How to Model Glycogenolysis in Computational Studies
AUG 28, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Glycogenolysis Modeling Background and Objectives
Glycogenolysis, the biochemical process of glycogen breakdown into glucose-1-phosphate and glucose, represents a critical metabolic pathway for maintaining blood glucose homeostasis. This process has been studied extensively since the mid-20th century, with significant breakthroughs in understanding its enzymatic mechanisms, regulatory factors, and physiological significance. The computational modeling of glycogenolysis has evolved from simple mathematical representations to sophisticated multi-scale simulations incorporating molecular dynamics, systems biology approaches, and machine learning techniques.
The evolution of glycogenolysis modeling reflects broader trends in computational biology, moving from reductionist approaches focusing on individual reactions to holistic models capturing the complexity of metabolic networks. Early models in the 1960s-1970s primarily employed basic differential equations to describe enzyme kinetics. The 1980s-1990s saw the integration of regulatory mechanisms and spatial considerations. Recent advances have enabled whole-cell metabolic models incorporating glycogenolysis within broader energy metabolism frameworks.
Current research aims to develop increasingly accurate and predictive computational models of glycogenolysis that can simulate this process across multiple biological scales—from molecular interactions to tissue-level responses. These models seek to capture the dynamic nature of glycogen metabolism under various physiological and pathological conditions, including exercise, fasting, and metabolic disorders such as glycogen storage diseases.
The primary technical objectives for glycogenolysis modeling include: developing multi-scale frameworks that integrate molecular, cellular, and tissue-level processes; incorporating realistic representations of enzyme kinetics and regulatory mechanisms; accounting for spatial heterogeneity in glycogen distribution and metabolism; and enabling predictive capabilities for personalized medicine applications.
Significant challenges remain in accurately representing the complex structural dynamics of glycogen particles, which feature branched structures affecting enzyme accessibility and reaction rates. Additionally, modeling the intricate regulatory networks involving hormonal control, allosteric regulation, and post-translational modifications presents substantial computational hurdles.
The field is moving toward integrating glycogenolysis models with broader metabolic networks, particularly focusing on connections with glycolysis, gluconeogenesis, and the pentose phosphate pathway. This integration aims to provide a comprehensive understanding of glucose homeostasis and energy metabolism, with applications ranging from sports physiology to diabetes management and rare disease therapeutics.
Future directions point toward incorporating patient-specific data into glycogenolysis models, enabling personalized predictions of metabolic responses and potential therapeutic interventions. This represents a significant step toward precision medicine approaches for metabolic disorders and optimized nutrition strategies.
The evolution of glycogenolysis modeling reflects broader trends in computational biology, moving from reductionist approaches focusing on individual reactions to holistic models capturing the complexity of metabolic networks. Early models in the 1960s-1970s primarily employed basic differential equations to describe enzyme kinetics. The 1980s-1990s saw the integration of regulatory mechanisms and spatial considerations. Recent advances have enabled whole-cell metabolic models incorporating glycogenolysis within broader energy metabolism frameworks.
Current research aims to develop increasingly accurate and predictive computational models of glycogenolysis that can simulate this process across multiple biological scales—from molecular interactions to tissue-level responses. These models seek to capture the dynamic nature of glycogen metabolism under various physiological and pathological conditions, including exercise, fasting, and metabolic disorders such as glycogen storage diseases.
The primary technical objectives for glycogenolysis modeling include: developing multi-scale frameworks that integrate molecular, cellular, and tissue-level processes; incorporating realistic representations of enzyme kinetics and regulatory mechanisms; accounting for spatial heterogeneity in glycogen distribution and metabolism; and enabling predictive capabilities for personalized medicine applications.
Significant challenges remain in accurately representing the complex structural dynamics of glycogen particles, which feature branched structures affecting enzyme accessibility and reaction rates. Additionally, modeling the intricate regulatory networks involving hormonal control, allosteric regulation, and post-translational modifications presents substantial computational hurdles.
The field is moving toward integrating glycogenolysis models with broader metabolic networks, particularly focusing on connections with glycolysis, gluconeogenesis, and the pentose phosphate pathway. This integration aims to provide a comprehensive understanding of glucose homeostasis and energy metabolism, with applications ranging from sports physiology to diabetes management and rare disease therapeutics.
