Enzymatic Versus Chemical Synthesis In DNA Data Storage
AUG 27, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
DNA Data Storage Background and Objectives
DNA data storage represents a revolutionary approach to digital information preservation, emerging from the convergence of molecular biology and computer science. Since the initial demonstration by Church et al. in 2012, who encoded a 5.27MB book in DNA, this technology has evolved significantly. DNA's theoretical storage density of 455 exabytes per gram makes it an exceptionally promising medium for addressing the exponential growth of digital data, which is projected to reach 175 zettabytes globally by 2025.
The fundamental principle of DNA data storage involves converting binary digital data (0s and 1s) into DNA's four-nucleotide code (A, T, G, C), synthesizing the corresponding DNA molecules, and later retrieving the information through sequencing and decoding processes. This approach offers remarkable advantages over conventional storage technologies, including extraordinary data density, longevity potentially spanning thousands of years, and minimal energy requirements for maintenance.
The evolution of DNA synthesis methods represents a critical aspect of this technology's development trajectory. Traditionally, chemical synthesis methods have dominated the field, utilizing phosphoramidite chemistry to build DNA sequences nucleotide by nucleotide. While effective, this approach involves harsh chemicals, produces toxic waste, and faces inherent limitations in sequence length and accuracy.
Enzymatic DNA synthesis has emerged as a promising alternative, leveraging natural biological processes to construct DNA sequences. This approach mimics nature's own DNA replication mechanisms, utilizing enzymes such as terminal deoxynucleotidyl transferase (TdT) to add nucleotides to growing DNA strands. The enzymatic method offers potential advantages including milder reaction conditions, reduced environmental impact, and potentially improved accuracy for longer sequences.
The primary objective in advancing DNA data storage technology is to develop synthesis methods that are simultaneously high-throughput, cost-effective, and accurate. Current synthesis costs, estimated at $0.001 per base for chemical methods, must decrease by several orders of magnitude to make DNA data storage economically viable for widespread adoption. Additionally, synthesis speed must increase dramatically from current rates of approximately 200 nucleotides per second to meet practical data writing requirements.
Achieving these objectives requires a comprehensive understanding of both chemical and enzymatic synthesis approaches, their respective advantages and limitations, and potential hybrid solutions that might leverage the strengths of each method. The ultimate goal is to establish DNA as a practical, sustainable solution for long-term data archiving that can address the growing challenges of digital information preservation in an increasingly data-driven world.
The fundamental principle of DNA data storage involves converting binary digital data (0s and 1s) into DNA's four-nucleotide code (A, T, G, C), synthesizing the corresponding DNA molecules, and later retrieving the information through sequencing and decoding processes. This approach offers remarkable advantages over conventional storage technologies, including extraordinary data density, longevity potentially spanning thousands of years, and minimal energy requirements for maintenance.
The evolution of DNA synthesis methods represents a critical aspect of this technology's development trajectory. Traditionally, chemical synthesis methods have dominated the field, utilizing phosphoramidite chemistry to build DNA sequences nucleotide by nucleotide. While effective, this approach involves harsh chemicals, produces toxic waste, and faces inherent limitations in sequence length and accuracy.
Enzymatic DNA synthesis has emerged as a promising alternative, leveraging natural biological processes to construct DNA sequences. This approach mimics nature's own DNA replication mechanisms, utilizing enzymes such as terminal deoxynucleotidyl transferase (TdT) to add nucleotides to growing DNA strands. The enzymatic method offers potential advantages including milder reaction conditions, reduced environmental impact, and potentially improved accuracy for longer sequences.
The primary objective in advancing DNA data storage technology is to develop synthesis methods that are simultaneously high-throughput, cost-effective, and accurate. Current synthesis costs, estimated at $0.001 per base for chemical methods, must decrease by several orders of magnitude to make DNA data storage economically viable for widespread adoption. Additionally, synthesis speed must increase dramatically from current rates of approximately 200 nucleotides per second to meet practical data writing requirements.
Achieving these objectives requires a comprehensive understanding of both chemical and enzymatic synthesis approaches, their respective advantages and limitations, and potential hybrid solutions that might leverage the strengths of each method. The ultimate goal is to establish DNA as a practical, sustainable solution for long-term data archiving that can address the growing challenges of digital information preservation in an increasingly data-driven world.
