Cell-free Protein Synthesis for Site-Specific Labeling
OCT 13, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
Cell-free Protein Synthesis Evolution and Objectives
Cell-free protein synthesis (CFPS) has evolved significantly since its inception in the 1960s when Nirenberg and Matthaei first demonstrated protein synthesis outside living cells. Initially developed as a research tool to decipher the genetic code, CFPS systems have transformed into sophisticated platforms for protein production and engineering. The evolution of CFPS technology has been marked by continuous improvements in extract preparation methods, energy regeneration systems, and reaction conditions, leading to enhanced protein yields and extended reaction durations.
The early CFPS systems derived from E. coli extracts suffered from limited productivity due to rapid depletion of energy resources and accumulation of inhibitory byproducts. Significant breakthroughs occurred in the 1990s and early 2000s with the development of continuous-exchange cell-free (CECF) systems and improved energy regeneration pathways, which substantially increased protein yields from micrograms to milligrams per milliliter of reaction volume.
Further advancements came with the diversification of cell extract sources beyond E. coli to include wheat germ, rabbit reticulocytes, insect cells, and human cell lines, each offering unique advantages for specific applications. The integration of non-canonical amino acids (ncAAs) into CFPS systems represented another pivotal development, enabling site-specific labeling of proteins with various functional groups, fluorophores, and affinity tags.
The primary objective of CFPS for site-specific labeling is to develop efficient and versatile methods for incorporating specific labels at defined positions within protein structures. This capability is crucial for studying protein dynamics, interactions, and functions in complex biological systems. Site-specific labeling aims to overcome the limitations of traditional labeling approaches that often result in heterogeneous products or compromise protein functionality.
Technical objectives include optimizing orthogonal translation systems for efficient ncAA incorporation, developing novel bioorthogonal chemistries for post-translational modifications, and enhancing the scalability of CFPS reactions for industrial applications. Additionally, researchers aim to expand the repertoire of incorporable ncAAs and compatible labeling chemistries to increase the diversity of functional groups that can be introduced into proteins.
The long-term vision encompasses the development of fully automated, high-throughput CFPS platforms capable of producing libraries of site-specifically labeled proteins for applications in structural biology, drug discovery, diagnostics, and therapeutic development. Achieving these objectives would significantly advance our understanding of protein function and accelerate the development of novel protein-based technologies and therapeutics.
The early CFPS systems derived from E. coli extracts suffered from limited productivity due to rapid depletion of energy resources and accumulation of inhibitory byproducts. Significant breakthroughs occurred in the 1990s and early 2000s with the development of continuous-exchange cell-free (CECF) systems and improved energy regeneration pathways, which substantially increased protein yields from micrograms to milligrams per milliliter of reaction volume.
Further advancements came with the diversification of cell extract sources beyond E. coli to include wheat germ, rabbit reticulocytes, insect cells, and human cell lines, each offering unique advantages for specific applications. The integration of non-canonical amino acids (ncAAs) into CFPS systems represented another pivotal development, enabling site-specific labeling of proteins with various functional groups, fluorophores, and affinity tags.
The primary objective of CFPS for site-specific labeling is to develop efficient and versatile methods for incorporating specific labels at defined positions within protein structures. This capability is crucial for studying protein dynamics, interactions, and functions in complex biological systems. Site-specific labeling aims to overcome the limitations of traditional labeling approaches that often result in heterogeneous products or compromise protein functionality.
Technical objectives include optimizing orthogonal translation systems for efficient ncAA incorporation, developing novel bioorthogonal chemistries for post-translational modifications, and enhancing the scalability of CFPS reactions for industrial applications. Additionally, researchers aim to expand the repertoire of incorporable ncAAs and compatible labeling chemistries to increase the diversity of functional groups that can be introduced into proteins.
The long-term vision encompasses the development of fully automated, high-throughput CFPS platforms capable of producing libraries of site-specifically labeled proteins for applications in structural biology, drug discovery, diagnostics, and therapeutic development. Achieving these objectives would significantly advance our understanding of protein function and accelerate the development of novel protein-based technologies and therapeutics.
