Phosphorylation in Biochemical Pathway Architecture: Evaluation
SEP 23, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Phosphorylation Biochemical Background and Objectives
Phosphorylation represents one of the most fundamental post-translational modifications in biological systems, playing a pivotal role in cellular signaling, metabolic regulation, and protein function modulation. The historical trajectory of phosphorylation research dates back to the 1950s when Fischer and Krebs first discovered reversible protein phosphorylation during their studies on glycogen metabolism, a breakthrough that eventually earned them the Nobel Prize in Physiology or Medicine in 1992.
The evolution of phosphorylation research has progressed through several distinct phases, beginning with basic biochemical characterization, advancing through the identification of kinases and phosphatases, and culminating in our current understanding of complex phosphorylation networks and cascades. Modern technological developments, particularly in mass spectrometry and phosphoproteomics, have revolutionized our ability to identify and quantify phosphorylation events on a global scale.
Current trends in phosphorylation research focus on systems-level understanding of phosphorylation networks, the temporal dynamics of phosphorylation events, and the integration of phosphorylation with other post-translational modifications. The emergence of artificial intelligence and machine learning approaches has further enhanced our ability to predict phosphorylation sites and interpret their functional significance within biochemical pathways.
The architectural principles governing phosphorylation in biochemical pathways reveal sophisticated design features, including amplification mechanisms, feedback and feedforward loops, and scaffold-mediated organization. These architectural elements confer specific functional properties to signaling networks, such as ultrasensitivity, bistability, and adaptation, which are essential for robust cellular responses to environmental stimuli.
The primary objectives of this technical evaluation are multifaceted. First, we aim to comprehensively assess the current understanding of phosphorylation mechanisms within the context of biochemical pathway architecture. Second, we seek to identify emerging technologies and methodologies that enable more precise manipulation and measurement of phosphorylation events. Third, we intend to evaluate the potential applications of phosphorylation engineering in biotechnology, medicine, and synthetic biology.
Additionally, this evaluation will explore the computational frameworks and mathematical models that have been developed to predict and simulate phosphorylation dynamics in complex biological systems. The integration of experimental data with computational models represents a particularly promising approach for advancing our understanding of phosphorylation networks and their functional implications.
Finally, we will establish clear metrics for evaluating the efficiency, specificity, and physiological relevance of phosphorylation events within biochemical pathways, with the ultimate goal of developing strategies to modulate these events for therapeutic or biotechnological applications.
The evolution of phosphorylation research has progressed through several distinct phases, beginning with basic biochemical characterization, advancing through the identification of kinases and phosphatases, and culminating in our current understanding of complex phosphorylation networks and cascades. Modern technological developments, particularly in mass spectrometry and phosphoproteomics, have revolutionized our ability to identify and quantify phosphorylation events on a global scale.
Current trends in phosphorylation research focus on systems-level understanding of phosphorylation networks, the temporal dynamics of phosphorylation events, and the integration of phosphorylation with other post-translational modifications. The emergence of artificial intelligence and machine learning approaches has further enhanced our ability to predict phosphorylation sites and interpret their functional significance within biochemical pathways.
The architectural principles governing phosphorylation in biochemical pathways reveal sophisticated design features, including amplification mechanisms, feedback and feedforward loops, and scaffold-mediated organization. These architectural elements confer specific functional properties to signaling networks, such as ultrasensitivity, bistability, and adaptation, which are essential for robust cellular responses to environmental stimuli.
The primary objectives of this technical evaluation are multifaceted. First, we aim to comprehensively assess the current understanding of phosphorylation mechanisms within the context of biochemical pathway architecture. Second, we seek to identify emerging technologies and methodologies that enable more precise manipulation and measurement of phosphorylation events. Third, we intend to evaluate the potential applications of phosphorylation engineering in biotechnology, medicine, and synthetic biology.
