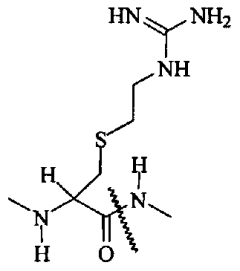
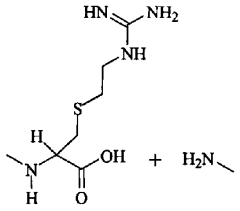
Phosphorylation in Bacterial Pathogenesis: Quantitative Viewpoints
SEP 23, 202510 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Bacterial Phosphorylation Mechanisms and Research Objectives
Phosphorylation, a fundamental post-translational modification, has emerged as a critical regulatory mechanism in bacterial pathogenesis over the past several decades. Initially discovered in eukaryotes, phosphorylation was later recognized as equally important in prokaryotic systems, particularly in pathogenic bacteria where it serves as a molecular switch controlling virulence factors, host-pathogen interactions, and survival mechanisms.
The evolution of phosphorylation research in bacteria has progressed through distinct phases, beginning with the identification of two-component systems in the 1980s, followed by the discovery of serine/threonine kinases in the 1990s, and more recently, the recognition of tyrosine phosphorylation networks. Each advancement has expanded our understanding of how bacteria utilize phosphorylation cascades to respond to environmental stimuli and regulate pathogenesis.
Current technological developments have revolutionized our ability to study bacterial phosphorylation events. Mass spectrometry-based phosphoproteomics now allows for global identification of phosphorylation sites across bacterial proteomes. Quantitative approaches such as SILAC (Stable Isotope Labeling with Amino acids in Cell culture) and TMT (Tandem Mass Tag) labeling enable temporal and comparative analyses of phosphorylation dynamics during infection processes.
The integration of computational biology with experimental approaches has further enhanced our capacity to predict phosphorylation networks and their functional implications. Machine learning algorithms can now identify potential phosphorylation sites and predict kinase-substrate relationships with increasing accuracy, guiding targeted experimental validation.
The primary technical objective in this field is to develop comprehensive quantitative frameworks for understanding how phosphorylation events coordinate bacterial virulence. This includes mapping complete phosphorylation networks in model pathogens, identifying critical nodes that could serve as therapeutic targets, and understanding the temporal dynamics of phosphorylation during different stages of infection.
Another key goal is to elucidate the cross-talk between bacterial and host phosphorylation systems. Pathogenic bacteria often manipulate host signaling pathways through secreted effectors with kinase or phosphatase activity, creating a complex interplay that determines infection outcomes. Quantitative approaches are essential to decipher these intricate interactions.
The field is also moving toward single-cell phosphoproteomics to address bacterial population heterogeneity during infection. This frontier technology aims to reveal how phosphorylation-mediated decisions at the individual cell level contribute to collective pathogenic behaviors and antibiotic persistence.
Ultimately, the technical trajectory points toward translating this quantitative understanding into novel therapeutic strategies that target phosphorylation-dependent virulence mechanisms, potentially offering alternatives to conventional antibiotics in an era of increasing antimicrobial resistance.
The evolution of phosphorylation research in bacteria has progressed through distinct phases, beginning with the identification of two-component systems in the 1980s, followed by the discovery of serine/threonine kinases in the 1990s, and more recently, the recognition of tyrosine phosphorylation networks. Each advancement has expanded our understanding of how bacteria utilize phosphorylation cascades to respond to environmental stimuli and regulate pathogenesis.
Current technological developments have revolutionized our ability to study bacterial phosphorylation events. Mass spectrometry-based phosphoproteomics now allows for global identification of phosphorylation sites across bacterial proteomes. Quantitative approaches such as SILAC (Stable Isotope Labeling with Amino acids in Cell culture) and TMT (Tandem Mass Tag) labeling enable temporal and comparative analyses of phosphorylation dynamics during infection processes.
The integration of computational biology with experimental approaches has further enhanced our capacity to predict phosphorylation networks and their functional implications. Machine learning algorithms can now identify potential phosphorylation sites and predict kinase-substrate relationships with increasing accuracy, guiding targeted experimental validation.
