Proteome analysis with cell-free expression systems.
SEP 5, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Cell-free Expression Systems Background and Objectives
Cell-free expression systems represent a revolutionary approach in proteomics research, emerging from the convergence of biochemistry and molecular biology. These systems have evolved from rudimentary extracts in the 1960s to sophisticated platforms capable of producing complex proteins without the constraints of living cells. The technology leverages cellular machinery extracted from various organisms—primarily E. coli, wheat germ, rabbit reticulocytes, and insect cells—while eliminating cellular barriers that traditionally limit protein expression studies.
The historical trajectory of cell-free systems began with their use in deciphering the genetic code and has progressively expanded to address increasingly complex proteomics challenges. Recent advancements have transformed these systems from academic curiosities into powerful tools for high-throughput protein production and analysis, enabling unprecedented insights into protein structure, function, and interaction networks.
The primary objective of cell-free expression systems in proteome analysis is to overcome the limitations inherent in traditional cell-based methods. These limitations include cellular toxicity of certain proteins, formation of inclusion bodies, and difficulties in expressing membrane proteins. By providing an open and highly controllable environment, cell-free systems aim to facilitate the expression of challenging proteins and enable direct manipulation of the translation machinery.
Technical goals include enhancing expression yields, improving post-translational modification capabilities, and developing specialized systems for particular protein classes. The field is moving toward creating more defined and reproducible systems that can reliably express proteins with native-like structures and functions, essential for accurate proteome characterization.
Current research trends focus on miniaturization and automation to enable high-throughput proteomics applications. The integration of cell-free systems with microfluidic platforms and advanced detection methods represents a significant direction, potentially revolutionizing protein biomarker discovery and personalized medicine approaches.
The technology aims to bridge fundamental proteomics research with practical applications in drug discovery, diagnostics, and synthetic biology. By providing rapid protein production capabilities, cell-free systems are positioned to accelerate the development cycle from protein identification to functional characterization and therapeutic application.
As proteomics increasingly moves toward systems-level understanding, cell-free expression systems are expected to play a crucial role in mapping comprehensive protein interaction networks and elucidating the functional significance of post-translational modifications across the proteome. The ultimate objective is to establish these systems as standard tools in proteomics research, complementing traditional approaches and enabling new experimental paradigms previously considered unfeasible.
The historical trajectory of cell-free systems began with their use in deciphering the genetic code and has progressively expanded to address increasingly complex proteomics challenges. Recent advancements have transformed these systems from academic curiosities into powerful tools for high-throughput protein production and analysis, enabling unprecedented insights into protein structure, function, and interaction networks.
The primary objective of cell-free expression systems in proteome analysis is to overcome the limitations inherent in traditional cell-based methods. These limitations include cellular toxicity of certain proteins, formation of inclusion bodies, and difficulties in expressing membrane proteins. By providing an open and highly controllable environment, cell-free systems aim to facilitate the expression of challenging proteins and enable direct manipulation of the translation machinery.
Technical goals include enhancing expression yields, improving post-translational modification capabilities, and developing specialized systems for particular protein classes. The field is moving toward creating more defined and reproducible systems that can reliably express proteins with native-like structures and functions, essential for accurate proteome characterization.
Current research trends focus on miniaturization and automation to enable high-throughput proteomics applications. The integration of cell-free systems with microfluidic platforms and advanced detection methods represents a significant direction, potentially revolutionizing protein biomarker discovery and personalized medicine approaches.
The technology aims to bridge fundamental proteomics research with practical applications in drug discovery, diagnostics, and synthetic biology. By providing rapid protein production capabilities, cell-free systems are positioned to accelerate the development cycle from protein identification to functional characterization and therapeutic application.
As proteomics increasingly moves toward systems-level understanding, cell-free expression systems are expected to play a crucial role in mapping comprehensive protein interaction networks and elucidating the functional significance of post-translational modifications across the proteome. The ultimate objective is to establish these systems as standard tools in proteomics research, complementing traditional approaches and enabling new experimental paradigms previously considered unfeasible.
Market Analysis for Proteome Analysis Technologies
The global market for proteome analysis technologies has witnessed substantial growth in recent years, driven by increasing research activities in proteomics and advancements in analytical techniques. The market was valued at approximately $21.6 billion in 2022 and is projected to reach $49.8 billion by 2028, growing at a CAGR of 14.9% during the forecast period.
