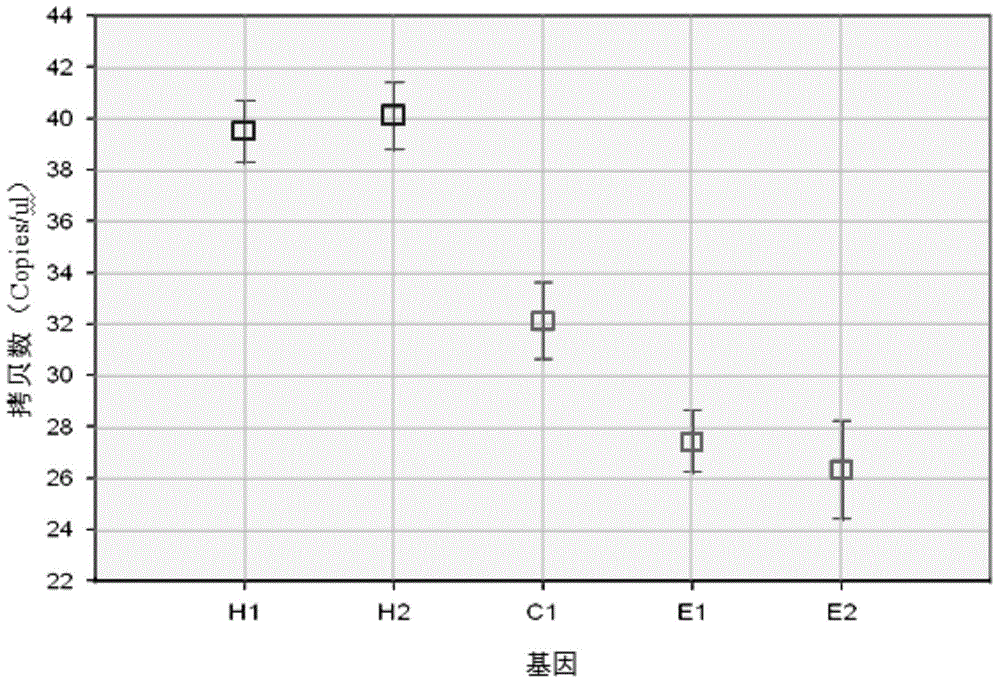
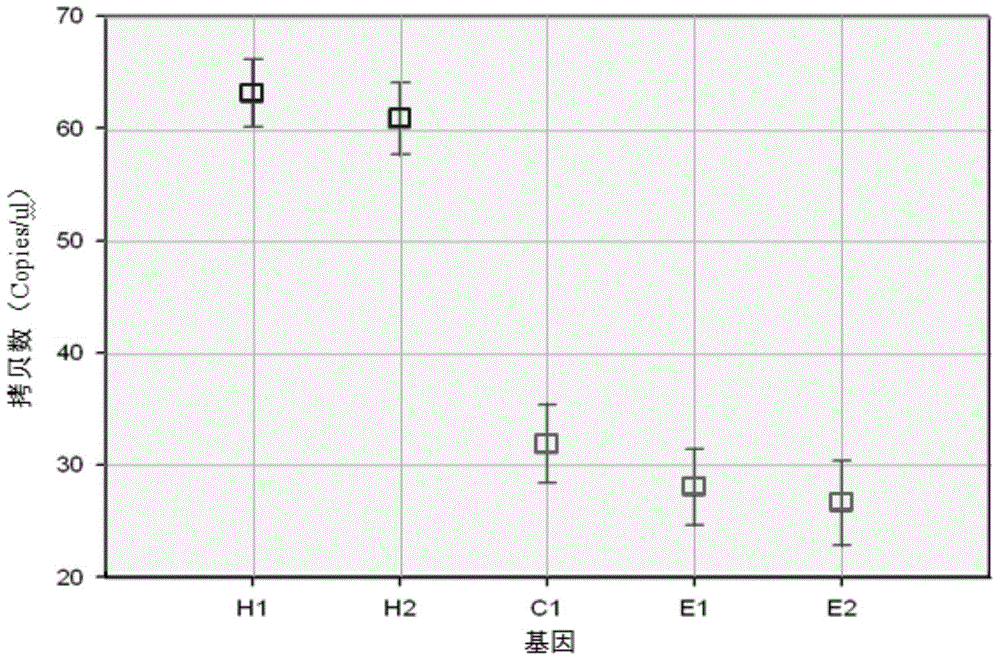
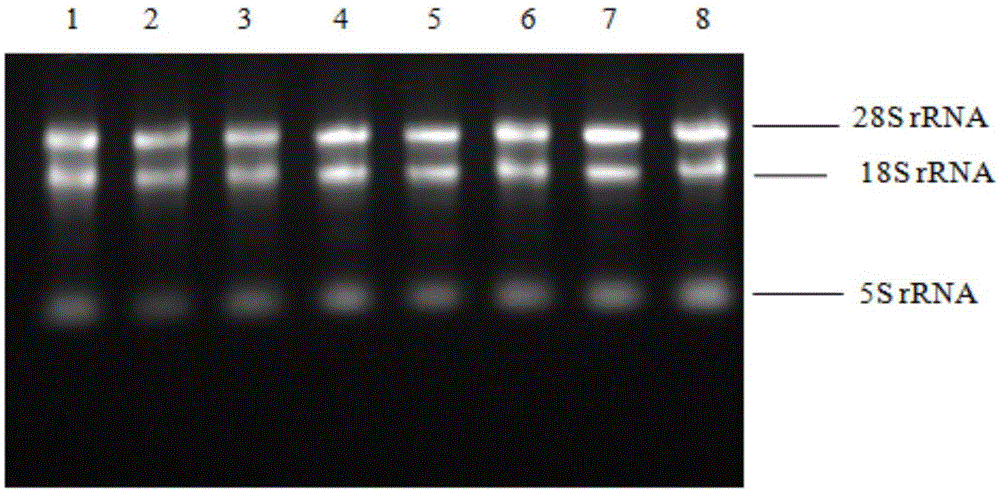
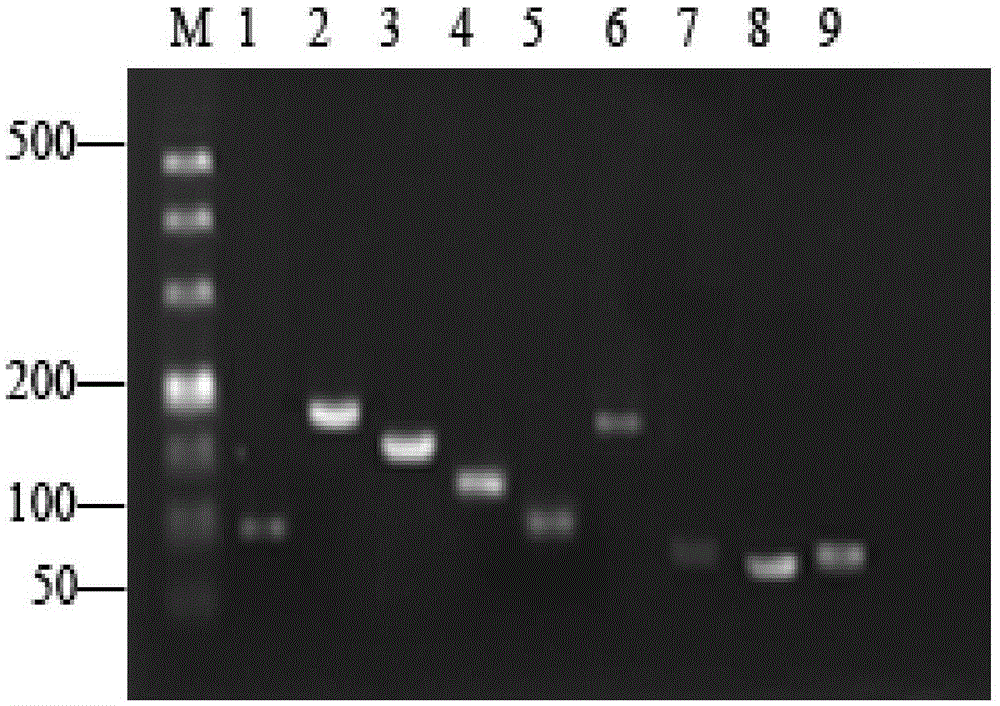
Patents
Literature
863 results about "Reference genes" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
This article discusses the specific topic of reference genes. Reference genes are expressed in all cells of an organism under normal and patho-physiological conditions. Although some housekeeping genes (such as LDHA, NONO, PGK1 and PPIH,) are expressed at relatively constant levels in most non-pathological situations, other housekeeping genes may vary depending on experimental conditions. Although the terms "housekeeping gene" and "reference gene" are used somewhat interchangeably, caution must be used in selecting genes for reference purposes.
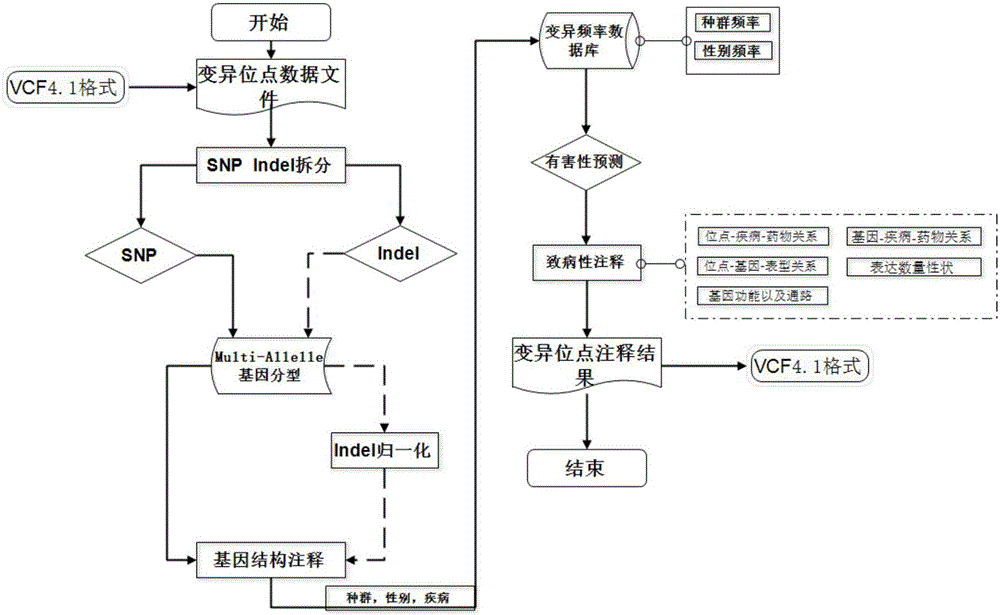
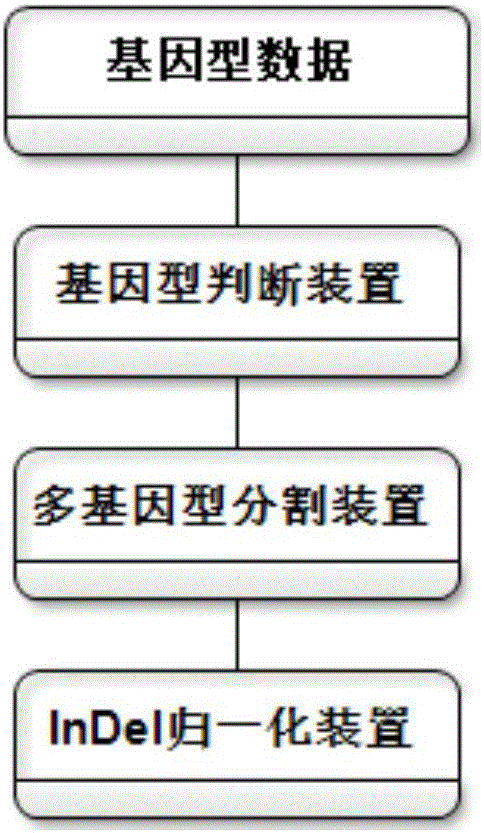
Method for genotype determination
InactiveUS20040115684A1Enabling detectionMicrobiological testing/measurementFermentationReference genesMedicine
The present invention is directed to a new method for genotype determination at a specific gene locus of an individual or a fetus comprising (i) amplifying a first sequence of said gene locus and a second sequence of a second reference gene locus from DNA originating from a sample containing biological material of said individual or fetus (ii) Monitoring both amplifications preferably in real time and determining the amount of amplification products after each cycle, and (iii) Calculating the ratio between the amount of DNA from the first gene locus and the amount of DNA from the second gene locus. The new method is useful for a variety of applications, especially for detection of chromosomal abnormalities in fetal cells.
