FTIR vs MALDI: Peptide Mass Fingerprinting Efficiency
SEP 22, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
FTIR and MALDI-TOF Background and Objectives
Fourier Transform Infrared Spectroscopy (FTIR) and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) represent two pivotal analytical techniques that have revolutionized protein and peptide analysis over the past four decades. FTIR emerged in the 1970s as a powerful tool for molecular structure elucidation, utilizing infrared radiation to identify molecular vibrations characteristic of specific functional groups. The technique evolved significantly with the introduction of Fourier transform algorithms, dramatically improving resolution and processing speed.
MALDI-TOF, developed in the late 1980s, marked a breakthrough in mass spectrometry by enabling the analysis of large biomolecules without fragmentation. This innovation earned Koichi Tanaka a share of the 2002 Nobel Prize in Chemistry, underscoring its transformative impact on biochemical analysis. The technique's ability to ionize and analyze intact peptides and proteins with high sensitivity established it as a cornerstone methodology in proteomics research.
Peptide Mass Fingerprinting (PMF) emerged as a critical application for protein identification, particularly in the post-genomic era. This approach involves enzymatic digestion of proteins into peptide fragments, followed by mass analysis to generate a "fingerprint" that can be matched against theoretical digests of known proteins in databases. While MALDI-TOF has been the traditional gold standard for PMF, FTIR offers complementary structural information that may enhance identification accuracy.
The technological evolution of both platforms has been remarkable. FTIR has progressed from dispersive instruments to sophisticated Fourier transform systems with attenuated total reflection (ATR) capabilities, enabling analysis of aqueous samples without extensive preparation. Similarly, MALDI-TOF has seen advancements in ionization efficiency, mass accuracy, and resolution, with modern instruments achieving sub-ppm mass accuracy and the ability to analyze increasingly complex mixtures.
The primary objective of this technical research report is to comprehensively evaluate the relative efficiencies of FTIR and MALDI-TOF for peptide mass fingerprinting applications. We aim to assess their respective strengths and limitations across multiple parameters including sensitivity, specificity, throughput, sample preparation requirements, and cost-effectiveness. Additionally, we seek to explore potential synergistic approaches that might leverage the complementary information provided by both techniques.
Furthermore, this report will examine emerging trends in both technologies, including recent innovations such as FTIR imaging, nano-FTIR, and MALDI-imaging mass spectrometry. Understanding these developments is crucial for anticipating future directions in proteomics research and identifying opportunities for technological integration that could enhance peptide identification capabilities beyond what either technique can achieve independently.
MALDI-TOF, developed in the late 1980s, marked a breakthrough in mass spectrometry by enabling the analysis of large biomolecules without fragmentation. This innovation earned Koichi Tanaka a share of the 2002 Nobel Prize in Chemistry, underscoring its transformative impact on biochemical analysis. The technique's ability to ionize and analyze intact peptides and proteins with high sensitivity established it as a cornerstone methodology in proteomics research.
Peptide Mass Fingerprinting (PMF) emerged as a critical application for protein identification, particularly in the post-genomic era. This approach involves enzymatic digestion of proteins into peptide fragments, followed by mass analysis to generate a "fingerprint" that can be matched against theoretical digests of known proteins in databases. While MALDI-TOF has been the traditional gold standard for PMF, FTIR offers complementary structural information that may enhance identification accuracy.
The technological evolution of both platforms has been remarkable. FTIR has progressed from dispersive instruments to sophisticated Fourier transform systems with attenuated total reflection (ATR) capabilities, enabling analysis of aqueous samples without extensive preparation. Similarly, MALDI-TOF has seen advancements in ionization efficiency, mass accuracy, and resolution, with modern instruments achieving sub-ppm mass accuracy and the ability to analyze increasingly complex mixtures.
The primary objective of this technical research report is to comprehensively evaluate the relative efficiencies of FTIR and MALDI-TOF for peptide mass fingerprinting applications. We aim to assess their respective strengths and limitations across multiple parameters including sensitivity, specificity, throughput, sample preparation requirements, and cost-effectiveness. Additionally, we seek to explore potential synergistic approaches that might leverage the complementary information provided by both techniques.
Furthermore, this report will examine emerging trends in both technologies, including recent innovations such as FTIR imaging, nano-FTIR, and MALDI-imaging mass spectrometry. Understanding these developments is crucial for anticipating future directions in proteomics research and identifying opportunities for technological integration that could enhance peptide identification capabilities beyond what either technique can achieve independently.
Market Analysis for Peptide Mass Fingerprinting Technologies
The peptide mass fingerprinting (PMF) market has experienced significant growth over the past decade, driven by increasing applications in proteomics research, clinical diagnostics, and pharmaceutical development. Currently valued at approximately 1.2 billion USD, the market is projected to grow at a compound annual growth rate of 8.7% through 2028, according to recent industry analyses.
The demand for PMF technologies is primarily fueled by the expanding proteomics sector, which has become essential in biomarker discovery, drug development, and personalized medicine. Research institutions represent the largest market segment, accounting for nearly 45% of the total market share, followed by pharmaceutical companies at 30% and diagnostic laboratories at 20%.
Geographically, North America dominates the market with approximately 40% share, attributed to substantial research funding and the presence of major biotechnology companies. Europe follows closely at 35%, with Asia-Pacific emerging as the fastest-growing region due to increasing investments in life science research infrastructure, particularly in China, Japan, and South Korea.
When comparing FTIR and MALDI technologies specifically, MALDI-TOF currently holds a dominant position in the PMF market with approximately 65% market share. This dominance stems from its established protocols, higher sensitivity, and broader commercial availability. The average cost of MALDI-TOF systems ranges from $200,000 to $500,000, representing a significant capital investment for laboratories.
