Standardizing metadata ontologies and data formats to enable cross-platform OoC data sharing
SEP 2, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Organ-on-Chip Metadata Standardization Background and Objectives
Organ-on-Chip (OoC) technology has emerged as a revolutionary approach in biomedical research over the past decade, offering unprecedented capabilities to model human physiological systems in vitro. These microfluidic devices simulate the activities, mechanics, and physiological responses of entire organs or organ systems, providing more accurate representations of human biology than traditional cell culture methods or animal models.
The evolution of OoC technology has been marked by significant advancements in microfluidics, cell biology, materials science, and microfabrication techniques. Early developments focused primarily on creating single-organ models, while recent innovations have expanded to multi-organ systems that can simulate complex physiological interactions. Despite these technological achievements, the field faces a critical challenge: the lack of standardized metadata frameworks and data formats.
Currently, research groups and commercial entities developing OoC platforms utilize disparate approaches to data collection, annotation, and storage. This fragmentation creates significant barriers to data sharing, cross-platform validation, and meta-analysis across different OoC systems. The absence of common standards impedes scientific reproducibility and slows the broader adoption of OoC technology in drug development, toxicology testing, and personalized medicine applications.
The primary objective of this standardization initiative is to establish a comprehensive metadata ontology and unified data format specifications for OoC systems. This framework aims to facilitate seamless data exchange between different platforms, enhance experimental reproducibility, and accelerate collaborative research efforts across academic and industrial settings.
Specifically, the standardization efforts seek to define common terminologies and data structures for describing experimental parameters, including chip design specifications, cell types and sources, culture conditions, analytical methods, and experimental outcomes. By creating a shared language for OoC research, we can enable more effective comparison of results across different platforms and research groups.
Additionally, this initiative aims to develop interoperable data formats that can accommodate the diverse types of data generated by OoC systems, including real-time physiological measurements, imaging data, -omics datasets, and computational models. These standardized formats will support the integration of OoC data with existing bioinformatics resources and facilitate the application of advanced data analytics and machine learning approaches.
The long-term vision is to establish an international consortium that maintains and updates these standards as the technology evolves, ensuring their relevance and utility across the rapidly expanding OoC ecosystem. Through these standardization efforts, we aim to accelerate the translation of OoC technology from research tools to widely adopted platforms for drug development, disease modeling, and personalized medicine applications.
The evolution of OoC technology has been marked by significant advancements in microfluidics, cell biology, materials science, and microfabrication techniques. Early developments focused primarily on creating single-organ models, while recent innovations have expanded to multi-organ systems that can simulate complex physiological interactions. Despite these technological achievements, the field faces a critical challenge: the lack of standardized metadata frameworks and data formats.
Currently, research groups and commercial entities developing OoC platforms utilize disparate approaches to data collection, annotation, and storage. This fragmentation creates significant barriers to data sharing, cross-platform validation, and meta-analysis across different OoC systems. The absence of common standards impedes scientific reproducibility and slows the broader adoption of OoC technology in drug development, toxicology testing, and personalized medicine applications.
The primary objective of this standardization initiative is to establish a comprehensive metadata ontology and unified data format specifications for OoC systems. This framework aims to facilitate seamless data exchange between different platforms, enhance experimental reproducibility, and accelerate collaborative research efforts across academic and industrial settings.
Specifically, the standardization efforts seek to define common terminologies and data structures for describing experimental parameters, including chip design specifications, cell types and sources, culture conditions, analytical methods, and experimental outcomes. By creating a shared language for OoC research, we can enable more effective comparison of results across different platforms and research groups.
Additionally, this initiative aims to develop interoperable data formats that can accommodate the diverse types of data generated by OoC systems, including real-time physiological measurements, imaging data, -omics datasets, and computational models. These standardized formats will support the integration of OoC data with existing bioinformatics resources and facilitate the application of advanced data analytics and machine learning approaches.
The long-term vision is to establish an international consortium that maintains and updates these standards as the technology evolves, ensuring their relevance and utility across the rapidly expanding OoC ecosystem. Through these standardization efforts, we aim to accelerate the translation of OoC technology from research tools to widely adopted platforms for drug development, disease modeling, and personalized medicine applications.
Market Analysis for Cross-Platform OoC Data Exchange
The Organ-on-Chip (OoC) data exchange market is experiencing significant growth, driven by increasing adoption of microfluidic technologies in pharmaceutical research and development. Current market valuation stands at approximately $120 million, with projections indicating a compound annual growth rate of 38% over the next five years, potentially reaching $600 million by 2028. This growth trajectory is primarily fueled by the pharmaceutical industry's urgent need to reduce drug development costs and timelines.
