layer removal of material. This technique is particularly valuable for analyzing biological tissues, polymers, and thin films. By adjusting laser parameters or using specialized sample preparation methods, researchers can obtain depth-resolved molecular information. Recent advancements include 3D MALDI imaging, which combines lateral and depth information for comprehensive sample characterization.Expand Specific Solutions03 Combined Raman and MALDI-TOF analysis systems
Integrated systems that combine Raman spectroscopy and MALDI-TOF mass spectrometry provide complementary information about sample composition and structure at various depths. These hybrid approaches allow for correlation between molecular vibration data from Raman and precise molecular identification from mass spectrometry. Such combined systems offer enhanced analytical capabilities for complex samples, enabling researchers to obtain comprehensive chemical information with spatial resolution in both lateral and depth dimensions.Expand Specific Solutions04 Sample preparation techniques for depth analysis
Specialized sample preparation methods enhance the depth analysis capabilities of both Raman spectroscopy and MALDI-TOF. These include sectioning techniques, optical clearing methods for biological samples, and controlled etching processes for materials science applications. Proper sample preparation is crucial for achieving optimal depth resolution and accurate chemical characterization. Advanced preparation protocols can minimize artifacts and improve the signal-to-noise ratio for subsurface measurements.Expand Specific Solutions05 Data processing algorithms for depth profile interpretation
Sophisticated data processing algorithms are essential for interpreting depth profiles obtained from Raman spectroscopy and MALDI-TOF analysis. These algorithms address challenges such as signal attenuation with depth, scattering effects, and matrix interference. Machine learning approaches can be applied to extract meaningful information from complex depth-resolved spectral data. Advanced computational methods enable deconvolution of overlapping signals and enhance the accuracy of depth measurements in heterogeneous samples.Expand Specific Solutions
Key Industry Players in Analytical Instrumentation
The Raman Spectroscopy versus MALDI-TOF proteomic analysis market is currently in a growth phase, with an estimated global market size of $2-3 billion and expanding at 8-10% annually. The competitive landscape features established analytical instrument manufacturers like Shimadzu Corp. and Thermo Finnigan (Thermo Fisher Scientific) dominating with mature MALDI-TOF technologies, while specialized players such as Bioyong Technology, Rongzhi Biotechnology, and Zhejiang Digena are advancing innovative applications. Technical maturity varies significantly between platforms - MALDI-TOF has reached commercial maturity with widespread clinical adoption, particularly in microbial identification, while Raman spectroscopy remains in earlier commercialization stages despite offering complementary proteomic insights with non-destructive sampling advantages.
Shimadzu Corp.
Technical Solution: Shimadzu has developed advanced Raman spectroscopy systems with their RAMAN-11 platform that offers high-resolution molecular fingerprinting capabilities. Their technology integrates confocal Raman microscopy with automated sampling for rapid proteomic analysis. For comparative proteomics, Shimadzu has engineered hybrid systems that combine Raman spectroscopy with their MALDI-TOF platforms (such as the AXIMA series), allowing researchers to leverage both technologies' strengths. Their proprietary software algorithms enable direct comparison between Raman spectral data and MALDI-TOF mass spectra, providing complementary structural and mass information. Shimadzu's systems demonstrate particular strength in analyzing intact proteins and maintaining sample integrity through non-destructive Raman analysis followed by MALDI-TOF for precise mass determination.
Strengths: Superior optical engineering providing exceptional spectral resolution in Raman systems; integrated workflows that allow sequential or parallel analysis using both technologies; comprehensive software for comparative data analysis. Weaknesses: Higher initial investment costs compared to standalone systems; complex calibration requirements when switching between modalities; steeper learning curve for operators.
Thermo Finnigan Corp.