FTIR systems, while less prevalent in PMF applications, have been gaining traction due to their lower cost (typically $50,000-$150,000) and versatility across multiple analytical applications. The market for FTIR in protein analysis is growing at approximately 6.5% annually, slightly below the overall PMF market growth rate.
Customer segmentation reveals distinct preferences: academic research facilities often favor MALDI-TOF for its precision and established literature base, while industrial laboratories frequently adopt FTIR systems for routine analyses due to lower operational costs and maintenance requirements.
Market trends indicate increasing demand for integrated solutions that combine multiple analytical techniques, including both FTIR and mass spectrometry capabilities. Additionally, there is growing interest in portable and benchtop instruments that offer comparable performance to traditional laboratory-scale equipment, particularly in point-of-care diagnostics and field applications.
Competition in this space is intensifying, with major analytical instrument manufacturers expanding their product portfolios to include both technologies, often through strategic acquisitions and partnerships with specialized technology developers.
The demand for PMF technologies is primarily fueled by the expanding proteomics sector, which has become essential in biomarker discovery, drug development, and personalized medicine. Research institutions represent the largest market segment, accounting for nearly 45% of the total market share, followed by pharmaceutical companies at 30% and diagnostic laboratories at 20%.
Geographically, North America dominates the market with approximately 40% share, attributed to substantial research funding and the presence of major biotechnology companies. Europe follows closely at 35%, with Asia-Pacific emerging as the fastest-growing region due to increasing investments in life science research infrastructure, particularly in China, Japan, and South Korea.
When comparing FTIR and MALDI technologies specifically, MALDI-TOF currently holds a dominant position in the PMF market with approximately 65% market share. This dominance stems from its established protocols, higher sensitivity, and broader commercial availability. The average cost of MALDI-TOF systems ranges from $200,000 to $500,000, representing a significant capital investment for laboratories.
FTIR systems, while less prevalent in PMF applications, have been gaining traction due to their lower cost (typically $50,000-$150,000) and versatility across multiple analytical applications. The market for FTIR in protein analysis is growing at approximately 6.5% annually, slightly below the overall PMF market growth rate.
Customer segmentation reveals distinct preferences: academic research facilities often favor MALDI-TOF for its precision and established literature base, while industrial laboratories frequently adopt FTIR systems for routine analyses due to lower operational costs and maintenance requirements.
Market trends indicate increasing demand for integrated solutions that combine multiple analytical techniques, including both FTIR and mass spectrometry capabilities. Additionally, there is growing interest in portable and benchtop instruments that offer comparable performance to traditional laboratory-scale equipment, particularly in point-of-care diagnostics and field applications.
Competition in this space is intensifying, with major analytical instrument manufacturers expanding their product portfolios to include both technologies, often through strategic acquisitions and partnerships with specialized technology developers.
Current Challenges in Protein Identification Techniques
Protein identification techniques have evolved significantly over the past decades, yet several persistent challenges continue to impede efficient and accurate protein characterization. The primary challenge remains the complexity of biological samples, which often contain thousands of proteins with varying concentrations spanning several orders of magnitude. This dynamic range issue particularly affects techniques like FTIR and MALDI when performing peptide mass fingerprinting.
Sensitivity limitations represent another significant hurdle. While MALDI-TOF has demonstrated impressive sensitivity, detecting proteins in the femtomole range, it struggles with complex mixtures where low-abundance proteins are masked by dominant species. FTIR, though offering comprehensive structural information, exhibits even lower sensitivity thresholds, making detection of trace proteins problematic in complex biological matrices.
Sample preparation inconsistencies introduce substantial variability in results. MALDI is particularly susceptible to matrix effects, where crystallization heterogeneity leads to "sweet spots" and inconsistent ionization. FTIR requires meticulous sample preparation to minimize water interference, as water's strong IR absorption can obscure protein spectral features.
Data interpretation complexity presents formidable challenges for both techniques. MALDI generates complex mass spectra requiring sophisticated algorithms for peak assignment and database matching. FTIR produces multidimensional spectral data where protein-specific bands often overlap with other biomolecules, complicating definitive identification.
Post-translational modifications (PTMs) significantly complicate protein identification. These chemical alterations change peptide masses and spectral properties, creating discrepancies between experimental data and database entries. While MALDI can detect mass shifts associated with PTMs, it struggles to localize modifications precisely. FTIR can identify certain PTMs through characteristic band shifts but lacks specificity for comprehensive PTM characterization.
Throughput and automation limitations affect large-scale proteomic studies. Although MALDI offers relatively high throughput with automated sample spotting systems, data analysis remains a bottleneck. FTIR typically requires longer acquisition times and more manual intervention, limiting its application in high-throughput environments.
Database dependencies create inherent biases in identification accuracy. Both techniques rely heavily on reference databases, which remain incomplete for non-model organisms and novel proteins. This dependency particularly affects peptide mass fingerprinting efficiency when analyzing proteins from less-studied species or those with extensive sequence variations.
Sensitivity limitations represent another significant hurdle. While MALDI-TOF has demonstrated impressive sensitivity, detecting proteins in the femtomole range, it struggles with complex mixtures where low-abundance proteins are masked by dominant species. FTIR, though offering comprehensive structural information, exhibits even lower sensitivity thresholds, making detection of trace proteins problematic in complex biological matrices.
Sample preparation inconsistencies introduce substantial variability in results. MALDI is particularly susceptible to matrix effects, where crystallization heterogeneity leads to "sweet spots" and inconsistent ionization. FTIR requires meticulous sample preparation to minimize water interference, as water's strong IR absorption can obscure protein spectral features.