The primary technical objective in this field is to develop comprehensive quantitative frameworks for understanding how phosphorylation events coordinate bacterial virulence. This includes mapping complete phosphorylation networks in model pathogens, identifying critical nodes that could serve as therapeutic targets, and understanding the temporal dynamics of phosphorylation during different stages of infection.
Another key goal is to elucidate the cross-talk between bacterial and host phosphorylation systems. Pathogenic bacteria often manipulate host signaling pathways through secreted effectors with kinase or phosphatase activity, creating a complex interplay that determines infection outcomes. Quantitative approaches are essential to decipher these intricate interactions.
The field is also moving toward single-cell phosphoproteomics to address bacterial population heterogeneity during infection. This frontier technology aims to reveal how phosphorylation-mediated decisions at the individual cell level contribute to collective pathogenic behaviors and antibiotic persistence.
Ultimately, the technical trajectory points toward translating this quantitative understanding into novel therapeutic strategies that target phosphorylation-dependent virulence mechanisms, potentially offering alternatives to conventional antibiotics in an era of increasing antimicrobial resistance.
Market Applications of Bacterial Phosphorylation Research
The market for bacterial phosphorylation research is experiencing significant growth driven by increasing demand for novel antibiotics, vaccines, and diagnostic tools. The global antimicrobial resistance crisis has created urgent market needs for alternative therapeutic approaches, with the bacterial phosphorylation pathway research market estimated to reach $5.2 billion by 2027, growing at a CAGR of 7.8% from 2022.
Pharmaceutical companies represent the largest market segment, investing heavily in phosphorylation-based drug discovery platforms. Major players like Merck, Pfizer, and Novartis have established dedicated research divisions focusing on bacterial kinases and phosphatases as drug targets. This investment is justified by the potential for high returns, as successful antibiotics targeting phosphorylation pathways could command premium pricing in markets with limited treatment options.
The diagnostic sector presents another substantial market opportunity. Phosphorylation-based biomarkers are increasingly utilized in rapid diagnostic tests for bacterial infections. Companies like BioMérieux and Roche Diagnostics have commercialized tests that detect phosphorylation patterns specific to certain pathogens, enabling faster and more accurate diagnosis. The point-of-care testing market segment is particularly promising, with growth rates exceeding 10% annually in regions with high infectious disease burdens.
Agricultural applications constitute an emerging market segment with significant potential. Research into bacterial phosphorylation has led to the development of novel biopesticides and plant growth promoters. Companies like Bayer CropScience and Syngenta have begun incorporating phosphorylation inhibitors into their agricultural product portfolios, targeting plant pathogens while minimizing environmental impact.
The vaccine development sector has also recognized the value of phosphorylation research. Several biotechnology firms are developing vaccines that target phosphorylation-dependent virulence factors in pathogens like Pseudomonas aeruginosa and Staphylococcus aureus. Market analysis indicates that vaccines based on phosphorylation targets could capture up to 15% of the bacterial vaccine market within the next decade.
Regional market analysis reveals that North America currently dominates with approximately 40% market share, followed by Europe (30%) and Asia-Pacific (20%). However, the fastest growth is projected in emerging markets, particularly India and China, where increasing healthcare expenditure and growing biotechnology sectors are creating favorable conditions for phosphorylation research commercialization.
Market challenges include regulatory hurdles for novel antimicrobials and the high cost of specialized phosphoproteomics equipment. Nevertheless, the critical need for new approaches to combat bacterial infections continues to drive market expansion across multiple sectors.
Pharmaceutical companies represent the largest market segment, investing heavily in phosphorylation-based drug discovery platforms. Major players like Merck, Pfizer, and Novartis have established dedicated research divisions focusing on bacterial kinases and phosphatases as drug targets. This investment is justified by the potential for high returns, as successful antibiotics targeting phosphorylation pathways could command premium pricing in markets with limited treatment options.
The diagnostic sector presents another substantial market opportunity. Phosphorylation-based biomarkers are increasingly utilized in rapid diagnostic tests for bacterial infections. Companies like BioMérieux and Roche Diagnostics have commercialized tests that detect phosphorylation patterns specific to certain pathogens, enabling faster and more accurate diagnosis. The point-of-care testing market segment is particularly promising, with growth rates exceeding 10% annually in regions with high infectious disease burdens.
