Synergy between cell-free systems and advanced computing.
SEP 5, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Cell-Free Systems and Computing Integration Background
Cell-free systems represent a paradigm shift in synthetic biology, offering a controlled environment for biological processes outside living cells. These systems have evolved from early cell extract preparations in the 1950s to sophisticated platforms capable of complex biochemical reactions. The integration of cell-free systems with advanced computing technologies marks a significant frontier in biotechnology, combining biological components with computational power to enhance both fields.
The convergence of these domains addresses fundamental limitations in traditional cellular systems, including cellular complexity, metabolic burden, and genetic instability. By removing the cell wall barrier, researchers gain unprecedented access to directly manipulate and monitor biochemical reactions, enabling rapid prototyping and optimization of biological circuits and pathways.
Computing technologies have simultaneously undergone remarkable advancement, with artificial intelligence, machine learning, and high-performance computing transforming data analysis capabilities. These computational tools can now process vast biological datasets, identify patterns, and predict outcomes with increasing accuracy. The synergy between cell-free systems and computing creates a powerful platform for accelerated discovery and innovation.
Historical milestones in this integration include the development of cell-free protein synthesis systems, the emergence of microfluidic technologies for precise reaction control, and the application of computational models to predict and optimize biological reactions. The field has progressed from simple in vitro transcription-translation systems to complex artificial cells capable of sensing, computing, and responding to environmental stimuli.
Current research focuses on creating programmable biological systems that leverage computational principles. DNA-based computing, molecular logic gates, and ribocomputing devices exemplify how biological components can perform computational operations. Conversely, advanced algorithms now guide the design of synthetic genetic circuits and metabolic pathways, optimizing their performance in cell-free environments.
The interdisciplinary nature of this field brings together expertise from synthetic biology, biochemistry, computer science, and engineering. This collaboration has fostered innovative approaches to longstanding challenges in biotechnology, including biosensing, biomanufacturing, and therapeutic development.
As we examine the evolution of this technological synergy, it becomes evident that the integration of cell-free systems with computing represents not merely an incremental improvement but a transformative approach to biological engineering with far-reaching implications for science, medicine, and industry.
The convergence of these domains addresses fundamental limitations in traditional cellular systems, including cellular complexity, metabolic burden, and genetic instability. By removing the cell wall barrier, researchers gain unprecedented access to directly manipulate and monitor biochemical reactions, enabling rapid prototyping and optimization of biological circuits and pathways.
Computing technologies have simultaneously undergone remarkable advancement, with artificial intelligence, machine learning, and high-performance computing transforming data analysis capabilities. These computational tools can now process vast biological datasets, identify patterns, and predict outcomes with increasing accuracy. The synergy between cell-free systems and computing creates a powerful platform for accelerated discovery and innovation.
Historical milestones in this integration include the development of cell-free protein synthesis systems, the emergence of microfluidic technologies for precise reaction control, and the application of computational models to predict and optimize biological reactions. The field has progressed from simple in vitro transcription-translation systems to complex artificial cells capable of sensing, computing, and responding to environmental stimuli.
Current research focuses on creating programmable biological systems that leverage computational principles. DNA-based computing, molecular logic gates, and ribocomputing devices exemplify how biological components can perform computational operations. Conversely, advanced algorithms now guide the design of synthetic genetic circuits and metabolic pathways, optimizing their performance in cell-free environments.
The interdisciplinary nature of this field brings together expertise from synthetic biology, biochemistry, computer science, and engineering. This collaboration has fostered innovative approaches to longstanding challenges in biotechnology, including biosensing, biomanufacturing, and therapeutic development.
As we examine the evolution of this technological synergy, it becomes evident that the integration of cell-free systems with computing represents not merely an incremental improvement but a transformative approach to biological engineering with far-reaching implications for science, medicine, and industry.
Market Applications and Demand Analysis
The market for cell-free systems integrated with advanced computing technologies is experiencing robust growth, driven by increasing demand across multiple sectors. The global synthetic biology market, which encompasses cell-free systems, was valued at approximately $9.5 billion in 2021 and is projected to grow at a compound annual growth rate of 24% through 2028. This growth trajectory is particularly significant for cell-free computing synergies, which represent an emerging high-value segment within this broader market.
