Correlate Biological Activity with Protein Chemical Shifts in NMR
SEP 22, 20259 MIN READ
Generate Your Research Report Instantly with AI Agent
Patsnap Eureka helps you evaluate technical feasibility & market potential.
NMR Protein Shift Analysis Background and Objectives
Nuclear Magnetic Resonance (NMR) spectroscopy has evolved as a pivotal analytical technique in structural biology since its discovery in the 1940s. The correlation between protein chemical shifts and biological activity represents a frontier in biophysical research with significant implications for drug discovery, protein engineering, and understanding fundamental biological processes. This technological domain has witnessed remarkable advancements over the past three decades, transitioning from simple one-dimensional spectra to sophisticated multi-dimensional experiments capable of elucidating complex protein structures and dynamics.
The evolution of NMR technology for protein analysis has been characterized by several key milestones: the development of superconducting magnets enabling higher field strengths, the introduction of multi-dimensional pulse sequences, and the advancement of computational methods for spectral analysis. These innovations have collectively enhanced the resolution and information content obtainable from NMR experiments, making it possible to correlate subtle changes in chemical shifts with specific biological functions.
Current technological trends in this field include the integration of artificial intelligence for spectral interpretation, the development of hyperpolarization techniques to enhance sensitivity, and the combination of NMR with other analytical methods such as mass spectrometry and cryo-electron microscopy for comprehensive structural characterization. These developments are expanding the applicability of NMR beyond traditional structural determination to include dynamic analyses of protein-ligand interactions and conformational changes associated with biological activity.
The primary objective of correlating biological activity with protein chemical shifts is to establish quantitative structure-activity relationships that can inform rational drug design and protein engineering efforts. This involves developing robust methodologies for detecting and interpreting chemical shift perturbations induced by ligand binding, post-translational modifications, or environmental changes that modulate protein function.
Secondary objectives include enhancing the throughput and accessibility of NMR-based screening approaches, reducing the sample requirements for analysis, and improving the integration of NMR data with computational modeling to predict biological outcomes. These goals align with the broader aim of positioning NMR as a central tool in systems biology and personalized medicine, where understanding protein behavior at the molecular level is essential for developing targeted therapeutic interventions.
The technical challenges that must be addressed to achieve these objectives include improving signal-to-noise ratios for detecting subtle chemical shift changes, developing standardized protocols for data acquisition and analysis, and creating comprehensive databases that link chemical shift patterns to specific biological activities across diverse protein families. Overcoming these challenges requires interdisciplinary collaboration between spectroscopists, structural biologists, computational scientists, and medicinal chemists.
The evolution of NMR technology for protein analysis has been characterized by several key milestones: the development of superconducting magnets enabling higher field strengths, the introduction of multi-dimensional pulse sequences, and the advancement of computational methods for spectral analysis. These innovations have collectively enhanced the resolution and information content obtainable from NMR experiments, making it possible to correlate subtle changes in chemical shifts with specific biological functions.
Current technological trends in this field include the integration of artificial intelligence for spectral interpretation, the development of hyperpolarization techniques to enhance sensitivity, and the combination of NMR with other analytical methods such as mass spectrometry and cryo-electron microscopy for comprehensive structural characterization. These developments are expanding the applicability of NMR beyond traditional structural determination to include dynamic analyses of protein-ligand interactions and conformational changes associated with biological activity.
The primary objective of correlating biological activity with protein chemical shifts is to establish quantitative structure-activity relationships that can inform rational drug design and protein engineering efforts. This involves developing robust methodologies for detecting and interpreting chemical shift perturbations induced by ligand binding, post-translational modifications, or environmental changes that modulate protein function.
Secondary objectives include enhancing the throughput and accessibility of NMR-based screening approaches, reducing the sample requirements for analysis, and improving the integration of NMR data with computational modeling to predict biological outcomes. These goals align with the broader aim of positioning NMR as a central tool in systems biology and personalized medicine, where understanding protein behavior at the molecular level is essential for developing targeted therapeutic interventions.