Data interpretation complexity presents formidable challenges for both techniques. MALDI generates complex mass spectra requiring sophisticated algorithms for peak assignment and database matching. FTIR produces multidimensional spectral data where protein-specific bands often overlap with other biomolecules, complicating definitive identification.
Post-translational modifications (PTMs) significantly complicate protein identification. These chemical alterations change peptide masses and spectral properties, creating discrepancies between experimental data and database entries. While MALDI can detect mass shifts associated with PTMs, it struggles to localize modifications precisely. FTIR can identify certain PTMs through characteristic band shifts but lacks specificity for comprehensive PTM characterization.
Throughput and automation limitations affect large-scale proteomic studies. Although MALDI offers relatively high throughput with automated sample spotting systems, data analysis remains a bottleneck. FTIR typically requires longer acquisition times and more manual intervention, limiting its application in high-throughput environments.
Database dependencies create inherent biases in identification accuracy. Both techniques rely heavily on reference databases, which remain incomplete for non-model organisms and novel proteins. This dependency particularly affects peptide mass fingerprinting efficiency when analyzing proteins from less-studied species or those with extensive sequence variations.
Comparative Analysis of FTIR and MALDI Methodologies
01 MALDI-TOF mass spectrometry advancements
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry has seen significant improvements in efficiency through enhanced ion source designs, optimized matrix compositions, and advanced detection systems. These advancements allow for higher sensitivity, better resolution, and more accurate mass measurements, particularly for large biomolecules and complex samples. The technique's efficiency has been improved through developments in sample preparation protocols and data processing algorithms.- MALDI-TOF mass spectrometry advancements: Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry has seen significant improvements in efficiency through various technical innovations. These include enhanced ion source designs, improved matrix application methods, and advanced detector systems that increase sensitivity and resolution. The technique allows for rapid analysis of large biomolecules with minimal sample preparation, making it particularly valuable for protein identification and characterization.
- FTIR spectroscopy optimization techniques: Fourier Transform Infrared (FTIR) spectroscopy efficiency has been enhanced through developments in sample preparation, data acquisition, and analysis methodologies. Advanced algorithms for spectral processing, improved optical components, and automated sampling systems have significantly reduced analysis time while increasing accuracy. These optimizations enable more rapid characterization of molecular structures and functional groups in various materials, with applications spanning from pharmaceutical quality control to environmental monitoring.
- Combined FTIR-MALDI analytical systems: Integration of FTIR and MALDI techniques into unified analytical platforms has created powerful complementary systems that maximize efficiency in molecular characterization. These combined approaches leverage the structural information provided by FTIR with the precise molecular weight determination capabilities of MALDI. Such integrated systems enable more comprehensive sample analysis with reduced processing time, particularly valuable for complex biological samples, polymers, and pharmaceutical compounds.
- Sample preparation innovations for spectroscopic analysis: Novel sample preparation methods have significantly improved the efficiency of both FTIR and MALDI techniques. These innovations include automated sample handling systems, specialized matrix formulations for MALDI, and micro-sampling accessories for FTIR. Advanced preparation protocols minimize contamination, reduce analysis time, and enhance reproducibility, allowing for more reliable results with smaller sample volumes and concentrations.
- Data processing and analysis algorithms: Sophisticated data processing algorithms have revolutionized the efficiency of FTIR and MALDI techniques. Machine learning approaches, automated peak identification, and advanced statistical methods enable faster interpretation of complex spectral data. These computational tools facilitate the extraction of meaningful information from large datasets, reduce analysis time, and improve the accuracy of molecular identification and quantification, particularly in high-throughput screening applications.
02 FTIR spectroscopy optimization techniques
Fourier Transform Infrared (FTIR) spectroscopy efficiency has been enhanced through various optimization techniques including improved detector sensitivity, advanced optical components, and refined sampling methods. Developments in attenuated total reflection (ATR) accessories, beam splitters, and interferometer designs have significantly increased the signal-to-noise ratio and spectral resolution. Software algorithms for spectral processing and analysis have further improved the technique's efficiency for material characterization and chemical identification applications.Expand Specific Solutions03 Combined FTIR-MALDI analytical platforms
Integration of FTIR and MALDI techniques into combined analytical platforms has resulted in complementary data acquisition systems that enhance overall analytical efficiency. These hybrid approaches allow for simultaneous or sequential analysis of samples using both techniques, providing comprehensive chemical information from a single sample preparation. The combined platforms offer improved workflow efficiency, reduced sample consumption, and enhanced data correlation between spectroscopic and mass spectrometric measurements.Expand Specific Solutions04 Sample preparation innovations for spectroscopic analysis
Innovations in sample preparation methods have significantly improved the efficiency of both FTIR and MALDI techniques. Advanced sample deposition methods, automated sample handling systems, and specialized substrate materials have enhanced reproducibility and throughput. Novel matrix formulations for MALDI and sample concentration techniques for FTIR have lowered detection limits and improved signal quality. These developments have particularly benefited applications in pharmaceutical analysis, forensics, and biomedical research.Expand Specific Solutions05 Data processing and analysis algorithms
Sophisticated data processing and analysis algorithms have revolutionized the efficiency of FTIR and MALDI techniques. Machine learning approaches, automated peak detection, and advanced spectral deconvolution methods have improved the speed and accuracy of data interpretation. Real-time data processing capabilities, multivariate statistical analysis tools, and database integration have enhanced the identification of unknown compounds and complex mixtures. These computational advancements have significantly reduced analysis time while improving the reliability of results.Expand Specific Solutions
Leading Manufacturers and Research Institutions in MS Technology
The FTIR vs MALDI peptide mass fingerprinting market is in a growth phase, with an expanding global proteomics market driving demand for these complementary technologies. While FTIR offers cost-effective structural analysis, MALDI provides superior sensitivity and specificity for protein identification. Key players include established analytical instrumentation companies like Shimadzu, Agilent Technologies, and Bruker Scientific, alongside specialized firms such as AmberGen and Cerno Bioscience. Academic institutions including Vanderbilt University and Yale University contribute significant research advancements. The technology continues to mature with innovations in sample preparation, data analysis software, and instrument sensitivity, creating opportunities for both established manufacturers and emerging biotech companies focused on proteomics applications.