Owner:COSTA JEAN MARC
Method for single cell classification and screening and device therefor
ActiveCN102952854AAvoid marked actionsImprove accuracyBioreactor/fermenter combinationsBiological substance pretreatmentsReference genesData set
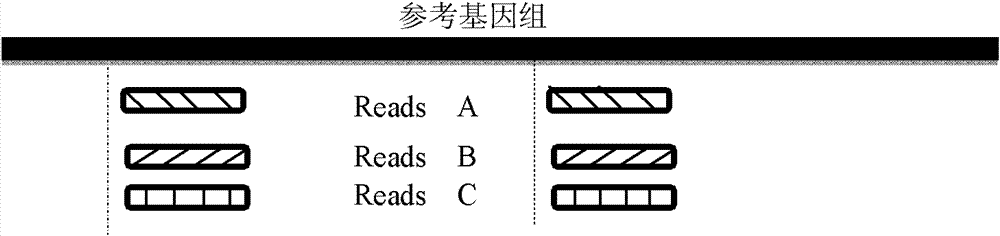
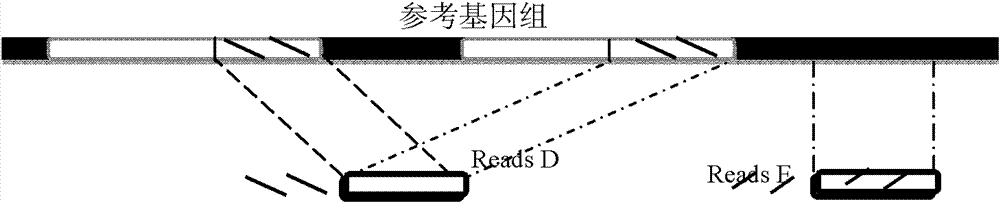
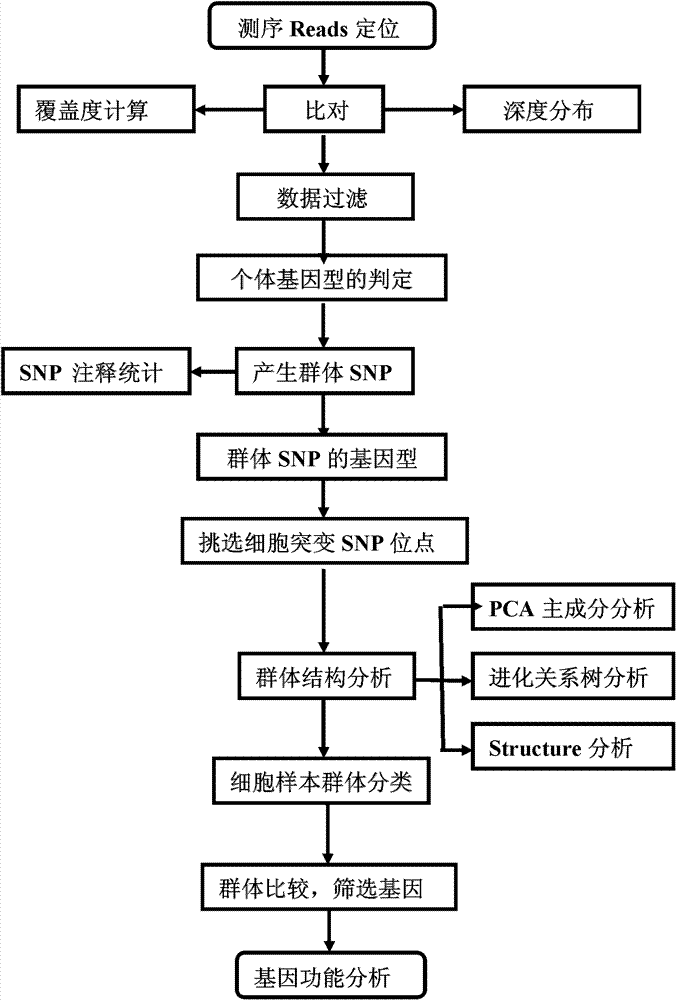
The invention provides a method for single cell classification and screening and a device therefor. The method comprises the following steps of carrying out comparison of reads obtained by sample sequencing and a reference genome, carrying out data filtration of a comparison result, determining a consistent genotype of all single cell samples according to filtered data, saving the consistent genotype of the all single cell samples into a SNP data set, extracting genotype files of loci corresponding to reference genome SNP data set positions from the saved SNP data set, selecting a cell mutation SNP locus, and carrying out cell classification and functional gene screening according to a genotype file of the cell mutation SNP locus. The method and the device avoid cell marking, solve the problem that the traditional single cell classification method can not realize classification of a certain cell subset having no corresponding specific markers, realize complete analysis of genetic variation information of a single cell genome, and greatly improve the accuracy of cell subset classification.
Owner:BGI SHENZHEN CO LTD +1
Method for detecting mutation information in multiplex amplification sequencing product of genome
ActiveCN106202991AEfficient identificationQuick identificationHybridisationSpecial data processing applicationsReference genesNatural abundance
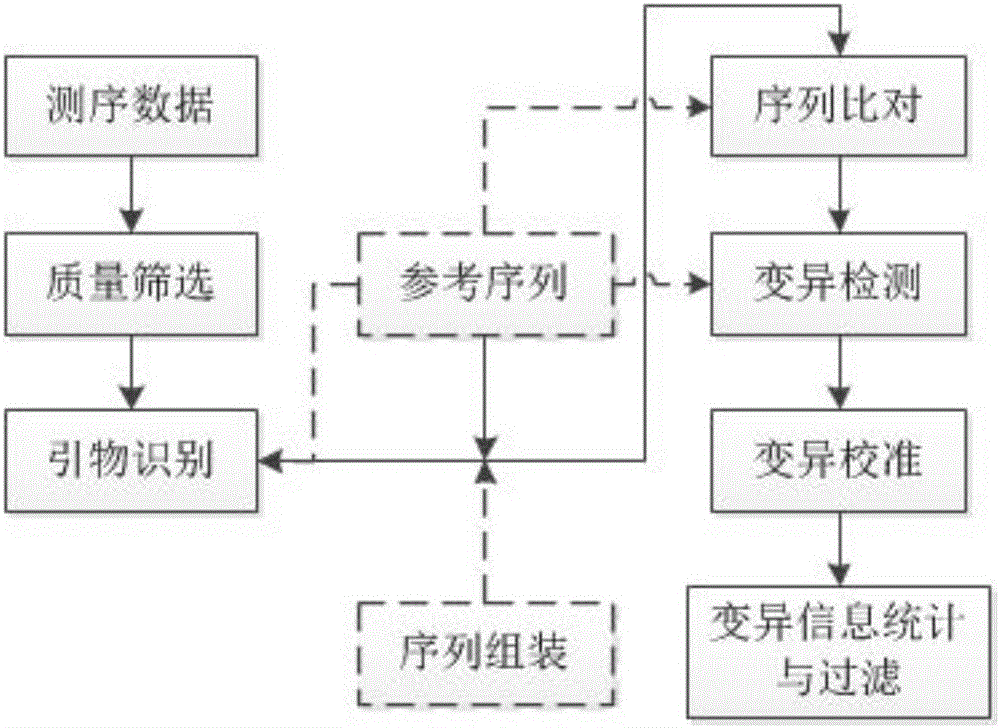
The invention discloses a method for detecting mutation information in a multiplex amplification sequencing product of a genome. The method comprises steps as follows: sequencing data are subjected to quality assessment and preprocessing; a recognizable sequencing sequence is selected for sequence assembling; the recognizable sequencing sequence or a sequence obtained through assembling is compared with a reference gene sequence, and preliminary variation information is obtained; fine calibration of sequence variation is performed according to different types of conditions; a calibrated sequencing fragment is obtained; the homozygosis or heterozygosis state of a target fragment is obtained according to the type of the sequencing fragment with the highest abundance; finally, the mutation information in the multiplex amplification sequencing product of the genome is obtained. By means of the method, the amplification product can be rapidly, efficiently and accurately recognized, and the calculation resources are saved; the sequence assembling process is compatible, and the problem of reduction of the quality value of basic groups produced in the sequencing process can be effectively solved; the homozygosis / heterozygosis state of variation information can be more effectively and stably judged, and random errors introduced in the PCR (polymerase chain reaction) process and the sequencing process are eliminated.
Owner:AMOY DIAGNOSTICS CO LTD +1
Annotation method and annotation system of whole-genome variant data
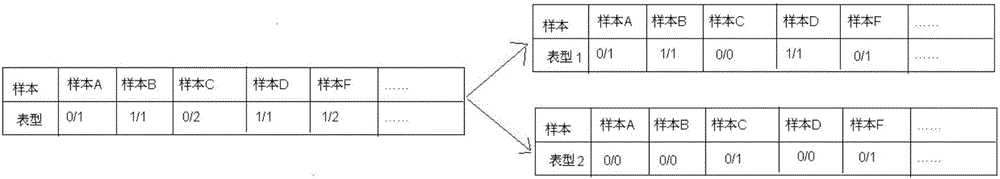
InactiveCN106156538AAccurate NotesSolve the screening puzzleProteomicsGenomicsReference genesAllele frequency
The invention discloses an annotation method and an annotation system of whole-genome variant data. The method comprises the following steps of S1, creating a variant data file, wherein the variant data are stored according to a national standard VCF format as an input file; S2, performing multi-allele genotyping, firstly performing genotype judgment, representing a basic group which is consistent with a reference genome by zero, and representing the basic groups which are inconsistent with the reference gene group by 1, 2, 3,..., then performing SNP and InDel multi-allele type resolution so that the allele type is represented by zero and one; S3, causing InDel generation position normalization, namely performing InDel generation position normalization according to a left justification and simplification normalization method; and S4, performing annotation, namely performing gene structure annotation, allele frequency annotation, variable site harm prediction and pathogenicity annotation. The annotation method and the annotation system improve integrity and accuracy of annotation information.
