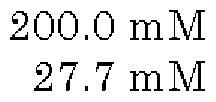Analysis of Ribosome Efficiency in Cell-free Protein Synthesis
OCT 13, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Ribosome Efficiency Background and Objectives
Cell-free protein synthesis (CFPS) has emerged as a powerful biotechnological platform over the past several decades, evolving from a research tool for understanding fundamental translation mechanisms to a versatile technology with applications spanning from protein production to synthetic biology. At the heart of this system lies the ribosome, the cellular machinery responsible for translating messenger RNA into proteins, whose efficiency directly impacts the overall productivity of CFPS systems.
The historical trajectory of ribosome research began with their discovery in the mid-20th century, followed by structural elucidation through X-ray crystallography and cryo-electron microscopy in subsequent decades. These advances have provided unprecedented insights into ribosomal architecture and function, enabling researchers to better understand the molecular mechanisms underlying protein synthesis.
Recent technological developments have shifted focus toward optimizing ribosomal performance in cell-free environments, where traditional cellular regulatory mechanisms are absent. This represents both a challenge and an opportunity for enhancing protein production efficiency through targeted engineering approaches.
The evolution of CFPS systems has seen significant improvements in extract preparation methods, energy regeneration systems, and reaction conditions. However, ribosome functionality remains a critical bottleneck in achieving industrial-scale production capabilities. Current research indicates that ribosomes in cell-free systems operate at approximately 20-30% of their theoretical maximum efficiency, highlighting substantial room for improvement.
Global research efforts are increasingly directed toward understanding and enhancing ribosomal performance parameters, including translation initiation rates, elongation efficiency, recycling dynamics, and overall stability in cell-free environments. These efforts span multiple disciplines, incorporating insights from structural biology, biochemistry, systems biology, and computational modeling.
The primary objectives of current ribosome efficiency research include: quantifying and characterizing factors limiting ribosomal performance in cell-free systems; developing novel methodologies for ribosome isolation, modification, and stabilization; engineering ribosomes with enhanced catalytic properties or expanded capabilities; and integrating optimized ribosomes into next-generation CFPS platforms for improved productivity and functionality.
Achieving these objectives would significantly advance the field toward realizing the full potential of CFPS technology for applications ranging from rapid protein production for structural studies to on-demand synthesis of therapeutics, biosensors, and novel biomaterials. The ultimate goal remains developing highly efficient, scalable, and economically viable cell-free protein synthesis systems where ribosomal performance approaches its theoretical maximum.
The historical trajectory of ribosome research began with their discovery in the mid-20th century, followed by structural elucidation through X-ray crystallography and cryo-electron microscopy in subsequent decades. These advances have provided unprecedented insights into ribosomal architecture and function, enabling researchers to better understand the molecular mechanisms underlying protein synthesis.
Recent technological developments have shifted focus toward optimizing ribosomal performance in cell-free environments, where traditional cellular regulatory mechanisms are absent. This represents both a challenge and an opportunity for enhancing protein production efficiency through targeted engineering approaches.
The evolution of CFPS systems has seen significant improvements in extract preparation methods, energy regeneration systems, and reaction conditions. However, ribosome functionality remains a critical bottleneck in achieving industrial-scale production capabilities. Current research indicates that ribosomes in cell-free systems operate at approximately 20-30% of their theoretical maximum efficiency, highlighting substantial room for improvement.
Global research efforts are increasingly directed toward understanding and enhancing ribosomal performance parameters, including translation initiation rates, elongation efficiency, recycling dynamics, and overall stability in cell-free environments. These efforts span multiple disciplines, incorporating insights from structural biology, biochemistry, systems biology, and computational modeling.
The primary objectives of current ribosome efficiency research include: quantifying and characterizing factors limiting ribosomal performance in cell-free systems; developing novel methodologies for ribosome isolation, modification, and stabilization; engineering ribosomes with enhanced catalytic properties or expanded capabilities; and integrating optimized ribosomes into next-generation CFPS platforms for improved productivity and functionality.
