Optimize Analytical Techniques for Phosphorylation Studies
SEP 23, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Phosphorylation Analysis Background and Objectives
Phosphorylation, a post-translational modification involving the addition of phosphate groups to proteins, has been studied extensively since its discovery in the late 19th century. This critical cellular process regulates numerous biological functions including signal transduction, cell cycle progression, metabolism, and gene expression. The evolution of phosphorylation analysis techniques has progressed from basic radioactive labeling methods in the 1950s to sophisticated mass spectrometry-based approaches in the 21st century.
The technological trajectory has been marked by significant breakthroughs, including the development of phospho-specific antibodies in the 1980s, the emergence of phosphoproteomics in the early 2000s, and the recent integration of artificial intelligence for data analysis. These advancements have transformed our understanding of cellular signaling networks and their dysregulation in disease states, particularly in cancer, neurodegenerative disorders, and metabolic syndromes.
Current analytical techniques face several limitations that impede comprehensive phosphorylation studies. These include challenges in detecting low-abundance phosphoproteins, difficulties in distinguishing closely related phosphorylation sites, and complications in quantifying dynamic phosphorylation events in complex biological samples. Additionally, existing methods often require substantial sample amounts and involve time-consuming protocols that limit throughput capabilities.
The primary objective of optimizing analytical techniques for phosphorylation studies is to develop more sensitive, specific, and high-throughput methodologies that can accurately identify and quantify phosphorylation events across diverse biological contexts. This includes enhancing detection limits to capture transient and low-abundance phosphorylation events that may be functionally significant but currently evade analysis.
Another crucial goal is to improve spatial and temporal resolution in phosphorylation analysis, enabling researchers to track phosphorylation dynamics in real-time within living cells and tissues. This would provide unprecedented insights into the kinetics of signaling cascades and their regulation under physiological and pathological conditions.
Furthermore, optimization efforts aim to establish standardized protocols and reference materials that ensure reproducibility and comparability of phosphorylation data across different laboratories and experimental platforms. This standardization is essential for building comprehensive phosphorylation databases that can serve as valuable resources for systems biology approaches and computational modeling of cellular signaling networks.
Ultimately, these technological improvements seek to bridge the gap between basic phosphorylation research and clinical applications, facilitating the development of phosphorylation-based biomarkers for disease diagnosis and monitoring, as well as enabling the rational design of kinase inhibitors and other therapeutics targeting phosphorylation-dependent pathways.
The technological trajectory has been marked by significant breakthroughs, including the development of phospho-specific antibodies in the 1980s, the emergence of phosphoproteomics in the early 2000s, and the recent integration of artificial intelligence for data analysis. These advancements have transformed our understanding of cellular signaling networks and their dysregulation in disease states, particularly in cancer, neurodegenerative disorders, and metabolic syndromes.
Current analytical techniques face several limitations that impede comprehensive phosphorylation studies. These include challenges in detecting low-abundance phosphoproteins, difficulties in distinguishing closely related phosphorylation sites, and complications in quantifying dynamic phosphorylation events in complex biological samples. Additionally, existing methods often require substantial sample amounts and involve time-consuming protocols that limit throughput capabilities.
The primary objective of optimizing analytical techniques for phosphorylation studies is to develop more sensitive, specific, and high-throughput methodologies that can accurately identify and quantify phosphorylation events across diverse biological contexts. This includes enhancing detection limits to capture transient and low-abundance phosphorylation events that may be functionally significant but currently evade analysis.
Another crucial goal is to improve spatial and temporal resolution in phosphorylation analysis, enabling researchers to track phosphorylation dynamics in real-time within living cells and tissues. This would provide unprecedented insights into the kinetics of signaling cascades and their regulation under physiological and pathological conditions.
Furthermore, optimization efforts aim to establish standardized protocols and reference materials that ensure reproducibility and comparability of phosphorylation data across different laboratories and experimental platforms. This standardization is essential for building comprehensive phosphorylation databases that can serve as valuable resources for systems biology approaches and computational modeling of cellular signaling networks.
Ultimately, these technological improvements seek to bridge the gap between basic phosphorylation research and clinical applications, facilitating the development of phosphorylation-based biomarkers for disease diagnosis and monitoring, as well as enabling the rational design of kinase inhibitors and other therapeutics targeting phosphorylation-dependent pathways.
Market Demand for Advanced Phosphoproteomic Technologies
The global market for advanced phosphoproteomic technologies is experiencing robust growth, driven primarily by increasing research activities in proteomics and the rising prevalence of chronic diseases. Current market valuations indicate that the phosphoproteomics market reached approximately 1.8 billion USD in 2022 and is projected to grow at a compound annual growth rate of 7.5% through 2030, reflecting the expanding application scope of these technologies.
Pharmaceutical and biotechnology companies represent the largest market segment, accounting for nearly 45% of the total market share. These organizations are increasingly investing in phosphorylation studies to accelerate drug discovery processes and develop targeted therapies. The ability to precisely map phosphorylation sites and understand their regulatory functions has become critical for identifying novel drug targets and biomarkers.
Academic research institutions constitute the second-largest market segment, with growing demand for high-throughput phosphoproteomic technologies to support fundamental research in cell signaling pathways. Government funding for proteomics research has seen a notable increase in recent years, particularly in North America and Europe, further stimulating market growth in these regions.