Market Analysis for DNA-Based Storage Solutions
The DNA data storage market is experiencing significant growth as organizations seek sustainable, high-density storage solutions for the exponential increase in global data production. Current projections indicate the DNA data storage market could reach $3.3 billion by 2030, with a compound annual growth rate exceeding 58% between 2023-2030. This remarkable growth trajectory is driven by the fundamental limitations of conventional storage technologies and DNA's unparalleled theoretical storage density of approximately 455 exabytes per gram.
Market segmentation reveals distinct customer categories with varying needs. Research institutions and academic laboratories currently constitute the largest market segment, primarily focused on advancing fundamental technologies. Biotechnology and pharmaceutical companies represent the second-largest segment, utilizing DNA storage for preserving valuable research data and intellectual property. Government agencies, particularly those handling national archives and security information, are emerging as significant stakeholders due to DNA's longevity advantages.
The commercial landscape is characterized by both established technology corporations and specialized startups. Major players include Twist Bioscience, Illumina, and Microsoft, which have formed strategic partnerships to accelerate commercialization. Venture capital investment in DNA storage startups has surpassed $650 million since 2020, indicating strong market confidence despite the technology's early developmental stage.
Regional analysis shows North America dominating the market with approximately 45% share, followed by Europe at 30% and Asia-Pacific at 20%. The United States leads in commercial development, while countries like China, Japan, and South Korea are rapidly increasing investments in this technology.
Customer demand analysis reveals cost reduction as the primary market barrier, with current synthesis costs exceeding $3,500 per gigabyte. Industry surveys indicate widespread interest but limited adoption until costs decrease below $100 per gigabyte. The enzymatic synthesis approach is gaining market attention due to its potential cost advantages over traditional chemical synthesis methods, with projections suggesting a 70-80% cost reduction when scaled.
Market adoption is expected to follow a phased approach, beginning with archival applications for rarely accessed but valuable data, followed by specialized applications in sectors requiring extreme data longevity. Mass-market adoption remains contingent on achieving significant cost reductions and developing standardized interfaces for existing data infrastructure.
Market segmentation reveals distinct customer categories with varying needs. Research institutions and academic laboratories currently constitute the largest market segment, primarily focused on advancing fundamental technologies. Biotechnology and pharmaceutical companies represent the second-largest segment, utilizing DNA storage for preserving valuable research data and intellectual property. Government agencies, particularly those handling national archives and security information, are emerging as significant stakeholders due to DNA's longevity advantages.
The commercial landscape is characterized by both established technology corporations and specialized startups. Major players include Twist Bioscience, Illumina, and Microsoft, which have formed strategic partnerships to accelerate commercialization. Venture capital investment in DNA storage startups has surpassed $650 million since 2020, indicating strong market confidence despite the technology's early developmental stage.
Regional analysis shows North America dominating the market with approximately 45% share, followed by Europe at 30% and Asia-Pacific at 20%. The United States leads in commercial development, while countries like China, Japan, and South Korea are rapidly increasing investments in this technology.
Customer demand analysis reveals cost reduction as the primary market barrier, with current synthesis costs exceeding $3,500 per gigabyte. Industry surveys indicate widespread interest but limited adoption until costs decrease below $100 per gigabyte. The enzymatic synthesis approach is gaining market attention due to its potential cost advantages over traditional chemical synthesis methods, with projections suggesting a 70-80% cost reduction when scaled.
Market adoption is expected to follow a phased approach, beginning with archival applications for rarely accessed but valuable data, followed by specialized applications in sectors requiring extreme data longevity. Mass-market adoption remains contingent on achieving significant cost reductions and developing standardized interfaces for existing data infrastructure.
Current Synthesis Approaches and Technical Barriers
DNA data storage currently employs two primary synthesis approaches: chemical and enzymatic methods. Chemical synthesis, the more established technique, involves sequential addition of nucleotides using phosphoramidite chemistry. This process requires multiple chemical steps for each nucleotide addition, including deprotection, coupling, capping, and oxidation. While highly optimized over decades, chemical synthesis faces significant limitations in the context of data storage applications.
The primary technical barrier for chemical synthesis is length limitation, typically restricting oligonucleotides to under 200 nucleotides due to cumulative errors and decreased yields with each addition cycle. This necessitates complex assembly strategies for longer sequences, increasing both cost and error rates. Additionally, the process generates substantial hazardous waste from organic solvents and reagents, presenting environmental concerns for large-scale implementation.