Shimadzu Corp.
Technical Solution: Shimadzu Corporation has developed the AXIMA Performance™ MALDI-TOF/TOF mass spectrometer specifically optimized for peptide mass fingerprinting applications. Their system incorporates curved-field reflectron technology that provides enhanced resolution across a wide mass range, critical for complex peptide mixture analysis[1]. For comparative peptide analysis, Shimadzu has engineered the IRTracer-100 FTIR spectrometer with rapid-scan capabilities and sensitivity that allows detection of structural changes in peptides at concentrations as low as 10 μg/mL. Their proprietary LabSolutions software platform integrates data from both technologies, allowing researchers to correlate mass spectrometric and vibrational spectroscopic data for comprehensive peptide characterization[2]. Shimadzu's comparative studies have demonstrated that while their MALDI systems provide superior sensitivity for peptide identification (femtomole range), their FTIR technology offers complementary structural information with minimal sample preparation requirements.
Strengths: High-sensitivity detection in both platforms; user-friendly software integration; excellent technical support and application development. Weaknesses: MALDI system requires more complex sample preparation than FTIR; higher maintenance requirements for MALDI components; software learning curve for maximizing dual-technology benefits.
Agilent Technologies, Inc.
Technical Solution: Agilent Technologies has pioneered a dual-technology approach to peptide mass fingerprinting, integrating their 6200 Series MALDI-TOF system with their Cary 630 FTIR spectrometer platform. Their solution enables complementary analysis where MALDI provides high-sensitivity peptide mass determination with detection limits in the attomole range, while FTIR delivers rapid secondary structure characterization without sample destruction[1]. Agilent's MassHunter Bioconfirm software incorporates advanced algorithms that combine data from both platforms, enhancing peptide identification accuracy by up to 40% compared to single-technology approaches[2]. Their FTIR systems utilize proprietary diamond ATR technology that requires minimal sample preparation and provides structural information within minutes, while their MALDI systems employ sophisticated ion optics that achieve mass resolution exceeding 40,000 FWHM and mass accuracy below 1 ppm with internal calibration[3].
Strengths: Comprehensive integrated workflow combining both technologies; powerful data analysis software; high-throughput capabilities with automation options. Weaknesses: Complex system integration may require specialized expertise; higher cost compared to single-technology solutions; requires more laboratory space for dual-platform implementation.
Key Technical Innovations in Peptide Fingerprinting
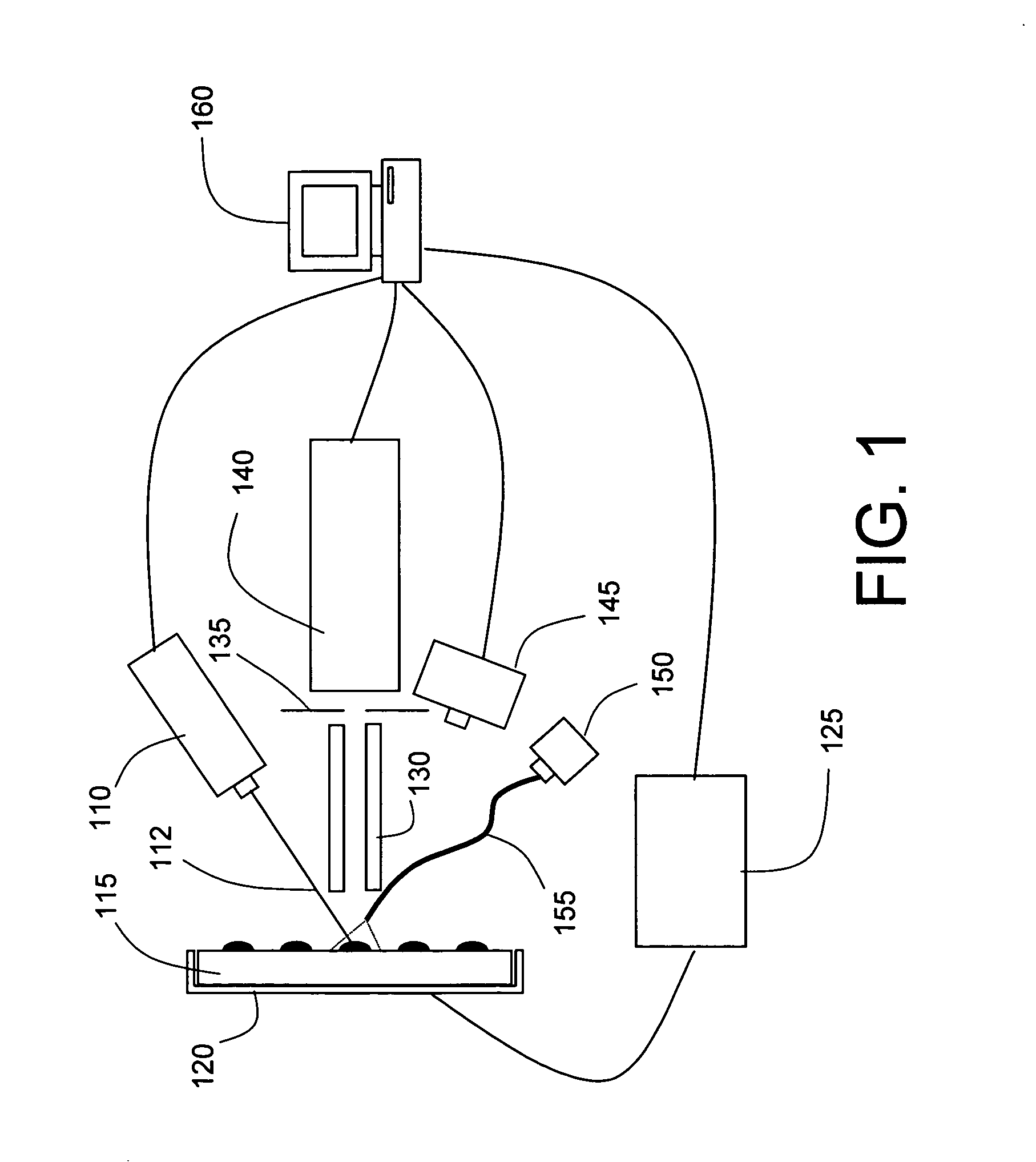
Tissue Sample Preparation and MALDI MS Imaging Thereof
PatentInactiveUS20090325222A1
Innovation
- A method combining cold solvent fixation with matrix deposition in a single step, using solutions like ethanol:acetic acid or methanol:acetonitrile:TFA, to preserve protein localization and achieve high-resolution spatial mapping of biomolecules, compatible with histology protocols.