Geographically, North America dominates the market with approximately 40% share, followed by Europe and Asia-Pacific. However, the Asia-Pacific region is expected to witness the fastest growth rate due to increasing investments in life sciences research infrastructure, particularly in China, Japan, and South Korea.
The clinical diagnostics sector represents an emerging application area with significant growth potential. There is increasing recognition of phosphorylation-based biomarkers for early disease detection and personalized medicine approaches. Healthcare providers are showing growing interest in phosphoproteomic technologies that can enable more precise patient stratification and treatment selection.
Industry surveys indicate that end-users are primarily demanding improvements in three key areas: analytical sensitivity for low-abundance phosphoproteins, throughput capabilities for large-scale studies, and data analysis solutions that can effectively interpret complex phosphorylation patterns. The ability to detect and quantify phosphorylation events in limited sample quantities, such as patient biopsies or rare cell populations, represents a particularly strong market need.
Integration capabilities with other omics technologies, especially genomics and metabolomics, is another emerging demand trend as researchers increasingly adopt multi-omics approaches to understand complex biological systems. This has created market opportunities for comprehensive analytical platforms that can provide integrated solutions rather than standalone technologies.
Pharmaceutical and biotechnology companies represent the largest market segment, accounting for nearly 45% of the total market share. These organizations are increasingly investing in phosphorylation studies to accelerate drug discovery processes and develop targeted therapies. The ability to precisely map phosphorylation sites and understand their regulatory functions has become critical for identifying novel drug targets and biomarkers.
Academic research institutions constitute the second-largest market segment, with growing demand for high-throughput phosphoproteomic technologies to support fundamental research in cell signaling pathways. Government funding for proteomics research has seen a notable increase in recent years, particularly in North America and Europe, further stimulating market growth in these regions.
Geographically, North America dominates the market with approximately 40% share, followed by Europe and Asia-Pacific. However, the Asia-Pacific region is expected to witness the fastest growth rate due to increasing investments in life sciences research infrastructure, particularly in China, Japan, and South Korea.
The clinical diagnostics sector represents an emerging application area with significant growth potential. There is increasing recognition of phosphorylation-based biomarkers for early disease detection and personalized medicine approaches. Healthcare providers are showing growing interest in phosphoproteomic technologies that can enable more precise patient stratification and treatment selection.
Industry surveys indicate that end-users are primarily demanding improvements in three key areas: analytical sensitivity for low-abundance phosphoproteins, throughput capabilities for large-scale studies, and data analysis solutions that can effectively interpret complex phosphorylation patterns. The ability to detect and quantify phosphorylation events in limited sample quantities, such as patient biopsies or rare cell populations, represents a particularly strong market need.
Integration capabilities with other omics technologies, especially genomics and metabolomics, is another emerging demand trend as researchers increasingly adopt multi-omics approaches to understand complex biological systems. This has created market opportunities for comprehensive analytical platforms that can provide integrated solutions rather than standalone technologies.
Current Challenges in Phosphorylation Detection Methods
Despite significant advancements in phosphorylation analysis techniques, researchers continue to face substantial challenges that limit comprehensive phosphoproteome characterization. Current detection methods struggle with sensitivity issues, particularly when identifying low-abundance phosphoproteins that often play crucial regulatory roles in cellular signaling pathways. Even state-of-the-art mass spectrometry approaches typically capture only a fraction of the phosphoproteome, leaving many biologically significant modifications undetected.
Sample preparation remains a critical bottleneck, with phosphopeptide enrichment techniques like titanium dioxide (TiO2) and immobilized metal affinity chromatography (IMAC) showing bias toward certain phosphopeptide classes. These methods often preferentially enrich multiply phosphorylated peptides or those with specific amino acid compositions, creating systematic gaps in phosphoproteome coverage. Additionally, the labile nature of phosphorylation modifications frequently leads to their loss during sample processing and ionization.
Quantification accuracy presents another significant challenge. Label-free quantification methods suffer from run-to-run variability, while isotope labeling approaches like SILAC and TMT have limitations in multiplexing capacity and dynamic range. The transient nature of phosphorylation events, which can change within seconds to minutes, further complicates accurate temporal profiling, as sample collection and processing times often exceed the biological timescale of these modifications.
Site localization ambiguity remains problematic, particularly when multiple potential phosphorylation sites exist in close proximity within a peptide sequence. Current algorithms for site localization assignment still produce false positives and negatives, complicating downstream functional analysis. This challenge is exacerbated in highly phosphorylated regions where multiple modifications may influence fragmentation patterns during mass spectrometry.
Data analysis workflows present computational challenges due to the enormous complexity of phosphoproteomics datasets. Existing software tools struggle with the integration of phosphorylation data with other omics datasets, limiting systems-level understanding. Furthermore, the biological interpretation of phosphorylation events remains difficult, as the functional consequences of many phosphorylation sites remain unknown or poorly characterized in databases.
Reproducibility issues plague the field, with different laboratories often reporting varying phosphorylation profiles from identical samples. This stems from differences in sample handling, instrument sensitivity, and data analysis pipelines. The lack of standardized protocols and quality control metrics further exacerbates this problem, making cross-study comparisons challenging and hindering the establishment of reliable phosphorylation signatures for various biological states.