The technical challenges that must be addressed to achieve these objectives include improving signal-to-noise ratios for detecting subtle chemical shift changes, developing standardized protocols for data acquisition and analysis, and creating comprehensive databases that link chemical shift patterns to specific biological activities across diverse protein families. Overcoming these challenges requires interdisciplinary collaboration between spectroscopists, structural biologists, computational scientists, and medicinal chemists.
Market Applications for Protein-Activity Correlation Methods
The protein-activity correlation market is experiencing significant growth, driven by the pharmaceutical and biotechnology sectors' increasing focus on structure-based drug design. The global market for protein analysis technologies, including NMR-based methods, was valued at approximately $32 billion in 2022 and is projected to reach $58 billion by 2028, with a compound annual growth rate of 10.4%.
Drug discovery represents the largest application segment, where correlating biological activity with protein chemical shifts provides critical insights into protein-ligand interactions. Pharmaceutical companies are leveraging these technologies to accelerate lead compound identification and optimization, potentially reducing drug development timelines by 20-30%. Companies like Merck, Pfizer, and Novartis have integrated NMR-based protein activity correlation methods into their drug discovery pipelines.
The diagnostic sector presents another substantial market opportunity. NMR-based protein activity correlation techniques are being developed for early disease detection, particularly for neurodegenerative disorders like Alzheimer's and Parkinson's. These methods can identify subtle changes in protein structure and function before clinical symptoms appear, potentially creating a market estimated at $5 billion by 2027.
Academic research institutions constitute a stable market segment, with universities and research centers investing in advanced NMR technologies to study protein dynamics and function. This segment accounts for approximately 25% of the total market and serves as an innovation hub for new methodological developments.
The agricultural biotechnology sector is emerging as a promising application area, using protein activity correlation to develop enhanced crop varieties and bio-pesticides. This segment is expected to grow at 12% annually over the next five years, reaching a market value of $3.5 billion by 2028.
Personalized medicine represents perhaps the most promising future application, with protein activity correlation methods being developed to predict individual patient responses to treatments. This approach could revolutionize therapeutic strategies for cancer, autoimmune disorders, and metabolic diseases, with a potential market exceeding $10 billion by 2030.
Geographically, North America dominates the market with a 45% share, followed by Europe (30%) and Asia-Pacific (20%). However, the Asia-Pacific region is experiencing the fastest growth rate at 15% annually, driven by increasing R&D investments in China, Japan, and South Korea.
Drug discovery represents the largest application segment, where correlating biological activity with protein chemical shifts provides critical insights into protein-ligand interactions. Pharmaceutical companies are leveraging these technologies to accelerate lead compound identification and optimization, potentially reducing drug development timelines by 20-30%. Companies like Merck, Pfizer, and Novartis have integrated NMR-based protein activity correlation methods into their drug discovery pipelines.
The diagnostic sector presents another substantial market opportunity. NMR-based protein activity correlation techniques are being developed for early disease detection, particularly for neurodegenerative disorders like Alzheimer's and Parkinson's. These methods can identify subtle changes in protein structure and function before clinical symptoms appear, potentially creating a market estimated at $5 billion by 2027.
Academic research institutions constitute a stable market segment, with universities and research centers investing in advanced NMR technologies to study protein dynamics and function. This segment accounts for approximately 25% of the total market and serves as an innovation hub for new methodological developments.
The agricultural biotechnology sector is emerging as a promising application area, using protein activity correlation to develop enhanced crop varieties and bio-pesticides. This segment is expected to grow at 12% annually over the next five years, reaching a market value of $3.5 billion by 2028.
Personalized medicine represents perhaps the most promising future application, with protein activity correlation methods being developed to predict individual patient responses to treatments. This approach could revolutionize therapeutic strategies for cancer, autoimmune disorders, and metabolic diseases, with a potential market exceeding $10 billion by 2030.