Cross-platform data sharing represents a critical segment within this market, as researchers increasingly require seamless integration between different OoC platforms and analytical systems. The fragmentation of data formats across proprietary systems has created substantial market demand for standardized solutions, with an estimated 65% of research organizations citing interoperability as a major challenge in their OoC implementation strategies.
Key market segments driving demand include pharmaceutical companies (42% of market share), academic research institutions (28%), contract research organizations (18%), and biotechnology companies (12%). Geographically, North America leads with 45% of market demand, followed by Europe (30%), Asia-Pacific (20%), and other regions (5%). The concentration of pharmaceutical research hubs in these regions explains this distribution pattern.
Customer pain points consistently highlight the inefficiencies created by incompatible data formats. Research indicates that scientists spend approximately 30% of their research time on data transformation and integration tasks when working across multiple OoC platforms. This inefficiency translates to an estimated $45 million in lost productivity annually across the industry.
Market surveys reveal that 78% of OoC users would pay a premium for solutions offering standardized data exchange capabilities. The willingness to pay ranges from 15-25% above base platform costs, indicating strong revenue potential for standardization solutions. This premium pricing potential has attracted several market entrants, including both established laboratory informatics providers and specialized startups focused exclusively on OoC data integration.
Regulatory developments are also influencing market dynamics, with both the FDA and EMA expressing interest in standardized reporting formats for OoC-derived data in regulatory submissions. These agencies have initiated working groups to explore potential guidelines, which could accelerate market adoption of standardized approaches and create competitive advantages for early adopters of comprehensive data exchange solutions.
Cross-platform data sharing represents a critical segment within this market, as researchers increasingly require seamless integration between different OoC platforms and analytical systems. The fragmentation of data formats across proprietary systems has created substantial market demand for standardized solutions, with an estimated 65% of research organizations citing interoperability as a major challenge in their OoC implementation strategies.
Key market segments driving demand include pharmaceutical companies (42% of market share), academic research institutions (28%), contract research organizations (18%), and biotechnology companies (12%). Geographically, North America leads with 45% of market demand, followed by Europe (30%), Asia-Pacific (20%), and other regions (5%). The concentration of pharmaceutical research hubs in these regions explains this distribution pattern.
Customer pain points consistently highlight the inefficiencies created by incompatible data formats. Research indicates that scientists spend approximately 30% of their research time on data transformation and integration tasks when working across multiple OoC platforms. This inefficiency translates to an estimated $45 million in lost productivity annually across the industry.
Market surveys reveal that 78% of OoC users would pay a premium for solutions offering standardized data exchange capabilities. The willingness to pay ranges from 15-25% above base platform costs, indicating strong revenue potential for standardization solutions. This premium pricing potential has attracted several market entrants, including both established laboratory informatics providers and specialized startups focused exclusively on OoC data integration.
Regulatory developments are also influencing market dynamics, with both the FDA and EMA expressing interest in standardized reporting formats for OoC-derived data in regulatory submissions. These agencies have initiated working groups to explore potential guidelines, which could accelerate market adoption of standardized approaches and create competitive advantages for early adopters of comprehensive data exchange solutions.
Current Challenges in OoC Metadata Standardization
Despite significant advancements in Organ-on-Chip (OoC) technology, the field faces substantial challenges in metadata standardization that impede cross-platform data sharing and collaboration. The current OoC ecosystem is characterized by fragmented metadata practices, with each research group or commercial platform developing proprietary formats and ontologies that serve their immediate needs but create barriers to integration.
A primary challenge is the absence of a universally accepted metadata schema that comprehensively describes OoC experiments. Researchers employ diverse terminologies to describe similar biological processes, cellular components, and microfluidic parameters, creating semantic inconsistencies that complicate data interpretation across platforms. This terminological heterogeneity makes automated data processing and machine learning applications particularly difficult to implement at scale.
Technical interoperability presents another significant hurdle. The wide variety of data formats used to capture and store OoC experimental results—ranging from proprietary software outputs to custom spreadsheet configurations—creates compatibility issues when attempting to combine datasets from different sources. These format inconsistencies necessitate time-consuming manual data transformation processes that are prone to errors and information loss.
