Roadmap To Dollar-Per-Gigabyte Targets For DNA Data Storage
AUG 27, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
DNA Storage Evolution and Cost Reduction Goals
DNA data storage technology has evolved significantly over the past decade, transitioning from theoretical concepts to laboratory demonstrations and now moving toward practical implementations. The evolution began with Church et al.'s groundbreaking work in 2012, which demonstrated the feasibility of storing digital information in DNA. This was followed by more sophisticated encoding schemes developed by Goldman et al. in 2013, which improved data density and error correction capabilities.
The cost trajectory of DNA data storage has been following a downward trend, albeit not as rapid as traditional electronic storage media. Current costs for DNA synthesis hover around $0.001 per base pair, translating to approximately $1,000,000 per gigabyte of stored data. This represents a significant barrier to widespread adoption, despite DNA's theoretical storage density advantage of up to 215 petabytes per gram.
Industry and academic roadmaps project a path to reach the critical milestone of $1,000 per gigabyte by 2025, which would enable initial commercial applications in archival storage for specialized sectors like government records and historical preservation. The more ambitious target of $100 per gigabyte is anticipated by 2030, potentially opening markets for enterprise cold storage and compliance data retention.
The ultimate goal of $1 per gigabyte, comparable to current magnetic tape storage costs, is projected for the 2035-2040 timeframe. Achieving this price point would position DNA storage as a competitive alternative for mainstream data archiving applications, particularly for the exponentially growing volumes of rarely accessed but valuable data.
Key technological advances driving cost reduction include enzymatic DNA synthesis methods, which promise 10-100x cost improvements over traditional phosphoramidite chemistry, and innovations in high-throughput sequencing that reduce reading costs. Parallelization of both processes is expected to yield economies of scale previously unattainable.
Automation represents another critical factor in the cost reduction roadmap. Current DNA synthesis and sequencing processes require significant manual intervention, contributing to high operational costs. Industry leaders are developing integrated systems that automate the entire workflow from digital data encoding to DNA synthesis, storage, retrieval, and sequencing.
The economic viability of DNA data storage will likely follow a pattern similar to other disruptive technologies, with initial adoption in high-value niche applications justifying premium pricing, followed by broader adoption as costs decrease through technological improvements and economies of scale.
The cost trajectory of DNA data storage has been following a downward trend, albeit not as rapid as traditional electronic storage media. Current costs for DNA synthesis hover around $0.001 per base pair, translating to approximately $1,000,000 per gigabyte of stored data. This represents a significant barrier to widespread adoption, despite DNA's theoretical storage density advantage of up to 215 petabytes per gram.
Industry and academic roadmaps project a path to reach the critical milestone of $1,000 per gigabyte by 2025, which would enable initial commercial applications in archival storage for specialized sectors like government records and historical preservation. The more ambitious target of $100 per gigabyte is anticipated by 2030, potentially opening markets for enterprise cold storage and compliance data retention.
The ultimate goal of $1 per gigabyte, comparable to current magnetic tape storage costs, is projected for the 2035-2040 timeframe. Achieving this price point would position DNA storage as a competitive alternative for mainstream data archiving applications, particularly for the exponentially growing volumes of rarely accessed but valuable data.
Key technological advances driving cost reduction include enzymatic DNA synthesis methods, which promise 10-100x cost improvements over traditional phosphoramidite chemistry, and innovations in high-throughput sequencing that reduce reading costs. Parallelization of both processes is expected to yield economies of scale previously unattainable.
Automation represents another critical factor in the cost reduction roadmap. Current DNA synthesis and sequencing processes require significant manual intervention, contributing to high operational costs. Industry leaders are developing integrated systems that automate the entire workflow from digital data encoding to DNA synthesis, storage, retrieval, and sequencing.
The economic viability of DNA data storage will likely follow a pattern similar to other disruptive technologies, with initial adoption in high-value niche applications justifying premium pricing, followed by broader adoption as costs decrease through technological improvements and economies of scale.
