Retrieval Accuracy And QA Pipelines In DNA Data Storage
AUG 27, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
DNA Data Storage Background and Objectives
DNA data storage represents a revolutionary approach to digital information preservation, leveraging the biological molecule's exceptional data density and longevity. Since the initial demonstration by Church et al. in 2012, who encoded a 659-kB book in DNA, this technology has evolved significantly. DNA's theoretical storage density of 455 exabytes per gram makes it an attractive solution for the exponentially growing global data, projected to reach 175 zettabytes by 2025.
The fundamental principle involves translating binary data into DNA nucleotide sequences (A, T, G, C), synthesizing these sequences, and later retrieving the information through sequencing and decoding. This process offers remarkable advantages: DNA can potentially store data for thousands of years under proper conditions, compared to conventional media's typical lifespan of 5-30 years, and achieves storage densities orders of magnitude greater than current technologies.
However, retrieval accuracy remains a critical challenge in DNA data storage systems. Current DNA synthesis and sequencing technologies introduce errors including insertions, deletions, and substitutions at rates that necessitate robust error correction mechanisms. The error rates in DNA storage systems typically range from 0.1% to 3%, depending on the specific technologies employed, significantly higher than electronic storage media's error rates of 10^-15 to 10^-18.
Quality assurance pipelines for DNA data storage must address multiple challenges across the workflow. During encoding, algorithms must generate DNA sequences that avoid problematic motifs like homopolymers and extreme GC content. Synthesis quality control requires methods to verify the accuracy of manufactured DNA sequences. Storage conditions must be optimized to minimize degradation, while sequencing processes need to maximize read accuracy and coverage depth.
The objective of current research is to develop integrated QA pipelines that can achieve bit error rates below 10^-9, making DNA storage practical for commercial applications. This involves innovations in encoding schemes, error correction codes specifically designed for the DNA channel, improved synthesis and sequencing technologies, and advanced computational methods for sequence reconstruction.
Achieving these objectives would position DNA data storage as a viable solution for archival storage needs, particularly for "cold" data that requires long-term preservation but infrequent access. The ultimate goal is to create systems that combine DNA's inherent advantages with retrieval reliability comparable to current digital storage technologies, enabling a new paradigm in information preservation.
The fundamental principle involves translating binary data into DNA nucleotide sequences (A, T, G, C), synthesizing these sequences, and later retrieving the information through sequencing and decoding. This process offers remarkable advantages: DNA can potentially store data for thousands of years under proper conditions, compared to conventional media's typical lifespan of 5-30 years, and achieves storage densities orders of magnitude greater than current technologies.
However, retrieval accuracy remains a critical challenge in DNA data storage systems. Current DNA synthesis and sequencing technologies introduce errors including insertions, deletions, and substitutions at rates that necessitate robust error correction mechanisms. The error rates in DNA storage systems typically range from 0.1% to 3%, depending on the specific technologies employed, significantly higher than electronic storage media's error rates of 10^-15 to 10^-18.
Quality assurance pipelines for DNA data storage must address multiple challenges across the workflow. During encoding, algorithms must generate DNA sequences that avoid problematic motifs like homopolymers and extreme GC content. Synthesis quality control requires methods to verify the accuracy of manufactured DNA sequences. Storage conditions must be optimized to minimize degradation, while sequencing processes need to maximize read accuracy and coverage depth.
The objective of current research is to develop integrated QA pipelines that can achieve bit error rates below 10^-9, making DNA storage practical for commercial applications. This involves innovations in encoding schemes, error correction codes specifically designed for the DNA channel, improved synthesis and sequencing technologies, and advanced computational methods for sequence reconstruction.
Achieving these objectives would position DNA data storage as a viable solution for archival storage needs, particularly for "cold" data that requires long-term preservation but infrequent access. The ultimate goal is to create systems that combine DNA's inherent advantages with retrieval reliability comparable to current digital storage technologies, enabling a new paradigm in information preservation.