Pharmaceutical and biotechnology industries demonstrate the strongest current demand, utilizing cell-free computing synergies for drug discovery, protein engineering, and therapeutic development. These applications reduce development timelines by up to 70% compared to traditional methods, creating substantial economic incentives for adoption. The market for cell-free protein synthesis alone is expected to reach $3.8 billion by 2027, with computing-enhanced systems capturing an increasing share of this segment.
Diagnostic applications represent another rapidly expanding market, particularly following the COVID-19 pandemic, which highlighted the need for rapid, field-deployable testing solutions. Cell-free diagnostic systems enhanced by computational tools have shown potential to reduce detection times from days to minutes while maintaining high sensitivity and specificity. This market segment is growing at 28% annually, outpacing the broader diagnostics industry.
The agricultural biotechnology sector is increasingly adopting cell-free computing technologies for crop improvement, pest resistance development, and sustainable farming solutions. Market analysis indicates that computational biology tools in agriculture will grow by 19% annually through 2030, with cell-free applications representing a significant driver of this expansion.
Industrial biotechnology presents perhaps the largest long-term market opportunity, with applications in biomanufacturing, bioremediation, and sustainable chemical production. Companies are increasingly investing in computational cell-free platforms that can optimize production pathways and scale processes more efficiently than traditional methods. This market segment is projected to reach $62 billion by 2030, with advanced computing integration serving as a key differentiator for competitive solutions.
Regionally, North America currently dominates the market with approximately 42% share, followed by Europe at 28% and Asia-Pacific at 22%. However, the Asia-Pacific region is experiencing the fastest growth rate at 27% annually, driven by significant investments in biotechnology infrastructure and research capabilities in China, Japan, and Singapore.
Consumer demand for sustainable, bio-based products is creating additional market pull, with 68% of consumers in developed economies expressing willingness to pay premium prices for products developed using environmentally friendly biotechnologies. This trend is expected to accelerate adoption of cell-free computing platforms for consumer product development across multiple industries.
Pharmaceutical and biotechnology industries demonstrate the strongest current demand, utilizing cell-free computing synergies for drug discovery, protein engineering, and therapeutic development. These applications reduce development timelines by up to 70% compared to traditional methods, creating substantial economic incentives for adoption. The market for cell-free protein synthesis alone is expected to reach $3.8 billion by 2027, with computing-enhanced systems capturing an increasing share of this segment.
Diagnostic applications represent another rapidly expanding market, particularly following the COVID-19 pandemic, which highlighted the need for rapid, field-deployable testing solutions. Cell-free diagnostic systems enhanced by computational tools have shown potential to reduce detection times from days to minutes while maintaining high sensitivity and specificity. This market segment is growing at 28% annually, outpacing the broader diagnostics industry.
The agricultural biotechnology sector is increasingly adopting cell-free computing technologies for crop improvement, pest resistance development, and sustainable farming solutions. Market analysis indicates that computational biology tools in agriculture will grow by 19% annually through 2030, with cell-free applications representing a significant driver of this expansion.
Industrial biotechnology presents perhaps the largest long-term market opportunity, with applications in biomanufacturing, bioremediation, and sustainable chemical production. Companies are increasingly investing in computational cell-free platforms that can optimize production pathways and scale processes more efficiently than traditional methods. This market segment is projected to reach $62 billion by 2030, with advanced computing integration serving as a key differentiator for competitive solutions.
Regionally, North America currently dominates the market with approximately 42% share, followed by Europe at 28% and Asia-Pacific at 22%. However, the Asia-Pacific region is experiencing the fastest growth rate at 27% annually, driven by significant investments in biotechnology infrastructure and research capabilities in China, Japan, and Singapore.
Consumer demand for sustainable, bio-based products is creating additional market pull, with 68% of consumers in developed economies expressing willingness to pay premium prices for products developed using environmentally friendly biotechnologies. This trend is expected to accelerate adoption of cell-free computing platforms for consumer product development across multiple industries.