Technical Solution: Thermo Finnigan (now part of Thermo Fisher Scientific) has pioneered integrated analytical solutions comparing Raman spectroscopy with MALDI-TOF for comprehensive proteomic analysis. Their DXR Raman microscope systems provide spatial resolution down to 1μm with spectral resolution of 0.5cm⁻¹, enabling detailed protein structural analysis. When combined with their high-performance MALDI-TOF systems like the LTQ Orbitrap, researchers can achieve multi-dimensional protein characterization. Their proprietary software platform enables direct correlation between Raman-derived structural information and MALDI-identified peptide fragments. Thermo's approach emphasizes complementary data acquisition, where Raman spectroscopy provides detailed secondary structure information (α-helices, β-sheets) while MALDI-TOF delivers precise mass identification and post-translational modification mapping. This dual-technology approach has demonstrated particular value in analyzing complex protein mixtures where traditional single-method approaches fail to provide complete characterization.
Strengths: Exceptional mass accuracy in MALDI-TOF systems (sub-ppm); sophisticated data integration software; comprehensive workflow solutions from sample preparation through analysis. Weaknesses: Higher operational costs; requires significant expertise to fully leverage both technologies; limited throughput compared to standalone MALDI-TOF for high-volume applications.
Core Innovations in Spectroscopic Protein Detection
method
PatentInactiveUS20200072839A1
Innovation
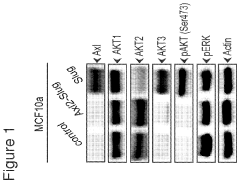
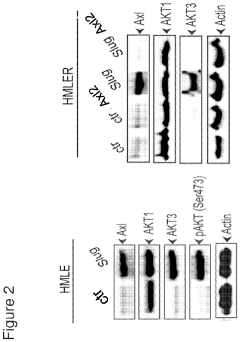
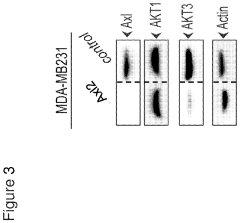
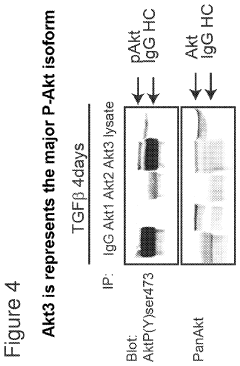
- Identifying Akt3 as a central regulator of EMT and cancer stem cell traits, with methods to inhibit Akt3 activity using pharmaceutical compounds, thereby reversing EMT and CSC traits, and using Akt3 as a biomarker for Axl signaling to predict cancer outcomes and treatment responses.
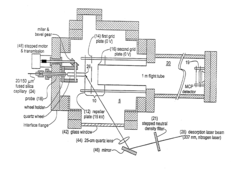
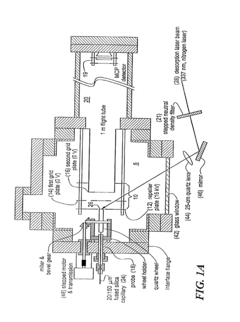
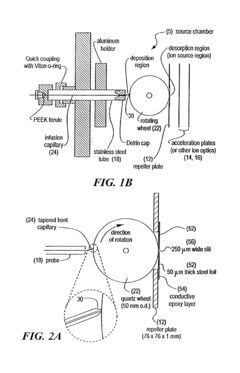
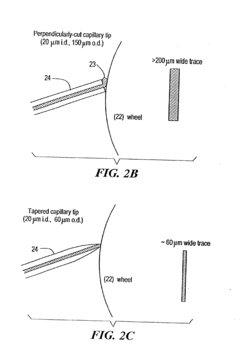
On-line and off-line deposition of liquid samples for matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectroscopy
PatentInactiveUS6825463B2
Innovation
- A universal interface and sample load mechanism for continuous on-line liquid sample introduction into a time-of-flight mass spectrometer at subatmospheric pressure, using a rotating quartz wheel for sample deposition and desorption, allowing for high-throughput analysis with improved reproducibility and sensitivity.