Market Analysis for DNA Storage Solutions
The DNA data storage market is experiencing significant growth, driven by the exponential increase in global data production and the limitations of conventional storage technologies. Current projections estimate the DNA data storage market to reach approximately $3.3 billion by 2030, with a compound annual growth rate exceeding 55% between 2023 and 2030. This remarkable growth trajectory is fueled by the inherent advantages of DNA storage, including exceptional data density, longevity, and sustainability compared to traditional electronic media.
Key market segments for DNA data storage include archival storage for government agencies, scientific research institutions, healthcare organizations, and large technology companies with massive data retention requirements. These sectors generate enormous volumes of cold data that must be preserved for extended periods, making them ideal early adopters of DNA storage technology.
The market landscape is characterized by two distinct customer segments: technology pioneers willing to pay premium prices for cutting-edge solutions, and mainstream enterprises awaiting cost reductions before adoption. Current DNA synthesis and sequencing costs remain prohibitively expensive for widespread commercial deployment, with estimates suggesting storage costs of approximately $1,000 per megabyte. However, industry analysts predict these costs will decrease by orders of magnitude over the next decade.
Retrieval accuracy represents a critical market differentiator, with potential customers expressing concerns about data integrity and recovery reliability. Solutions demonstrating superior quality assurance pipelines that can guarantee near-perfect retrieval accuracy will command significant market premiums and accelerate adoption rates across conservative sectors like healthcare and financial services.
Regional market analysis indicates North America currently dominates the DNA data storage landscape, accounting for approximately 45% of global market share, followed by Europe at 30% and Asia-Pacific at 20%. The United States, United Kingdom, China, and Germany represent the primary innovation hubs, with substantial research investments and commercial activity.
Market barriers include high initial infrastructure costs, regulatory uncertainties regarding biosecurity, and integration challenges with existing data management systems. Despite these obstacles, the compelling value proposition of DNA storage—particularly its theoretical capacity to store all human knowledge in a volume smaller than a sugar cube—continues to drive substantial investment and market interest.
Key market segments for DNA data storage include archival storage for government agencies, scientific research institutions, healthcare organizations, and large technology companies with massive data retention requirements. These sectors generate enormous volumes of cold data that must be preserved for extended periods, making them ideal early adopters of DNA storage technology.
The market landscape is characterized by two distinct customer segments: technology pioneers willing to pay premium prices for cutting-edge solutions, and mainstream enterprises awaiting cost reductions before adoption. Current DNA synthesis and sequencing costs remain prohibitively expensive for widespread commercial deployment, with estimates suggesting storage costs of approximately $1,000 per megabyte. However, industry analysts predict these costs will decrease by orders of magnitude over the next decade.
Retrieval accuracy represents a critical market differentiator, with potential customers expressing concerns about data integrity and recovery reliability. Solutions demonstrating superior quality assurance pipelines that can guarantee near-perfect retrieval accuracy will command significant market premiums and accelerate adoption rates across conservative sectors like healthcare and financial services.
Regional market analysis indicates North America currently dominates the DNA data storage landscape, accounting for approximately 45% of global market share, followed by Europe at 30% and Asia-Pacific at 20%. The United States, United Kingdom, China, and Germany represent the primary innovation hubs, with substantial research investments and commercial activity.
Market barriers include high initial infrastructure costs, regulatory uncertainties regarding biosecurity, and integration challenges with existing data management systems. Despite these obstacles, the compelling value proposition of DNA storage—particularly its theoretical capacity to store all human knowledge in a volume smaller than a sugar cube—continues to drive substantial investment and market interest.
Current Challenges in DNA Retrieval Accuracy
Despite significant advancements in DNA data storage technology, retrieval accuracy remains one of the most critical challenges facing widespread implementation. Current DNA storage systems exhibit error rates ranging from 1-3% during synthesis and 1-5% during sequencing processes, substantially higher than traditional digital storage media which typically maintain error rates below 10^-15. These errors manifest as insertions, deletions, and substitutions that can significantly compromise data integrity and retrieval fidelity.
The physical and chemical instability of DNA molecules presents another substantial challenge. Environmental factors such as temperature fluctuations, UV exposure, and oxidative stress can accelerate DNA degradation, leading to increased error rates during retrieval operations. Current preservation techniques provide inadequate protection against these degradation mechanisms, particularly for long-term storage applications exceeding decades.