Owner:天津诺禾医学检验所有限公司
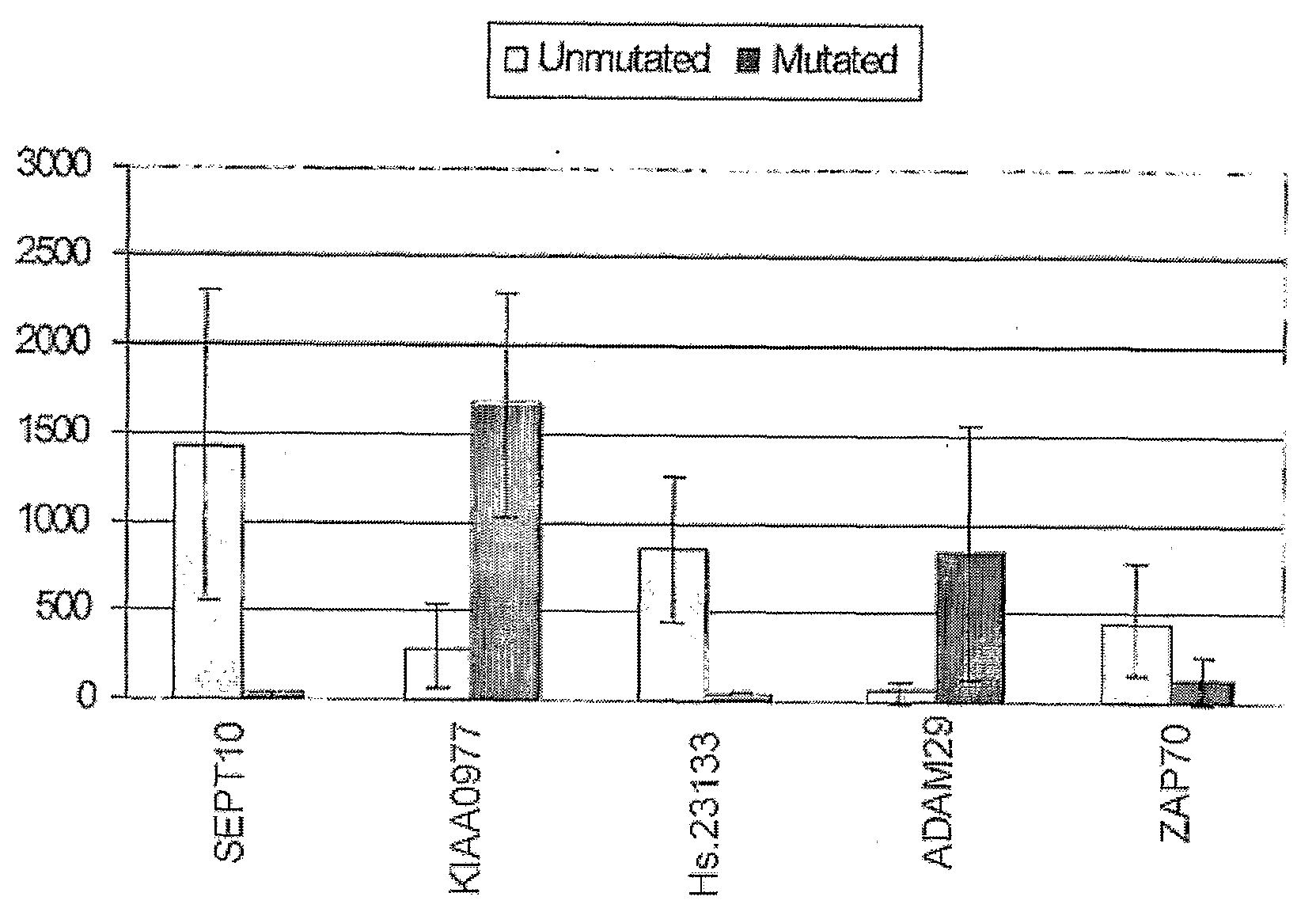
Compositions and Methods for Differential Diagnosis of Chronic Lymphocytic Leukemia
InactiveUS20080280297A1Microbiological testing/measurementImmunoglobulinsReference genesLymphocytic cell
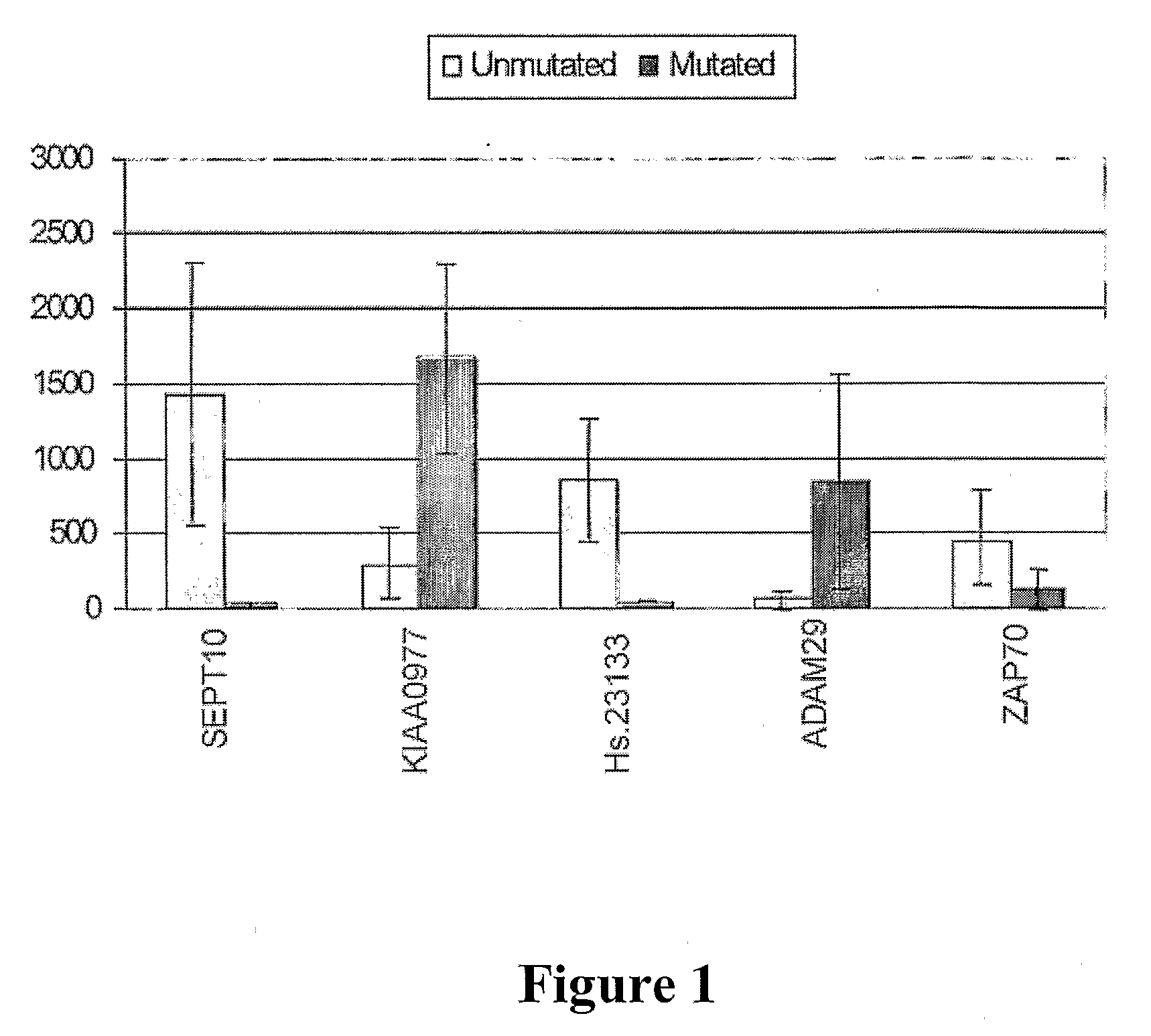
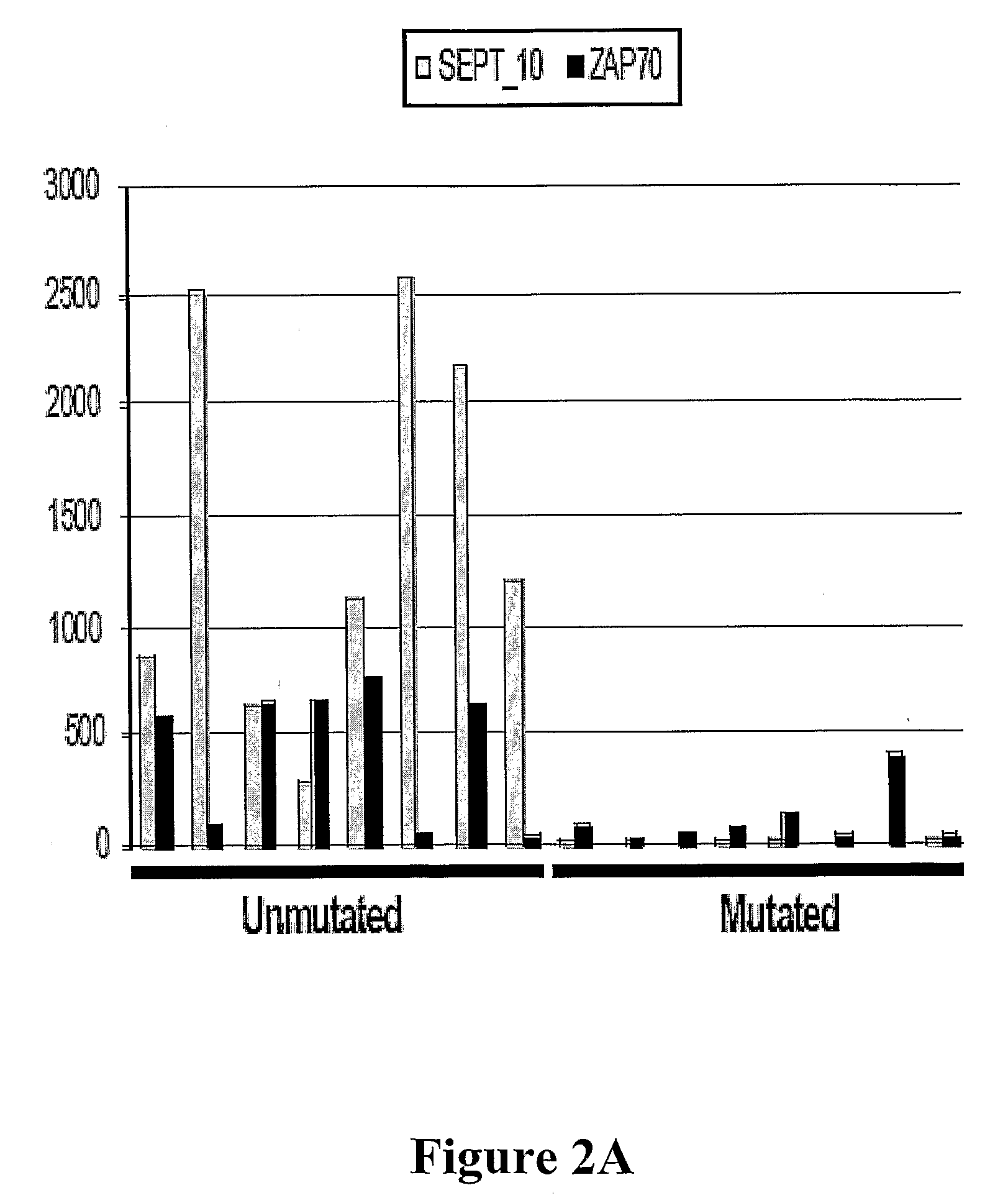
The invention provides compositions and methods for determining a prognosis of a B cell chronic lymphocytic leukemia (CLL) in a subject based on the level of expression of at least one marker gene. Marker genes provided by the invention are SEPTlO, KIAA0799, Hs.23133, and ADAM29. The marker genes can be used to differentially diagnose CLL in a subject based on relative gene expression levels in the subject compared to reference gene expression levels established from a clinically characterized population of patients. The invention also provides diagnostic reagents and compositions and kits based on the marker genes.