Additionally, this evaluation will explore the computational frameworks and mathematical models that have been developed to predict and simulate phosphorylation dynamics in complex biological systems. The integration of experimental data with computational models represents a particularly promising approach for advancing our understanding of phosphorylation networks and their functional implications.
Finally, we will establish clear metrics for evaluating the efficiency, specificity, and physiological relevance of phosphorylation events within biochemical pathways, with the ultimate goal of developing strategies to modulate these events for therapeutic or biotechnological applications.
Market Applications of Phosphorylation Pathway Analysis
The phosphorylation pathway analysis market has experienced significant growth across multiple sectors, driven by increasing demand for precision medicine and advanced diagnostic tools. Pharmaceutical companies represent the largest market segment, utilizing phosphorylation analysis to develop targeted therapies that modulate specific signaling pathways implicated in diseases. This application has revolutionized cancer treatment through the development of kinase inhibitors that target aberrant phosphorylation events, with the global market for such inhibitors reaching $27 billion in 2022.
Clinical diagnostics constitutes another major application area, where phosphorylation biomarkers serve as valuable indicators for disease diagnosis, progression monitoring, and treatment response assessment. Phosphoproteomic profiling is increasingly integrated into companion diagnostic platforms, enabling personalized treatment selection based on a patient's unique pathway activation patterns. Several FDA-approved diagnostic tests now incorporate phosphorylation markers, particularly in oncology.
The agricultural biotechnology sector has adopted phosphorylation pathway analysis to develop crops with enhanced stress resistance and improved nutritional profiles. By understanding plant signaling networks, companies have engineered varieties with optimized phosphorylation-dependent responses to environmental stressors, contributing to sustainable agriculture initiatives and food security efforts.
Research institutions and academic laboratories represent a stable market segment, utilizing phosphorylation analysis tools for fundamental biological research. This sector drives innovation in analytical techniques and contributes to the discovery of novel pathway architectures that eventually translate into commercial applications.
Emerging applications include neurodegenerative disease research, where abnormal protein phosphorylation plays a critical role in conditions like Alzheimer's and Parkinson's disease. Several biotechnology companies are developing diagnostic tools and therapeutic approaches targeting these phosphorylation events, with the first such products expected to reach market within 3-5 years.
The cosmetics and anti-aging industry has begun exploring phosphorylation pathways related to cellular senescence and skin aging. Products targeting specific phosphorylation events in dermal cells are being developed to promote skin rejuvenation and address age-related changes at the molecular level.
Industrial biotechnology represents a nascent but promising application area, with companies investigating phosphorylation-based biosensors for environmental monitoring and bioremediation. These technologies leverage natural phosphorylation cascades to detect pollutants or trigger remediation processes in contaminated environments.
Clinical diagnostics constitutes another major application area, where phosphorylation biomarkers serve as valuable indicators for disease diagnosis, progression monitoring, and treatment response assessment. Phosphoproteomic profiling is increasingly integrated into companion diagnostic platforms, enabling personalized treatment selection based on a patient's unique pathway activation patterns. Several FDA-approved diagnostic tests now incorporate phosphorylation markers, particularly in oncology.
The agricultural biotechnology sector has adopted phosphorylation pathway analysis to develop crops with enhanced stress resistance and improved nutritional profiles. By understanding plant signaling networks, companies have engineered varieties with optimized phosphorylation-dependent responses to environmental stressors, contributing to sustainable agriculture initiatives and food security efforts.
Research institutions and academic laboratories represent a stable market segment, utilizing phosphorylation analysis tools for fundamental biological research. This sector drives innovation in analytical techniques and contributes to the discovery of novel pathway architectures that eventually translate into commercial applications.
Emerging applications include neurodegenerative disease research, where abnormal protein phosphorylation plays a critical role in conditions like Alzheimer's and Parkinson's disease. Several biotechnology companies are developing diagnostic tools and therapeutic approaches targeting these phosphorylation events, with the first such products expected to reach market within 3-5 years.