Achieving these objectives would significantly advance the field toward realizing the full potential of CFPS technology for applications ranging from rapid protein production for structural studies to on-demand synthesis of therapeutics, biosensors, and novel biomaterials. The ultimate goal remains developing highly efficient, scalable, and economically viable cell-free protein synthesis systems where ribosomal performance approaches its theoretical maximum.
Market Analysis for Cell-free Protein Synthesis
The global market for cell-free protein synthesis (CFPS) has experienced significant growth in recent years, driven by increasing demand for rapid protein production methods across various industries. The market was valued at approximately $250 million in 2021 and is projected to reach $500 million by 2026, representing a compound annual growth rate (CAGR) of 14.9%. This growth trajectory reflects the expanding applications of CFPS technology beyond traditional research settings.
Pharmaceutical and biotechnology sectors currently dominate the CFPS market, accounting for nearly 60% of the total market share. These industries leverage CFPS for drug discovery, protein engineering, and therapeutic protein production. The ability to rapidly synthesize proteins without the constraints of living cells makes CFPS particularly valuable for high-throughput screening and optimization of protein-based therapeutics.
Academic and research institutions constitute the second-largest market segment, utilizing CFPS systems for fundamental research in protein science, synthetic biology, and metabolic engineering. The diagnostic sector represents a rapidly growing segment, with CFPS enabling the development of point-of-care diagnostic tools and biosensors that can detect pathogens or biomarkers with high sensitivity and specificity.
Geographically, North America leads the CFPS market with approximately 45% market share, followed by Europe (30%) and Asia-Pacific (20%). The Asia-Pacific region, particularly China, Japan, and South Korea, is expected to witness the fastest growth due to increasing investments in biotechnology research and development.
Key market drivers include the rising demand for personalized medicine, advancements in synthetic biology, and the need for rapid protein production methods in vaccine development—a need highlighted during the COVID-19 pandemic. The ability of CFPS to produce complex proteins with post-translational modifications has expanded its application potential in therapeutic protein production.
Market challenges include high costs associated with reagents and extract preparation, scalability issues for industrial production, and regulatory uncertainties surrounding CFPS-derived products. The cost per reaction remains significantly higher than traditional cell-based expression systems, limiting widespread adoption in cost-sensitive applications.
Customer segments are diversifying beyond traditional research laboratories to include pharmaceutical companies, diagnostic manufacturers, and synthetic biology startups. These emerging customers are particularly interested in CFPS platforms that offer improved ribosome efficiency, as this directly impacts protein yield and production costs—critical factors for commercial viability.
Pharmaceutical and biotechnology sectors currently dominate the CFPS market, accounting for nearly 60% of the total market share. These industries leverage CFPS for drug discovery, protein engineering, and therapeutic protein production. The ability to rapidly synthesize proteins without the constraints of living cells makes CFPS particularly valuable for high-throughput screening and optimization of protein-based therapeutics.
Academic and research institutions constitute the second-largest market segment, utilizing CFPS systems for fundamental research in protein science, synthetic biology, and metabolic engineering. The diagnostic sector represents a rapidly growing segment, with CFPS enabling the development of point-of-care diagnostic tools and biosensors that can detect pathogens or biomarkers with high sensitivity and specificity.
Geographically, North America leads the CFPS market with approximately 45% market share, followed by Europe (30%) and Asia-Pacific (20%). The Asia-Pacific region, particularly China, Japan, and South Korea, is expected to witness the fastest growth due to increasing investments in biotechnology research and development.
Key market drivers include the rising demand for personalized medicine, advancements in synthetic biology, and the need for rapid protein production methods in vaccine development—a need highlighted during the COVID-19 pandemic. The ability of CFPS to produce complex proteins with post-translational modifications has expanded its application potential in therapeutic protein production.
