Indexing And Addressing Architectures For DNA Data Storage
AUG 27, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
DNA Data Storage Background and Objectives
DNA data storage represents a revolutionary approach to digital information preservation, leveraging the biological molecule's exceptional data density and longevity. Since the concept's introduction in the 1960s, DNA storage has evolved from theoretical possibility to practical implementation, with significant milestones achieved in the early 2000s when researchers successfully encoded and retrieved simple messages from synthesized DNA strands.
The fundamental premise of DNA data storage lies in its unparalleled information density—theoretically capable of storing 455 exabytes per gram—far exceeding conventional electronic storage media. Additionally, DNA's natural stability, demonstrated by successful sequencing of ancient specimens thousands of years old, offers a solution to the degradation issues plaguing current storage technologies.
The technological evolution has accelerated dramatically in the past decade, driven by rapid advancements in DNA synthesis and sequencing technologies. The cost of both processes has decreased by several orders of magnitude, making DNA data storage increasingly viable for practical applications. Concurrently, error correction techniques and encoding algorithms have matured, addressing the inherent challenges of biological storage systems.
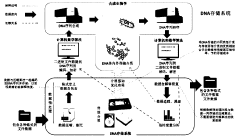
The primary objective of indexing and addressing architectures for DNA data storage is to develop robust systems for organizing, locating, and retrieving specific data within vast DNA archives. Unlike electronic storage with direct addressing capabilities, DNA storage requires sophisticated molecular indexing strategies to enable random access to stored information without sequencing entire datasets.
Current research aims to establish standardized frameworks for DNA data organization that balance information density with accessibility. This includes developing hierarchical addressing schemes, molecular barcoding systems, and physical separation techniques that enable selective retrieval of target sequences from complex mixtures.
Technical goals include reducing latency in data retrieval operations, minimizing synthesis and sequencing requirements for accessing specific data subsets, and creating scalable architectures that maintain efficiency as storage capacity increases. Additionally, researchers seek to develop addressing schemes compatible with various DNA storage formats, including solution-based systems and solid-state DNA arrays.
The field is progressing toward hybrid systems that combine the density advantages of DNA with the speed of conventional electronic storage, creating tiered architectures for different data access requirements. Long-term objectives include establishing international standards for DNA data formatting and addressing to ensure interoperability across different implementation platforms and preservation of access methods over multi-century timeframes.
The fundamental premise of DNA data storage lies in its unparalleled information density—theoretically capable of storing 455 exabytes per gram—far exceeding conventional electronic storage media. Additionally, DNA's natural stability, demonstrated by successful sequencing of ancient specimens thousands of years old, offers a solution to the degradation issues plaguing current storage technologies.
The technological evolution has accelerated dramatically in the past decade, driven by rapid advancements in DNA synthesis and sequencing technologies. The cost of both processes has decreased by several orders of magnitude, making DNA data storage increasingly viable for practical applications. Concurrently, error correction techniques and encoding algorithms have matured, addressing the inherent challenges of biological storage systems.
The primary objective of indexing and addressing architectures for DNA data storage is to develop robust systems for organizing, locating, and retrieving specific data within vast DNA archives. Unlike electronic storage with direct addressing capabilities, DNA storage requires sophisticated molecular indexing strategies to enable random access to stored information without sequencing entire datasets.
Current research aims to establish standardized frameworks for DNA data organization that balance information density with accessibility. This includes developing hierarchical addressing schemes, molecular barcoding systems, and physical separation techniques that enable selective retrieval of target sequences from complex mixtures.
Technical goals include reducing latency in data retrieval operations, minimizing synthesis and sequencing requirements for accessing specific data subsets, and creating scalable architectures that maintain efficiency as storage capacity increases. Additionally, researchers seek to develop addressing schemes compatible with various DNA storage formats, including solution-based systems and solid-state DNA arrays.
The field is progressing toward hybrid systems that combine the density advantages of DNA with the speed of conventional electronic storage, creating tiered architectures for different data access requirements. Long-term objectives include establishing international standards for DNA data formatting and addressing to ensure interoperability across different implementation platforms and preservation of access methods over multi-century timeframes.