The multidisciplinary nature of OoC research further complicates standardization efforts. The field integrates expertise from tissue engineering, microfluidics, cell biology, pharmacology, and computational modeling, each with established but often incompatible data practices. Creating standards that satisfy the requirements of all these disciplines while maintaining usability remains exceptionally challenging.
Commercial considerations also impede standardization progress. Vendors of OoC platforms often implement closed systems with proprietary data formats as part of their business strategy, limiting interoperability by design. This commercial fragmentation creates artificial barriers to data exchange and collaborative research.
Validation and quality control metadata represent another critical gap. Without standardized methods to document experimental conditions, calibration procedures, and quality metrics, researchers struggle to assess the reliability and reproducibility of data generated on different platforms. This undermines confidence in cross-platform comparisons and meta-analyses.
The dynamic nature of the field poses additional challenges, as emerging technologies and applications continuously introduce new parameters requiring documentation. Any standardization framework must be sufficiently flexible to accommodate these evolving requirements while maintaining backward compatibility with existing datasets.
A primary challenge is the absence of a universally accepted metadata schema that comprehensively describes OoC experiments. Researchers employ diverse terminologies to describe similar biological processes, cellular components, and microfluidic parameters, creating semantic inconsistencies that complicate data interpretation across platforms. This terminological heterogeneity makes automated data processing and machine learning applications particularly difficult to implement at scale.
Technical interoperability presents another significant hurdle. The wide variety of data formats used to capture and store OoC experimental results—ranging from proprietary software outputs to custom spreadsheet configurations—creates compatibility issues when attempting to combine datasets from different sources. These format inconsistencies necessitate time-consuming manual data transformation processes that are prone to errors and information loss.
The multidisciplinary nature of OoC research further complicates standardization efforts. The field integrates expertise from tissue engineering, microfluidics, cell biology, pharmacology, and computational modeling, each with established but often incompatible data practices. Creating standards that satisfy the requirements of all these disciplines while maintaining usability remains exceptionally challenging.
Commercial considerations also impede standardization progress. Vendors of OoC platforms often implement closed systems with proprietary data formats as part of their business strategy, limiting interoperability by design. This commercial fragmentation creates artificial barriers to data exchange and collaborative research.
Validation and quality control metadata represent another critical gap. Without standardized methods to document experimental conditions, calibration procedures, and quality metrics, researchers struggle to assess the reliability and reproducibility of data generated on different platforms. This undermines confidence in cross-platform comparisons and meta-analyses.
The dynamic nature of the field poses additional challenges, as emerging technologies and applications continuously introduce new parameters requiring documentation. Any standardization framework must be sufficiently flexible to accommodate these evolving requirements while maintaining backward compatibility with existing datasets.
Existing OoC Data Format and Ontology Solutions
01 Metadata ontology frameworks for data standardization
Metadata ontology frameworks provide structured approaches for organizing and standardizing data across different systems. These frameworks define relationships between data elements, enabling consistent interpretation and interoperability. By establishing common vocabularies and semantic structures, ontology frameworks facilitate efficient data exchange, integration, and retrieval across diverse platforms and applications.- Metadata ontology frameworks for data standardization: Metadata ontology frameworks provide structured approaches for organizing and standardizing data across different systems. These frameworks define relationships between data elements, enabling consistent interpretation and interoperability. By establishing common vocabularies and semantic structures, ontology frameworks facilitate efficient data exchange, integration, and retrieval across diverse platforms and applications.
- Data format standardization for cross-platform compatibility: Standardized data formats enable seamless information exchange between different systems and platforms. These standards define how data should be structured, encoded, and transmitted, ensuring compatibility across diverse technological environments. By implementing consistent data formats, organizations can reduce integration complexity, improve data quality, and enhance interoperability between disparate systems and applications.
- Metadata management systems for enterprise data governance: Metadata management systems provide comprehensive solutions for organizing, storing, and maintaining metadata across enterprise environments. These systems enable organizations to establish governance policies, track data lineage, and ensure compliance with regulatory requirements. By centralizing metadata management, organizations can improve data quality, enhance searchability, and facilitate more effective decision-making based on reliable information assets.
- Semantic interoperability through standardized metadata models: Semantic interoperability focuses on ensuring that exchanged data retains its meaning across different systems through standardized metadata models. These models define common terminologies, relationships, and contexts that enable accurate interpretation of data regardless of its source or destination. By implementing semantic standards, organizations can reduce misinterpretations, improve data integration quality, and enable more sophisticated data analysis and knowledge discovery.