Market challenges include high costs associated with reagents and extract preparation, scalability issues for industrial production, and regulatory uncertainties surrounding CFPS-derived products. The cost per reaction remains significantly higher than traditional cell-based expression systems, limiting widespread adoption in cost-sensitive applications.
Customer segments are diversifying beyond traditional research laboratories to include pharmaceutical companies, diagnostic manufacturers, and synthetic biology startups. These emerging customers are particularly interested in CFPS platforms that offer improved ribosome efficiency, as this directly impacts protein yield and production costs—critical factors for commercial viability.
Current Challenges in Ribosome Performance
Despite significant advancements in cell-free protein synthesis (CFPS) systems, ribosome performance remains a critical bottleneck limiting overall system efficiency. Current CFPS platforms typically utilize ribosomes extracted from cellular lysates, which often exhibit suboptimal activity in the artificial environment. These ribosomes, evolved for functioning within the complex cellular milieu, face numerous challenges when operating in cell-free conditions that lack the sophisticated regulatory mechanisms present in living cells.
One major challenge is the rapid degradation of ribosomal components during the CFPS reaction. Studies have shown that ribosomal RNA and associated proteins can degrade within hours, significantly reducing the productive lifetime of the system. This degradation is particularly problematic for industrial applications requiring extended production periods and has been linked to the presence of nucleases and proteases that remain active in cell extracts despite purification efforts.
Energy limitations represent another significant constraint on ribosome performance. Translation is an energy-intensive process, consuming approximately 4 ATP equivalents per peptide bond formed. In cell-free systems, energy resources are finite and quickly depleted, leading to premature termination of protein synthesis. Current energy regeneration systems often fail to maintain optimal ATP/GTP levels throughout the reaction duration, resulting in declining ribosome activity over time.
The coupling efficiency between transcription and translation presents additional challenges. In living cells, these processes are tightly coordinated, with ribosomes often initiating translation on nascent mRNA transcripts. This coupling is frequently disrupted in cell-free systems, leading to reduced efficiency and increased resource wastage as ribosomes struggle to locate and engage with mRNA templates effectively.
Ribosome heterogeneity further complicates performance optimization. Extracted ribosomes exhibit variable activity levels depending on their source cells' growth phase and extraction methods. Recent research has revealed that only a fraction of ribosomes in typical CFPS reactions (approximately 30-50%) actively participate in translation, with the remainder remaining functionally inactive despite consuming valuable resources.
The lack of proper ribosome recycling mechanisms represents another significant challenge. In cellular environments, specialized factors facilitate ribosome disassembly and recycling after termination. These mechanisms are often compromised in cell-free systems, leading to ribosome sequestration on terminated mRNAs and reducing the pool of available ribosomes for new translation initiation events.
Additionally, the artificial buffer conditions of CFPS systems rarely provide the optimal ionic environment for ribosome function. Ribosomes have evolved to operate within the precise ionic conditions maintained by cellular homeostasis, and deviations in magnesium, potassium, and other ion concentrations can dramatically impact their structural integrity and catalytic efficiency.
One major challenge is the rapid degradation of ribosomal components during the CFPS reaction. Studies have shown that ribosomal RNA and associated proteins can degrade within hours, significantly reducing the productive lifetime of the system. This degradation is particularly problematic for industrial applications requiring extended production periods and has been linked to the presence of nucleases and proteases that remain active in cell extracts despite purification efforts.
Energy limitations represent another significant constraint on ribosome performance. Translation is an energy-intensive process, consuming approximately 4 ATP equivalents per peptide bond formed. In cell-free systems, energy resources are finite and quickly depleted, leading to premature termination of protein synthesis. Current energy regeneration systems often fail to maintain optimal ATP/GTP levels throughout the reaction duration, resulting in declining ribosome activity over time.
The coupling efficiency between transcription and translation presents additional challenges. In living cells, these processes are tightly coordinated, with ribosomes often initiating translation on nascent mRNA transcripts. This coupling is frequently disrupted in cell-free systems, leading to reduced efficiency and increased resource wastage as ribosomes struggle to locate and engage with mRNA templates effectively.