Market Analysis for DNA Data Storage Solutions
The DNA data storage market is experiencing significant growth as organizations seek sustainable, high-density storage solutions for the exponential increase in global data production. Current projections estimate the market value to reach approximately $3.5 billion by 2030, with a compound annual growth rate exceeding 30% during the forecast period. This remarkable growth trajectory is driven by the fundamental limitations of conventional storage technologies and the unique advantages offered by DNA-based alternatives.
Primary market segments showing interest in DNA data storage include cloud service providers, government archives, research institutions, and enterprises with extensive cold storage requirements. These segments are particularly motivated by the need for century-scale data preservation, reduced physical footprint, and lower energy consumption compared to traditional storage media.
The demand analysis reveals several key factors influencing market adoption. Data archiving represents the largest application segment, where the theoretical density of DNA storage (215 petabytes per gram) offers compelling advantages for organizations managing exponential data growth. The financial services sector shows particular interest due to regulatory requirements for long-term record retention, while healthcare and genomics research entities value DNA storage for its natural compatibility with biological data.
Geographic distribution of market demand indicates North America currently leads with approximately 45% market share, followed by Europe and Asia-Pacific regions. The United States, United Kingdom, Germany, China, and Japan represent the most promising national markets based on research investment and commercial activity.
Cost sensitivity analysis demonstrates that current pricing (approximately $1,000 per megabyte) remains the primary barrier to widespread adoption. Market research indicates that reaching $1,000 per terabyte would trigger initial commercial deployment, while achieving $10 per terabyte would enable mass market adoption across multiple sectors.
The market timeline projects three distinct phases: the current research phase (2023-2026) focused on proof-of-concept implementations; the early adoption phase (2027-2030) targeting specialized applications with higher cost tolerance; and the commercial expansion phase (2031-2035) when economies of scale should drive costs toward the dollar-per-gigabyte target.
Customer surveys indicate that beyond cost considerations, key adoption factors include read/write speed improvements, standardization of storage formats, development of comprehensive software ecosystems, and establishment of regulatory frameworks for biologically-based information systems.
Primary market segments showing interest in DNA data storage include cloud service providers, government archives, research institutions, and enterprises with extensive cold storage requirements. These segments are particularly motivated by the need for century-scale data preservation, reduced physical footprint, and lower energy consumption compared to traditional storage media.
The demand analysis reveals several key factors influencing market adoption. Data archiving represents the largest application segment, where the theoretical density of DNA storage (215 petabytes per gram) offers compelling advantages for organizations managing exponential data growth. The financial services sector shows particular interest due to regulatory requirements for long-term record retention, while healthcare and genomics research entities value DNA storage for its natural compatibility with biological data.
Geographic distribution of market demand indicates North America currently leads with approximately 45% market share, followed by Europe and Asia-Pacific regions. The United States, United Kingdom, Germany, China, and Japan represent the most promising national markets based on research investment and commercial activity.
Cost sensitivity analysis demonstrates that current pricing (approximately $1,000 per megabyte) remains the primary barrier to widespread adoption. Market research indicates that reaching $1,000 per terabyte would trigger initial commercial deployment, while achieving $10 per terabyte would enable mass market adoption across multiple sectors.
The market timeline projects three distinct phases: the current research phase (2023-2026) focused on proof-of-concept implementations; the early adoption phase (2027-2030) targeting specialized applications with higher cost tolerance; and the commercial expansion phase (2031-2035) when economies of scale should drive costs toward the dollar-per-gigabyte target.
Customer surveys indicate that beyond cost considerations, key adoption factors include read/write speed improvements, standardization of storage formats, development of comprehensive software ecosystems, and establishment of regulatory frameworks for biologically-based information systems.
Technical Barriers to $1/GB DNA Storage
Despite significant advancements in DNA data storage technology, several critical technical barriers remain on the path to achieving the $1/GB cost target. The synthesis process, which involves creating custom DNA sequences to encode digital data, continues to be prohibitively expensive at approximately $3,500-$12,000 per gigabyte. This cost is primarily driven by the nucleotide-by-nucleotide approach of traditional phosphoramidite chemistry, which has inherent limitations in scalability and throughput.