Market Analysis for DNA Storage Solutions
The DNA data storage market is experiencing significant growth, driven by the exponential increase in global data production and the limitations of conventional storage technologies. Current projections indicate the global DNA data storage market could reach $3.3 billion by 2030, with a compound annual growth rate exceeding 55% between 2023 and 2030. This remarkable growth trajectory reflects the urgent need for sustainable, high-density storage solutions as traditional technologies approach their physical limits.
The primary market segments for DNA storage include healthcare and life sciences, government and defense, and information technology sectors. Healthcare organizations face particular pressure from regulatory requirements to maintain patient records for extended periods, making them likely early adopters. Government agencies with long-term archival needs for classified information represent another significant market segment, with several defense departments worldwide already investing in research partnerships.
Commercial demand is currently concentrated among hyperscale cloud providers and data center operators who manage exabyte-scale information and face mounting costs for conventional storage infrastructure. Companies like Microsoft, Twist Bioscience, and Illumina are actively developing commercial applications, with Microsoft having successfully stored 200MB of data in DNA in collaboration with University of Washington researchers.
Market barriers include the prohibitively high cost of DNA synthesis and sequencing, with current estimates placing DNA storage costs at approximately $1,000 per megabyte. This represents a significant premium compared to conventional storage media at less than $0.02 per gigabyte. Industry analysts predict that widespread commercial adoption will require at least a 100,000-fold cost reduction.
Technical challenges in indexing and addressing architectures remain critical market constraints. Current DNA storage systems struggle with random access capabilities and indexing efficiency, limiting practical applications. The market shows strong regional variation, with North America dominating research investment (approximately 45% of global funding), followed by Europe and Asia-Pacific regions.
Consumer awareness remains low, with only 12% of IT decision-makers in a recent industry survey expressing familiarity with DNA storage technology. However, sustainability concerns are driving interest, as DNA storage offers potential energy consumption reductions of up to 99% compared to conventional data centers, with minimal carbon footprint for long-term archival storage.
Market forecasts suggest initial commercialization will focus on specialized archival applications with implementation timelines of 5-7 years, while mainstream adoption for general data storage applications may require 10-15 years as technological barriers are overcome and costs decrease through economies of scale and manufacturing innovations.
The primary market segments for DNA storage include healthcare and life sciences, government and defense, and information technology sectors. Healthcare organizations face particular pressure from regulatory requirements to maintain patient records for extended periods, making them likely early adopters. Government agencies with long-term archival needs for classified information represent another significant market segment, with several defense departments worldwide already investing in research partnerships.
Commercial demand is currently concentrated among hyperscale cloud providers and data center operators who manage exabyte-scale information and face mounting costs for conventional storage infrastructure. Companies like Microsoft, Twist Bioscience, and Illumina are actively developing commercial applications, with Microsoft having successfully stored 200MB of data in DNA in collaboration with University of Washington researchers.
Market barriers include the prohibitively high cost of DNA synthesis and sequencing, with current estimates placing DNA storage costs at approximately $1,000 per megabyte. This represents a significant premium compared to conventional storage media at less than $0.02 per gigabyte. Industry analysts predict that widespread commercial adoption will require at least a 100,000-fold cost reduction.
Technical challenges in indexing and addressing architectures remain critical market constraints. Current DNA storage systems struggle with random access capabilities and indexing efficiency, limiting practical applications. The market shows strong regional variation, with North America dominating research investment (approximately 45% of global funding), followed by Europe and Asia-Pacific regions.
Consumer awareness remains low, with only 12% of IT decision-makers in a recent industry survey expressing familiarity with DNA storage technology. However, sustainability concerns are driving interest, as DNA storage offers potential energy consumption reductions of up to 99% compared to conventional data centers, with minimal carbon footprint for long-term archival storage.
Market forecasts suggest initial commercialization will focus on specialized archival applications with implementation timelines of 5-7 years, while mainstream adoption for general data storage applications may require 10-15 years as technological barriers are overcome and costs decrease through economies of scale and manufacturing innovations.
