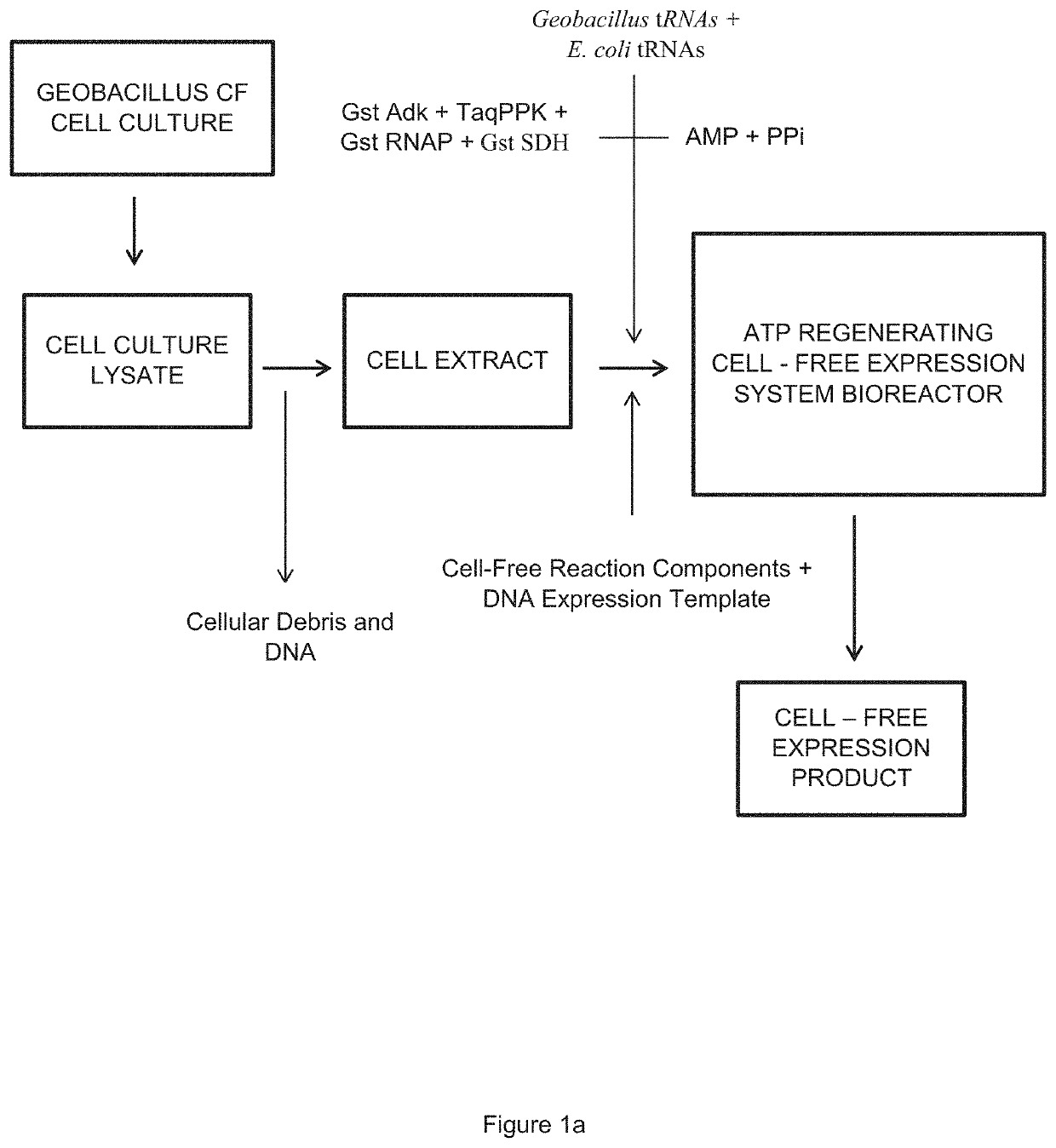
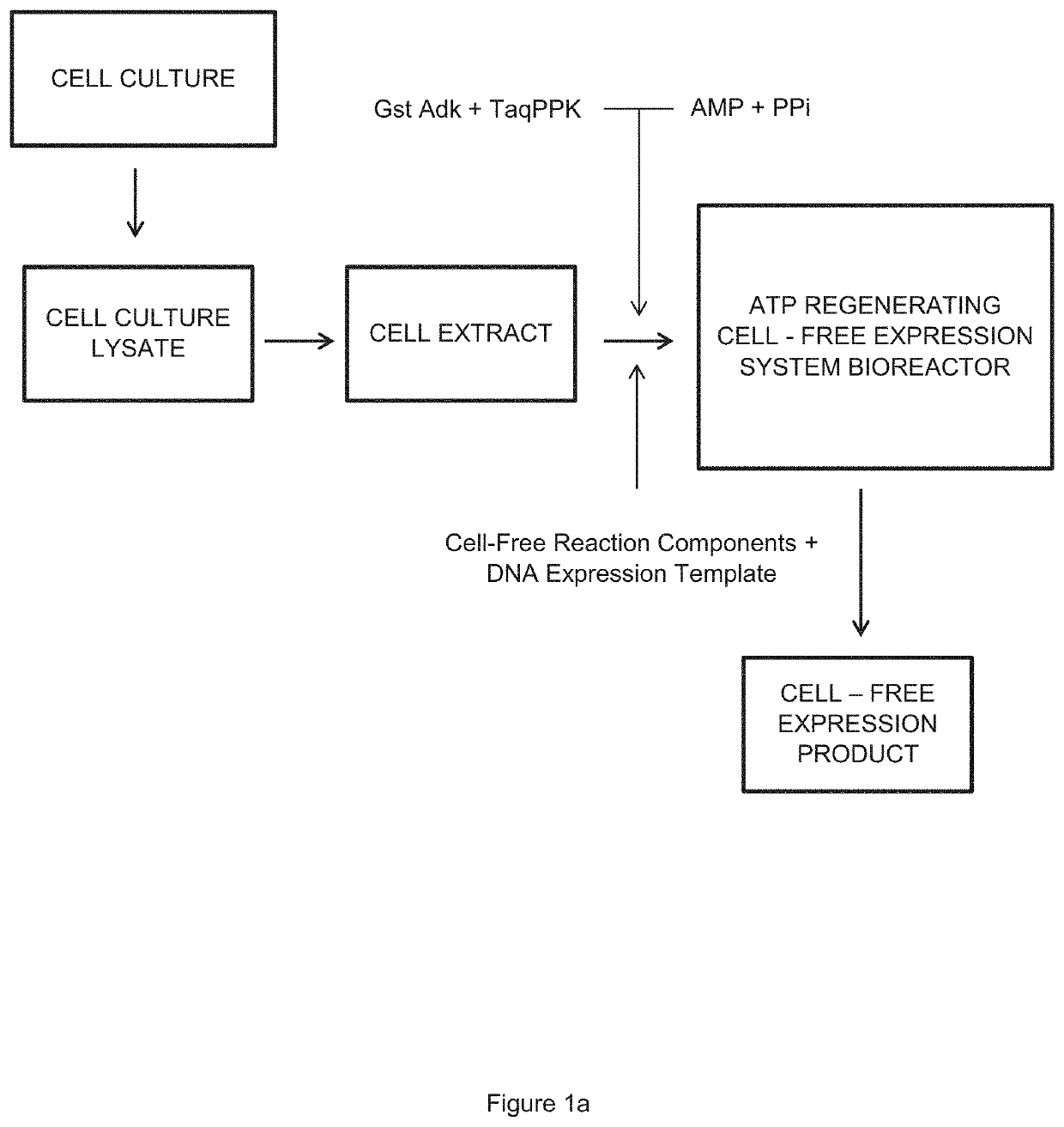
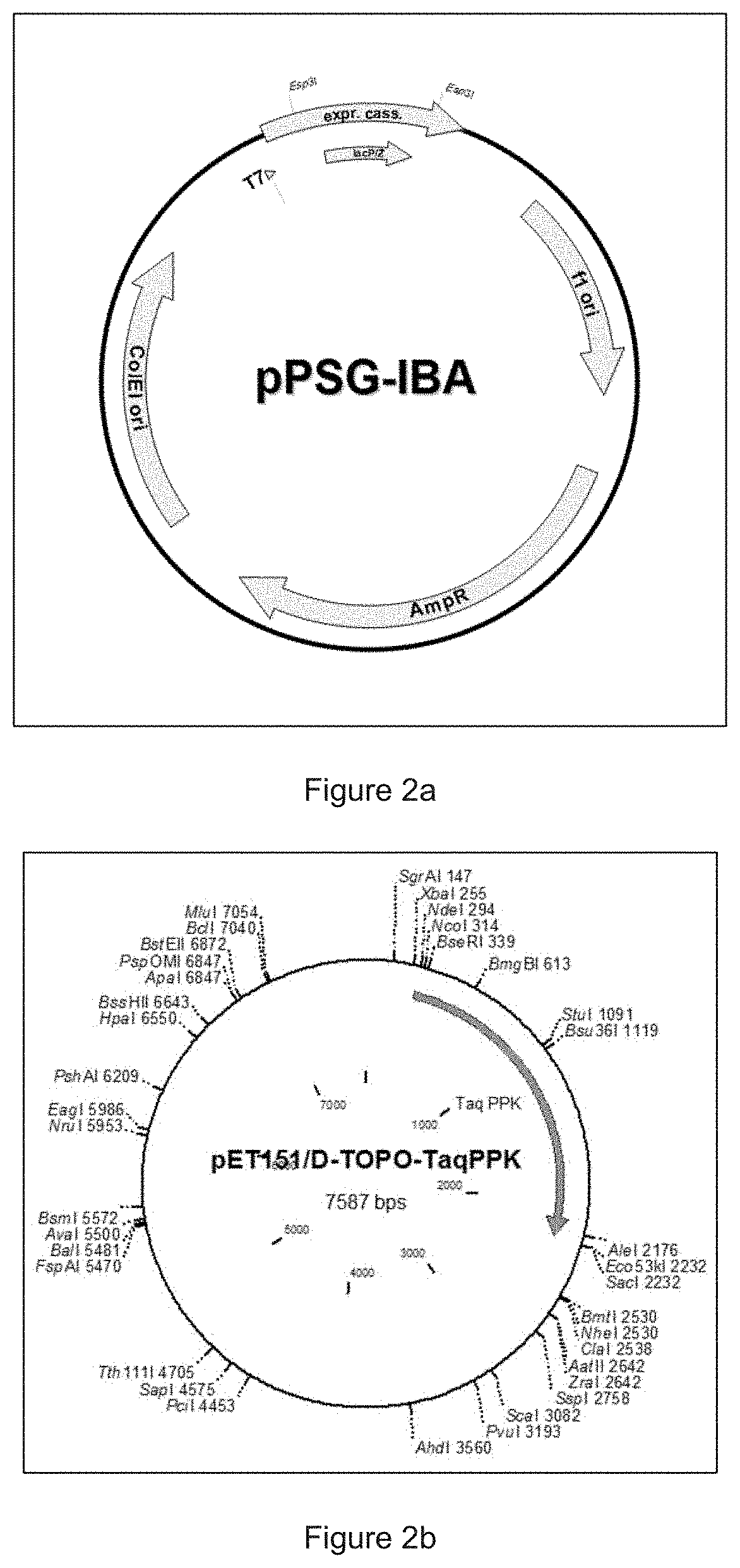
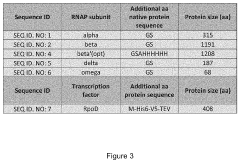
Kinetic Modeling of Protein Expression in Cell-free Systems
OCT 13, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Cell-free Protein Expression Background and Objectives
Cell-free protein expression systems have evolved significantly since their inception in the 1960s, originating from the groundbreaking work of Nirenberg and Matthaei who utilized cell extracts to decipher the genetic code. These systems represent a paradigm shift from traditional in vivo protein production methods by eliminating the constraints of cell viability and growth. The technology has progressed from simple batch reactions to sophisticated continuous-exchange systems capable of sustained protein synthesis over extended periods.
The field has witnessed remarkable technological advancements in recent decades, transitioning from academic curiosity to practical biotechnological tools. Modern cell-free systems derive from various organisms including Escherichia coli, wheat germ, rabbit reticulocytes, and insect cells, each offering distinct advantages for specific applications. The development trajectory indicates a clear trend toward more efficient, scalable, and customizable platforms for protein production.