Enzymatic DNA synthesis has emerged as a promising alternative, leveraging natural biological processes. This approach utilizes DNA polymerases or terminal deoxynucleotidyl transferase (TdT) to add nucleotides to growing DNA strands. The enzymatic method operates in aqueous solutions at moderate temperatures, offering significant environmental advantages over chemical methods.
Recent innovations in enzymatic synthesis include reversible terminator nucleotides that allow controlled, single-nucleotide additions. Companies like DNA Script, Ansa Biotechnologies, and Molecular Assemblies have developed proprietary enzymatic synthesis platforms achieving improved accuracy and length capabilities. These systems can potentially produce longer DNA sequences with fewer errors, addressing a critical need for DNA data storage.
Despite progress, enzymatic synthesis faces its own technical challenges. Current enzymatic methods exhibit lower fidelity compared to optimized chemical approaches, resulting in higher error rates. Reaction speed remains significantly slower than required for commercial-scale data storage applications, with current throughput falling orders of magnitude below what would be economically viable.
Cross-talk between adjacent synthesis sites represents another significant barrier, particularly in high-density array formats necessary for parallel synthesis. Additionally, enzymatic methods currently struggle with homopolymer regions (consecutive identical nucleotides), often resulting in insertion or deletion errors that compromise data integrity.
The cost structure also presents challenges for both approaches. Chemical synthesis benefits from decades of industrial optimization but remains expensive at approximately $0.05-0.15 per nucleotide. Enzymatic methods currently cost more but show potential for significant cost reduction through process improvements and scale economies, particularly as they eliminate expensive and hazardous chemical waste management requirements.
The primary technical barrier for chemical synthesis is length limitation, typically restricting oligonucleotides to under 200 nucleotides due to cumulative errors and decreased yields with each addition cycle. This necessitates complex assembly strategies for longer sequences, increasing both cost and error rates. Additionally, the process generates substantial hazardous waste from organic solvents and reagents, presenting environmental concerns for large-scale implementation.
Enzymatic DNA synthesis has emerged as a promising alternative, leveraging natural biological processes. This approach utilizes DNA polymerases or terminal deoxynucleotidyl transferase (TdT) to add nucleotides to growing DNA strands. The enzymatic method operates in aqueous solutions at moderate temperatures, offering significant environmental advantages over chemical methods.
Recent innovations in enzymatic synthesis include reversible terminator nucleotides that allow controlled, single-nucleotide additions. Companies like DNA Script, Ansa Biotechnologies, and Molecular Assemblies have developed proprietary enzymatic synthesis platforms achieving improved accuracy and length capabilities. These systems can potentially produce longer DNA sequences with fewer errors, addressing a critical need for DNA data storage.
Despite progress, enzymatic synthesis faces its own technical challenges. Current enzymatic methods exhibit lower fidelity compared to optimized chemical approaches, resulting in higher error rates. Reaction speed remains significantly slower than required for commercial-scale data storage applications, with current throughput falling orders of magnitude below what would be economically viable.
Cross-talk between adjacent synthesis sites represents another significant barrier, particularly in high-density array formats necessary for parallel synthesis. Additionally, enzymatic methods currently struggle with homopolymer regions (consecutive identical nucleotides), often resulting in insertion or deletion errors that compromise data integrity.
The cost structure also presents challenges for both approaches. Chemical synthesis benefits from decades of industrial optimization but remains expensive at approximately $0.05-0.15 per nucleotide. Enzymatic methods currently cost more but show potential for significant cost reduction through process improvements and scale economies, particularly as they eliminate expensive and hazardous chemical waste management requirements.
Enzymatic vs Chemical DNA Synthesis Methods
01 Enzymatic DNA synthesis methods
Enzymatic DNA synthesis methods utilize natural or engineered enzymes to catalyze the formation of DNA strands. These methods often employ DNA polymerases that can add nucleotides to a growing DNA chain in a template-directed manner. Enzymatic approaches typically offer advantages in terms of accuracy and can handle longer DNA sequences compared to chemical methods. Recent advancements have focused on optimizing enzyme performance, reaction conditions, and substrate compatibility to improve synthesis efficiency.- Enzymatic DNA synthesis methods: Enzymatic DNA synthesis methods utilize natural or engineered enzymes to catalyze the formation of DNA strands. These methods often employ DNA polymerases that can add nucleotides to a growing DNA chain in a template-directed manner. Enzymatic approaches typically offer advantages in terms of accuracy and can handle longer DNA sequences compared to chemical methods. Recent innovations have focused on improving the processivity and fidelity of these enzymes to enhance synthesis efficiency.