Geographically, North America dominates the market with a 45% share, followed by Europe (30%) and Asia-Pacific (20%). However, the Asia-Pacific region is experiencing the fastest growth rate at 15% annually, driven by increasing R&D investments in China, Japan, and South Korea.
Current Challenges in Biological Activity-Chemical Shift Correlation
Despite significant advancements in NMR spectroscopy techniques, correlating biological activity with protein chemical shifts remains challenging due to several fundamental limitations. The relationship between structural changes detected by chemical shifts and functional biological activity is often indirect and complex, making straightforward correlations difficult to establish. This complexity stems from the multifaceted nature of protein function, which involves dynamic conformational changes, allosteric effects, and interactions with various binding partners.
One major challenge is the inherent sensitivity limitations of NMR spectroscopy. While chemical shifts provide atomic-level information, they may not capture subtle conformational changes that significantly impact biological activity. Additionally, the signal-to-noise ratio in NMR experiments often requires high protein concentrations that may not reflect physiological conditions, potentially leading to artifacts or misinterpretations when correlating with biological activity.
The time scale discrepancy between NMR measurements and biological processes presents another significant obstacle. Many biological activities occur on microsecond to millisecond timescales, while NMR chemical shift measurements typically represent time-averaged values. This temporal mismatch can obscure critical dynamic processes that drive biological function, resulting in incomplete correlations between observed shifts and actual activity.
Environmental factors further complicate these correlations. In vitro NMR conditions (buffer composition, pH, temperature, absence of cellular components) often differ substantially from the native cellular environment where biological activity naturally occurs. These differences can significantly alter protein behavior, making direct extrapolation from NMR data to biological activity problematic.
Data interpretation challenges also persist in the field. Chemical shift perturbations can result from various factors beyond those directly related to biological activity, including non-specific binding events, minor structural adjustments, or experimental artifacts. Distinguishing relevant shifts from background variations requires sophisticated statistical approaches and validation through complementary techniques.
The lack of standardized protocols for correlating chemical shifts with biological activity represents a methodological barrier. Different research groups employ varied approaches for data collection, processing, and analysis, making cross-study comparisons difficult and hindering the development of robust correlation models. This methodological heterogeneity impedes progress toward establishing reliable structure-activity relationships based on chemical shift data.
Finally, computational challenges remain significant. While molecular dynamics simulations and machine learning approaches show promise for bridging chemical shift data with biological activity, current computational methods still struggle with accurately modeling the complex relationship between protein structure, dynamics, and function across different timescales and environmental conditions.
One major challenge is the inherent sensitivity limitations of NMR spectroscopy. While chemical shifts provide atomic-level information, they may not capture subtle conformational changes that significantly impact biological activity. Additionally, the signal-to-noise ratio in NMR experiments often requires high protein concentrations that may not reflect physiological conditions, potentially leading to artifacts or misinterpretations when correlating with biological activity.
The time scale discrepancy between NMR measurements and biological processes presents another significant obstacle. Many biological activities occur on microsecond to millisecond timescales, while NMR chemical shift measurements typically represent time-averaged values. This temporal mismatch can obscure critical dynamic processes that drive biological function, resulting in incomplete correlations between observed shifts and actual activity.
Environmental factors further complicate these correlations. In vitro NMR conditions (buffer composition, pH, temperature, absence of cellular components) often differ substantially from the native cellular environment where biological activity naturally occurs. These differences can significantly alter protein behavior, making direct extrapolation from NMR data to biological activity problematic.
Data interpretation challenges also persist in the field. Chemical shift perturbations can result from various factors beyond those directly related to biological activity, including non-specific binding events, minor structural adjustments, or experimental artifacts. Distinguishing relevant shifts from background variations requires sophisticated statistical approaches and validation through complementary techniques.
The lack of standardized protocols for correlating chemical shifts with biological activity represents a methodological barrier. Different research groups employ varied approaches for data collection, processing, and analysis, making cross-study comparisons difficult and hindering the development of robust correlation models. This methodological heterogeneity impedes progress toward establishing reliable structure-activity relationships based on chemical shift data.