Sample preparation remains a critical bottleneck, with phosphopeptide enrichment techniques like titanium dioxide (TiO2) and immobilized metal affinity chromatography (IMAC) showing bias toward certain phosphopeptide classes. These methods often preferentially enrich multiply phosphorylated peptides or those with specific amino acid compositions, creating systematic gaps in phosphoproteome coverage. Additionally, the labile nature of phosphorylation modifications frequently leads to their loss during sample processing and ionization.
Quantification accuracy presents another significant challenge. Label-free quantification methods suffer from run-to-run variability, while isotope labeling approaches like SILAC and TMT have limitations in multiplexing capacity and dynamic range. The transient nature of phosphorylation events, which can change within seconds to minutes, further complicates accurate temporal profiling, as sample collection and processing times often exceed the biological timescale of these modifications.
Site localization ambiguity remains problematic, particularly when multiple potential phosphorylation sites exist in close proximity within a peptide sequence. Current algorithms for site localization assignment still produce false positives and negatives, complicating downstream functional analysis. This challenge is exacerbated in highly phosphorylated regions where multiple modifications may influence fragmentation patterns during mass spectrometry.
Data analysis workflows present computational challenges due to the enormous complexity of phosphoproteomics datasets. Existing software tools struggle with the integration of phosphorylation data with other omics datasets, limiting systems-level understanding. Furthermore, the biological interpretation of phosphorylation events remains difficult, as the functional consequences of many phosphorylation sites remain unknown or poorly characterized in databases.
Reproducibility issues plague the field, with different laboratories often reporting varying phosphorylation profiles from identical samples. This stems from differences in sample handling, instrument sensitivity, and data analysis pipelines. The lack of standardized protocols and quality control metrics further exacerbates this problem, making cross-study comparisons challenging and hindering the establishment of reliable phosphorylation signatures for various biological states.
Mainstream Phosphopeptide Enrichment Strategies
01 Mass spectrometry-based phosphorylation analysis techniques
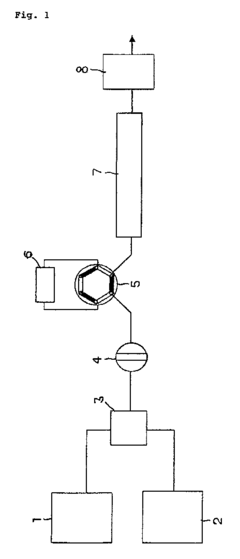
Mass spectrometry (MS) has become a powerful analytical tool for phosphorylation studies, enabling high-throughput identification and quantification of phosphorylated proteins and peptides. Advanced MS techniques allow for precise detection of phosphorylation sites, measurement of phosphorylation dynamics, and characterization of complex phosphoproteomes. These methods can be optimized through improved sample preparation protocols, enrichment strategies for phosphopeptides, and specialized data acquisition methods to enhance sensitivity and coverage of phosphorylation events.- Mass Spectrometry-Based Phosphorylation Analysis: Mass spectrometry techniques are widely used for the identification and quantification of phosphorylation sites in proteins. These methods allow for high-throughput analysis of phosphoproteomes, enabling researchers to detect multiple phosphorylation events simultaneously. Advanced MS techniques include phosphopeptide enrichment strategies, targeted MS approaches, and data-dependent acquisition methods that enhance sensitivity and specificity for phosphorylation site detection.
- Computational Methods for Phosphorylation Data Analysis: Computational tools and algorithms play a crucial role in analyzing complex phosphorylation data. These include machine learning approaches, statistical models, and bioinformatics software designed specifically for phosphoproteomics data interpretation. Such computational methods help in predicting phosphorylation sites, understanding kinase-substrate relationships, and integrating phosphorylation data with other omics datasets to gain comprehensive insights into cellular signaling networks.
- Phosphoprotein Enrichment and Purification Techniques: Various techniques have been developed to enrich and purify phosphorylated proteins or peptides from complex biological samples. These include immobilized metal affinity chromatography (IMAC), titanium dioxide chromatography, phospho-specific antibody immunoprecipitation, and chemical derivatization methods. These enrichment strategies are essential for increasing the detection sensitivity of low-abundance phosphoproteins and improving the overall coverage of phosphorylation site analysis.
- Kinase Activity Assays and Inhibitor Screening: Specialized assays have been developed to measure kinase activity and screen for kinase inhibitors in phosphorylation studies. These include fluorescence-based assays, bioluminescence resonance energy transfer (BRET) assays, and high-throughput screening platforms. These methods enable researchers to evaluate the efficiency of phosphorylation reactions, identify specific kinase inhibitors, and optimize conditions for studying phosphorylation events in various biological contexts.
- In Vivo Phosphorylation Monitoring Systems: Advanced technologies for monitoring phosphorylation events in living cells and organisms have been developed. These include genetically encoded biosensors, phosphorylation-specific fluorescent probes, and real-time imaging techniques. Such systems allow researchers to track dynamic phosphorylation events in their native cellular environment, providing insights into the spatial and temporal regulation of phosphorylation-dependent signaling pathways under physiological conditions.