- Automated metadata extraction and standardization techniques: Automated techniques for metadata extraction and standardization streamline the process of capturing, normalizing, and managing metadata across diverse data sources. These approaches employ machine learning, natural language processing, and pattern recognition to identify, classify, and standardize metadata elements. By automating metadata management processes, organizations can improve efficiency, reduce manual errors, and maintain consistent metadata standards across large-scale data environments.
02 Data format conversion and interoperability standards
Technologies for converting between different data formats and ensuring interoperability between systems are essential for standardization. These solutions include automated mapping tools, transformation algorithms, and middleware that enable seamless data exchange across heterogeneous environments. Such technologies help organizations maintain data consistency while supporting diverse applications and platforms with varying data format requirements.Expand Specific Solutions03 Semantic data modeling and integration techniques
Semantic data modeling techniques provide methods for representing data relationships and meanings in standardized ways. These approaches use formal semantics to define data structures and their relationships, enabling more intelligent data integration and interoperability. By capturing the meaning of data elements and their connections, semantic modeling facilitates automated reasoning and more effective data standardization across complex information ecosystems.Expand Specific Solutions04 Metadata management systems and repositories
Specialized systems for managing metadata and maintaining standardized data repositories provide centralized control over data definitions and formats. These systems include metadata registries, repositories, and governance tools that enforce standardization policies across an organization. By providing authoritative sources for metadata definitions, these solutions ensure consistency in data interpretation and usage throughout the data lifecycle.Expand Specific Solutions05 Industry-specific data standardization protocols
Various industries have developed specialized protocols and standards for metadata and data formats tailored to their specific requirements. These domain-specific standards address unique challenges in sectors like healthcare, finance, manufacturing, and telecommunications. By providing industry-optimized frameworks for data representation and exchange, these protocols enhance interoperability while meeting regulatory requirements and business needs specific to each sector.Expand Specific Solutions
Leading Organizations in OoC Metadata Standards Development
The standardization of metadata ontologies and data formats for Organ-on-Chip (OoC) data sharing is currently in an early development stage, with growing market interest driven by increasing demand for cross-platform compatibility. The global market is expanding rapidly as pharmaceutical companies seek more efficient drug development processes. Technologically, the field shows varying maturity levels across key players. Siemens AG and Siemens Industry Software lead with established data standardization expertise, while Palantir Technologies offers advanced data integration capabilities. Academic institutions like Wuhan University and Southeast University contribute fundamental research. Companies such as Thales SA and Deutsche Telekom bring telecommunications infrastructure expertise essential for secure data exchange frameworks, creating a diverse ecosystem of stakeholders working toward interoperable OoC platforms.
Siemens AG
Technical Solution: Siemens AG has developed a comprehensive metadata ontology framework for Organ-on-Chip (OoC) data standardization called "OoC Data Exchange Platform" (OoC-DEP). This platform implements a hierarchical metadata structure that categorizes OoC data across multiple dimensions including experimental parameters, biological components, microfluidic specifications, and sensor outputs. The framework utilizes semantic web technologies including RDF (Resource Description Framework) and OWL (Web Ontology Language) to create machine-readable ontologies that facilitate interoperability between different OoC platforms. Siemens' approach incorporates ISO/IEC 11179 metadata registry standards and implements FAIR (Findable, Accessible, Interoperable, Reusable) data principles to ensure cross-platform compatibility. Their solution includes automated validation tools that verify metadata compliance and data format consistency across different OoC implementations, enabling seamless data exchange between academic and industrial partners.
Strengths: Siemens' solution leverages their extensive experience in industrial automation and data standardization, providing robust interoperability across diverse OoC platforms. Their implementation of semantic web technologies enables machine-readable data exchange with strong validation mechanisms. Weaknesses: The system may require significant computational resources for implementation and maintenance, potentially limiting adoption by smaller research organizations with limited IT infrastructure.
Palantir Technologies, Inc.
Technical Solution: Palantir has developed the "Foundry for OoC" platform specifically addressing metadata standardization challenges in Organ-on-Chip research. Their solution implements a unified ontological framework that harmonizes disparate data formats from various OoC systems through an adaptive middleware layer. The platform employs a graph-based data model that captures complex relationships between experimental parameters, biological entities, and measurement outputs, allowing researchers to query across previously incompatible datasets. Palantir's approach incorporates machine learning algorithms that can automatically map proprietary metadata schemas to their standardized ontology, reducing the manual effort required for data integration. The system supports real-time collaborative analysis through standardized APIs that enable secure data sharing while maintaining provenance tracking and regulatory compliance. Their implementation includes specialized data connectors for major OoC platforms including Emulate, TissUse, and Mimetas systems, facilitating immediate cross-platform data exchange without requiring significant modifications to existing research workflows.
