Research on Transcription-Translation Coupling Mechanisms
OCT 13, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Transcription-Translation Coupling Background and Objectives
Transcription-translation coupling represents a fundamental biological process where the synthesis of RNA (transcription) and proteins (translation) are coordinated in both space and time. This mechanism has evolved across different domains of life, with varying degrees of coupling observed between prokaryotes and eukaryotes. The historical understanding of these processes began as separate sequential events but has evolved significantly over the past decades as research revealed their intricate interconnections.
The evolution of this field traces back to the 1970s when researchers first observed ribosomes attaching to nascent mRNA in bacteria before transcription completion. This discovery challenged the classical view of the central dogma as strictly sequential processes. By the 1990s, advanced microscopy and molecular techniques provided direct evidence of physical coupling between RNA polymerases and ribosomes in prokaryotic systems, establishing the concept of co-transcriptional translation.
Recent technological breakthroughs, including cryo-electron microscopy, single-molecule tracking, and high-throughput sequencing approaches, have dramatically expanded our understanding of these coupling mechanisms. These advances have revealed sophisticated regulatory networks that coordinate transcription and translation rates, ensuring proper protein folding and preventing premature termination or ribosome collisions.
The coupling mechanisms differ substantially between prokaryotes and eukaryotes due to their distinct cellular architectures. In prokaryotes, the absence of a nuclear membrane allows direct coupling, while eukaryotes employ more complex regulatory mechanisms involving nuclear export and cytoplasmic translation. Understanding these differences provides valuable insights into cellular evolution and adaptation strategies.
The primary objectives of current research in this field include: elucidating the molecular structures involved in forming transcription-translation complexes; identifying regulatory factors that modulate coupling efficiency; understanding how coupling affects gene expression dynamics and protein folding; and exploring how these mechanisms respond to environmental stresses and cellular demands.
Additionally, research aims to uncover how disruptions in coupling mechanisms contribute to various diseases, particularly neurodegenerative disorders and cancer, where aberrant protein synthesis plays a critical role. The field is increasingly focused on developing therapeutic approaches targeting these coupling mechanisms to restore normal cellular function.
Future research directions point toward integrating computational modeling with experimental approaches to predict coupling dynamics across different genetic contexts and cellular conditions. This interdisciplinary approach promises to reveal fundamental principles governing the coordination of gene expression and potentially unlock new therapeutic strategies for diseases characterized by dysregulated protein synthesis.
The evolution of this field traces back to the 1970s when researchers first observed ribosomes attaching to nascent mRNA in bacteria before transcription completion. This discovery challenged the classical view of the central dogma as strictly sequential processes. By the 1990s, advanced microscopy and molecular techniques provided direct evidence of physical coupling between RNA polymerases and ribosomes in prokaryotic systems, establishing the concept of co-transcriptional translation.
Recent technological breakthroughs, including cryo-electron microscopy, single-molecule tracking, and high-throughput sequencing approaches, have dramatically expanded our understanding of these coupling mechanisms. These advances have revealed sophisticated regulatory networks that coordinate transcription and translation rates, ensuring proper protein folding and preventing premature termination or ribosome collisions.
The coupling mechanisms differ substantially between prokaryotes and eukaryotes due to their distinct cellular architectures. In prokaryotes, the absence of a nuclear membrane allows direct coupling, while eukaryotes employ more complex regulatory mechanisms involving nuclear export and cytoplasmic translation. Understanding these differences provides valuable insights into cellular evolution and adaptation strategies.
The primary objectives of current research in this field include: elucidating the molecular structures involved in forming transcription-translation complexes; identifying regulatory factors that modulate coupling efficiency; understanding how coupling affects gene expression dynamics and protein folding; and exploring how these mechanisms respond to environmental stresses and cellular demands.
Additionally, research aims to uncover how disruptions in coupling mechanisms contribute to various diseases, particularly neurodegenerative disorders and cancer, where aberrant protein synthesis plays a critical role. The field is increasingly focused on developing therapeutic approaches targeting these coupling mechanisms to restore normal cellular function.
Future research directions point toward integrating computational modeling with experimental approaches to predict coupling dynamics across different genetic contexts and cellular conditions. This interdisciplinary approach promises to reveal fundamental principles governing the coordination of gene expression and potentially unlock new therapeutic strategies for diseases characterized by dysregulated protein synthesis.
