Compare Phosphorylation and Transcriptional Dynamics
SEP 23, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Phosphorylation and Transcription Background and Objectives
Phosphorylation and transcription represent two fundamental cellular processes that orchestrate the complex symphony of gene expression and protein function regulation. Historically, these processes were studied separately, with phosphorylation research dating back to the 1950s when Edmond Fischer and Edwin Krebs discovered protein kinases. Transcriptional regulation studies gained momentum in the 1960s with the elucidation of the lac operon model by Jacob and Monod.
The evolution of these fields has been marked by significant technological breakthroughs. The development of radioactive labeling techniques in the 1970s enabled researchers to track phosphorylation events, while the advent of DNA sequencing and molecular cloning revolutionized transcription studies. More recently, high-throughput technologies such as mass spectrometry for phosphoproteomics and RNA-seq for transcriptomics have dramatically accelerated our understanding of both processes.
Current research increasingly recognizes the intricate interplay between phosphorylation and transcriptional dynamics. Phosphorylation events can directly modulate transcription factor activity, alter chromatin structure, and influence RNA polymerase function. Conversely, transcriptional outputs can regulate kinase expression, creating complex feedback loops that maintain cellular homeostasis or drive cellular transitions.
The temporal aspects of these processes represent a particularly fascinating area of investigation. While phosphorylation can occur within seconds to minutes, transcriptional changes typically manifest over minutes to hours. This temporal disconnect creates complex signaling cascades where rapid phosphorylation events trigger more sustained transcriptional responses, allowing cells to integrate immediate environmental signals into longer-term adaptive strategies.
The technical objective of this research is to develop comprehensive models that capture the dynamic relationship between phosphorylation networks and transcriptional programs across different time scales and cellular contexts. This includes mapping kinase-substrate relationships that influence transcription, identifying phosphorylation-dependent transcriptional regulators, and elucidating how transcriptional outputs feedback to modulate phosphorylation networks.
Understanding these dynamics has profound implications for multiple fields, including developmental biology, immunology, and cancer research. For instance, aberrant phosphorylation-transcription coupling underlies many pathological conditions, making this interface an attractive target for therapeutic intervention. The emergence of single-cell multi-omics technologies now enables unprecedented resolution in tracking these processes simultaneously, promising new insights into cellular decision-making mechanisms.
The evolution of these fields has been marked by significant technological breakthroughs. The development of radioactive labeling techniques in the 1970s enabled researchers to track phosphorylation events, while the advent of DNA sequencing and molecular cloning revolutionized transcription studies. More recently, high-throughput technologies such as mass spectrometry for phosphoproteomics and RNA-seq for transcriptomics have dramatically accelerated our understanding of both processes.
Current research increasingly recognizes the intricate interplay between phosphorylation and transcriptional dynamics. Phosphorylation events can directly modulate transcription factor activity, alter chromatin structure, and influence RNA polymerase function. Conversely, transcriptional outputs can regulate kinase expression, creating complex feedback loops that maintain cellular homeostasis or drive cellular transitions.
The temporal aspects of these processes represent a particularly fascinating area of investigation. While phosphorylation can occur within seconds to minutes, transcriptional changes typically manifest over minutes to hours. This temporal disconnect creates complex signaling cascades where rapid phosphorylation events trigger more sustained transcriptional responses, allowing cells to integrate immediate environmental signals into longer-term adaptive strategies.
The technical objective of this research is to develop comprehensive models that capture the dynamic relationship between phosphorylation networks and transcriptional programs across different time scales and cellular contexts. This includes mapping kinase-substrate relationships that influence transcription, identifying phosphorylation-dependent transcriptional regulators, and elucidating how transcriptional outputs feedback to modulate phosphorylation networks.
Understanding these dynamics has profound implications for multiple fields, including developmental biology, immunology, and cancer research. For instance, aberrant phosphorylation-transcription coupling underlies many pathological conditions, making this interface an attractive target for therapeutic intervention. The emergence of single-cell multi-omics technologies now enables unprecedented resolution in tracking these processes simultaneously, promising new insights into cellular decision-making mechanisms.