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
Method and device for quick contrast and analysis of short sequence for second-generation sequencing
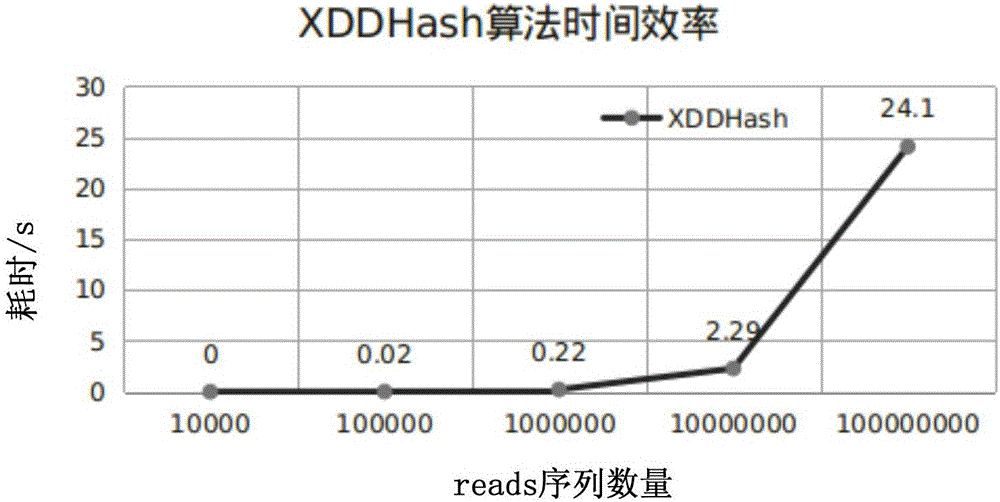
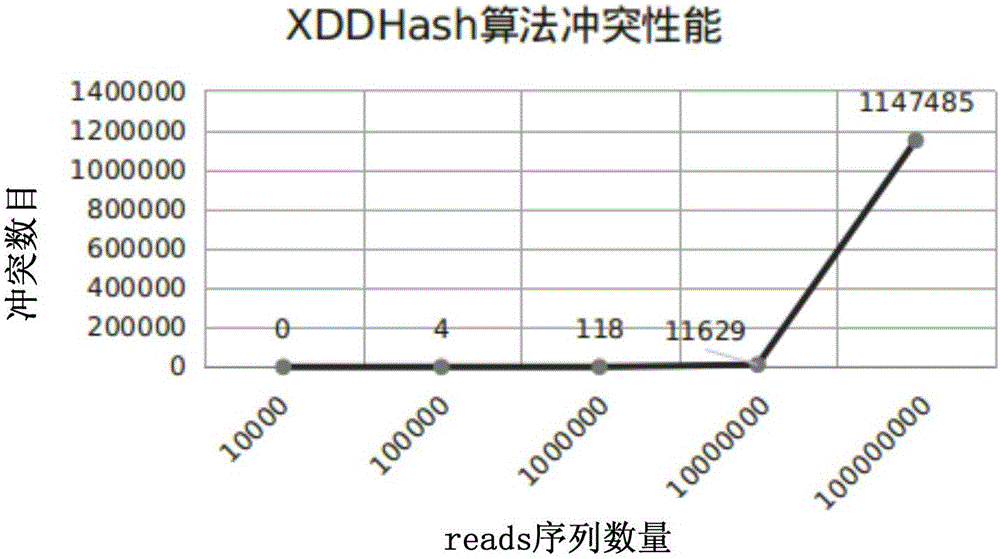
ActiveCN106295250AImprove time and efficiencyFaster thanSequence analysisSpecial data processing applicationsReference genesArray data structure
The invention discloses a method and a device for quick contrast and analysis of a short sequence for second-generation sequencing, which can solve the problems of low contrast efficiency and high memory occupation ratio of sequencing data. The method comprises the following steps of obtaining a DNA (deoxyribonucleic acid) short sequence obtained by sequencing, and respectively mapping and encoding the DNA short sequence by a first hash algorithm and a second hash algorithm, so as to respectively obtain a first index and a second index; according to a preset index query library, the first index and the second index, contrasting the DNA short sequence and a reference gene group, wherein the index query library consists of an unit structure array, and each unit structure comprises value and index 2; storing the array index offset of each unit structure as the corresponding index 1, namely the index value corresponding to the structure array, wherein K is the length of segment sequence; according to the contrast result, when the contrast result is correct, obtaining the value of the K-mer segment contrasted with the corresponding DNA short sequence, and determining the chromosome number of the corresponding DNA short sequence and the site on the chromosome.
Owner:北京普康瑞仁医学检验所有限公司
Endpoint taqman methods for determining zygosity of corn comprising tc1507 events
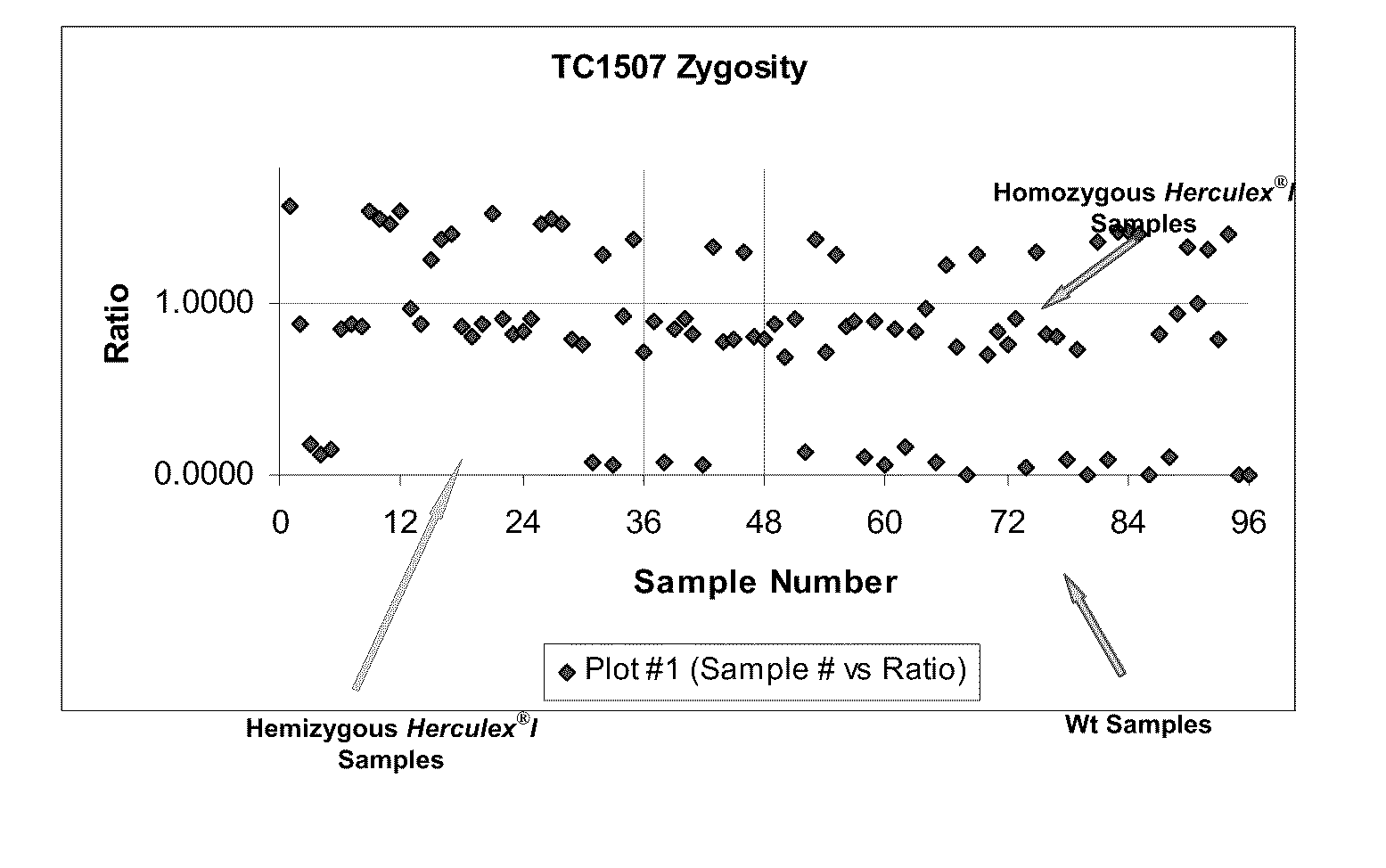
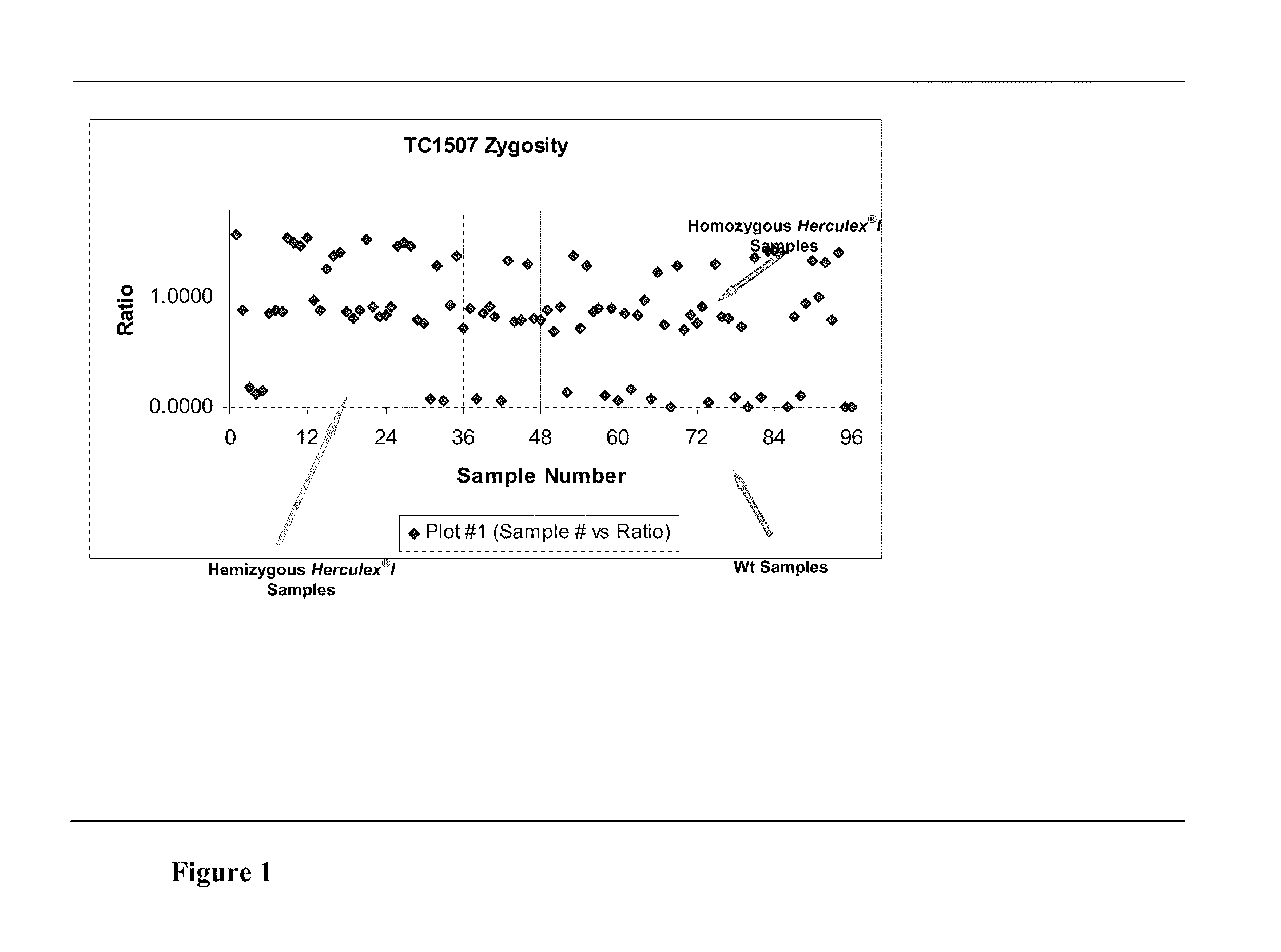
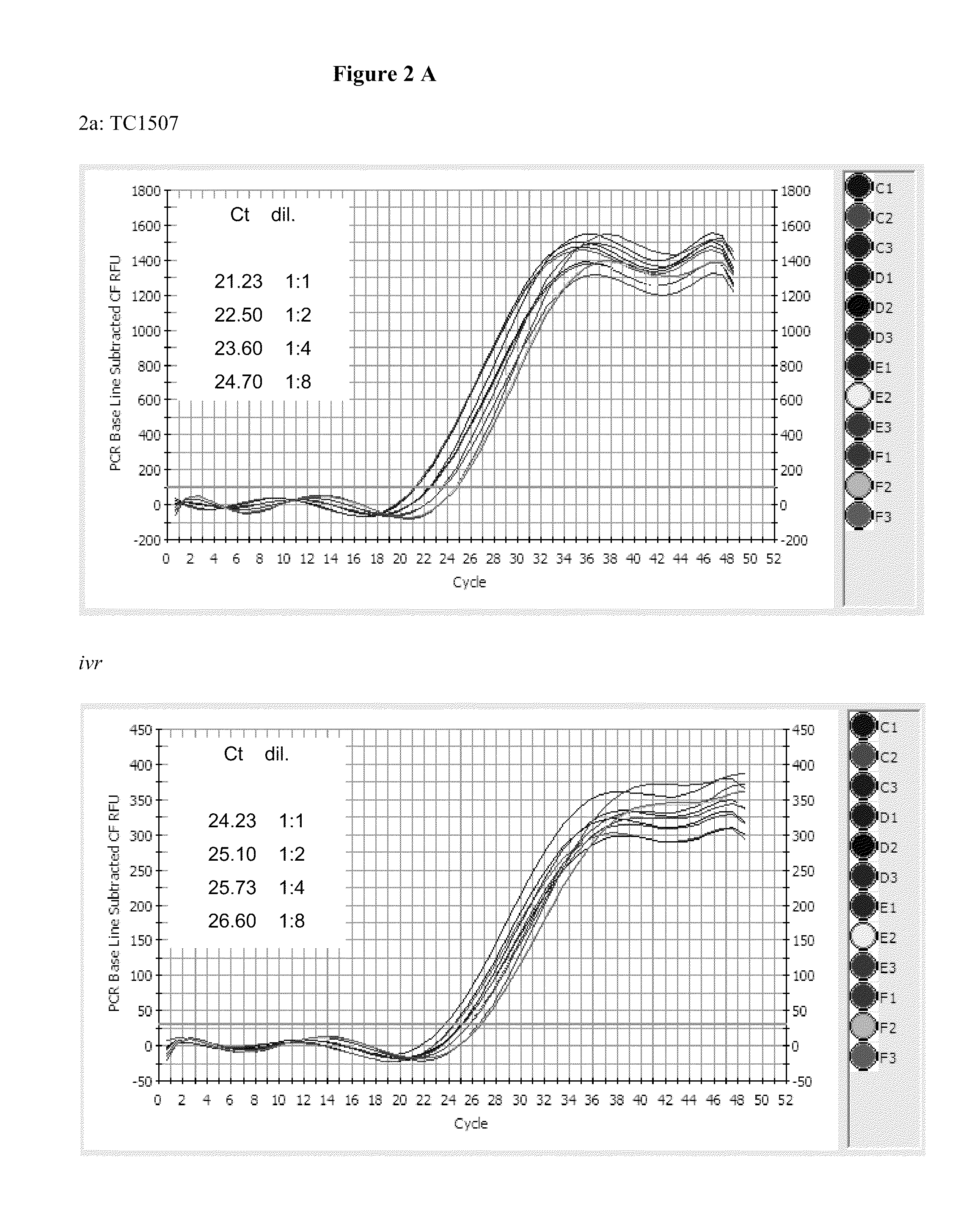
InactiveUS20110151441A1High throughput zygosity analysisSugar derivativesMicrobiological testing/measurementReference genesPcr assay
A method for zygosity analysis of the maize Cry1F event TC1507 is provided. The method provides TC1507 event-specific and maize endogenous reference gene-specific primers and TaqMan probe combinations for use in an endpoint biplex TaqMan PCR assay capable of producing robust genotype calls for assisting in molecular breeding of TC1507.