02 Computational methods for phosphorylation data analysis
Computational approaches play a crucial role in analyzing and interpreting phosphorylation data. These include algorithms for phosphorylation site prediction, statistical methods for quantitative phosphoproteomics, network analysis tools to understand phosphorylation-mediated signaling pathways, and machine learning approaches to identify patterns in phosphorylation datasets. Optimization of these computational methods involves improving prediction accuracy, developing more efficient data processing workflows, and integrating multiple data types to gain comprehensive insights into phosphorylation-regulated cellular processes.Expand Specific Solutions03 Phosphoprotein enrichment and purification strategies
Effective enrichment and purification of phosphorylated proteins and peptides are essential for successful phosphorylation studies. Various techniques have been developed, including immobilized metal affinity chromatography (IMAC), metal oxide affinity chromatography (MOAC), phospho-specific antibody-based immunoprecipitation, and chemical derivatization methods. Optimization of these strategies focuses on improving specificity, reducing non-specific binding, enhancing recovery rates, and developing protocols suitable for different sample types and phosphorylation patterns.Expand Specific Solutions04 Kinase activity assays and phosphorylation dynamics monitoring
Methods for measuring kinase activity and monitoring phosphorylation dynamics in real-time provide valuable insights into cellular signaling mechanisms. These include fluorescence-based kinase assays, bioluminescence resonance energy transfer (BRET) techniques, phospho-specific antibody-based assays, and label-free approaches. Optimization of these methods involves improving temporal resolution, enhancing sensitivity for detecting low-abundance phosphorylation events, developing multiplexed assays for simultaneous monitoring of multiple kinases, and adapting techniques for in vivo applications.Expand Specific Solutions05 Phosphorylation site mapping and structural analysis
Techniques for precise mapping of phosphorylation sites and analyzing structural changes induced by phosphorylation are critical for understanding the functional consequences of these modifications. These include site-directed mutagenesis approaches, hydrogen/deuterium exchange mass spectrometry, X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy. Optimization strategies focus on improving spatial resolution, developing methods for analyzing phosphorylation in native protein complexes, and integrating structural data with functional assays to establish structure-function relationships of phosphorylated proteins.Expand Specific Solutions
Key Industry Players in Phosphoproteomics Research
Phosphorylation studies are currently in a mature development phase, with the analytical techniques market showing steady growth driven by increasing applications in proteomics and drug development. The global market size for phosphorylation analysis is estimated at $2-3 billion annually, expanding at approximately 8-10% CAGR. Leading companies like Agilent Technologies, Life Technologies, and Illumina have established robust technological platforms, while academic institutions including Harvard, University of California, and University of Michigan contribute significant research advancements. Emerging players such as Toyobo and Dalian Institute of Chemical Physics are developing novel approaches to improve sensitivity and throughput. The technology landscape shows high maturity in mass spectrometry-based techniques, with growing innovation in antibody-free detection methods and high-throughput screening platforms for phosphorylation site identification.
Revvity Health Sciences, Inc.
Technical Solution: Revvity (formerly PerkinElmer) has developed the AlphaLISA SureFire Ultra phosphorylation assay platform, which utilizes their proprietary Alpha (Amplified Luminescent Proximity Homogeneous Assay) technology for highly sensitive detection of protein phosphorylation. This no-wash assay format employs donor and acceptor beads that generate a luminescent signal when brought into proximity by phosphorylated protein binding. Their technology enables detection of phosphorylation events at endogenous protein levels without overexpression systems, with sensitivity down to femtogram levels. Revvity has expanded their phosphorylation analysis portfolio with the Phospho-Explorer Antibody Array, which simultaneously profiles over 1,300 phosphorylation sites across 400 human proteins. Their latest innovation combines microfluidics with their established Alpha technology to enable real-time monitoring of phosphorylation dynamics in living cells with temporal resolution under 30 seconds.
Strengths: Exceptional sensitivity for detecting low-abundance phosphorylation events with minimal sample requirements and high-throughput capabilities. Their homogeneous assay format eliminates washing steps, reducing variability. Weaknesses: Some of their assays have limited dynamic range compared to mass spectrometry-based approaches, and their antibody-dependent methods may face specificity challenges with closely related phosphorylation sites.
Agilent Technologies, Inc.
Technical Solution: Agilent has developed advanced analytical platforms specifically optimized for phosphorylation studies, including their Phosphoproteomics Solution that combines high-resolution liquid chromatography with triple quadrupole mass spectrometry. Their technology employs titanium dioxide (TiO2) and immobilized metal affinity chromatography (IMAC) enrichment strategies to selectively capture phosphopeptides prior to LC-MS/MS analysis. Agilent's AssayMAP Bravo platform automates phosphopeptide enrichment workflows, significantly reducing manual handling errors and improving reproducibility. Their latest innovations include the integration of ion mobility separation with high-resolution mass spectrometry, enabling detection of low-abundance phosphopeptides in complex biological matrices with up to 30% increased phosphosite identification compared to conventional methods.
Strengths: Comprehensive end-to-end workflow solutions with high automation capabilities and reproducibility. Their integrated hardware-software ecosystem provides seamless data acquisition and analysis. Weaknesses: Higher initial investment costs compared to some competitors, and their proprietary software ecosystems may limit flexibility for custom analytical workflows.