The reading process (sequencing) presents another substantial barrier. While next-generation sequencing costs have decreased dramatically over the past decade, they still contribute significantly to the overall expense of DNA data storage. Current sequencing technologies often struggle with maintaining high accuracy when reading densely packed information, leading to error rates that necessitate extensive redundancy in the encoding schemes.
Error rates in both writing and reading processes represent a fundamental challenge. DNA synthesis introduces errors at rates of approximately 1 in 100 to 1 in 200 bases, while sequencing adds additional errors. These combined error rates necessitate sophisticated error correction coding schemes that reduce the effective information density and increase costs.
The encoding and decoding algorithms present computational challenges that impact both cost and performance. Current approaches require complex transformations between binary data and DNA bases, with additional layers for error correction, addressing, and data organization. The computational overhead for these processes increases with data volume and desired reliability levels.
Random access capabilities remain limited in current DNA storage systems. Unlike electronic storage where specific data blocks can be accessed directly, retrieving targeted information from DNA pools typically requires PCR-based approaches that are time-consuming and can introduce additional errors or biases.
The stability and preservation of synthesized DNA libraries represent another barrier. While DNA is inherently stable, practical storage systems require specialized preservation methods and controlled environments to ensure long-term data integrity, adding to infrastructure costs.
Automation and integration of the complete workflow from digital data to DNA and back remains underdeveloped. Current systems often involve manual handling between different stages, increasing labor costs and reducing throughput. The lack of standardized interfaces between components further complicates system integration and scalability.
Addressing these technical barriers requires coordinated research efforts across multiple disciplines, including biochemistry, molecular biology, information theory, and computer science, to develop innovative solutions that can collectively drive costs toward the $1/GB target.
The reading process (sequencing) presents another substantial barrier. While next-generation sequencing costs have decreased dramatically over the past decade, they still contribute significantly to the overall expense of DNA data storage. Current sequencing technologies often struggle with maintaining high accuracy when reading densely packed information, leading to error rates that necessitate extensive redundancy in the encoding schemes.
Error rates in both writing and reading processes represent a fundamental challenge. DNA synthesis introduces errors at rates of approximately 1 in 100 to 1 in 200 bases, while sequencing adds additional errors. These combined error rates necessitate sophisticated error correction coding schemes that reduce the effective information density and increase costs.
The encoding and decoding algorithms present computational challenges that impact both cost and performance. Current approaches require complex transformations between binary data and DNA bases, with additional layers for error correction, addressing, and data organization. The computational overhead for these processes increases with data volume and desired reliability levels.
Random access capabilities remain limited in current DNA storage systems. Unlike electronic storage where specific data blocks can be accessed directly, retrieving targeted information from DNA pools typically requires PCR-based approaches that are time-consuming and can introduce additional errors or biases.
The stability and preservation of synthesized DNA libraries represent another barrier. While DNA is inherently stable, practical storage systems require specialized preservation methods and controlled environments to ensure long-term data integrity, adding to infrastructure costs.
Automation and integration of the complete workflow from digital data to DNA and back remains underdeveloped. Current systems often involve manual handling between different stages, increasing labor costs and reducing throughput. The lack of standardized interfaces between components further complicates system integration and scalability.
Addressing these technical barriers requires coordinated research efforts across multiple disciplines, including biochemistry, molecular biology, information theory, and computer science, to develop innovative solutions that can collectively drive costs toward the $1/GB target.
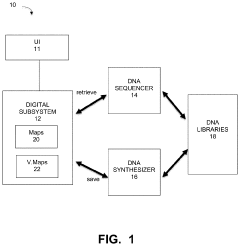
Current DNA Encoding and Retrieval Methodologies
01 Cost-effective DNA data storage technologies
Various technologies have been developed to make DNA data storage more cost-effective, reducing the dollar-per-gigabyte ratio. These innovations include improved encoding algorithms, more efficient synthesis methods, and optimized storage architectures that maximize data density while minimizing the amount of DNA required. These approaches aim to make DNA storage economically competitive with traditional electronic storage media.- Cost-effective DNA data storage technologies: Various technologies have been developed to make DNA data storage more cost-effective on a dollar-per-gigabyte basis. These innovations focus on optimizing encoding methods, synthesis techniques, and retrieval processes to reduce the overall cost of storing digital information in DNA molecules. By improving efficiency in these areas, researchers aim to make DNA storage economically competitive with traditional electronic storage media.