Owner:DOW AGROSCIENCES LLC
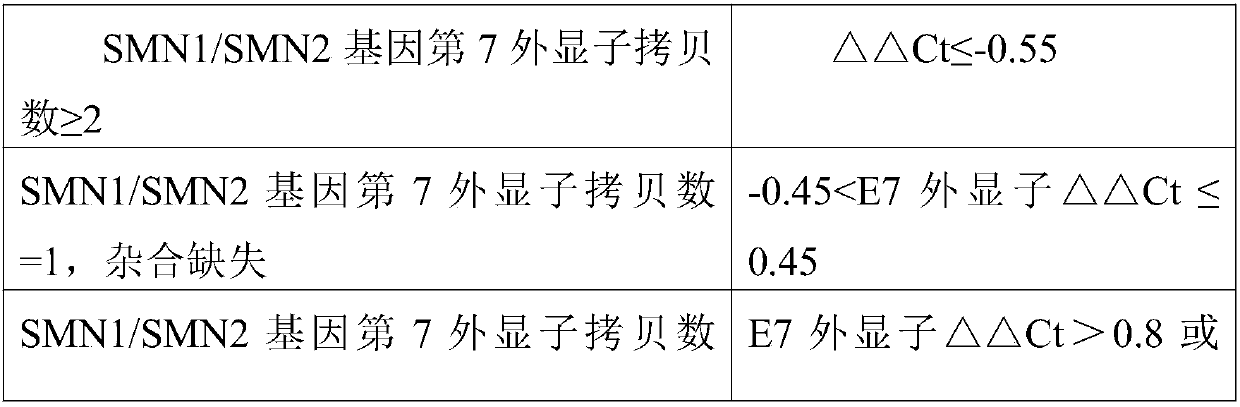
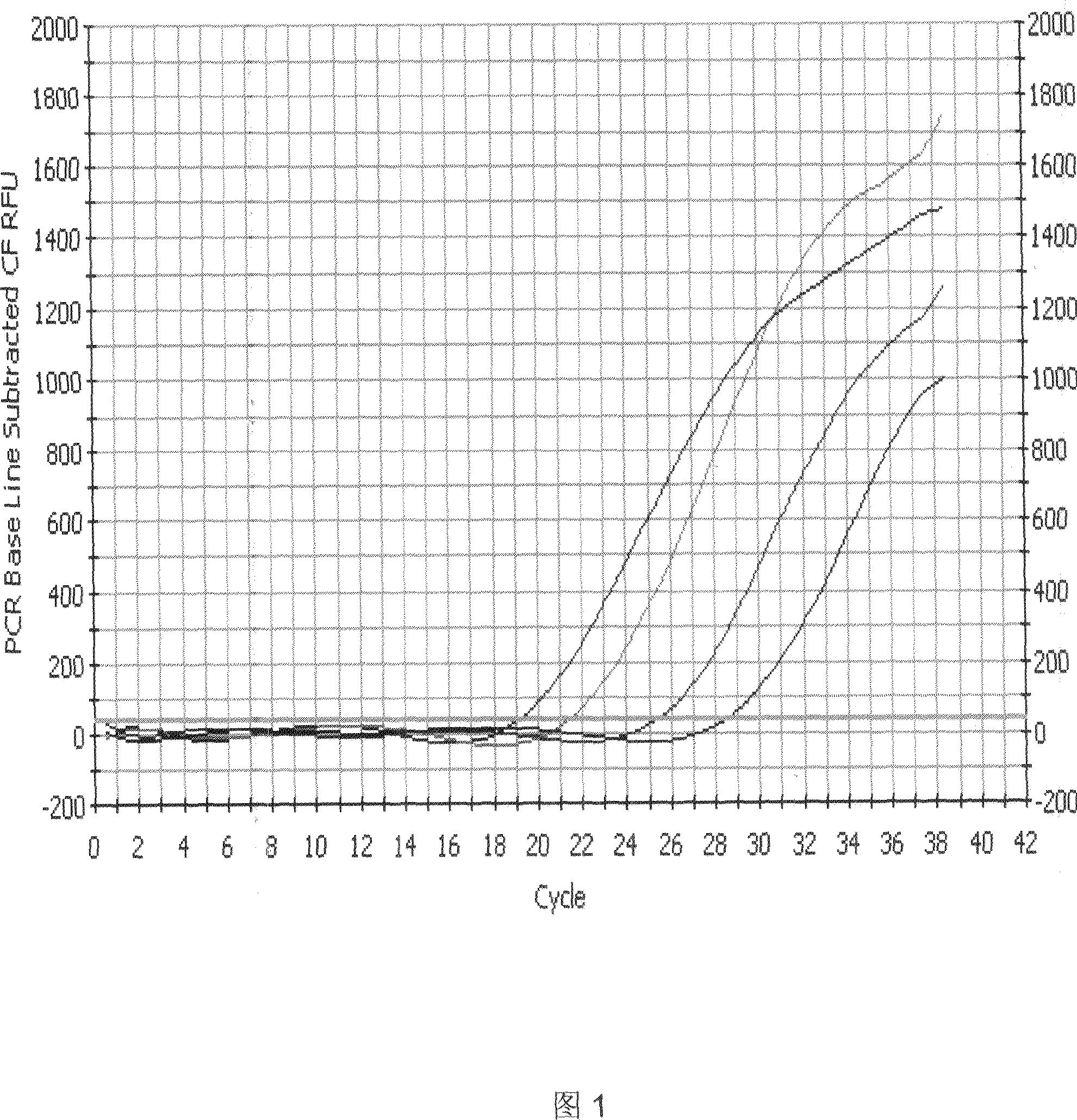
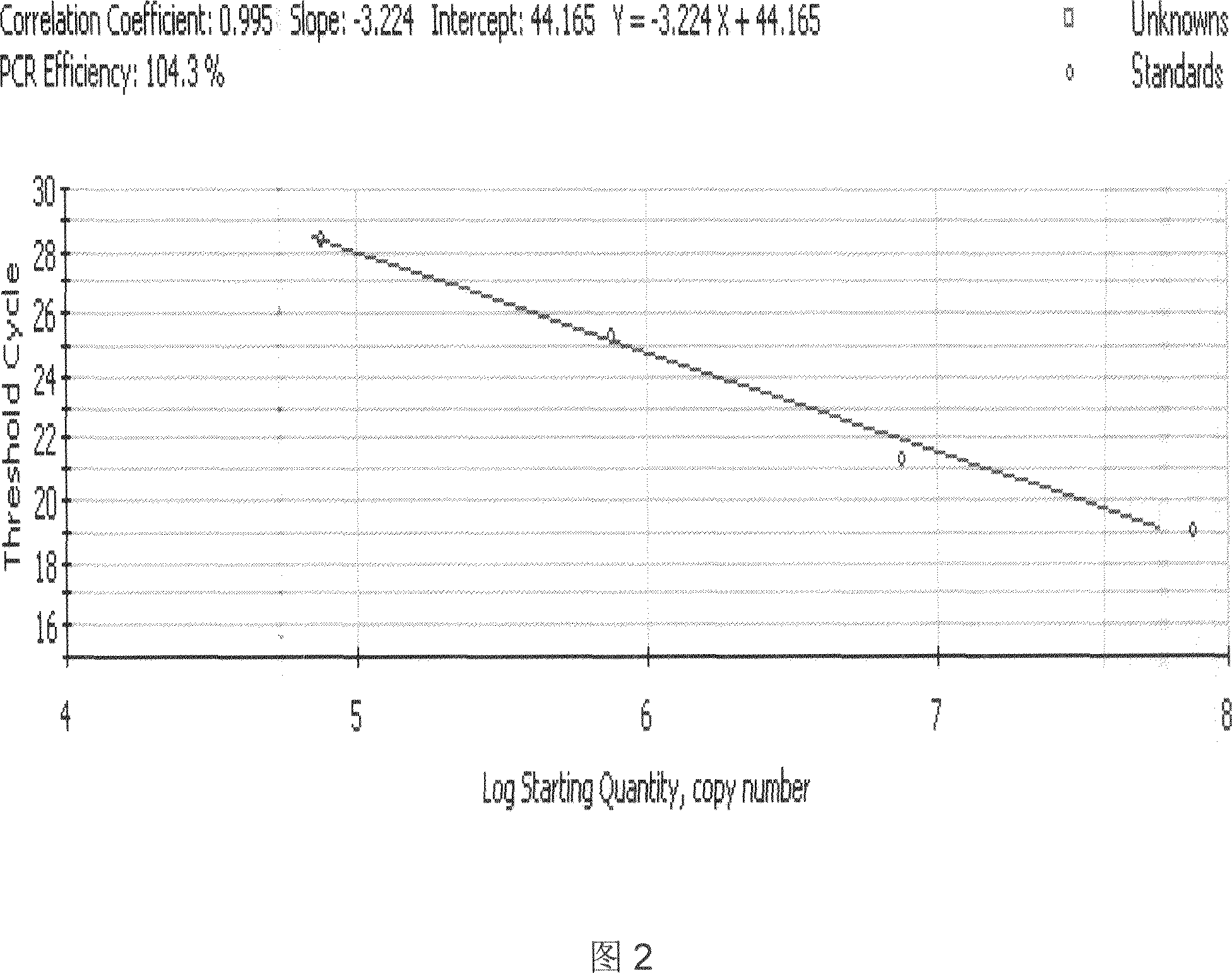
Fluorescent quantitative PCR kit for detecting copy number of virulence gene of human spinal muscular atrophy
InactiveCN108048548AAvoid interferenceEliminate interfering fluorescent signalsMicrobiological testing/measurementDNA/RNA fragmentationReference genesVirulent characteristics
The invention discloses a fluorescent quantitative PCR kit for detecting the copy number of a virulence gene of human spinal muscular atrophy. The fluorescent quantitative PCR kit comprises amplimersand fluorescent probes, wherein the amplimers consist of a pair of shared primers for specific amplification of the seventh exon of SMN1 and SMN2 genes, a pair of shared primers for specific amplification of the eighth exon of the SMN1 and SMN2 genes, and a pair of specific primers for specific amplification of a reference gene, i.e., a CFTR gene; and the fluorescent probes consist of a fluorescent probe for specific detection of the seventh exon of the SMN1 gene, a fluorescent probe for specific detection of the eighth exon of the SMN1 gene, a fluorescent probe for specific detection of the seventh exon of the SMN2 gene, a fluorescent probe for specific detection of the eighth exon of the SMN2 gene and a fluorescent probe for specific detection of the reference gene CFTR. The fluorescentquantitative PCR kit for detecting the copy number of the virulence gene of human spinal muscular atrophy is directed at problems and insufficiencies in quantitative detection of the copy numbers of human motoneuron genes and is rapid and simple in detection and reliable in detection results.