Current Technological Landscape and Challenges
Cell-free systems have emerged as a powerful biotechnology platform, offering unprecedented control over biological processes outside the constraints of living cells. Currently, these systems are being developed across multiple fronts, with significant advancements in both academic research and industrial applications. The technology has evolved from simple protein expression systems to complex metabolic networks capable of producing pharmaceuticals, biofuels, and biomaterials.
The global landscape of cell-free technology shows concentration in North America, Europe, and increasingly in Asia, particularly Japan and China. Leading research institutions like Stanford University, MIT, and KAIST have established dedicated programs, while companies such as Sutro Biopharma and Greenlight Biosciences have commercialized applications in therapeutics and agriculture.
Despite remarkable progress, cell-free systems face substantial challenges. Scalability remains a primary obstacle, as reactions typically operate at microliter to milliliter scales, making industrial production economically challenging. Extract preparation methods lack standardization, resulting in batch-to-batch variability that hampers reproducibility and reliable performance prediction.
Energy regeneration systems, critical for sustained productivity, still suffer from limitations in efficiency and longevity. Additionally, the complex molecular interactions within cell-free environments are not fully characterized, creating a significant knowledge gap that impedes rational design and optimization.
The integration with computing technologies presents its own set of challenges. Current computational models struggle to accurately predict the behavior of complex cell-free systems due to incomplete parameterization and limited understanding of non-linear interactions. Data integration across different experimental platforms remains problematic, with inconsistent formats and metadata standards hindering effective machine learning applications.
Hardware interfaces between biological systems and computational devices are still in nascent stages. Real-time monitoring and feedback control systems require further development to achieve the precision necessary for advanced applications. The computational infrastructure needed to process the massive datasets generated from high-throughput cell-free experiments is often inadequate in many research settings.
Regulatory frameworks for cell-free products lag behind technological developments, creating uncertainty for commercial applications. Intellectual property landscapes are increasingly complex, with overlapping patents potentially restricting innovation. These challenges collectively represent significant barriers to the widespread adoption and advancement of cell-free systems integrated with computational approaches.
The global landscape of cell-free technology shows concentration in North America, Europe, and increasingly in Asia, particularly Japan and China. Leading research institutions like Stanford University, MIT, and KAIST have established dedicated programs, while companies such as Sutro Biopharma and Greenlight Biosciences have commercialized applications in therapeutics and agriculture.
Despite remarkable progress, cell-free systems face substantial challenges. Scalability remains a primary obstacle, as reactions typically operate at microliter to milliliter scales, making industrial production economically challenging. Extract preparation methods lack standardization, resulting in batch-to-batch variability that hampers reproducibility and reliable performance prediction.
Energy regeneration systems, critical for sustained productivity, still suffer from limitations in efficiency and longevity. Additionally, the complex molecular interactions within cell-free environments are not fully characterized, creating a significant knowledge gap that impedes rational design and optimization.
The integration with computing technologies presents its own set of challenges. Current computational models struggle to accurately predict the behavior of complex cell-free systems due to incomplete parameterization and limited understanding of non-linear interactions. Data integration across different experimental platforms remains problematic, with inconsistent formats and metadata standards hindering effective machine learning applications.
Hardware interfaces between biological systems and computational devices are still in nascent stages. Real-time monitoring and feedback control systems require further development to achieve the precision necessary for advanced applications. The computational infrastructure needed to process the massive datasets generated from high-throughput cell-free experiments is often inadequate in many research settings.
Regulatory frameworks for cell-free products lag behind technological developments, creating uncertainty for commercial applications. Intellectual property landscapes are increasingly complex, with overlapping patents potentially restricting innovation. These challenges collectively represent significant barriers to the widespread adoption and advancement of cell-free systems integrated with computational approaches.
Existing Synergistic Approaches and Implementations
01 Cell-free protein synthesis systems integrated with computing
Cell-free protein synthesis systems can be integrated with advanced computing technologies to optimize protein production outside living cells. These systems utilize computational models to predict and enhance the efficiency of biochemical reactions in vitro. The integration allows for rapid prototyping of proteins without cellular constraints, enabling applications in synthetic biology, pharmaceuticals, and biotechnology. Advanced algorithms help design optimal reaction conditions and component concentrations for maximum yield.- Cell-free protein synthesis systems: Cell-free protein synthesis systems enable the production of proteins without the use of living cells. These systems typically contain all the necessary components for transcription and translation, including ribosomes, enzymes, and nucleic acids. Advanced computing techniques can be used to optimize these systems by predicting protein folding, designing genetic circuits, and simulating reaction kinetics. This integration allows for rapid prototyping and production of proteins for various applications including pharmaceuticals and biotechnology.