Market Analysis for Site-Specific Labeling Applications
The site-specific labeling market has experienced significant growth in recent years, driven by increasing demand for precision in protein research and therapeutic applications. The global market for protein labeling technologies was valued at approximately $2.1 billion in 2022 and is projected to reach $3.5 billion by 2027, representing a compound annual growth rate (CAGR) of 10.8%. Within this broader market, site-specific labeling applications are emerging as a particularly dynamic segment.
Pharmaceutical and biotechnology companies represent the largest market segment, accounting for roughly 45% of the total market share. These organizations utilize site-specific labeling technologies primarily for drug discovery, protein-protein interaction studies, and the development of antibody-drug conjugates (ADCs). The ADC market alone is expected to grow from $7.9 billion in 2022 to $16.4 billion by 2026, creating substantial demand for advanced site-specific labeling methods.
Academic and research institutions constitute the second-largest market segment at approximately 30%, where site-specific labeling techniques are employed for fundamental protein research, structural biology studies, and method development. Government funding for proteomics research has increased by 15% over the past five years, further stimulating market growth in this sector.
Geographically, North America dominates the market with a 40% share, followed by Europe (30%) and Asia-Pacific (25%). The Asia-Pacific region, particularly China and India, is expected to witness the fastest growth rate of 14.2% annually due to increasing investment in life sciences research infrastructure and growing biotechnology sectors.
Cell-free protein synthesis (CFPS) for site-specific labeling represents a specialized but rapidly growing sub-segment. The CFPS market was valued at $246 million in 2022 and is projected to reach $372 million by 2026. The integration of CFPS with site-specific labeling technologies offers significant advantages in terms of speed, flexibility, and precision, driving adoption across various application areas.
Key application areas showing strong market demand include proteomics (35% of applications), diagnostics (25%), therapeutics development (20%), and structural biology (15%). The diagnostics segment is expected to grow at the highest rate due to increasing demand for highly sensitive protein detection methods in clinical settings.
Customer pain points driving market growth include the need for higher labeling specificity, reduced background signal, compatibility with complex biological environments, and scalability for industrial applications. Companies addressing these challenges through innovative CFPS-based site-specific labeling technologies are positioned to capture significant market share in this evolving landscape.
Pharmaceutical and biotechnology companies represent the largest market segment, accounting for roughly 45% of the total market share. These organizations utilize site-specific labeling technologies primarily for drug discovery, protein-protein interaction studies, and the development of antibody-drug conjugates (ADCs). The ADC market alone is expected to grow from $7.9 billion in 2022 to $16.4 billion by 2026, creating substantial demand for advanced site-specific labeling methods.
Academic and research institutions constitute the second-largest market segment at approximately 30%, where site-specific labeling techniques are employed for fundamental protein research, structural biology studies, and method development. Government funding for proteomics research has increased by 15% over the past five years, further stimulating market growth in this sector.
Geographically, North America dominates the market with a 40% share, followed by Europe (30%) and Asia-Pacific (25%). The Asia-Pacific region, particularly China and India, is expected to witness the fastest growth rate of 14.2% annually due to increasing investment in life sciences research infrastructure and growing biotechnology sectors.
Cell-free protein synthesis (CFPS) for site-specific labeling represents a specialized but rapidly growing sub-segment. The CFPS market was valued at $246 million in 2022 and is projected to reach $372 million by 2026. The integration of CFPS with site-specific labeling technologies offers significant advantages in terms of speed, flexibility, and precision, driving adoption across various application areas.
Key application areas showing strong market demand include proteomics (35% of applications), diagnostics (25%), therapeutics development (20%), and structural biology (15%). The diagnostics segment is expected to grow at the highest rate due to increasing demand for highly sensitive protein detection methods in clinical settings.
Customer pain points driving market growth include the need for higher labeling specificity, reduced background signal, compatibility with complex biological environments, and scalability for industrial applications. Companies addressing these challenges through innovative CFPS-based site-specific labeling technologies are positioned to capture significant market share in this evolving landscape.
Technical Barriers in Cell-free Protein Labeling
Despite significant advancements in cell-free protein synthesis (CFPS) for site-specific labeling, several technical barriers continue to impede broader implementation and commercial viability. The primary challenge remains the limited efficiency of incorporation of non-canonical amino acids (ncAAs), with success rates typically ranging from 20-80% depending on the specific ncAA and target protein. This variability creates significant obstacles for applications requiring high purity and homogeneity, particularly in therapeutic protein development.