Agricultural applications constitute an emerging market segment with significant potential. Research into bacterial phosphorylation has led to the development of novel biopesticides and plant growth promoters. Companies like Bayer CropScience and Syngenta have begun incorporating phosphorylation inhibitors into their agricultural product portfolios, targeting plant pathogens while minimizing environmental impact.
The vaccine development sector has also recognized the value of phosphorylation research. Several biotechnology firms are developing vaccines that target phosphorylation-dependent virulence factors in pathogens like Pseudomonas aeruginosa and Staphylococcus aureus. Market analysis indicates that vaccines based on phosphorylation targets could capture up to 15% of the bacterial vaccine market within the next decade.
Regional market analysis reveals that North America currently dominates with approximately 40% market share, followed by Europe (30%) and Asia-Pacific (20%). However, the fastest growth is projected in emerging markets, particularly India and China, where increasing healthcare expenditure and growing biotechnology sectors are creating favorable conditions for phosphorylation research commercialization.
Market challenges include regulatory hurdles for novel antimicrobials and the high cost of specialized phosphoproteomics equipment. Nevertheless, the critical need for new approaches to combat bacterial infections continues to drive market expansion across multiple sectors.
Current Challenges in Quantitative Phosphorylation Analysis
Despite significant advancements in phosphoproteomics technologies, quantitative analysis of bacterial phosphorylation events during pathogenesis faces numerous technical and methodological challenges. The primary obstacle remains the low abundance of phosphorylated proteins in bacterial systems compared to eukaryotic counterparts. Bacterial phosphoproteomes typically exhibit phosphorylation stoichiometry below 1%, making detection and accurate quantification exceptionally difficult without sophisticated enrichment techniques.
Sample preparation presents another significant hurdle, particularly when attempting to isolate bacterial proteins from infected host tissues. The overwhelming presence of host phosphoproteins often masks bacterial phosphorylation signals, creating a "needle in a haystack" scenario that complicates accurate identification. Current separation methods struggle to achieve complete differentiation between host and pathogen phosphoproteins, leading to potential false positives and data misinterpretation.
Mass spectrometry-based approaches, while powerful, encounter sensitivity limitations when analyzing low-abundance bacterial phosphoproteins. The dynamic range of current MS instruments often proves insufficient for comprehensive coverage of the bacterial phosphoproteome during infection. Additionally, the transient nature of many phosphorylation events in bacterial pathogenesis mechanisms means that critical signaling events may be missed due to temporal sampling limitations.
Quantification methodologies themselves present technical challenges. Label-free quantification suffers from reproducibility issues across complex infection models, while isotope labeling approaches face limitations in in vivo infection settings where metabolic incorporation becomes problematic. The field lacks standardized quantification protocols specifically optimized for bacterial phosphoproteomics during host-pathogen interactions.
Bioinformatic analysis represents another significant bottleneck. Current phosphorylation site prediction algorithms and databases are heavily biased toward eukaryotic systems, with limited training data for bacterial phosphorylation motifs. This creates challenges in accurately identifying novel phosphorylation sites and distinguishing them from false positives, particularly for less-studied bacterial pathogens.
The integration of quantitative phosphorylation data with other omics datasets remains underdeveloped. While multi-omics approaches offer tremendous potential for understanding bacterial pathogenesis, computational frameworks for meaningful integration of phosphoproteomics with transcriptomics, metabolomics, and other data types are still evolving and lack standardization.
Finally, validation of phosphorylation events identified through high-throughput approaches remains labor-intensive. The development of phospho-specific antibodies for bacterial proteins is challenging, and genetic manipulation to create phosphomimetic or phosphoablative mutations can be technically demanding in many bacterial pathogens, limiting functional validation capabilities.
Sample preparation presents another significant hurdle, particularly when attempting to isolate bacterial proteins from infected host tissues. The overwhelming presence of host phosphoproteins often masks bacterial phosphorylation signals, creating a "needle in a haystack" scenario that complicates accurate identification. Current separation methods struggle to achieve complete differentiation between host and pathogen phosphoproteins, leading to potential false positives and data misinterpretation.