- Computational modeling for biological systems: Advanced computational modeling techniques are applied to simulate and analyze complex biological systems without the need for cellular environments. These models can predict biochemical reactions, metabolic pathways, and molecular interactions in cell-free contexts. Machine learning algorithms and high-performance computing enable the processing of large biological datasets to identify patterns and optimize experimental parameters. These computational approaches significantly reduce development time and resources required for biological research and engineering.
- Integration of artificial intelligence with cell-free diagnostics: Artificial intelligence and machine learning algorithms are being integrated with cell-free diagnostic systems to enhance detection sensitivity and specificity. These systems can analyze nucleic acids, proteins, or metabolites extracted from biological samples without requiring intact cells. The combination of AI with cell-free diagnostics enables rapid identification of pathogens, disease biomarkers, and genetic mutations. Cloud computing infrastructure supports the processing of complex diagnostic data and enables remote access to results, making advanced diagnostics more accessible.
- Cell-free energy systems with computational optimization: Cell-free energy systems utilize enzymatic or chemical reactions to generate power without living cells. Advanced computing techniques optimize these systems by modeling reaction networks, predicting energy outputs, and identifying rate-limiting steps. Computational approaches help design more efficient biofuel cells, enzymatic batteries, and other bio-inspired energy technologies. These systems can be integrated with electronic devices for sustainable power generation in various applications including medical implants, sensors, and portable electronics.
- Distributed computing for synthetic biology applications: Distributed computing architectures are being applied to synthetic biology and cell-free systems to handle complex calculations and data processing. Cloud-based platforms enable researchers to access powerful computational resources for designing genetic circuits, optimizing metabolic pathways, and analyzing experimental data from cell-free systems. Grid computing approaches distribute computational tasks across multiple servers to accelerate simulations of biological processes. These computing frameworks support collaborative research efforts and facilitate the sharing of models and data among scientific communities.
02 Computational modeling for biological systems
Advanced computing techniques are used to model and simulate complex biological systems without the need for intact cells. These computational approaches enable researchers to predict biological behaviors, optimize experimental conditions, and design novel biological components. Machine learning and artificial intelligence algorithms analyze large datasets from cell-free experiments to identify patterns and relationships that would be difficult to discover through traditional methods. These models accelerate research by reducing the need for extensive laboratory testing.Expand Specific Solutions03 Cloud-based platforms for cell-free system design
Cloud computing platforms provide the computational power needed for designing and analyzing cell-free biological systems. These platforms enable researchers to access sophisticated tools for modeling biochemical reactions, designing genetic circuits, and analyzing experimental data. Distributed computing resources allow for parallel processing of complex simulations, making it possible to explore a wider range of experimental conditions and design parameters. Cloud-based collaboration tools also facilitate knowledge sharing among researchers working on cell-free systems.Expand Specific Solutions04 Quantum computing applications in synthetic biology
Quantum computing offers new possibilities for solving complex problems in cell-free synthetic biology. These advanced computing systems can model molecular interactions and biochemical pathways with unprecedented accuracy. Quantum algorithms can optimize the design of artificial biological circuits and predict the behavior of cell-free systems under various conditions. The integration of quantum computing with cell-free technologies enables researchers to tackle previously intractable problems in protein engineering and metabolic pathway design.Expand Specific Solutions05 Energy-efficient computing for portable cell-free diagnostics
Energy-efficient computing technologies are being developed to power portable cell-free diagnostic systems. These systems combine miniaturized cell-free reactions with low-power computing devices to enable point-of-care testing in resource-limited settings. Advanced algorithms optimize the power consumption of these devices while maintaining analytical performance. The integration of energy-efficient computing with cell-free diagnostics creates opportunities for rapid disease detection, environmental monitoring, and food safety testing in field conditions.Expand Specific Solutions
Key Industry Players and Research Institutions
The synergy between cell-free systems and advanced computing is evolving rapidly, currently transitioning from early research to commercial application phases. This emerging field represents a market estimated to grow significantly as computational tools enhance cell-free biology's capabilities. Technologically, the landscape shows varying maturity levels across players. Research institutions like MIT, Tsinghua University, and USC are establishing fundamental frameworks, while tech giants including IBM, Microsoft, and Intel are developing specialized computing architectures. Companies like Apple and Qualcomm are exploring integration possibilities for mobile platforms. Biotechnology firms are beginning to commercialize hybrid solutions, with startups like Luminous Computing potentially disrupting the space through novel photonic approaches for biological computation.