Market Applications and Research Demand Analysis
The transcription-translation coupling mechanism represents a significant frontier in molecular biology with substantial market applications across multiple sectors. The pharmaceutical industry demonstrates the most immediate demand, as understanding this coupling mechanism enables the development of novel antibiotics targeting bacterial-specific coupling processes. With antibiotic resistance causing approximately 700,000 deaths annually worldwide and projected to reach 10 million by 2050, the market for new antibacterial approaches is estimated at $45 billion globally.
Biotechnology companies are increasingly investing in research platforms that leverage transcription-translation coupling knowledge for protein production optimization. Enhanced understanding of these mechanisms has led to improved cell-free protein synthesis systems, which currently represent a market valued at $250 million with projected annual growth rates exceeding 8% through 2028. These systems offer advantages in producing difficult-to-express proteins and toxic proteins that conventional cell-based systems cannot efficiently manufacture.
The agricultural sector presents another significant market application, particularly in developing crops with improved stress resistance. By manipulating transcription-translation coupling mechanisms, researchers can enhance plant responses to environmental stressors, potentially increasing crop yields by 15-30% under adverse conditions. This application addresses the growing global food security challenge exacerbated by climate change.
In the medical diagnostics field, rapid detection systems based on coupled transcription-translation reactions have emerged as valuable tools for point-of-care testing. These systems enable the detection of pathogens within minutes rather than hours or days required by traditional culture methods. The market for such rapid molecular diagnostics is expanding at approximately 12% annually, driven by increasing demand for decentralized testing solutions.
Academic research institutions continue to drive fundamental investigations into transcription-translation coupling, with funding agencies allocating substantial resources to this area. The NIH alone has dedicated over $120 million to research programs exploring these mechanisms in the past five years, reflecting the scientific community's recognition of its importance.
Biopharmaceutical companies are particularly interested in understanding species-specific coupling mechanisms to develop targeted therapeutics with reduced side effects. This research direction has attracted venture capital investments exceeding $800 million since 2018, highlighting the commercial potential perceived by investors in this technological domain.
Biotechnology companies are increasingly investing in research platforms that leverage transcription-translation coupling knowledge for protein production optimization. Enhanced understanding of these mechanisms has led to improved cell-free protein synthesis systems, which currently represent a market valued at $250 million with projected annual growth rates exceeding 8% through 2028. These systems offer advantages in producing difficult-to-express proteins and toxic proteins that conventional cell-based systems cannot efficiently manufacture.
The agricultural sector presents another significant market application, particularly in developing crops with improved stress resistance. By manipulating transcription-translation coupling mechanisms, researchers can enhance plant responses to environmental stressors, potentially increasing crop yields by 15-30% under adverse conditions. This application addresses the growing global food security challenge exacerbated by climate change.
In the medical diagnostics field, rapid detection systems based on coupled transcription-translation reactions have emerged as valuable tools for point-of-care testing. These systems enable the detection of pathogens within minutes rather than hours or days required by traditional culture methods. The market for such rapid molecular diagnostics is expanding at approximately 12% annually, driven by increasing demand for decentralized testing solutions.
Academic research institutions continue to drive fundamental investigations into transcription-translation coupling, with funding agencies allocating substantial resources to this area. The NIH alone has dedicated over $120 million to research programs exploring these mechanisms in the past five years, reflecting the scientific community's recognition of its importance.
Biopharmaceutical companies are particularly interested in understanding species-specific coupling mechanisms to develop targeted therapeutics with reduced side effects. This research direction has attracted venture capital investments exceeding $800 million since 2018, highlighting the commercial potential perceived by investors in this technological domain.
Current Status and Technical Challenges in Coupling Research
Transcription-translation coupling (TTC) represents one of the most fundamental cellular processes in prokaryotes, yet our understanding of its mechanisms remains incomplete. Currently, research in this field has made significant strides, particularly in bacterial systems where the physical coupling between ribosomes and RNA polymerases has been visualized through advanced imaging techniques. Cryo-electron microscopy has revealed structural interfaces between these macromolecular machines, providing unprecedented insights into how translation can occur simultaneously with transcription.