Market Applications and Research Demand Analysis
The market for phosphorylation and transcriptional dynamics research tools and technologies has experienced significant growth over the past decade, driven primarily by advances in systems biology, precision medicine, and drug discovery. Current market estimates place the global proteomics market, which encompasses phosphorylation analysis tools, at approximately $21 billion, with a compound annual growth rate of 14% projected through 2028. Meanwhile, the transcriptomics market, covering transcriptional dynamics technologies, is valued at around $8.7 billion with a projected growth rate of 9.3% annually.
Pharmaceutical and biotechnology companies represent the largest market segment, accounting for nearly 45% of the total demand. These organizations leverage phosphorylation and transcriptional dynamics research to develop targeted therapies, particularly for cancer, neurodegenerative diseases, and inflammatory conditions. The ability to understand post-translational modifications and gene expression changes simultaneously provides crucial insights for drug development pipelines.
Academic and research institutions constitute the second-largest market segment, driven by fundamental research funding and collaborative industry partnerships. Government initiatives supporting precision medicine, such as the NIH's Precision Medicine Initiative in the United States and similar programs in Europe and Asia, have allocated substantial resources toward understanding molecular mechanisms of disease, further stimulating market growth.
Clinical diagnostics represents an emerging application area with significant growth potential. The integration of phosphorylation and transcriptional analysis into diagnostic workflows enables more precise patient stratification and treatment selection. This is particularly evident in oncology, where phosphorylation patterns of specific proteins combined with gene expression signatures are increasingly used to guide therapeutic decisions.
Regionally, North America dominates the market with approximately 40% share, followed by Europe at 30% and Asia-Pacific at 22%. However, the Asia-Pacific region is experiencing the fastest growth rate, driven by increasing research investments in China, Japan, and South Korea, along with the expansion of contract research organizations in these regions.
The demand for integrated multi-omics approaches that combine phosphorylation and transcriptional analysis has grown substantially, with researchers increasingly recognizing the limitations of studying these processes in isolation. This has created market opportunities for companies developing integrated analytical platforms and computational tools that can correlate post-translational modifications with transcriptional changes across time and cellular contexts.
Key market challenges include the high cost of advanced instrumentation, complexity of data integration and analysis, and the need for specialized expertise. These factors have created demand for more accessible, user-friendly technologies and turnkey solutions that can democratize access to these powerful analytical approaches across different research settings.
Pharmaceutical and biotechnology companies represent the largest market segment, accounting for nearly 45% of the total demand. These organizations leverage phosphorylation and transcriptional dynamics research to develop targeted therapies, particularly for cancer, neurodegenerative diseases, and inflammatory conditions. The ability to understand post-translational modifications and gene expression changes simultaneously provides crucial insights for drug development pipelines.
Academic and research institutions constitute the second-largest market segment, driven by fundamental research funding and collaborative industry partnerships. Government initiatives supporting precision medicine, such as the NIH's Precision Medicine Initiative in the United States and similar programs in Europe and Asia, have allocated substantial resources toward understanding molecular mechanisms of disease, further stimulating market growth.
Clinical diagnostics represents an emerging application area with significant growth potential. The integration of phosphorylation and transcriptional analysis into diagnostic workflows enables more precise patient stratification and treatment selection. This is particularly evident in oncology, where phosphorylation patterns of specific proteins combined with gene expression signatures are increasingly used to guide therapeutic decisions.
Regionally, North America dominates the market with approximately 40% share, followed by Europe at 30% and Asia-Pacific at 22%. However, the Asia-Pacific region is experiencing the fastest growth rate, driven by increasing research investments in China, Japan, and South Korea, along with the expansion of contract research organizations in these regions.
The demand for integrated multi-omics approaches that combine phosphorylation and transcriptional analysis has grown substantially, with researchers increasingly recognizing the limitations of studying these processes in isolation. This has created market opportunities for companies developing integrated analytical platforms and computational tools that can correlate post-translational modifications with transcriptional changes across time and cellular contexts.
Key market challenges include the high cost of advanced instrumentation, complexity of data integration and analysis, and the need for specialized expertise. These factors have created demand for more accessible, user-friendly technologies and turnkey solutions that can democratize access to these powerful analytical approaches across different research settings.
Current Technical Challenges in Comparative Dynamics
The comparative analysis of phosphorylation and transcriptional dynamics presents several significant technical challenges that currently limit our comprehensive understanding of these interconnected cellular processes. One primary obstacle is the vast difference in temporal scales between these two regulatory mechanisms. Phosphorylation events occur within seconds to minutes, while transcriptional changes typically manifest over hours to days, making simultaneous measurement technically demanding.