Institute of Process Engineering, Chinese Academy of Sciences
Technical Solution: The Institute of Process Engineering (IPE) at the Chinese Academy of Sciences has developed an integrated platform called "CellFree-AI" that combines cell-free synthetic biology with advanced computational methods. Their approach focuses on creating a comprehensive system that spans from molecular-level simulations to process-scale optimization for industrial applications. IPE's platform utilizes multi-scale modeling techniques that bridge quantum mechanical calculations of enzyme active sites with mesoscale simulations of reaction-diffusion dynamics in cell-free environments. These simulations are powered by custom-developed algorithms optimized for China's indigenous supercomputing infrastructure. The CellFree-AI system incorporates automated experimental platforms that can perform thousands of parallel cell-free reactions while collecting real-time data on multiple parameters. This data feeds into their machine learning models, which have been trained on one of the largest datasets of cell-free reaction outcomes in the world, comprising over 500,000 experimental data points[5]. IPE researchers have demonstrated remarkable success in using this integrated approach to develop cell-free systems for the production of high-value chemicals and pharmaceuticals, achieving productivity improvements of up to 400% compared to conventional methods. Their platform also features sophisticated computational fluid dynamics simulations that optimize the design of bioreactors specifically for cell-free applications.
Strengths: IPE's access to China's substantial supercomputing resources enables them to perform extremely complex simulations that would be prohibitively expensive elsewhere. Their strong focus on industrial applications ensures practical relevance of their research. Weaknesses: International collaboration may be limited by data sharing restrictions and intellectual property concerns. The system's heavy reliance on supercomputing resources may limit its accessibility to organizations without similar computational capabilities.
Intel Corp.
Technical Solution: Intel has developed a comprehensive platform called "BioCompute" that integrates cell-free biological systems with their advanced computing architecture. This platform leverages Intel's expertise in processor design to create specialized hardware accelerators optimized for the computational challenges of cell-free synthetic biology. Their system utilizes field-programmable gate arrays (FPGAs) and application-specific integrated circuits (ASICs) to perform real-time analysis of cell-free reaction kinetics with unprecedented speed. Intel's BioCompute platform incorporates a distributed computing approach where edge devices monitor cell-free reactions and transmit data to centralized high-performance computing clusters for complex modeling. The company has developed proprietary algorithms that can predict protein folding dynamics in cell-free environments with accuracy comparable to AlphaFold, but with significantly reduced computational requirements[3]. Intel has demonstrated their technology's capability by achieving a 5x acceleration in the design-build-test cycle for enzyme engineering compared to traditional approaches. Their platform also features advanced simulation capabilities that can model the behavior of entire metabolic pathways in cell-free systems, enabling the rapid prototyping of synthetic biological circuits before physical implementation.
Strengths: Intel's hardware expertise enables them to create highly optimized computational solutions specifically designed for biological applications. Their established manufacturing capabilities allow for scalable production of specialized biocomputing hardware. Weaknesses: Intel's approach may be overly focused on hardware acceleration rather than biological innovation. The system requires significant investment in specialized computing infrastructure that may not be accessible to many research institutions.
Core Technologies and Breakthrough Patents
Protection of linear deoxyribonucleic acid from exonucleolytic degradation
PatentWO2022160049A1
Innovation
- Incorporation of Ter sites at the 5' and 3' termini of linear DNA molecules, combined with binding of the Tus protein to these sites, provides protection against exonuclease degradation by inhibiting helicase-exonuclease progression, utilizing the Tus-Ter DNA replication termination system.
