Research on Ribosome Stalling and Rescue Mechanisms
OCT 13, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Ribosomal Stalling Background and Research Objectives
Ribosomal stalling represents a critical regulatory mechanism in protein synthesis that has evolved across all domains of life. This phenomenon occurs when the ribosome, the cellular machinery responsible for translating messenger RNA (mRNA) into proteins, temporarily or permanently halts during the elongation phase of translation. The study of ribosome stalling has gained significant attention over the past two decades as researchers have uncovered its importance in protein quality control, gene expression regulation, and cellular stress responses.
Historically, ribosome stalling was first observed in the 1970s, but the molecular mechanisms and physiological significance remained poorly understood until advanced techniques in structural biology and molecular genetics emerged in the early 2000s. The field has since experienced exponential growth, with breakthrough discoveries revealing that stalling is not merely a translation error but often a programmed event with regulatory functions.
Ribosome stalling can be triggered by various factors, including rare codons, specific amino acid sequences, mRNA secondary structures, and insufficient availability of charged tRNAs. Additionally, certain antibiotics and toxins specifically target the ribosome to induce stalling, highlighting its importance as a potential therapeutic target. The SecM, TnaC, and ErmCL leader peptides represent well-characterized examples of sequences that induce programmed ribosome stalling for regulatory purposes.
The rescue mechanisms that resolve stalled ribosomes are equally important for cellular homeostasis. These include specialized factors such as tmRNA in bacteria (which employs trans-translation), ArfA, ArfB, and the recently discovered bacterial ribosome rescue factor C (RRF-C). In eukaryotes, mechanisms involving Dom34/Hbs1, Ski7, and the ribosome quality control complex (RQC) play crucial roles in resolving stalled translation complexes.
The primary objectives of current research in this field include: (1) elucidating the molecular mechanisms of ribosome stalling at atomic resolution; (2) identifying and characterizing novel stalling sequences and their regulatory functions; (3) understanding the interplay between ribosome stalling and cellular stress responses; (4) developing therapeutic strategies targeting ribosome stalling or rescue mechanisms; and (5) exploring the evolutionary conservation and divergence of these processes across species.
Recent technological advances, particularly in cryo-electron microscopy, ribosome profiling, and computational modeling, have revolutionized our ability to study ribosome stalling at unprecedented resolution. These techniques have revealed intricate details of how nascent peptides interact with the ribosomal exit tunnel to induce conformational changes that lead to stalling.
Understanding ribosome stalling and rescue mechanisms has profound implications for biotechnology, medicine, and fundamental biology, potentially leading to novel antibiotics, improved protein production systems, and insights into neurodegenerative diseases associated with protein misfolding and aggregation.
Historically, ribosome stalling was first observed in the 1970s, but the molecular mechanisms and physiological significance remained poorly understood until advanced techniques in structural biology and molecular genetics emerged in the early 2000s. The field has since experienced exponential growth, with breakthrough discoveries revealing that stalling is not merely a translation error but often a programmed event with regulatory functions.
Ribosome stalling can be triggered by various factors, including rare codons, specific amino acid sequences, mRNA secondary structures, and insufficient availability of charged tRNAs. Additionally, certain antibiotics and toxins specifically target the ribosome to induce stalling, highlighting its importance as a potential therapeutic target. The SecM, TnaC, and ErmCL leader peptides represent well-characterized examples of sequences that induce programmed ribosome stalling for regulatory purposes.
The rescue mechanisms that resolve stalled ribosomes are equally important for cellular homeostasis. These include specialized factors such as tmRNA in bacteria (which employs trans-translation), ArfA, ArfB, and the recently discovered bacterial ribosome rescue factor C (RRF-C). In eukaryotes, mechanisms involving Dom34/Hbs1, Ski7, and the ribosome quality control complex (RQC) play crucial roles in resolving stalled translation complexes.