Future directions point toward incorporating patient-specific data into glycogenolysis models, enabling personalized predictions of metabolic responses and potential therapeutic interventions. This represents a significant step toward precision medicine approaches for metabolic disorders and optimized nutrition strategies.
Market Applications for Glycogenolysis Computational Models
Computational models of glycogenolysis have significant market applications across multiple sectors, driven by the fundamental role this metabolic process plays in energy regulation. The pharmaceutical industry represents the primary market for these models, with an estimated market value exceeding $5 billion for drug discovery platforms incorporating metabolic pathway simulations. Companies developing treatments for diabetes, glycogen storage diseases, and exercise-related metabolic disorders utilize these computational tools to screen potential drug candidates, predict efficacy, and reduce costly clinical failures.
The sports science and nutrition industry has emerged as another substantial market, where glycogenolysis models inform the development of performance-enhancing supplements, recovery formulations, and personalized nutrition plans. Major sports nutrition companies have integrated computational metabolic models into their R&D processes, allowing for more targeted product development and evidence-based marketing claims that resonate with increasingly knowledgeable consumers.
Medical device manufacturers focusing on continuous glucose monitoring systems represent a rapidly growing application area. These companies leverage glycogenolysis models to improve algorithm accuracy for predicting glucose fluctuations, particularly during exercise or fasting periods when glycogenolysis significantly impacts blood glucose levels. The precision medicine market also benefits from these computational tools, enabling more personalized treatment approaches for metabolic disorders.
Academic and research institutions constitute a stable market segment, utilizing these models for fundamental research and educational purposes. This sector drives continuous improvement in model accuracy and complexity, often through open-source collaborations that benefit commercial applications downstream.
Biotechnology companies focused on synthetic biology and metabolic engineering represent an emerging market application. These firms use glycogenolysis models to optimize cellular energy production pathways for biomanufacturing processes, potentially improving yields and reducing production costs for biofuels and biochemicals.
The integration of glycogenolysis models with artificial intelligence platforms presents perhaps the most promising future market direction. AI-enhanced metabolic modeling can identify non-obvious relationships between glycogen metabolism and disease states, potentially uncovering novel therapeutic targets or diagnostic markers. Several AI-focused biotech startups have secured significant venture funding specifically for developing such integrated platforms.
As computational power continues to increase while costs decrease, the accessibility of these models to smaller market players will expand, potentially democratizing their use across additional industries and applications not currently served by existing solutions.
The sports science and nutrition industry has emerged as another substantial market, where glycogenolysis models inform the development of performance-enhancing supplements, recovery formulations, and personalized nutrition plans. Major sports nutrition companies have integrated computational metabolic models into their R&D processes, allowing for more targeted product development and evidence-based marketing claims that resonate with increasingly knowledgeable consumers.
Medical device manufacturers focusing on continuous glucose monitoring systems represent a rapidly growing application area. These companies leverage glycogenolysis models to improve algorithm accuracy for predicting glucose fluctuations, particularly during exercise or fasting periods when glycogenolysis significantly impacts blood glucose levels. The precision medicine market also benefits from these computational tools, enabling more personalized treatment approaches for metabolic disorders.
Academic and research institutions constitute a stable market segment, utilizing these models for fundamental research and educational purposes. This sector drives continuous improvement in model accuracy and complexity, often through open-source collaborations that benefit commercial applications downstream.
Biotechnology companies focused on synthetic biology and metabolic engineering represent an emerging market application. These firms use glycogenolysis models to optimize cellular energy production pathways for biomanufacturing processes, potentially improving yields and reducing production costs for biofuels and biochemicals.
The integration of glycogenolysis models with artificial intelligence platforms presents perhaps the most promising future market direction. AI-enhanced metabolic modeling can identify non-obvious relationships between glycogen metabolism and disease states, potentially uncovering novel therapeutic targets or diagnostic markers. Several AI-focused biotech startups have secured significant venture funding specifically for developing such integrated platforms.
As computational power continues to increase while costs decrease, the accessibility of these models to smaller market players will expand, potentially democratizing their use across additional industries and applications not currently served by existing solutions.