Ribosome heterogeneity further complicates performance optimization. Extracted ribosomes exhibit variable activity levels depending on their source cells' growth phase and extraction methods. Recent research has revealed that only a fraction of ribosomes in typical CFPS reactions (approximately 30-50%) actively participate in translation, with the remainder remaining functionally inactive despite consuming valuable resources.
The lack of proper ribosome recycling mechanisms represents another significant challenge. In cellular environments, specialized factors facilitate ribosome disassembly and recycling after termination. These mechanisms are often compromised in cell-free systems, leading to ribosome sequestration on terminated mRNAs and reducing the pool of available ribosomes for new translation initiation events.
Additionally, the artificial buffer conditions of CFPS systems rarely provide the optimal ionic environment for ribosome function. Ribosomes have evolved to operate within the precise ionic conditions maintained by cellular homeostasis, and deviations in magnesium, potassium, and other ion concentrations can dramatically impact their structural integrity and catalytic efficiency.
Current Ribosome Engineering Solutions
01 Optimization of ribosome binding sites
Enhancing ribosome binding site (RBS) sequences can significantly improve translation efficiency. By optimizing the Shine-Dalgarno sequence and adjusting the spacing between the RBS and start codon, researchers can increase the affinity of ribosomes to mRNA, leading to more efficient protein synthesis. These optimizations can be achieved through computational modeling and experimental validation to create ideal ribosome binding conditions.- Optimization of ribosome binding sites: Enhancing ribosome efficiency through optimization of ribosome binding sites (RBS) is a key approach in biotechnology. By modifying the sequence and structure of RBS, researchers can improve the initiation of translation, which is often a rate-limiting step in protein synthesis. This optimization can lead to increased protein production yields and more efficient use of cellular resources. Various computational tools and experimental methods have been developed to design optimal RBS sequences for specific applications.
- Modification of tRNA and translation factors: Ribosome efficiency can be enhanced through modifications of transfer RNA (tRNA) and translation factors. By engineering tRNAs with improved anticodon recognition or stability, or by modifying translation factors that assist in the various stages of protein synthesis, researchers can overcome bottlenecks in the translation process. These approaches can lead to faster elongation rates, reduced mistranslation, and overall improved ribosomal performance in both natural and synthetic biological systems.
- Genetic engineering for improved ribosomal proteins: Genetic engineering techniques can be used to modify ribosomal proteins and rRNA to enhance ribosome efficiency. By introducing specific mutations or modifications to the ribosomal components, researchers can create ribosomes with improved catalytic activity, stability, or specificity. These engineered ribosomes can exhibit enhanced translation rates, better tolerance to environmental stresses, or the ability to preferentially translate specific mRNAs, leading to applications in biotechnology, medicine, and synthetic biology.
- Codon optimization strategies: Codon optimization is a powerful strategy to enhance ribosome efficiency by adapting the codon usage in heterologous genes to match the preferred codons of the host organism. This approach minimizes ribosome stalling caused by rare codons and optimizes the translation elongation rate. Advanced algorithms and experimental methods have been developed to design codon-optimized sequences that consider factors such as mRNA secondary structure, GC content, and codon context, resulting in significantly improved protein expression levels.
- Cell-free translation systems for enhanced ribosome performance: Cell-free translation systems provide a controlled environment for studying and enhancing ribosome efficiency. By removing cellular constraints and allowing direct manipulation of translation components, researchers can optimize conditions for maximal ribosomal activity. These systems enable the addition of specific factors to enhance translation initiation, elongation, or termination, as well as the incorporation of non-natural amino acids. Cell-free systems have applications in protein production, synthetic biology, and the development of novel therapeutics.