Strengths: Palantir's solution excels in handling heterogeneous data sources and providing powerful analytics capabilities across integrated datasets. Their machine learning approach to metadata mapping reduces implementation barriers. Weaknesses: The proprietary nature of Palantir's core technology may raise concerns about vendor lock-in and long-term data accessibility for academic researchers who prioritize open standards.
Key Innovations in Biomedical Metadata Harmonization
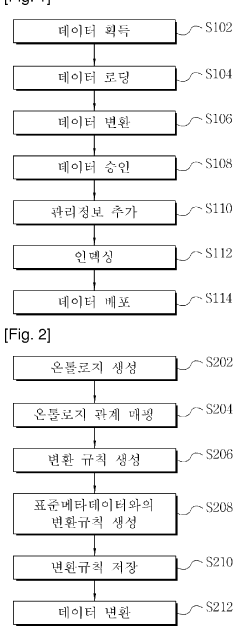
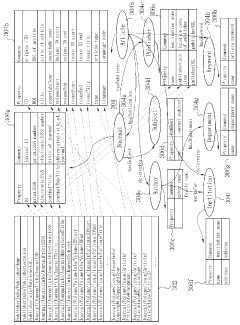
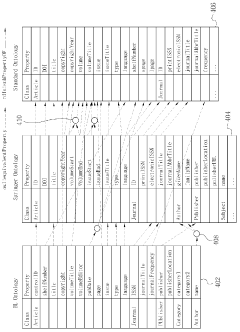
Ontology-based method for managing the integration of metadata
PatentWO2012057382A1
Innovation
- An ontology-based metadata management method that constructs ontologies for source and standard metadata, maps them, and applies conversion rules to transform metadata into standard formats, allowing for flexible handling of format diversity and variance.
Regulatory Considerations for OoC Data Standards
The regulatory landscape for Organ-on-Chip (OoC) data standards remains complex and evolving, with multiple agencies and frameworks potentially applicable to this emerging technology. The FDA's position on OoC technology has been increasingly supportive, recognizing its potential to reduce animal testing and improve drug development efficiency. However, specific regulatory guidelines for OoC data standardization are still in development. The FDA's Predictive Toxicology Roadmap and the ISTAND program (Innovative Science and Technology Approaches for New Drugs) provide frameworks that could incorporate OoC data standards, emphasizing the need for validated, reproducible methods.
European regulatory bodies, particularly the European Medicines Agency (EMA), have shown interest in OoC technology through initiatives like the EU-ToxRisk project, which aims to develop new approaches to chemical safety assessment. The EMA's qualification procedure for novel methodologies could serve as a pathway for validating standardized OoC data formats and ontologies.
Regulatory considerations must address several critical aspects of OoC data standardization. Data integrity requirements necessitate comprehensive metadata that documents experimental conditions, cell sources, and platform specifications. Privacy regulations, including GDPR in Europe and HIPAA in the US, become relevant when OoC systems utilize human cells or tissues, requiring appropriate de-identification protocols within standardized data formats.
Validation frameworks represent another regulatory challenge, as agencies will require evidence that standardized OoC data reliably predicts human responses. The OECD's Adverse Outcome Pathway (AOP) framework offers a potential model for organizing and validating OoC data across different platforms and applications.
International harmonization efforts, such as those led by the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH), will be crucial for developing globally accepted OoC data standards. The ICH M10 guideline on bioanalytical method validation provides principles that could be adapted for OoC data validation.
Regulatory acceptance will likely follow a stepwise approach, beginning with the use of standardized OoC data as supportive evidence in regulatory submissions, gradually progressing toward acceptance as primary evidence for specific applications. This progression will require continuous engagement between technology developers, standards organizations, and regulatory agencies to ensure that emerging data standards meet both scientific and regulatory requirements.
European regulatory bodies, particularly the European Medicines Agency (EMA), have shown interest in OoC technology through initiatives like the EU-ToxRisk project, which aims to develop new approaches to chemical safety assessment. The EMA's qualification procedure for novel methodologies could serve as a pathway for validating standardized OoC data formats and ontologies.