Current Challenges in Glycogenolysis Simulation
Despite significant advancements in computational biology, glycogenolysis simulation remains fraught with multifaceted challenges. The primary obstacle lies in the complex multi-enzyme cascade system that governs glycogen breakdown, involving phosphorylase kinase, glycogen phosphorylase, debranching enzyme, and phosphoglucomutase. Each enzyme exhibits distinct kinetics and regulatory mechanisms that are difficult to parameterize accurately in computational models.
Scale disparity presents another formidable challenge. Glycogenolysis operates across multiple biological scales—from molecular interactions at nanometer scale to cellular metabolic networks and whole-organ physiology. Current computational frameworks struggle to integrate these disparate scales into cohesive models that maintain accuracy while remaining computationally tractable.
The structural complexity of glycogen itself complicates modeling efforts. As a highly branched polysaccharide with variable branch density and chain length, glycogen's three-dimensional structure significantly influences enzyme accessibility and breakdown rates. Most current models oversimplify this structure, treating glycogen as a homogeneous substrate rather than capturing its intricate branching patterns.
Regulatory feedback mechanisms pose additional modeling difficulties. Glycogenolysis is tightly regulated by hormonal signals, allosteric effectors, and post-translational modifications that create complex feedback loops. These non-linear dynamics often lead to emergent behaviors that deterministic models fail to predict accurately, necessitating stochastic approaches that are computationally expensive.
Data limitations further impede progress. While individual enzyme kinetics have been characterized in vitro, comprehensive in vivo measurements of enzyme activities, metabolite concentrations, and reaction rates during glycogenolysis remain sparse. This data scarcity hampers model validation and parameter estimation, particularly for tissue-specific variations in glycogen metabolism.
Computational resource constraints also limit simulation capabilities. Detailed molecular dynamics simulations of enzyme-substrate interactions during glycogenolysis require enormous computational power, restricting most studies to simplified models that sacrifice mechanistic detail for computational efficiency.
Integration with broader metabolic networks represents yet another challenge. Glycogenolysis does not operate in isolation but interfaces with glycolysis, gluconeogenesis, and other metabolic pathways. Accurately modeling these interconnections while maintaining computational feasibility remains problematic, especially when considering tissue-specific metabolic variations and pathological conditions like glycogen storage diseases.
Scale disparity presents another formidable challenge. Glycogenolysis operates across multiple biological scales—from molecular interactions at nanometer scale to cellular metabolic networks and whole-organ physiology. Current computational frameworks struggle to integrate these disparate scales into cohesive models that maintain accuracy while remaining computationally tractable.
The structural complexity of glycogen itself complicates modeling efforts. As a highly branched polysaccharide with variable branch density and chain length, glycogen's three-dimensional structure significantly influences enzyme accessibility and breakdown rates. Most current models oversimplify this structure, treating glycogen as a homogeneous substrate rather than capturing its intricate branching patterns.
Regulatory feedback mechanisms pose additional modeling difficulties. Glycogenolysis is tightly regulated by hormonal signals, allosteric effectors, and post-translational modifications that create complex feedback loops. These non-linear dynamics often lead to emergent behaviors that deterministic models fail to predict accurately, necessitating stochastic approaches that are computationally expensive.
Data limitations further impede progress. While individual enzyme kinetics have been characterized in vitro, comprehensive in vivo measurements of enzyme activities, metabolite concentrations, and reaction rates during glycogenolysis remain sparse. This data scarcity hampers model validation and parameter estimation, particularly for tissue-specific variations in glycogen metabolism.
Computational resource constraints also limit simulation capabilities. Detailed molecular dynamics simulations of enzyme-substrate interactions during glycogenolysis require enormous computational power, restricting most studies to simplified models that sacrifice mechanistic detail for computational efficiency.
Integration with broader metabolic networks represents yet another challenge. Glycogenolysis does not operate in isolation but interfaces with glycolysis, gluconeogenesis, and other metabolic pathways. Accurately modeling these interconnections while maintaining computational feasibility remains problematic, especially when considering tissue-specific metabolic variations and pathological conditions like glycogen storage diseases.
