How to Conduct High-Throughput Phosphorylation Screening
SEP 23, 202510 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Phosphorylation Screening Background and Objectives
Protein phosphorylation, a fundamental post-translational modification, has been extensively studied since its discovery in the late 1950s. This reversible biochemical process involves the addition of a phosphate group to specific amino acid residues, primarily serine, threonine, and tyrosine, catalyzed by protein kinases. The significance of phosphorylation in cellular signaling pathways, metabolic regulation, and disease mechanisms has driven continuous technological advancement in detection and analysis methods over the past decades.
The evolution of phosphorylation screening techniques has progressed from low-throughput radioactive labeling methods to sophisticated high-throughput platforms integrating mass spectrometry, protein microarrays, and computational approaches. Early techniques in the 1980s and 1990s focused on individual protein analysis, while the post-genomic era has witnessed a paradigm shift toward comprehensive phosphoproteome analysis, enabling the simultaneous examination of thousands of phosphorylation events.
Current technological trends in phosphorylation screening emphasize increased sensitivity, higher throughput, reduced sample requirements, and enhanced quantitative accuracy. The integration of artificial intelligence and machine learning algorithms for data analysis represents a significant advancement, allowing researchers to extract meaningful patterns from complex phosphoproteomic datasets and predict functional consequences of phosphorylation events with greater precision.
The primary objective of high-throughput phosphorylation screening is to systematically identify and characterize phosphorylation sites across the proteome under various physiological and pathological conditions. This comprehensive approach aims to elucidate the dynamic nature of phosphorylation networks, revealing how these networks respond to stimuli, regulate cellular processes, and contribute to disease mechanisms when dysregulated.
Secondary objectives include the identification of novel kinase-substrate relationships, the discovery of phosphorylation-dependent protein-protein interactions, and the elucidation of temporal dynamics in phosphorylation cascades. These insights are crucial for understanding cellular signaling architecture and developing targeted therapeutic interventions for diseases characterized by aberrant phosphorylation patterns, such as cancer, neurodegenerative disorders, and inflammatory conditions.
The technical goals of modern phosphorylation screening methodologies focus on achieving comprehensive coverage of the phosphoproteome while maintaining high specificity and reproducibility. Researchers aim to develop platforms capable of detecting low-abundance phosphoproteins, distinguishing between closely related phosphorylation sites, and providing accurate quantitative measurements across diverse sample types and experimental conditions.
Looking forward, the field is moving toward integrating phosphorylation data with other omics datasets to construct multi-dimensional models of cellular regulation. This systems biology approach promises to reveal emergent properties of biological networks that cannot be discerned from phosphorylation analysis alone, ultimately advancing our understanding of complex biological processes and facilitating the development of precision medicine approaches.
The evolution of phosphorylation screening techniques has progressed from low-throughput radioactive labeling methods to sophisticated high-throughput platforms integrating mass spectrometry, protein microarrays, and computational approaches. Early techniques in the 1980s and 1990s focused on individual protein analysis, while the post-genomic era has witnessed a paradigm shift toward comprehensive phosphoproteome analysis, enabling the simultaneous examination of thousands of phosphorylation events.
Current technological trends in phosphorylation screening emphasize increased sensitivity, higher throughput, reduced sample requirements, and enhanced quantitative accuracy. The integration of artificial intelligence and machine learning algorithms for data analysis represents a significant advancement, allowing researchers to extract meaningful patterns from complex phosphoproteomic datasets and predict functional consequences of phosphorylation events with greater precision.
The primary objective of high-throughput phosphorylation screening is to systematically identify and characterize phosphorylation sites across the proteome under various physiological and pathological conditions. This comprehensive approach aims to elucidate the dynamic nature of phosphorylation networks, revealing how these networks respond to stimuli, regulate cellular processes, and contribute to disease mechanisms when dysregulated.