Kinetic modeling of these systems represents a critical frontier in optimizing their performance and predictability. Understanding the complex interplay of transcription, translation, energy metabolism, and resource consumption requires sophisticated mathematical frameworks that can capture both the dynamic behavior and steady-state characteristics of these multi-component reactions.
The primary objectives of kinetic modeling in cell-free protein expression include: establishing quantitative relationships between system components and protein yield; identifying rate-limiting steps in the expression process; optimizing reaction conditions for maximum productivity; and developing predictive tools for rational design of synthetic biological circuits. These models aim to transform cell-free systems from empirically-driven platforms to precisely engineered production systems with predictable outcomes.
Current modeling approaches range from simplified black-box models to comprehensive mechanistic representations incorporating hundreds of parameters. The evolution of computational capabilities has enabled increasingly complex simulations that account for resource limitations, metabolic fluctuations, and molecular crowding effects that influence expression kinetics.
The ultimate goal of this technical research is to develop robust, validated kinetic models that can accurately predict protein expression dynamics across different scales and configurations of cell-free systems. Such models would significantly accelerate the development cycle for biopharmaceuticals, enable rapid prototyping of synthetic biological circuits, and advance fundamental understanding of translation processes outside the cellular context.
As the field progresses, integration of machine learning approaches with mechanistic modeling presents promising opportunities for creating hybrid models that combine theoretical rigor with data-driven insights, potentially revolutionizing our ability to design and control cell-free protein expression systems.
The field has witnessed remarkable technological advancements in recent decades, transitioning from academic curiosity to practical biotechnological tools. Modern cell-free systems derive from various organisms including Escherichia coli, wheat germ, rabbit reticulocytes, and insect cells, each offering distinct advantages for specific applications. The development trajectory indicates a clear trend toward more efficient, scalable, and customizable platforms for protein production.
Kinetic modeling of these systems represents a critical frontier in optimizing their performance and predictability. Understanding the complex interplay of transcription, translation, energy metabolism, and resource consumption requires sophisticated mathematical frameworks that can capture both the dynamic behavior and steady-state characteristics of these multi-component reactions.