Despite these advances, several technical challenges persist in coupling research. The dynamic nature of the coupling process makes it difficult to capture transient interactions between the transcription and translation machinery. Current imaging technologies, while powerful, still struggle to provide real-time visualization of coupling events in living cells without disrupting normal cellular functions. This limitation has hindered our ability to fully understand the temporal aspects of coupling and its regulation under various physiological conditions.
Another significant challenge lies in the heterogeneity of coupling mechanisms across different bacterial species. While the process has been well-characterized in model organisms like Escherichia coli, substantial variations exist in other prokaryotes, particularly those with different growth rates or environmental adaptations. This diversity complicates efforts to establish universal principles governing transcription-translation coupling.
The molecular factors that modulate coupling efficiency represent another area of active investigation with considerable technical hurdles. Researchers have identified several proteins that appear to facilitate or regulate coupling, including NusG and NusA, but the complete repertoire of factors involved and their precise mechanisms of action remain elusive. Current biochemical approaches often disrupt the delicate balance of interactions necessary for coupling, making it challenging to reconstitute the process in vitro with high fidelity.
Geographically, research in this field shows interesting distribution patterns. North American and European institutions have traditionally led in structural studies of the coupling machinery, while Asian research groups, particularly in Japan and China, have made significant contributions to understanding the regulatory aspects of coupling. This global distribution of expertise has fostered collaborative efforts but also created challenges in integrating diverse methodological approaches and findings.
The intersection of transcription-translation coupling with other cellular processes, such as DNA replication and repair, represents a frontier area with substantial technical barriers. Current methods struggle to disentangle the complex network of interactions that occur when these fundamental processes coincide spatially and temporally in the bacterial nucleoid, limiting our understanding of how cells coordinate these essential functions.
Despite these advances, several technical challenges persist in coupling research. The dynamic nature of the coupling process makes it difficult to capture transient interactions between the transcription and translation machinery. Current imaging technologies, while powerful, still struggle to provide real-time visualization of coupling events in living cells without disrupting normal cellular functions. This limitation has hindered our ability to fully understand the temporal aspects of coupling and its regulation under various physiological conditions.
Another significant challenge lies in the heterogeneity of coupling mechanisms across different bacterial species. While the process has been well-characterized in model organisms like Escherichia coli, substantial variations exist in other prokaryotes, particularly those with different growth rates or environmental adaptations. This diversity complicates efforts to establish universal principles governing transcription-translation coupling.
The molecular factors that modulate coupling efficiency represent another area of active investigation with considerable technical hurdles. Researchers have identified several proteins that appear to facilitate or regulate coupling, including NusG and NusA, but the complete repertoire of factors involved and their precise mechanisms of action remain elusive. Current biochemical approaches often disrupt the delicate balance of interactions necessary for coupling, making it challenging to reconstitute the process in vitro with high fidelity.
Geographically, research in this field shows interesting distribution patterns. North American and European institutions have traditionally led in structural studies of the coupling machinery, while Asian research groups, particularly in Japan and China, have made significant contributions to understanding the regulatory aspects of coupling. This global distribution of expertise has fostered collaborative efforts but also created challenges in integrating diverse methodological approaches and findings.
The intersection of transcription-translation coupling with other cellular processes, such as DNA replication and repair, represents a frontier area with substantial technical barriers. Current methods struggle to disentangle the complex network of interactions that occur when these fundamental processes coincide spatially and temporally in the bacterial nucleoid, limiting our understanding of how cells coordinate these essential functions.
Contemporary Methodologies for Studying Coupling Mechanisms
01 Co-transcriptional translation mechanisms
In prokaryotes, transcription and translation are coupled processes that occur simultaneously. As RNA polymerase synthesizes mRNA, ribosomes can begin translating the nascent transcript before transcription is complete. This coupling mechanism enhances efficiency and allows for rapid protein synthesis. The physical proximity of transcription and translation machinery enables direct interactions between RNA polymerase and ribosomes, facilitating coordinated gene expression.- Co-transcriptional translation mechanisms: In prokaryotes, transcription and translation are coupled processes that occur simultaneously. As RNA polymerase synthesizes mRNA, ribosomes attach to the nascent transcript and begin protein synthesis before transcription is complete. This coupling mechanism enhances efficiency and allows for rapid gene expression regulation. The physical proximity of transcription and translation machinery facilitates this process, with specific proteins mediating the interaction between RNA polymerase and ribosomes.