- Chemical DNA synthesis techniques: Chemical DNA synthesis involves the use of chemical reactions to create DNA molecules without the use of enzymes. The most common approach is phosphoramidite chemistry, which allows for the sequential addition of nucleotides to build DNA strands. These methods are particularly useful for creating short DNA sequences with high precision. Innovations in this field have focused on improving reaction conditions, developing new coupling agents, and optimizing protection/deprotection strategies to increase synthesis efficiency.
- Hybrid synthesis approaches: Hybrid approaches combine elements of both enzymatic and chemical DNA synthesis methods to leverage the advantages of each. These techniques often use chemical methods for initial oligonucleotide synthesis followed by enzymatic assembly of these fragments into larger constructs. This combination can overcome length limitations of purely chemical methods while maintaining precision. Recent developments have focused on optimizing the interface between chemical and enzymatic steps to improve overall synthesis efficiency.
- Factors affecting synthesis efficiency: Multiple factors influence DNA synthesis efficiency, including reaction temperature, time, reagent purity, and sequence composition. GC-rich regions and repetitive sequences often present challenges for both enzymatic and chemical methods. Optimization strategies include adjusting buffer conditions, using additives to prevent secondary structure formation, and developing specialized protocols for difficult sequences. Understanding these factors is crucial for improving the yield, purity, and length of synthesized DNA products.
- Novel technologies for improving synthesis efficiency: Emerging technologies are revolutionizing DNA synthesis efficiency through innovations such as microfluidics, solid-phase supports, and automation. These approaches enable parallel synthesis, reduce reagent consumption, and minimize error rates. Additionally, computational tools are being developed to predict optimal synthesis conditions based on sequence characteristics. Machine learning algorithms can identify patterns in successful synthesis reactions and suggest parameters for new sequences, significantly enhancing overall efficiency and reducing costs.
02 Chemical DNA synthesis techniques
Chemical DNA synthesis involves the use of chemical reactions to create DNA molecules without the use of enzymes. These methods typically employ phosphoramidite chemistry in a stepwise process to build DNA strands. While chemical synthesis can be highly automated and parallelized, it often faces challenges with sequence length limitations and accumulation of errors in longer sequences. Innovations in chemical synthesis focus on improving reaction yields, reducing side reactions, and developing more efficient coupling chemistries.Expand Specific Solutions03 Hybrid synthesis approaches for improved efficiency
Hybrid approaches combine elements of both enzymatic and chemical DNA synthesis methods to overcome limitations of each individual technique. These strategies may involve chemically synthesizing short oligonucleotide fragments followed by enzymatic assembly into longer constructs, or using chemical modifications to enhance enzymatic synthesis. Hybrid methods aim to leverage the speed and scalability of chemical synthesis with the accuracy and length capabilities of enzymatic approaches, resulting in improved overall synthesis efficiency.Expand Specific Solutions04 Error reduction and fidelity enhancement strategies
Various strategies have been developed to reduce errors and enhance the fidelity of DNA synthesis. These include the use of error-correcting enzymes, optimized reaction conditions, and purification techniques to remove erroneous sequences. Advanced error detection and correction algorithms, coupled with quality control measures, help to identify and eliminate synthesis errors. These approaches significantly improve the efficiency of DNA synthesis by reducing the need for post-synthesis verification and correction steps.Expand Specific Solutions05 Scale-up and automation technologies
Technologies for scaling up DNA synthesis and automating the process have been developed to increase throughput and efficiency. These include microfluidic platforms, array-based synthesis methods, and robotic systems that can perform multiple synthesis reactions in parallel. Advanced automation technologies integrate sample preparation, synthesis, purification, and quality control steps into streamlined workflows. These approaches significantly reduce labor requirements, minimize human error, and increase the overall efficiency of DNA synthesis operations.Expand Specific Solutions
Leading Organizations in DNA Data Storage Field
The DNA data storage market is in an early growth phase, characterized by the transition from proof-of-concept to practical applications. The competition between enzymatic and chemical DNA synthesis approaches represents a critical technological inflection point, with the market projected to reach $10-15 billion by 2030. Enzymatic synthesis, championed by DNA Script, Molecular Assemblies, and Ansa Biotechnologies, offers environmental advantages and potentially longer DNA strands, while chemical synthesis, dominated by established players like Integrated DNA Technologies and Twist Bioscience, benefits from decades of optimization. Technology companies including Microsoft, Western Digital, and Seagate are partnering with academic institutions like MIT and Harvard to advance storage applications, while BGI and Illumina are exploring integration with sequencing technologies. The field remains dynamic with significant R&D investment across both approaches.