Owner:北京华瑞康源生物科技发展有限公司
BCR-ABL gene fluorescence quantitative RT-PCR primer and probe and reagent kit
InactiveCN1995386AEasy to detectImprove featuresMicrobiological testing/measurementReference genesAgricultural science
The invention discloses a quantitative RT-PCR primer and probe and agent box of BCR-ABL fusing gene mRNA fluorescence with BCR-ABL fusing gene primer and probe sequence as SEQ ID NO1-4 and internal reference gene primer and probe sequence as SEQ ID NO5-7, wherein the agent box contains cell cracking liquid, water, RT-PCR reacting liquid, internal reference TBP RT-PCR reacting liquid, BCR-ABL fusing gene detecting probe, TBP internal reference gene testing probe, composite enzyme, standard material and comparing material; the box can test the expressive level of mRNA of P210BCR / ABL and P190BCR / ABL in the specimen, which provides the reference to diagnose, recurrent and treat chronic granulocytic leukemia and acute lymphocyte leukemia.
Owner:SHANGHAI FOSUN PHARMA (GROUP) CO LTD +1
Screening method for real-time fluorescent quantitative PCR reference genes of Hibiscus hamabo Sieb.et Zucc
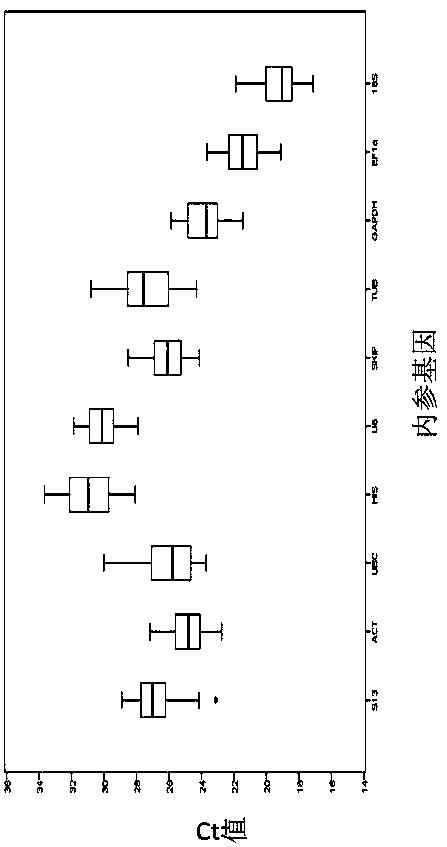
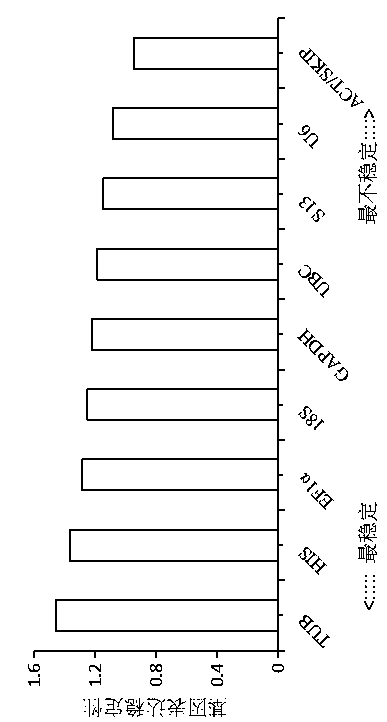
The invention belongs to the technical field of plant gene engineering, and discloses a screening method for real-time fluorescent quantitative PCR reference genes of Hibiscus hamabo Sieb.et Zucc. Thescreening method utilizes a transcriptome database of Hibiscus hamabo Sieb.et Zucc, selects 10 candidate reference genes, and takes 10 candidate reference gene sequences as templates to design real-time fluorescent quantitative PCR specific primers, selects different tissues of Hibiscus hamabo Sieb.et Zucc and Hibiscus hamabo Sieb.et Zucc leaves under various abiotic stresses and hormone treatments as experimental materials for performing a fluorescent quantitative PCR experiment, and finally, performs data analysis by applying geNorm, NormFinder and BestKeeper software. The most stable reference genes in the screening method are ACT and SKIP. The screening method for real-time fluorescent quantitative PCR reference genes of Hibiscus hamabo Sieb.et Zucc establishes a screening system forstable reference genes of hibiscus hamabo, and provides stable reference genes for developing gene function research of Hibiscus hamabo Sieb.et Zucc.
Owner:INST OF BOTANY JIANGSU PROVINCE & CHINESE ACADEMY OF SCI
Method for diagnosing lung cancers using gene expression profiles in peripheral blood mononuclear cells
ActiveUS8476420B2Bioreactor/fermenter combinationsBiological substance pretreatmentsDiseaseReference genes
Owner:THE WISTAR INST OF ANATOMY & BIOLOGY
Multi-real-time fluorescent quantitative PCR kit, method and primer probe composition for detecting 2019 novel coronavirus
InactiveCN111270013AShort detection cycleStrong specificityMicrobiological testing/measurementAgainst vector-borne diseasesReference genesBlood specimen
Owner:NINGBO HEALTH GENE TECHNOLOGIES CO LTD +1
Method for detecting HER 2 positivity of breast cancer through ddPCR technology
The invention provides a method for detecting HER 2 positivity of breast cancer through the ddPCR technology and specifically provides an HER 2 copy number variation detection kit. The kit is characterized by comprising two pairs of primers for detecting HER 2 genes and one or more pairs of primers for detecting reference genes of CEP 17 or EFTUD2 or a combination of CEP 17 and EFTUD2. By combining the ddPCR technology with a combination of multiple reference genes, whether a sample with suspended positivity determined through a conventional method has HER 2 positivity or not can be effectively judged, and therefore assistance and guide can be provided for breast cancer medication.
Owner:SHANGHAI BIOTECAN PHARMA +2
Tea tree miRNA fluorescent quantitative PCR reference gene under low temperature stress as well as screening method and application of reference gene
ActiveCN105132417AMicrobiological testing/measurementDNA/RNA fragmentationReference genesGene selection
The invention relates to a tea tree miRNA fluorescent quantitative PCR reference gene Pc-222-3p, which has a nucleotide sequence as shown in SEQ ID NO: 1, as well as a screening method of the reference gene. According to the method, 6 common plant miRNA reference genes and 3 tea tree miRNAs are selected as candidate reference genes, and the expression stabilities of the candidate reference genes under different low temperature stresses and in different tea tree tissues are evaluated by virtue of qRT-PCR technology through two software, namely BestKeeper and geNorm, so as to provide a reference for the selection of reference genes for tea tree miRNA differential expression analysis under low temperature stress. The obtained tea tree miRNA fluorescent quantitative PCR reference gene Pc-222-3p is stable in gene expression under different temperatures of treatment and in different tea tree tissues; therefore, the reference gene Pc-222-3p can be used as a reference gene for tea tree miRNA fluorescent quantitative PCR under low temperature stress.