The primary objectives of kinetic modeling in cell-free protein expression include: establishing quantitative relationships between system components and protein yield; identifying rate-limiting steps in the expression process; optimizing reaction conditions for maximum productivity; and developing predictive tools for rational design of synthetic biological circuits. These models aim to transform cell-free systems from empirically-driven platforms to precisely engineered production systems with predictable outcomes.
Current modeling approaches range from simplified black-box models to comprehensive mechanistic representations incorporating hundreds of parameters. The evolution of computational capabilities has enabled increasingly complex simulations that account for resource limitations, metabolic fluctuations, and molecular crowding effects that influence expression kinetics.
The ultimate goal of this technical research is to develop robust, validated kinetic models that can accurately predict protein expression dynamics across different scales and configurations of cell-free systems. Such models would significantly accelerate the development cycle for biopharmaceuticals, enable rapid prototyping of synthetic biological circuits, and advance fundamental understanding of translation processes outside the cellular context.
As the field progresses, integration of machine learning approaches with mechanistic modeling presents promising opportunities for creating hybrid models that combine theoretical rigor with data-driven insights, potentially revolutionizing our ability to design and control cell-free protein expression systems.
Market Analysis for Cell-free Protein Production Systems
The cell-free protein production systems market has witnessed significant growth in recent years, driven by increasing applications in synthetic biology, personalized medicine, and biopharmaceutical development. The global market value reached approximately $103 million in 2022 and is projected to grow at a compound annual growth rate (CAGR) of 6.8% through 2028, potentially reaching $153 million by the end of the forecast period.
Research institutions currently dominate the market demand, accounting for roughly 45% of the total market share, followed by pharmaceutical and biotechnology companies at 35%. This distribution reflects the technology's current positioning primarily as a research tool rather than a mainstream production platform. However, the transition from research to commercial applications is accelerating, particularly in therapeutic protein production and diagnostic applications.
Geographically, North America leads the market with approximately 40% share, followed by Europe (30%) and Asia-Pacific (20%). The dominance of these regions correlates with their advanced biotechnology infrastructure and substantial R&D investments. Notably, China and South Korea are emerging as rapidly growing markets, with annual growth rates exceeding 9%, driven by increasing government funding for biotechnology research.
The market segmentation reveals that E. coli-based cell-free systems hold the largest market share (55%), followed by wheat germ (20%) and rabbit reticulocyte lysate systems (15%). This preference stems from E. coli systems' cost-effectiveness and high protein yield capabilities. However, insect and mammalian cell-free systems are gaining traction due to their superior post-translational modification capabilities, essential for producing complex therapeutic proteins.
Key market drivers include the rising demand for rapid protein production methods, increasing focus on personalized medicine, and the growing need for efficient drug discovery tools. The COVID-19 pandemic has further accelerated market growth by highlighting the importance of rapid vaccine and therapeutic development platforms.
Challenges limiting market expansion include high production costs, scalability issues, and regulatory uncertainties surrounding products manufactured using cell-free systems. The average cost per reaction remains significantly higher than traditional cell-based methods, creating a barrier to widespread commercial adoption.
Customer needs analysis indicates growing demand for integrated systems that combine cell-free protein expression with real-time monitoring capabilities, allowing for kinetic modeling and optimization of protein production parameters. This trend aligns with the broader industry movement toward process analytical technology (PAT) and quality by design (QbD) approaches in biomanufacturing.
Research institutions currently dominate the market demand, accounting for roughly 45% of the total market share, followed by pharmaceutical and biotechnology companies at 35%. This distribution reflects the technology's current positioning primarily as a research tool rather than a mainstream production platform. However, the transition from research to commercial applications is accelerating, particularly in therapeutic protein production and diagnostic applications.
Geographically, North America leads the market with approximately 40% share, followed by Europe (30%) and Asia-Pacific (20%). The dominance of these regions correlates with their advanced biotechnology infrastructure and substantial R&D investments. Notably, China and South Korea are emerging as rapidly growing markets, with annual growth rates exceeding 9%, driven by increasing government funding for biotechnology research.
The market segmentation reveals that E. coli-based cell-free systems hold the largest market share (55%), followed by wheat germ (20%) and rabbit reticulocyte lysate systems (15%). This preference stems from E. coli systems' cost-effectiveness and high protein yield capabilities. However, insect and mammalian cell-free systems are gaining traction due to their superior post-translational modification capabilities, essential for producing complex therapeutic proteins.
Key market drivers include the rising demand for rapid protein production methods, increasing focus on personalized medicine, and the growing need for efficient drug discovery tools. The COVID-19 pandemic has further accelerated market growth by highlighting the importance of rapid vaccine and therapeutic development platforms.
Challenges limiting market expansion include high production costs, scalability issues, and regulatory uncertainties surrounding products manufactured using cell-free systems. The average cost per reaction remains significantly higher than traditional cell-based methods, creating a barrier to widespread commercial adoption.