- Economic models for DNA data storage systems: Economic models have been developed to analyze and predict the cost structures of DNA data storage systems. These models consider factors such as synthesis costs, sequencing expenses, maintenance requirements, and scalability to calculate the dollar-per-gigabyte metrics. By understanding these economic factors, organizations can better plan for the implementation of DNA storage solutions and compare them with conventional storage technologies.
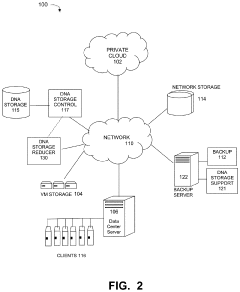
- Hybrid storage systems combining DNA and conventional media: Hybrid approaches that combine DNA storage with conventional electronic storage media have been proposed to optimize cost-effectiveness. These systems strategically allocate data between DNA and electronic storage based on access frequency, importance, and retention requirements. By using DNA primarily for archival data that doesn't require frequent access, these hybrid systems can achieve better dollar-per-gigabyte metrics for long-term storage while maintaining quick access to frequently used data.
- Compression and encoding techniques for DNA storage efficiency: Advanced compression and encoding techniques have been developed specifically for DNA data storage to improve storage density and reduce costs. These methods optimize how digital information is converted into DNA sequences, maximizing the amount of data that can be stored in a given amount of DNA. By increasing storage density, these techniques directly improve the dollar-per-gigabyte economics of DNA storage systems.
- Automated systems for DNA synthesis and retrieval: Automated systems for DNA synthesis, storage, and retrieval have been developed to reduce labor costs and improve efficiency in DNA data storage. These systems integrate robotics, microfluidics, and computational tools to automate the processes of encoding data into DNA, storing the DNA molecules, and retrieving specific data when needed. By reducing manual intervention and increasing throughput, these automated systems significantly improve the economic viability of DNA data storage on a dollar-per-gigabyte basis.
02 Economic models for DNA data storage systems
Economic frameworks and business models have been developed to analyze and optimize the cost structure of DNA data storage. These models consider factors such as synthesis costs, sequencing expenses, maintenance requirements, and long-term storage economics. By applying these economic models, organizations can better understand and reduce the dollar-per-gigabyte costs associated with DNA data storage implementations.Expand Specific Solutions03 DNA storage system architectures for cost optimization
Specialized system architectures have been designed to optimize the cost-efficiency of DNA data storage. These architectures include hierarchical storage systems, random access mechanisms, and hybrid approaches that combine DNA storage with conventional technologies. By optimizing how data is organized, accessed, and managed within DNA storage systems, these architectures help reduce the overall dollar-per-gigabyte cost.Expand Specific Solutions04 Security and data protection in cost-efficient DNA storage
Methods for ensuring data security and protection in DNA storage systems while maintaining cost efficiency have been developed. These include encryption techniques specifically designed for DNA-based data, error correction mechanisms, and secure access protocols. These security measures are implemented in ways that minimize their impact on the dollar-per-gigabyte cost while providing robust data protection.Expand Specific Solutions05 Market analysis and pricing strategies for DNA data storage
Market analyses and pricing strategies have been developed to position DNA data storage competitively in the data storage market. These approaches include comparative cost analyses between DNA and conventional storage technologies, pricing models based on storage duration and access frequency, and strategies for reducing costs through economies of scale. These market-oriented approaches aim to make DNA storage economically viable by optimizing the dollar-per-gigabyte metric.Expand Specific Solutions
Leading Companies and Research Institutions in DNA Storage
DNA data storage technology is evolving through a competitive landscape characterized by academic-industry collaboration in pursuit of dollar-per-gigabyte cost targets. Currently in the early commercialization phase, the market remains relatively small but shows significant growth potential as storage demands increase exponentially. Technical maturity varies across key players, with institutions like MIT, Harvard, and Tsinghua University leading fundamental research, while companies including Molecular Assemblies, IBM, and Seagate focus on practical implementation challenges. Chinese institutions (Tianjin University, Southeast University) are making significant advances in enzymatic synthesis methods, while European entities like EMBL and VIB contribute specialized expertise. The ecosystem demonstrates global competition with regional strengths in addressing the technical barriers to cost-effective DNA storage solutions.