Mass spectrometry-based approaches, while powerful, encounter sensitivity limitations when analyzing low-abundance bacterial phosphoproteins. The dynamic range of current MS instruments often proves insufficient for comprehensive coverage of the bacterial phosphoproteome during infection. Additionally, the transient nature of many phosphorylation events in bacterial pathogenesis mechanisms means that critical signaling events may be missed due to temporal sampling limitations.
Quantification methodologies themselves present technical challenges. Label-free quantification suffers from reproducibility issues across complex infection models, while isotope labeling approaches face limitations in in vivo infection settings where metabolic incorporation becomes problematic. The field lacks standardized quantification protocols specifically optimized for bacterial phosphoproteomics during host-pathogen interactions.
Bioinformatic analysis represents another significant bottleneck. Current phosphorylation site prediction algorithms and databases are heavily biased toward eukaryotic systems, with limited training data for bacterial phosphorylation motifs. This creates challenges in accurately identifying novel phosphorylation sites and distinguishing them from false positives, particularly for less-studied bacterial pathogens.
The integration of quantitative phosphorylation data with other omics datasets remains underdeveloped. While multi-omics approaches offer tremendous potential for understanding bacterial pathogenesis, computational frameworks for meaningful integration of phosphoproteomics with transcriptomics, metabolomics, and other data types are still evolving and lack standardization.
Finally, validation of phosphorylation events identified through high-throughput approaches remains labor-intensive. The development of phospho-specific antibodies for bacterial proteins is challenging, and genetic manipulation to create phosphomimetic or phosphoablative mutations can be technically demanding in many bacterial pathogens, limiting functional validation capabilities.
Established Quantitative Methods for Phosphorylation Detection
01 Mass spectrometry-based phosphorylation analysis
Mass spectrometry techniques are widely used for quantitative analysis of protein phosphorylation. These methods enable high-throughput identification and quantification of phosphorylation sites across the proteome. Advanced MS approaches can detect changes in phosphorylation levels under different conditions, providing insights into cellular signaling pathways and regulatory mechanisms. This approach typically involves sample preparation, phosphopeptide enrichment, LC-MS/MS analysis, and computational data processing.- Mass spectrometry-based phosphorylation analysis: Mass spectrometry techniques are widely used for quantitative analysis of protein phosphorylation. These methods enable high-throughput identification and quantification of phosphorylation sites across the proteome. Advanced MS approaches can detect changes in phosphorylation levels under different conditions, providing insights into cellular signaling pathways and regulatory mechanisms. The techniques often involve sample preparation steps such as phosphopeptide enrichment, followed by LC-MS/MS analysis and computational data processing.
- Antibody-based phosphorylation detection methods: Antibody-based approaches provide specific detection of phosphorylated proteins and peptides. These methods include Western blotting, ELISA, immunoprecipitation, and phospho-specific antibody arrays for quantitative analysis of phosphorylation events. The techniques rely on antibodies that specifically recognize phosphorylated amino acid residues (primarily phospho-serine, phospho-threonine, and phospho-tyrosine) or specific phosphorylated sequences within proteins. These approaches allow for targeted analysis of known phosphorylation sites and can be applied to clinical samples.
- Imaging and microscopy techniques for phosphorylation analysis: Advanced imaging techniques enable visualization and quantification of protein phosphorylation in cells and tissues. These methods include fluorescence microscopy, phospho-specific immunohistochemistry, and live-cell imaging approaches. The techniques allow for spatial and temporal resolution of phosphorylation events within cellular compartments and can track dynamic changes in phosphorylation status. Image analysis algorithms and computational tools are used to quantify phosphorylation signals and correlate them with cellular functions.
- Phosphoproteomic data analysis and computational methods: Computational approaches for analyzing phosphorylation data include specialized software tools, statistical methods, and machine learning algorithms. These methods enable processing of large-scale phosphoproteomic datasets, identification of phosphorylation sites, quantification of phosphorylation levels, and prediction of kinase-substrate relationships. Bioinformatic tools can integrate phosphorylation data with other omics datasets to provide comprehensive understanding of cellular signaling networks and identify potential biomarkers or therapeutic targets.