Owner:ANHUI QIMEN COUNTY QIHONG TEA
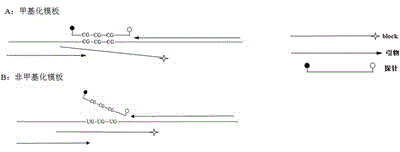
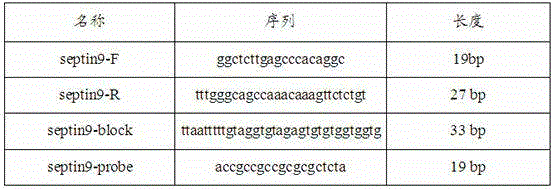
Primer and kit for detecting septin9 gene methylation
InactiveCN106244724AEasy to detectEliminate distractionsMicrobiological testing/measurementDNA/RNA fragmentationForward primerReference genes
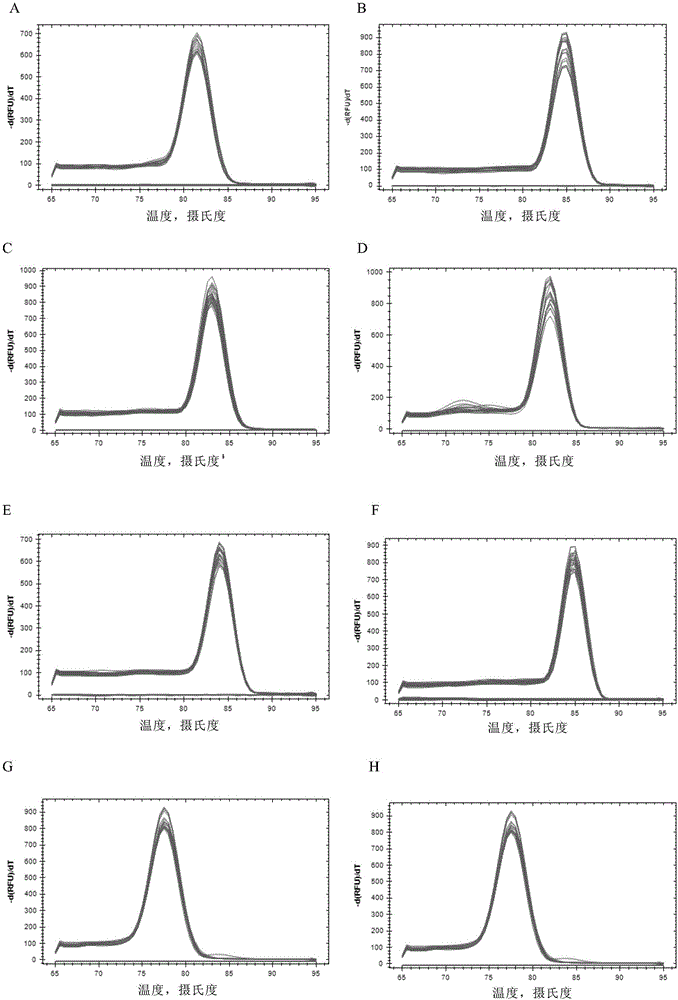
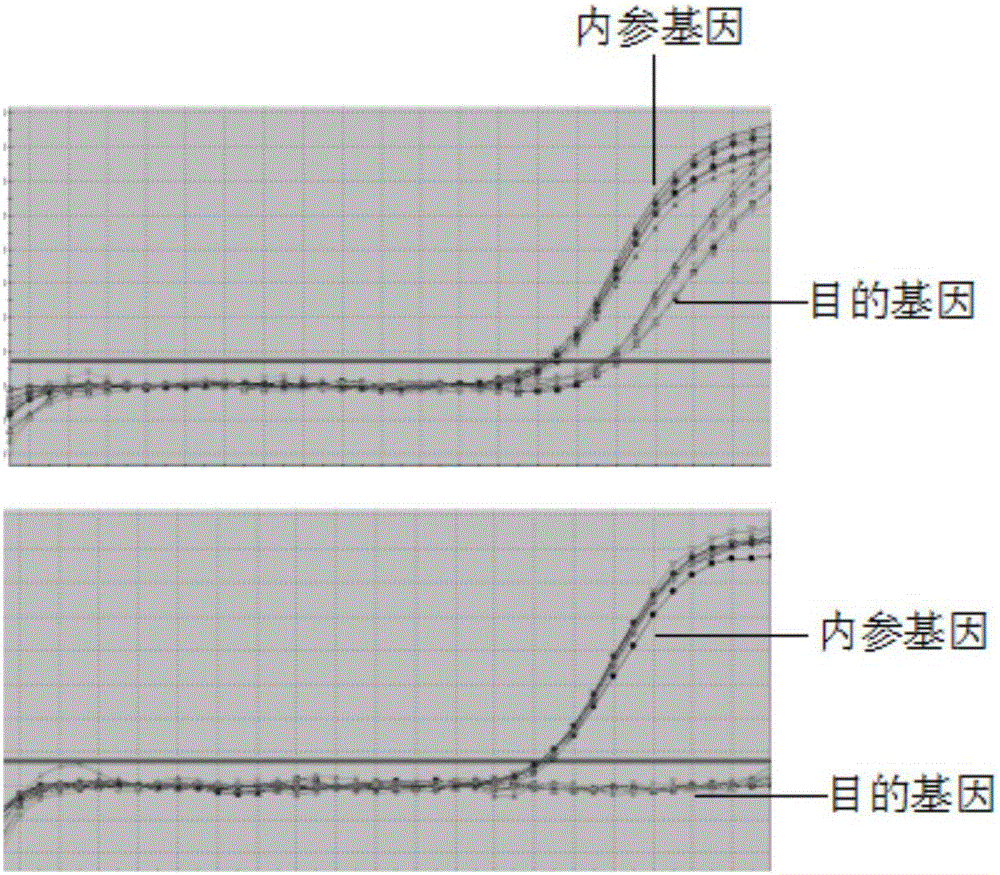
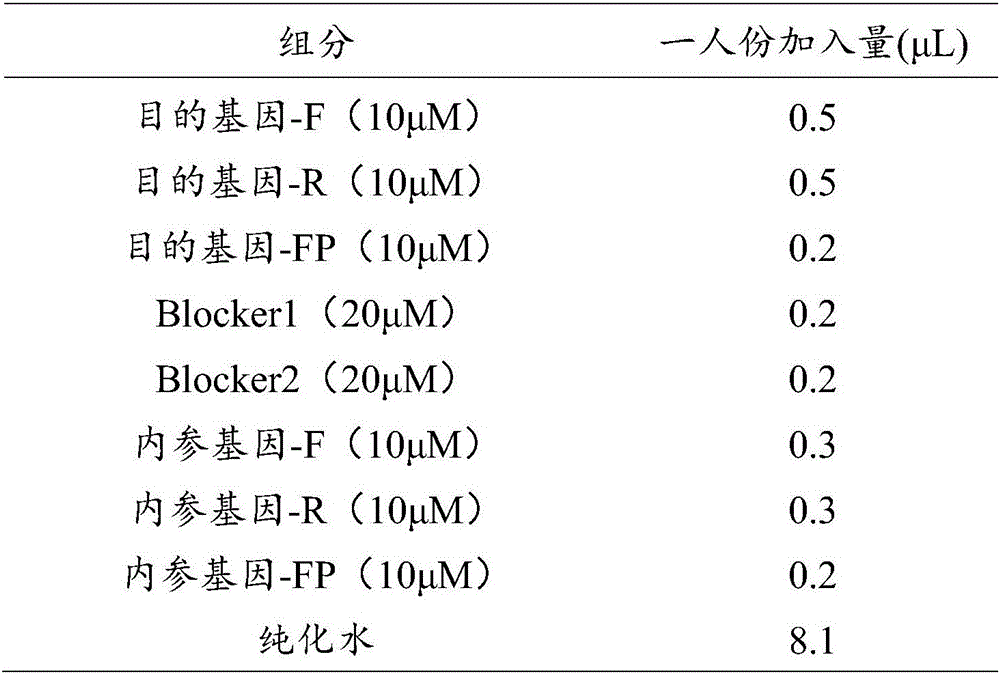
The invention discloses a primer and a kit for detecting septin9 gene methylation. The primer comprises a target gene primer pair Septin9 forward primer F and an Septin9 reverse primer R. The kit comprises the primer and optimally further comprises a reference gene primer, a TaqMan-MGB probe and two Blocker primers. The kit is simple and convenient to operate, is easy to judge and is low in requirement for instruments; the whole PCR process is fully closed; the cross contamination possibility is avoided; the result is more accurate. Compared with the method of detecting by adopting PCR specific primer, the method of identifying the methylated site by utilizing the probe has higher flexibility and accuracy. Two Blocker primers are utilized to strictly control the amplification of the non-specific stripe, so that the kit has higher detection sensitivity and specificity and is especially fit for early screening of free DNA of colorectal cancer plasma.
Owner:北京鑫诺美迪基因检测技术有限公司
A method and system for quickly comparing gene data
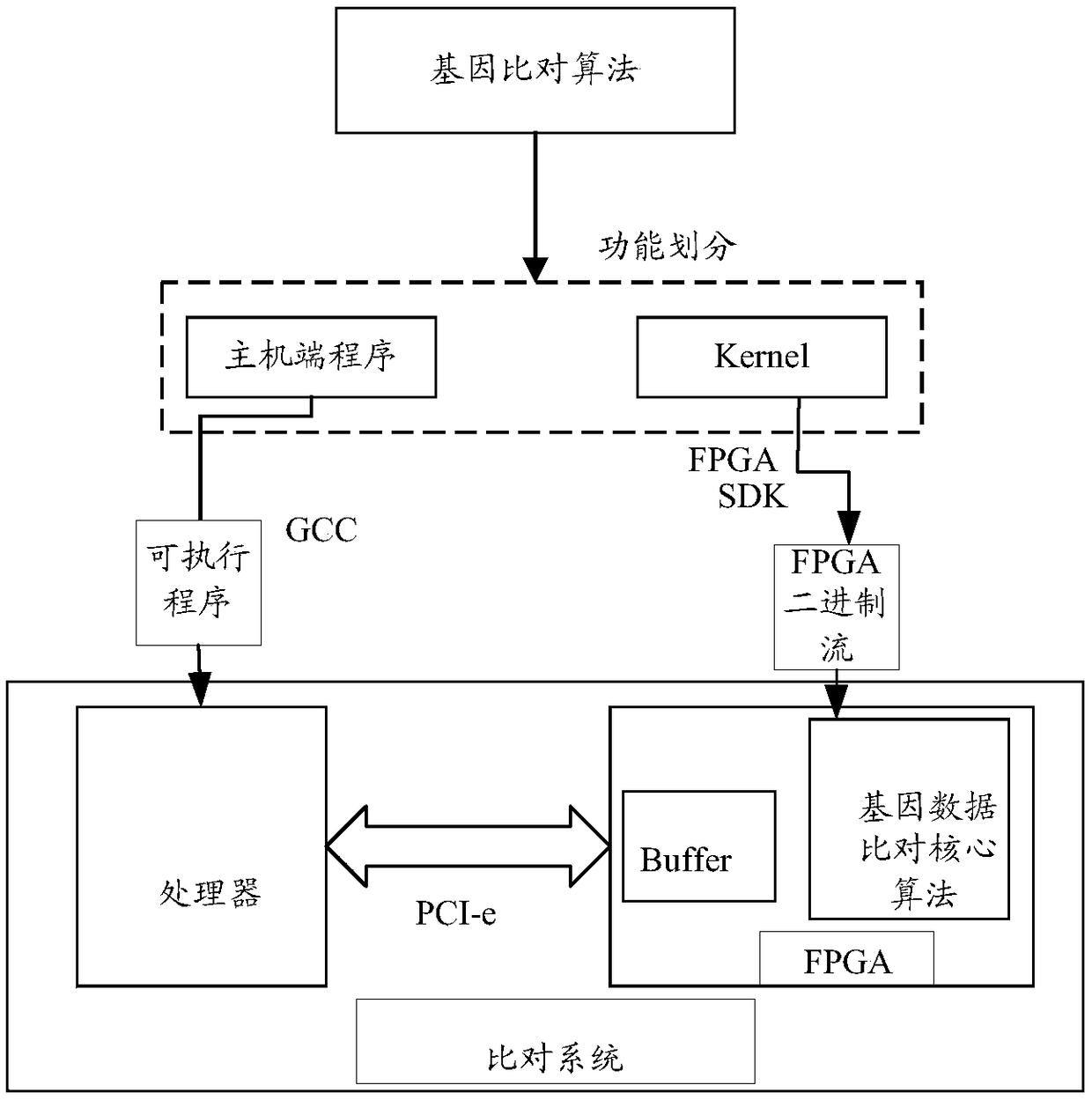
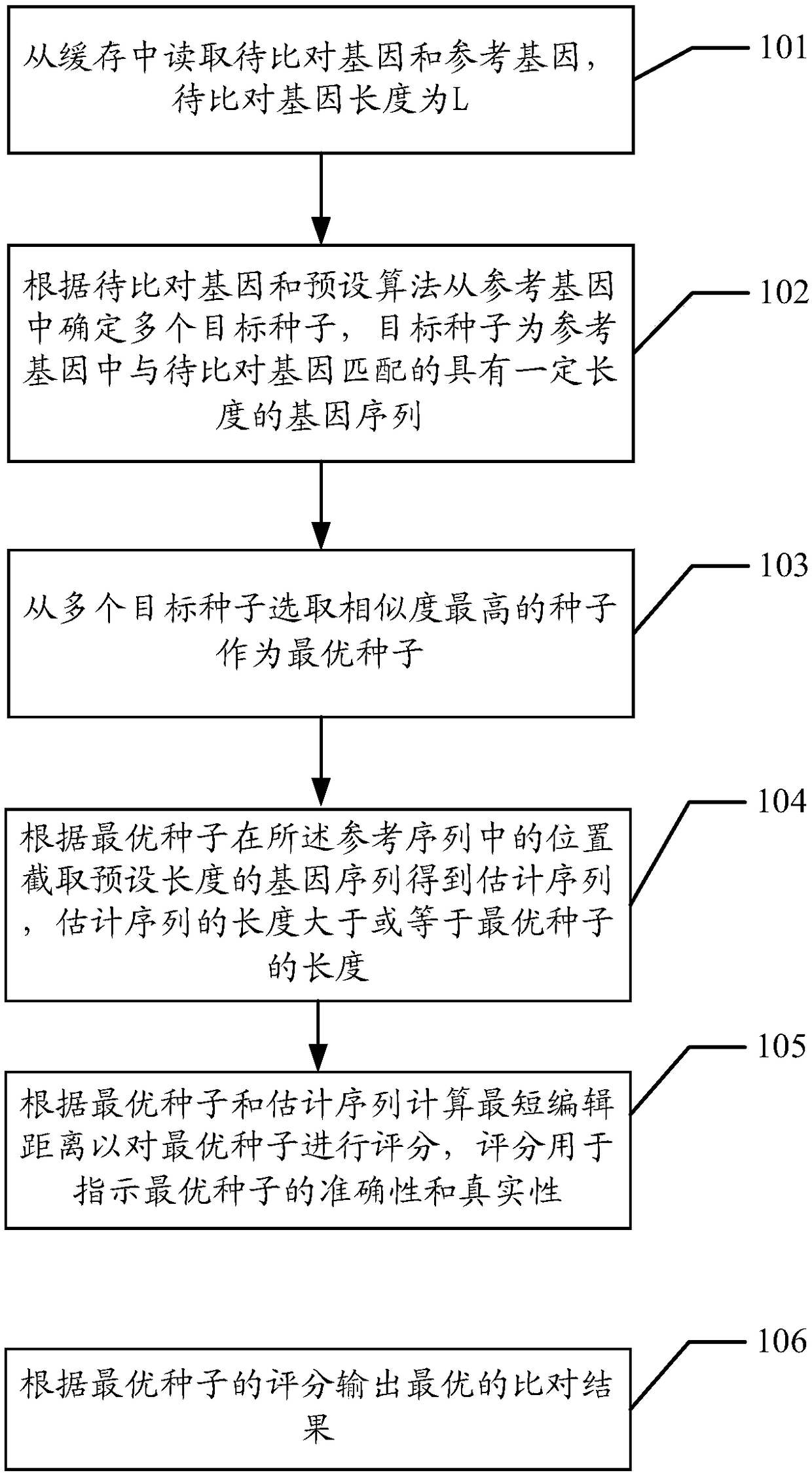
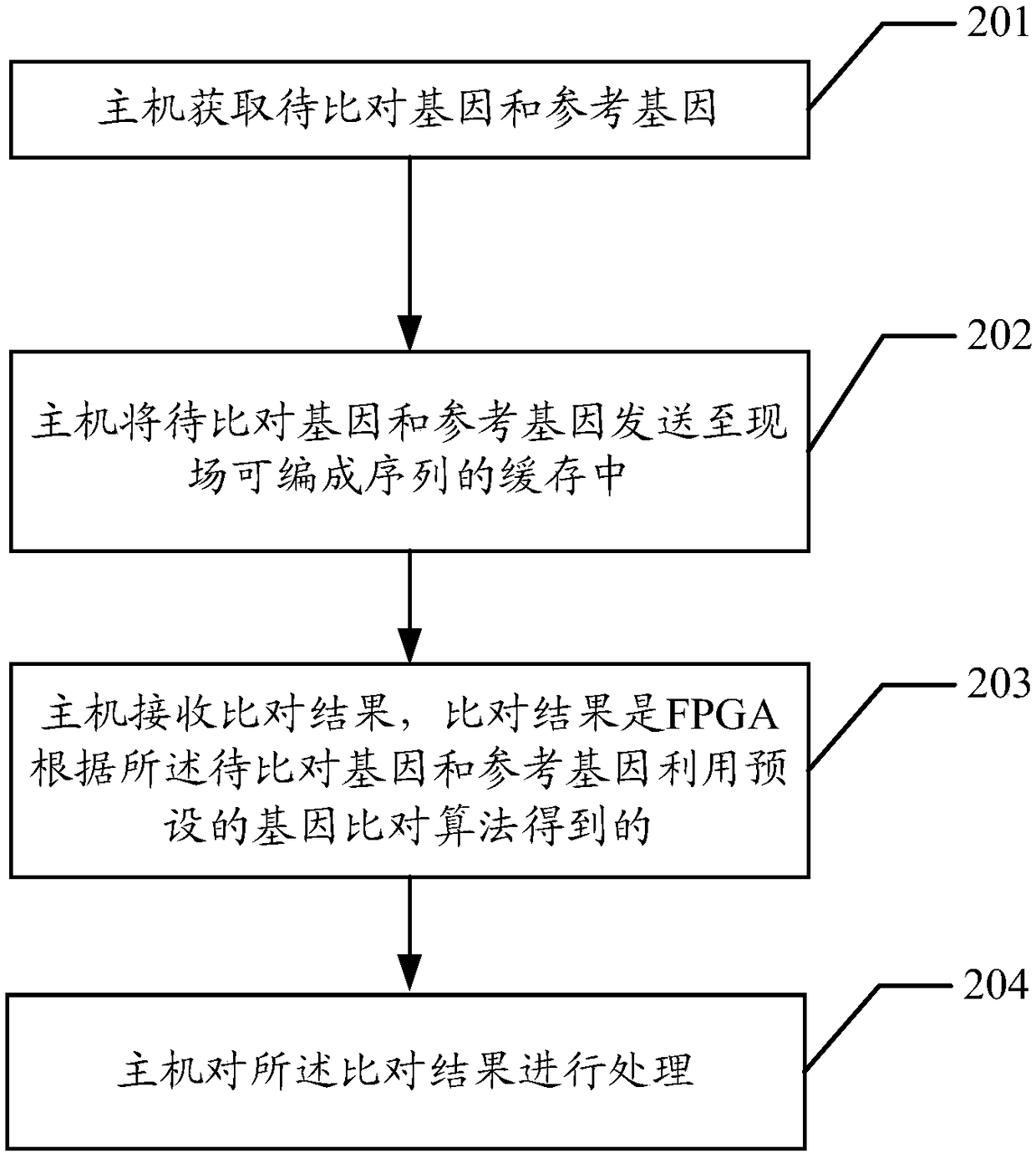
ActiveCN108985008AImprove parallelismImprove throughput performanceSpecial data processing applicationsReference genesEdit distance
The embodiment of the invention discloses a method for quickly comparing gene data, which comprises the following steps: an FPGA reads genes to be compared and a reference gene from a buffer, and thelength of the genes to be compared is L; the FPGA determines a plurality of target seeds from a reference gene accord to a gene to be aligned and a preset algorithm, and the target seeds are gene sequences match with the gene to be aligned for a certain length; the FPGA selects the seeds with the highest similarity from multiple target seeds as the optimal seeds; according to the position of the best seed in the reference sequence, the FPGA obtains the gene sequence with preset length, and the length of the gene sequence is greater than or equal to the length of the best seed; the FPGA scoresthe best seeds by calculating the shortest editing distance between the best seeds and the estimated sequence, and the score is used to indicate the accuracy and authenticity of the best seeds; the FPGA outputs an optimal comparison result according to the scoring of the optimal seeds. The throughput performance of the algorithm can be improved and the efficiency of gene alignment can be improved.
Owner:ZHENGZHOU YUNHAI INFORMATION TECH CO LTD
Kit for detecting methylation level of lung cancer associated SHOX2 gene promoter region
ActiveCN105316421AHigh sensitivityImprove throughputMicrobiological testing/measurementReference genesForward primer