Customer needs analysis indicates growing demand for integrated systems that combine cell-free protein expression with real-time monitoring capabilities, allowing for kinetic modeling and optimization of protein production parameters. This trend aligns with the broader industry movement toward process analytical technology (PAT) and quality by design (QbD) approaches in biomanufacturing.
Current Challenges in Kinetic Modeling of Cell-free Systems
Despite significant advancements in cell-free protein synthesis (CFPS) systems, kinetic modeling of these systems faces several substantial challenges that impede their broader application and optimization. One fundamental challenge is the inherent complexity of biochemical reactions occurring simultaneously in cell-free environments. Unlike cellular systems with compartmentalization and regulatory mechanisms, cell-free systems contain numerous components interacting in a less controlled manner, making comprehensive modeling extremely difficult.
The lack of standardized experimental protocols represents another significant obstacle. Different research groups employ varying preparation methods for cell extracts and reaction conditions, leading to inconsistent results across studies. This variability complicates the development of universally applicable kinetic models and hinders comparative analysis between different modeling approaches.
Data acquisition limitations further constrain modeling efforts. Real-time measurement of multiple reaction components simultaneously remains technically challenging. Current analytical techniques often provide only endpoint measurements or focus on a limited subset of components, resulting in incomplete datasets for model development and validation. This data scarcity forces modelers to make numerous assumptions, potentially reducing model accuracy.
Parameter estimation presents another formidable challenge. Cell-free systems involve dozens to hundreds of parameters, many of which cannot be directly measured. Statistical approaches for parameter estimation often struggle with identifiability issues when applied to such complex systems. The interdependence of parameters further complicates this process, as small variations in one parameter can significantly impact others.
Scale-up issues also plague kinetic modeling efforts. Models developed for small-scale laboratory reactions frequently fail to predict behavior in larger reaction volumes due to emerging phenomena like diffusion limitations, temperature gradients, and resource depletion patterns that differ between scales. This scale-dependence limits the industrial applicability of many current models.
Resource depletion dynamics remain poorly understood in kinetic models. As reactions progress, the consumption of energy sources, amino acids, and cofactors significantly alters reaction kinetics. Current models often inadequately capture these dynamic changes, particularly for extended reaction durations.
Finally, integrating stochastic effects into deterministic models presents a methodological challenge. Protein expression in cell-free systems exhibits inherent variability, especially at low concentrations of key components. Most current models rely on deterministic approaches that fail to capture this stochasticity, potentially missing important system behaviors at critical operational boundaries.
The lack of standardized experimental protocols represents another significant obstacle. Different research groups employ varying preparation methods for cell extracts and reaction conditions, leading to inconsistent results across studies. This variability complicates the development of universally applicable kinetic models and hinders comparative analysis between different modeling approaches.
Data acquisition limitations further constrain modeling efforts. Real-time measurement of multiple reaction components simultaneously remains technically challenging. Current analytical techniques often provide only endpoint measurements or focus on a limited subset of components, resulting in incomplete datasets for model development and validation. This data scarcity forces modelers to make numerous assumptions, potentially reducing model accuracy.
Parameter estimation presents another formidable challenge. Cell-free systems involve dozens to hundreds of parameters, many of which cannot be directly measured. Statistical approaches for parameter estimation often struggle with identifiability issues when applied to such complex systems. The interdependence of parameters further complicates this process, as small variations in one parameter can significantly impact others.
Scale-up issues also plague kinetic modeling efforts. Models developed for small-scale laboratory reactions frequently fail to predict behavior in larger reaction volumes due to emerging phenomena like diffusion limitations, temperature gradients, and resource depletion patterns that differ between scales. This scale-dependence limits the industrial applicability of many current models.
Resource depletion dynamics remain poorly understood in kinetic models. As reactions progress, the consumption of energy sources, amino acids, and cofactors significantly alters reaction kinetics. Current models often inadequately capture these dynamic changes, particularly for extended reaction durations.
Finally, integrating stochastic effects into deterministic models presents a methodological challenge. Protein expression in cell-free systems exhibits inherent variability, especially at low concentrations of key components. Most current models rely on deterministic approaches that fail to capture this stochasticity, potentially missing important system behaviors at critical operational boundaries.
Existing Kinetic Modeling Approaches and Frameworks
01 Mathematical modeling of protein expression kinetics
Mathematical models are developed to simulate and predict protein expression dynamics over time. These models incorporate various parameters such as transcription rates, translation efficiency, and protein degradation to accurately represent the kinetic behavior of protein expression systems. Advanced computational algorithms are used to analyze experimental data and optimize model parameters for improved predictive capabilities.- Mathematical modeling of protein expression kinetics: Mathematical models are developed to predict and analyze protein expression kinetics in biological systems. These models incorporate various parameters such as transcription rates, translation efficiency, and protein degradation to simulate the dynamic behavior of protein expression over time. Advanced computational algorithms are used to solve these mathematical models, enabling researchers to understand complex biological processes and optimize protein production.