Finally, computational challenges remain significant. While molecular dynamics simulations and machine learning approaches show promise for bridging chemical shift data with biological activity, current computational methods still struggle with accurately modeling the complex relationship between protein structure, dynamics, and function across different timescales and environmental conditions.
Established Methods for Correlating Structure and Function
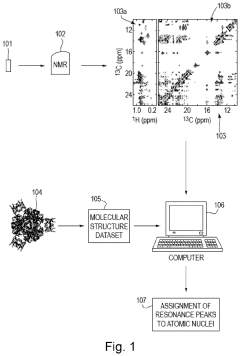
01 NMR techniques for protein structure determination
Nuclear Magnetic Resonance (NMR) spectroscopy is used to determine protein structures by analyzing chemical shifts in protein molecules. These chemical shifts provide valuable information about the three-dimensional structure of proteins, including secondary structure elements, backbone conformations, and folding patterns. Advanced NMR techniques allow researchers to map protein structures with high resolution, which is essential for understanding their biological functions and activities.- NMR techniques for protein structure determination: Nuclear Magnetic Resonance (NMR) spectroscopy is used to determine protein structures by analyzing chemical shifts in protein molecules. These chemical shifts provide valuable information about the three-dimensional structure of proteins, including secondary structure elements, backbone conformations, and folding patterns. Advanced NMR techniques allow researchers to map protein structures with high resolution, which is crucial for understanding their biological functions and activities.
- Correlation of protein chemical shifts with biological activity: Chemical shifts observed in NMR spectra can be directly correlated with the biological activity of proteins. By analyzing changes in chemical shifts upon ligand binding or protein-protein interactions, researchers can identify active sites and understand the mechanisms of biological functions. This approach enables the study of structure-activity relationships and helps in predicting how structural changes might affect a protein's biological role in cellular processes.
- Automated analysis systems for NMR protein data: Automated systems have been developed to analyze the complex data generated by NMR studies of proteins. These systems use computational algorithms to process chemical shift data, identify patterns, and correlate them with biological activities. Such automation allows for high-throughput screening of protein-ligand interactions and facilitates drug discovery by rapidly identifying compounds that interact with target proteins based on chemical shift perturbations.
- NMR-based screening methods for drug discovery: Chemical shift analysis in NMR provides a powerful tool for screening potential drug candidates. By monitoring changes in protein chemical shifts upon compound binding, researchers can identify molecules that interact with target proteins. This approach allows for the identification of lead compounds and helps in optimizing their binding properties. NMR-based screening is particularly valuable for identifying weak binders that might be missed by other screening methods.
- Enhanced NMR hardware for protein analysis: Specialized NMR hardware has been developed to enhance the sensitivity and resolution of protein chemical shift measurements. These advancements include improved magnet designs, cryogenic probe technology, and pulse sequence innovations that allow for better detection of subtle chemical shift changes. Such technological improvements enable the study of larger proteins, lower concentration samples, and more complex biological systems, expanding the applicability of NMR in biological research.