Current detection technologies also present limitations. While mass spectrometry offers high-throughput phosphoproteomic analysis, it struggles with temporal resolution and requires substantial sample amounts. RNA sequencing provides excellent transcriptome coverage but faces challenges in capturing rapid transcriptional changes. The integration of these disparate data types remains problematic due to differences in measurement units, dynamic ranges, and noise characteristics.
Single-cell analysis introduces additional complexity. Technologies for simultaneous measurement of phosphorylation states and transcriptional profiles at single-cell resolution are still emerging. Current approaches often require cell fixation, preventing true dynamic studies in living systems. The trade-off between spatial resolution and temporal dynamics continues to challenge researchers attempting to correlate these processes within cellular compartments.
Computational challenges further complicate comparative analyses. Existing algorithms struggle to integrate multi-omics data with different temporal characteristics. Network inference methods often fail to capture the complex feedback loops between phosphorylation cascades and transcriptional regulation. Statistical frameworks for determining causality versus correlation between these processes remain underdeveloped.
Biological variability adds another layer of complexity. Cell-to-cell heterogeneity in both phosphorylation states and gene expression patterns makes population-level measurements potentially misleading. Environmental factors and cellular states significantly influence both processes, requiring careful experimental design to control for confounding variables.
Technical standardization issues persist across laboratories. Differences in sample preparation, instrument calibration, and data processing pipelines make cross-study comparisons difficult. The lack of universal standards for quantifying phosphorylation stoichiometry and absolute transcript abundance further complicates comparative analyses.
Emerging areas like spatial proteomics and transcriptomics offer promising approaches but face technical limitations in resolution and throughput. The development of biosensors for real-time monitoring of phosphorylation events has advanced significantly but remains limited to a small subset of kinase activities, while parallel monitoring of transcriptional dynamics still relies heavily on reporter constructs with inherent limitations.
Current detection technologies also present limitations. While mass spectrometry offers high-throughput phosphoproteomic analysis, it struggles with temporal resolution and requires substantial sample amounts. RNA sequencing provides excellent transcriptome coverage but faces challenges in capturing rapid transcriptional changes. The integration of these disparate data types remains problematic due to differences in measurement units, dynamic ranges, and noise characteristics.
Single-cell analysis introduces additional complexity. Technologies for simultaneous measurement of phosphorylation states and transcriptional profiles at single-cell resolution are still emerging. Current approaches often require cell fixation, preventing true dynamic studies in living systems. The trade-off between spatial resolution and temporal dynamics continues to challenge researchers attempting to correlate these processes within cellular compartments.
Computational challenges further complicate comparative analyses. Existing algorithms struggle to integrate multi-omics data with different temporal characteristics. Network inference methods often fail to capture the complex feedback loops between phosphorylation cascades and transcriptional regulation. Statistical frameworks for determining causality versus correlation between these processes remain underdeveloped.
Biological variability adds another layer of complexity. Cell-to-cell heterogeneity in both phosphorylation states and gene expression patterns makes population-level measurements potentially misleading. Environmental factors and cellular states significantly influence both processes, requiring careful experimental design to control for confounding variables.
Technical standardization issues persist across laboratories. Differences in sample preparation, instrument calibration, and data processing pipelines make cross-study comparisons difficult. The lack of universal standards for quantifying phosphorylation stoichiometry and absolute transcript abundance further complicates comparative analyses.
Emerging areas like spatial proteomics and transcriptomics offer promising approaches but face technical limitations in resolution and throughput. The development of biosensors for real-time monitoring of phosphorylation events has advanced significantly but remains limited to a small subset of kinase activities, while parallel monitoring of transcriptional dynamics still relies heavily on reporter constructs with inherent limitations.