02 Modification of tRNA and aminoacyl-tRNA synthetases
Modifications to transfer RNA (tRNA) and aminoacyl-tRNA synthetases can enhance ribosomal efficiency by improving the delivery of amino acids to the ribosome during translation. These modifications can include altering tRNA abundance, optimizing codon-anticodon interactions, and engineering synthetases with improved catalytic properties. Such approaches lead to faster and more accurate protein synthesis, particularly when incorporating non-standard amino acids.Expand Specific Solutions03 Genetic engineering of ribosomal components
Direct modification of ribosomal RNA (rRNA) and ribosomal proteins through genetic engineering can enhance translational efficiency. By altering specific nucleotides in rRNA or amino acids in ribosomal proteins, researchers can create ribosomes with improved catalytic activity, better mRNA binding capabilities, or enhanced resistance to inhibitory factors. These engineered ribosomes demonstrate increased protein synthesis rates and can be tailored for specific translation tasks.Expand Specific Solutions04 Regulation of ribosome biogenesis and assembly
Controlling the processes of ribosome biogenesis and assembly can optimize ribosomal efficiency. This includes regulating the expression of ribosomal proteins, modifying ribosome assembly factors, and optimizing the cellular environment for ribosome formation. Improved assembly leads to structurally sound ribosomes with enhanced functional capabilities, resulting in more efficient protein synthesis and cellular growth.Expand Specific Solutions05 Development of translation enhancers and inhibitor removal
Various compounds and molecular strategies can enhance ribosomal efficiency by either directly promoting translation or removing inhibitory factors. These include small molecules that stabilize ribosome-mRNA interactions, peptides that facilitate ribosome recycling, and methods to eliminate translation inhibitors. Additionally, certain RNA elements can be incorporated into mRNA to enhance ribosome recruitment and translation initiation, leading to overall improved protein synthesis efficiency.Expand Specific Solutions
Key Industry Players in CFPS Technology
The cell-free protein synthesis (CFPS) market is currently in a growth phase, with an estimated global market size of $250-300 million and projected annual growth of 8-10%. The technology has evolved from early research stages to commercial applications, particularly in pharmaceutical development. Technical maturity varies across applications, with companies demonstrating different specialization levels. Sutro Biopharma leads with its proprietary XpressCF platform for biopharmaceutical development, while Shimadzu and QIAGEN provide supporting analytical technologies. Academic institutions like Cornell, Tsinghua, and Kyoto University contribute fundamental research advancements. Emerging players like Spiber and Kangma are applying CFPS to novel areas including synthetic proteins and diagnostic tools, indicating the technology's expanding commercial potential beyond traditional pharmaceutical applications.
Riken Corp.
Technical Solution: Riken has developed the PURE (Protein synthesis Using Recombinant Elements) system, a reconstituted cell-free protein synthesis platform that provides unprecedented control over ribosomal efficiency. Unlike extract-based systems, PURE consists of individually purified and reconstituted components of the E. coli translation machinery, including ribosomes, translation factors, and enzymes. This approach allows precise manipulation of each component's concentration to optimize ribosomal performance. Riken's researchers have systematically analyzed how different translation factors affect ribosome processivity and fidelity, leading to optimized formulations for different protein classes. Their system includes modified release factors that enhance termination efficiency and reduce premature translation termination. Riken has also developed computational models that predict ribosome loading rates and elongation dynamics, allowing rational design of mRNA templates with optimized codon usage and secondary structures to maximize ribosomal throughput.
Strengths: Offers unparalleled control over the translation process by using defined components rather than crude extracts. The system provides exceptional reproducibility and consistency between batches. Weaknesses: The PURE system typically yields lower protein amounts compared to extract-based systems. The requirement for multiple purified components makes it significantly more expensive than extract-based alternatives.
Sutro Biopharma, Inc.