The primary objectives of current research in this field include: (1) elucidating the molecular mechanisms of ribosome stalling at atomic resolution; (2) identifying and characterizing novel stalling sequences and their regulatory functions; (3) understanding the interplay between ribosome stalling and cellular stress responses; (4) developing therapeutic strategies targeting ribosome stalling or rescue mechanisms; and (5) exploring the evolutionary conservation and divergence of these processes across species.
Recent technological advances, particularly in cryo-electron microscopy, ribosome profiling, and computational modeling, have revolutionized our ability to study ribosome stalling at unprecedented resolution. These techniques have revealed intricate details of how nascent peptides interact with the ribosomal exit tunnel to induce conformational changes that lead to stalling.
Understanding ribosome stalling and rescue mechanisms has profound implications for biotechnology, medicine, and fundamental biology, potentially leading to novel antibiotics, improved protein production systems, and insights into neurodegenerative diseases associated with protein misfolding and aggregation.
Market Applications of Ribosome Stalling Research
The research on ribosome stalling and rescue mechanisms presents significant market applications across multiple industries, particularly in pharmaceuticals, biotechnology, and agriculture. Understanding these cellular processes offers unprecedented opportunities for developing novel therapeutic approaches and enhancing protein production systems.
In the pharmaceutical sector, ribosome stalling research enables the development of new antibiotics targeting bacterial translation machinery. Companies like Roche and Novartis are investing in translation-targeting compounds that exploit differences between prokaryotic and eukaryotic ribosomes. The global antibiotic market, facing resistance challenges, views ribosome-targeting drugs as a promising avenue with reduced resistance potential.
Biotechnology companies are leveraging ribosome stalling knowledge to optimize recombinant protein production. By manipulating ribosome pause sites, manufacturers can enhance protein folding efficiency and increase yields in biopharmaceutical production. This application directly addresses the growing demand for biologics, which require precise translational control during manufacturing.
The agricultural sector benefits from ribosome stalling research through the development of safer, more selective pesticides. Compounds that induce stalling in pest ribosomes while sparing beneficial organisms represent an environmentally friendly alternative to broad-spectrum chemicals. Companies like Bayer and Syngenta have research programs exploring translation-specific targeting in agricultural applications.
Diagnostic applications represent another emerging market. Biomarkers based on ribosome stalling patterns are being investigated for early detection of neurodegenerative diseases where protein misfolding plays a central role. Several diagnostics companies are developing platforms to detect aberrant ribosome activity as disease indicators.
Gene therapy and mRNA therapeutics benefit substantially from ribosome rescue mechanism research. Companies like Moderna and BioNTech are incorporating findings on ribosome rescue pathways to optimize mRNA stability and translation efficiency in their vaccine and therapeutic platforms. Understanding how to prevent premature ribosome stalling on therapeutic mRNAs directly impacts product efficacy.
Research tools and reagents for studying translation represent a specialized but growing market segment. Companies providing ribosome profiling kits, translation inhibitors, and related research tools serve academic and industrial laboratories investigating protein synthesis mechanisms.
The market for custom protein design also benefits from ribosome stalling research. By strategically introducing controlled stalling sites, protein engineers can influence co-translational folding pathways, enabling novel protein functionalities for industrial enzymes and therapeutic proteins with enhanced stability profiles.
In the pharmaceutical sector, ribosome stalling research enables the development of new antibiotics targeting bacterial translation machinery. Companies like Roche and Novartis are investing in translation-targeting compounds that exploit differences between prokaryotic and eukaryotic ribosomes. The global antibiotic market, facing resistance challenges, views ribosome-targeting drugs as a promising avenue with reduced resistance potential.
Biotechnology companies are leveraging ribosome stalling knowledge to optimize recombinant protein production. By manipulating ribosome pause sites, manufacturers can enhance protein folding efficiency and increase yields in biopharmaceutical production. This application directly addresses the growing demand for biologics, which require precise translational control during manufacturing.
The agricultural sector benefits from ribosome stalling research through the development of safer, more selective pesticides. Compounds that induce stalling in pest ribosomes while sparing beneficial organisms represent an environmentally friendly alternative to broad-spectrum chemicals. Companies like Bayer and Syngenta have research programs exploring translation-specific targeting in agricultural applications.