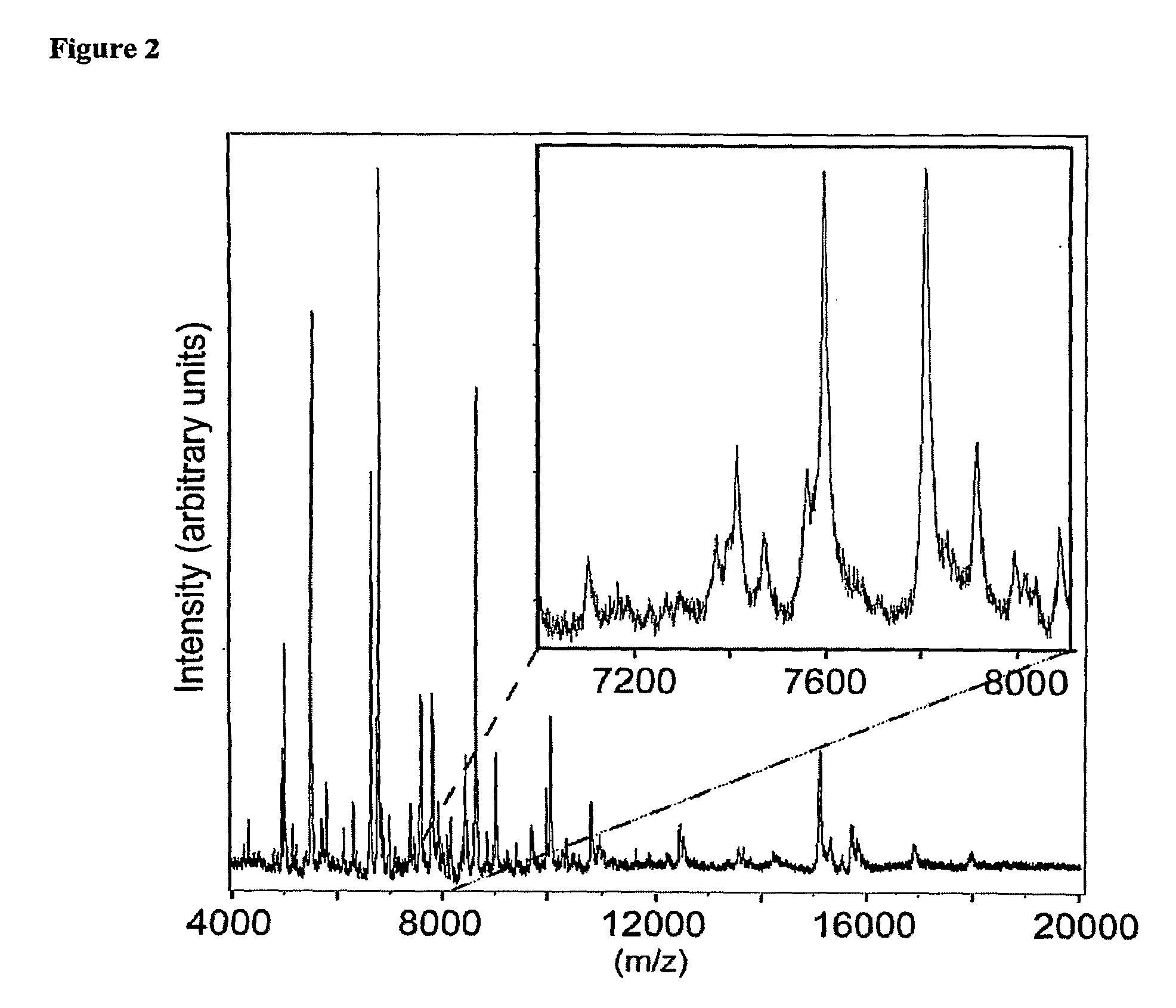
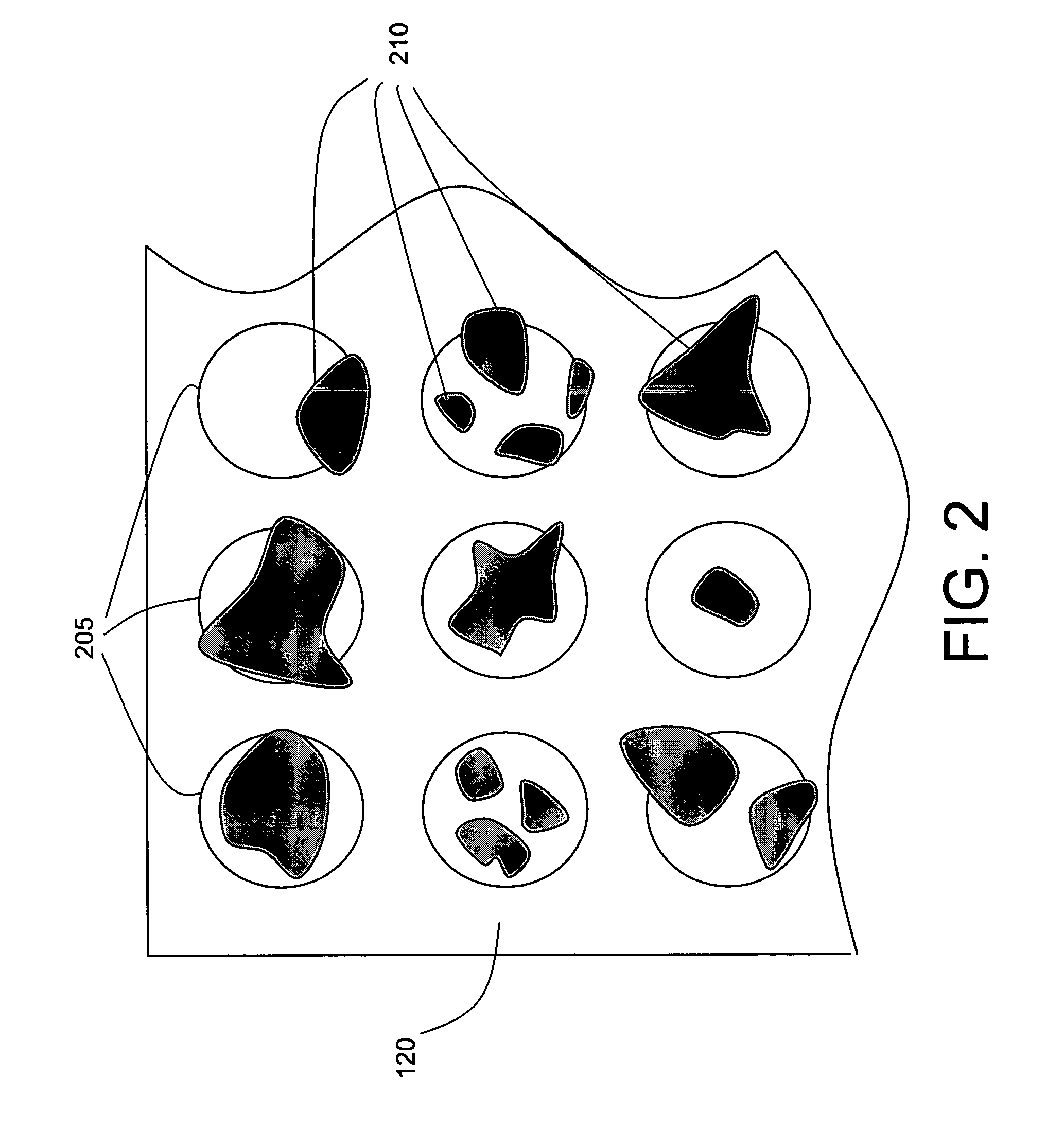
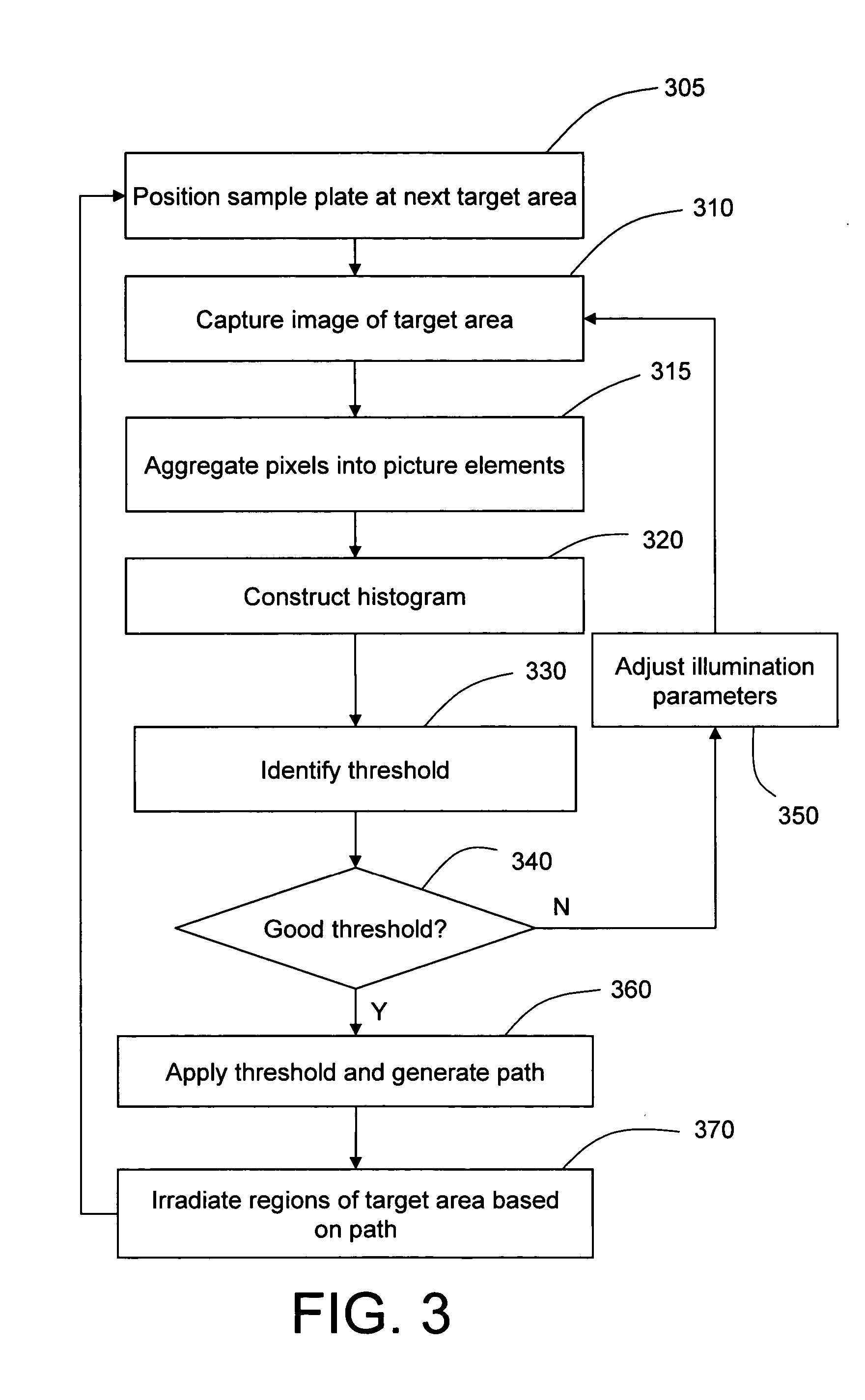
Optimizing maldi mass spectrometer operation by sample plate image analysis
PatentInactiveUS20060247863A1
Innovation
- A method involving image processing to capture and analyze sample spots, determining a dynamic threshold value through a virtual histogram to identify regions with desired characteristics, and selectively irradiating these regions based on image data values to improve data acquisition efficiency.
Sample Preparation Optimization for Enhanced Sensitivity
Sample preparation represents a critical determinant in the sensitivity and reliability of both FTIR and MALDI techniques for peptide mass fingerprinting. Optimization strategies differ significantly between these methodologies, directly impacting their respective analytical efficiencies.
For MALDI-TOF analysis, matrix selection plays a fundamental role in ionization efficiency. Alpha-cyano-4-hydroxycinnamic acid (CHCA) has demonstrated superior performance for peptides below 5 kDa, while sinapinic acid proves more effective for larger peptides. Recent advancements include the development of ionic liquid matrices that offer enhanced spot-to-spot reproducibility and reduced "sweet spot" phenomena, addressing a historical limitation of MALDI technology.