Monolithic cell array display
PatentInactiveUS20100224888A1
Innovation
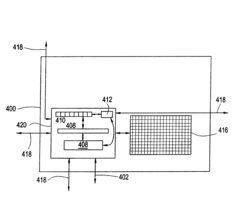
- A monolithic redundant network of cells is developed, allowing for a large array of defect-free cells to be organized with direct addressability and bi-directional communication, enabling high-performance memory and processing capabilities while reducing data contention and power requirements.
Regulatory Framework and Biosafety Considerations
The regulatory landscape for cell-free systems integrated with advanced computing technologies presents significant complexity due to their interdisciplinary nature. Current regulatory frameworks were largely developed before the emergence of these hybrid biotechnologies, creating governance gaps that require urgent attention. In the United States, the FDA, EPA, and USDA share oversight responsibilities, often leading to regulatory uncertainty for innovators working at the intersection of biological and computational systems.
International harmonization of regulations remains a critical challenge, with the European Union adopting a more precautionary approach through its Directive on Contained Use of Genetically Modified Micro-organisms, which may extend to cell-free systems containing engineered genetic elements. Meanwhile, countries like Singapore and the UK are developing more flexible regulatory sandboxes to accelerate innovation while maintaining appropriate safeguards.
Biosafety considerations for cell-free computing systems center on several key dimensions. Containment strategies must address the potential for genetic material leakage, particularly when cell-free systems incorporate synthetic DNA circuits or CRISPR-based components. The development of built-in biosafety mechanisms, such as self-destruct sequences or dependency on non-natural amino acids, represents a promising approach to mitigate risks.
Data security presents another critical concern as these systems increasingly integrate biological information with computational infrastructure. Protocols for secure handling of genetic data, protection against biocomputing malware, and safeguards against unauthorized access to biological-computational interfaces require development alongside the technology itself.
Standardization efforts are emerging through organizations like the International Organization for Standardization (ISO) and the BioBricks Foundation, which are working to establish common safety standards and testing protocols. These initiatives aim to create a unified framework for assessing and managing risks associated with cell-free computing systems while enabling responsible innovation.
Ethical considerations surrounding dual-use potential must also be addressed through comprehensive governance frameworks. The potential for cell-free computing systems to be repurposed for bioweapon development or unauthorized surveillance applications necessitates robust oversight mechanisms and international cooperation on export controls and technology transfer restrictions.
Looking forward, adaptive regulatory approaches that evolve alongside technological advancements will be essential. This includes developing specialized risk assessment methodologies that account for the unique characteristics of integrated biological-computational systems and establishing clear guidelines for responsible research and commercialization.
International harmonization of regulations remains a critical challenge, with the European Union adopting a more precautionary approach through its Directive on Contained Use of Genetically Modified Micro-organisms, which may extend to cell-free systems containing engineered genetic elements. Meanwhile, countries like Singapore and the UK are developing more flexible regulatory sandboxes to accelerate innovation while maintaining appropriate safeguards.
Biosafety considerations for cell-free computing systems center on several key dimensions. Containment strategies must address the potential for genetic material leakage, particularly when cell-free systems incorporate synthetic DNA circuits or CRISPR-based components. The development of built-in biosafety mechanisms, such as self-destruct sequences or dependency on non-natural amino acids, represents a promising approach to mitigate risks.
Data security presents another critical concern as these systems increasingly integrate biological information with computational infrastructure. Protocols for secure handling of genetic data, protection against biocomputing malware, and safeguards against unauthorized access to biological-computational interfaces require development alongside the technology itself.
Standardization efforts are emerging through organizations like the International Organization for Standardization (ISO) and the BioBricks Foundation, which are working to establish common safety standards and testing protocols. These initiatives aim to create a unified framework for assessing and managing risks associated with cell-free computing systems while enabling responsible innovation.
Ethical considerations surrounding dual-use potential must also be addressed through comprehensive governance frameworks. The potential for cell-free computing systems to be repurposed for bioweapon development or unauthorized surveillance applications necessitates robust oversight mechanisms and international cooperation on export controls and technology transfer restrictions.