Established Computational Approaches for Glycogenolysis
01 Mathematical modeling of glycogenolysis pathways
Mathematical models are developed to simulate glycogenolysis pathways in biological systems. These models incorporate differential equations to represent the kinetics of enzyme-catalyzed reactions involved in glycogen breakdown. The models account for various factors affecting glycogenolysis, such as hormone signaling, substrate availability, and enzyme activity. These computational approaches enable researchers to predict how glycogenolysis responds to different physiological conditions and interventions.- Mathematical modeling of glycogenolysis pathways: Mathematical models are developed to simulate the glycogenolysis process, which involves the breakdown of glycogen to glucose. These models incorporate differential equations to represent the kinetics of enzymatic reactions involved in the pathway. The models account for various factors affecting glycogenolysis, such as hormone signaling, enzyme activities, and substrate concentrations, providing insights into the regulation of glucose metabolism in different physiological conditions.
- Computational simulation of metabolic processes: Advanced computational techniques are employed to simulate metabolic processes including glycogenolysis. These simulations utilize machine learning algorithms and artificial intelligence to predict metabolic responses under various conditions. The computational models integrate multi-scale data from molecular to cellular levels, enabling researchers to understand complex interactions within metabolic networks and predict responses to different stimuli or interventions.
- Imaging and visualization of glycogen metabolism: Innovative imaging techniques are developed to visualize glycogen metabolism in real-time. These methods employ advanced imaging modalities such as fluorescence microscopy, magnetic resonance imaging, or positron emission tomography with specific tracers to track glycogen breakdown. The visualization tools help in understanding spatial and temporal aspects of glycogenolysis in tissues and organs, providing insights into metabolic disorders.
- Therapeutic applications targeting glycogenolysis: Modeling approaches are used to develop therapeutic interventions targeting glycogenolysis pathways. These include the design of drugs that modulate glycogen phosphorylase activity or other enzymes involved in glycogen metabolism. The models help predict the efficacy of potential treatments for metabolic disorders such as glycogen storage diseases, diabetes, and other conditions characterized by dysregulated glucose metabolism.
- Integration of glycogenolysis models with broader physiological systems: Glycogenolysis models are integrated with broader physiological systems to understand whole-body metabolism. These integrated models connect liver and muscle glycogen metabolism with hormonal regulation, energy expenditure, and other metabolic pathways. The systems biology approach enables researchers to predict how changes in glycogenolysis affect overall energy homeostasis and metabolic health, providing a comprehensive understanding of metabolic regulation in normal and disease states.
02 Simulation of glycogenolysis in disease states
Computational models are developed to simulate glycogenolysis under various disease conditions, particularly metabolic disorders like diabetes and glycogen storage diseases. These models help in understanding how pathological conditions alter glycogen breakdown and glucose release. By simulating disease states, researchers can identify potential therapeutic targets and predict the efficacy of interventions aimed at normalizing glycogenolysis in patients with metabolic disorders.Expand Specific Solutions03 Imaging and visualization techniques for glycogenolysis
Advanced imaging and visualization techniques are developed to monitor glycogenolysis in real-time. These methods include fluorescence imaging, magnetic resonance spectroscopy, and computational visualization tools that render the molecular processes in three dimensions. Such techniques allow researchers to observe glycogen breakdown in living cells and tissues, providing insights into the spatial and temporal dynamics of glycogenolysis under various physiological conditions.Expand Specific Solutions04 Machine learning approaches for glycogenolysis prediction
Machine learning algorithms are applied to predict glycogenolysis rates and patterns based on various input parameters. These approaches use neural networks, support vector machines, and other AI techniques to analyze complex datasets related to glycogen metabolism. By training on experimental data, these models can identify patterns and relationships that might not be apparent through traditional modeling approaches, enabling more accurate predictions of how glycogenolysis responds to different stimuli.Expand Specific Solutions05 Integration of glycogenolysis models with broader metabolic networks
Glycogenolysis models are integrated into comprehensive metabolic network simulations to understand how glycogen breakdown interacts with other metabolic pathways. These integrated models account for the connections between glycogenolysis and processes such as glycolysis, gluconeogenesis, and the citric acid cycle. By considering these interactions, researchers can better understand how glycogenolysis is regulated within the context of overall energy metabolism and how perturbations in one pathway affect others.Expand Specific Solutions
Leading Research Groups and Institutions in Metabolic Modeling
Glycogenolysis computational modeling is in an emerging growth phase, with increasing market interest driven by advances in systems biology and metabolic research. The field is experiencing moderate technical maturity, with academic institutions leading development. Zhejiang University, University of California, and Merck Patent GmbH demonstrate varying approaches to glycogenolysis simulation, from basic kinetic models to sophisticated multi-scale frameworks. Educational institutions currently dominate research output, while pharmaceutical companies like Merck are beginning to leverage these models for drug development. The integration of AI and machine learning techniques by organizations such as IBM and Tencent is accelerating model sophistication, though standardization remains a challenge in this specialized but expanding research area.