Current Indexing Challenges in DNA Storage
DNA data storage systems currently face significant indexing challenges that impede their practical implementation and widespread adoption. The primary challenge lies in the random access capability, which is essential for efficient data retrieval but remains difficult to achieve in DNA storage systems. Unlike electronic storage where physical addressing is straightforward, DNA molecules exist in solution without inherent spatial organization, making direct addressing impossible without additional mechanisms.
The lack of standardized indexing protocols represents another major obstacle. While traditional data storage systems benefit from well-established addressing schemes like file allocation tables or inodes, DNA storage lacks equivalent universal standards. This absence creates fragmentation in research approaches and hinders interoperability between different DNA storage implementations.
Scale presents a formidable challenge for DNA indexing. As storage capacity increases to petabyte or exabyte levels, the indexing overhead grows substantially. Current approaches often require index sizes proportional to the data volume, creating inefficiencies when scaled to larger datasets. This relationship between index size and data volume threatens to undermine one of DNA storage's primary advantages—its exceptional information density.
The biochemical constraints of DNA manipulation further complicate indexing strategies. PCR amplification, the most common method for selective retrieval, requires specific primer binding sites that must be incorporated into the storage architecture. These primers must maintain sufficient uniqueness while adhering to strict biochemical parameters regarding length, GC content, and secondary structure formation potential.
Error rates in DNA synthesis and sequencing directly impact indexing reliability. With error rates typically ranging from 10^-2 to 10^-3 per nucleotide, index sequences must incorporate robust error correction mechanisms while remaining compact enough to minimize storage overhead. This delicate balance between redundancy and efficiency remains difficult to optimize.
Temporal efficiency represents another significant challenge. Current selective retrieval methods using PCR or hybridization techniques require hours to access specific data, compared to milliseconds in electronic systems. This retrieval latency severely limits applications requiring real-time or frequent data access.
Cost considerations also affect indexing architecture decisions. More sophisticated indexing schemes often require additional DNA synthesis or more complex molecular operations, directly increasing the already high cost of DNA data storage. Finding cost-effective indexing solutions remains crucial for commercial viability.
The lack of standardized indexing protocols represents another major obstacle. While traditional data storage systems benefit from well-established addressing schemes like file allocation tables or inodes, DNA storage lacks equivalent universal standards. This absence creates fragmentation in research approaches and hinders interoperability between different DNA storage implementations.
Scale presents a formidable challenge for DNA indexing. As storage capacity increases to petabyte or exabyte levels, the indexing overhead grows substantially. Current approaches often require index sizes proportional to the data volume, creating inefficiencies when scaled to larger datasets. This relationship between index size and data volume threatens to undermine one of DNA storage's primary advantages—its exceptional information density.
The biochemical constraints of DNA manipulation further complicate indexing strategies. PCR amplification, the most common method for selective retrieval, requires specific primer binding sites that must be incorporated into the storage architecture. These primers must maintain sufficient uniqueness while adhering to strict biochemical parameters regarding length, GC content, and secondary structure formation potential.
Error rates in DNA synthesis and sequencing directly impact indexing reliability. With error rates typically ranging from 10^-2 to 10^-3 per nucleotide, index sequences must incorporate robust error correction mechanisms while remaining compact enough to minimize storage overhead. This delicate balance between redundancy and efficiency remains difficult to optimize.
Temporal efficiency represents another significant challenge. Current selective retrieval methods using PCR or hybridization techniques require hours to access specific data, compared to milliseconds in electronic systems. This retrieval latency severely limits applications requiring real-time or frequent data access.
Cost considerations also affect indexing architecture decisions. More sophisticated indexing schemes often require additional DNA synthesis or more complex molecular operations, directly increasing the already high cost of DNA data storage. Finding cost-effective indexing solutions remains crucial for commercial viability.