Technical Solution: Sutro Biopharma has developed a proprietary cell-free protein synthesis platform called XpressCF that specifically addresses ribosome efficiency challenges. Their system utilizes an E. coli extract optimized for high-yield protein production with modified ribosomes that demonstrate enhanced translation efficiency. The platform incorporates specialized RNA polymerases and optimized reaction conditions that maintain ribosomal activity for extended periods. Sutro's technology enables site-specific incorporation of non-natural amino acids through engineered orthogonal tRNA-synthetase pairs that work efficiently with their modified ribosomes. Their approach includes real-time monitoring of ribosomal activity using fluorescent reporters to optimize reaction conditions dynamically, resulting in protein yields up to 2 mg/mL in batch reactions.
Strengths: Proprietary technology allows for precise control of the translation machinery, enabling incorporation of non-canonical amino acids with high specificity. Their system maintains ribosomal activity for extended periods (up to 24 hours) compared to conventional systems. Weaknesses: The platform requires specialized reagents and equipment, making it less accessible for general research applications. The system is primarily optimized for therapeutic protein production rather than general research applications.
Critical Patents in Ribosome Optimization
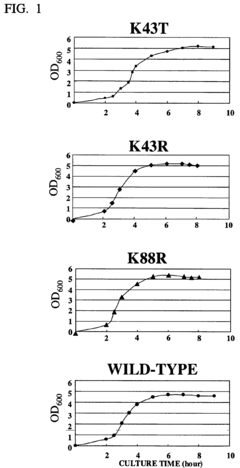
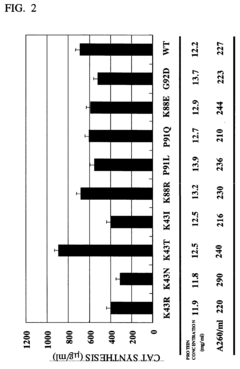
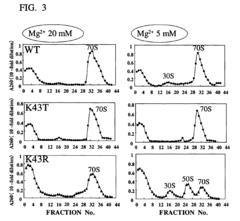
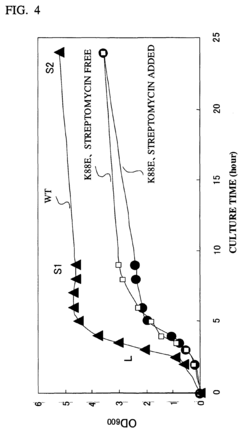
CELL EXTRACT OF i ESCHERICHIA COLI /i HAVING MUTATION IN S12 RIBOSOMAL PROTEIN AND PROCESS FOR PRODUCING PROTEIN IN CELL-FREE SYSTEM USING THE SAME
PatentInactiveEP1582582A1
Innovation
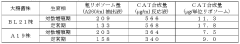
- A cell extract is prepared from E. coli with a mutation in the ribosomal protein S12 gene, specifically a substitution at position 43 from lysine to threonine, which enhances protein synthesis activity and resistance to streptomycin, allowing for improved codon recognition accuracy and increased protein production.
Process for producing extract for cell-free protein synthesis and cell extract produced thereby
PatentWO2004104209A1
Innovation
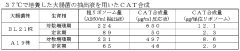
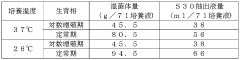
- Culturing Escherichia coli under suppressed growth conditions, specifically at temperatures between 20-32°C, to produce a cell extract with improved ribosome stability and protein synthesis activity, which is then used in a cell-free protein synthesis system with linear DNA as a template.
Scalability Factors in CFPS Systems
Scaling up Cell-Free Protein Synthesis (CFPS) systems represents a critical challenge for transitioning this technology from laboratory research to industrial applications. The scalability of CFPS is influenced by multiple interconnected factors that must be carefully optimized to maintain or enhance protein production efficiency at larger volumes.
Reaction vessel design emerges as a primary consideration, with surface-to-volume ratios significantly impacting oxygen transfer rates and heat distribution. As reaction volumes increase, traditional laboratory vessels become inadequate, necessitating specialized bioreactors with enhanced mixing capabilities and precise temperature control systems. Research indicates that continuous-flow reactors may offer superior performance for large-scale operations compared to batch processes.