Cell-free expression systems represent a rapidly expanding segment within this market, offering significant advantages over traditional cell-based methods for protein production and analysis. This segment is expected to grow at a higher rate than the overall proteomics market, with projections indicating a CAGR of 18.7% through 2028.
North America currently dominates the proteome analysis market, accounting for about 42% of the global market share, followed by Europe (28%) and Asia-Pacific (22%). The Asia-Pacific region, particularly China and India, is experiencing the fastest growth due to increasing investments in life sciences research infrastructure and rising pharmaceutical R&D activities.
Key market drivers include the growing prevalence of chronic diseases necessitating protein biomarker discovery, increasing pharmaceutical and biotechnology R&D expenditure, and technological advancements in proteomics tools. The application of proteomics in precision medicine and personalized healthcare is creating substantial market opportunities.
The COVID-19 pandemic has further accelerated market growth, highlighting the importance of rapid protein analysis technologies in understanding disease mechanisms and developing therapeutic interventions. This has led to increased funding for proteomics research and development of more efficient analytical platforms.
Major end-users of proteome analysis technologies include academic and research institutions (38%), pharmaceutical and biotechnology companies (34%), clinical research organizations (18%), and hospitals and diagnostic centers (10%). The pharmaceutical sector is expected to witness the highest growth rate due to increasing applications in drug discovery and development processes.
Challenges facing the market include the high cost of advanced proteomics instruments, complexity of data analysis, and shortage of skilled professionals. Additionally, standardization issues and regulatory hurdles present significant barriers to market entry and expansion.
Emerging trends shaping the market include the integration of artificial intelligence and machine learning in proteomics data analysis, development of high-throughput proteomics platforms, and increasing adoption of multi-omics approaches combining proteomics with genomics, transcriptomics, and metabolomics for comprehensive biological insights.
Cell-free expression systems represent a rapidly expanding segment within this market, offering significant advantages over traditional cell-based methods for protein production and analysis. This segment is expected to grow at a higher rate than the overall proteomics market, with projections indicating a CAGR of 18.7% through 2028.
North America currently dominates the proteome analysis market, accounting for about 42% of the global market share, followed by Europe (28%) and Asia-Pacific (22%). The Asia-Pacific region, particularly China and India, is experiencing the fastest growth due to increasing investments in life sciences research infrastructure and rising pharmaceutical R&D activities.
Key market drivers include the growing prevalence of chronic diseases necessitating protein biomarker discovery, increasing pharmaceutical and biotechnology R&D expenditure, and technological advancements in proteomics tools. The application of proteomics in precision medicine and personalized healthcare is creating substantial market opportunities.
The COVID-19 pandemic has further accelerated market growth, highlighting the importance of rapid protein analysis technologies in understanding disease mechanisms and developing therapeutic interventions. This has led to increased funding for proteomics research and development of more efficient analytical platforms.
Major end-users of proteome analysis technologies include academic and research institutions (38%), pharmaceutical and biotechnology companies (34%), clinical research organizations (18%), and hospitals and diagnostic centers (10%). The pharmaceutical sector is expected to witness the highest growth rate due to increasing applications in drug discovery and development processes.
Challenges facing the market include the high cost of advanced proteomics instruments, complexity of data analysis, and shortage of skilled professionals. Additionally, standardization issues and regulatory hurdles present significant barriers to market entry and expansion.
Emerging trends shaping the market include the integration of artificial intelligence and machine learning in proteomics data analysis, development of high-throughput proteomics platforms, and increasing adoption of multi-omics approaches combining proteomics with genomics, transcriptomics, and metabolomics for comprehensive biological insights.
Current Challenges in Cell-free Proteomics
Despite significant advancements in cell-free proteomics, several critical challenges continue to impede the full realization of this technology's potential. One of the most persistent obstacles is the limited yield of protein expression in cell-free systems compared to cellular environments. Current cell-free systems typically produce protein yields in the microgram to milligram range, which remains insufficient for comprehensive proteome-wide analyses requiring substantial sample quantities.