Secondary objectives include the identification of novel kinase-substrate relationships, the discovery of phosphorylation-dependent protein-protein interactions, and the elucidation of temporal dynamics in phosphorylation cascades. These insights are crucial for understanding cellular signaling architecture and developing targeted therapeutic interventions for diseases characterized by aberrant phosphorylation patterns, such as cancer, neurodegenerative disorders, and inflammatory conditions.
The technical goals of modern phosphorylation screening methodologies focus on achieving comprehensive coverage of the phosphoproteome while maintaining high specificity and reproducibility. Researchers aim to develop platforms capable of detecting low-abundance phosphoproteins, distinguishing between closely related phosphorylation sites, and providing accurate quantitative measurements across diverse sample types and experimental conditions.
Looking forward, the field is moving toward integrating phosphorylation data with other omics datasets to construct multi-dimensional models of cellular regulation. This systems biology approach promises to reveal emergent properties of biological networks that cannot be discerned from phosphorylation analysis alone, ultimately advancing our understanding of complex biological processes and facilitating the development of precision medicine approaches.
Market Demand Analysis for High-Throughput Phosphorylation Assays
The global market for high-throughput phosphorylation screening technologies has experienced significant growth over the past decade, driven primarily by advances in drug discovery and personalized medicine. Current market valuations indicate that the high-throughput screening (HTS) market reached approximately 22 billion USD in 2022, with phosphorylation assays representing a substantial segment estimated at 3.5 billion USD.
Pharmaceutical and biotechnology companies constitute the largest consumer base, accounting for nearly 60% of the market demand. These organizations increasingly rely on phosphorylation screening to identify novel drug candidates that target kinase pathways, which are implicated in numerous diseases including cancer, inflammatory disorders, and neurological conditions. The oncology segment alone generates over 40% of the demand for phosphorylation assays due to the critical role of kinase dysregulation in cancer progression.
Academic research institutions represent the second-largest market segment, contributing approximately 25% of the demand. This sector's growth is fueled by substantial research grants focused on understanding fundamental cellular signaling mechanisms and disease pathways. Contract research organizations (CROs) account for another 15% of the market as pharmaceutical outsourcing continues to expand.
Regionally, North America dominates the market with approximately 45% share, followed by Europe (30%) and Asia-Pacific (20%). However, the Asia-Pacific region is projected to witness the highest compound annual growth rate of 12% through 2028, primarily due to increasing R&D investments in China, Japan, and South Korea.
The demand for more efficient and cost-effective phosphorylation screening methods continues to rise as drug discovery pipelines expand. Industry surveys indicate that 78% of pharmaceutical companies plan to increase their investment in high-throughput screening technologies over the next five years. This trend is particularly evident in the development of targeted therapies, where precise understanding of phosphorylation events is crucial.
Market analysis reveals growing demand for integrated solutions that combine phosphorylation screening with other post-translational modification analyses. Customers increasingly seek platforms that offer multiplexed capabilities, allowing simultaneous assessment of multiple phosphorylation sites and pathways. Additionally, there is rising interest in technologies that can perform phosphorylation screening in more physiologically relevant models, including 3D cell cultures and patient-derived samples.
The emergence of artificial intelligence and machine learning applications in data analysis represents another significant market trend, with companies willing to pay premium prices for solutions that incorporate advanced analytics capabilities. This reflects the growing complexity of phosphorylation data and the need for sophisticated interpretation tools to extract meaningful biological insights.
Pharmaceutical and biotechnology companies constitute the largest consumer base, accounting for nearly 60% of the market demand. These organizations increasingly rely on phosphorylation screening to identify novel drug candidates that target kinase pathways, which are implicated in numerous diseases including cancer, inflammatory disorders, and neurological conditions. The oncology segment alone generates over 40% of the demand for phosphorylation assays due to the critical role of kinase dysregulation in cancer progression.
Academic research institutions represent the second-largest market segment, contributing approximately 25% of the demand. This sector's growth is fueled by substantial research grants focused on understanding fundamental cellular signaling mechanisms and disease pathways. Contract research organizations (CROs) account for another 15% of the market as pharmaceutical outsourcing continues to expand.