Critical Technologies in Mass Spectrometry-Based Phosphorylation Analysis
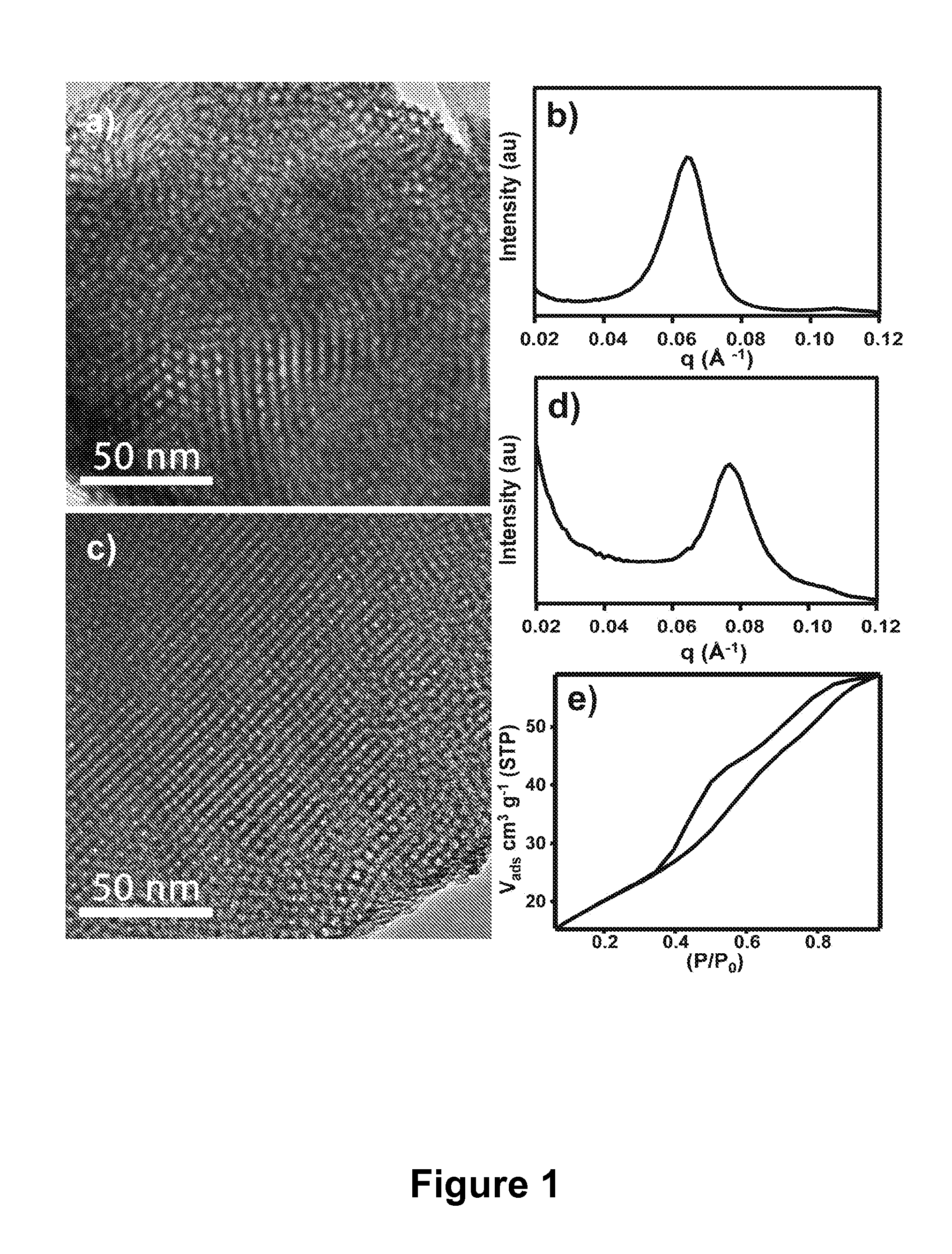
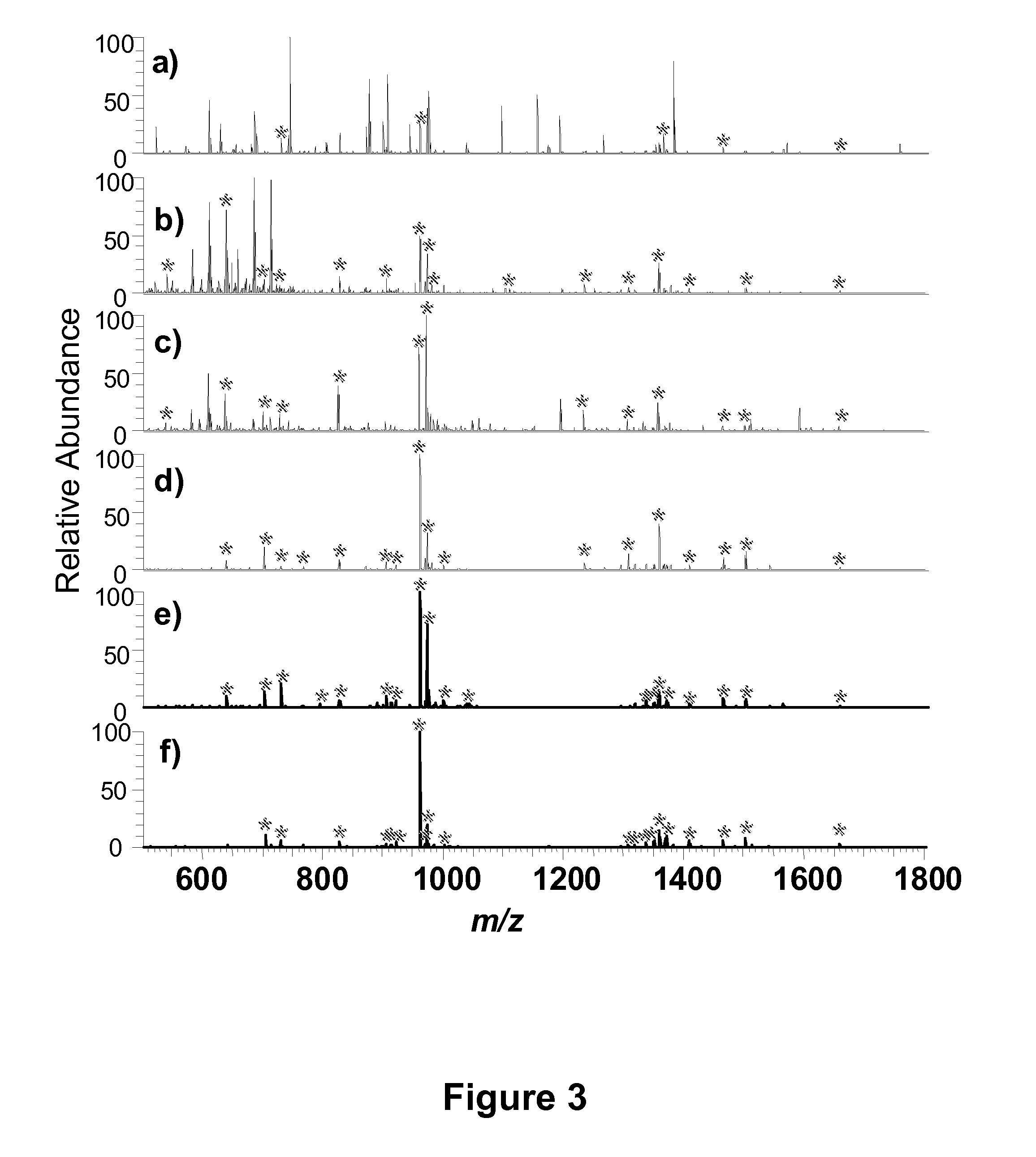
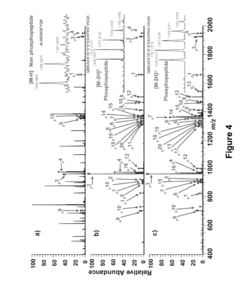
Mesoporous metal oxide materials for phosphoproteomics
PatentActiveUS20100093102A1
Innovation
- The use of nanostructured metal oxide mesoporous materials, such as transition metal oxides and Group IIIA metal oxides, which provide high specificity and reversible binding for phosphorylated compounds, allowing for efficient enrichment and purification through controlled release mechanisms, compatible with various separation platforms.
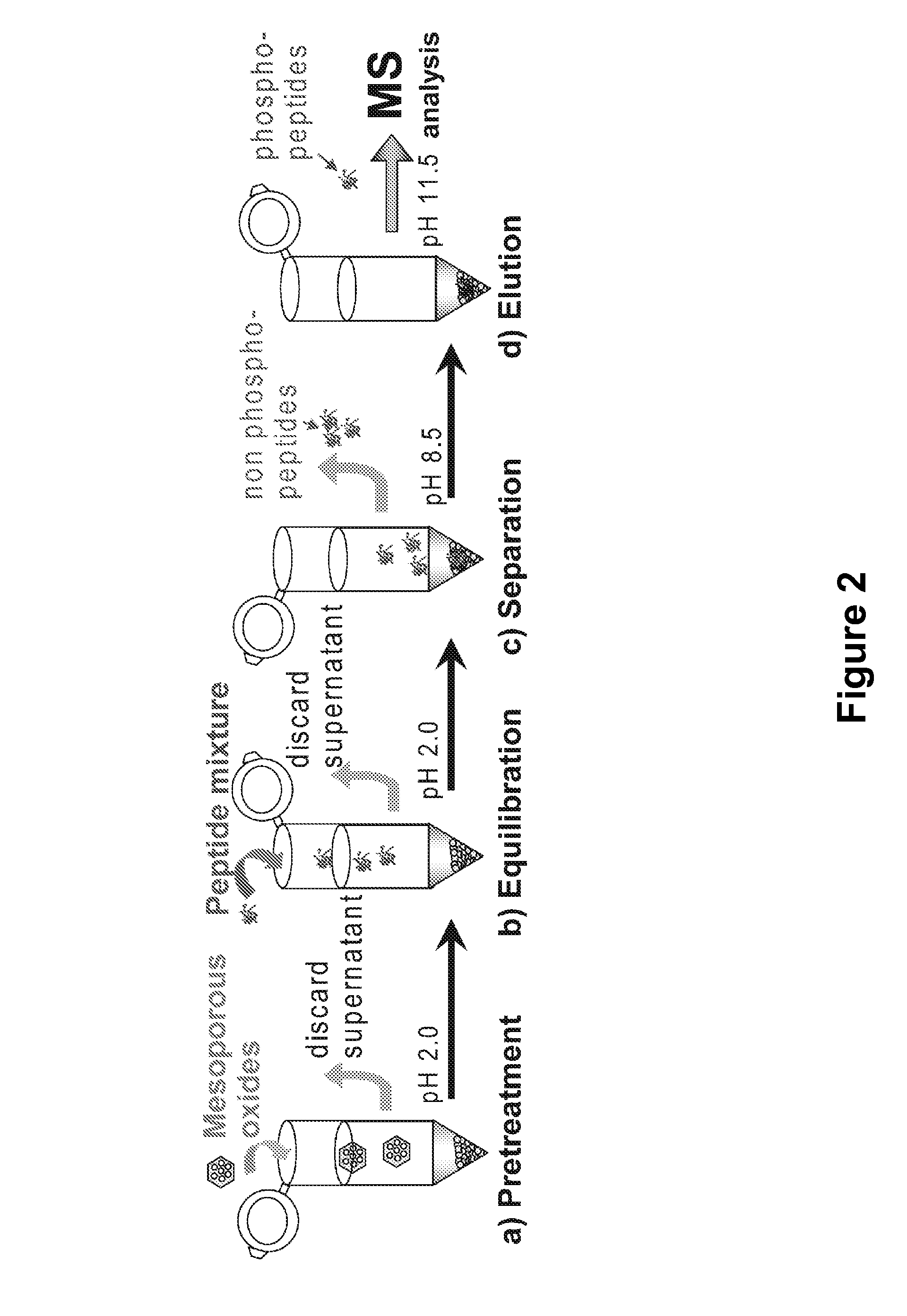
Method and apparatus for analyzing amino acid, peptide, protein, saccharide or lipid
PatentInactiveEP1477800A8
Innovation
- A method utilizing a titanium dioxide column for selective extraction and separation of phosphorylated compounds under acidic conditions, allowing for simple, rapid, and cost-effective analysis by retaining phosphorylated compounds and eluting non-phosphorylated ones with phosphate or alkali solutions, thereby simplifying experimental operations and eliminating the need for detection steps in some cases.
Bioinformatics Tools for Phosphoproteome Data Analysis
The phosphoproteome data analysis landscape has evolved significantly with the development of sophisticated bioinformatics tools designed specifically for phosphorylation studies. These tools address the complex challenges of processing large-scale phosphoproteomics datasets, from raw mass spectrometry data to biological interpretation.