Diagnostic applications represent another emerging market. Biomarkers based on ribosome stalling patterns are being investigated for early detection of neurodegenerative diseases where protein misfolding plays a central role. Several diagnostics companies are developing platforms to detect aberrant ribosome activity as disease indicators.
Gene therapy and mRNA therapeutics benefit substantially from ribosome rescue mechanism research. Companies like Moderna and BioNTech are incorporating findings on ribosome rescue pathways to optimize mRNA stability and translation efficiency in their vaccine and therapeutic platforms. Understanding how to prevent premature ribosome stalling on therapeutic mRNAs directly impacts product efficacy.
Research tools and reagents for studying translation represent a specialized but growing market segment. Companies providing ribosome profiling kits, translation inhibitors, and related research tools serve academic and industrial laboratories investigating protein synthesis mechanisms.
The market for custom protein design also benefits from ribosome stalling research. By strategically introducing controlled stalling sites, protein engineers can influence co-translational folding pathways, enabling novel protein functionalities for industrial enzymes and therapeutic proteins with enhanced stability profiles.
Current Challenges in Ribosome Stalling Research
Despite significant advances in understanding ribosome stalling mechanisms, several critical challenges persist in this field. One of the primary obstacles is the transient nature of stalled ribosomes, which makes their detection and characterization exceptionally difficult. Current methodologies often capture only snapshots of these events, failing to provide a comprehensive temporal understanding of the stalling process and subsequent rescue mechanisms.
Technical limitations in real-time monitoring of translation at the single-molecule level continue to impede progress. While techniques such as ribosome profiling have revolutionized our ability to identify stalling sites genome-wide, they lack the temporal resolution necessary to distinguish between different types of stalling events and their durations. This limitation creates significant gaps in our understanding of how cells prioritize different rescue pathways.
The complexity of ribosome stalling is further compounded by its context-dependent nature. Stalling can be induced by various factors including rare codons, mRNA secondary structures, amino acid limitations, and nascent peptide-mediated effects. Each mechanism may trigger different cellular responses, yet current research struggles to differentiate between these various causes in vivo, particularly when multiple factors operate simultaneously.
Another significant challenge lies in understanding the regulatory networks that govern rescue pathway selection. Cells possess multiple rescue mechanisms including no-go decay (NGD), non-stop decay (NSD), and ribosome quality control (RQC), but the decision-making process that determines which pathway activates under specific conditions remains poorly understood. This knowledge gap hampers efforts to manipulate these pathways for therapeutic purposes.
Cross-species variations in stalling and rescue mechanisms present additional complications. While core components are conserved across eukaryotes, significant differences exist in regulatory mechanisms and pathway preferences. These variations limit the translational potential of findings from model organisms to human systems, particularly for developing therapeutics targeting ribosome-associated diseases.
Methodological inconsistencies across research groups further complicate the field. Variations in experimental conditions, cell types, and analytical approaches make direct comparisons between studies challenging. This fragmentation of knowledge impedes the development of unified models that could accelerate progress in understanding ribosome stalling and rescue mechanisms.
The integration of ribosome stalling research with other cellular processes represents another frontier challenge. Stalling events do not occur in isolation but interact with various cellular pathways including stress responses, protein quality control, and metabolic regulation. Elucidating these complex interactions requires interdisciplinary approaches that are still developing.
Technical limitations in real-time monitoring of translation at the single-molecule level continue to impede progress. While techniques such as ribosome profiling have revolutionized our ability to identify stalling sites genome-wide, they lack the temporal resolution necessary to distinguish between different types of stalling events and their durations. This limitation creates significant gaps in our understanding of how cells prioritize different rescue pathways.
The complexity of ribosome stalling is further compounded by its context-dependent nature. Stalling can be induced by various factors including rare codons, mRNA secondary structures, amino acid limitations, and nascent peptide-mediated effects. Each mechanism may trigger different cellular responses, yet current research struggles to differentiate between these various causes in vivo, particularly when multiple factors operate simultaneously.
