Method and system for efficient indexed storage for unstructured content
a technology of unstructured content and indexing techniques, applied in the field of storage, can solve the problems of unstructured content, multimedia, lack of robust, scalable indexing techniques, and inability to insert and query through efficient indexes, and achieve the effect of distinguishing unstructured content from structured content, and avoiding the loss of indexing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example # 1
Token Sequence Example #1
Text
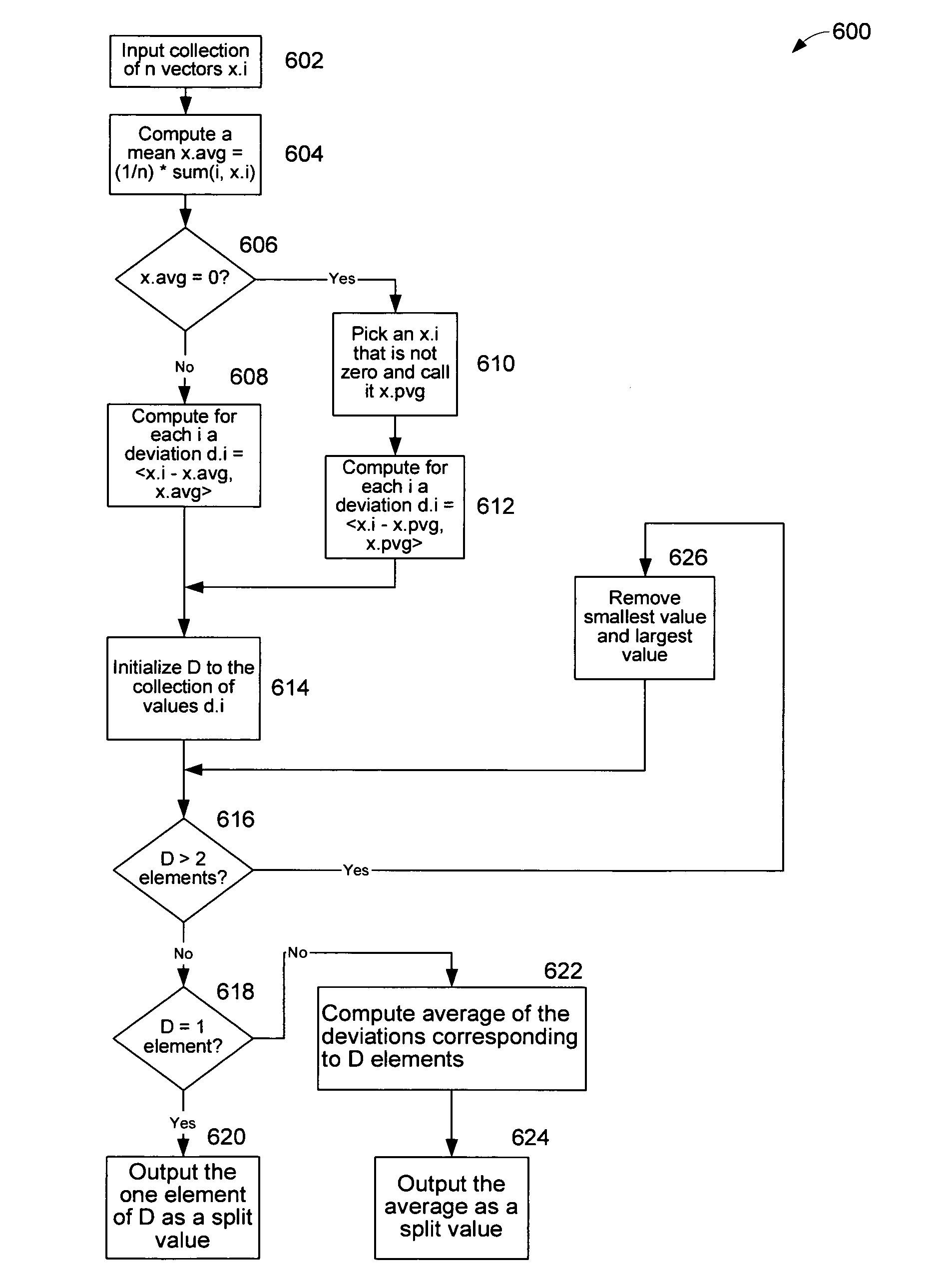
[0025]Given a block of text, tokenize it by mapping words, word breaks, punctuation, and other formatting into a sequence of tokens. The tokenizer performs the desired mapping from text to tokens. From the above discussion, it follows that a block of text corresponds to a probability transition matrix. In other words, associate a block of text with its corresponding vector in an inner product space. A distance function expressed between two vectors as,
Distance(A,B)=sqrt(A−B,A−B>)
[0026]This distance is also known as the “norm,”
Distance(A,B)=∥A−B∥=sqrt(A−B,A−B>)
[0027]According to this metric, we say that two text blocks are similar if the distance between their corresponding probability transition matrices is small. In practice, we have found that this numerical measure corroborates our own ordinary concept of “similar” text. For example, when the distance between two text blocks is small, the text is satisfyingly similar to a human reader. Empirically, we...
example # 2
Token Sequence Example #2
Genomic Data
[0029]Another kind of unstructured content is genomic data. Using an embodiment of the present invention, DNA sequences of nucleotides can be parsed and transformed into sparse probabilistic tuple-to-token transition matrices. For example, the VIIth and XIVth chromosomes of Baker's yeast (Saccharomyces cerevisiae) consist of 563,000 and 784,000 bases, respectively. In one embodiment of the invention, a tuple width of 8, 15, and 30 have been used to perform the “splitting” operation. The operations to compute the average and deviation each take 40 seconds to 3 minutes, depending on the tuple width, for each chromosome on a vintage 2002 desktop PC. This confirms that long token sequences associated with raw nucleotide data can be mapped to an inner product vector space.
[0030]Image Blocks
[0031]The techniques of the present invention may be used on image blocks, as similarity between two images can be represented by computing an inner product over “s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com