Extract preparation represents another substantial hurdle, as current methods yield inconsistent quality across batches. The presence of proteases, nucleases, and other degradative enzymes in crude extracts can compromise both the stability of the template DNA/RNA and the synthesized protein products. Furthermore, the shelf-life of these extracts rarely exceeds 6-12 months even under optimal storage conditions, limiting their practical utility in industrial settings.
Energy regeneration systems constitute a critical bottleneck in sustained CFPS reactions. Most current systems can maintain protein synthesis for only 4-8 hours before energy depletion occurs, whereas optimal labeling often requires extended reaction times. The cost of energy substrates like phosphoenolpyruvate (PEP) or creatine phosphate also contributes significantly to the overall expense of CFPS reactions, often accounting for 30-40% of total material costs.
Scale-up challenges present formidable barriers to industrial implementation. Most successful CFPS labeling protocols operate efficiently at microliter to milliliter scales, but encounter significant performance decreases when scaled to industrial volumes. Factors including oxygen transfer limitations, heat dissipation issues, and mixing inefficiencies contribute to this scale-dependent decline in performance.
The orthogonal translation machinery required for site-specific incorporation of ncAAs introduces additional complexity. Current orthogonal aminoacyl-tRNA synthetase/tRNA pairs often exhibit cross-reactivity with endogenous components, leading to misincorporation and reduced specificity. Engineering these components for improved specificity while maintaining activity remains technically challenging.
Post-translational modifications present another significant barrier, as many CFPS systems lack the enzymatic machinery necessary for glycosylation, phosphorylation, and other modifications essential for protein functionality. This limitation particularly affects the production of therapeutic proteins and complex enzymes where such modifications are crucial for biological activity.
Regulatory and quality control challenges also persist, as standardized protocols for validating CFPS-produced labeled proteins remain underdeveloped. The absence of comprehensive analytical methods to verify site-specific incorporation efficiency and to detect potential side reactions or misincorporations hampers regulatory approval processes for CFPS-derived products.
Extract preparation represents another substantial hurdle, as current methods yield inconsistent quality across batches. The presence of proteases, nucleases, and other degradative enzymes in crude extracts can compromise both the stability of the template DNA/RNA and the synthesized protein products. Furthermore, the shelf-life of these extracts rarely exceeds 6-12 months even under optimal storage conditions, limiting their practical utility in industrial settings.
Energy regeneration systems constitute a critical bottleneck in sustained CFPS reactions. Most current systems can maintain protein synthesis for only 4-8 hours before energy depletion occurs, whereas optimal labeling often requires extended reaction times. The cost of energy substrates like phosphoenolpyruvate (PEP) or creatine phosphate also contributes significantly to the overall expense of CFPS reactions, often accounting for 30-40% of total material costs.
Scale-up challenges present formidable barriers to industrial implementation. Most successful CFPS labeling protocols operate efficiently at microliter to milliliter scales, but encounter significant performance decreases when scaled to industrial volumes. Factors including oxygen transfer limitations, heat dissipation issues, and mixing inefficiencies contribute to this scale-dependent decline in performance.
The orthogonal translation machinery required for site-specific incorporation of ncAAs introduces additional complexity. Current orthogonal aminoacyl-tRNA synthetase/tRNA pairs often exhibit cross-reactivity with endogenous components, leading to misincorporation and reduced specificity. Engineering these components for improved specificity while maintaining activity remains technically challenging.
Post-translational modifications present another significant barrier, as many CFPS systems lack the enzymatic machinery necessary for glycosylation, phosphorylation, and other modifications essential for protein functionality. This limitation particularly affects the production of therapeutic proteins and complex enzymes where such modifications are crucial for biological activity.
Regulatory and quality control challenges also persist, as standardized protocols for validating CFPS-produced labeled proteins remain underdeveloped. The absence of comprehensive analytical methods to verify site-specific incorporation efficiency and to detect potential side reactions or misincorporations hampers regulatory approval processes for CFPS-derived products.