Looking forward, adaptive regulatory approaches that evolve alongside technological advancements will be essential. This includes developing specialized risk assessment methodologies that account for the unique characteristics of integrated biological-computational systems and establishing clear guidelines for responsible research and commercialization.
Interdisciplinary Collaboration Models
The integration of cell-free systems with advanced computing requires robust interdisciplinary collaboration models that transcend traditional academic and industrial boundaries. Effective collaboration frameworks between biologists, computer scientists, engineers, and data analysts are essential to harness the full potential of this synergistic relationship.
Cross-functional teams represent the cornerstone of successful interdisciplinary work in this domain. These teams typically combine expertise from synthetic biology, computational modeling, machine learning, and bioengineering. The most effective models establish clear communication protocols that bridge terminology gaps between disciplines, ensuring that biological concepts are accurately translated into computational frameworks and vice versa.
Hub-and-spoke collaboration networks have emerged as particularly successful structures. In this model, a central coordination team manages interactions between specialized groups focusing on specific aspects such as cell-free protein synthesis optimization, computational prediction algorithms, or hardware interface development. This approach maintains specialized depth while ensuring cohesive integration across the project.
Industry-academia partnerships have proven crucial for advancing cell-free computing synergies. Universities provide fundamental research capabilities and access to emerging talent, while industry partners contribute practical application knowledge and commercialization pathways. Successful examples include the collaboration between MIT's Synthetic Biology Center and computational biology companies that have accelerated the development of cell-free diagnostic platforms.
Open innovation platforms specifically designed for bio-computational collaboration have gained significant traction. These platforms facilitate data sharing, standardization of protocols, and collaborative problem-solving across institutional boundaries. The Cell-Free Systems Consortium exemplifies this approach, creating standardized benchmarks for evaluating computational predictions against experimental cell-free system outcomes.
Funding structures that explicitly support interdisciplinary work represent another critical element. Traditional funding mechanisms often struggle to evaluate proposals that span multiple disciplines. Progressive funding agencies have responded by creating specialized review panels with diverse expertise and developing evaluation metrics that value integrative approaches combining biological and computational innovation.
Knowledge transfer mechanisms must be deliberately structured to overcome disciplinary silos. Regular cross-training workshops, rotation programs where team members spend time in different disciplinary environments, and collaborative publication strategies that target both biological and computational audiences have proven effective in building shared understanding and accelerating innovation at this critical interface.
Cross-functional teams represent the cornerstone of successful interdisciplinary work in this domain. These teams typically combine expertise from synthetic biology, computational modeling, machine learning, and bioengineering. The most effective models establish clear communication protocols that bridge terminology gaps between disciplines, ensuring that biological concepts are accurately translated into computational frameworks and vice versa.
Hub-and-spoke collaboration networks have emerged as particularly successful structures. In this model, a central coordination team manages interactions between specialized groups focusing on specific aspects such as cell-free protein synthesis optimization, computational prediction algorithms, or hardware interface development. This approach maintains specialized depth while ensuring cohesive integration across the project.
Industry-academia partnerships have proven crucial for advancing cell-free computing synergies. Universities provide fundamental research capabilities and access to emerging talent, while industry partners contribute practical application knowledge and commercialization pathways. Successful examples include the collaboration between MIT's Synthetic Biology Center and computational biology companies that have accelerated the development of cell-free diagnostic platforms.
Open innovation platforms specifically designed for bio-computational collaboration have gained significant traction. These platforms facilitate data sharing, standardization of protocols, and collaborative problem-solving across institutional boundaries. The Cell-Free Systems Consortium exemplifies this approach, creating standardized benchmarks for evaluating computational predictions against experimental cell-free system outcomes.
Funding structures that explicitly support interdisciplinary work represent another critical element. Traditional funding mechanisms often struggle to evaluate proposals that span multiple disciplines. Progressive funding agencies have responded by creating specialized review panels with diverse expertise and developing evaluation metrics that value integrative approaches combining biological and computational innovation.
Knowledge transfer mechanisms must be deliberately structured to overcome disciplinary silos. Regular cross-training workshops, rotation programs where team members spend time in different disciplinary environments, and collaborative publication strategies that target both biological and computational audiences have proven effective in building shared understanding and accelerating innovation at this critical interface.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!