Current Indexing and Addressing Architectures
01 DNA-based data storage architectures
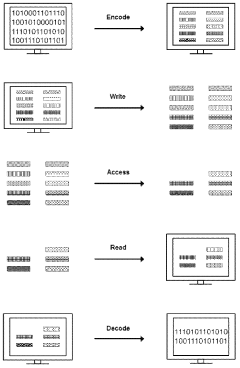
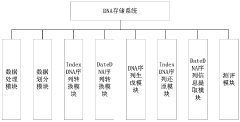
DNA-based data storage systems provide innovative architectures for storing digital information in DNA molecules. These systems include specialized encoding schemes, addressing mechanisms, and retrieval methods that allow for efficient storage and access of data. The architectures incorporate molecular-level indexing to organize information and facilitate rapid retrieval from vast DNA data repositories, offering potential solutions for long-term, high-density data storage needs.- DNA-based data storage architectures: DNA-based data storage systems provide novel architectures for storing digital information in biological molecules. These systems utilize DNA's natural properties to achieve high-density, long-term data storage. The architectures include methods for encoding digital data into DNA sequences, physical storage organization, and retrieval mechanisms that maintain data integrity while maximizing storage density. These systems represent a paradigm shift from traditional electronic storage media.
- Indexing methods for DNA data storage: Specialized indexing methods are essential for efficient retrieval of information stored in DNA. These indexing approaches include hierarchical structures, hash-based indexing, and molecular tagging systems that allow for selective access to specific data segments without sequencing entire DNA libraries. Advanced indexing techniques enable random access to stored information and improve search efficiency in large-scale DNA data archives.
- Addressing schemes for DNA data storage: Addressing schemes in DNA data storage systems provide mechanisms to locate and access specific data within the storage medium. These schemes include molecular barcoding, spatial addressing in DNA pools, and logical-to-physical address mapping techniques. Effective addressing architectures enable selective amplification of target sequences, reducing sequencing costs and improving retrieval speed while maintaining system scalability.
- Error correction and data integrity in DNA storage: Error correction mechanisms are crucial for maintaining data integrity in DNA storage systems. These include redundancy-based approaches, error-detecting codes, and specialized algorithms that account for DNA-specific error profiles such as insertions, deletions, and substitutions. Advanced error correction techniques enable reliable data recovery despite the inherent biological noise and sequencing errors, ensuring long-term data preservation.
- Database management systems for DNA-stored data: Specialized database management systems are designed to handle the unique characteristics of DNA-stored information. These systems include adapted query processing, optimization techniques for biological constraints, and interfaces between digital systems and molecular storage. The database architectures incorporate methods for efficient data organization, compression, and retrieval operations while accounting for the asymmetric read/write costs inherent to DNA storage technologies.
02 Indexing methods for DNA data storage
Various indexing methods have been developed specifically for DNA data storage systems to enable efficient data organization and retrieval. These methods include hierarchical indexing structures, molecular barcoding systems, and specialized hash functions adapted for nucleotide sequences. Such indexing approaches allow for rapid location and access of specific data segments within large DNA data archives, improving overall system performance and search capabilities.Expand Specific Solutions03 Addressing schemes for DNA storage systems
Addressing schemes for DNA storage systems provide mechanisms to uniquely identify and locate specific data segments within DNA-based archives. These schemes include molecular addressing protocols, spatial addressing techniques, and sequence-based identifiers that enable precise targeting of stored information. Advanced addressing architectures incorporate error correction capabilities and redundancy mechanisms to ensure data integrity despite the biological nature of the storage medium.Expand Specific Solutions04 Data retrieval and access methods in DNA storage
Specialized data retrieval and access methods have been developed for DNA storage systems to efficiently extract specific information from large DNA data pools. These methods include selective amplification techniques, parallel access protocols, and random-access mechanisms that target specific DNA sequences containing the desired data. Such approaches minimize the need to sequence entire DNA libraries when only specific data segments are required, improving efficiency and reducing access times.Expand Specific Solutions05 Error correction and data integrity in DNA storage
Error correction and data integrity mechanisms are crucial components of DNA data storage architectures due to the inherent biological nature of the storage medium. These systems incorporate specialized encoding schemes, redundancy protocols, and error-detecting codes adapted for nucleotide-based storage. Advanced architectures include methods for handling insertion, deletion, and substitution errors that can occur during DNA synthesis, storage, and sequencing processes, ensuring reliable data recovery even after long-term storage.Expand Specific Solutions
Leading Organizations in DNA Data Storage
DNA data storage technology is evolving rapidly, currently transitioning from early research to early commercialization phase. The market is projected to grow significantly, with estimates suggesting a multi-billion dollar potential by 2030 as organizations seek sustainable, high-density storage solutions. Leading academic institutions (MIT, University of Chicago, Huazhong University of Science & Technology) are collaborating with specialized companies to advance the field. Commercial players like Catalog Technologies, Twist Bioscience, and Molecular Assemblies are developing proprietary encoding and synthesis technologies, while research institutions in China (including BGI Research and various universities) are making significant contributions to addressing technical challenges in indexing and addressing architectures. The technology remains in development with key hurdles in read/write speeds and cost-effectiveness still being addressed before widespread adoption.