The cosmetics and anti-aging industry has begun exploring phosphorylation pathways related to cellular senescence and skin aging. Products targeting specific phosphorylation events in dermal cells are being developed to promote skin rejuvenation and address age-related changes at the molecular level.
Industrial biotechnology represents a nascent but promising application area, with companies investigating phosphorylation-based biosensors for environmental monitoring and bioremediation. These technologies leverage natural phosphorylation cascades to detect pollutants or trigger remediation processes in contaminated environments.
Current Phosphorylation Research Challenges
Despite significant advancements in phosphorylation research, several critical challenges continue to impede comprehensive understanding of this fundamental biochemical process. The complexity of phosphorylation networks presents a formidable obstacle, with thousands of kinases, phosphatases, and substrates forming intricate regulatory webs that defy simple mapping approaches. Current technologies struggle to capture the dynamic nature of these networks, particularly the temporal aspects of phosphorylation events that occur on millisecond timescales.
Methodological limitations persist in phosphorylation detection and quantification. While mass spectrometry has revolutionized phosphoproteomic analysis, sensitivity issues remain when dealing with low-abundance phosphoproteins or those with poor ionization properties. Additionally, the field lacks standardized protocols for sample preparation and data analysis, leading to inconsistencies across research groups and hindering comparative studies.
Spatial regulation of phosphorylation events represents another significant challenge. Current techniques provide limited insight into the subcellular localization of phosphorylation reactions, despite growing evidence that spatial organization critically influences signaling outcomes. Technologies capable of visualizing phosphorylation events with high spatial and temporal resolution in living cells remain underdeveloped.
The functional significance of identified phosphorylation sites poses a persistent question. High-throughput studies have revealed hundreds of thousands of phosphorylation sites across proteomes, yet determining which sites are physiologically relevant versus which represent "noise" in the system remains problematic. Computational approaches for predicting functional importance show promise but require further refinement.
Integration of phosphorylation data with other post-translational modifications presents another frontier challenge. Phosphorylation often works in concert with other modifications like acetylation, methylation, and ubiquitination, creating a complex "modification code" that current analytical frameworks struggle to decipher comprehensively.
Translating phosphorylation research into clinical applications faces significant hurdles. While kinase inhibitors represent a major class of therapeutic agents, developing drugs that target specific phosphorylation events with minimal off-target effects remains challenging. Furthermore, identifying reliable phosphorylation-based biomarkers for disease diagnosis and monitoring requires more robust validation approaches.
Computational modeling of phosphorylation networks represents both a challenge and opportunity. Current models often fail to capture the full complexity of these networks, particularly feedback and feedforward loops that create non-linear behaviors. Advanced mathematical frameworks and machine learning approaches show promise but require extensive experimental data for validation and refinement.
Methodological limitations persist in phosphorylation detection and quantification. While mass spectrometry has revolutionized phosphoproteomic analysis, sensitivity issues remain when dealing with low-abundance phosphoproteins or those with poor ionization properties. Additionally, the field lacks standardized protocols for sample preparation and data analysis, leading to inconsistencies across research groups and hindering comparative studies.
Spatial regulation of phosphorylation events represents another significant challenge. Current techniques provide limited insight into the subcellular localization of phosphorylation reactions, despite growing evidence that spatial organization critically influences signaling outcomes. Technologies capable of visualizing phosphorylation events with high spatial and temporal resolution in living cells remain underdeveloped.
The functional significance of identified phosphorylation sites poses a persistent question. High-throughput studies have revealed hundreds of thousands of phosphorylation sites across proteomes, yet determining which sites are physiologically relevant versus which represent "noise" in the system remains problematic. Computational approaches for predicting functional importance show promise but require further refinement.