Methodological Approaches for Comparative Dynamic Analysis
01 Phosphorylation mechanisms in gene expression regulation
Phosphorylation plays a critical role in regulating gene expression by modifying transcription factors and other regulatory proteins. This post-translational modification can activate or inhibit transcription factor binding to DNA, affecting their ability to recruit RNA polymerase and initiate transcription. The dynamic nature of phosphorylation allows for rapid responses to cellular signals and environmental changes, providing a mechanism for temporal control of gene expression.- Phosphorylation mechanisms in gene expression regulation: Phosphorylation plays a crucial role in regulating gene expression by modifying transcription factors and other regulatory proteins. This post-translational modification can activate or inhibit transcription factor activity, affecting their DNA binding capabilities, nuclear localization, and protein-protein interactions. The dynamic nature of phosphorylation allows for rapid responses to cellular signals, creating a sophisticated control mechanism for transcriptional dynamics.
- Kinase-mediated signaling pathways in transcriptional control: Various kinase-mediated signaling pathways are involved in transcriptional regulation through phosphorylation events. These pathways transmit external signals to the nucleus, where they influence gene expression patterns. Specific kinases target different transcription factors, creating complex regulatory networks that control cellular responses to environmental stimuli. Understanding these pathways provides insights into how cells coordinate transcriptional dynamics in response to changing conditions.
- Methods for detecting phosphorylation-dependent transcriptional changes: Advanced techniques have been developed to detect and analyze phosphorylation-dependent changes in transcriptional activity. These methods include phospho-specific antibodies, mass spectrometry, reporter gene assays, and high-throughput screening approaches. Such techniques enable researchers to monitor the temporal dynamics of phosphorylation events and their correlation with changes in gene expression, providing valuable tools for studying the relationship between phosphorylation and transcriptional regulation.
- Phosphorylation-dependent transcription factor complexes: Phosphorylation can regulate the assembly and disassembly of transcription factor complexes, affecting their functionality. These complexes often include multiple proteins whose interactions are modulated by phosphorylation states. The dynamic formation of these complexes allows for precise control of gene expression in response to cellular signals. The composition of these complexes can determine which genes are activated or repressed, contributing to the specificity of transcriptional responses.
- Therapeutic applications targeting phosphorylation in transcriptional regulation: Targeting phosphorylation events involved in transcriptional regulation has emerged as a promising therapeutic strategy for various diseases. Inhibitors of specific kinases or phosphatases can modulate aberrant transcriptional activities associated with cancer, inflammatory disorders, and neurodegenerative diseases. Understanding the relationship between phosphorylation and transcriptional dynamics has led to the development of novel drugs that can restore normal gene expression patterns in pathological conditions.
02 Kinase-mediated signaling pathways in transcriptional control
Various kinase-mediated signaling pathways are involved in transcriptional regulation through phosphorylation events. These pathways transmit signals from the cell membrane to the nucleus, where they influence transcription factor activity. Specific kinases target different transcription factors, creating complex networks of regulation. Understanding these pathways provides insights into how cells respond to stimuli by altering gene expression patterns and can reveal potential therapeutic targets for diseases involving dysregulated transcription.Expand Specific Solutions03 Methods for detecting phosphorylation-dependent transcriptional changes
Advanced techniques have been developed to detect and analyze phosphorylation-dependent changes in transcriptional activity. These methods include phospho-specific antibodies, mass spectrometry, and reporter gene assays that can monitor real-time changes in transcription following phosphorylation events. High-throughput approaches allow for comprehensive mapping of phosphorylation sites and their effects on transcriptional dynamics, enabling researchers to understand the temporal relationship between phosphorylation and subsequent gene expression changes.Expand Specific Solutions04 Phosphorylation dynamics in chromatin remodeling and epigenetic regulation
Phosphorylation events influence transcriptional dynamics by affecting chromatin structure and epigenetic modifications. Histone phosphorylation can alter chromatin accessibility, while phosphorylation of chromatin remodeling complexes can change their activity or targeting. These modifications create a dynamic interplay between phosphorylation signaling and chromatin states, contributing to both short-term and long-term regulation of gene expression patterns. This mechanism is particularly important during development and cellular differentiation processes.Expand Specific Solutions05 Dysregulation of phosphorylation in transcriptional diseases
Abnormal phosphorylation patterns can lead to dysregulated transcriptional dynamics associated with various diseases, including cancer and neurodegenerative disorders. Mutations in kinases, phosphatases, or their substrates can disrupt normal transcriptional control mechanisms. Understanding these aberrant phosphorylation events provides opportunities for developing targeted therapeutics that can restore normal transcriptional dynamics. Phosphorylation biomarkers also offer diagnostic potential for identifying disease states characterized by altered transcriptional regulation.Expand Specific Solutions
Leading Research Institutions and Biotechnology Companies
The phosphorylation and transcriptional dynamics field is currently in a growth phase, with an estimated market size of $3-5 billion and expanding at 8-10% annually. The competitive landscape features academic powerhouses like University of California, Yale University, and Institut Pasteur leading fundamental research, while pharmaceutical giants including Roche, Novartis, and Merck are advancing clinical applications. Technology companies such as Illumina and IBM are developing analytical platforms to study these complex cellular processes. The field's technical maturity varies significantly across subdomains, with phosphorylation analysis being more established than integrated multi-omics approaches. Research institutions are increasingly partnering with industry players to bridge the gap between basic science discoveries and therapeutic applications targeting cancer, neurodegenerative diseases, and metabolic disorders.