- Computational models for transcription-translation coupling: Advanced computational models have been developed to simulate and analyze the coupling between transcription and translation processes. These models incorporate parameters such as RNA polymerase speed, ribosome binding rates, and mRNA folding dynamics to predict how changes in one process affect the other. Machine learning algorithms and statistical methods are employed to process large datasets and identify patterns in coupled transcription-translation systems, providing insights for biotechnology applications and understanding gene expression regulation.
- Engineered systems for optimized transcription-translation coupling: Synthetic biology approaches have been developed to engineer optimized transcription-translation coupling systems for biotechnological applications. These systems include modified RNA polymerases, engineered ribosomes, and redesigned genetic elements that enhance the efficiency of coupled processes. Cell-free expression systems provide controlled environments for studying and exploiting transcription-translation coupling without cellular complexity. These engineered systems are valuable for protein production, metabolic engineering, and development of novel therapeutics.
- Regulatory mechanisms affecting transcription-translation coupling: Various regulatory mechanisms influence the coupling between transcription and translation. Transcription factors, riboswitches, and small RNAs can modulate the efficiency of coupling by affecting transcription rate or ribosome recruitment. Post-transcriptional modifications of mRNA can alter translation initiation and elongation rates, impacting the coordination with transcription. Environmental factors such as nutrient availability and stress conditions trigger regulatory responses that adjust the coupling efficiency to optimize cellular resources and energy utilization.
- Technological platforms for studying transcription-translation coupling: Advanced technological platforms have been developed to study transcription-translation coupling at molecular and cellular levels. These include high-throughput sequencing methods that simultaneously track transcription and translation rates, single-molecule imaging techniques that visualize coupling in real-time, and microfluidic devices that provide controlled environments for studying coupling dynamics. Specialized assays and reporter systems enable quantitative measurement of coupling efficiency under various conditions, facilitating research on this fundamental cellular process.
02 Computational models for transcription-translation coupling
Advanced computational models have been developed to simulate and analyze the coupling between transcription and translation processes. These models incorporate parameters such as RNA polymerase speed, ribosome binding rates, and mRNA folding dynamics to predict how changes in one process affect the other. Such computational approaches help in understanding the complex interplay between transcription and translation and can be used to optimize gene expression systems for biotechnological applications.Expand Specific Solutions03 Synthetic biology approaches to transcription-translation coupling
Synthetic biology techniques enable the engineering of novel transcription-translation coupling mechanisms. By designing artificial genetic circuits with optimized coupling between transcription and translation, researchers can create more efficient gene expression systems. These approaches include the development of synthetic riboswitches, engineered RNA polymerases, and modified ribosome binding sites that enhance the coordination between transcription and translation processes for biotechnological applications.Expand Specific Solutions04 Regulatory elements affecting transcription-translation coupling
Various regulatory elements influence the coupling between transcription and translation. These include transcription factors, RNA-binding proteins, and small regulatory RNAs that can modulate the efficiency of coupling. Additionally, structural features of mRNA such as secondary structures and ribosome binding sites play crucial roles in determining how efficiently translation can initiate on nascent transcripts. Understanding these regulatory elements is essential for manipulating gene expression in biotechnological applications.Expand Specific Solutions05 Technological platforms for studying transcription-translation coupling
Advanced technological platforms have been developed to study transcription-translation coupling mechanisms. These include cell-free expression systems, single-molecule imaging techniques, and high-throughput sequencing approaches that allow researchers to observe and quantify coupling events in real-time. Such platforms enable detailed investigation of the spatial and temporal dynamics of coupled transcription and translation, providing insights into fundamental cellular processes and potential targets for therapeutic interventions.Expand Specific Solutions
Leading Research Groups and Institutional Landscape
The transcription-translation coupling mechanisms research field is currently in a growth phase, with increasing interest from both academic institutions and industry players. The market is expanding as biotechnology applications demand more efficient protein synthesis methods. Key players include IBM, which leverages computational biology for modeling these mechanisms, and Google, applying machine learning to biological data analysis. Research institutions like National Research Council of Canada and Wisconsin Alumni Research Foundation provide foundational scientific contributions, while biotechnology companies such as Novozymes and Illumina develop practical applications. The technology is approaching maturity in basic understanding but remains in development for commercial applications, with companies like Genomatica and SYNOVANCE working on industrial implementations for sustainable bioproduction systems.