Cost-Benefit Analysis of Competing Technologies
When evaluating Raman Spectroscopy versus MALDI-TOF for proteomic analysis, cost-benefit considerations are crucial for research institutions and clinical laboratories making technology investment decisions. Initial acquisition costs represent a significant difference between these technologies, with Raman systems typically ranging from $50,000 to $150,000, while MALDI-TOF systems generally command $200,000 to $500,000, depending on specifications and capabilities.
Operational expenses also differ substantially. Raman spectroscopy offers lower per-sample costs, estimated at $2-5 per analysis, primarily due to minimal sample preparation requirements and reusable substrates. In contrast, MALDI-TOF incurs costs of approximately $10-20 per sample, driven by matrix reagents, target plates, and calibration standards. This cost differential becomes particularly significant in high-throughput environments processing thousands of samples annually.
Maintenance requirements present another important consideration. Raman systems typically require annual maintenance contracts of $5,000-10,000, with laser components needing replacement every 3-5 years at approximately $15,000. MALDI-TOF systems demand more extensive maintenance, with annual service contracts ranging from $20,000-40,000 and vacuum systems requiring periodic attention.
Personnel training investments vary between technologies. Raman spectroscopy generally requires 1-2 weeks of operator training, while MALDI-TOF demands 2-4 weeks of specialized training due to its more complex sample preparation protocols and data interpretation requirements. This translates to different staffing costs and implementation timelines.
Return on investment calculations must consider throughput capabilities. MALDI-TOF demonstrates superior processing capacity, handling 96-384 samples per batch with analysis times of seconds per sample. Raman systems typically process samples individually with 1-5 minutes per analysis. For high-volume laboratories, MALDI-TOF's throughput advantage may offset its higher initial investment through operational efficiency.
Data quality and analytical depth represent the ultimate value proposition. While MALDI-TOF excels in protein identification with detection limits in the femtomole range and comprehensive database support, Raman spectroscopy offers unique structural insights and non-destructive analysis capabilities. The optimal technology choice ultimately depends on specific application requirements, sample volumes, and available resources.
Operational expenses also differ substantially. Raman spectroscopy offers lower per-sample costs, estimated at $2-5 per analysis, primarily due to minimal sample preparation requirements and reusable substrates. In contrast, MALDI-TOF incurs costs of approximately $10-20 per sample, driven by matrix reagents, target plates, and calibration standards. This cost differential becomes particularly significant in high-throughput environments processing thousands of samples annually.
Maintenance requirements present another important consideration. Raman systems typically require annual maintenance contracts of $5,000-10,000, with laser components needing replacement every 3-5 years at approximately $15,000. MALDI-TOF systems demand more extensive maintenance, with annual service contracts ranging from $20,000-40,000 and vacuum systems requiring periodic attention.
Personnel training investments vary between technologies. Raman spectroscopy generally requires 1-2 weeks of operator training, while MALDI-TOF demands 2-4 weeks of specialized training due to its more complex sample preparation protocols and data interpretation requirements. This translates to different staffing costs and implementation timelines.
Return on investment calculations must consider throughput capabilities. MALDI-TOF demonstrates superior processing capacity, handling 96-384 samples per batch with analysis times of seconds per sample. Raman systems typically process samples individually with 1-5 minutes per analysis. For high-volume laboratories, MALDI-TOF's throughput advantage may offset its higher initial investment through operational efficiency.
Data quality and analytical depth represent the ultimate value proposition. While MALDI-TOF excels in protein identification with detection limits in the femtomole range and comprehensive database support, Raman spectroscopy offers unique structural insights and non-destructive analysis capabilities. The optimal technology choice ultimately depends on specific application requirements, sample volumes, and available resources.
Integration Potential with Existing Laboratory Workflows
The integration of advanced analytical technologies into existing laboratory workflows represents a critical consideration for research facilities and clinical laboratories. When evaluating Raman spectroscopy and MALDI-TOF for proteomic analysis, their compatibility with established protocols and infrastructure significantly impacts adoption decisions. Both technologies offer distinct integration pathways that merit careful assessment.