Regionally, North America dominates the market with approximately 45% share, followed by Europe (30%) and Asia-Pacific (20%). However, the Asia-Pacific region is projected to witness the highest compound annual growth rate of 12% through 2028, primarily due to increasing R&D investments in China, Japan, and South Korea.
The demand for more efficient and cost-effective phosphorylation screening methods continues to rise as drug discovery pipelines expand. Industry surveys indicate that 78% of pharmaceutical companies plan to increase their investment in high-throughput screening technologies over the next five years. This trend is particularly evident in the development of targeted therapies, where precise understanding of phosphorylation events is crucial.
Market analysis reveals growing demand for integrated solutions that combine phosphorylation screening with other post-translational modification analyses. Customers increasingly seek platforms that offer multiplexed capabilities, allowing simultaneous assessment of multiple phosphorylation sites and pathways. Additionally, there is rising interest in technologies that can perform phosphorylation screening in more physiologically relevant models, including 3D cell cultures and patient-derived samples.
The emergence of artificial intelligence and machine learning applications in data analysis represents another significant market trend, with companies willing to pay premium prices for solutions that incorporate advanced analytics capabilities. This reflects the growing complexity of phosphorylation data and the need for sophisticated interpretation tools to extract meaningful biological insights.
Current Phosphorylation Screening Technologies and Limitations
Phosphorylation screening technologies have evolved significantly over the past decades, transitioning from low-throughput methods to sophisticated high-throughput platforms. Currently, several established technologies dominate the field, each with distinct advantages and limitations that influence their applicability in different research contexts.
Mass spectrometry (MS) represents the gold standard for phosphorylation analysis, offering unparalleled depth in phosphoproteome coverage. Advanced MS techniques like SILAC, TMT, and iTRAQ enable quantitative phosphoproteomic analysis across multiple samples simultaneously. However, MS-based approaches require expensive instrumentation, specialized expertise, and complex sample preparation protocols that can introduce variability. Additionally, the detection of low-abundance phosphorylation events remains challenging despite recent technological improvements.
Protein microarrays have emerged as powerful tools for high-throughput phosphorylation screening, allowing simultaneous analysis of thousands of potential substrates. These platforms require minimal sample volume and provide rapid results. Nevertheless, they suffer from limited dynamic range, potential cross-reactivity issues, and challenges in detecting phosphorylation events in their native cellular context. The production of high-quality protein arrays with properly folded proteins also presents significant technical hurdles.
Bead-based multiplexing technologies, such as Luminex xMAP, offer another approach for high-throughput phosphorylation analysis. These systems provide good sensitivity and multiplexing capabilities but are constrained by the availability of specific antibodies and potential cross-reactivity issues that can compromise data quality.
Cell-based assays utilizing phospho-specific antibodies in formats such as ELISA, AlphaScreen, or TR-FRET provide valuable information on cellular phosphorylation dynamics. While these approaches maintain physiological relevance, they typically offer lower throughput compared to other screening methods and are heavily dependent on antibody quality and specificity.
Kinase activity assays using peptide substrates in microplate formats enable direct measurement of kinase function across many samples. However, these assays often utilize artificial substrates that may not accurately reflect in vivo phosphorylation preferences, potentially leading to misleading results when translated to cellular contexts.
A significant limitation across all current technologies is the challenge of temporal resolution. Phosphorylation events occur dynamically within millisecond to second timeframes, yet most current technologies provide only static snapshots rather than continuous monitoring of phosphorylation dynamics.
Data analysis represents another major bottleneck, as high-throughput phosphorylation screening generates massive datasets requiring sophisticated computational approaches for meaningful interpretation. The integration of phosphoproteomic data with other omics datasets remains challenging but essential for comprehensive understanding of cellular signaling networks.