Another significant challenge lies in understanding the regulatory networks that govern rescue pathway selection. Cells possess multiple rescue mechanisms including no-go decay (NGD), non-stop decay (NSD), and ribosome quality control (RQC), but the decision-making process that determines which pathway activates under specific conditions remains poorly understood. This knowledge gap hampers efforts to manipulate these pathways for therapeutic purposes.
Cross-species variations in stalling and rescue mechanisms present additional complications. While core components are conserved across eukaryotes, significant differences exist in regulatory mechanisms and pathway preferences. These variations limit the translational potential of findings from model organisms to human systems, particularly for developing therapeutics targeting ribosome-associated diseases.
Methodological inconsistencies across research groups further complicate the field. Variations in experimental conditions, cell types, and analytical approaches make direct comparisons between studies challenging. This fragmentation of knowledge impedes the development of unified models that could accelerate progress in understanding ribosome stalling and rescue mechanisms.
The integration of ribosome stalling research with other cellular processes represents another frontier challenge. Stalling events do not occur in isolation but interact with various cellular pathways including stress responses, protein quality control, and metabolic regulation. Elucidating these complex interactions requires interdisciplinary approaches that are still developing.
Established Methodologies for Studying Ribosome Rescue
01 Mechanisms of ribosome stalling during translation
Ribosome stalling occurs during protein synthesis when the translation process is interrupted. This can happen due to various factors including rare codons, mRNA secondary structures, or amino acid limitations. The stalling mechanism involves the ribosome pausing at specific sites on the mRNA, which can lead to incomplete protein synthesis. Understanding these mechanisms is crucial for developing strategies to address translation issues in both natural and engineered biological systems.- Mechanisms of ribosome stalling: Ribosome stalling occurs when the translation process is interrupted, often due to specific mRNA sequences, amino acid limitations, or structural obstacles. These stalling events can be triggered by rare codons, strong mRNA secondary structures, or specific peptide sequences within the nascent chain. Understanding these mechanisms is crucial for developing strategies to address translation issues in both natural and engineered biological systems.
- Ribosome rescue pathways: Cells have evolved specialized rescue mechanisms to resolve stalled ribosomes and prevent cellular toxicity. These pathways include trans-translation mediated by tmRNA, ArfA (Alternative Rescue Factor A), ArfB, and other factors that recognize and resolve stalled translation complexes. These rescue systems are essential for maintaining cellular homeostasis by recycling stalled ribosomes and preventing the accumulation of incomplete proteins.
- Pharmaceutical applications targeting ribosome stalling: Therapeutic approaches have been developed that either induce or resolve ribosome stalling for medical applications. These include antibiotics that target bacterial translation machinery, compounds that can rescue disease-related ribosome stalling, and strategies to modulate protein synthesis in specific disease contexts. Understanding ribosome stalling mechanisms has led to novel drug development strategies for treating various diseases including genetic disorders and infections.
- Genetic factors affecting ribosome stalling: Various genetic elements influence ribosome stalling, including specific mRNA sequences, rare codons, and regulatory elements. These genetic factors can be natural control mechanisms or pathological features in certain diseases. Research has identified specific sequence motifs that induce programmed ribosome stalling, which can serve regulatory functions in gene expression or contribute to disease pathology when dysregulated.
- Detection and analysis methods for ribosome stalling: Advanced techniques have been developed to detect and analyze ribosome stalling events in cells. These include ribosome profiling, computational prediction tools, and biochemical assays that can identify stalling sites with high precision. These methodologies enable researchers to study translation dynamics in real-time and understand how ribosome stalling affects overall protein synthesis and cellular function under various conditions.