Database-centric platforms such as PhosphoSitePlus, PHOSIDA, and PhosphoPep serve as comprehensive repositories for experimentally verified phosphorylation sites across multiple species. These resources enable researchers to validate their findings against established phosphorylation events and explore conservation patterns across evolutionary boundaries.
For data processing and phosphopeptide identification, specialized software suites like MaxQuant, Proteome Discoverer, and PEAKS have incorporated phosphorylation-specific algorithms that account for the unique characteristics of phosphopeptide fragmentation patterns. These tools implement advanced features such as phosphosite localization scoring, which addresses the critical challenge of pinpointing the exact amino acid residue that carries the phosphate group.
Network analysis tools represent another crucial category, with platforms like Cytoscape and its phosphorylation-specific plugins enabling visualization and analysis of phosphorylation-mediated signaling networks. These tools facilitate the integration of phosphoproteomics data with protein-protein interaction networks, revealing functional modules and regulatory circuits within cellular signaling pathways.
Machine learning approaches have recently transformed phosphoproteomics data analysis. Tools like PhosphoPredict and MusiteDeep employ deep learning algorithms to predict phosphorylation sites and their kinase-specific associations with increasing accuracy. These computational methods help overcome experimental limitations by predicting potential phosphorylation events that might be missed in experimental settings.
Kinase-substrate prediction tools such as NetPhorest, GPS, and KinasePhos leverage position-specific scoring matrices and structural information to predict kinase-specific phosphorylation sites. These predictions provide valuable insights into the regulatory mechanisms governing phosphorylation events and help identify potential therapeutic targets in disease contexts.
Pathway enrichment analysis tools like DAVID, GSEA, and IPA have been adapted to accommodate phosphoproteomics data, enabling researchers to identify significantly enriched biological pathways and functions within their phosphoproteomic datasets. These analyses provide a systems-level understanding of phosphorylation-mediated cellular processes.
Database-centric platforms such as PhosphoSitePlus, PHOSIDA, and PhosphoPep serve as comprehensive repositories for experimentally verified phosphorylation sites across multiple species. These resources enable researchers to validate their findings against established phosphorylation events and explore conservation patterns across evolutionary boundaries.