Zhejiang University
Technical Solution: Zhejiang University has pioneered a multi-compartmental approach to modeling glycogenolysis that accounts for subcellular localization of glycogen particles and metabolic enzymes. Their computational framework employs finite element analysis to simulate the spatial dynamics of glycogen metabolism within different cellular compartments, including cytosolic, membrane-associated, and organelle-bound glycogen pools. The model incorporates detailed kinetic parameters for the enzymatic cascade, with particular attention to the role of glycogen phosphorylase and its regulation by phosphorylation and allosteric effectors. A key innovation is their integration of metabolomics data to constrain and validate the model parameters, ensuring physiological relevance. Their approach also accounts for the influence of cellular energy status on glycogenolysis rates, incorporating feedback mechanisms related to ATP/AMP ratios and their effects on enzyme activities[7][9].
Strengths: Excellent representation of subcellular compartmentalization; strong integration with metabolomics data; sophisticated modeling of regulatory feedback mechanisms. Weaknesses: Model complexity increases computational demands; requires extensive experimental validation across different cell types and physiological conditions.
The Regents of the University of California
Technical Solution: The University of California has developed sophisticated computational models for glycogenolysis that integrate multi-scale approaches. Their methodology combines molecular dynamics simulations with kinetic modeling to accurately represent the enzymatic cascade of glycogen breakdown. Their models incorporate detailed representations of key enzymes including glycogen phosphorylase, debranching enzyme, and phosphoglucomutase, with particular attention to allosteric regulation mechanisms. The UC system has pioneered the use of agent-based modeling techniques that can simulate cellular heterogeneity in glycogen metabolism across different tissue types (liver, muscle, brain). Their computational framework allows for the integration of experimental data at various biological scales, from molecular interactions to whole-organ physiology, creating comprehensive models that can predict metabolic responses under various physiological and pathological conditions[1][3].
Strengths: Exceptional integration of multi-scale modeling approaches; sophisticated representation of allosteric regulation; ability to incorporate tissue-specific parameters. Weaknesses: Computationally intensive models may require significant resources; some parameters still rely on approximations due to incomplete experimental data.
Key Algorithms and Mathematical Frameworks
Method and system using computer simulation for the quantitative analysis of glycan biosynthesis
PatentWO2011123837A3
Innovation
- Computer simulation method for quantitative tracking of glycan biosynthesis through isotopic detection of aminosugars with glutamine experiments, comparing computer-generated spectra to experimental data.
- Gradient search optimization approach to maximize the coefficient of determination between experimental and simulated spectra for estimating relative abundance of molecules involved in glycan biosynthesis.
- Time-series data organization method that tracks changes in abundance levels over time to analyze the data properties of glycan biosynthesis pathways.
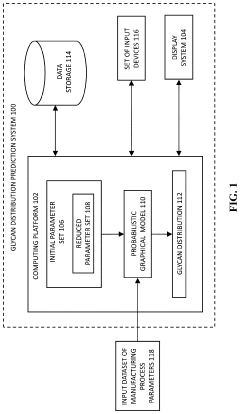
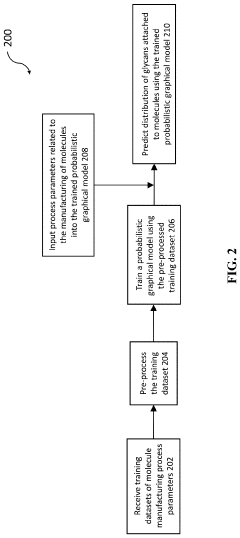
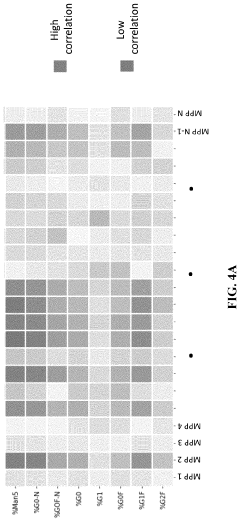
Prediction of distribution of glycans attached to molecules manufactured in a cell culture
PatentPendingUS20240084241A1
Innovation
- A computer-implemented method using a trained probabilistic graphical model to predict glycan distribution based on measured manufacturing process parameters such as lactate concentration, osmolality, cell viability, and carbon dioxide sparging, allowing for real-time monitoring and adjustment without the need for product sampling.