02 Ribosome rescue pathways and factors
When ribosomes stall during translation, cellular rescue mechanisms are activated to resolve the stalled complexes. These rescue pathways involve specialized factors such as tmRNA, ArfA, and ArfB in bacteria, and PELO, HBS1, and ABCE1 in eukaryotes. These factors recognize stalled ribosomes and facilitate their recycling, allowing the translation machinery to be reused. The rescue mechanisms are essential for maintaining cellular protein synthesis capacity and preventing the accumulation of incomplete proteins.Expand Specific Solutions03 Drug development targeting ribosome stalling
Pharmaceutical approaches have been developed to either induce or resolve ribosome stalling for therapeutic purposes. Some antibiotics work by causing ribosome stalling in pathogenic bacteria, while other compounds are designed to overcome stalling in genetic diseases where premature termination occurs. These drug development strategies focus on modulating the interaction between the ribosome and specific mRNA sequences or affecting the function of rescue factors to achieve the desired therapeutic outcome.Expand Specific Solutions04 Genetic and molecular tools for studying ribosome stalling
Advanced molecular techniques have been developed to investigate ribosome stalling and rescue mechanisms. These include ribosome profiling, cryo-electron microscopy, and genetic screening approaches that allow researchers to identify stalling sequences and characterize the structural basis of ribosome stalling. These tools provide insights into the molecular interactions that occur during translation pausing and the subsequent rescue processes, contributing to our understanding of protein synthesis regulation.Expand Specific Solutions05 Biotechnological applications of ribosome stalling
Controlled ribosome stalling has been harnessed for various biotechnological applications. These include the production of difficult-to-express proteins, the development of biosensors based on translation efficiency, and the creation of synthetic gene circuits that respond to specific cellular conditions. By engineering sequences that induce regulated stalling, researchers can control protein expression levels or create novel biological functions that have applications in synthetic biology and protein manufacturing.Expand Specific Solutions
Leading Research Institutions and Biotechnology Companies
The ribosome stalling and rescue mechanisms research field is currently in a growth phase, with increasing market interest driven by its potential applications in drug development and disease treatment. The global market size for this niche is expanding, estimated to reach significant value as pharmaceutical companies recognize its importance in protein synthesis regulation. Technologically, the field shows moderate maturity with established research foundations but substantial room for innovation. Leading players include academic powerhouses like Johns Hopkins University, Yale University, and Washington University in St. Louis conducting fundamental research, while pharmaceutical companies such as Regeneron, F. Hoffmann-La Roche, and FibroGen are translating findings into therapeutic applications. Government entities and research institutions like Fred Hutchinson Cancer Research Center provide critical funding and infrastructure support, creating a diverse competitive landscape spanning both public and private sectors.
Washington University in St. Louis
Technical Solution: Washington University in St. Louis has developed advanced molecular biology techniques to study ribosome stalling and rescue mechanisms. Their research focuses on the role of specific mRNA sequences and nascent peptide interactions that cause ribosomal pausing. They have pioneered ribosome profiling methods that allow genome-wide identification of stalling sites with single-codon resolution[1]. Their technology employs deep sequencing of ribosome-protected mRNA fragments to create detailed maps of ribosome occupancy across transcripts. Additionally, they've developed in vitro reconstitution systems to study the molecular interactions between stalled ribosomes and rescue factors such as ArfA, ArfB, and tmRNA-SmpB[3]. Their approach combines structural biology with biochemical assays to elucidate the precise mechanisms by which these factors recognize and resolve stalled translation complexes.
Strengths: Their ribosome profiling technology provides unprecedented resolution for identifying stalling sites across the entire transcriptome. Their integrated structural and biochemical approach offers comprehensive insights into rescue factor mechanisms. Weaknesses: Their in vitro systems may not fully recapitulate the complexity of cellular environments, and their techniques require specialized equipment and expertise that limits widespread adoption.