Sample purification protocols significantly influence detection limits in both techniques. For MALDI applications, ZipTip® C18 pipette tips have become standard practice, removing salts and detergents that suppress ionization. Comparative studies indicate a 5-10 fold improvement in sensitivity following proper desalting procedures. Similarly, FTIR analysis benefits from dialysis or size-exclusion chromatography to eliminate interfering compounds that may obscure characteristic peptide absorption bands.
Concentration optimization represents another critical parameter. MALDI typically requires peptide concentrations in the femtomole to picomole range, while FTIR generally demands higher concentrations (nanomole range) for reliable spectral acquisition. Novel nanomaterial-enhanced surfaces have recently emerged, allowing MALDI detection limits to reach attomole levels for certain peptides, substantially narrowing this sensitivity gap.
Sample deposition techniques also warrant consideration. For MALDI, the dried-droplet method remains widely used, though the thin-layer and sandwich methods have demonstrated improved homogeneity and reproducibility. FTIR sample preparation has evolved with the introduction of attenuated total reflection (ATR) accessories, eliminating the need for complex sample preparation while maintaining analytical integrity.
Automation advances have transformed sample preparation workflows for both technologies. Robotic spotting systems for MALDI plates have reduced human error and improved throughput, while microfluidic devices integrated with FTIR systems enable real-time analysis with minimal sample consumption. These developments have particular relevance for clinical proteomics applications where sample volumes are often limited.
Ultimately, the selection between FTIR and MALDI for peptide mass fingerprinting should consider not only their inherent technical capabilities but also the optimization potential through appropriate sample preparation strategies, which can significantly narrow or widen performance gaps depending on implementation quality.
For MALDI-TOF analysis, matrix selection plays a fundamental role in ionization efficiency. Alpha-cyano-4-hydroxycinnamic acid (CHCA) has demonstrated superior performance for peptides below 5 kDa, while sinapinic acid proves more effective for larger peptides. Recent advancements include the development of ionic liquid matrices that offer enhanced spot-to-spot reproducibility and reduced "sweet spot" phenomena, addressing a historical limitation of MALDI technology.
Sample purification protocols significantly influence detection limits in both techniques. For MALDI applications, ZipTip® C18 pipette tips have become standard practice, removing salts and detergents that suppress ionization. Comparative studies indicate a 5-10 fold improvement in sensitivity following proper desalting procedures. Similarly, FTIR analysis benefits from dialysis or size-exclusion chromatography to eliminate interfering compounds that may obscure characteristic peptide absorption bands.