Current Methodologies for Site-Specific Labeling
01 Incorporation of unnatural amino acids for site-specific labeling
Cell-free protein synthesis systems can be engineered to incorporate unnatural amino acids at specific sites in proteins. These unnatural amino acids often contain unique functional groups that can be selectively labeled with various probes. This approach allows for precise control over the location of labels in the protein structure, enabling studies of protein structure, function, and interactions with minimal disruption to the native protein.- Incorporation of unnatural amino acids for site-specific labeling: Cell-free protein synthesis systems can be engineered to incorporate unnatural amino acids at specific sites in proteins. These unnatural amino acids often contain unique functional groups that can be selectively labeled with various probes. This approach allows for precise control over the location of labels in the protein structure, enabling studies of protein structure, function, and interactions with minimal disruption to the native protein.
- tRNA-mediated labeling strategies: Modified tRNAs can be used in cell-free protein synthesis systems to introduce labels at specific sites. These tRNAs are often chemically aminoacylated with labeled amino acids or amino acid analogs. When these charged tRNAs are added to the cell-free system, they can incorporate the labeled amino acids at specific codons, allowing for site-specific labeling of proteins during translation.
- Enzymatic approaches for post-translational labeling: After cell-free protein synthesis, enzymatic methods can be used to introduce labels at specific sites in proteins. These methods often involve engineered proteins with tags that can be recognized by specific enzymes, which then catalyze the attachment of labels to these tags. This approach allows for site-specific labeling without the need to modify the translation machinery and can be performed under mild conditions that preserve protein structure and function.
- Click chemistry and bioorthogonal reactions: Cell-free protein synthesis can be combined with click chemistry and other bioorthogonal reactions for site-specific labeling. In this approach, proteins are first synthesized with reactive handles at specific sites, and then these handles are selectively modified with labels using highly specific chemical reactions. These reactions are compatible with biological systems and can be performed in aqueous solutions under mild conditions, making them ideal for labeling proteins produced in cell-free systems.
- Fluorescent and affinity tag labeling methods: Various fluorescent and affinity tags can be incorporated into proteins during cell-free synthesis for detection and purification purposes. These tags can be genetically encoded and fused to the protein of interest, or they can be introduced through modified amino acids. Fluorescent tags enable visualization and tracking of proteins, while affinity tags facilitate purification and immobilization. Both types of tags can be introduced at specific sites to minimize interference with protein function.
02 tRNA-mediated site-specific labeling methods
Modified tRNAs can be used in cell-free protein synthesis systems to introduce labeled amino acids at specific sites. These tRNAs are often chemically aminoacylated with the desired labeled amino acid and then added to the cell-free system. The specificity is achieved through the use of suppressor tRNAs that recognize specific codons, such as amber stop codons, allowing for site-directed incorporation of labels at predetermined positions in the protein sequence.Expand Specific Solutions03 Enzymatic approaches for post-translational labeling
After cell-free protein synthesis, enzymatic methods can be employed to achieve site-specific labeling. These methods utilize enzymes that recognize specific amino acid sequences or structural motifs in the synthesized protein and catalyze the attachment of labels at these sites. This approach offers high specificity and can be performed under mild conditions that preserve protein structure and function.Expand Specific Solutions04 Click chemistry and bioorthogonal reactions for labeling
Cell-free protein synthesis can be combined with click chemistry and other bioorthogonal reactions to achieve site-specific labeling. This approach involves incorporating amino acids with reactive handles during protein synthesis, followed by selective chemical reactions with complementary labeled probes. These reactions are highly specific and can occur in complex biological environments without interfering with native biochemical processes.Expand Specific Solutions05 Optimization of cell-free systems for enhanced labeling efficiency
Various modifications to cell-free protein synthesis systems have been developed to improve the efficiency of site-specific labeling. These include engineering of the translation machinery, optimization of reaction conditions, and development of specialized cell extracts. Such optimizations can increase the yield of labeled protein, improve the specificity of labeling, and expand the range of labels that can be incorporated.Expand Specific Solutions
Leading Research Groups and Companies
Cell-free Protein Synthesis for Site-Specific Labeling is in an early growth phase, characterized by increasing research interest but limited commercial applications. The global market is estimated at $100-150 million, with projected annual growth of 15-20% as applications expand in pharmaceutical research and diagnostics. Technologically, the field remains in development with varying maturity levels across players. Academic institutions (Rutgers, Tsinghua, ETH Zurich) lead fundamental research, while specialized companies like Cellfree Sciences and Anima Cell Metrology offer commercial platforms. Larger corporations (Novartis, Shimadzu, Olympus) are investing in the technology for drug development applications. Research collaborations between institutions like RIKEN and commercial entities are accelerating technological advancement toward broader market adoption.