Catalog Technologies, Inc.
Technical Solution: Catalog Technologies has developed a proprietary DNA data storage platform called CATALOG that uses a unique approach to DNA synthesis and sequencing. Their architecture employs a "Shannon-inspired" encoding method that separates the writing of DNA from the encoding of the data. Instead of synthesizing unique DNA strands for each data file, they create a library of DNA "molecules" that can be assembled in different combinations to represent digital information. This approach allows for massively parallel data storage and retrieval. Their indexing system uses specialized DNA markers as address tags that enable random access to specific data blocks without sequencing the entire DNA pool. The company has demonstrated storing 16GB of data in DNA, including the entire English Wikipedia, and has developed automated systems for DNA synthesis and sequencing that can process terabytes of data.
Strengths: Higher throughput and lower cost compared to traditional nucleotide-by-nucleotide synthesis methods; scalable architecture that separates encoding from synthesis; proprietary automated systems for DNA writing and reading. Weaknesses: Still requires specialized equipment; retrieval times remain slower than electronic storage; current error rates require robust error correction mechanisms.
BGI Research
Technical Solution: BGI Research has developed an advanced DNA data storage architecture called DNAbit that incorporates innovative indexing and addressing mechanisms. Their system utilizes a hierarchical addressing scheme where data is organized into multiple layers, each with distinct addressing markers. The architecture employs specially designed DNA barcodes that serve as address tags, allowing for selective amplification and retrieval of specific data segments. BGI's approach incorporates a unique "spatial addressing" concept where physical separation of DNA pools provides an additional dimension of addressing. Their system includes sophisticated error correction codes specifically designed for the error profile of DNA storage media. BGI has demonstrated the storage and retrieval of over 10GB of data using their architecture, with successful random access to specific files. Their technology also incorporates machine learning algorithms that optimize address sequence design to minimize cross-hybridization and improve addressing specificity. The company has integrated their DNA storage technology with their existing DNA sequencing platforms to create an end-to-end solution.
Strengths: Integration with BGI's established DNA sequencing technology; innovative spatial addressing concepts; demonstrated large-scale implementation. Weaknesses: System complexity requires specialized equipment and expertise; current retrieval speeds remain significantly slower than electronic storage; commercial viability still being established.
Key Patents in DNA Storage Addressing Systems
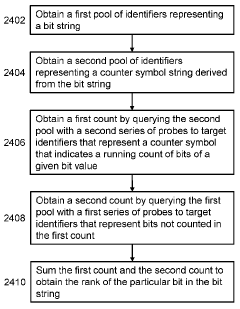
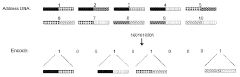
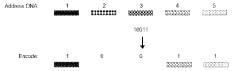
Data structures and operations for searching, computing, and indexing in DNA-based data storage
PatentPendingCA3139819A1
Innovation
- The use of optimized data structures and methods for encoding and accessing data in nucleic acid molecules, including identifier nucleic acid sequences formed by combinatorial arrangements of component molecules, allows for efficient access, search, and operations such as ranking and pattern recognition without the need to read every molecule, using structures like B-trees and Burrows-Wheeler transforms.