- High-throughput methods for protein expression analysis: High-throughput technologies enable rapid analysis of protein expression kinetics across multiple conditions or samples simultaneously. These methods include automated systems for protein expression monitoring, data collection, and analysis. By integrating advanced detection technologies with computational tools, researchers can efficiently characterize protein expression patterns, identify optimal expression conditions, and accelerate the development of biopharmaceuticals and other protein-based products.
- Recombinant protein expression optimization: Techniques for optimizing recombinant protein expression focus on manipulating expression kinetics to maximize yield and quality. These approaches involve modifying expression vectors, selecting appropriate host cells, optimizing culture conditions, and controlling induction timing. By understanding the kinetics of protein expression, researchers can design strategies to overcome bottlenecks in protein production, reduce protein aggregation, and enhance the functionality of expressed proteins.
- Real-time monitoring of protein expression kinetics: Real-time monitoring systems allow continuous observation of protein expression kinetics throughout the production process. These systems utilize fluorescent reporters, biosensors, and spectroscopic techniques to track protein synthesis, folding, and accumulation in living cells. Real-time data enables immediate adjustments to expression conditions, facilitating process control and quality assurance in protein production for research and industrial applications.
- Machine learning approaches for protein expression prediction: Machine learning algorithms are applied to predict protein expression kinetics based on sequence features, expression conditions, and historical data. These computational approaches can identify patterns and relationships that influence protein expression, enabling more accurate predictions of expression outcomes. By leveraging large datasets and advanced analytics, researchers can develop models that guide experimental design, reduce trial-and-error approaches, and accelerate the development of efficient protein expression systems.
02 Real-time monitoring of protein expression kinetics
Systems and methods for real-time monitoring of protein expression kinetics enable continuous tracking of protein production in various expression systems. These approaches utilize fluorescent markers, biosensors, or other detection technologies to measure protein levels dynamically without disrupting the expression process. The real-time data collection allows for immediate adjustments to expression conditions and provides valuable insights into expression kinetics.Expand Specific Solutions03 Optimization of expression conditions based on kinetic models
Kinetic modeling approaches are used to optimize protein expression conditions in various host systems. By understanding the rate-limiting steps in protein production through kinetic analysis, researchers can modify culture conditions, genetic elements, or process parameters to enhance expression yields. These optimization strategies consider factors such as temperature, pH, nutrient availability, and induction timing to maximize protein production efficiency.Expand Specific Solutions04 Recombinant protein expression kinetics in different host systems
Comparative analysis of protein expression kinetics across different host systems helps in selecting the optimal expression platform for specific proteins. The kinetic parameters of protein expression vary significantly between bacterial, yeast, insect, and mammalian cell systems. Understanding these differences enables researchers to predict expression outcomes and choose the most suitable host system for particular protein production requirements.Expand Specific Solutions05 Machine learning approaches for protein expression kinetic prediction
Advanced machine learning algorithms are applied to predict protein expression kinetics based on sequence information and experimental data. These computational approaches can identify patterns and relationships between protein sequence features and expression parameters that are not apparent through traditional analysis methods. Machine learning models improve prediction accuracy for protein expression outcomes and help in designing expression systems with desired kinetic properties.Expand Specific Solutions
Leading Research Groups and Companies in Cell-free Systems
The cell-free protein expression market is currently in a growth phase, characterized by increasing adoption across pharmaceutical and research sectors. The global market size is expanding rapidly, driven by advantages in speed, scalability, and flexibility over traditional expression systems. Technologically, this field shows moderate maturity with ongoing innovations. Leading players include Sutro Biopharma and Nature's Toolbox, which have developed proprietary cell-free platforms for therapeutic applications, while academic institutions like Cornell University and Swiss Federal Institute of Technology contribute significant research advances. Established companies like Shimadzu and Roche provide supporting technologies and instrumentation. Emerging players such as Nuclera and Invitris are introducing novel approaches to democratize protein synthesis technology, indicating a dynamic competitive landscape with substantial room for innovation and market expansion.
Sutro Biopharma, Inc.
Technical Solution: Sutro Biopharma has developed a proprietary cell-free protein synthesis platform called XpressCF® specifically designed for kinetic modeling of protein expression. Their system utilizes an E. coli extract-based approach that allows for precise control over reaction conditions and real-time monitoring of protein synthesis kinetics. The platform incorporates a mathematical framework that accounts for transcription, translation, and post-translational modifications in a coupled reaction environment. Sutro's technology enables the incorporation of non-natural amino acids at precise positions through their XpressCF+™ technology, allowing for site-specific conjugation of various molecules to the expressed proteins[1]. Their kinetic modeling approach includes detailed characterization of reaction rates, resource consumption, and inhibitory byproduct accumulation, which enables predictive optimization of protein yield and quality. The company has successfully scaled this system from microliter to 100L scale while maintaining consistent kinetic parameters[2].