Integration of phosphorylation data with other post-translational modifications presents another frontier challenge. Phosphorylation often works in concert with other modifications like acetylation, methylation, and ubiquitination, creating a complex "modification code" that current analytical frameworks struggle to decipher comprehensively.
Translating phosphorylation research into clinical applications faces significant hurdles. While kinase inhibitors represent a major class of therapeutic agents, developing drugs that target specific phosphorylation events with minimal off-target effects remains challenging. Furthermore, identifying reliable phosphorylation-based biomarkers for disease diagnosis and monitoring requires more robust validation approaches.
Computational modeling of phosphorylation networks represents both a challenge and opportunity. Current models often fail to capture the full complexity of these networks, particularly feedback and feedforward loops that create non-linear behaviors. Advanced mathematical frameworks and machine learning approaches show promise but require extensive experimental data for validation and refinement.
Established Phosphorylation Detection and Analysis Methods
01 Computational modeling of phosphorylation pathways
Computational methods and systems for modeling phosphorylation biochemical pathways, including simulation algorithms that predict protein interactions and signal transduction cascades. These models incorporate kinetic parameters of enzymatic reactions and provide visual representations of pathway architecture to better understand cellular signaling networks and their regulation mechanisms.- Computational modeling of phosphorylation pathways: Computational methods and systems for modeling phosphorylation biochemical pathways, including simulation tools that predict protein interactions and signal transduction cascades. These models help visualize and analyze complex phosphorylation networks, enabling researchers to understand pathway architecture and dynamics. Advanced algorithms are used to integrate experimental data and create predictive models of phosphorylation events in cellular signaling.
- Detection and analysis of phosphorylation events: Methods and systems for detecting and analyzing phosphorylation events in biochemical pathways, including assays and diagnostic tools that identify phosphorylated proteins and quantify phosphorylation levels. These technologies enable researchers to map phosphorylation sites, determine kinase activities, and elucidate pathway architectures. High-throughput screening approaches allow for comprehensive analysis of phosphorylation networks and their regulation in various biological contexts.
- Therapeutic targeting of phosphorylation pathways: Therapeutic approaches targeting phosphorylation pathways for disease treatment, particularly in cancer and neurological disorders. These include development of kinase inhibitors and phosphatase modulators that can regulate specific nodes within phosphorylation cascades. By understanding the architecture of phosphorylation pathways, researchers can identify critical intervention points and develop targeted therapies that modify aberrant signaling with minimal side effects.
- Visualization tools for phosphorylation networks: Visualization technologies and software tools designed to represent complex phosphorylation pathway architectures in comprehensible formats. These tools enable researchers to map intricate signaling networks, visualize protein interactions, and identify regulatory hubs within phosphorylation cascades. Advanced graphical interfaces allow for dynamic representation of phosphorylation events, facilitating the understanding of temporal and spatial aspects of signal transduction.
- Engineered phosphorylation systems: Engineered biological systems with modified phosphorylation pathways for biotechnological applications. These include synthetic biology approaches to create novel signaling circuits, biosensors based on phosphorylation events, and engineered cellular systems with customized phosphorylation architectures. By redesigning phosphorylation networks, researchers can develop cells with new functionalities for bioproduction, environmental sensing, or therapeutic applications.