Cellfree Sciences Co., Ltd.
Technical Solution: Cellfree Sciences has developed a proprietary WEPRO® cell-free protein synthesis system based on wheat germ extract, specifically optimized for site-specific labeling applications. Their technology enables the incorporation of non-canonical amino acids at predetermined positions using modified tRNA synthetases and orthogonal tRNA pairs. The company's approach allows for high-yield production (up to 5 mg/ml) of labeled proteins with minimal background incorporation, making it ideal for structural biology and protein interaction studies. Their system maintains protein folding fidelity while accommodating various labeling chemistries including fluorescent dyes, biotin tags, and isotopic labels. The wheat germ-based system offers advantages over E. coli systems by reducing endogenous amino acid competition and providing a eukaryotic-like environment for complex protein synthesis. Recent innovations include microfluidic integration for high-throughput applications and optimization for membrane protein labeling.
Strengths: Superior yield compared to many competing systems; excellent compatibility with eukaryotic proteins; reduced background labeling. Weaknesses: Higher cost than bacterial systems; more complex setup requirements; limited scalability for industrial applications compared to traditional expression systems.
Jiangsu Fulcrum Biotechnology Co., Ltd.
Technical Solution: Jiangsu Fulcrum Biotechnology has developed the FlexiCFPS™ platform for cell-free protein synthesis with enhanced site-specific labeling capabilities. Their technology combines optimized cell extracts from multiple sources (E. coli, wheat germ, and CHO cells) with proprietary reaction components to achieve high-yield production of labeled proteins. The company's approach utilizes a dual-plasmid system: one encoding the target protein with amber codons at desired labeling positions, and another expressing the orthogonal tRNA/synthetase pair optimized for specific non-canonical amino acids. This system achieves incorporation efficiencies of 60-75% at designated sites while maintaining protein yields of 50-300 μg/ml depending on the extract source. Fulcrum has further enhanced their technology by developing specialized microfluidic devices that enable continuous-exchange cell-free protein synthesis, extending reaction lifetimes and improving overall yields by approximately 2-3 fold compared to batch reactions. Their platform is particularly suitable for producing labeled membrane proteins and toxic proteins that are challenging to express in living cells.
Strengths: Flexibility in choosing optimal extract source for different protein types; good scalability from microliter to milliliter reactions; compatible with continuous-flow systems for extended reactions. Weaknesses: Variable performance across different protein targets; requires optimization for each new labeling application; moderate cost compared to simpler systems.
Key Patents in Cell-free Labeling Technology
Cell-free synthesis of isotopic labelled proteins from amino-acids precursors
PatentInactiveEP3589740A1
Innovation
- Development of a cell-free protein synthesis system that utilizes alpha-keto acid precursors (excluding 2-ketobutyric acid) for site-specific isotopic labeling of proteins.
- Integration of precursor transformation mechanisms directly within the cell-free extract, creating a one-pot system for converting precursors to amino acids and subsequently incorporating them into target proteins.
- Enabling improved NMR spectroscopy analysis of large proteins through selective isotopic labeling, enhancing both sensitivity and resolution compared to conventional methods.
Process for producing protein by cell-free protein synthesis system and proten synthesis reagent kit
PatentWO2004104210A1
Innovation
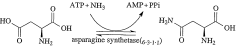
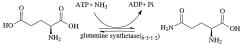
- Using a cell-free protein synthesis system with an amino acid mixture of 18 kinds, excluding asparagine and glutamine, and adding an ammonium salt to metabolically produce these amino acids, allowing specific labeling of their side chains or main chains without them, reducing costs and improving labeling efficiency.