Strengths: Highly controllable reaction environment allowing precise manipulation of expression parameters; capability to incorporate non-natural amino acids; scalable from small to large volumes while maintaining kinetic properties. Weaknesses: Relatively high cost compared to in vivo systems; limited post-translational modification capabilities compared to mammalian cell-based systems; potential challenges in modeling complex multi-domain proteins.
Nature's Toolbox, Inc.
Technical Solution: Nature's Toolbox (NTx) has developed a cell-free protein expression platform called CellFree™ with sophisticated kinetic modeling capabilities specifically designed for industrial applications. Their system utilizes a proprietary extract preparation method that enhances energy efficiency and extends reaction lifetimes, critical factors for accurate kinetic modeling over extended timeframes. NTx's approach incorporates a systems biology framework that accounts for the complex interplay between transcription, translation, and metabolic processes in cell-free environments. Their kinetic models integrate differential equation-based descriptions of reaction networks with empirical data collected through real-time monitoring technologies[5]. The company has developed specialized software tools that enable simulation of protein expression under various conditions, allowing for in silico optimization before experimental validation. NTx's platform includes capabilities for modeling the effects of different energy regeneration systems, buffer compositions, and inhibitory byproduct removal strategies on protein expression kinetics. Their technology has been successfully applied to the production of complex therapeutic proteins, vaccines, and enzymes with predictable yields and quality attributes[6].
Strengths: Extended reaction lifetimes enabling more complete kinetic characterization; integrated systems biology approach; specialized software tools for simulation and optimization. Weaknesses: Proprietary nature limits academic adoption and cross-validation; potential challenges in modeling membrane proteins; higher complexity may increase development time for new applications.
Key Mathematical Models and Computational Methods
Cell-Free Expression System Having Novel Inorganic Polyphosphate-Based Energy Regeneration
PatentActiveUS20210024912A1
Innovation
- A novel inorganic polyphosphate-based energy regeneration system using synergistic high-efficiency kinase proteins from select bacterial strains, such as AdK and PPK, to drive the chemical regeneration of ATP, extending reaction time and increasing protein yields.
Cell-free protein expression systems and methods of use thereof
PatentWO2007044894A3
Innovation
- Development of specialized in vitro translation (IVT) systems optimized for the expression of protein tyrosine kinases (PTKs), including both receptor and cytoplasmic tyrosine kinases.
- Ability to express functional fragments of kinases (e.g., isolated kinase domains or active fragments) in a cell-free environment, enabling studies of specific functional domains without interference from cellular components.
- Creation of a panel-based approach for simultaneous expression of multiple tyrosine kinases in a cell-free system, facilitating comparative studies and screening applications.
Scalability and Industrial Applications of Cell-free Systems
Cell-free protein synthesis (CFPS) systems have evolved from laboratory research tools to promising platforms for industrial applications. The scalability of these systems represents a critical factor in their commercial viability. Recent advancements have demonstrated successful scale-up from microliter to liter volumes while maintaining productivity, addressing one of the major historical limitations of cell-free systems.
Industrial applications of cell-free systems span multiple sectors, with biopharmaceuticals leading adoption. Companies like Sutro Biopharma and Greenlight Biosciences have pioneered commercial applications for rapid production of therapeutic proteins and vaccines. The COVID-19 pandemic accelerated interest in cell-free systems as rapid-response platforms for diagnostic reagents and vaccine components, highlighting their advantage in speed-to-market scenarios.
The economic considerations for industrial implementation have improved significantly. Production costs have decreased from approximately $1000/mg to under $100/mg for many proteins over the past decade. This cost reduction stems from improved extract preparation methods, enhanced energy regeneration systems, and optimized reaction conditions derived from kinetic modeling insights.
Technical challenges for industrial scale-up include maintaining reaction homogeneity, managing heat transfer in larger volumes, and ensuring consistent quality across batches. Continuous-flow cell-free systems represent a promising approach to overcome volume limitations while enabling steady-state production. These systems have demonstrated production runs exceeding 24 hours, compared to typical batch reactions that terminate within 4-6 hours.
Regulatory frameworks for cell-free produced products are still evolving. The absence of living cells in final products potentially simplifies regulatory approval processes compared to traditional cell-based manufacturing. Several companies have cell-free produced products in clinical trials, establishing precedents for regulatory pathways.
Future industrial applications may extend beyond protein production to include on-demand biosensors, distributed manufacturing of biologics, and personalized medicine applications. The integration of kinetic modeling with process engineering principles will be crucial for optimizing large-scale operations and ensuring economic viability across these diverse applications.
Industrial applications of cell-free systems span multiple sectors, with biopharmaceuticals leading adoption. Companies like Sutro Biopharma and Greenlight Biosciences have pioneered commercial applications for rapid production of therapeutic proteins and vaccines. The COVID-19 pandemic accelerated interest in cell-free systems as rapid-response platforms for diagnostic reagents and vaccine components, highlighting their advantage in speed-to-market scenarios.
The economic considerations for industrial implementation have improved significantly. Production costs have decreased from approximately $1000/mg to under $100/mg for many proteins over the past decade. This cost reduction stems from improved extract preparation methods, enhanced energy regeneration systems, and optimized reaction conditions derived from kinetic modeling insights.
