Molecular markers, internal reference genes and their applications, detection kits and methods for constructing detection models
A molecular marker, internal reference gene technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. Excessive treatment and inappropriate treatment, satisfying individualized precision treatment, and improving the effect of technical methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
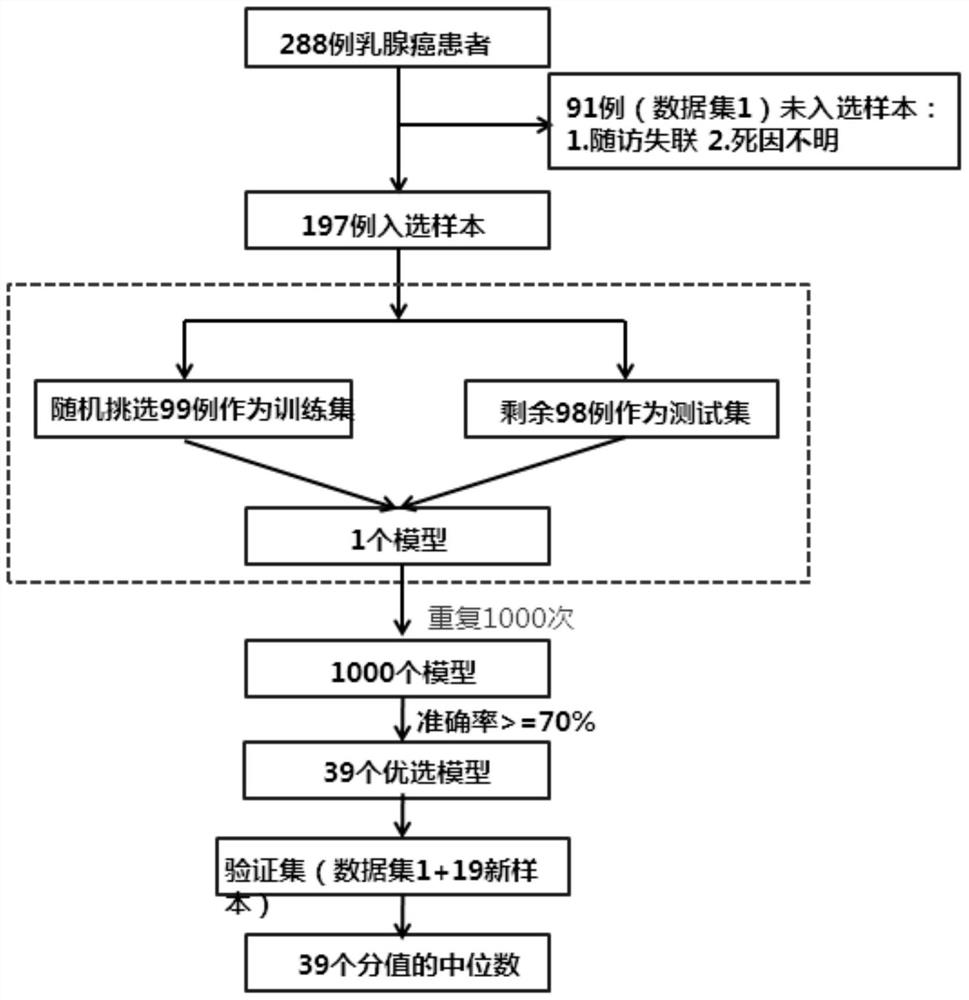
[0062] Example 1 Collection of Samples, Sample Data Consolidation
[0063] The system collects complete clinical follow-up information without treatment of the initial breast cancer patient, and the inventors have selected 341 samples as TLDA (TAQman Lowdeysity Array, TLDA) chip detection. And subsequent series of QRT-PCR verification experimental samples:
[0064] (1) Unexpected early, medium-term breast cancer first diagnosis;
[0065] (2) Patients with Uneavenated early, medium ER or (and) PR positive breast cancer;
[0066] (3) LN or (and) no transfer, and the number of LN transfer;
[0067] (4) It has accurate and detailed follow-up information.
Embodiment 2
[0068] Example 2TLDA chip Screening molecular marker and interference gene
[0069] Two-six cases of breast cancer prognosis in compliance with the above conditions were detected by TLDA chip detection for TLDA chip detection. The specific steps are:
[0070] (1) Extraction of RNA in the FFPE sample: 8 tablets of 20 μm per sample 4 or 10 μm slices are taken, and RNA is extracted according to the highpure ffpet rna isolation kit (roche), and the extraction RNA is quantitatively controlled by NanoDrop-2000 After the downstream reverse transcription experiment.
[0071] (2) Total RNA is reactive to CDNA samples: Take 1μg total RNA according to Vilo TM The Master Mix Kit (Invitrogen) manual is reversed.
[0072] (3) CDNA sample for TLDA test: above CDNA products and After the Universal PCR Mastermix is fully mixed, the test experiment was performed on the ABI 7900 fluorescent quantitative PCR instrument in accordance with the TLDA standard program.
[0073] (4) Data analysis and p...