Mass spectrometry (MS) represents the gold standard for phosphorylation analysis, offering unparalleled depth in phosphoproteome coverage. Advanced MS techniques like SILAC, TMT, and iTRAQ enable quantitative phosphoproteomic analysis across multiple samples simultaneously. However, MS-based approaches require expensive instrumentation, specialized expertise, and complex sample preparation protocols that can introduce variability. Additionally, the detection of low-abundance phosphorylation events remains challenging despite recent technological improvements.
Protein microarrays have emerged as powerful tools for high-throughput phosphorylation screening, allowing simultaneous analysis of thousands of potential substrates. These platforms require minimal sample volume and provide rapid results. Nevertheless, they suffer from limited dynamic range, potential cross-reactivity issues, and challenges in detecting phosphorylation events in their native cellular context. The production of high-quality protein arrays with properly folded proteins also presents significant technical hurdles.
Bead-based multiplexing technologies, such as Luminex xMAP, offer another approach for high-throughput phosphorylation analysis. These systems provide good sensitivity and multiplexing capabilities but are constrained by the availability of specific antibodies and potential cross-reactivity issues that can compromise data quality.
Cell-based assays utilizing phospho-specific antibodies in formats such as ELISA, AlphaScreen, or TR-FRET provide valuable information on cellular phosphorylation dynamics. While these approaches maintain physiological relevance, they typically offer lower throughput compared to other screening methods and are heavily dependent on antibody quality and specificity.
Kinase activity assays using peptide substrates in microplate formats enable direct measurement of kinase function across many samples. However, these assays often utilize artificial substrates that may not accurately reflect in vivo phosphorylation preferences, potentially leading to misleading results when translated to cellular contexts.
A significant limitation across all current technologies is the challenge of temporal resolution. Phosphorylation events occur dynamically within millisecond to second timeframes, yet most current technologies provide only static snapshots rather than continuous monitoring of phosphorylation dynamics.
Data analysis represents another major bottleneck, as high-throughput phosphorylation screening generates massive datasets requiring sophisticated computational approaches for meaningful interpretation. The integration of phosphoproteomic data with other omics datasets remains challenging but essential for comprehensive understanding of cellular signaling networks.
Established High-Throughput Phosphorylation Screening Approaches
01 Microfluidic platforms for high-throughput phosphorylation screening
Microfluidic devices enable high-throughput phosphorylation screening by providing miniaturized reaction environments that require minimal sample volumes. These platforms integrate multiple analytical steps including sample preparation, reaction, separation, and detection into a single system. The miniaturization allows for parallel processing of numerous samples simultaneously, significantly increasing throughput while reducing reagent consumption and analysis time. Advanced microfluidic systems incorporate automated fluid handling and can be coupled with various detection methods for real-time monitoring of phosphorylation events.- Microarray-based phosphorylation screening methods: Microarray technology enables high-throughput screening of protein phosphorylation events by immobilizing substrates on solid surfaces. These platforms allow for parallel analysis of multiple kinase reactions simultaneously, significantly increasing throughput compared to traditional methods. The technology incorporates detection systems such as fluorescence or chemiluminescence to identify phosphorylation events across thousands of potential substrates in a single experiment.
- Automated systems for phosphorylation analysis: Automated platforms have been developed to streamline phosphorylation screening workflows, incorporating robotic liquid handling, automated sample preparation, and integrated detection systems. These systems minimize manual intervention, reduce human error, and enable continuous operation for processing large sample batches. The integration of computational tools for data analysis further enhances the efficiency of phosphorylation screening, allowing for rapid identification of kinase-substrate relationships.
- Mass spectrometry-based phosphoproteomic approaches: Mass spectrometry has revolutionized high-throughput phosphorylation screening by enabling the identification and quantification of thousands of phosphorylation sites in complex biological samples. Advanced techniques such as phosphopeptide enrichment, multiplexed labeling strategies, and data-independent acquisition methods have significantly increased the coverage and throughput of phosphoproteomic analyses. These approaches provide comprehensive insights into cellular signaling networks and kinase activities in various biological contexts.