The Johns Hopkins University
Technical Solution: Johns Hopkins University has developed innovative approaches to study ribosome stalling and rescue mechanisms, focusing particularly on the role of collided ribosomes in triggering quality control pathways. Their research employs cryo-electron microscopy (cryo-EM) to visualize the structural conformations of stalled ribosomes at near-atomic resolution[2]. They have pioneered techniques to capture transient interactions between stalled ribosomes and rescue factors such as Pelota, Hbs1, and ABCE1. Their technology platform includes ribosome profiling coupled with selective drug treatments to induce specific types of stalling, allowing them to map the cellular response to different translation obstacles[4]. Additionally, they've developed genetic screening methods to identify novel factors involved in ribosome-associated quality control (RQC) pathways that detect and resolve stalled translation complexes, revealing the intricate connections between ribosome stalling and cellular stress responses.
Strengths: Their cryo-EM approach provides detailed structural insights into the molecular mechanisms of stalling and rescue. Their integrated genetic and biochemical methods offer a comprehensive view of quality control pathways. Weaknesses: The high technical complexity of their approaches limits accessibility, and their focus on model systems may not fully capture the diversity of stalling mechanisms across different organisms and cellular conditions.
Key Molecular Mechanisms of Ribosomal Quality Control
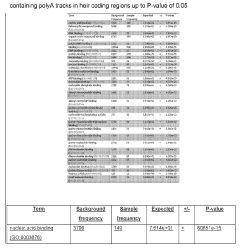
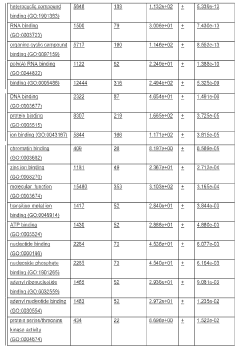
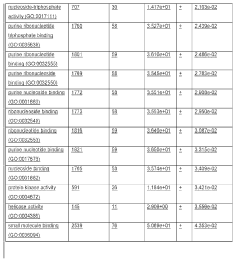
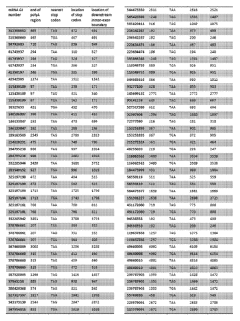
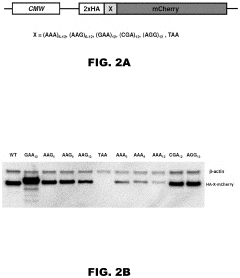
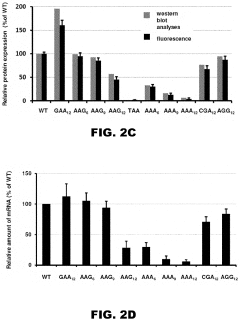
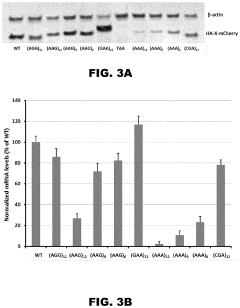
Incorporation of internal polya-encoded poly-lysine sequence tags and their variations for the tunable control of protein synthesis in bacterial and eukaryotic cells
PatentWO2018013720A1
Innovation
- Incorporation of internal polyA-encoded poly-lysine sequence tags and their variations in open reading frames to modulate protein synthesis by altering the number of consecutive adenine nucleotides in lysine codons, using CRISPR, ZFN, or TALEN systems to introduce synonymous mutations, thereby controlling protein expression levels.
Incorporation of internal polya-encoded poly-lysine sequence tags and their variations for the tunable control of protein synthesis in bacterial and eukaryotic cells
PatentActiveUS20230265442A1
Innovation
- The method involves modulating consecutive adenine nucleotides in lysine codons within a polynucleotide sequence to control protein expression, using expression vectors with engineered synonymous mutations in AAA or AAG codons to increase or decrease protein production, and employing CRISPR or zinc-finger nucleases for genomic DNA modification.
Therapeutic Implications for Protein Misfolding Diseases
Recent advances in understanding ribosome stalling and rescue mechanisms have opened promising avenues for therapeutic interventions in protein misfolding diseases. These disorders, including Alzheimer's, Parkinson's, and Huntington's diseases, are characterized by the accumulation of misfolded proteins that form toxic aggregates. The connection between ribosomal function and protein folding provides a novel target for therapeutic development.