Technical challenges for industrial scale-up include maintaining reaction homogeneity, managing heat transfer in larger volumes, and ensuring consistent quality across batches. Continuous-flow cell-free systems represent a promising approach to overcome volume limitations while enabling steady-state production. These systems have demonstrated production runs exceeding 24 hours, compared to typical batch reactions that terminate within 4-6 hours.
Regulatory frameworks for cell-free produced products are still evolving. The absence of living cells in final products potentially simplifies regulatory approval processes compared to traditional cell-based manufacturing. Several companies have cell-free produced products in clinical trials, establishing precedents for regulatory pathways.
Future industrial applications may extend beyond protein production to include on-demand biosensors, distributed manufacturing of biologics, and personalized medicine applications. The integration of kinetic modeling with process engineering principles will be crucial for optimizing large-scale operations and ensuring economic viability across these diverse applications.
Regulatory Considerations for Cell-free Protein Products
The regulatory landscape for cell-free protein products presents unique challenges compared to traditional biopharmaceuticals. As these technologies advance from laboratory research to commercial applications, manufacturers must navigate complex regulatory frameworks that were primarily designed for cell-based production systems. The U.S. Food and Drug Administration (FDA) and European Medicines Agency (EMA) have begun developing specific guidance for cell-free expression systems, focusing on quality control parameters that address the distinct characteristics of these production methods.
Key regulatory considerations include the sourcing and quality of cell extracts, which must be thoroughly characterized and consistently produced. Regulatory bodies require comprehensive documentation of extract preparation methods, component origins, and batch-to-batch reproducibility. The absence of cellular containment in these systems necessitates stringent purity standards and robust contaminant removal processes, particularly for endotoxins and nucleic acid fragments that may remain from cell lysates.
Quality by Design (QbD) principles are increasingly emphasized for cell-free protein products, requiring manufacturers to establish critical quality attributes (CQAs) specific to cell-free expression systems. These include kinetic parameters that influence protein folding, post-translational modifications, and functional activity. Regulatory submissions must demonstrate how these parameters are monitored and controlled throughout the production process.
Stability testing protocols for cell-free protein products require adaptation from traditional guidelines, as these products may exhibit different degradation pathways and shelf-life characteristics. Regulators expect comprehensive stability data that accounts for the unique composition of cell-free reaction environments and their impact on product integrity over time.
Scale-up considerations present another regulatory challenge, as the kinetic modeling that proves effective at laboratory scale must demonstrate consistent performance at commercial production volumes. Regulatory agencies increasingly request computational models that predict how reaction kinetics will behave across different scales, supported by experimental validation data.
International harmonization of regulatory requirements remains incomplete for cell-free protein products, creating compliance challenges for global manufacturers. Industry consortia and regulatory science initiatives are working to establish standardized approaches to safety assessment, quality control, and validation methodologies specifically tailored to cell-free expression systems.
As the field evolves, regulatory frameworks are expected to incorporate more sophisticated understanding of the kinetic modeling that underlies cell-free protein expression, potentially leading to streamlined approval pathways for products that demonstrate well-characterized and controlled production processes.
Key regulatory considerations include the sourcing and quality of cell extracts, which must be thoroughly characterized and consistently produced. Regulatory bodies require comprehensive documentation of extract preparation methods, component origins, and batch-to-batch reproducibility. The absence of cellular containment in these systems necessitates stringent purity standards and robust contaminant removal processes, particularly for endotoxins and nucleic acid fragments that may remain from cell lysates.
Quality by Design (QbD) principles are increasingly emphasized for cell-free protein products, requiring manufacturers to establish critical quality attributes (CQAs) specific to cell-free expression systems. These include kinetic parameters that influence protein folding, post-translational modifications, and functional activity. Regulatory submissions must demonstrate how these parameters are monitored and controlled throughout the production process.
Stability testing protocols for cell-free protein products require adaptation from traditional guidelines, as these products may exhibit different degradation pathways and shelf-life characteristics. Regulators expect comprehensive stability data that accounts for the unique composition of cell-free reaction environments and their impact on product integrity over time.
Scale-up considerations present another regulatory challenge, as the kinetic modeling that proves effective at laboratory scale must demonstrate consistent performance at commercial production volumes. Regulatory agencies increasingly request computational models that predict how reaction kinetics will behave across different scales, supported by experimental validation data.
International harmonization of regulatory requirements remains incomplete for cell-free protein products, creating compliance challenges for global manufacturers. Industry consortia and regulatory science initiatives are working to establish standardized approaches to safety assessment, quality control, and validation methodologies specifically tailored to cell-free expression systems.
As the field evolves, regulatory frameworks are expected to incorporate more sophisticated understanding of the kinetic modeling that underlies cell-free protein expression, potentially leading to streamlined approval pathways for products that demonstrate well-characterized and controlled production processes.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!