- Microfluidic and chip-based phosphorylation assays: Miniaturized platforms and microfluidic devices enable high-throughput, sensitive detection of protein phosphorylation with minimal sample requirements. These technologies include protein microarrays, lab-on-a-chip devices, and microfluidic systems integrated with various detection methods. The approaches allow for multiplexed analysis of multiple phosphorylation sites simultaneously and can be automated for increased throughput. These platforms are particularly valuable for clinical applications and personalized medicine approaches where sample quantities are limited.
02 Phosphopeptide enrichment strategies
Various enrichment strategies have been developed to isolate phosphorylated peptides prior to quantitative analysis. These include immobilized metal affinity chromatography (IMAC), titanium dioxide (TiO2) chromatography, and phospho-specific antibody-based methods. These enrichment techniques are crucial for increasing the detection sensitivity of phosphorylated proteins, which are often present at low abundance in complex biological samples. Combining multiple enrichment approaches can further enhance the coverage of phosphoproteome analysis.Expand Specific Solutions03 Imaging-based phosphorylation quantification
Imaging techniques provide spatial information about phosphorylation events within cells and tissues. These methods include fluorescence microscopy with phospho-specific antibodies, phospho-sensors, and FRET-based approaches. Advanced imaging algorithms and software tools enable quantitative analysis of phosphorylation signals in single cells or specific subcellular compartments. These techniques are particularly valuable for studying the dynamics and localization of phosphorylation events in living systems.Expand Specific Solutions04 Computational methods for phosphoproteomics data analysis
Specialized computational tools and algorithms have been developed for processing and interpreting phosphoproteomics data. These include software for phosphorylation site identification, quantification of phosphorylation stoichiometry, and statistical analysis of differential phosphorylation. Machine learning approaches are increasingly being applied to predict functional consequences of phosphorylation events and to integrate phosphoproteomics data with other omics datasets. These computational methods help researchers extract meaningful biological insights from complex phosphorylation data.Expand Specific Solutions05 Multiplexed assays for phosphorylation analysis
Multiplexed technologies enable simultaneous quantification of multiple phosphorylation sites or phosphorylated proteins. These include protein microarrays, bead-based assays, and multiplexed mass spectrometry approaches using isobaric labeling reagents (e.g., TMT, iTRAQ). These methods increase throughput and allow for comparative analysis across multiple samples, time points, or treatment conditions. Multiplexed approaches are particularly valuable for studying complex signaling networks and phosphorylation-mediated cellular responses.Expand Specific Solutions
Leading Research Institutions and Biotechnology Companies
Phosphorylation in bacterial pathogenesis represents a dynamic research area currently in its growth phase, with an estimated market size of $2-3 billion and expanding at 7-9% annually. The competitive landscape features established pharmaceutical companies like Novartis AG and Vertex Pharmaceuticals alongside specialized research institutions such as the German Cancer Research Center and Institut Pasteur. Academic institutions including University of Michigan and University of California contribute significant research advancements. Technology maturity varies across applications, with diagnostic tools reaching commercial viability while therapeutic interventions remain largely experimental. Illumina and Life Technologies lead in providing research tools, while companies like Antabio SAS focus on developing novel antibacterial resistance-breakers targeting phosphorylation pathways in pathogens.
The Regents of the University of California
Technical Solution: The University of California system has developed an integrated multi-omics approach to bacterial phosphorylation analysis that combines phosphoproteomics with transcriptomics and metabolomics. Their technical solution employs titanium dioxide (TiO2) and immobilized metal affinity chromatography (IMAC) enrichment strategies optimized specifically for bacterial phosphopeptides, which typically occur at lower stoichiometry than eukaryotic counterparts. The UC researchers have created specialized bioinformatics tools that incorporate machine learning algorithms to predict functional consequences of phosphorylation events in bacterial virulence factors. Their quantitative approach utilizes both label-free and isobaric labeling techniques (TMT/iTRAQ) to achieve comprehensive coverage of the bacterial phosphoproteome during different stages of infection. The system has been validated across multiple bacterial pathogens including Pseudomonas aeruginosa, Salmonella, and Mycobacterium tuberculosis, demonstrating its versatility for studying diverse pathogenesis mechanisms.