02 Correlation of protein chemical shifts with biological activity
Chemical shifts observed in NMR spectra of proteins can be directly correlated with their biological activities. By analyzing changes in chemical shifts upon ligand binding, protein-protein interactions, or conformational changes, researchers can identify active sites and understand mechanisms of biological function. This approach enables the study of structure-activity relationships and helps in predicting biological functions based on NMR spectral data.Expand Specific Solutions03 High-throughput NMR screening for biological activity
High-throughput NMR screening methods have been developed to rapidly assess protein-ligand interactions and biological activity. These methods utilize chemical shift perturbations to identify binding events and characterize binding sites. Automated systems can analyze large libraries of compounds against target proteins, making this approach valuable for drug discovery and development. The chemical shift data obtained can be used to identify compounds that modulate protein function.Expand Specific Solutions04 Advanced NMR hardware and pulse sequences for protein analysis
Specialized NMR hardware and pulse sequences have been developed to enhance the detection and analysis of protein chemical shifts. These technological advancements include high-field magnets, cryogenic probes, and sophisticated pulse sequences that improve sensitivity and resolution. Such improvements allow for the detection of subtle chemical shift changes that correlate with biological activity, even in complex protein systems or at low concentrations.Expand Specific Solutions05 Computational methods for analyzing protein NMR data
Computational tools and algorithms have been developed to analyze and interpret protein chemical shift data from NMR experiments. These methods include automated assignment of chemical shifts, prediction of protein structures from chemical shift data, and simulation of NMR spectra. Machine learning approaches can identify patterns in chemical shift data that correlate with specific biological activities, enabling researchers to make predictions about protein function based on NMR spectral features.Expand Specific Solutions
Leading Research Groups and Companies in Protein NMR
The field of correlating biological activity with protein chemical shifts in NMR is currently in a growth phase, with an estimated market size of $500-700 million and expanding at 8-10% annually. The competitive landscape features a mix of academic institutions (Xiamen University, Peking University, The Scripps Research Institute, École Polytechnique Fédérale de Lausanne) and commercial entities (BASF Plant Science, CJ CheilJedang, INNOBIO). Technology maturity varies across applications, with pharmaceutical companies (Boehringer Ingelheim, Deciphera) focusing on drug discovery applications, while biotechnology firms (Shanghai Hongene Bioengineering, Heilongjiang Nuolu) are developing industrial applications. Academic-industry partnerships are accelerating technology development, with research institutions providing fundamental innovations that companies then commercialize for specific market applications.
The Scripps Research Institute
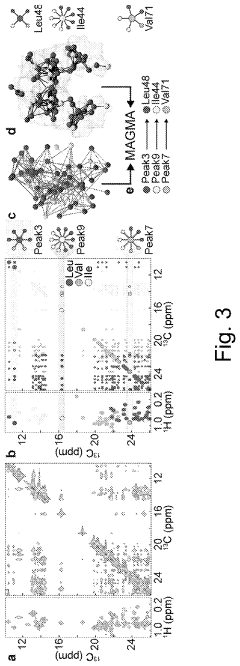
Technical Solution: The Scripps Research Institute has developed a comprehensive platform for correlating biological activity with protein chemical shifts in NMR. Their approach integrates high-resolution NMR spectroscopy with advanced computational modeling to establish structure-activity relationships. They've pioneered methods for automated assignment of protein resonances and chemical shift prediction algorithms that significantly improve the accuracy of structure determination. Their technology includes specialized pulse sequences for detecting protein-ligand interactions through chemical shift perturbation analysis, enabling drug discovery applications. The institute has also developed CHESCA (CHEmical Shift Covariance Analysis), a statistical approach that identifies networks of functionally linked residues by analyzing patterns of chemical shift changes across different protein states. This method has been particularly valuable for identifying allosteric networks in proteins and understanding how distant binding events propagate through protein structures to affect function. Additionally, they've implemented machine learning approaches to correlate chemical shift patterns with specific biological activities, creating predictive models for protein function based on NMR data.
Strengths: Their integrated approach combining experimental NMR with computational methods provides comprehensive insights into protein structure-function relationships. Their CHESCA method offers unique capabilities for identifying allosteric networks. Weaknesses: The complexity of their computational approaches requires significant expertise in both NMR spectroscopy and bioinformatics, potentially limiting widespread adoption.
Oxford University Innovation Ltd.