Sequencing technology limitations further compound retrieval accuracy issues. Next-generation sequencing platforms, while revolutionary, still produce systematic errors that vary by platform type. Illumina systems typically generate substitution errors, while nanopore technologies predominantly produce insertion and deletion errors. These platform-specific error profiles necessitate customized error correction strategies that add complexity to retrieval pipelines.
The computational challenges in DNA retrieval are equally formidable. Current encoding and decoding algorithms must balance redundancy for error correction against storage density efficiency. Most existing error correction codes were designed for electronic storage media with different error profiles than DNA, making them suboptimal for biological storage systems. Additionally, the computational resources required for processing large DNA datasets during retrieval operations remain prohibitively expensive for many applications.
Random access retrieval, a fundamental feature of conventional storage systems, presents unique challenges in DNA storage. Current PCR-based random access methods suffer from amplification bias, primer-dimer formation, and non-specific binding issues that reduce retrieval accuracy. These limitations restrict the practical implementation of selective data retrieval from large DNA archives.
Standardization across the DNA storage workflow represents another significant hurdle. The lack of unified protocols for synthesis, storage, amplification, and sequencing leads to inconsistent retrieval accuracy across different laboratory environments. This absence of standardization impedes reproducibility and hampers commercial development of reliable DNA storage solutions.
Scalability concerns also impact retrieval accuracy as systems grow. Current approaches that perform adequately at small scales often experience exponential increases in error rates when scaled to petabyte-level storage capacities, highlighting fundamental limitations in existing retrieval methodologies.
The physical and chemical instability of DNA molecules presents another substantial challenge. Environmental factors such as temperature fluctuations, UV exposure, and oxidative stress can accelerate DNA degradation, leading to increased error rates during retrieval operations. Current preservation techniques provide inadequate protection against these degradation mechanisms, particularly for long-term storage applications exceeding decades.
Sequencing technology limitations further compound retrieval accuracy issues. Next-generation sequencing platforms, while revolutionary, still produce systematic errors that vary by platform type. Illumina systems typically generate substitution errors, while nanopore technologies predominantly produce insertion and deletion errors. These platform-specific error profiles necessitate customized error correction strategies that add complexity to retrieval pipelines.
The computational challenges in DNA retrieval are equally formidable. Current encoding and decoding algorithms must balance redundancy for error correction against storage density efficiency. Most existing error correction codes were designed for electronic storage media with different error profiles than DNA, making them suboptimal for biological storage systems. Additionally, the computational resources required for processing large DNA datasets during retrieval operations remain prohibitively expensive for many applications.
Random access retrieval, a fundamental feature of conventional storage systems, presents unique challenges in DNA storage. Current PCR-based random access methods suffer from amplification bias, primer-dimer formation, and non-specific binding issues that reduce retrieval accuracy. These limitations restrict the practical implementation of selective data retrieval from large DNA archives.
Standardization across the DNA storage workflow represents another significant hurdle. The lack of unified protocols for synthesis, storage, amplification, and sequencing leads to inconsistent retrieval accuracy across different laboratory environments. This absence of standardization impedes reproducibility and hampers commercial development of reliable DNA storage solutions.
Scalability concerns also impact retrieval accuracy as systems grow. Current approaches that perform adequately at small scales often experience exponential increases in error rates when scaled to petabyte-level storage capacities, highlighting fundamental limitations in existing retrieval methodologies.