Strengths: Comprehensive multi-omics integration provides contextual understanding of phosphorylation events; robust bioinformatics infrastructure for data analysis; extensive validation across multiple bacterial pathogens. Weakness: The complexity of the integrated approach requires significant expertise across multiple technical domains, potentially limiting accessibility to specialized research groups.
Institut Pasteur
Technical Solution: Institut Pasteur has developed comprehensive phosphoproteomics platforms specifically optimized for bacterial pathogen analysis. Their technical approach combines mass spectrometry-based phosphopeptide enrichment techniques with custom bioinformatics pipelines to quantitatively measure phosphorylation events during host-pathogen interactions. The institute has pioneered methods for temporal phosphorylation profiling during infection cycles, allowing researchers to track dynamic changes in both bacterial effector proteins and host response mechanisms. Their platform integrates stable isotope labeling techniques (SILAC) with high-resolution mass spectrometry to achieve quantitative measurements of phosphorylation stoichiometry across thousands of sites simultaneously. Institut Pasteur researchers have specifically adapted these methods to overcome the technical challenges of bacterial phosphoproteomics, including the relatively low abundance of phosphorylated proteins in prokaryotes compared to eukaryotes.
Strengths: Exceptional expertise in bacterial pathogenesis mechanisms and host-pathogen interactions; established infrastructure for high-throughput phosphoproteomics; strong track record in method development for bacterial systems. Weakness: Their quantitative approaches may require specialized equipment not widely available in standard microbiology laboratories, potentially limiting broader adoption.
Key Phosphorylation Pathways in Bacterial Virulence
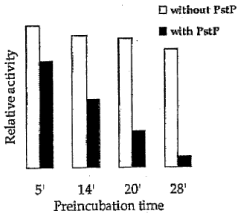
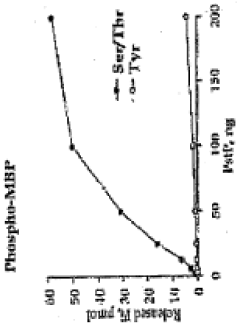
Pknb kinase and pstp phosphatase and methods of identifying inhibitory substances
PatentInactiveIN212DELNP2006A
Innovation
- The identification and characterization of the pknB kinase and pstP phosphatase, which are involved in signal transduction pathways, providing potential targets for developing antibacterial agents that can modulate their activity to treat Mycobacterium tuberculosis infections.
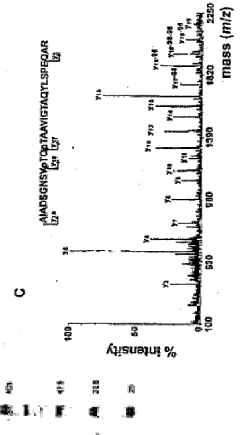
In-GEL tagging and in-GEL digestion for phosphoproteins analysis and phosphorylation site identification
PatentWO2006028310A1
Innovation
- An in-gel chemical tagging method using nucleophilic reagents like guanidinoethanethiol, which converts dephosphorylated amino acid residues into enzymatically recognizable sites, allowing for specific proteolysis and mass analysis of phosphopeptides, enhancing detectability and site identification.
Therapeutic Implications of Targeting Bacterial Phosphorylation
The therapeutic potential of targeting bacterial phosphorylation systems represents a promising frontier in antimicrobial development. Given the essential role of phosphorylation in bacterial virulence and survival, inhibiting these pathways offers several strategic advantages over conventional antibiotics. Phosphorylation inhibitors could potentially disrupt pathogen virulence without directly killing bacteria, thereby reducing selective pressure that drives resistance development.
Quantitative approaches have revealed that phosphorylation events often occur at specific thresholds during infection, suggesting that precisely calibrated interventions could effectively attenuate pathogenesis without complete pathway shutdown. This "tuning" rather than "switching off" approach may preserve commensal microbiota while selectively targeting pathogenic behavior.
Several classes of compounds have demonstrated efficacy in targeting bacterial kinases and phosphatases. Tyrosine kinase inhibitors, originally developed for cancer therapy, have shown cross-reactivity with bacterial counterparts. For instance, compounds targeting Staphylococcus aureus Stk1 have demonstrated reduced virulence in animal models without affecting bacterial viability in vitro, suggesting potential for anti-virulence applications.