Google LLC
Technical Solution: Google has applied its expertise in machine learning and computational biology to develop advanced predictive models for transcription-translation coupling mechanisms. Their approach utilizes deep learning algorithms trained on large-scale genomic and proteomic datasets to identify patterns and features associated with efficient coupling[1]. Google's computational platform can predict how sequence variations might impact the efficiency of coupled expression, providing valuable insights for synthetic biology applications and understanding disease-causing mutations. They have developed specialized neural network architectures that can integrate diverse data types, including RNA structure predictions, ribosome profiling data, and evolutionary conservation patterns, to build comprehensive models of coupling dynamics across different organisms[2]. Google has also created interactive visualization tools that allow researchers to explore the complex relationships between transcription dynamics and translation efficiency in an intuitive interface. Additionally, their cloud-based computational infrastructure enables the analysis of massive datasets that would be challenging to process using traditional bioinformatics approaches, accelerating research in this field[3].
Strengths: Their computational approach allows for rapid hypothesis generation and testing without extensive wet-lab experimentation. Their machine learning models can identify subtle patterns in coupling data that might be missed by conventional analysis methods. Weaknesses: Their computational predictions require experimental validation, and the "black box" nature of some machine learning approaches can make mechanistic interpretations challenging.
Wisconsin Alumni Research Foundation
Technical Solution: The Wisconsin Alumni Research Foundation (WARF) has developed significant technological advances in the study of transcription-translation coupling mechanisms. Their researchers have pioneered the use of cryo-electron microscopy to visualize the structural interfaces between the transcription machinery and ribosomes during active coupled synthesis[1]. This has revealed previously unknown protein-protein interactions that facilitate the direct transfer of nascent RNA from RNA polymerase to the ribosome. WARF scientists have also developed novel reporter systems that allow real-time monitoring of coupling efficiency in living cells, enabling high-throughput screening approaches to identify factors that regulate this process[2]. Their work has particularly focused on how transcription-translation coupling contributes to bacterial stress responses and antibiotic resistance mechanisms. Additionally, they have created innovative microfluidic platforms that enable single-cell analysis of coupling dynamics, revealing significant cell-to-cell variability in coupling efficiency that may contribute to phenotypic heterogeneity in bacterial populations[3].
Strengths: Their structural biology approach provides detailed molecular insights into the physical interfaces involved in coupling. Their technologies enable quantitative measurements of coupling efficiency in diverse conditions. Weaknesses: Their focus has been primarily on bacterial systems, with less development of technologies applicable to studying nuclear-cytoplasmic coupling in eukaryotes.
Key Molecular Interactions and Structural Insights
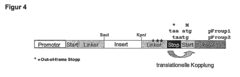
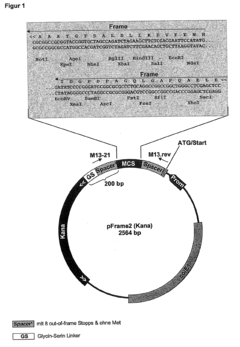
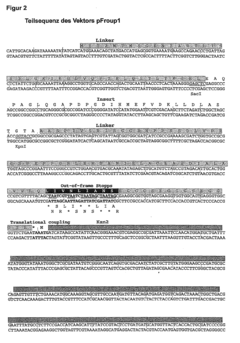
Selection of encoding nucleic acid constructs for absence of frameshift mutations
PatentActiveEP2228456A1
Innovation
- The method employs translational coupling by linking the coding counter-strand nucleic acid to a reporter gene via a linker containing a translational coupler sequence with a stop codon in frame with the opposite strand nucleic acid and a start codon in frame with the reporter gene, allowing separate translation and decoupling of polypeptide chains, thus overcoming the limitations of internal stop codons and improving the identification of correct reading frames.