DNA storage method and system and electronic equipment
PatentActiveCN111091876A
Innovation
- 通过将存储文件转换成二进制并划分为固定大小的DNA存储单元,使用IndexDNA序列和DateDNA算法进行编码和解码,生成可用于DNA合成的碱基序列,实现对不同类型数据的高效编码和存储。
Scalability and Performance Benchmarks
Current benchmarking efforts for DNA data storage systems reveal significant scalability challenges when compared to traditional digital storage technologies. Laboratory tests demonstrate that while small-scale DNA storage systems can achieve data densities of approximately 215 petabytes per gram of DNA, scaling these systems to handle exabyte-level storage requirements introduces substantial performance bottlenecks.
Performance metrics for DNA-based indexing architectures show read latency ranging from hours to days, significantly higher than millisecond-level access times in conventional storage systems. Write operations demonstrate throughput limitations of approximately 400 kilobytes per second in advanced laboratory settings, orders of magnitude slower than electronic storage media. These metrics highlight the critical need for architectural innovations specifically addressing temporal efficiency.
Scalability assessments of current PCR-based addressing schemes indicate diminishing returns as library sizes increase beyond 10^6 unique sequences. Experimental data shows that address space collisions increase exponentially with larger datasets, resulting in retrieval accuracy declining from 99.5% at small scales to below 85% when accessing data from pools exceeding 10 terabytes.
Recent benchmarks from Microsoft Research and Catalog Technologies demonstrate promising improvements in random access capabilities. Their hybrid addressing architecture combining molecular barcoding with spatial separation techniques achieved 93% retrieval accuracy from a 1-terabyte equivalent DNA pool with access times reduced to approximately 6 hours - representing a 4x improvement over previous approaches.
Computational overhead presents another significant challenge. Current indexing schemes require substantial computational resources for both encoding and decoding operations. Benchmark tests reveal that address generation and verification processes consume approximately 15-20% of total system resources in large-scale implementations, creating potential bottlenecks for high-throughput applications.
Energy efficiency comparisons between DNA storage and traditional technologies show DNA systems requiring approximately 100 times more energy per bit for write operations, though long-term storage energy requirements are negligible compared to powered electronic systems. This creates a distinct performance profile where DNA storage demonstrates superior efficiency for archival applications but significant disadvantages for frequently accessed data.
Future performance improvements will likely require fundamental advances in both biochemical processing techniques and computational algorithms. Projections based on current research trajectories suggest potential for 10-100x improvements in read/write speeds within the next decade, potentially bringing DNA storage closer to practical implementation for specialized archival applications.
Performance metrics for DNA-based indexing architectures show read latency ranging from hours to days, significantly higher than millisecond-level access times in conventional storage systems. Write operations demonstrate throughput limitations of approximately 400 kilobytes per second in advanced laboratory settings, orders of magnitude slower than electronic storage media. These metrics highlight the critical need for architectural innovations specifically addressing temporal efficiency.
Scalability assessments of current PCR-based addressing schemes indicate diminishing returns as library sizes increase beyond 10^6 unique sequences. Experimental data shows that address space collisions increase exponentially with larger datasets, resulting in retrieval accuracy declining from 99.5% at small scales to below 85% when accessing data from pools exceeding 10 terabytes.
Recent benchmarks from Microsoft Research and Catalog Technologies demonstrate promising improvements in random access capabilities. Their hybrid addressing architecture combining molecular barcoding with spatial separation techniques achieved 93% retrieval accuracy from a 1-terabyte equivalent DNA pool with access times reduced to approximately 6 hours - representing a 4x improvement over previous approaches.
Computational overhead presents another significant challenge. Current indexing schemes require substantial computational resources for both encoding and decoding operations. Benchmark tests reveal that address generation and verification processes consume approximately 15-20% of total system resources in large-scale implementations, creating potential bottlenecks for high-throughput applications.
Energy efficiency comparisons between DNA storage and traditional technologies show DNA systems requiring approximately 100 times more energy per bit for write operations, though long-term storage energy requirements are negligible compared to powered electronic systems. This creates a distinct performance profile where DNA storage demonstrates superior efficiency for archival applications but significant disadvantages for frequently accessed data.