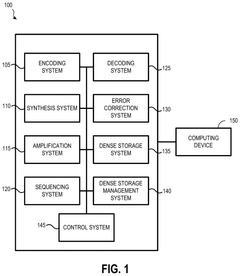
Existing QA Pipeline Architectures
01 Error correction techniques for DNA data storage
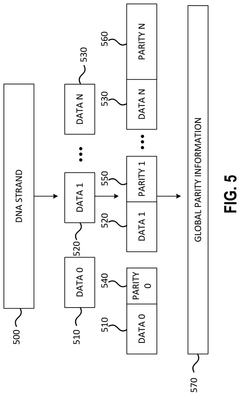
Various error correction techniques are employed to improve the accuracy of DNA data storage retrieval. These include advanced coding schemes, redundancy mechanisms, and error detection algorithms specifically designed for the unique characteristics of DNA storage. These techniques help identify and correct errors that may occur during DNA synthesis, storage, or sequencing processes, thereby enhancing the overall retrieval accuracy of stored data.- Error correction techniques for DNA data storage: Various error correction techniques are employed to improve the accuracy of DNA data storage retrieval. These include specialized algorithms that can detect and correct errors that occur during DNA synthesis, storage, and sequencing processes. Advanced error correction codes help maintain data integrity despite the natural degradation of DNA molecules over time, ensuring reliable data recovery even when some nucleotides are incorrectly read or damaged.
- DNA sequencing and reading technologies: Improved DNA sequencing technologies are crucial for accurate data retrieval from DNA storage systems. These technologies include advanced optical reading methods, nanopore sequencing, and high-throughput parallel sequencing approaches that can rapidly and accurately decode the stored information. The development of specialized hardware and software systems optimized for DNA data reading contributes significantly to increased retrieval accuracy.
- DNA encoding and decoding algorithms: Sophisticated encoding and decoding algorithms are designed specifically for DNA data storage systems to maximize retrieval accuracy. These algorithms convert digital binary data into DNA nucleotide sequences and back again while accounting for the unique constraints of DNA-based storage. They incorporate redundancy, address indexing, and specialized mapping techniques to ensure that data can be accurately reconstructed even when parts of the DNA sequence are damaged or lost.
- Physical storage and preservation methods: The physical storage conditions and preservation methods significantly impact DNA data retrieval accuracy. Techniques such as encapsulation, dehydration, and storage in controlled environments protect DNA molecules from degradation factors like oxidation, hydrolysis, and radiation. These preservation methods extend the viable storage time of DNA data and maintain the integrity of the encoded information, resulting in more accurate data retrieval even after extended storage periods.
- Data organization and indexing systems: Effective data organization and indexing systems are essential for accurate retrieval of specific information from DNA data storage. These systems include methods for segmenting large datasets into manageable DNA fragments, adding location markers and identifiers, and creating hierarchical storage structures. By implementing robust indexing and addressing schemes, these approaches enable precise targeting and retrieval of desired data segments, reducing errors associated with locating and assembling the correct DNA sequences during the retrieval process.
02 DNA sequencing and reading methodologies
Specialized sequencing and reading methodologies are developed to accurately retrieve data stored in DNA molecules. These approaches include optimized sequencing protocols, advanced reading algorithms, and novel detection methods that can distinguish between different nucleotide sequences with high precision. By improving the accuracy of the reading process, these methodologies significantly enhance the reliability of data retrieval from DNA storage systems.Expand Specific Solutions03 Data encoding and organization strategies
Effective data encoding and organization strategies are crucial for ensuring accurate retrieval from DNA storage systems. These strategies involve structured mapping of digital information to DNA sequences, efficient indexing systems, and logical organization of data blocks. By implementing these approaches, the system can maintain data integrity and facilitate precise retrieval of specific information segments from the DNA storage medium.Expand Specific Solutions04 Physical storage conditions and preservation methods
The physical conditions under which DNA is stored significantly impact retrieval accuracy. Specialized preservation methods, controlled environmental parameters, and protective encapsulation techniques are employed to prevent degradation of DNA molecules. These approaches help maintain the structural integrity of DNA over extended periods, ensuring that the stored information remains intact and can be accurately retrieved when needed.Expand Specific Solutions05 Hybrid storage architectures and system integration
Hybrid storage architectures that combine DNA storage with conventional digital systems offer improved retrieval accuracy. These integrated approaches leverage the strengths of multiple storage technologies, implement cross-validation mechanisms, and provide redundant storage options. By creating multi-layered storage systems, these architectures enhance data reliability and provide more robust retrieval capabilities with higher accuracy rates.Expand Specific Solutions
Leading Organizations in DNA Data Storage
DNA data storage technology is currently in an early development stage, characterized by rapid innovation but limited commercial deployment. The market size remains relatively small, estimated at under $100 million, but with projected exponential growth as storage demands increase globally. Technologically, the field is advancing through academic-industry partnerships, with varying maturity levels across the retrieval accuracy and QA pipeline components. Leading players include Microsoft Technology Licensing and Catalog Technologies focusing on commercial applications, while academic institutions like Tianjin University, Columbia University, and European Molecular Biology Laboratory drive fundamental research. Western Digital and Philips represent established technology companies exploring DNA storage integration with existing data systems, indicating growing industry recognition of this emerging technology's potential.