Phosphorylation-targeting therapeutics face distinct pharmacological challenges. The structural differences between bacterial and human kinases must be exploited to ensure specificity and minimize off-target effects. Quantitative structure-activity relationship (QSAR) models have identified promising scaffolds with selective activity against bacterial phosphorylation systems while sparing human homologs.
Combination therapies represent another promising approach. Sub-inhibitory concentrations of phosphorylation inhibitors can sensitize bacteria to conventional antibiotics, potentially revitalizing the efficacy of drugs to which resistance has developed. Quantitative studies have demonstrated synergistic effects when phosphorylation inhibitors are administered alongside β-lactams or aminoglycosides.
Delivery systems present both challenges and opportunities. Nanoparticle-based delivery platforms have shown promise in targeting phosphorylation inhibitors specifically to infection sites, potentially reducing systemic exposure and associated toxicity. Quantitative pharmacokinetic modeling suggests that such targeted delivery could achieve therapeutic concentrations at infection sites while maintaining sub-therapeutic levels in commensal communities.
Clinical translation remains the ultimate challenge. While preclinical data is promising, human trials of phosphorylation-targeting antimicrobials remain limited. Quantitative biomarkers that reliably indicate successful phosphorylation inhibition in vivo are needed to facilitate clinical development. Phosphoproteomic signatures may serve as surrogate endpoints in early-phase trials, potentially accelerating the path to regulatory approval.
Quantitative approaches have revealed that phosphorylation events often occur at specific thresholds during infection, suggesting that precisely calibrated interventions could effectively attenuate pathogenesis without complete pathway shutdown. This "tuning" rather than "switching off" approach may preserve commensal microbiota while selectively targeting pathogenic behavior.
Several classes of compounds have demonstrated efficacy in targeting bacterial kinases and phosphatases. Tyrosine kinase inhibitors, originally developed for cancer therapy, have shown cross-reactivity with bacterial counterparts. For instance, compounds targeting Staphylococcus aureus Stk1 have demonstrated reduced virulence in animal models without affecting bacterial viability in vitro, suggesting potential for anti-virulence applications.
Phosphorylation-targeting therapeutics face distinct pharmacological challenges. The structural differences between bacterial and human kinases must be exploited to ensure specificity and minimize off-target effects. Quantitative structure-activity relationship (QSAR) models have identified promising scaffolds with selective activity against bacterial phosphorylation systems while sparing human homologs.
Combination therapies represent another promising approach. Sub-inhibitory concentrations of phosphorylation inhibitors can sensitize bacteria to conventional antibiotics, potentially revitalizing the efficacy of drugs to which resistance has developed. Quantitative studies have demonstrated synergistic effects when phosphorylation inhibitors are administered alongside β-lactams or aminoglycosides.
Delivery systems present both challenges and opportunities. Nanoparticle-based delivery platforms have shown promise in targeting phosphorylation inhibitors specifically to infection sites, potentially reducing systemic exposure and associated toxicity. Quantitative pharmacokinetic modeling suggests that such targeted delivery could achieve therapeutic concentrations at infection sites while maintaining sub-therapeutic levels in commensal communities.
Clinical translation remains the ultimate challenge. While preclinical data is promising, human trials of phosphorylation-targeting antimicrobials remain limited. Quantitative biomarkers that reliably indicate successful phosphorylation inhibition in vivo are needed to facilitate clinical development. Phosphoproteomic signatures may serve as surrogate endpoints in early-phase trials, potentially accelerating the path to regulatory approval.
Regulatory Considerations for Phosphorylation-Based Antimicrobials
The development of phosphorylation-based antimicrobials represents a promising frontier in combating bacterial infections, yet navigating the regulatory landscape presents significant challenges. Regulatory bodies such as the FDA, EMA, and NMPA have established stringent frameworks for antimicrobial approval that must be carefully considered during development phases. These frameworks emphasize safety profiles, efficacy metrics, and resistance potential—all critical factors for phosphorylation-targeting therapeutics.