02 Detection and analysis of phosphorylation events
Methods and systems for detecting and analyzing phosphorylation events in biological samples, including high-throughput screening techniques, protein arrays, and mass spectrometry approaches. These technologies enable the identification of phosphorylation sites, quantification of phosphorylation levels, and characterization of kinase-substrate relationships within complex biochemical pathways.Expand Specific Solutions03 Therapeutic targeting of phosphorylation pathways
Therapeutic approaches targeting specific components of phosphorylation pathways for disease treatment, particularly in cancer and neurological disorders. These include kinase inhibitors, phosphatase modulators, and other compounds that can regulate abnormal phosphorylation events to restore normal cellular function or induce apoptosis in diseased cells.Expand Specific Solutions04 Visualization tools for phosphorylation pathway architecture
Software tools and visualization systems designed to represent complex phosphorylation pathway architectures in an intuitive and interactive manner. These tools help researchers understand the spatial and temporal dynamics of phosphorylation cascades, identify regulatory nodes, and predict the effects of perturbations on pathway function.Expand Specific Solutions05 Engineered phosphorylation systems for biotechnology applications
Engineered phosphorylation pathways and components for biotechnological applications, including biosensors, synthetic biology constructs, and bioproduction systems. These engineered systems utilize the specificity and reversibility of phosphorylation reactions to create novel cellular functions, control gene expression, or produce valuable compounds through modified metabolic pathways.Expand Specific Solutions
Leading Research Institutions and Biotech Companies
The phosphorylation biochemical pathway architecture market is currently in a growth phase, with increasing research focus and commercial applications. The global market size is expanding rapidly due to rising demand in drug discovery, personalized medicine, and biotechnology applications. Leading players include Illumina, Inc., which dominates the sequencing technology space essential for pathway analysis, and pharmaceutical giants like Novartis AG and Merck Patent GmbH that leverage phosphorylation insights for drug development. Research institutions such as the German Cancer Research Center and University of Michigan contribute significant academic advancements. Technology maturity varies across applications, with companies like Astex Therapeutics and Insilico Medicine pioneering computational approaches to pathway analysis, while established players like Samsung Electronics and Eppendorf SE provide supporting instrumentation and equipment for biochemical research.
German Cancer Research Center (Deutsches Krebsforschungszentrum, DKFZ)
Technical Solution: DKFZ has established a comprehensive phosphoproteomics platform for evaluating phosphorylation networks in cancer progression and treatment response. Their approach combines advanced mass spectrometry techniques with bioinformatic tools to map kinase-substrate relationships with high precision. DKFZ researchers have developed methods for analyzing phosphorylation dynamics in patient-derived xenograft models, enabling translational studies of pathway activation. Their technology includes specialized sample preparation protocols that maximize phosphopeptide recovery from limited clinical specimens. DKFZ has pioneered the integration of phosphoproteomic data with other omics platforms (genomics, transcriptomics) to create multi-dimensional views of signaling pathway architecture. Their phosphorylation evaluation framework has been particularly valuable in identifying resistance mechanisms to targeted therapies and developing rational drug combinations that overcome compensatory phosphorylation events. DKFZ has established international standards for phosphoproteomic data reporting and sharing, enhancing reproducibility across research centers.
Strengths: Exceptional technical infrastructure for phosphoproteomic analysis; strong integration of basic and clinical research; established collaborations with pharmaceutical companies for translational applications. Weaknesses: European-centric research network may limit global applicability; complex organizational structure can slow technology implementation; challenges in standardizing phosphoproteomic analyses across different tumor types.
President & Fellows of Harvard College
Technical Solution: Harvard has developed innovative technologies for evaluating phosphorylation dynamics in biochemical pathways. Their approach integrates multiplexed kinase activity profiling with systems biology modeling to create comprehensive phosphorylation network maps. Harvard researchers have pioneered the use of genetically encoded biosensors that enable real-time visualization of phosphorylation events in living cells, providing unprecedented temporal and spatial resolution. Their technology platform includes computational frameworks that predict how perturbations in phosphorylation networks propagate through cellular systems, allowing for in silico modeling of drug effects. Harvard has developed phosphoproteomic methods that require minimal sample input, enabling analysis of rare cell populations and clinical specimens. Their phosphorylation evaluation systems have been particularly valuable in understanding how post-translational modifications coordinate complex cellular processes like embryonic development, immune response, and neuronal signaling.
Strengths: Cutting-edge integration of experimental and computational approaches; access to diverse scientific expertise across disciplines; strong track record of developing novel phosphorylation analysis methods. Weaknesses: Academic focus sometimes limits translation to industrial applications; fragmented research efforts across different departments; challenges in commercializing complex technological platforms.