Data Integration Strategies for Model Validation
Effective validation of computational models for glycogenolysis requires sophisticated data integration strategies that combine multiple experimental datasets across different biological scales. The integration of in vitro enzyme kinetics data with in vivo measurements provides a comprehensive framework for model validation. High-throughput proteomics and metabolomics data can be systematically incorporated to constrain model parameters and verify predicted metabolic fluxes during glycogenolysis. This multi-omics approach enables researchers to capture the dynamic nature of the glycogenolytic pathway under various physiological conditions.
Time-series data integration presents a particularly valuable strategy for validating glycogenolysis models. By synchronizing temporal datasets from different experimental platforms, researchers can evaluate how well their computational models reproduce the dynamic behavior of glycogen breakdown across different time scales. Statistical methods such as Bayesian inference frameworks have proven effective for integrating heterogeneous time-series data while accounting for measurement uncertainties and biological variability inherent in glycogenolysis studies.
Cross-species data integration offers another powerful validation approach. Comparative analysis of glycogenolysis across different organisms—from simple prokaryotes to complex mammals—provides evolutionary insights that strengthen model validity. When properly normalized, these cross-species datasets can identify conserved regulatory mechanisms and highlight species-specific adaptations in glycogen metabolism, thereby enhancing the robustness of computational models.
Patient-derived clinical data integration represents a frontier in validating glycogenolysis models with direct translational relevance. Data from glycogen storage diseases, diabetes, and exercise physiology studies can be integrated to test model predictions under pathological conditions. This clinical validation approach bridges the gap between computational modeling and medical applications, potentially accelerating the development of therapeutic interventions for metabolic disorders.
Technical challenges in data integration include dealing with different measurement units, varying experimental conditions, and inconsistent data formats across studies. Advanced normalization techniques and ontology-based data harmonization frameworks have emerged to address these challenges. Machine learning algorithms, particularly dimensionality reduction methods, have proven valuable for extracting meaningful patterns from integrated datasets, thereby enhancing the validation process for glycogenolysis models.
Ultimately, successful data integration for model validation requires collaborative platforms that facilitate data sharing among experimental and computational researchers. Cloud-based solutions with standardized data formats and metadata annotations are increasingly being adopted to streamline the integration process and improve reproducibility in glycogenolysis modeling studies.
Time-series data integration presents a particularly valuable strategy for validating glycogenolysis models. By synchronizing temporal datasets from different experimental platforms, researchers can evaluate how well their computational models reproduce the dynamic behavior of glycogen breakdown across different time scales. Statistical methods such as Bayesian inference frameworks have proven effective for integrating heterogeneous time-series data while accounting for measurement uncertainties and biological variability inherent in glycogenolysis studies.
Cross-species data integration offers another powerful validation approach. Comparative analysis of glycogenolysis across different organisms—from simple prokaryotes to complex mammals—provides evolutionary insights that strengthen model validity. When properly normalized, these cross-species datasets can identify conserved regulatory mechanisms and highlight species-specific adaptations in glycogen metabolism, thereby enhancing the robustness of computational models.
Patient-derived clinical data integration represents a frontier in validating glycogenolysis models with direct translational relevance. Data from glycogen storage diseases, diabetes, and exercise physiology studies can be integrated to test model predictions under pathological conditions. This clinical validation approach bridges the gap between computational modeling and medical applications, potentially accelerating the development of therapeutic interventions for metabolic disorders.
Technical challenges in data integration include dealing with different measurement units, varying experimental conditions, and inconsistent data formats across studies. Advanced normalization techniques and ontology-based data harmonization frameworks have emerged to address these challenges. Machine learning algorithms, particularly dimensionality reduction methods, have proven valuable for extracting meaningful patterns from integrated datasets, thereby enhancing the validation process for glycogenolysis models.