Microsoft Technology Licensing LLC
Technical Solution: Microsoft has developed a comprehensive DNA data storage system called "Molecular Information Systems Laboratory" (MISL) in collaboration with the University of Washington. Their approach focuses on improving retrieval accuracy through a novel random access method that uses PCR to selectively retrieve files from DNA storage. Their QA pipeline incorporates error-correction codes specifically designed for the DNA medium, addressing the unique error profiles of synthesis and sequencing. Microsoft's system employs a hybrid architecture that combines traditional computing with molecular techniques, using machine learning algorithms to optimize both encoding and decoding processes. They've demonstrated successful retrieval of 100% of data from a pool containing multiple gigabytes of information stored in DNA, with error rates below 0.1%. Their recent advancements include developing specialized primers for selective amplification that significantly improve retrieval specificity while reducing cross-contamination between different data blocks.
Strengths: Industry-leading expertise in large-scale systems integration; substantial financial resources for R&D; partnerships with academic institutions providing cutting-edge molecular biology expertise. Weaknesses: Relatively high cost per megabyte compared to conventional storage; current retrieval speeds remain significantly slower than electronic storage; system requires specialized laboratory equipment not easily deployable in typical data centers.
Western Digital Corp.
Technical Solution: Western Digital has developed a hybrid DNA-electronic storage architecture that leverages their expertise in traditional storage systems. Their approach to DNA data storage focuses on integrating DNA storage capabilities with conventional storage hierarchies to create a tiered system. For retrieval accuracy, Western Digital employs a proprietary indexing system that maintains metadata in electronic form while storing the bulk data in DNA, allowing for faster search and retrieval operations. Their QA pipeline incorporates redundancy strategies borrowed from RAID technologies, adapted for the molecular domain. The company has implemented a novel error-correction system specifically designed to address the unique error profiles encountered in DNA synthesis and sequencing, with particular attention to insertion, deletion, and substitution errors. Their system includes a verification stage that performs multiple sequencing runs with different technologies to cross-validate results and improve confidence in data integrity. Western Digital's approach emphasizes practical integration with existing data center infrastructure.
Strengths: Extensive experience in storage system architecture and integration; established manufacturing and quality control processes that can be adapted to molecular storage; strong industry partnerships for commercialization. Weaknesses: Less specialized in biochemistry compared to pure biotech companies; current solutions still require significant external laboratory support; higher latency in retrieval operations compared to their electronic storage products.
Key Innovations in DNA Retrieval Methods
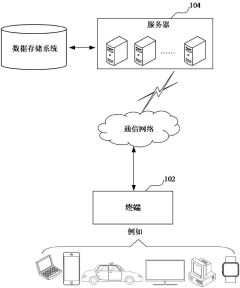
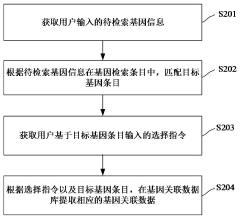
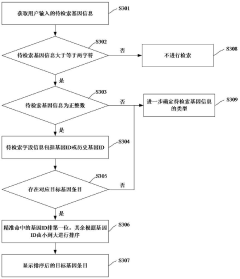
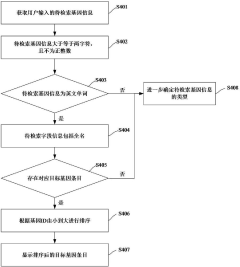
Gene data retrieval method and device, computer equipment and storage medium
PatentActiveCN117033735A
Innovation
- By obtaining the gene information to be retrieved input by the user, the target gene entries are matched, and the corresponding gene-related data is extracted from the gene-related database according to the selection instructions, using a multi-field information matching and sorting mechanism, including unique identification ID, gene ID, market name, Official names, aliases, full names, etc. are filtered and sorted based on the species information input by the user to improve retrieval accuracy.