Technical Solution: Oxford University Innovation has developed a sophisticated platform called "ShiftTrack" for correlating biological activity with protein chemical shifts in NMR. Their technology combines high-resolution multidimensional NMR experiments with proprietary computational algorithms to establish quantitative relationships between chemical shift changes and functional states of proteins. The platform incorporates machine learning approaches trained on extensive databases of protein chemical shifts to predict structural changes and biological activities from NMR data. Their methodology includes specialized pulse sequences that enhance sensitivity for detecting subtle conformational changes in proteins upon ligand binding or post-translational modifications. A key innovation is their "Chemical Shift Fingerprinting" technique, which creates unique spectral signatures for different functional states of proteins, allowing rapid screening of potential modulators of protein activity. The platform also integrates molecular dynamics simulations with experimental NMR data to provide mechanistic insights into how structural perturbations affect protein function. This comprehensive approach enables researchers to identify allosteric binding sites and design modulators of protein function based on chemical shift analysis.
Strengths: Their integrated platform combining experimental NMR with advanced computational methods provides comprehensive insights into protein structure-function relationships. Their machine learning approach leverages extensive databases to improve prediction accuracy. Weaknesses: The complexity of their system requires significant expertise in both NMR spectroscopy and computational biology, potentially limiting accessibility for smaller research groups.
Key Innovations in Chemical Shift-Activity Relationship Analysis
Method for processing nuclear magnetic resonance (NMR) spectroscopic data
PatentInactiveUS10866295B2
Innovation
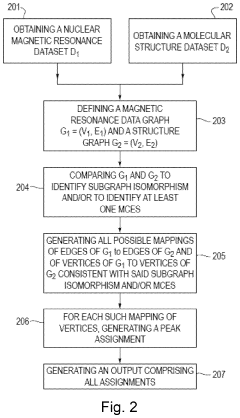
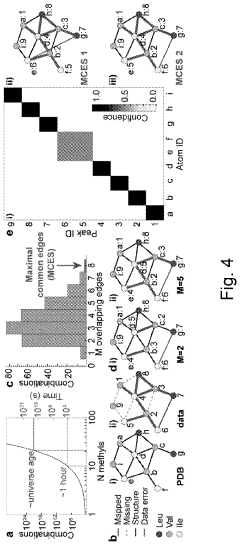
- A graph-matching algorithm that combines structural models with experimental multidimensional magnetic resonance data to accurately identify confident and ambiguous peak assignments by comparing experimental distance restraints with structural models, reducing the need for laborious experiments and providing exact sets of plausible assignments.
Method of using reduced dimensionality nuclear magnetic resonance spectroscopy for rapid chemical shift assignment and secondary structure determination of proteins
PatentInactiveUS7141432B2
Innovation
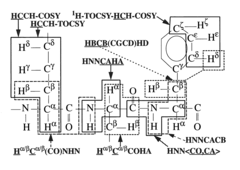
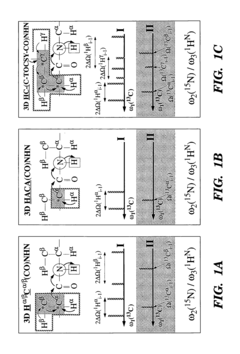
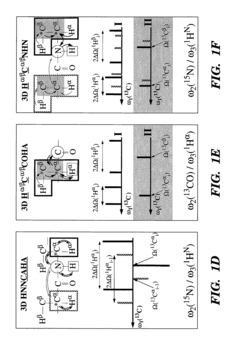
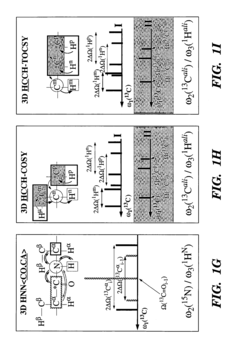
- The development of reduced dimensionality (RD) TR NMR experiments, such as 3D HA,CA,(CO),N,HN and H,C,(C-TOCSY—CO),N,HN, which apply specific radiofrequency pulses to encode chemical shift values in fewer dimensions, allowing for phase-sensitive measurement and cosine modulation to generate peak pairs that encode chemical shifts, enabling efficient data collection and analysis.