- Bead-based multiplexed phosphorylation assays: Bead-based technologies enable multiplexed analysis of phosphorylation events by coupling substrates to uniquely identifiable microspheres. These systems allow for simultaneous detection of multiple phosphorylation reactions in a single sample, significantly increasing throughput. The technology typically employs flow cytometry or imaging-based detection methods to quantify phosphorylation levels across numerous substrates, making it particularly valuable for screening kinase inhibitors and studying signaling pathways.
- Microfluidic platforms for kinase activity screening: Microfluidic devices have been developed for high-throughput phosphorylation screening, enabling miniaturized reactions with minimal reagent consumption. These platforms incorporate various detection modalities and can be designed for single-cell analysis or droplet-based assays. The integration of multiple functional components on a single chip allows for parallel processing of numerous samples, significantly increasing throughput while reducing analysis time and cost.
02 Mass spectrometry-based methods for phosphorylation analysis
Mass spectrometry techniques provide powerful tools for high-throughput phosphorylation screening by enabling the identification and quantification of phosphorylated proteins and peptides with high sensitivity and specificity. These methods can detect multiple phosphorylation sites simultaneously and determine their stoichiometry. Advanced mass spectrometry approaches incorporate stable isotope labeling, multiple reaction monitoring, and data-independent acquisition to enhance throughput and quantitative accuracy. Integration with automated sample preparation and chromatographic separation further increases the screening capacity for complex biological samples.Expand Specific Solutions03 Array-based technologies for phosphorylation detection
Protein and peptide arrays enable parallel analysis of multiple phosphorylation events in a high-throughput manner. These arrays consist of immobilized substrates or capture molecules arranged in a grid format that allows simultaneous screening of numerous kinase-substrate interactions or phosphorylation states. The technology incorporates various detection methods including fluorescence, chemiluminescence, or label-free approaches to visualize phosphorylation events. Advanced array platforms feature automated spotting, incubation, and washing steps to increase reproducibility and throughput while minimizing manual intervention.Expand Specific Solutions04 Automated systems for high-throughput kinase assays
Automated robotic systems significantly enhance phosphorylation screening throughput by performing repetitive tasks with high precision and minimal human intervention. These systems integrate liquid handling, incubation, washing, and detection steps into streamlined workflows that can process hundreds to thousands of samples per day. Advanced platforms incorporate machine learning algorithms for experimental design optimization and data analysis. The automation reduces experimental variability, increases reproducibility, and enables continuous operation for large-scale phosphorylation studies across multiple kinases and substrates.Expand Specific Solutions05 Fluorescence-based detection methods for phosphorylation screening
Fluorescence-based techniques provide sensitive and quantitative methods for high-throughput phosphorylation screening. These approaches utilize fluorescently labeled antibodies, phospho-specific dyes, or genetically encoded biosensors that change their spectral properties upon phosphorylation. Time-resolved fluorescence, fluorescence polarization, and Förster resonance energy transfer (FRET) enable real-time monitoring of phosphorylation reactions. When combined with plate reader technology or automated microscopy systems, these methods allow rapid analysis of thousands of samples with minimal reagent consumption and high sensitivity for detecting even subtle changes in phosphorylation states.Expand Specific Solutions
Key Industry Players in Phosphoproteomics Research
High-throughput phosphorylation screening technology is currently in a growth phase, with an estimated market size of $1.5-2 billion and expanding at 8-10% annually. The competitive landscape features pharmaceutical giants like Pfizer, Janssen, and Sanofi-Aventis alongside specialized research institutions such as MIT, Howard Hughes Medical Institute, and the Broad Institute. Academic-industry partnerships are accelerating technological maturity, with companies like Chugai Pharmaceutical and Inflazome developing proprietary screening platforms. Technology leaders are focusing on automation, AI integration, and miniaturization to increase throughput while reducing costs. The field is transitioning from early adoption to mainstream implementation, with significant opportunities for innovation in screening methodologies and data analysis tools.