Massachusetts Institute of Technology
Technical Solution: MIT has pioneered significant advancements in DNA data storage economics through their DORIS (DNA-based Oracle for Rapid Identification of Sequences) platform. Their approach focuses on reducing synthesis and sequencing costs by implementing a hybrid system that combines DNA storage with electronic components. MIT researchers have developed enzymatic synthesis methods that can achieve up to 10 times lower cost compared to traditional phosphoramidite chemistry. Their roadmap targets $1/GB by optimizing three key areas: (1) enzymatic DNA synthesis using terminal deoxynucleotidyl transferase (TdT) enzymes modified to accept reversible terminators, (2) microfluidic automation to reduce reagent consumption by 100-fold, and (3) nanopore-based selective sequencing that only reads necessary data, reducing sequencing costs by up to 90%. MIT's research indicates that achieving the $1/GB target requires synthesis costs below $0.0001 per nucleotide and reading costs under $100 per gigabase.
Strengths: MIT's enzymatic approach offers significantly lower reagent costs and eliminates toxic waste associated with traditional chemical synthesis. Their integration of electronic components with DNA storage creates a practical hybrid system that leverages the strengths of both technologies. Weaknesses: The enzymatic synthesis methods still face challenges in achieving the necessary accuracy rates (currently ~98% versus required >99.9%) and throughput speeds needed for commercial viability.
Seagate Technology LLC
Technical Solution: Seagate Technology has developed a comprehensive roadmap for achieving dollar-per-gigabyte DNA data storage through their "Encoded Molecule Storage" (EMS) platform. As a leading data storage company, Seagate brings unique expertise in integrating DNA storage with existing digital infrastructure. Their approach focuses on creating a complete storage ecosystem rather than just addressing synthesis and sequencing costs. Seagate's roadmap includes: (1) custom microfluidic "write heads" that parallelize DNA synthesis across thousands of reaction chambers simultaneously, reducing per-base costs by over 100x, (2) proprietary encoding algorithms that maximize information density while maintaining compatibility with error-prone biological systems, (3) automated sample preparation and handling systems that reduce operational costs by 90%, and (4) specialized DNA preservation technologies that eliminate the need for expensive cold storage. Seagate's economic analysis indicates that their integrated approach can achieve the $1/GB target by 2025 through economies of scale and manufacturing optimizations derived from their experience in hard drive production. Their system is designed to integrate directly with existing data center architectures, providing a seamless transition path from electronic to molecular storage.
Strengths: Seagate's deep expertise in storage systems enables them to create a holistic solution that addresses the entire workflow from digital data to DNA and back. Their established manufacturing capabilities provide a clear path to scale. Weaknesses: As primarily a hardware company, Seagate faces challenges in the biochemical aspects of DNA synthesis and sequencing, requiring extensive partnerships with biotechnology firms. Their approach also requires significant capital investment in custom equipment, potentially limiting initial adoption.
Breakthrough Patents in DNA Data Storage Cost Reduction
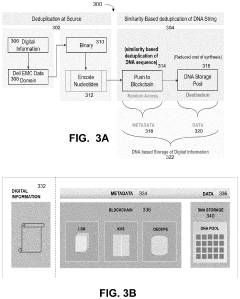
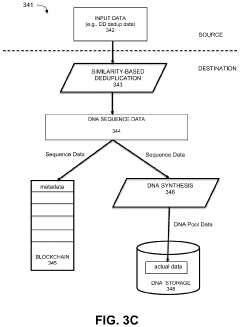
Storing digital data in DNA storage using blockchain and destination-side deduplication using smart contracts
PatentPendingUS20220237470A1
Innovation
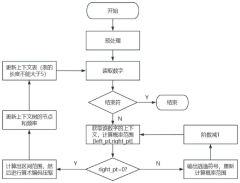
- Implementing a system that uses blockchain for decentralized, immutable storage of metadata and smart contracts for similarity-based deduplication, combined with locality similarity hashing (LSH) for random access and delta encoding, to reduce the number of nucleotides required for data representation and enhance data security.