Embodiment 3
[0077] Example 3 Molecular marker's large sample amount QRT-PCR verification
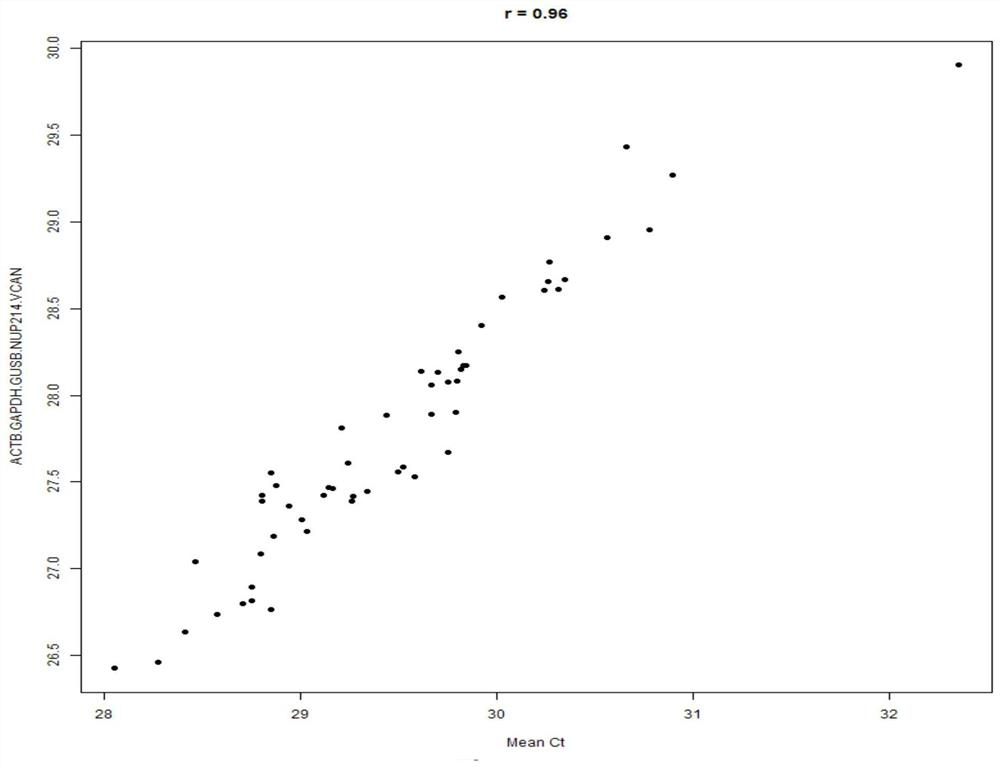
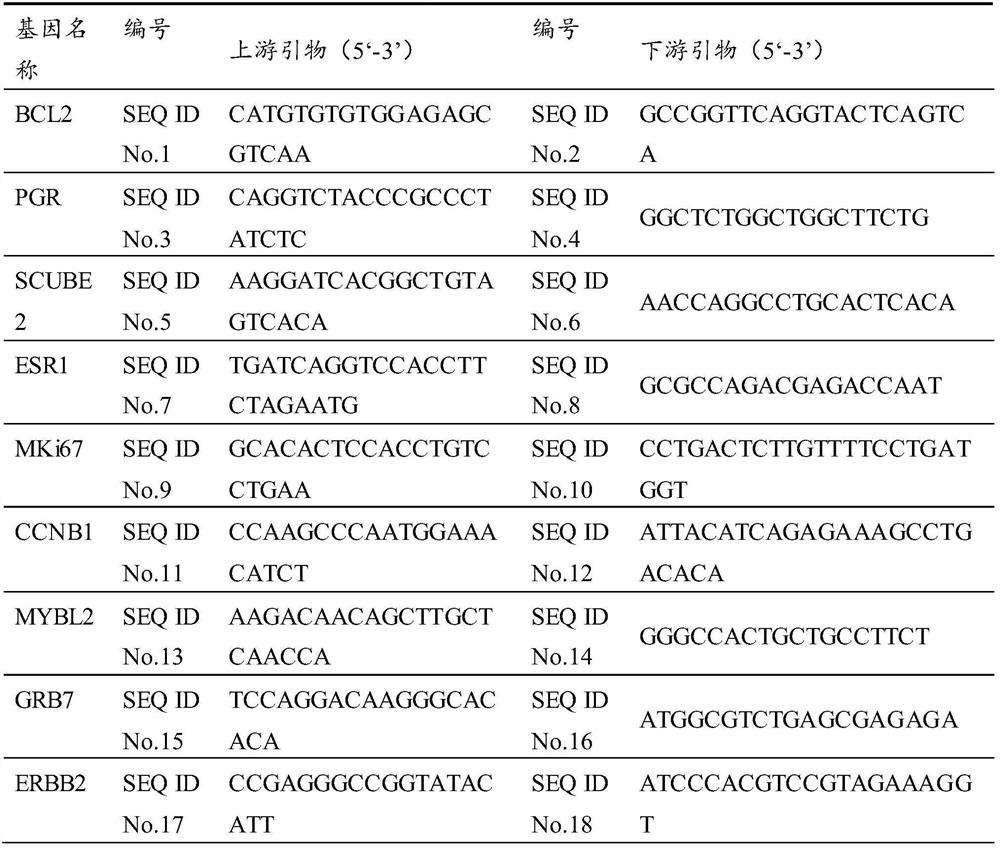
[0078] TLDA Screening 14 molecular markers and 5 innergic genes: BCl2, PGR, Scube2, ESR1, MKI67, CCNB1, MYBL2, GRB7, Erbb2, MMP11, CD68, BAG1, MAPT, MS4A1; ACTB, GAPDH, GUSB, NUP214 . The complete 289 FFPE samples in accordance with the above sample collection requirements and clinical follow-up information are used, and single tube QRT-PCR verification is performed.
[0079] (1) 289 cases of FFPE sample RNA extract: 8 μm slices per sample 4 or 10 μm sections are taken, and RNA is extracted according to the highpure ffpet RNA ISOLATION KIT (ROCHE). The extracted RNA is quantified by NanoDrop-2000 The downstream reverse transcription experiment is performed after the control.
[0080] (2) 289 cases of FFPE sample RNA reverse recorded cDNA: 1 μg total RNA according to Vilo TM Mastermix Kit (Invitrogen) Manual for reverse transcription.
[0081] (3) 289 cases of FFPE samples CDNA product for QPCR detectio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com