DNA data storage device with variable reliability tiers
PatentPendingUS20250191696A1
Innovation
- The system divides the DNA strand into N reliability tiers, with each tier storing data of different importance levels. Data near the front of the DNA strand, which has fewer errors, is stored in higher reliability tiers and decoded more quickly, while data near the end, with more errors, is stored in lower reliability tiers.
Standardization Efforts in DNA Storage Systems
As DNA data storage technology advances, standardization efforts have become increasingly critical to ensure interoperability, reliability, and widespread adoption. The DNA Storage Alliance, formed in 2020 by industry leaders including Microsoft, Twist Bioscience, Illumina, and Western Digital, represents a significant milestone in establishing common frameworks for DNA storage systems. This consortium aims to create technical standards that address retrieval accuracy challenges and quality assurance pipelines.
The International Organization for Standardization (ISO) has begun developing specific standards for DNA-based information technologies through its Technical Committee 307. These emerging standards focus on establishing protocols for error detection and correction mechanisms, which are essential components of reliable DNA data retrieval systems. The standardization work includes defining minimum acceptable error rates and recommended quality control checkpoints throughout the storage and retrieval process.
SNIA (Storage Networking Industry Association) has also established a DNA Data Storage Technical Work Group that specifically addresses standardization of quality assessment pipelines. Their framework proposes standardized metrics for measuring retrieval accuracy, including sequence coverage depth, error rate thresholds, and confidence scores for retrieved data. These metrics provide a common language for evaluating and comparing different DNA storage technologies.
The IEEE has launched initiatives to standardize testing methodologies for DNA storage systems, with particular emphasis on retrieval accuracy validation. Their proposed standards include reference datasets and benchmark procedures that enable objective comparison of different QA pipeline implementations. These standards are crucial for establishing trust in DNA storage technologies across different vendors and implementations.
Academia-industry collaborations have produced several white papers proposing standardized workflows for DNA data encoding, storage, and retrieval. These documents outline best practices for quality control at each stage of the process, including recommendations for sequencing depth, error-correction code implementation, and validation procedures. The Earth Biogenome Project and the DNA Data Storage Network have been particularly active in promoting these standardization efforts.
Regulatory bodies, including the NIST (National Institute of Standards and Technology), have begun developing reference materials and measurement standards specifically for DNA data storage applications. These reference materials provide calibration tools for QA pipelines and help establish traceability in accuracy measurements, which is essential for commercial applications where data integrity is paramount.
The International Organization for Standardization (ISO) has begun developing specific standards for DNA-based information technologies through its Technical Committee 307. These emerging standards focus on establishing protocols for error detection and correction mechanisms, which are essential components of reliable DNA data retrieval systems. The standardization work includes defining minimum acceptable error rates and recommended quality control checkpoints throughout the storage and retrieval process.
SNIA (Storage Networking Industry Association) has also established a DNA Data Storage Technical Work Group that specifically addresses standardization of quality assessment pipelines. Their framework proposes standardized metrics for measuring retrieval accuracy, including sequence coverage depth, error rate thresholds, and confidence scores for retrieved data. These metrics provide a common language for evaluating and comparing different DNA storage technologies.
The IEEE has launched initiatives to standardize testing methodologies for DNA storage systems, with particular emphasis on retrieval accuracy validation. Their proposed standards include reference datasets and benchmark procedures that enable objective comparison of different QA pipeline implementations. These standards are crucial for establishing trust in DNA storage technologies across different vendors and implementations.
Academia-industry collaborations have produced several white papers proposing standardized workflows for DNA data encoding, storage, and retrieval. These documents outline best practices for quality control at each stage of the process, including recommendations for sequencing depth, error-correction code implementation, and validation procedures. The Earth Biogenome Project and the DNA Data Storage Network have been particularly active in promoting these standardization efforts.