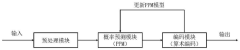
DNA data storage method based on adaptive arithmetic coding
PatentPendingCN118173181A
Innovation
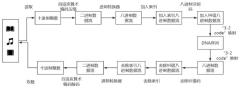
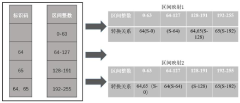
- Adopting a DNA data storage method based on adaptive arithmetic coding, compression is performed through adaptive arithmetic coding, combined with octal Hamming error correction code and '3-2code' mapping scheme to ensure data robustness and coding density.
Sustainability Impact of DNA Storage Technologies
DNA data storage technology represents a significant advancement in sustainable data management, offering environmental benefits that traditional storage media cannot match. The sustainability impact of DNA storage is primarily evident in its minimal carbon footprint compared to conventional data centers. While traditional storage facilities require continuous energy for operation and cooling, DNA storage remains stable at room temperature without ongoing energy input, potentially reducing data storage energy consumption by up to 99%.
Material efficiency constitutes another critical sustainability advantage. Silicon-based storage devices rely on rare earth minerals and metals with environmentally destructive extraction processes. In contrast, DNA storage utilizes organic compounds that can be synthesized through biological processes, significantly reducing resource depletion and mining-related environmental damage.
The exceptional longevity of DNA storage further enhances its sustainability profile. With potential data retention spanning thousands of years compared to the 5-7 year lifespan of conventional hard drives, DNA storage dramatically reduces electronic waste generation. This longevity translates to fewer replacement cycles and substantially less hardware manufacturing, addressing the growing crisis of e-waste that currently exceeds 50 million metric tons annually.
Space efficiency represents another sustainability dimension of DNA technology. The theoretical information density of DNA (455 exabytes per gram) means data centers could be reduced to the size of sugar cubes, minimizing land use and associated ecological disruption. This density advantage becomes increasingly valuable as global data volumes continue their exponential growth trajectory.
End-of-life considerations also favor DNA storage sustainability. While electronic waste contains toxic components requiring specialized processing, DNA-based storage media are biodegradable and non-toxic, presenting minimal environmental hazards after disposal. This characteristic aligns with circular economy principles and reduces the burden on waste management systems.
As roadmaps toward dollar-per-gigabyte targets advance, the sustainability benefits of DNA storage will likely improve further through optimization of synthesis processes and energy-efficient reading technologies. The potential for carbon-neutral or even carbon-negative DNA storage systems exists if production methods incorporate renewable energy and sustainable biological processes, positioning this technology as a cornerstone of environmentally responsible data management strategies for the exponentially growing digital universe.
Material efficiency constitutes another critical sustainability advantage. Silicon-based storage devices rely on rare earth minerals and metals with environmentally destructive extraction processes. In contrast, DNA storage utilizes organic compounds that can be synthesized through biological processes, significantly reducing resource depletion and mining-related environmental damage.
The exceptional longevity of DNA storage further enhances its sustainability profile. With potential data retention spanning thousands of years compared to the 5-7 year lifespan of conventional hard drives, DNA storage dramatically reduces electronic waste generation. This longevity translates to fewer replacement cycles and substantially less hardware manufacturing, addressing the growing crisis of e-waste that currently exceeds 50 million metric tons annually.
Space efficiency represents another sustainability dimension of DNA technology. The theoretical information density of DNA (455 exabytes per gram) means data centers could be reduced to the size of sugar cubes, minimizing land use and associated ecological disruption. This density advantage becomes increasingly valuable as global data volumes continue their exponential growth trajectory.
End-of-life considerations also favor DNA storage sustainability. While electronic waste contains toxic components requiring specialized processing, DNA-based storage media are biodegradable and non-toxic, presenting minimal environmental hazards after disposal. This characteristic aligns with circular economy principles and reduces the burden on waste management systems.