For data processing and phosphopeptide identification, specialized software suites like MaxQuant, Proteome Discoverer, and PEAKS have incorporated phosphorylation-specific algorithms that account for the unique characteristics of phosphopeptide fragmentation patterns. These tools implement advanced features such as phosphosite localization scoring, which addresses the critical challenge of pinpointing the exact amino acid residue that carries the phosphate group.
Network analysis tools represent another crucial category, with platforms like Cytoscape and its phosphorylation-specific plugins enabling visualization and analysis of phosphorylation-mediated signaling networks. These tools facilitate the integration of phosphoproteomics data with protein-protein interaction networks, revealing functional modules and regulatory circuits within cellular signaling pathways.
Machine learning approaches have recently transformed phosphoproteomics data analysis. Tools like PhosphoPredict and MusiteDeep employ deep learning algorithms to predict phosphorylation sites and their kinase-specific associations with increasing accuracy. These computational methods help overcome experimental limitations by predicting potential phosphorylation events that might be missed in experimental settings.
Kinase-substrate prediction tools such as NetPhorest, GPS, and KinasePhos leverage position-specific scoring matrices and structural information to predict kinase-specific phosphorylation sites. These predictions provide valuable insights into the regulatory mechanisms governing phosphorylation events and help identify potential therapeutic targets in disease contexts.
Pathway enrichment analysis tools like DAVID, GSEA, and IPA have been adapted to accommodate phosphoproteomics data, enabling researchers to identify significantly enriched biological pathways and functions within their phosphoproteomic datasets. These analyses provide a systems-level understanding of phosphorylation-mediated cellular processes.
Regulatory Considerations for Clinical Phosphoproteomics Applications
The implementation of phosphoproteomics technologies in clinical settings necessitates careful navigation of complex regulatory frameworks. In the United States, the FDA has established specific guidelines for the validation of mass spectrometry-based proteomics assays used in clinical decision-making. These guidelines require demonstration of analytical validity, clinical validity, and clinical utility before phosphoproteomic tests can be approved for patient care.
European regulatory bodies, including the European Medicines Agency (EMA), have developed parallel frameworks that emphasize CE marking requirements for in vitro diagnostic devices incorporating phosphoproteomics technologies. Manufacturers must demonstrate compliance with essential requirements related to safety, quality, and performance characteristics before market authorization.
Quality control standards represent a critical regulatory consideration, with organizations such as the Clinical and Laboratory Standards Institute (CLSI) providing guidelines for laboratory developed tests (LDTs) involving phosphoproteomic analyses. These standards address sample handling, instrument calibration, and data interpretation protocols to ensure reproducibility across different clinical laboratories.
Data privacy regulations, including HIPAA in the US and GDPR in Europe, impose strict requirements on the handling of patient data generated through phosphoproteomics studies. Researchers must implement robust data protection measures and obtain appropriate informed consent for the collection, storage, and analysis of phosphoproteome data from patient samples.
Reimbursement pathways present another regulatory challenge, as healthcare systems require evidence of cost-effectiveness before covering novel phosphoproteomics-based diagnostic tests. This necessitates comprehensive health economic analyses demonstrating improved patient outcomes relative to standard diagnostic approaches.
Harmonization of regulatory standards across international jurisdictions remains an ongoing challenge. The International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) has initiated efforts to develop globally consistent guidelines for the validation of biomarker assays, including those based on phosphoproteomics technologies.
Emerging regulatory considerations include the development of frameworks for AI-augmented phosphoproteomics data interpretation, with regulatory bodies increasingly focused on the validation of algorithms used to identify clinically relevant phosphorylation patterns. These frameworks aim to ensure the reliability and interpretability of computational approaches in clinical phosphoproteomics applications.
European regulatory bodies, including the European Medicines Agency (EMA), have developed parallel frameworks that emphasize CE marking requirements for in vitro diagnostic devices incorporating phosphoproteomics technologies. Manufacturers must demonstrate compliance with essential requirements related to safety, quality, and performance characteristics before market authorization.
Quality control standards represent a critical regulatory consideration, with organizations such as the Clinical and Laboratory Standards Institute (CLSI) providing guidelines for laboratory developed tests (LDTs) involving phosphoproteomic analyses. These standards address sample handling, instrument calibration, and data interpretation protocols to ensure reproducibility across different clinical laboratories.
Data privacy regulations, including HIPAA in the US and GDPR in Europe, impose strict requirements on the handling of patient data generated through phosphoproteomics studies. Researchers must implement robust data protection measures and obtain appropriate informed consent for the collection, storage, and analysis of phosphoproteome data from patient samples.
Reimbursement pathways present another regulatory challenge, as healthcare systems require evidence of cost-effectiveness before covering novel phosphoproteomics-based diagnostic tests. This necessitates comprehensive health economic analyses demonstrating improved patient outcomes relative to standard diagnostic approaches.
Harmonization of regulatory standards across international jurisdictions remains an ongoing challenge. The International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) has initiated efforts to develop globally consistent guidelines for the validation of biomarker assays, including those based on phosphoproteomics technologies.
Emerging regulatory considerations include the development of frameworks for AI-augmented phosphoproteomics data interpretation, with regulatory bodies increasingly focused on the validation of algorithms used to identify clinically relevant phosphorylation patterns. These frameworks aim to ensure the reliability and interpretability of computational approaches in clinical phosphoproteomics applications.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!