Energy regeneration systems face particular challenges during scale-up. The ATP regeneration pathways that function efficiently at microliter scales often encounter limitations in larger volumes due to byproduct accumulation and substrate depletion gradients. Secondary energy regeneration systems utilizing phosphoenolpyruvate or creatine phosphate demonstrate varying efficiency profiles across different scales, requiring recalibration of component concentrations when transitioning to larger volumes.
Ribosome performance represents another critical scalability factor. Studies reveal that ribosome activity can decrease by 15-30% when scaling from microliter to liter volumes, attributed to factors including oxygen limitation, pH drift, and accumulation of inhibitory byproducts. Maintaining optimal ribosomal efficiency at scale requires supplementation strategies and enhanced mixing protocols.
Resource distribution homogeneity becomes increasingly problematic at larger scales. Concentration gradients of amino acids, nucleotides, and cofactors can develop in poorly mixed systems, creating microenvironments with suboptimal conditions for protein synthesis. Advanced mixing technologies and feeding strategies have been developed to address these challenges, including fed-batch approaches that maintain critical component concentrations within optimal ranges.
Economic considerations ultimately determine industrial feasibility. The cost structure of CFPS changes dramatically with scale, as reagent costs decrease through bulk purchasing while equipment and operational expenses increase. Analysis of production economics indicates that medium-scale (10-100L) CFPS systems currently offer the most favorable balance between economies of scale and technical challenges, though continued innovation in bioreactor design and process engineering continues to push the boundaries of economically viable scales.
Reaction vessel design emerges as a primary consideration, with surface-to-volume ratios significantly impacting oxygen transfer rates and heat distribution. As reaction volumes increase, traditional laboratory vessels become inadequate, necessitating specialized bioreactors with enhanced mixing capabilities and precise temperature control systems. Research indicates that continuous-flow reactors may offer superior performance for large-scale operations compared to batch processes.
Energy regeneration systems face particular challenges during scale-up. The ATP regeneration pathways that function efficiently at microliter scales often encounter limitations in larger volumes due to byproduct accumulation and substrate depletion gradients. Secondary energy regeneration systems utilizing phosphoenolpyruvate or creatine phosphate demonstrate varying efficiency profiles across different scales, requiring recalibration of component concentrations when transitioning to larger volumes.
Ribosome performance represents another critical scalability factor. Studies reveal that ribosome activity can decrease by 15-30% when scaling from microliter to liter volumes, attributed to factors including oxygen limitation, pH drift, and accumulation of inhibitory byproducts. Maintaining optimal ribosomal efficiency at scale requires supplementation strategies and enhanced mixing protocols.
Resource distribution homogeneity becomes increasingly problematic at larger scales. Concentration gradients of amino acids, nucleotides, and cofactors can develop in poorly mixed systems, creating microenvironments with suboptimal conditions for protein synthesis. Advanced mixing technologies and feeding strategies have been developed to address these challenges, including fed-batch approaches that maintain critical component concentrations within optimal ranges.
Economic considerations ultimately determine industrial feasibility. The cost structure of CFPS changes dramatically with scale, as reagent costs decrease through bulk purchasing while equipment and operational expenses increase. Analysis of production economics indicates that medium-scale (10-100L) CFPS systems currently offer the most favorable balance between economies of scale and technical challenges, though continued innovation in bioreactor design and process engineering continues to push the boundaries of economically viable scales.
Bioethical Considerations in Synthetic Biology
The intersection of synthetic biology and cell-free protein synthesis raises significant bioethical considerations that warrant careful examination. As these technologies advance, particularly in ribosome efficiency optimization, society must address fundamental questions about the boundaries of human intervention in biological processes. The ability to create and manipulate biological systems outside natural contexts challenges traditional notions of life and biological integrity.
Concerns regarding biosafety and biosecurity are paramount when discussing ribosome efficiency in cell-free systems. Enhanced protein synthesis capabilities could potentially enable the production of harmful biological agents or toxins with greater efficiency. This dual-use potential necessitates robust regulatory frameworks and oversight mechanisms to prevent misuse while allowing beneficial research to proceed. The scientific community must establish standardized safety protocols specific to high-efficiency cell-free systems.