The Regents of the University of California
Technical Solution: The University of California has developed advanced mass spectrometry-based approaches for studying phosphorylation dynamics in cellular signaling networks. Their technology integrates stable isotope labeling with amino acids in cell culture (SILAC) and phosphopeptide enrichment techniques to quantitatively measure thousands of phosphorylation sites simultaneously. They've pioneered computational frameworks that integrate phosphoproteomic data with transcriptional profiling to establish causal relationships between kinase activity and downstream gene expression changes. Their recent work has focused on temporal analysis of phosphorylation cascades, demonstrating how rapid phosphorylation events (occurring within minutes) trigger subsequent waves of transcriptional changes (occurring over hours). This approach has been particularly valuable in understanding cancer signaling networks and drug resistance mechanisms.
Strengths: Exceptional integration of multi-omics data types; sophisticated computational modeling of signaling networks; access to cutting-edge mass spectrometry facilities. Weaknesses: Complex methodologies require specialized expertise; high cost of implementation; challenges in detecting low-abundance phosphorylation events.
Yale University
Technical Solution: Yale University has developed a pioneering approach called "Integrated Phospho-Transcriptional Analysis" (IPTA) that combines phosphoproteomics with RNA-seq and ChIP-seq technologies to create comprehensive models of cellular signaling networks. Their methodology employs a unique time-course experimental design that captures both rapid phosphorylation events (seconds to minutes) and subsequent transcriptional responses (minutes to hours). Yale researchers have created custom bioinformatics pipelines that identify statistically significant correlations between specific phosphorylation events and transcriptional changes, enabling the prediction of causal relationships. Their technology has been successfully applied to study immune cell activation, revealing previously unknown signaling nodes that connect T-cell receptor engagement to gene expression programs controlling T-cell differentiation and function.
Strengths: Sophisticated temporal resolution of signaling events; strong bioinformatics capabilities for network inference; innovative experimental designs for capturing rapid phosphorylation dynamics. Weaknesses: Requires substantial computational resources; limited throughput for large-scale studies; challenges in distinguishing direct versus indirect phosphorylation effects on transcription.
Key Technological Innovations in Temporal Profiling
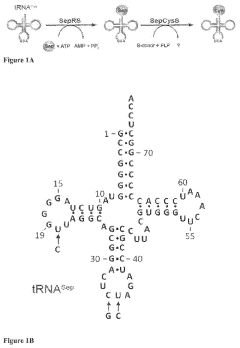
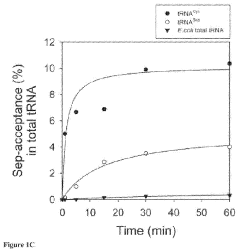
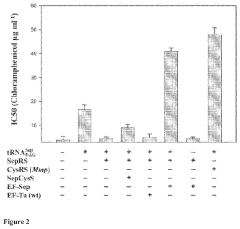
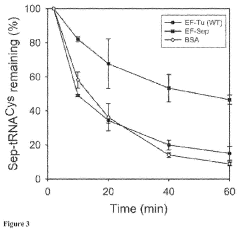
Site-specific incorporation of phosphoserine into proteins in escherichia coli
PatentInactiveUS20200385742A1
Innovation
- The use of mutant elongation factor proteins (EF-Sep) in conjunction with phosphoseryl-tRNA synthetase (SepRS) and phosphoseryl-tRNA (tRNASep) to specifically incorporate phosphoserine into proteins, where EF-Sep protects Sep-tRNASep from deacylation and catalyzes the covalent transfer of phosphoserine onto the polypeptide, allowing for site-specific phosphorylation indistinguishable from kinase-mediated phosphorylation.