Modulation of ribosome-associated quality control (RQC) pathways represents a significant therapeutic opportunity. By enhancing the efficiency of RQC mechanisms, it may be possible to reduce the burden of misfolded proteins before they accumulate and cause cellular damage. Small molecule compounds that selectively activate key components of the RQC pathway, such as Ltn1 E3 ubiquitin ligase or the Rqc2/NEMF protein, are currently under investigation in preclinical models.
Targeting ribosome rescue factors offers another promising approach. Compounds that modulate the activity of rescue factors like ArfA, ArfB, or eRF1 could potentially be used to fine-tune the balance between protein synthesis continuation and termination in cases where disease-causing mutations lead to ribosome stalling. This strategy could be particularly valuable for diseases caused by nonsense mutations or frameshift errors.
Translation speed modulation has emerged as a novel therapeutic concept. Research indicates that controlling the rate of translation can influence co-translational folding and reduce misfolding events. Compounds that slightly slow ribosomal progression through specific mRNA regions might allow more time for proper folding of challenging protein domains, potentially reducing the occurrence of misfolded species in diseases like cystic fibrosis.
Antisense oligonucleotides (ASOs) and RNA-based therapeutics represent another frontier in this field. These molecules can be designed to target specific mRNA sequences that cause ribosome stalling, either by promoting read-through of premature termination codons or by masking sequences that trigger ribosome pausing and subsequent protein misfolding.
CRISPR-based approaches are being explored to correct mutations that lead to ribosome stalling and protein misfolding at the genomic level. While still in early development stages, these techniques could potentially provide permanent solutions for genetic disorders where ribosome stalling contributes to pathology.
Clinical translation of these approaches faces significant challenges, including delivery to affected tissues, specificity of action, and potential side effects on global protein synthesis. However, the growing understanding of tissue-specific translation regulation mechanisms may enable the development of targeted therapies with acceptable safety profiles. Several pharmaceutical companies have established research programs focused on translation modulation, with early-stage clinical trials anticipated within the next 3-5 years.
Modulation of ribosome-associated quality control (RQC) pathways represents a significant therapeutic opportunity. By enhancing the efficiency of RQC mechanisms, it may be possible to reduce the burden of misfolded proteins before they accumulate and cause cellular damage. Small molecule compounds that selectively activate key components of the RQC pathway, such as Ltn1 E3 ubiquitin ligase or the Rqc2/NEMF protein, are currently under investigation in preclinical models.
Targeting ribosome rescue factors offers another promising approach. Compounds that modulate the activity of rescue factors like ArfA, ArfB, or eRF1 could potentially be used to fine-tune the balance between protein synthesis continuation and termination in cases where disease-causing mutations lead to ribosome stalling. This strategy could be particularly valuable for diseases caused by nonsense mutations or frameshift errors.
Translation speed modulation has emerged as a novel therapeutic concept. Research indicates that controlling the rate of translation can influence co-translational folding and reduce misfolding events. Compounds that slightly slow ribosomal progression through specific mRNA regions might allow more time for proper folding of challenging protein domains, potentially reducing the occurrence of misfolded species in diseases like cystic fibrosis.
Antisense oligonucleotides (ASOs) and RNA-based therapeutics represent another frontier in this field. These molecules can be designed to target specific mRNA sequences that cause ribosome stalling, either by promoting read-through of premature termination codons or by masking sequences that trigger ribosome pausing and subsequent protein misfolding.
CRISPR-based approaches are being explored to correct mutations that lead to ribosome stalling and protein misfolding at the genomic level. While still in early development stages, these techniques could potentially provide permanent solutions for genetic disorders where ribosome stalling contributes to pathology.