Scalability and Production Challenges
Despite the promising potential of cell-free protein synthesis (CFPS) for site-specific labeling, significant scalability and production challenges remain that hinder its widespread industrial adoption. The transition from laboratory-scale experiments to commercial production faces several technical hurdles. Current CFPS systems typically operate at microliter to milliliter scales, with production yields ranging from micrograms to milligrams of protein per milliliter of reaction mixture, which is insufficient for many commercial applications requiring gram to kilogram quantities.
Energy supply represents a critical bottleneck in scaling up CFPS reactions. As reaction volumes increase, maintaining consistent ATP regeneration throughout the mixture becomes increasingly difficult. Traditional energy regeneration systems like phosphoenolpyruvate (PEP) are expensive and impractical for large-scale operations, while more cost-effective alternatives such as glucose-based systems often suffer from reduced efficiency at larger scales due to byproduct accumulation.
Extract preparation methodology presents another significant challenge. The quality and consistency of cell extracts directly impact CFPS performance, yet current preparation methods show considerable batch-to-batch variation. This variability becomes more pronounced at larger scales, where maintaining homogeneity throughout the extraction process is technically demanding. Furthermore, the specialized equipment and expertise required for extract preparation represent substantial barriers to entry for many potential users.
Component stability during prolonged reaction times poses additional challenges for scaled-up production. Many CFPS components, including enzymes and nucleic acids, exhibit degradation over time, limiting the duration of productive synthesis. This is particularly problematic for site-specific labeling applications, where extended reaction times may be necessary to achieve sufficient incorporation of non-canonical amino acids or other labeling components.
Cost considerations remain paramount in scaling CFPS technology. Current estimates place the cost of CFPS reactions at $0.26-4.00 per milligram of protein produced—significantly higher than conventional in vivo expression systems. The expenses associated with specialized reagents for site-specific labeling, such as orthogonal tRNA/synthetase pairs and non-canonical amino acids, further compound this economic challenge.
Regulatory and quality control frameworks for CFPS-based products are still evolving, creating uncertainty for commercial applications. Unlike traditional biopharmaceutical production platforms, standardized protocols for validating the consistency, purity, and safety of CFPS-derived proteins with site-specific modifications are not yet well established, presenting additional hurdles for regulatory approval and commercial deployment.
Energy supply represents a critical bottleneck in scaling up CFPS reactions. As reaction volumes increase, maintaining consistent ATP regeneration throughout the mixture becomes increasingly difficult. Traditional energy regeneration systems like phosphoenolpyruvate (PEP) are expensive and impractical for large-scale operations, while more cost-effective alternatives such as glucose-based systems often suffer from reduced efficiency at larger scales due to byproduct accumulation.
Extract preparation methodology presents another significant challenge. The quality and consistency of cell extracts directly impact CFPS performance, yet current preparation methods show considerable batch-to-batch variation. This variability becomes more pronounced at larger scales, where maintaining homogeneity throughout the extraction process is technically demanding. Furthermore, the specialized equipment and expertise required for extract preparation represent substantial barriers to entry for many potential users.
Component stability during prolonged reaction times poses additional challenges for scaled-up production. Many CFPS components, including enzymes and nucleic acids, exhibit degradation over time, limiting the duration of productive synthesis. This is particularly problematic for site-specific labeling applications, where extended reaction times may be necessary to achieve sufficient incorporation of non-canonical amino acids or other labeling components.
Cost considerations remain paramount in scaling CFPS technology. Current estimates place the cost of CFPS reactions at $0.26-4.00 per milligram of protein produced—significantly higher than conventional in vivo expression systems. The expenses associated with specialized reagents for site-specific labeling, such as orthogonal tRNA/synthetase pairs and non-canonical amino acids, further compound this economic challenge.
Regulatory and quality control frameworks for CFPS-based products are still evolving, creating uncertainty for commercial applications. Unlike traditional biopharmaceutical production platforms, standardized protocols for validating the consistency, purity, and safety of CFPS-derived proteins with site-specific modifications are not yet well established, presenting additional hurdles for regulatory approval and commercial deployment.