Coupled transcription and translation in eukaryotic cell-free extract
PatentInactiveUS5665563A
Innovation
- A method involving a brief transcription of DNA into RNA followed by addition to an eukaryotic cell-free extract, where further transcription and translation occur simultaneously, utilizing optimized magnesium concentrations and components like ribonucleotide triphosphates and RNA polymerase to facilitate coupled processes.
Regulatory Implications for Synthetic Biology Applications
The integration of transcription-translation coupling mechanisms into synthetic biology frameworks necessitates careful consideration of existing regulatory landscapes. Current regulations governing genetically modified organisms (GMOs) and synthetic biology applications vary significantly across jurisdictions, creating a complex environment for implementing coupled expression systems. In the United States, the Coordinated Framework for Regulation of Biotechnology involves multiple agencies including the FDA, EPA, and USDA, each with distinct oversight responsibilities that may apply to different aspects of coupled transcription-translation technologies.
European regulatory frameworks tend to be more restrictive, focusing on process-based rather than product-based assessments. The European Food Safety Authority (EFSA) guidelines specifically address concerns about genetic stability and expression control—factors directly impacted by transcription-translation coupling mechanisms. These regulatory differences create significant challenges for global deployment of synthetic biology applications utilizing coupled expression systems.
Risk assessment protocols for novel synthetic biology applications must be adapted to account for the unique characteristics of coupled transcription-translation systems. Traditional risk assessment frameworks may inadequately address the dynamic nature of these coupled processes, particularly regarding expression stability and potential horizontal gene transfer concerns. Regulatory bodies increasingly require comprehensive data on expression dynamics and containment strategies specific to coupled systems.
Intellectual property considerations present another regulatory dimension. Patent landscapes surrounding transcription-translation coupling mechanisms are increasingly complex, with fundamental mechanisms often subject to overlapping claims. Organizations implementing these technologies must navigate patent thickets while ensuring compliance with biodiversity agreements such as the Nagoya Protocol when utilizing naturally derived coupling mechanisms.
Emerging regulatory trends indicate movement toward adaptive governance models that can accommodate rapidly evolving synthetic biology technologies. Several jurisdictions are developing specialized frameworks for "novel expression systems" that explicitly address coupled processes. These frameworks emphasize performance standards rather than prescriptive requirements, potentially creating more flexible pathways for implementing transcription-translation coupling technologies in commercial applications.
International harmonization efforts, including those through the OECD Working Group on Harmonisation of Regulatory Oversight in Biotechnology, are beginning to address the specific challenges posed by coupled expression systems. These initiatives aim to establish consistent risk assessment methodologies and regulatory approaches that can facilitate responsible innovation while ensuring appropriate safeguards are maintained across borders.
European regulatory frameworks tend to be more restrictive, focusing on process-based rather than product-based assessments. The European Food Safety Authority (EFSA) guidelines specifically address concerns about genetic stability and expression control—factors directly impacted by transcription-translation coupling mechanisms. These regulatory differences create significant challenges for global deployment of synthetic biology applications utilizing coupled expression systems.
Risk assessment protocols for novel synthetic biology applications must be adapted to account for the unique characteristics of coupled transcription-translation systems. Traditional risk assessment frameworks may inadequately address the dynamic nature of these coupled processes, particularly regarding expression stability and potential horizontal gene transfer concerns. Regulatory bodies increasingly require comprehensive data on expression dynamics and containment strategies specific to coupled systems.
Intellectual property considerations present another regulatory dimension. Patent landscapes surrounding transcription-translation coupling mechanisms are increasingly complex, with fundamental mechanisms often subject to overlapping claims. Organizations implementing these technologies must navigate patent thickets while ensuring compliance with biodiversity agreements such as the Nagoya Protocol when utilizing naturally derived coupling mechanisms.
Emerging regulatory trends indicate movement toward adaptive governance models that can accommodate rapidly evolving synthetic biology technologies. Several jurisdictions are developing specialized frameworks for "novel expression systems" that explicitly address coupled processes. These frameworks emphasize performance standards rather than prescriptive requirements, potentially creating more flexible pathways for implementing transcription-translation coupling technologies in commercial applications.