Massachusetts Institute of Technology
Technical Solution: MIT has developed a high-throughput phosphorylation screening platform that combines microfluidic technology with mass spectrometry. Their approach utilizes automated sample handling systems that can process thousands of phosphorylation reactions simultaneously. The platform incorporates a miniaturized reaction chamber design that requires minimal reagent volumes (nanoliter scale), significantly reducing costs while maintaining high sensitivity. MIT researchers have implemented machine learning algorithms to analyze the resulting phosphorylation patterns, enabling rapid identification of kinase-substrate relationships and phosphorylation site preferences. The system integrates with their proprietary database of known phosphorylation networks, allowing for contextual interpretation of results within cellular signaling pathways[1][3]. Their technology also features real-time monitoring capabilities, permitting kinetic analysis of phosphorylation events rather than just endpoint measurements.
Strengths: Exceptional integration of microfluidics with advanced computational analysis, providing both high throughput and deep mechanistic insights. The system requires minimal sample input, making it suitable for precious biological samples. Weaknesses: The sophisticated technology requires specialized expertise and expensive equipment, limiting accessibility to well-funded research institutions. The approach may have challenges with highly complex protein mixtures or membrane proteins.
The Howard Hughes Medical Institute
Technical Solution: HHMI has pioneered a comprehensive phosphorylation screening platform combining protein microarrays with advanced imaging techniques. Their system utilizes custom-designed protein chips containing thousands of potential substrate proteins in native or near-native conformations. The screening process employs fluorescently labeled ATP analogs that enable direct visualization of phosphorylation events without antibody detection steps, significantly increasing throughput. HHMI researchers have developed specialized surface chemistry that maintains protein activity while minimizing non-specific binding. Their platform incorporates parallel processing of multiple kinases against the same substrate array, allowing for comparative analysis of kinase specificity profiles[2]. The system also features an integrated computational pipeline that correlates phosphorylation patterns with protein structural features, enabling prediction of novel substrates based on structural homology. Additionally, they've implemented quantitative image analysis tools that can detect subtle differences in phosphorylation efficiency across experimental conditions.
Strengths: Exceptional substrate diversity on microarrays and direct visualization approach eliminates many false positives/negatives associated with antibody-based detection. The parallel processing capability dramatically increases screening efficiency. Weaknesses: The technology requires specialized chip manufacturing capabilities and advanced imaging systems. The approach may have limitations in detecting phosphorylation events that require specific cellular contexts or cofactors not present in the in vitro system.
Critical Technologies in Phosphoprotein Detection and Analysis
Method for screening compounds inhibiting signal transduction through inflammatory cytokines
PatentInactiveUS6989244B1
Innovation
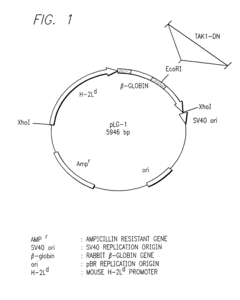
- A method for screening compounds that inhibit the binding between TAK1 and TAB1, using techniques such as contacting a test sample with TAK1 and TAB1, detecting their binding, and selecting compounds that inhibit this interaction, which can be used to develop inhibitors of signal transduction through inflammatory cytokines.
Methods of screening compounds inhibiting signal transduction through inflammatory cytokines
PatentInactiveUS8105799B2
Innovation
- A method for screening compounds that inhibit the binding or kinase activity of TAK1, using techniques such as contacting test samples with TAK1 and TAB1, detecting binding or phosphorylation, and selecting compounds that reduce biological activity associated with inflammatory cytokines, thereby inhibiting signal transduction through inflammatory cytokines.
Data Management Solutions for Large-Scale Phosphoproteomics
The exponential growth of phosphoproteomics data presents significant challenges for storage, processing, and analysis. Current high-throughput phosphorylation screening technologies generate terabytes of raw data that require sophisticated management solutions. Traditional database systems often struggle with the volume, velocity, and variety of phosphoproteomics data, necessitating specialized approaches.