Key Phosphorylation Regulatory Mechanisms and Patents
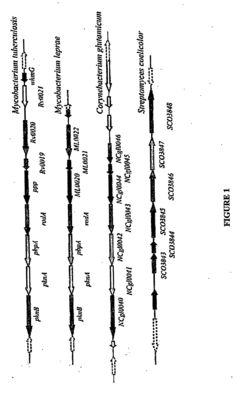
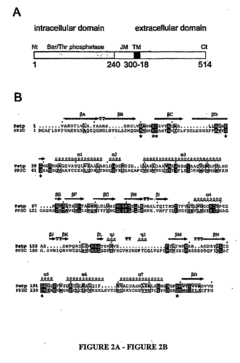
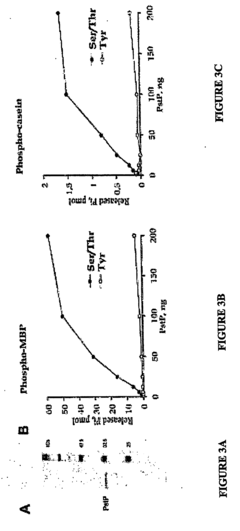
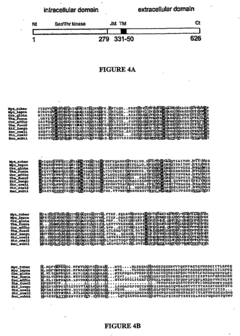
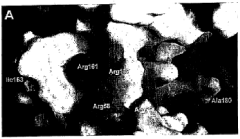
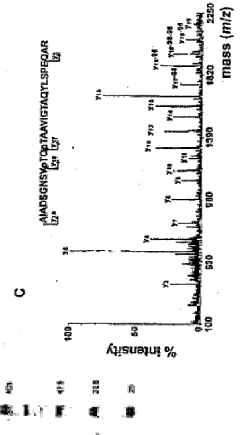
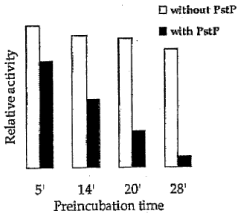
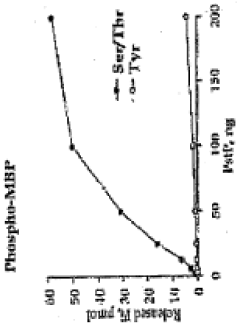
Pknb kinase and pstp phosphatase and methods of identifying inhibitory substances
PatentInactiveUS20070128678A1
Innovation
- The pknB kinase and pstP phosphatase, which are co-localized in a conserved operon with other signaling proteins, are identified and characterized for their role in regulating cell growth and latency, providing potential targets for antibacterial agents.
Pknb kinase and pstp phosphatase and methods of identifying inhibitory substances
PatentInactiveIN212DELNP2006A
Innovation
- The identification and characterization of the pknB kinase and pstP phosphatase, which are involved in signal transduction pathways, providing potential targets for developing antibacterial agents that can modulate their activity to treat Mycobacterium tuberculosis infections.
Computational Modeling of Phosphorylation Networks
Computational modeling of phosphorylation networks has emerged as a critical approach for understanding the complex dynamics of biochemical pathways. These models integrate mathematical frameworks with biological data to simulate how phosphorylation events propagate through cellular signaling networks, providing insights that would be difficult to obtain through experimental methods alone.
The development of computational models for phosphorylation networks typically involves several key components: kinetic parameters of enzymatic reactions, spatial distribution of signaling molecules, feedback and feedforward loops, and cross-talk between different pathways. Ordinary differential equation (ODE) models remain the most widely used approach, capturing the temporal dynamics of phosphorylation states with reasonable computational efficiency.