Clinical translation of these approaches faces significant challenges, including delivery to affected tissues, specificity of action, and potential side effects on global protein synthesis. However, the growing understanding of tissue-specific translation regulation mechanisms may enable the development of targeted therapies with acceptable safety profiles. Several pharmaceutical companies have established research programs focused on translation modulation, with early-stage clinical trials anticipated within the next 3-5 years.
Computational Approaches to Ribosome Stalling Prediction
Computational approaches to predicting ribosome stalling have evolved significantly in recent years, leveraging advances in machine learning and molecular dynamics simulations. These methods aim to identify sequence motifs and structural features that contribute to translational pausing, providing valuable insights into the fundamental mechanisms of protein synthesis regulation.
Traditional computational models have primarily focused on codon usage bias and mRNA secondary structures as predictors of ribosome stalling. However, these approaches often failed to capture the complex interplay between the nascent peptide chain, the ribosome exit tunnel, and translation factors that collectively influence stalling events.
More sophisticated algorithms now incorporate multiple parameters, including amino acid properties, peptide charge distribution, and hydrophobicity patterns. Deep learning frameworks have demonstrated particular promise, with convolutional neural networks capable of detecting subtle sequence patterns associated with stalling sites across diverse organisms.
Recent innovations include physics-based models that simulate the molecular interactions within the ribosome tunnel during translation. These models account for electrostatic forces, van der Waals interactions, and hydrogen bonding between the nascent peptide and ribosomal components, providing mechanistic explanations for experimentally observed stalling phenomena.
Integration of ribosome profiling data has substantially improved prediction accuracy. By training algorithms on genome-wide translational landscapes, researchers can now identify previously unrecognized stalling signatures with high sensitivity and specificity. Cross-species comparative analyses further enhance these models by distinguishing conserved stalling mechanisms from species-specific regulatory elements.
Several publicly available tools have emerged for stalling prediction, including RiboTempo, StallPred, and PausePred. These platforms employ different computational strategies but share the common goal of identifying potential stalling sites from primary sequence information. Benchmarking studies indicate accuracy rates of 70-85% for well-characterized stalling motifs, though performance varies considerably for novel sequences.
The computational prediction of ribosome stalling represents a crucial bridge between experimental observations and mechanistic understanding. As these tools continue to improve, they will facilitate the rational design of recombinant proteins with optimized translation efficiency and provide new targets for therapeutic interventions in diseases associated with translational dysregulation.
Traditional computational models have primarily focused on codon usage bias and mRNA secondary structures as predictors of ribosome stalling. However, these approaches often failed to capture the complex interplay between the nascent peptide chain, the ribosome exit tunnel, and translation factors that collectively influence stalling events.
More sophisticated algorithms now incorporate multiple parameters, including amino acid properties, peptide charge distribution, and hydrophobicity patterns. Deep learning frameworks have demonstrated particular promise, with convolutional neural networks capable of detecting subtle sequence patterns associated with stalling sites across diverse organisms.
Recent innovations include physics-based models that simulate the molecular interactions within the ribosome tunnel during translation. These models account for electrostatic forces, van der Waals interactions, and hydrogen bonding between the nascent peptide and ribosomal components, providing mechanistic explanations for experimentally observed stalling phenomena.
Integration of ribosome profiling data has substantially improved prediction accuracy. By training algorithms on genome-wide translational landscapes, researchers can now identify previously unrecognized stalling signatures with high sensitivity and specificity. Cross-species comparative analyses further enhance these models by distinguishing conserved stalling mechanisms from species-specific regulatory elements.
Several publicly available tools have emerged for stalling prediction, including RiboTempo, StallPred, and PausePred. These platforms employ different computational strategies but share the common goal of identifying potential stalling sites from primary sequence information. Benchmarking studies indicate accuracy rates of 70-85% for well-characterized stalling motifs, though performance varies considerably for novel sequences.
The computational prediction of ribosome stalling represents a crucial bridge between experimental observations and mechanistic understanding. As these tools continue to improve, they will facilitate the rational design of recombinant proteins with optimized translation efficiency and provide new targets for therapeutic interventions in diseases associated with translational dysregulation.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!