Data Management and Integration Strategies
The effective management and integration of NMR chemical shift data with biological activity information represents a critical challenge in modern bioinformatics. Current approaches typically involve disparate data storage systems that require significant manual curation to establish meaningful correlations. Leading research institutions have implemented specialized database architectures that combine spectral repositories with functional annotation frameworks, enabling more streamlined analysis workflows. These systems typically incorporate both structured databases for quantitative measurements and unstructured storage solutions for raw spectral data.
Integration strategies have evolved significantly in recent years, with API-driven approaches becoming increasingly prevalent. RESTful interfaces now commonly serve as the connective tissue between NMR data processing pipelines and biological activity databases. This architectural pattern facilitates real-time data exchange while maintaining the independence of specialized analytical platforms. Several open-source frameworks have emerged to standardize these integration patterns, with the Bio-NMR Exchange Format (BioNEF) gaining particular traction among academic research groups.
Data normalization presents persistent challenges when correlating chemical shifts with biological functions. Variations in experimental conditions, instrument calibration, and sample preparation can introduce significant noise into correlation analyses. Advanced preprocessing algorithms now employ machine learning techniques to identify and compensate for these experimental artifacts. Particularly promising are self-supervised models that can learn normalization parameters from large unlabeled datasets of protein NMR spectra.
Metadata management frameworks have become essential components of integrated NMR-activity correlation systems. These frameworks capture critical experimental parameters, sample provenance information, and analytical processing steps. The comprehensive documentation of the data lineage enables more robust statistical analysis and improves reproducibility. Industry leaders have adopted semantic web technologies to represent these complex relationships, with RDF-based knowledge graphs emerging as a flexible solution for capturing the multidimensional nature of structure-function relationships.
Cloud-based collaborative platforms are increasingly being deployed to facilitate multi-institutional research efforts. These systems provide secure access controls while enabling geographically distributed teams to contribute to shared datasets. Real-time synchronization mechanisms ensure that chemical shift annotations and biological activity measurements remain consistent across research sites. The adoption of containerized deployment models has significantly reduced the technical barriers to implementing these collaborative environments, accelerating the pace of discovery in this field.
Integration strategies have evolved significantly in recent years, with API-driven approaches becoming increasingly prevalent. RESTful interfaces now commonly serve as the connective tissue between NMR data processing pipelines and biological activity databases. This architectural pattern facilitates real-time data exchange while maintaining the independence of specialized analytical platforms. Several open-source frameworks have emerged to standardize these integration patterns, with the Bio-NMR Exchange Format (BioNEF) gaining particular traction among academic research groups.
Data normalization presents persistent challenges when correlating chemical shifts with biological functions. Variations in experimental conditions, instrument calibration, and sample preparation can introduce significant noise into correlation analyses. Advanced preprocessing algorithms now employ machine learning techniques to identify and compensate for these experimental artifacts. Particularly promising are self-supervised models that can learn normalization parameters from large unlabeled datasets of protein NMR spectra.
Metadata management frameworks have become essential components of integrated NMR-activity correlation systems. These frameworks capture critical experimental parameters, sample provenance information, and analytical processing steps. The comprehensive documentation of the data lineage enables more robust statistical analysis and improves reproducibility. Industry leaders have adopted semantic web technologies to represent these complex relationships, with RDF-based knowledge graphs emerging as a flexible solution for capturing the multidimensional nature of structure-function relationships.
Cloud-based collaborative platforms are increasingly being deployed to facilitate multi-institutional research efforts. These systems provide secure access controls while enabling geographically distributed teams to contribute to shared datasets. Real-time synchronization mechanisms ensure that chemical shift annotations and biological activity measurements remain consistent across research sites. The adoption of containerized deployment models has significantly reduced the technical barriers to implementing these collaborative environments, accelerating the pace of discovery in this field.
Regulatory Considerations for NMR-Based Drug Discovery
The regulatory landscape for NMR-based drug discovery involves complex considerations that pharmaceutical companies must navigate throughout the development process. The FDA, EMA, and other global regulatory bodies have established specific guidelines for analytical methods used in drug development, including NMR spectroscopy. These guidelines primarily focus on method validation, data integrity, and reproducibility of results when correlating biological activity with protein chemical shifts.