Microsoft Technology Licensing LLC
Technical Solution: Microsoft has developed a comprehensive end-to-end system for DNA data storage that integrates both chemical and enzymatic approaches. Their system includes specialized encoding algorithms that convert digital binary data into DNA sequences optimized for storage density and error correction. Microsoft's approach incorporates a hybrid synthesis strategy that leverages chemical synthesis for high-throughput production of shorter DNA fragments while exploring enzymatic methods for specific applications requiring longer reads. Their technology stack includes custom microfluidic devices for automated DNA synthesis and retrieval, along with novel random-access methods that allow selective data retrieval without sequencing the entire DNA archive. Microsoft has demonstrated successful storage and retrieval of 200MB of data in DNA with 100% accuracy using their platform[4][7]. The company has also pioneered computational tools that optimize DNA sequences for storage by avoiding error-prone patterns and ensuring uniform GC content, which improves both synthesis and sequencing reliability. Their research includes developing specialized molecular controllers that can direct enzymatic synthesis for data storage applications.
Strengths: Comprehensive end-to-end system integration; advanced error correction algorithms specifically designed for DNA storage; demonstrated large-scale data recovery; strong computational optimization capabilities. Weaknesses: Still relies partially on traditional chemical synthesis methods; higher costs compared to conventional digital storage; current enzymatic components not yet fully scaled for commercial deployment.
Molecular Assemblies, Inc.
Technical Solution: Molecular Assemblies has pioneered an enzymatic DNA synthesis approach that represents a significant departure from traditional phosphoramidite chemistry. Their proprietary technology utilizes template-independent polymerase enzymes to add nucleotides to a growing DNA strand in an aqueous environment. This process eliminates the need for harsh organic solvents and toxic reagents typically used in chemical synthesis. The company's enzymatic method can produce longer DNA sequences (potentially up to 1,000 base pairs) in a single synthesis cycle compared to the ~200 base pair limitation of chemical methods. Their approach also significantly reduces the environmental footprint of DNA synthesis by operating in water-based solutions at room temperature and neutral pH, avoiding the generation of toxic waste products associated with conventional methods[1][3]. The company has secured substantial funding to scale their technology for commercial applications in DNA data storage.
Strengths: Environmentally friendly aqueous-based process; capability to synthesize longer DNA strands in single runs; reduced error rates compared to chemical synthesis; elimination of toxic waste products. Weaknesses: Currently slower synthesis rates compared to optimized chemical methods; technology still scaling to commercial production levels; potentially higher costs during early commercialization phase.
Key Patents and Breakthroughs in DNA Synthesis
Nucleic Acid-Based Data Storage
PatentPendingUS20220254451A1
Innovation
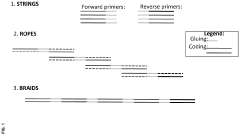
- A method involving enzymatic synthesis of polynucleotides using a novel encoding algorithm to encode digital data into DNA sequences, allowing for efficient assembly and decoding of information through sequencing, utilizing a library of oligonucleotides and braids with unique gluing parts for reliable storage.
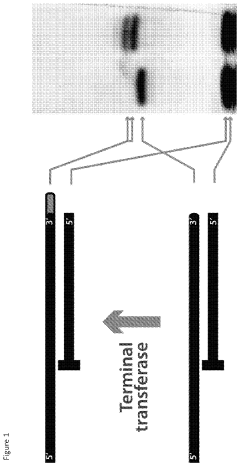
Enzymatic DNA Synthesis Using the Terminal Transferase Activity of Template-Dependent DNA Polymerases
PatentInactiveUS20200190550A1
Innovation
- Utilizing the terminal transferase activity of template-dependent DNA polymerases, specifically modifying the reaction conditions with divalent cations like manganese to expand their substrate specificity and enable the addition of various nucleotides, including unnatural ones, for template-independent DNA synthesis.