Access and equity issues emerge as critical considerations. As cell-free protein synthesis technologies become more sophisticated, disparities in access to these technologies may exacerbate existing inequalities in healthcare and biotechnology. Ensuring that benefits derived from improved ribosome efficiency are distributed globally requires intentional policies and international cooperation. This includes considerations for intellectual property rights and technology transfer mechanisms.
Environmental implications of synthetic biology applications utilizing enhanced ribosome efficiency demand attention. While cell-free systems potentially reduce environmental footprints compared to whole-cell approaches, questions remain about waste management, resource consumption, and potential ecological impacts of synthetic products. Life cycle assessments of these technologies should be conducted to evaluate their true environmental sustainability.
The concept of "playing god" or fundamentally altering nature's processes raises philosophical and religious concerns. Cell-free systems that achieve unprecedented efficiency in protein production represent a significant step toward human design of biological processes. These developments necessitate ongoing dialogue between scientists, ethicists, religious leaders, and the public to navigate complex questions about human limits and responsibilities.
Informed consent and public engagement present additional challenges. As research advances, ensuring transparent communication about risks, benefits, and uncertainties becomes increasingly important. The technical complexity of ribosome efficiency optimization makes meaningful public discourse difficult but essential. Scientists must develop effective communication strategies that enable non-specialists to participate in decision-making about these technologies.
Governance frameworks for synthetic biology must evolve alongside technical capabilities. International coordination is necessary to address transboundary risks while avoiding regulatory fragmentation that could impede beneficial innovation. Adaptive governance approaches that can respond to rapidly evolving technologies while upholding ethical principles will be crucial for responsible advancement of ribosome efficiency in cell-free protein synthesis.
Concerns regarding biosafety and biosecurity are paramount when discussing ribosome efficiency in cell-free systems. Enhanced protein synthesis capabilities could potentially enable the production of harmful biological agents or toxins with greater efficiency. This dual-use potential necessitates robust regulatory frameworks and oversight mechanisms to prevent misuse while allowing beneficial research to proceed. The scientific community must establish standardized safety protocols specific to high-efficiency cell-free systems.
Access and equity issues emerge as critical considerations. As cell-free protein synthesis technologies become more sophisticated, disparities in access to these technologies may exacerbate existing inequalities in healthcare and biotechnology. Ensuring that benefits derived from improved ribosome efficiency are distributed globally requires intentional policies and international cooperation. This includes considerations for intellectual property rights and technology transfer mechanisms.
Environmental implications of synthetic biology applications utilizing enhanced ribosome efficiency demand attention. While cell-free systems potentially reduce environmental footprints compared to whole-cell approaches, questions remain about waste management, resource consumption, and potential ecological impacts of synthetic products. Life cycle assessments of these technologies should be conducted to evaluate their true environmental sustainability.
The concept of "playing god" or fundamentally altering nature's processes raises philosophical and religious concerns. Cell-free systems that achieve unprecedented efficiency in protein production represent a significant step toward human design of biological processes. These developments necessitate ongoing dialogue between scientists, ethicists, religious leaders, and the public to navigate complex questions about human limits and responsibilities.
Informed consent and public engagement present additional challenges. As research advances, ensuring transparent communication about risks, benefits, and uncertainties becomes increasingly important. The technical complexity of ribosome efficiency optimization makes meaningful public discourse difficult but essential. Scientists must develop effective communication strategies that enable non-specialists to participate in decision-making about these technologies.
Governance frameworks for synthetic biology must evolve alongside technical capabilities. International coordination is necessary to address transboundary risks while avoiding regulatory fragmentation that could impede beneficial innovation. Adaptive governance approaches that can respond to rapidly evolving technologies while upholding ethical principles will be crucial for responsible advancement of ribosome efficiency in cell-free protein synthesis.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!