Raman spectroscopy demonstrates considerable flexibility in laboratory integration, requiring minimal sample preparation compared to many proteomic techniques. Modern Raman systems can analyze samples in various states—solid, liquid, or gas—often without destructive processing, allowing for subsequent analysis by complementary methods. This non-destructive nature creates valuable workflow efficiencies where sample conservation is paramount. Additionally, recent developments in portable and benchtop Raman instruments facilitate point-of-need analysis, enabling decentralized testing protocols that can reduce bottlenecks in centralized laboratory facilities.
MALDI-TOF systems, while requiring more specialized infrastructure, have achieved significant integration into clinical microbiology workflows, establishing precedent for broader proteomic applications. The technology's high-throughput capabilities align well with batch processing environments, and automated sample preparation platforms have substantially reduced hands-on time requirements. Many clinical laboratories have already invested in MALDI-TOF infrastructure for microbial identification, creating potential synergies for expanded proteomic applications without significant additional capital investment.
Informatics integration represents another crucial dimension for both technologies. MALDI-TOF benefits from mature bioinformatic pipelines and extensive spectral libraries, particularly for protein identification applications. These established databases facilitate straightforward integration with laboratory information management systems (LIMS) and electronic health records. Conversely, Raman spectroscopy often requires more customized data processing workflows, though recent advances in machine learning algorithms have improved automated spectral interpretation capabilities.
Regulatory considerations also influence integration potential. MALDI-TOF has achieved regulatory clearance for specific clinical applications, establishing pathways for laboratory adoption within regulated environments. Raman spectroscopy, while widely used in research settings, has fewer cleared clinical applications, potentially requiring additional validation steps for certain workflow integrations.
Cost-effectiveness calculations must account for both initial implementation expenses and ongoing operational requirements. Raman systems typically demand lower consumable costs and maintenance requirements, potentially offering advantages for laboratories with limited recurring budgets. MALDI-TOF systems, while often requiring higher consumable expenses, may leverage existing infrastructure in laboratories already utilizing the technology for microbial identification.
Raman spectroscopy demonstrates considerable flexibility in laboratory integration, requiring minimal sample preparation compared to many proteomic techniques. Modern Raman systems can analyze samples in various states—solid, liquid, or gas—often without destructive processing, allowing for subsequent analysis by complementary methods. This non-destructive nature creates valuable workflow efficiencies where sample conservation is paramount. Additionally, recent developments in portable and benchtop Raman instruments facilitate point-of-need analysis, enabling decentralized testing protocols that can reduce bottlenecks in centralized laboratory facilities.
MALDI-TOF systems, while requiring more specialized infrastructure, have achieved significant integration into clinical microbiology workflows, establishing precedent for broader proteomic applications. The technology's high-throughput capabilities align well with batch processing environments, and automated sample preparation platforms have substantially reduced hands-on time requirements. Many clinical laboratories have already invested in MALDI-TOF infrastructure for microbial identification, creating potential synergies for expanded proteomic applications without significant additional capital investment.
Informatics integration represents another crucial dimension for both technologies. MALDI-TOF benefits from mature bioinformatic pipelines and extensive spectral libraries, particularly for protein identification applications. These established databases facilitate straightforward integration with laboratory information management systems (LIMS) and electronic health records. Conversely, Raman spectroscopy often requires more customized data processing workflows, though recent advances in machine learning algorithms have improved automated spectral interpretation capabilities.
Regulatory considerations also influence integration potential. MALDI-TOF has achieved regulatory clearance for specific clinical applications, establishing pathways for laboratory adoption within regulated environments. Raman spectroscopy, while widely used in research settings, has fewer cleared clinical applications, potentially requiring additional validation steps for certain workflow integrations.
Cost-effectiveness calculations must account for both initial implementation expenses and ongoing operational requirements. Raman systems typically demand lower consumable costs and maintenance requirements, potentially offering advantages for laboratories with limited recurring budgets. MALDI-TOF systems, while often requiring higher consumable expenses, may leverage existing infrastructure in laboratories already utilizing the technology for microbial identification.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!