Cloud-based storage solutions have emerged as the primary infrastructure for large-scale phosphoproteomics data management. Platforms such as Amazon Web Services, Google Cloud, and Microsoft Azure offer scalable storage options with built-in redundancy and security features. These platforms provide flexible computing resources that can be dynamically allocated based on processing demands, making them ideal for handling periodic intensive computational tasks common in phosphoproteomics analysis.
Specialized database architectures optimized for phosphoproteomics data have been developed to address the unique requirements of this field. NoSQL databases like MongoDB and Cassandra have gained popularity due to their ability to handle heterogeneous data types and schema flexibility. These systems efficiently store and retrieve complex phosphorylation site information, experimental metadata, and quantitative measurements across multiple conditions and replicates.
Data integration frameworks represent another critical component of phosphoproteomics data management. Tools like PhosphoSitePlus, PHOSIDA, and PhosphoPep facilitate the integration of experimental data with existing knowledge bases, enabling researchers to contextualize their findings within the broader phosphoproteome landscape. These frameworks implement standardized data formats and ontologies to ensure interoperability between different phosphoproteomics datasets and analysis platforms.
Automated data processing pipelines have revolutionized phosphoproteomics data handling. Workflow management systems such as Nextflow, Snakemake, and Galaxy enable the creation of reproducible analysis pipelines that can be deployed across different computing environments. These pipelines typically incorporate quality control metrics, normalization procedures, and statistical analysis tools specifically designed for phosphoproteomics data, ensuring consistent processing across large datasets.
Machine learning approaches are increasingly being applied to phosphoproteomics data management challenges. Dimensionality reduction techniques like principal component analysis and t-SNE help visualize complex phosphorylation patterns, while clustering algorithms identify co-regulated phosphorylation events. Advanced deep learning models can predict functional consequences of phosphorylation events, aiding in the biological interpretation of large-scale screening results.
Cloud-based storage solutions have emerged as the primary infrastructure for large-scale phosphoproteomics data management. Platforms such as Amazon Web Services, Google Cloud, and Microsoft Azure offer scalable storage options with built-in redundancy and security features. These platforms provide flexible computing resources that can be dynamically allocated based on processing demands, making them ideal for handling periodic intensive computational tasks common in phosphoproteomics analysis.
Specialized database architectures optimized for phosphoproteomics data have been developed to address the unique requirements of this field. NoSQL databases like MongoDB and Cassandra have gained popularity due to their ability to handle heterogeneous data types and schema flexibility. These systems efficiently store and retrieve complex phosphorylation site information, experimental metadata, and quantitative measurements across multiple conditions and replicates.
Data integration frameworks represent another critical component of phosphoproteomics data management. Tools like PhosphoSitePlus, PHOSIDA, and PhosphoPep facilitate the integration of experimental data with existing knowledge bases, enabling researchers to contextualize their findings within the broader phosphoproteome landscape. These frameworks implement standardized data formats and ontologies to ensure interoperability between different phosphoproteomics datasets and analysis platforms.
Automated data processing pipelines have revolutionized phosphoproteomics data handling. Workflow management systems such as Nextflow, Snakemake, and Galaxy enable the creation of reproducible analysis pipelines that can be deployed across different computing environments. These pipelines typically incorporate quality control metrics, normalization procedures, and statistical analysis tools specifically designed for phosphoproteomics data, ensuring consistent processing across large datasets.
Machine learning approaches are increasingly being applied to phosphoproteomics data management challenges. Dimensionality reduction techniques like principal component analysis and t-SNE help visualize complex phosphorylation patterns, while clustering algorithms identify co-regulated phosphorylation events. Advanced deep learning models can predict functional consequences of phosphorylation events, aiding in the biological interpretation of large-scale screening results.