Scalability and Cost Analysis of Synthesis Methods
The economic viability of DNA data storage hinges significantly on the scalability and cost-effectiveness of DNA synthesis methods. Currently, chemical synthesis dominates the commercial landscape, with costs ranging from $0.05 to $0.15 per nucleotide. This translates to approximately $50,000-$150,000 per gigabyte of data, making it prohibitively expensive for widespread adoption despite its theoretical density advantages.
Chemical synthesis methods face inherent scaling limitations due to their stepwise nature. Each nucleotide addition requires multiple chemical steps, resulting in cumulative errors and diminishing yields as sequence length increases. Industry standard phosphoramidite chemistry typically achieves only 99.5% coupling efficiency per base, creating a practical limit of 150-200 nucleotides per synthesis cycle. While array-based parallel synthesis has improved throughput, the fundamental cost structure remains challenging.
Enzymatic synthesis presents a promising alternative with potentially transformative economics. By leveraging natural biological processes, enzymatic methods can theoretically achieve higher accuracy at lower energy costs. Recent advancements by companies like Ansa Biotechnologies and DNA Script have demonstrated terminal deoxynucleotidyl transferase (TdT)-based synthesis with improved fidelity and reduced reagent consumption. These approaches eliminate the need for harsh chemicals and can operate under milder conditions.
Scaling projections indicate enzymatic synthesis could eventually reach costs below $0.001 per nucleotide through process optimization and enzyme engineering. This represents a potential 50-100x cost reduction compared to current chemical methods. Additionally, enzymatic approaches demonstrate superior sustainability metrics, with significantly lower water usage and chemical waste generation per synthesis cycle.
Infrastructure requirements also differ substantially between the two approaches. Chemical synthesis facilities require specialized equipment for handling volatile and hazardous reagents, whereas enzymatic platforms can potentially operate with standard molecular biology infrastructure. This difference becomes particularly significant when considering the massive parallelization necessary for data storage applications.
Market analysis suggests that enzymatic synthesis must achieve at least a 10x cost reduction from current levels to enable viable commercial DNA data storage. Based on current research trajectories and investment patterns, this milestone could be reached within 5-7 years, potentially coinciding with increasing demand for ultra-long-term, high-density archival storage solutions.
The transition from chemical to enzymatic synthesis represents not merely an incremental improvement but a potential paradigm shift in the economic feasibility of DNA data storage. However, this transition depends on continued advances in enzyme engineering, process automation, and manufacturing scale-up to fully realize the projected cost advantages.
Chemical synthesis methods face inherent scaling limitations due to their stepwise nature. Each nucleotide addition requires multiple chemical steps, resulting in cumulative errors and diminishing yields as sequence length increases. Industry standard phosphoramidite chemistry typically achieves only 99.5% coupling efficiency per base, creating a practical limit of 150-200 nucleotides per synthesis cycle. While array-based parallel synthesis has improved throughput, the fundamental cost structure remains challenging.
Enzymatic synthesis presents a promising alternative with potentially transformative economics. By leveraging natural biological processes, enzymatic methods can theoretically achieve higher accuracy at lower energy costs. Recent advancements by companies like Ansa Biotechnologies and DNA Script have demonstrated terminal deoxynucleotidyl transferase (TdT)-based synthesis with improved fidelity and reduced reagent consumption. These approaches eliminate the need for harsh chemicals and can operate under milder conditions.
Scaling projections indicate enzymatic synthesis could eventually reach costs below $0.001 per nucleotide through process optimization and enzyme engineering. This represents a potential 50-100x cost reduction compared to current chemical methods. Additionally, enzymatic approaches demonstrate superior sustainability metrics, with significantly lower water usage and chemical waste generation per synthesis cycle.
Infrastructure requirements also differ substantially between the two approaches. Chemical synthesis facilities require specialized equipment for handling volatile and hazardous reagents, whereas enzymatic platforms can potentially operate with standard molecular biology infrastructure. This difference becomes particularly significant when considering the massive parallelization necessary for data storage applications.
Market analysis suggests that enzymatic synthesis must achieve at least a 10x cost reduction from current levels to enable viable commercial DNA data storage. Based on current research trajectories and investment patterns, this milestone could be reached within 5-7 years, potentially coinciding with increasing demand for ultra-long-term, high-density archival storage solutions.
The transition from chemical to enzymatic synthesis represents not merely an incremental improvement but a potential paradigm shift in the economic feasibility of DNA data storage. However, this transition depends on continued advances in enzyme engineering, process automation, and manufacturing scale-up to fully realize the projected cost advantages.