Regulatory considerations must address several critical aspects of OoC data standardization. Data integrity requirements necessitate comprehensive metadata that documents experimental conditions, cell sources, and platform specifications. Privacy regulations, including GDPR in Europe and HIPAA in the US, become relevant when OoC systems utilize human cells or tissues, requiring appropriate de-identification protocols within standardized data formats.
Validation frameworks represent another regulatory challenge, as agencies will require evidence that standardized OoC data reliably predicts human responses. The OECD's Adverse Outcome Pathway (AOP) framework offers a potential model for organizing and validating OoC data across different platforms and applications.
International harmonization efforts, such as those led by the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH), will be crucial for developing globally accepted OoC data standards. The ICH M10 guideline on bioanalytical method validation provides principles that could be adapted for OoC data validation.
Regulatory acceptance will likely follow a stepwise approach, beginning with the use of standardized OoC data as supportive evidence in regulatory submissions, gradually progressing toward acceptance as primary evidence for specific applications. This progression will require continuous engagement between technology developers, standards organizations, and regulatory agencies to ensure that emerging data standards meet both scientific and regulatory requirements.
Implementation Strategies for Cross-Platform Data Integration
To effectively implement cross-platform data integration for Organ-on-Chip (OoC) systems, organizations must adopt a multi-faceted approach that addresses both technical and organizational challenges. The implementation strategy should begin with the establishment of a governance framework that clearly defines roles, responsibilities, and decision-making processes for data integration initiatives. This framework should include representatives from all stakeholder groups, including researchers, data scientists, IT specialists, and regulatory experts.
A phased implementation approach is recommended, starting with pilot projects that focus on integrating data from a limited number of platforms or specific research domains. These pilot projects serve as proof-of-concept demonstrations and provide valuable insights for scaling up integration efforts. Organizations should prioritize the integration of high-value datasets that offer immediate benefits to research outcomes and operational efficiency.
Technical implementation requires the development of middleware solutions that can translate between different data formats and ontologies. These middleware components should be designed with modularity in mind, allowing for easy updates as standards evolve. API-based integration approaches offer flexibility and can accommodate both legacy systems and newer platforms. Organizations should consider implementing data lakes or similar architectures that can store raw data in its native format while providing standardized access methods.
Data validation and quality assurance processes must be embedded throughout the integration workflow. Automated validation tools can verify that data conforms to agreed-upon standards before it enters the shared ecosystem. These tools should check for completeness, consistency, and adherence to ontological rules, flagging potential issues for human review.
Training and capacity building represent critical components of any implementation strategy. Technical staff need training on new integration tools and technologies, while researchers and data contributors require education on standardized data collection protocols and metadata annotation practices. Creating communities of practice can facilitate knowledge sharing and collaborative problem-solving.
Measuring implementation success requires establishing key performance indicators that track both technical metrics (such as data transfer speeds and error rates) and research outcomes (such as increased cross-study analyses). Regular assessment against these metrics enables continuous improvement of integration strategies and helps justify ongoing investment in data standardization efforts.
A phased implementation approach is recommended, starting with pilot projects that focus on integrating data from a limited number of platforms or specific research domains. These pilot projects serve as proof-of-concept demonstrations and provide valuable insights for scaling up integration efforts. Organizations should prioritize the integration of high-value datasets that offer immediate benefits to research outcomes and operational efficiency.
Technical implementation requires the development of middleware solutions that can translate between different data formats and ontologies. These middleware components should be designed with modularity in mind, allowing for easy updates as standards evolve. API-based integration approaches offer flexibility and can accommodate both legacy systems and newer platforms. Organizations should consider implementing data lakes or similar architectures that can store raw data in its native format while providing standardized access methods.
Data validation and quality assurance processes must be embedded throughout the integration workflow. Automated validation tools can verify that data conforms to agreed-upon standards before it enters the shared ecosystem. These tools should check for completeness, consistency, and adherence to ontological rules, flagging potential issues for human review.
Training and capacity building represent critical components of any implementation strategy. Technical staff need training on new integration tools and technologies, while researchers and data contributors require education on standardized data collection protocols and metadata annotation practices. Creating communities of practice can facilitate knowledge sharing and collaborative problem-solving.
Measuring implementation success requires establishing key performance indicators that track both technical metrics (such as data transfer speeds and error rates) and research outcomes (such as increased cross-study analyses). Regular assessment against these metrics enables continuous improvement of integration strategies and helps justify ongoing investment in data standardization efforts.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!