Pknb kinase and pstp phosphatase and methods of identifying inhibitory substances
PatentInactiveUS20070128678A1
Innovation
- The pknB kinase and pstP phosphatase, which are co-localized in a conserved operon with other signaling proteins, are identified and characterized for their role in regulating cell growth and latency, providing potential targets for antibacterial agents.
Computational Tools for Dynamic Systems Biology
The field of systems biology has witnessed significant advancements in computational tools designed to analyze and model dynamic biological processes. When comparing phosphorylation and transcriptional dynamics, researchers require sophisticated computational approaches that can handle the distinct temporal scales and regulatory mechanisms of these processes.
Differential equation-based models have emerged as powerful tools for capturing the kinetics of phosphorylation events, which typically occur on timescales of seconds to minutes. These models, including ordinary differential equations (ODEs) and partial differential equations (PDEs), can effectively represent the rapid state changes in signaling cascades and provide quantitative predictions about system behavior under various conditions.
For transcriptional dynamics, which generally unfold over hours to days, stochastic simulation algorithms have proven particularly valuable. These approaches account for the inherent randomness in gene expression and can reveal how noise propagates through genetic networks. Tools such as the Gillespie algorithm and its variants enable researchers to simulate discrete molecular events that characterize transcriptional regulation.
Network-based analytical frameworks offer another computational approach for studying both processes simultaneously. These tools, including Bayesian networks and Boolean networks, can integrate diverse data types and identify causal relationships between phosphorylation events and subsequent transcriptional changes, thereby elucidating the complex interplay between these regulatory layers.
Machine learning techniques have revolutionized the analysis of high-throughput data generated from phosphoproteomic and transcriptomic experiments. Supervised learning algorithms can predict phosphorylation sites and transcription factor binding, while unsupervised methods like clustering and dimensionality reduction help identify patterns in complex datasets that span both regulatory domains.
Visualization tools specifically designed for temporal data have become essential for interpreting the dynamic behavior of biological systems. These include heatmaps with time-course clustering, trajectory plots, and interactive dashboards that allow researchers to explore the temporal relationships between phosphorylation cascades and downstream transcriptional responses.
Multi-scale modeling approaches are increasingly important for bridging the gap between phosphorylation and transcriptional dynamics. These hybrid computational frameworks can integrate fast signaling events with slower gene regulatory processes, providing a more comprehensive understanding of cellular decision-making across different time scales and biological contexts.
Differential equation-based models have emerged as powerful tools for capturing the kinetics of phosphorylation events, which typically occur on timescales of seconds to minutes. These models, including ordinary differential equations (ODEs) and partial differential equations (PDEs), can effectively represent the rapid state changes in signaling cascades and provide quantitative predictions about system behavior under various conditions.
For transcriptional dynamics, which generally unfold over hours to days, stochastic simulation algorithms have proven particularly valuable. These approaches account for the inherent randomness in gene expression and can reveal how noise propagates through genetic networks. Tools such as the Gillespie algorithm and its variants enable researchers to simulate discrete molecular events that characterize transcriptional regulation.
Network-based analytical frameworks offer another computational approach for studying both processes simultaneously. These tools, including Bayesian networks and Boolean networks, can integrate diverse data types and identify causal relationships between phosphorylation events and subsequent transcriptional changes, thereby elucidating the complex interplay between these regulatory layers.
Machine learning techniques have revolutionized the analysis of high-throughput data generated from phosphoproteomic and transcriptomic experiments. Supervised learning algorithms can predict phosphorylation sites and transcription factor binding, while unsupervised methods like clustering and dimensionality reduction help identify patterns in complex datasets that span both regulatory domains.
Visualization tools specifically designed for temporal data have become essential for interpreting the dynamic behavior of biological systems. These include heatmaps with time-course clustering, trajectory plots, and interactive dashboards that allow researchers to explore the temporal relationships between phosphorylation cascades and downstream transcriptional responses.
Multi-scale modeling approaches are increasingly important for bridging the gap between phosphorylation and transcriptional dynamics. These hybrid computational frameworks can integrate fast signaling events with slower gene regulatory processes, providing a more comprehensive understanding of cellular decision-making across different time scales and biological contexts.