Phosphorylation inhibitors face unique regulatory scrutiny due to their novel mechanisms of action. Unlike conventional antibiotics that target cell wall synthesis or protein translation, these compounds interfere with bacterial signaling pathways, raising questions about specificity and potential off-target effects in human cells. Regulatory agencies typically require extensive cross-reactivity studies demonstrating minimal interference with human kinases and phosphatases.
Clinical trial designs for phosphorylation-based antimicrobials must address regulatory expectations for both traditional antibacterial endpoints and mechanism-specific biomarkers. Quantitative assessment of phosphorylation states in bacterial populations during infection presents methodological challenges that regulators increasingly expect developers to overcome. Recent guidance documents suggest incorporating phosphoproteomic analyses as supportive evidence in regulatory submissions.
The accelerated approval pathways available for novel antimicrobials offer potential advantages for phosphorylation-targeting compounds, particularly those addressing multidrug-resistant pathogens. The GAIN Act in the US and similar initiatives in Europe provide extended market exclusivity and expedited review processes. However, qualifying for these pathways requires demonstrating significant advantages over existing therapies—a hurdle requiring robust quantitative data on phosphorylation dynamics during infection.
Regulatory considerations also extend to companion diagnostics for phosphorylation-based therapeutics. As these compounds often target specific phosphorylation patterns in particular bacterial species, diagnostic tools identifying suitable patients may require parallel regulatory approval. The FDA's combined diagnostic-therapeutic review process offers a streamlined approach but demands coordinated development strategies.
Manufacturing controls present another regulatory dimension, with agencies requiring validated analytical methods for assessing the purity and stability of phosphorylation-targeting compounds. The complex chemical structures of many kinase inhibitors necessitate sophisticated quality control measures that meet increasingly stringent regulatory standards for impurity profiling and stability testing.
Post-approval regulatory requirements will likely include enhanced pharmacovigilance for monitoring resistance development. Regulatory agencies have signaled interest in real-time surveillance of bacterial phosphorylation patterns in clinical settings, potentially mandating periodic susceptibility testing and reporting mechanisms for developers of these novel antimicrobials.
Phosphorylation inhibitors face unique regulatory scrutiny due to their novel mechanisms of action. Unlike conventional antibiotics that target cell wall synthesis or protein translation, these compounds interfere with bacterial signaling pathways, raising questions about specificity and potential off-target effects in human cells. Regulatory agencies typically require extensive cross-reactivity studies demonstrating minimal interference with human kinases and phosphatases.
Clinical trial designs for phosphorylation-based antimicrobials must address regulatory expectations for both traditional antibacterial endpoints and mechanism-specific biomarkers. Quantitative assessment of phosphorylation states in bacterial populations during infection presents methodological challenges that regulators increasingly expect developers to overcome. Recent guidance documents suggest incorporating phosphoproteomic analyses as supportive evidence in regulatory submissions.
The accelerated approval pathways available for novel antimicrobials offer potential advantages for phosphorylation-targeting compounds, particularly those addressing multidrug-resistant pathogens. The GAIN Act in the US and similar initiatives in Europe provide extended market exclusivity and expedited review processes. However, qualifying for these pathways requires demonstrating significant advantages over existing therapies—a hurdle requiring robust quantitative data on phosphorylation dynamics during infection.
Regulatory considerations also extend to companion diagnostics for phosphorylation-based therapeutics. As these compounds often target specific phosphorylation patterns in particular bacterial species, diagnostic tools identifying suitable patients may require parallel regulatory approval. The FDA's combined diagnostic-therapeutic review process offers a streamlined approach but demands coordinated development strategies.
Manufacturing controls present another regulatory dimension, with agencies requiring validated analytical methods for assessing the purity and stability of phosphorylation-targeting compounds. The complex chemical structures of many kinase inhibitors necessitate sophisticated quality control measures that meet increasingly stringent regulatory standards for impurity profiling and stability testing.
Post-approval regulatory requirements will likely include enhanced pharmacovigilance for monitoring resistance development. Regulatory agencies have signaled interest in real-time surveillance of bacterial phosphorylation patterns in clinical settings, potentially mandating periodic susceptibility testing and reporting mechanisms for developers of these novel antimicrobials.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!