Protein folding and post-translational modifications (PTMs) present another significant hurdle. Cell-free systems often lack the complete cellular machinery necessary for proper protein folding and modification. This limitation is particularly problematic for complex eukaryotic proteins that require extensive chaperone assistance or specific PTMs such as glycosylation, phosphorylation, or disulfide bond formation, which are essential for proper function and structural integrity.
Scalability and cost-effectiveness remain substantial barriers to widespread adoption. The reagents required for cell-free expression, including nucleotides, amino acids, and energy regeneration components, are expensive, making large-scale proteome analysis financially prohibitive for many research institutions. Additionally, the preparation of cell extracts is labor-intensive and subject to batch-to-batch variations, compromising reproducibility across experiments.
The complexity of proteome analysis itself poses technical challenges. Unlike genomics, where amplification techniques can overcome sample limitations, proteins cannot be amplified, making the detection of low-abundance proteins particularly difficult. Current cell-free systems struggle to express the full dynamic range of cellular proteomes, often favoring abundant and soluble proteins while missing critical low-abundance regulatory proteins.
Integration with downstream analytical techniques presents additional complications. Compatibility issues between cell-free expression products and mass spectrometry or other analytical platforms can result in data loss or misinterpretation. The presence of components from the cell-free system itself can interfere with accurate protein identification and quantification.
Temporal control and stability issues further complicate proteome analysis. Cell-free reactions typically have limited lifespans (hours rather than days), restricting the expression window for slow-folding or complex proteins. Additionally, the lack of cellular compartmentalization means that expressed proteins are exposed to degradative enzymes, potentially reducing yield and complicating analysis of the complete proteome.
Addressing these challenges requires interdisciplinary approaches combining synthetic biology, biochemical engineering, and analytical chemistry to develop next-generation cell-free systems capable of more accurately recapitulating the cellular environment while maintaining the advantages of cell-free methodologies.
Protein folding and post-translational modifications (PTMs) present another significant hurdle. Cell-free systems often lack the complete cellular machinery necessary for proper protein folding and modification. This limitation is particularly problematic for complex eukaryotic proteins that require extensive chaperone assistance or specific PTMs such as glycosylation, phosphorylation, or disulfide bond formation, which are essential for proper function and structural integrity.
Scalability and cost-effectiveness remain substantial barriers to widespread adoption. The reagents required for cell-free expression, including nucleotides, amino acids, and energy regeneration components, are expensive, making large-scale proteome analysis financially prohibitive for many research institutions. Additionally, the preparation of cell extracts is labor-intensive and subject to batch-to-batch variations, compromising reproducibility across experiments.
The complexity of proteome analysis itself poses technical challenges. Unlike genomics, where amplification techniques can overcome sample limitations, proteins cannot be amplified, making the detection of low-abundance proteins particularly difficult. Current cell-free systems struggle to express the full dynamic range of cellular proteomes, often favoring abundant and soluble proteins while missing critical low-abundance regulatory proteins.
Integration with downstream analytical techniques presents additional complications. Compatibility issues between cell-free expression products and mass spectrometry or other analytical platforms can result in data loss or misinterpretation. The presence of components from the cell-free system itself can interfere with accurate protein identification and quantification.
Temporal control and stability issues further complicate proteome analysis. Cell-free reactions typically have limited lifespans (hours rather than days), restricting the expression window for slow-folding or complex proteins. Additionally, the lack of cellular compartmentalization means that expressed proteins are exposed to degradative enzymes, potentially reducing yield and complicating analysis of the complete proteome.
Addressing these challenges requires interdisciplinary approaches combining synthetic biology, biochemical engineering, and analytical chemistry to develop next-generation cell-free systems capable of more accurately recapitulating the cellular environment while maintaining the advantages of cell-free methodologies.
Current Methodologies for Cell-free Proteomics
01 Cell-free protein expression systems for high-throughput applications
Cell-free protein expression systems enable rapid and efficient production of proteins without the need for living cells, making them ideal for high-throughput proteome analysis. These systems utilize cellular extracts containing the necessary machinery for transcription and translation, allowing for direct protein synthesis from DNA templates. This approach facilitates large-scale protein production for comprehensive proteome studies and can be optimized for various analytical techniques.- Cell-free protein expression systems for proteome analysis: Cell-free protein expression systems provide a platform for rapid and efficient synthesis of proteins without the need for living cells. These systems typically contain all the necessary components for transcription and translation, including ribosomes, enzymes, and cofactors. They are particularly useful for proteome analysis as they allow for high-throughput protein production and characterization, enabling researchers to study protein function, structure, and interactions on a large scale.