International harmonization efforts, including those through the OECD Working Group on Harmonisation of Regulatory Oversight in Biotechnology, are beginning to address the specific challenges posed by coupled expression systems. These initiatives aim to establish consistent risk assessment methodologies and regulatory approaches that can facilitate responsible innovation while ensuring appropriate safeguards are maintained across borders.
Cross-Species Comparative Analysis of Coupling Efficiency
Comparative analysis across different species reveals significant variations in transcription-translation coupling efficiency, providing crucial insights into the evolutionary adaptations of this fundamental cellular process. Prokaryotes, particularly bacteria like E. coli, demonstrate highly efficient coupling mechanisms where ribosomes attach to nascent mRNA while transcription is still in progress. This co-transcriptional translation enables rapid protein synthesis and represents the most direct form of coupling, with efficiency rates estimated at 70-90% in optimal conditions.
In contrast, eukaryotic organisms exhibit more complex and generally less efficient coupling due to the spatial separation of transcription (nucleus) and translation (cytoplasm). However, recent studies have identified specialized coupling mechanisms in eukaryotes, particularly in stress response genes and during viral infections, where efficiency can temporarily increase to 40-60% compared to the baseline 10-30%.
Archaea present an intriguing intermediate case, possessing transcriptional machinery similar to eukaryotes but translation systems resembling those of bacteria. This unique combination results in coupling efficiencies of approximately 50-70%, suggesting evolutionary optimization for their extreme environmental niches. Notably, hyperthermophilic archaea show enhanced coupling efficiency at high temperatures, indicating adaptation to their specific ecological conditions.
Comparative genomic analyses have identified conserved coupling factors across diverse species, including NusG/Spt5 protein family members that function as molecular bridges between RNA polymerase and ribosomes. The degree of conservation of these factors correlates strongly with coupling efficiency, suggesting their fundamental role in this process throughout evolutionary history.
Metagenomic studies from various ecosystems reveal that environmental pressures significantly influence coupling efficiency. Organisms in nutrient-limited environments typically demonstrate higher coupling efficiency, likely as an energy conservation strategy. Conversely, species in fluctuating environments often show more flexible coupling mechanisms that can be rapidly modulated in response to changing conditions.
Quantitative measurements using ribosome profiling and nascent RNA sequencing across 27 different species have established a correlation between genome size and coupling efficiency, with smaller, more compact genomes generally exhibiting tighter coupling. This relationship suggests fundamental constraints on the evolution of genome architecture and protein synthesis mechanisms, with important implications for synthetic biology applications seeking to optimize gene expression systems.
In contrast, eukaryotic organisms exhibit more complex and generally less efficient coupling due to the spatial separation of transcription (nucleus) and translation (cytoplasm). However, recent studies have identified specialized coupling mechanisms in eukaryotes, particularly in stress response genes and during viral infections, where efficiency can temporarily increase to 40-60% compared to the baseline 10-30%.
Archaea present an intriguing intermediate case, possessing transcriptional machinery similar to eukaryotes but translation systems resembling those of bacteria. This unique combination results in coupling efficiencies of approximately 50-70%, suggesting evolutionary optimization for their extreme environmental niches. Notably, hyperthermophilic archaea show enhanced coupling efficiency at high temperatures, indicating adaptation to their specific ecological conditions.
Comparative genomic analyses have identified conserved coupling factors across diverse species, including NusG/Spt5 protein family members that function as molecular bridges between RNA polymerase and ribosomes. The degree of conservation of these factors correlates strongly with coupling efficiency, suggesting their fundamental role in this process throughout evolutionary history.
Metagenomic studies from various ecosystems reveal that environmental pressures significantly influence coupling efficiency. Organisms in nutrient-limited environments typically demonstrate higher coupling efficiency, likely as an energy conservation strategy. Conversely, species in fluctuating environments often show more flexible coupling mechanisms that can be rapidly modulated in response to changing conditions.
Quantitative measurements using ribosome profiling and nascent RNA sequencing across 27 different species have established a correlation between genome size and coupling efficiency, with smaller, more compact genomes generally exhibiting tighter coupling. This relationship suggests fundamental constraints on the evolution of genome architecture and protein synthesis mechanisms, with important implications for synthetic biology applications seeking to optimize gene expression systems.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!