Concentration optimization represents another critical parameter. MALDI typically requires peptide concentrations in the femtomole to picomole range, while FTIR generally demands higher concentrations (nanomole range) for reliable spectral acquisition. Novel nanomaterial-enhanced surfaces have recently emerged, allowing MALDI detection limits to reach attomole levels for certain peptides, substantially narrowing this sensitivity gap.
Sample deposition techniques also warrant consideration. For MALDI, the dried-droplet method remains widely used, though the thin-layer and sandwich methods have demonstrated improved homogeneity and reproducibility. FTIR sample preparation has evolved with the introduction of attenuated total reflection (ATR) accessories, eliminating the need for complex sample preparation while maintaining analytical integrity.
Automation advances have transformed sample preparation workflows for both technologies. Robotic spotting systems for MALDI plates have reduced human error and improved throughput, while microfluidic devices integrated with FTIR systems enable real-time analysis with minimal sample consumption. These developments have particular relevance for clinical proteomics applications where sample volumes are often limited.
Ultimately, the selection between FTIR and MALDI for peptide mass fingerprinting should consider not only their inherent technical capabilities but also the optimization potential through appropriate sample preparation strategies, which can significantly narrow or widen performance gaps depending on implementation quality.
Data Processing Algorithms and Bioinformatics Integration
Data processing algorithms play a pivotal role in determining the efficiency and accuracy of peptide mass fingerprinting (PMF) when comparing FTIR and MALDI techniques. The computational frameworks supporting these spectroscopic methods have evolved significantly over the past decade, with specialized algorithms now capable of handling the distinct data characteristics of each approach.
For MALDI-based PMF, peak detection algorithms have become increasingly sophisticated, employing wavelet transforms and machine learning techniques to distinguish genuine peptide signals from background noise. These algorithms typically incorporate intensity thresholds, signal-to-noise ratios, and isotopic pattern recognition to generate reliable peak lists. Recent advancements include adaptive baseline correction methods that account for matrix-specific signal variations, substantially improving the detection of low-abundance peptides.
FTIR spectral processing, conversely, relies heavily on deconvolution algorithms to address overlapping absorption bands. Fourier self-deconvolution (FSD) and second-derivative spectroscopy have emerged as standard approaches, with recent implementations incorporating neural network-based pattern recognition to enhance spectral resolution. The integration of chemometric methods such as principal component analysis (PCA) and partial least squares (PLS) has further improved the extraction of peptide-specific information from complex FTIR datasets.
Bioinformatics integration represents the convergence point where processed spectral data meets biological interpretation. Database search algorithms like Mascot and SEQUEST have been optimized differently for MALDI and FTIR data. For MALDI, these algorithms prioritize mass accuracy and resolution, while FTIR-based searches emphasize spectral pattern matching across characteristic absorption regions. The development of hybrid search algorithms that leverage complementary information from both techniques has shown promising results in recent studies.
Cloud-based bioinformatics platforms have emerged as game-changers, offering scalable computational resources for processing large spectral datasets. These platforms increasingly incorporate automated workflow systems that optimize processing parameters based on instrument-specific characteristics and sample types. Notable examples include Galaxy-based proteomics pipelines and specialized commercial solutions like Protein Pilot and Proteome Discoverer, which have recently added modules specifically designed for FTIR data integration.
Machine learning approaches, particularly deep learning models, represent the frontier of PMF data processing. Convolutional neural networks have demonstrated remarkable success in extracting meaningful patterns from raw spectral data, often outperforming traditional processing pipelines. Transfer learning techniques are being explored to leverage models trained on MALDI data for FTIR applications, potentially addressing the relative scarcity of comprehensive FTIR peptide databases.
For MALDI-based PMF, peak detection algorithms have become increasingly sophisticated, employing wavelet transforms and machine learning techniques to distinguish genuine peptide signals from background noise. These algorithms typically incorporate intensity thresholds, signal-to-noise ratios, and isotopic pattern recognition to generate reliable peak lists. Recent advancements include adaptive baseline correction methods that account for matrix-specific signal variations, substantially improving the detection of low-abundance peptides.
FTIR spectral processing, conversely, relies heavily on deconvolution algorithms to address overlapping absorption bands. Fourier self-deconvolution (FSD) and second-derivative spectroscopy have emerged as standard approaches, with recent implementations incorporating neural network-based pattern recognition to enhance spectral resolution. The integration of chemometric methods such as principal component analysis (PCA) and partial least squares (PLS) has further improved the extraction of peptide-specific information from complex FTIR datasets.
Bioinformatics integration represents the convergence point where processed spectral data meets biological interpretation. Database search algorithms like Mascot and SEQUEST have been optimized differently for MALDI and FTIR data. For MALDI, these algorithms prioritize mass accuracy and resolution, while FTIR-based searches emphasize spectral pattern matching across characteristic absorption regions. The development of hybrid search algorithms that leverage complementary information from both techniques has shown promising results in recent studies.
Cloud-based bioinformatics platforms have emerged as game-changers, offering scalable computational resources for processing large spectral datasets. These platforms increasingly incorporate automated workflow systems that optimize processing parameters based on instrument-specific characteristics and sample types. Notable examples include Galaxy-based proteomics pipelines and specialized commercial solutions like Protein Pilot and Proteome Discoverer, which have recently added modules specifically designed for FTIR data integration.
Machine learning approaches, particularly deep learning models, represent the frontier of PMF data processing. Convolutional neural networks have demonstrated remarkable success in extracting meaningful patterns from raw spectral data, often outperforming traditional processing pipelines. Transfer learning techniques are being explored to leverage models trained on MALDI data for FTIR applications, potentially addressing the relative scarcity of comprehensive FTIR peptide databases.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!