- High-throughput proteomics using cell-free systems: Cell-free expression systems enable high-throughput proteomics by allowing parallel synthesis of multiple proteins simultaneously. These systems can be adapted for automation and miniaturization, making them suitable for large-scale proteome analysis. By eliminating the constraints of cell viability, researchers can express proteins that might be toxic to living cells and can rapidly produce proteins for functional and structural studies, accelerating proteome-wide investigations.
- Modified cell-free systems for improved protein expression: Various modifications to cell-free expression systems have been developed to enhance protein yield and functionality for proteome analysis. These include the addition of chaperones to assist protein folding, incorporation of post-translational modification enzymes, optimization of energy regeneration systems, and supplementation with specific cofactors. Such modifications enable the production of proteins with native-like structures and functions, which is crucial for accurate proteome characterization.
- Integration of cell-free expression with analytical techniques: Cell-free expression systems can be directly integrated with various analytical techniques for comprehensive proteome analysis. These include mass spectrometry for protein identification and quantification, surface plasmon resonance for interaction studies, and microarrays for high-throughput functional screening. This integration streamlines the workflow from protein synthesis to analysis, reducing sample handling and potential contamination while increasing throughput and reproducibility.
- Applications of cell-free proteomics in biomedical research: Cell-free expression systems coupled with proteome analysis have numerous applications in biomedical research. These include drug discovery through target protein expression and screening, biomarker identification for disease diagnosis, vaccine development through rapid antigen production, and personalized medicine through patient-specific protein expression profiling. The flexibility and speed of cell-free systems make them valuable tools for addressing various biomedical challenges that require proteome-level investigations.
02 Advanced analytical techniques for cell-free proteome analysis
Various analytical techniques have been developed specifically for proteome analysis in cell-free systems. These include mass spectrometry-based approaches, protein microarrays, and fluorescence-based detection methods that enable comprehensive characterization of proteins produced in cell-free expression systems. These techniques allow researchers to analyze protein-protein interactions, post-translational modifications, and functional properties of expressed proteins, providing deeper insights into proteome dynamics.Expand Specific Solutions03 Optimization of cell-free expression conditions for proteome studies
Optimizing cell-free expression conditions is crucial for effective proteome analysis. This includes adjusting factors such as energy regeneration systems, cofactor concentrations, and reaction environments to enhance protein yield and quality. Specialized formulations have been developed to improve the expression of difficult proteins, including membrane proteins and those with complex folding requirements, enabling more comprehensive proteome coverage in analytical studies.Expand Specific Solutions04 Integration of cell-free systems with computational proteomics
The integration of cell-free expression systems with computational proteomics has revolutionized proteome analysis. Bioinformatic tools and algorithms have been developed to predict protein expression outcomes, analyze large-scale proteomics data, and model protein interactions in cell-free environments. This computational approach enhances the interpretation of experimental results and enables more targeted proteome studies, improving the efficiency and accuracy of proteome analysis using cell-free systems.Expand Specific Solutions05 Miniaturized and automated cell-free proteome analysis platforms
Miniaturized and automated platforms for cell-free proteome analysis have been developed to increase throughput and reduce sample requirements. These include microfluidic devices, droplet-based systems, and automated workstations that integrate sample preparation, protein expression, and analytical detection. Such platforms enable parallel processing of multiple samples, real-time monitoring of protein synthesis, and rapid screening of proteome components, significantly accelerating proteome research and discovery.Expand Specific Solutions
Key Technical Innovations in Cell-free Expression
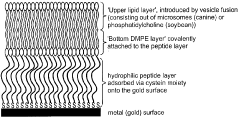
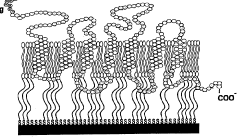
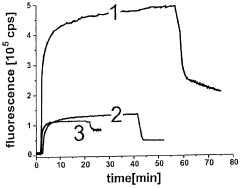
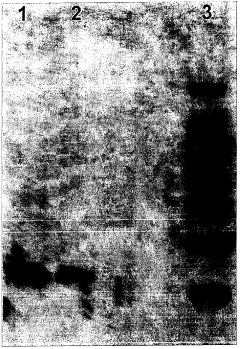
Cell-free in vitro transcription and translation of membrane proteins into tethered planar lipid layers
PatentWO2007048459A1
Innovation
- A process involving a cell-free expression system and nucleic acid coding for membrane proteins is applied directly to synthetic membranes, allowing for the expression and directed incorporation of membrane proteins in their native functional form without prior isolation.