For NMR data to be accepted in regulatory submissions, pharmaceutical companies must demonstrate that their methodologies for correlating chemical shifts with biological activity are scientifically sound and reproducible. This includes validation of the NMR experimental conditions, data processing protocols, and statistical methods used to establish structure-activity relationships. The ICH Q2(R1) guideline on validation of analytical procedures provides a framework that can be adapted for NMR-based studies.
Data integrity represents another critical regulatory consideration. Complete audit trails for NMR data acquisition, processing, and analysis must be maintained in compliance with 21 CFR Part 11 (for FDA submissions) or equivalent regulations in other jurisdictions. This includes proper documentation of software versions, processing parameters, and any mathematical transformations applied to the raw data when establishing correlations between chemical shifts and biological activity.
The use of NMR as a biomarker qualification tool requires additional regulatory considerations. If chemical shift changes are proposed as biomarkers for drug efficacy or safety, the FDA's Biomarker Qualification Program or the EMA's qualification procedure for novel methodologies may apply. These pathways require robust evidence linking the observed NMR parameters to clinically relevant biological outcomes.
Quality control measures for NMR instrumentation and sample preparation must also meet regulatory standards. This includes regular calibration of NMR spectrometers, validation of reference standards, and implementation of quality management systems that ensure consistent performance across multiple experiments and sites. The variability in chemical shift measurements must be quantified and demonstrated to be within acceptable limits for regulatory acceptance.
For advanced applications like in-cell NMR or real-time monitoring of drug-target interactions, regulatory frameworks are still evolving. Companies pioneering these approaches should engage in early discussions with regulatory agencies through programs like the FDA's Critical Path Initiative or the EMA's Innovation Task Force to establish appropriate validation criteria for these novel methodologies.
For NMR data to be accepted in regulatory submissions, pharmaceutical companies must demonstrate that their methodologies for correlating chemical shifts with biological activity are scientifically sound and reproducible. This includes validation of the NMR experimental conditions, data processing protocols, and statistical methods used to establish structure-activity relationships. The ICH Q2(R1) guideline on validation of analytical procedures provides a framework that can be adapted for NMR-based studies.
Data integrity represents another critical regulatory consideration. Complete audit trails for NMR data acquisition, processing, and analysis must be maintained in compliance with 21 CFR Part 11 (for FDA submissions) or equivalent regulations in other jurisdictions. This includes proper documentation of software versions, processing parameters, and any mathematical transformations applied to the raw data when establishing correlations between chemical shifts and biological activity.
The use of NMR as a biomarker qualification tool requires additional regulatory considerations. If chemical shift changes are proposed as biomarkers for drug efficacy or safety, the FDA's Biomarker Qualification Program or the EMA's qualification procedure for novel methodologies may apply. These pathways require robust evidence linking the observed NMR parameters to clinically relevant biological outcomes.
Quality control measures for NMR instrumentation and sample preparation must also meet regulatory standards. This includes regular calibration of NMR spectrometers, validation of reference standards, and implementation of quality management systems that ensure consistent performance across multiple experiments and sites. The variability in chemical shift measurements must be quantified and demonstrated to be within acceptable limits for regulatory acceptance.
For advanced applications like in-cell NMR or real-time monitoring of drug-target interactions, regulatory frameworks are still evolving. Companies pioneering these approaches should engage in early discussions with regulatory agencies through programs like the FDA's Critical Path Initiative or the EMA's Innovation Task Force to establish appropriate validation criteria for these novel methodologies.
Unlock deeper insights with Patsnap Eureka Quick Research — get a full tech report to explore trends and direct your research. Try now!
Generate Your Research Report Instantly with AI Agent
Supercharge your innovation with Patsnap Eureka AI Agent Platform!