More sophisticated approaches include partial differential equation (PDE) models that incorporate spatial dimensions, stochastic models that account for the inherent randomness in biochemical reactions, and agent-based models that simulate individual molecule behaviors. Each modeling paradigm offers distinct advantages depending on the specific research questions and available data.
Recent advances in machine learning have significantly enhanced our ability to model phosphorylation networks. Deep learning algorithms can now predict phosphorylation sites with high accuracy, while reinforcement learning approaches help optimize model parameters against experimental data. These AI-driven methods are particularly valuable when dealing with incomplete knowledge of network architecture.
Multi-scale modeling represents another frontier, connecting molecular-level phosphorylation events to cellular phenotypes and even tissue-level responses. Such models integrate data across different biological scales, from protein structures to cellular behaviors, providing a more comprehensive understanding of how phosphorylation networks influence physiological and pathological states.
Challenges in computational modeling of phosphorylation networks include parameter identifiability, model validation, and computational complexity. The high dimensionality of these networks often leads to underdetermined systems where multiple parameter sets can produce similar outputs. Sensitivity analysis and parameter estimation techniques help address these challenges but require sophisticated computational resources.
The integration of phosphoproteomics data with computational models has revolutionized our understanding of signaling networks. Mass spectrometry-based approaches now provide quantitative measurements of thousands of phosphorylation sites simultaneously, creating rich datasets for model calibration and validation. This data-driven modeling approach has proven particularly valuable for identifying novel regulatory mechanisms and potential therapeutic targets in diseases characterized by dysregulated phosphorylation.
The development of computational models for phosphorylation networks typically involves several key components: kinetic parameters of enzymatic reactions, spatial distribution of signaling molecules, feedback and feedforward loops, and cross-talk between different pathways. Ordinary differential equation (ODE) models remain the most widely used approach, capturing the temporal dynamics of phosphorylation states with reasonable computational efficiency.
More sophisticated approaches include partial differential equation (PDE) models that incorporate spatial dimensions, stochastic models that account for the inherent randomness in biochemical reactions, and agent-based models that simulate individual molecule behaviors. Each modeling paradigm offers distinct advantages depending on the specific research questions and available data.
Recent advances in machine learning have significantly enhanced our ability to model phosphorylation networks. Deep learning algorithms can now predict phosphorylation sites with high accuracy, while reinforcement learning approaches help optimize model parameters against experimental data. These AI-driven methods are particularly valuable when dealing with incomplete knowledge of network architecture.
Multi-scale modeling represents another frontier, connecting molecular-level phosphorylation events to cellular phenotypes and even tissue-level responses. Such models integrate data across different biological scales, from protein structures to cellular behaviors, providing a more comprehensive understanding of how phosphorylation networks influence physiological and pathological states.
Challenges in computational modeling of phosphorylation networks include parameter identifiability, model validation, and computational complexity. The high dimensionality of these networks often leads to underdetermined systems where multiple parameter sets can produce similar outputs. Sensitivity analysis and parameter estimation techniques help address these challenges but require sophisticated computational resources.
The integration of phosphoproteomics data with computational models has revolutionized our understanding of signaling networks. Mass spectrometry-based approaches now provide quantitative measurements of thousands of phosphorylation sites simultaneously, creating rich datasets for model calibration and validation. This data-driven modeling approach has proven particularly valuable for identifying novel regulatory mechanisms and potential therapeutic targets in diseases characterized by dysregulated phosphorylation.
Therapeutic Implications of Phosphorylation Research
The therapeutic potential of phosphorylation research represents a cornerstone in modern drug development strategies. Kinase inhibitors have emerged as one of the most successful classes of targeted therapeutics, with over 60 FDA-approved drugs currently in clinical use for cancer, inflammatory disorders, and neurological conditions. These pharmaceuticals function by disrupting aberrant phosphorylation events that drive disease pathogenesis, demonstrating the direct translation of basic phosphorylation research into clinical applications.