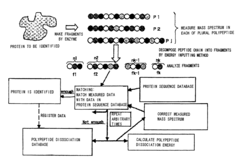
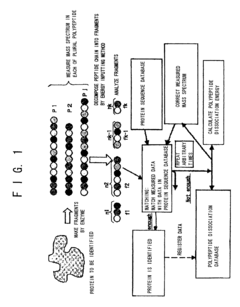
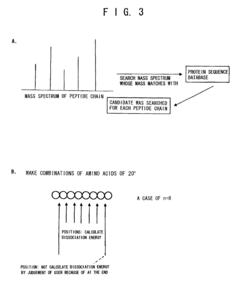
Proteome analysis method and proteome analysis system
PatentInactiveUS7523026B2
Innovation
- A proteome analysis method using a tandem type mass spectrometer that calculates bond dissociation energy via molecular simulation, predicts cleavage patterns and peak positions, and corrects mass spectra to enhance protein identification accuracy and throughput by integrating predicted information with experimental data.
Scalability and High-throughput Applications
Cell-free expression systems have demonstrated remarkable potential for scalability in proteome analysis applications. The transition from traditional small-scale analytical methods to industrial-scale production represents a significant advancement in this field. Current systems can be scaled from microliter volumes in 96-well plates to multi-liter bioreactors, enabling both high-throughput screening and large-scale protein production within the same technological framework.
The implementation of automation technologies has dramatically enhanced the throughput capabilities of cell-free proteome analysis. Robotic liquid handling systems, when integrated with cell-free expression platforms, can process thousands of protein samples simultaneously. This automation reduces human error, increases reproducibility, and significantly decreases the time required for comprehensive proteome studies from months to days.
Microfluidic technologies have emerged as a revolutionary approach for miniaturizing cell-free expression systems. These platforms require minimal sample volumes (nanoliters to picoliters) while maintaining analytical sensitivity. The integration of microfluidic chips with detection systems enables real-time monitoring of protein synthesis, providing dynamic information about expression kinetics that was previously unattainable in traditional batch processes.
Parallel processing capabilities represent another dimension of scalability in cell-free proteome analysis. Modern systems can simultaneously express and analyze thousands of different proteins under identical conditions, facilitating comparative studies across entire proteomes. This parallelization is particularly valuable for identifying protein interaction networks and conducting systematic structure-function relationship studies.
Cost considerations remain critical for widespread adoption of high-throughput cell-free proteome analysis. Recent innovations in extract preparation and reaction component recycling have reduced costs by up to 70% compared to early systems. Additionally, the development of lyophilized cell-free reagents has extended shelf-life and simplified logistics, making these technologies accessible to laboratories worldwide without specialized equipment.
Data management infrastructure has evolved to handle the massive datasets generated by high-throughput proteome analysis. Cloud-based platforms with machine learning algorithms now facilitate automated data processing, pattern recognition, and integration with existing proteome databases. These computational tools are essential for extracting meaningful biological insights from the terabytes of data generated in large-scale proteome studies.
The implementation of automation technologies has dramatically enhanced the throughput capabilities of cell-free proteome analysis. Robotic liquid handling systems, when integrated with cell-free expression platforms, can process thousands of protein samples simultaneously. This automation reduces human error, increases reproducibility, and significantly decreases the time required for comprehensive proteome studies from months to days.
Microfluidic technologies have emerged as a revolutionary approach for miniaturizing cell-free expression systems. These platforms require minimal sample volumes (nanoliters to picoliters) while maintaining analytical sensitivity. The integration of microfluidic chips with detection systems enables real-time monitoring of protein synthesis, providing dynamic information about expression kinetics that was previously unattainable in traditional batch processes.