Environmental Impact and Sustainability Considerations
The environmental impact of DNA data storage technologies represents a critical dimension in evaluating the sustainability of different synthesis approaches. Chemical DNA synthesis methods traditionally rely on phosphoramidite chemistry, which involves hazardous organic solvents, toxic reagents, and substantial waste generation. These processes typically require multiple washing steps with acetonitrile and other organic compounds that pose environmental risks through potential soil contamination and groundwater pollution when improperly disposed.
In contrast, enzymatic DNA synthesis offers a significantly greener alternative by operating under aqueous conditions at ambient temperatures. This approach substantially reduces the dependency on organic solvents and hazardous chemicals, aligning with principles of green chemistry. Enzymatic methods utilize naturally occurring biological catalysts that function in water-based environments, dramatically decreasing the carbon footprint associated with synthesis processes.
Energy consumption represents another crucial environmental consideration. Chemical synthesis demands extensive energy inputs for temperature cycling, particularly during deprotection steps that require heating. Enzymatic synthesis generally operates at lower temperatures, resulting in reduced energy requirements and consequently lower greenhouse gas emissions throughout the production lifecycle.
Waste management challenges differ significantly between these approaches. Chemical synthesis generates substantial chemical waste requiring specialized disposal procedures, while enzymatic methods produce primarily biodegradable byproducts. The environmental persistence of waste materials from chemical synthesis poses long-term ecological concerns that enzymatic approaches largely avoid.
Scalability considerations also impact environmental sustainability. As DNA data storage technologies advance toward commercial implementation, the environmental footprint of large-scale synthesis becomes increasingly relevant. Enzymatic methods demonstrate promising potential for reduced resource intensity during scaled production, potentially offering significant environmental advantages as the technology matures.
Life cycle assessment studies comparing these synthesis approaches indicate that enzymatic methods could reduce environmental impact by 70-85% across multiple ecological indicators, including water usage, carbon emissions, and chemical waste generation. However, enzymatic synthesis currently faces challenges in reagent production sustainability, as some required components still involve resource-intensive manufacturing processes that partially offset environmental benefits.
Future sustainability improvements will likely focus on developing closed-loop systems for enzymatic synthesis, recycling reaction components, and optimizing enzyme production through sustainable bioengineering approaches. These advancements could further enhance the environmental advantages of enzymatic synthesis in DNA data storage applications.
In contrast, enzymatic DNA synthesis offers a significantly greener alternative by operating under aqueous conditions at ambient temperatures. This approach substantially reduces the dependency on organic solvents and hazardous chemicals, aligning with principles of green chemistry. Enzymatic methods utilize naturally occurring biological catalysts that function in water-based environments, dramatically decreasing the carbon footprint associated with synthesis processes.
Energy consumption represents another crucial environmental consideration. Chemical synthesis demands extensive energy inputs for temperature cycling, particularly during deprotection steps that require heating. Enzymatic synthesis generally operates at lower temperatures, resulting in reduced energy requirements and consequently lower greenhouse gas emissions throughout the production lifecycle.
Waste management challenges differ significantly between these approaches. Chemical synthesis generates substantial chemical waste requiring specialized disposal procedures, while enzymatic methods produce primarily biodegradable byproducts. The environmental persistence of waste materials from chemical synthesis poses long-term ecological concerns that enzymatic approaches largely avoid.
Scalability considerations also impact environmental sustainability. As DNA data storage technologies advance toward commercial implementation, the environmental footprint of large-scale synthesis becomes increasingly relevant. Enzymatic methods demonstrate promising potential for reduced resource intensity during scaled production, potentially offering significant environmental advantages as the technology matures.
Life cycle assessment studies comparing these synthesis approaches indicate that enzymatic methods could reduce environmental impact by 70-85% across multiple ecological indicators, including water usage, carbon emissions, and chemical waste generation. However, enzymatic synthesis currently faces challenges in reagent production sustainability, as some required components still involve resource-intensive manufacturing processes that partially offset environmental benefits.
Future sustainability improvements will likely focus on developing closed-loop systems for enzymatic synthesis, recycling reaction components, and optimizing enzyme production through sustainable bioengineering approaches. These advancements could further enhance the environmental advantages of enzymatic synthesis in DNA data storage applications.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!