Phosphorylation-based therapies offer distinct advantages in precision medicine approaches. By targeting specific kinases or phosphatases that are dysregulated in particular disease states, these interventions can achieve higher specificity and reduced off-target effects compared to conventional treatments. The development of isoform-selective inhibitors has further refined this approach, allowing for modulation of specific signaling pathways while preserving essential cellular functions mediated by related enzymes.
Recent advances in phosphoproteomics have revolutionized the identification of disease-specific phosphorylation signatures, enabling the development of novel therapeutic strategies. These phospho-signatures serve as both diagnostic biomarkers and therapeutic targets, facilitating patient stratification for clinical trials and treatment selection. The integration of phosphoproteomics with other omics technologies has created comprehensive disease models that reveal previously unrecognized therapeutic opportunities within phosphorylation networks.
Combination therapies targeting multiple nodes within phosphorylation cascades have shown promise in overcoming resistance mechanisms that frequently develop with single-agent approaches. By simultaneously inhibiting complementary or redundant phosphorylation pathways, these strategies can achieve synergistic effects and prevent compensatory signaling that often limits therapeutic efficacy. This network-based approach represents a paradigm shift from traditional single-target drug development.
Emerging therapeutic modalities, including proteolysis-targeting chimeras (PROTACs) and allosteric modulators, are expanding the druggable phosphoproteome beyond conventional active-site inhibition. These technologies enable the targeting of previously "undruggable" components of phosphorylation networks, including scaffold proteins and regulatory domains that lack enzymatic activity but play critical roles in signal transduction.
The development of phosphorylation-responsive drug delivery systems represents another innovative therapeutic application. These smart delivery platforms utilize disease-specific phosphorylation events to trigger controlled release of therapeutic agents at target sites, improving efficacy while reducing systemic toxicity. Such approaches exemplify how fundamental understanding of phosphorylation dynamics can inspire novel therapeutic strategies beyond direct enzyme inhibition.
Phosphorylation-based therapies offer distinct advantages in precision medicine approaches. By targeting specific kinases or phosphatases that are dysregulated in particular disease states, these interventions can achieve higher specificity and reduced off-target effects compared to conventional treatments. The development of isoform-selective inhibitors has further refined this approach, allowing for modulation of specific signaling pathways while preserving essential cellular functions mediated by related enzymes.
Recent advances in phosphoproteomics have revolutionized the identification of disease-specific phosphorylation signatures, enabling the development of novel therapeutic strategies. These phospho-signatures serve as both diagnostic biomarkers and therapeutic targets, facilitating patient stratification for clinical trials and treatment selection. The integration of phosphoproteomics with other omics technologies has created comprehensive disease models that reveal previously unrecognized therapeutic opportunities within phosphorylation networks.
Combination therapies targeting multiple nodes within phosphorylation cascades have shown promise in overcoming resistance mechanisms that frequently develop with single-agent approaches. By simultaneously inhibiting complementary or redundant phosphorylation pathways, these strategies can achieve synergistic effects and prevent compensatory signaling that often limits therapeutic efficacy. This network-based approach represents a paradigm shift from traditional single-target drug development.
Emerging therapeutic modalities, including proteolysis-targeting chimeras (PROTACs) and allosteric modulators, are expanding the druggable phosphoproteome beyond conventional active-site inhibition. These technologies enable the targeting of previously "undruggable" components of phosphorylation networks, including scaffold proteins and regulatory domains that lack enzymatic activity but play critical roles in signal transduction.
The development of phosphorylation-responsive drug delivery systems represents another innovative therapeutic application. These smart delivery platforms utilize disease-specific phosphorylation events to trigger controlled release of therapeutic agents at target sites, improving efficacy while reducing systemic toxicity. Such approaches exemplify how fundamental understanding of phosphorylation dynamics can inspire novel therapeutic strategies beyond direct enzyme inhibition.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!