Parallel processing capabilities represent another dimension of scalability in cell-free proteome analysis. Modern systems can simultaneously express and analyze thousands of different proteins under identical conditions, facilitating comparative studies across entire proteomes. This parallelization is particularly valuable for identifying protein interaction networks and conducting systematic structure-function relationship studies.
Cost considerations remain critical for widespread adoption of high-throughput cell-free proteome analysis. Recent innovations in extract preparation and reaction component recycling have reduced costs by up to 70% compared to early systems. Additionally, the development of lyophilized cell-free reagents has extended shelf-life and simplified logistics, making these technologies accessible to laboratories worldwide without specialized equipment.
Data management infrastructure has evolved to handle the massive datasets generated by high-throughput proteome analysis. Cloud-based platforms with machine learning algorithms now facilitate automated data processing, pattern recognition, and integration with existing proteome databases. These computational tools are essential for extracting meaningful biological insights from the terabytes of data generated in large-scale proteome studies.
Biosafety and Regulatory Considerations
Cell-free expression systems for proteome analysis operate outside traditional biosafety containment frameworks, necessitating careful consideration of regulatory requirements and safety protocols. These systems involve handling biological materials including DNA templates, cell extracts, and potentially hazardous reagents, requiring adherence to laboratory biosafety levels (BSL) appropriate to the risk classification of materials used. Most cell-free proteomics work falls under BSL-1 or BSL-2 categories, though work with pathogenic proteins may require higher containment levels.
Regulatory oversight varies significantly across jurisdictions, with agencies such as the FDA in the United States, the EMA in Europe, and the NMPA in China establishing different frameworks for cell-free technologies. These regulations particularly impact diagnostic applications and therapeutic protein production, where Good Manufacturing Practice (GMP) compliance becomes essential for clinical translation.
Environmental considerations must address the disposal of reaction components containing recombinant DNA and expressed proteins. While cell-free systems generally present reduced environmental risks compared to living organisms, proper waste management protocols remain necessary to prevent unintended environmental exposure or contamination.
Intellectual property landscapes surrounding cell-free expression technologies present additional regulatory challenges. Many fundamental techniques are patent-protected, requiring licensing agreements for commercial applications. This complex IP environment necessitates thorough freedom-to-operate analyses before implementing these systems in industrial settings.
Ethical considerations emerge when cell-free systems express proteins from human origin or those with dual-use potential. Institutional review boards and ethics committees should evaluate research protocols involving human genetic material, while international biosecurity frameworks govern work with potentially dangerous proteins.
Looking forward, regulatory frameworks are evolving to accommodate cell-free technologies' unique characteristics. Industry stakeholders are collaborating with regulatory bodies to develop standardized approaches for safety assessment and quality control. The establishment of international standards for cell-free expression systems would significantly facilitate cross-border research collaboration and commercialization efforts while ensuring appropriate biosafety measures remain in place.
Regulatory oversight varies significantly across jurisdictions, with agencies such as the FDA in the United States, the EMA in Europe, and the NMPA in China establishing different frameworks for cell-free technologies. These regulations particularly impact diagnostic applications and therapeutic protein production, where Good Manufacturing Practice (GMP) compliance becomes essential for clinical translation.
Environmental considerations must address the disposal of reaction components containing recombinant DNA and expressed proteins. While cell-free systems generally present reduced environmental risks compared to living organisms, proper waste management protocols remain necessary to prevent unintended environmental exposure or contamination.
Intellectual property landscapes surrounding cell-free expression technologies present additional regulatory challenges. Many fundamental techniques are patent-protected, requiring licensing agreements for commercial applications. This complex IP environment necessitates thorough freedom-to-operate analyses before implementing these systems in industrial settings.
Ethical considerations emerge when cell-free systems express proteins from human origin or those with dual-use potential. Institutional review boards and ethics committees should evaluate research protocols involving human genetic material, while international biosecurity frameworks govern work with potentially dangerous proteins.
Looking forward, regulatory frameworks are evolving to accommodate cell-free technologies' unique characteristics. Industry stakeholders are collaborating with regulatory bodies to develop standardized approaches for safety assessment and quality control. The establishment of international standards for cell-free expression systems would significantly facilitate cross-border research collaboration and commercialization efforts while ensuring appropriate biosafety measures remain in place.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!