Regulatory Considerations for Phosphorylation-Based Diagnostics
The regulatory landscape for phosphorylation-based diagnostics presents significant challenges for developers implementing high-throughput phosphorylation screening technologies in clinical settings. In the United States, the FDA regulates these diagnostics primarily through the Clinical Laboratory Improvement Amendments (CLIA) and the medical device regulatory framework. Phosphorylation-based tests may be classified as Laboratory Developed Tests (LDTs) or In Vitro Diagnostic devices (IVDs), with different regulatory pathways depending on their intended use and risk classification.
For high-throughput phosphorylation screening technologies seeking clinical application, developers must demonstrate analytical validity, clinical validity, and clinical utility. Analytical validity requirements include specificity, sensitivity, reproducibility, and robustness of phosphorylation detection across diverse sample types. The FDA typically requires extensive validation data showing consistent performance across different laboratories and operators.
European regulations under the In Vitro Diagnostic Regulation (IVDR) impose additional requirements, including more stringent clinical evidence standards and post-market surveillance. The IVDR specifically addresses multiplex assays and high-throughput technologies, requiring validation of each biomarker in the panel and demonstration of clinical relevance for the combined results.
Quality control considerations are particularly critical for phosphorylation-based diagnostics due to the dynamic and transient nature of phosphorylation events. Regulatory bodies require implementation of rigorous quality management systems, including standardized protocols for sample collection, handling, and storage to preserve phosphorylation states. Reference standards and calibrators specific to phosphorylated proteins must be developed and validated.
Data privacy regulations present another layer of complexity, especially for high-throughput screening generating large datasets. Compliance with GDPR in Europe and HIPAA in the US requires robust data management systems and clear consent procedures for patient samples used in test development and validation.
Reimbursement pathways for novel phosphorylation-based diagnostics remain challenging, with payers requiring strong evidence of clinical utility and cost-effectiveness. Developers must engage early with both regulatory agencies and payers to align evidence generation strategies that satisfy both regulatory approval and coverage determination requirements.
International harmonization efforts through organizations like the International Medical Device Regulators Forum (IMDRF) are working to standardize requirements for novel biomarker-based diagnostics, potentially streamlining global market access for phosphorylation-based tests. Companies developing high-throughput phosphorylation screening technologies should monitor these evolving frameworks and engage proactively with regulatory agencies through pre-submission consultations.
For high-throughput phosphorylation screening technologies seeking clinical application, developers must demonstrate analytical validity, clinical validity, and clinical utility. Analytical validity requirements include specificity, sensitivity, reproducibility, and robustness of phosphorylation detection across diverse sample types. The FDA typically requires extensive validation data showing consistent performance across different laboratories and operators.
European regulations under the In Vitro Diagnostic Regulation (IVDR) impose additional requirements, including more stringent clinical evidence standards and post-market surveillance. The IVDR specifically addresses multiplex assays and high-throughput technologies, requiring validation of each biomarker in the panel and demonstration of clinical relevance for the combined results.
Quality control considerations are particularly critical for phosphorylation-based diagnostics due to the dynamic and transient nature of phosphorylation events. Regulatory bodies require implementation of rigorous quality management systems, including standardized protocols for sample collection, handling, and storage to preserve phosphorylation states. Reference standards and calibrators specific to phosphorylated proteins must be developed and validated.
Data privacy regulations present another layer of complexity, especially for high-throughput screening generating large datasets. Compliance with GDPR in Europe and HIPAA in the US requires robust data management systems and clear consent procedures for patient samples used in test development and validation.
Reimbursement pathways for novel phosphorylation-based diagnostics remain challenging, with payers requiring strong evidence of clinical utility and cost-effectiveness. Developers must engage early with both regulatory agencies and payers to align evidence generation strategies that satisfy both regulatory approval and coverage determination requirements.
International harmonization efforts through organizations like the International Medical Device Regulators Forum (IMDRF) are working to standardize requirements for novel biomarker-based diagnostics, potentially streamlining global market access for phosphorylation-based tests. Companies developing high-throughput phosphorylation screening technologies should monitor these evolving frameworks and engage proactively with regulatory agencies through pre-submission consultations.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!