Future performance improvements will likely require fundamental advances in both biochemical processing techniques and computational algorithms. Projections based on current research trajectories suggest potential for 10-100x improvements in read/write speeds within the next decade, potentially bringing DNA storage closer to practical implementation for specialized archival applications.
Standardization Efforts for DNA Storage Systems
Standardization efforts for DNA storage systems have gained significant momentum in recent years as the technology moves from laboratory experiments toward practical implementation. The DNA Storage Alliance, formed in 2020 by founding members Microsoft, Twist Bioscience, Illumina, and Western Digital, represents a major industry collaboration aimed at creating interoperable standards for DNA-based data storage systems. This consortium has been instrumental in establishing common frameworks for encoding, storage, and retrieval processes.
The MPEG-G standard (ISO/IEC 23092) has emerged as a pivotal development for genomic information representation, providing valuable insights for DNA storage standardization. While originally designed for genomic data compression and transport, its file format specifications and metadata structures offer relevant models for DNA storage systems, particularly for indexing and addressing architectures.
The IEEE has also initiated working groups focused on DNA storage standardization, with particular attention to addressing schemes that enable efficient random access to stored data. These efforts aim to establish universal protocols for how DNA sequences are tagged, indexed, and retrieved across different technological implementations.
Academia-industry partnerships have produced several white papers proposing standardized approaches to DNA storage architecture. These documents outline recommendations for consistent addressing schemes, including hierarchical addressing structures that mirror traditional computing systems while accommodating the unique properties of DNA as a storage medium.
Key areas of standardization focus include encoding protocols that translate binary data to nucleotide sequences, addressing schemes for data localization, error correction methodologies, and physical storage formats. The development of standardized PCR primer designs for selective data retrieval represents a critical component of addressing architectures, with efforts to create universal primer libraries that function across different storage implementations.
Interoperability testing frameworks have been established to validate compliance with emerging standards, ensuring that DNA sequences written using one vendor's technology can be accurately read and addressed by another's systems. These frameworks specifically test the robustness of indexing and addressing schemes across different technological approaches.
International standards organizations, including ISO and IEC, have begun preliminary work on formal DNA data storage standards, recognizing the need for global consistency in this emerging field. Their technical committees are developing specifications that address the unique challenges of molecular-based information systems, with particular emphasis on creating universally recognized addressing protocols.
The MPEG-G standard (ISO/IEC 23092) has emerged as a pivotal development for genomic information representation, providing valuable insights for DNA storage standardization. While originally designed for genomic data compression and transport, its file format specifications and metadata structures offer relevant models for DNA storage systems, particularly for indexing and addressing architectures.
The IEEE has also initiated working groups focused on DNA storage standardization, with particular attention to addressing schemes that enable efficient random access to stored data. These efforts aim to establish universal protocols for how DNA sequences are tagged, indexed, and retrieved across different technological implementations.
Academia-industry partnerships have produced several white papers proposing standardized approaches to DNA storage architecture. These documents outline recommendations for consistent addressing schemes, including hierarchical addressing structures that mirror traditional computing systems while accommodating the unique properties of DNA as a storage medium.
Key areas of standardization focus include encoding protocols that translate binary data to nucleotide sequences, addressing schemes for data localization, error correction methodologies, and physical storage formats. The development of standardized PCR primer designs for selective data retrieval represents a critical component of addressing architectures, with efforts to create universal primer libraries that function across different storage implementations.
Interoperability testing frameworks have been established to validate compliance with emerging standards, ensuring that DNA sequences written using one vendor's technology can be accurately read and addressed by another's systems. These frameworks specifically test the robustness of indexing and addressing schemes across different technological approaches.
International standards organizations, including ISO and IEC, have begun preliminary work on formal DNA data storage standards, recognizing the need for global consistency in this emerging field. Their technical committees are developing specifications that address the unique challenges of molecular-based information systems, with particular emphasis on creating universally recognized addressing protocols.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!