Bioethical Implications of Synthetic Biology
The rapid advancement of cell-free protein synthesis (CFPS) for site-specific labeling raises significant bioethical considerations within the broader context of synthetic biology. This technology, which enables the production of proteins outside living cells with precise control over amino acid incorporation, intersects with fundamental questions about human intervention in biological processes and the boundaries of artificial life creation.
The ability to engineer proteins with site-specific modifications represents a profound level of control over biological molecules that was previously unattainable. This capability raises questions about the appropriate limits of human manipulation of biological systems. While CFPS offers tremendous benefits for research and medicine, it also contributes to the blurring distinction between natural and artificial biological entities, challenging traditional ontological categories that inform ethical frameworks.
Concerns regarding biosafety and biosecurity are particularly relevant as CFPS technologies become more accessible. The reduced biological containment requirements compared to whole-cell systems may facilitate wider adoption but simultaneously create potential for misuse. The dual-use nature of this technology—capable of producing both therapeutic proteins and potentially harmful biological agents—necessitates robust governance frameworks that balance innovation with appropriate safeguards.
Intellectual property considerations surrounding CFPS and site-specific labeling technologies present another ethical dimension. Patent landscapes in this field may create access inequities, particularly for researchers and institutions in developing countries. This raises questions about global justice in scientific advancement and whether essential biotechnologies should be subject to different intellectual property regimes to ensure equitable access.
Environmental implications must also be considered, as the chemical components and energy requirements of CFPS systems may present sustainability challenges. While CFPS eliminates concerns about genetically modified organisms escaping into ecosystems, the disposal of reaction components and potential for novel protein interactions with environmental systems require careful assessment.
The democratization of protein engineering through CFPS technologies also raises questions about appropriate governance. As these technologies become more accessible to biohackers and DIY biology communities, ensuring responsible use while preserving innovation becomes increasingly complex. Participatory governance approaches that engage diverse stakeholders may be necessary to navigate these challenges effectively.
Finally, CFPS for site-specific labeling contributes to broader philosophical questions about human relationships with biological systems. As we gain unprecedented control over the fundamental building blocks of life, we must reconsider what constitutes responsible stewardship of biological technologies and how these advances align with diverse cultural and religious perspectives on the appropriate boundaries of biotechnological intervention.
The ability to engineer proteins with site-specific modifications represents a profound level of control over biological molecules that was previously unattainable. This capability raises questions about the appropriate limits of human manipulation of biological systems. While CFPS offers tremendous benefits for research and medicine, it also contributes to the blurring distinction between natural and artificial biological entities, challenging traditional ontological categories that inform ethical frameworks.
Concerns regarding biosafety and biosecurity are particularly relevant as CFPS technologies become more accessible. The reduced biological containment requirements compared to whole-cell systems may facilitate wider adoption but simultaneously create potential for misuse. The dual-use nature of this technology—capable of producing both therapeutic proteins and potentially harmful biological agents—necessitates robust governance frameworks that balance innovation with appropriate safeguards.
Intellectual property considerations surrounding CFPS and site-specific labeling technologies present another ethical dimension. Patent landscapes in this field may create access inequities, particularly for researchers and institutions in developing countries. This raises questions about global justice in scientific advancement and whether essential biotechnologies should be subject to different intellectual property regimes to ensure equitable access.
Environmental implications must also be considered, as the chemical components and energy requirements of CFPS systems may present sustainability challenges. While CFPS eliminates concerns about genetically modified organisms escaping into ecosystems, the disposal of reaction components and potential for novel protein interactions with environmental systems require careful assessment.
The democratization of protein engineering through CFPS technologies also raises questions about appropriate governance. As these technologies become more accessible to biohackers and DIY biology communities, ensuring responsible use while preserving innovation becomes increasingly complex. Participatory governance approaches that engage diverse stakeholders may be necessary to navigate these challenges effectively.
Finally, CFPS for site-specific labeling contributes to broader philosophical questions about human relationships with biological systems. As we gain unprecedented control over the fundamental building blocks of life, we must reconsider what constitutes responsible stewardship of biological technologies and how these advances align with diverse cultural and religious perspectives on the appropriate boundaries of biotechnological intervention.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!