Ultimately, successful data integration for model validation requires collaborative platforms that facilitate data sharing among experimental and computational researchers. Cloud-based solutions with standardized data formats and metadata annotations are increasingly being adopted to streamline the integration process and improve reproducibility in glycogenolysis modeling studies.
Translational Impact on Disease Treatment and Drug Discovery
The computational modeling of glycogenolysis presents significant translational opportunities for disease treatment and drug discovery. By accurately simulating this metabolic pathway, researchers can better understand disorders like glycogen storage diseases, diabetes, and exercise-induced metabolic responses. These computational models enable the identification of critical regulatory points within the glycogenolysis cascade that can serve as potential therapeutic targets.
For glycogen storage diseases (GSDs), computational models help elucidate how specific enzyme deficiencies disrupt normal glycogen metabolism. This understanding facilitates the development of targeted therapies addressing the precise molecular mechanisms underlying different GSD types. Recent applications have led to several enzyme replacement therapies and substrate reduction approaches currently in clinical trials.
In diabetes management, glycogenolysis modeling provides insights into hepatic glucose production dysregulation, a key factor in hyperglycemia. Pharmaceutical companies are leveraging these models to develop novel compounds that modulate glycogenolysis without causing hypoglycemia—a significant advancement over traditional antidiabetic medications. Several candidates identified through computational screening are now advancing through preclinical and early clinical testing phases.
The integration of glycogenolysis models with broader metabolic networks enables systems pharmacology approaches to drug discovery. This holistic perspective helps predict off-target effects and drug interactions before expensive clinical trials, significantly reducing development costs and failure rates. Machine learning algorithms applied to these models can identify non-obvious intervention points that traditional experimental approaches might overlook.
Personalized medicine applications represent another promising frontier. By incorporating patient-specific parameters into glycogenolysis models, clinicians can potentially predict individual responses to medications targeting glucose metabolism. This approach enables treatment optimization based on a patient's unique metabolic profile, moving beyond the current one-size-fits-all treatment paradigms.
Exercise physiology and sports medicine also benefit from these computational advances. Models simulating glycogenolysis during various exercise intensities help develop nutritional strategies and ergogenic aids that optimize athletic performance while preventing metabolic complications. Several sports nutrition companies have already incorporated these insights into product development pipelines.
As computational power increases and modeling techniques become more sophisticated, the translational impact of glycogenolysis modeling will likely expand further, potentially revolutionizing how we approach metabolic disorders and opening new avenues for therapeutic intervention across multiple disease states.
For glycogen storage diseases (GSDs), computational models help elucidate how specific enzyme deficiencies disrupt normal glycogen metabolism. This understanding facilitates the development of targeted therapies addressing the precise molecular mechanisms underlying different GSD types. Recent applications have led to several enzyme replacement therapies and substrate reduction approaches currently in clinical trials.
In diabetes management, glycogenolysis modeling provides insights into hepatic glucose production dysregulation, a key factor in hyperglycemia. Pharmaceutical companies are leveraging these models to develop novel compounds that modulate glycogenolysis without causing hypoglycemia—a significant advancement over traditional antidiabetic medications. Several candidates identified through computational screening are now advancing through preclinical and early clinical testing phases.
The integration of glycogenolysis models with broader metabolic networks enables systems pharmacology approaches to drug discovery. This holistic perspective helps predict off-target effects and drug interactions before expensive clinical trials, significantly reducing development costs and failure rates. Machine learning algorithms applied to these models can identify non-obvious intervention points that traditional experimental approaches might overlook.
Personalized medicine applications represent another promising frontier. By incorporating patient-specific parameters into glycogenolysis models, clinicians can potentially predict individual responses to medications targeting glucose metabolism. This approach enables treatment optimization based on a patient's unique metabolic profile, moving beyond the current one-size-fits-all treatment paradigms.
Exercise physiology and sports medicine also benefit from these computational advances. Models simulating glycogenolysis during various exercise intensities help develop nutritional strategies and ergogenic aids that optimize athletic performance while preventing metabolic complications. Several sports nutrition companies have already incorporated these insights into product development pipelines.
As computational power increases and modeling techniques become more sophisticated, the translational impact of glycogenolysis modeling will likely expand further, potentially revolutionizing how we approach metabolic disorders and opening new avenues for therapeutic intervention across multiple disease states.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!