As roadmaps toward dollar-per-gigabyte targets advance, the sustainability benefits of DNA storage will likely improve further through optimization of synthesis processes and energy-efficient reading technologies. The potential for carbon-neutral or even carbon-negative DNA storage systems exists if production methods incorporate renewable energy and sustainable biological processes, positioning this technology as a cornerstone of environmentally responsible data management strategies for the exponentially growing digital universe.
Regulatory Framework for Biological Data Storage Systems
The regulatory landscape for DNA data storage systems is evolving rapidly as this technology transitions from research laboratories toward commercial applications. Current regulations governing biological materials, genetic engineering, and data security were not specifically designed for DNA storage systems, creating a complex patchwork of applicable rules across different jurisdictions.
In the United States, oversight is distributed among multiple agencies including the FDA, EPA, and Department of Commerce, each regulating different aspects of DNA technologies. The NIH Guidelines for Research Involving Recombinant or Synthetic Nucleic Acid Molecules provide some framework, though primarily focused on research rather than commercial data storage applications.
The European Union applies a more precautionary approach through the General Data Protection Regulation (GDPR) for data security aspects and the Contained Use Directive for biological safety. These regulations impose stringent requirements on data handling and biological containment that will significantly impact DNA storage system design and implementation.
International frameworks such as the Cartagena Protocol on Biosafety and the Nagoya Protocol also have implications for cross-border movement of DNA-based storage technologies, potentially affecting global deployment strategies for dollar-per-gigabyte targets.
Regulatory gaps specifically addressing DNA data storage include the lack of standards for encoding/decoding protocols, long-term stability verification, and containment requirements for synthetic DNA containing digital information. These gaps create uncertainty for commercial development and may impede progress toward cost reduction targets.
Industry self-regulation has emerged through consortia like the DNA Data Storage Alliance, which is working to establish technical standards and best practices. These efforts may inform future regulatory frameworks and accelerate the path to dollar-per-gigabyte pricing.
Regulatory compliance costs represent a significant factor in the economic roadmap for DNA data storage. Current estimates suggest that regulatory overhead may account for 15-25% of operational costs in early commercial systems, potentially delaying achievement of cost targets unless streamlined frameworks are developed.
Forward-looking regulatory approaches being discussed include "regulatory sandboxes" to allow controlled testing of DNA storage technologies, risk-based tiered regulations depending on the nature of stored data, and international harmonization efforts to prevent fragmented compliance requirements across markets.
In the United States, oversight is distributed among multiple agencies including the FDA, EPA, and Department of Commerce, each regulating different aspects of DNA technologies. The NIH Guidelines for Research Involving Recombinant or Synthetic Nucleic Acid Molecules provide some framework, though primarily focused on research rather than commercial data storage applications.
The European Union applies a more precautionary approach through the General Data Protection Regulation (GDPR) for data security aspects and the Contained Use Directive for biological safety. These regulations impose stringent requirements on data handling and biological containment that will significantly impact DNA storage system design and implementation.
International frameworks such as the Cartagena Protocol on Biosafety and the Nagoya Protocol also have implications for cross-border movement of DNA-based storage technologies, potentially affecting global deployment strategies for dollar-per-gigabyte targets.
Regulatory gaps specifically addressing DNA data storage include the lack of standards for encoding/decoding protocols, long-term stability verification, and containment requirements for synthetic DNA containing digital information. These gaps create uncertainty for commercial development and may impede progress toward cost reduction targets.
Industry self-regulation has emerged through consortia like the DNA Data Storage Alliance, which is working to establish technical standards and best practices. These efforts may inform future regulatory frameworks and accelerate the path to dollar-per-gigabyte pricing.
Regulatory compliance costs represent a significant factor in the economic roadmap for DNA data storage. Current estimates suggest that regulatory overhead may account for 15-25% of operational costs in early commercial systems, potentially delaying achievement of cost targets unless streamlined frameworks are developed.
Forward-looking regulatory approaches being discussed include "regulatory sandboxes" to allow controlled testing of DNA storage technologies, risk-based tiered regulations depending on the nature of stored data, and international harmonization efforts to prevent fragmented compliance requirements across markets.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!