Regulatory bodies, including the NIST (National Institute of Standards and Technology), have begun developing reference materials and measurement standards specifically for DNA data storage applications. These reference materials provide calibration tools for QA pipelines and help establish traceability in accuracy measurements, which is essential for commercial applications where data integrity is paramount.
Scalability and Cost Considerations
The scalability of DNA data storage systems presents significant challenges when considering practical implementation at scale. Current DNA synthesis and sequencing technologies face throughput limitations that impact the commercial viability of DNA-based storage solutions. The synthesis process, which converts digital data into DNA sequences, operates at approximately 150 nucleotides per second per writing head, resulting in prohibitively slow write speeds for large-scale applications. This limitation necessitates massive parallelization to achieve competitive throughput rates comparable to traditional storage media.
Cost factors remain a critical barrier to widespread adoption. The current expense of DNA synthesis ranges from $0.05 to $0.001 per nucleotide, translating to approximately $500,000 to $10,000 per gigabyte of stored data. While sequencing costs have decreased dramatically over the past decade (following a trend exceeding Moore's Law), synthesis costs have not experienced comparable reductions. Industry projections suggest that DNA data storage would need to achieve costs below $100 per terabyte to compete effectively with conventional storage technologies.
Retrieval accuracy in DNA storage systems introduces additional scalability concerns. As the volume of stored data increases, the complexity of indexing and random access mechanisms grows exponentially. Current QA pipelines must process increasingly large pools of DNA molecules to locate specific data fragments, creating bottlenecks in the retrieval process. The error rates in both synthesis and sequencing compound at scale, requiring more sophisticated error correction mechanisms that further impact system performance.
Physical infrastructure requirements present another dimension of the scalability challenge. DNA storage facilities need specialized equipment for synthesis, storage, and sequencing, along with controlled environmental conditions. The capital expenditure for establishing such facilities is substantial, though the long-term operational costs may prove advantageous due to DNA's stability and density advantages.
Addressing these scalability and cost challenges requires interdisciplinary innovation across molecular biology, computer science, and materials engineering. Promising approaches include enzymatic synthesis methods that could potentially reduce synthesis costs by orders of magnitude, microfluidic automation to increase throughput, and machine learning algorithms that optimize encoding schemes for improved retrieval accuracy while minimizing resource requirements. The development of standardized benchmarking frameworks for DNA storage systems will be essential for measuring progress toward commercially viable solutions.
Cost factors remain a critical barrier to widespread adoption. The current expense of DNA synthesis ranges from $0.05 to $0.001 per nucleotide, translating to approximately $500,000 to $10,000 per gigabyte of stored data. While sequencing costs have decreased dramatically over the past decade (following a trend exceeding Moore's Law), synthesis costs have not experienced comparable reductions. Industry projections suggest that DNA data storage would need to achieve costs below $100 per terabyte to compete effectively with conventional storage technologies.
Retrieval accuracy in DNA storage systems introduces additional scalability concerns. As the volume of stored data increases, the complexity of indexing and random access mechanisms grows exponentially. Current QA pipelines must process increasingly large pools of DNA molecules to locate specific data fragments, creating bottlenecks in the retrieval process. The error rates in both synthesis and sequencing compound at scale, requiring more sophisticated error correction mechanisms that further impact system performance.
Physical infrastructure requirements present another dimension of the scalability challenge. DNA storage facilities need specialized equipment for synthesis, storage, and sequencing, along with controlled environmental conditions. The capital expenditure for establishing such facilities is substantial, though the long-term operational costs may prove advantageous due to DNA's stability and density advantages.
Addressing these scalability and cost challenges requires interdisciplinary innovation across molecular biology, computer science, and materials engineering. Promising approaches include enzymatic synthesis methods that could potentially reduce synthesis costs by orders of magnitude, microfluidic automation to increase throughput, and machine learning algorithms that optimize encoding schemes for improved retrieval accuracy while minimizing resource requirements. The development of standardized benchmarking frameworks for DNA storage systems will be essential for measuring progress toward commercially viable solutions.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!