Translational Impact on Drug Discovery and Therapeutics
The comparative analysis of phosphorylation and transcriptional dynamics has profound implications for drug discovery and therapeutic development. Understanding these molecular mechanisms enables pharmaceutical researchers to identify novel drug targets and develop more effective treatment strategies for various diseases, particularly cancer, neurodegenerative disorders, and inflammatory conditions.
Kinase inhibitors represent one of the most successful classes of targeted therapeutics developed based on phosphorylation dynamics research. By targeting specific kinases involved in dysregulated signaling pathways, these drugs have revolutionized cancer treatment. Notable examples include imatinib (Gleevec) for chronic myeloid leukemia and erlotinib for non-small cell lung cancer. The temporal dynamics of phosphorylation revealed through recent research provides opportunities for developing drugs with improved specificity and reduced off-target effects.
Transcription-based therapeutics have similarly emerged as promising approaches, with applications in gene therapy and RNA-based interventions. The integration of phosphorylation and transcriptional dynamics data has led to the development of dual-action compounds that can modulate both signaling cascades and gene expression patterns simultaneously. This approach shows particular promise for complex diseases where multiple molecular pathways contribute to pathogenesis.
High-throughput screening technologies that incorporate both phosphorylation and transcriptional readouts are transforming early-stage drug discovery. These platforms enable researchers to rapidly identify compounds that modulate specific cellular responses with greater precision. The temporal dimension of these molecular events provides critical insights for optimizing drug dosing schedules and combination therapies, potentially reducing treatment resistance and improving patient outcomes.
Personalized medicine approaches are increasingly leveraging phospho-proteomics and transcriptomics data to stratify patients and predict treatment responses. By analyzing individual phosphorylation and transcriptional profiles, clinicians can select therapies most likely to benefit specific patients, minimizing adverse effects and maximizing therapeutic efficacy. This precision medicine approach represents a significant advancement over traditional one-size-fits-all treatment paradigms.
Emerging therapeutic modalities, including proteolysis-targeting chimeras (PROTACs) and RNA-targeting small molecules, are being developed based on insights from integrated phosphorylation and transcriptional studies. These novel approaches offer potential solutions for previously "undruggable" targets, expanding the therapeutic landscape for challenging diseases with limited treatment options.
Kinase inhibitors represent one of the most successful classes of targeted therapeutics developed based on phosphorylation dynamics research. By targeting specific kinases involved in dysregulated signaling pathways, these drugs have revolutionized cancer treatment. Notable examples include imatinib (Gleevec) for chronic myeloid leukemia and erlotinib for non-small cell lung cancer. The temporal dynamics of phosphorylation revealed through recent research provides opportunities for developing drugs with improved specificity and reduced off-target effects.
Transcription-based therapeutics have similarly emerged as promising approaches, with applications in gene therapy and RNA-based interventions. The integration of phosphorylation and transcriptional dynamics data has led to the development of dual-action compounds that can modulate both signaling cascades and gene expression patterns simultaneously. This approach shows particular promise for complex diseases where multiple molecular pathways contribute to pathogenesis.
High-throughput screening technologies that incorporate both phosphorylation and transcriptional readouts are transforming early-stage drug discovery. These platforms enable researchers to rapidly identify compounds that modulate specific cellular responses with greater precision. The temporal dimension of these molecular events provides critical insights for optimizing drug dosing schedules and combination therapies, potentially reducing treatment resistance and improving patient outcomes.
Personalized medicine approaches are increasingly leveraging phospho-proteomics and transcriptomics data to stratify patients and predict treatment responses. By analyzing individual phosphorylation and transcriptional profiles, clinicians can select therapies most likely to benefit specific patients, minimizing adverse effects and maximizing therapeutic efficacy. This precision medicine approach represents a significant advancement over traditional one-size-fits-all treatment paradigms.
Emerging therapeutic modalities, including proteolysis-targeting chimeras (PROTACs) and RNA-targeting small molecules, are being developed based on insights from integrated phosphorylation and transcriptional studies. These novel approaches offer potential solutions for previously "undruggable" targets, expanding the therapeutic landscape for challenging diseases with limited treatment options.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!