The invention discloses a Kit for detecting the methylation level of a lung cancer associated SHOX2 gene promoter region, which comprises a target gene primer pair, a reference gene primer pair, Taqman probes, and blocking agents, wherein the Taqman probe is used for detecting target genes and reference genes, and the blocking agent has a blocking effect. The target gene primer pair comprises a target gene forward primer shown in a sequence SEQ ID NO.1 and a target gene reverse primer shown in a sequence SEQ ID NO.2; and the reference gene primer pair comprises a reference gene forward primer shown in a sequence SEQ ID NO.3 and a reference gene reverse primer shown in a sequence SEQ ID NO.4. The target gene Taqman probe for detecting is shown in a sequence SEQ ID NO.5, and the reference gene Taqman probe is shown in a sequence SEQ ID NO.6. The blocking agents are shown in a sequence SEQ ID NO.7 and a sequence SEQ ID NO.8.
Owner:湖南宏雅基因技术有限公司
GSTM3 (Glutathione S-Transferase M3) gene methylation quantitative detection method for hepatic failure prognosis and kit
InactiveCN103525907ASimple and fast operationHigh sensitivityMicrobiological testing/measurementReference genesDeoxyribose
The invention provides a GSTM3 (Glutathione S-Transferase M3) gene methylation quantitative detection method for hepatic failure prognosis and a kit, and provides a method and a device for quantitatively detecting the methylation degree of a glutathione S-transferase M3 promoter. The kit is internally provided with a reagent for extracting peripheral blood genome DNA (Deoxyribose Nucleic Acid), a reagent for modifying the genome DNA, a 5*premixing PCR (Polymerase Chain Reaction) system, a methylation specific primer pair and a Taqman fluorescence probe aiming at a target gene GSTM3 promoter, and a specific primer pair and a Taqman fluorescence probe of reference genes ALU-C4. According to the GSTM3 gene methylation quantitative detection method for the hepatic failure prognosis and the kit, the methylation specific primer pair and the probe of the target gene GSTM3 promoter and the specific primer pair and the probe of the reference genes can be used for carrying out a real-time quantitative polymerase chain reaction (PCR) on the peripheral blood genome DNA; a quantitative methylation specific PCR method is used for detecting a threshold value circulation value of the target gene GSTM3 and the reference genes ALU-C4 and calculating a gene GSTM3 methylation quantitative value, so as to be good for evaluating illness states, judging the prognosis and guiding the treatment.
Owner:SHANDONG UNIV QILU HOSPITAL
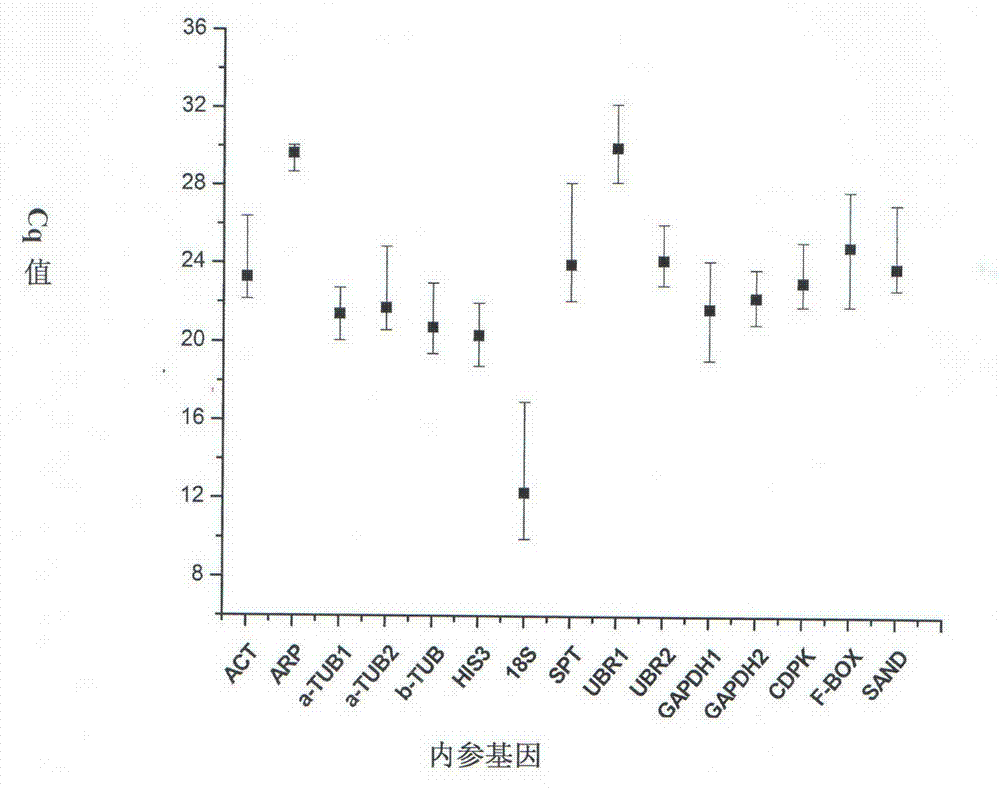
Method for screening reference genes for chilo suppressalis under temperature stress and application of reference genes
The invention discloses a method for screening reference genes for chilo suppressalis under different temperature stress conditions and application of the reference genes. The method includes utilizing the chilo suppressalis cultivated under room-temperature conditions as a control material; carrying out fluorescence quantitative PCR (polymerase chain reaction) detection by the aid of the chilo suppressalis as an experimental material after the chilo suppressalis is under high-temperature and low-temperature stress; inputting Ct values of all alternative reference genes into delta Ct, RefFinder, BestKeeper, GeNorm, NormFinder reference gene stability analysis software and analyzing the Ct values of all the alternative reference genes; screening the most stable in-vivo reference genes TUB for the chilo suppressalis under the temperature stress conditions. The method and the application have the advantage that expression of genes related to the temperature stress conditions can be accurately studied by the aid of genes, namely the reference genes.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Method and device for data classification of metagenome
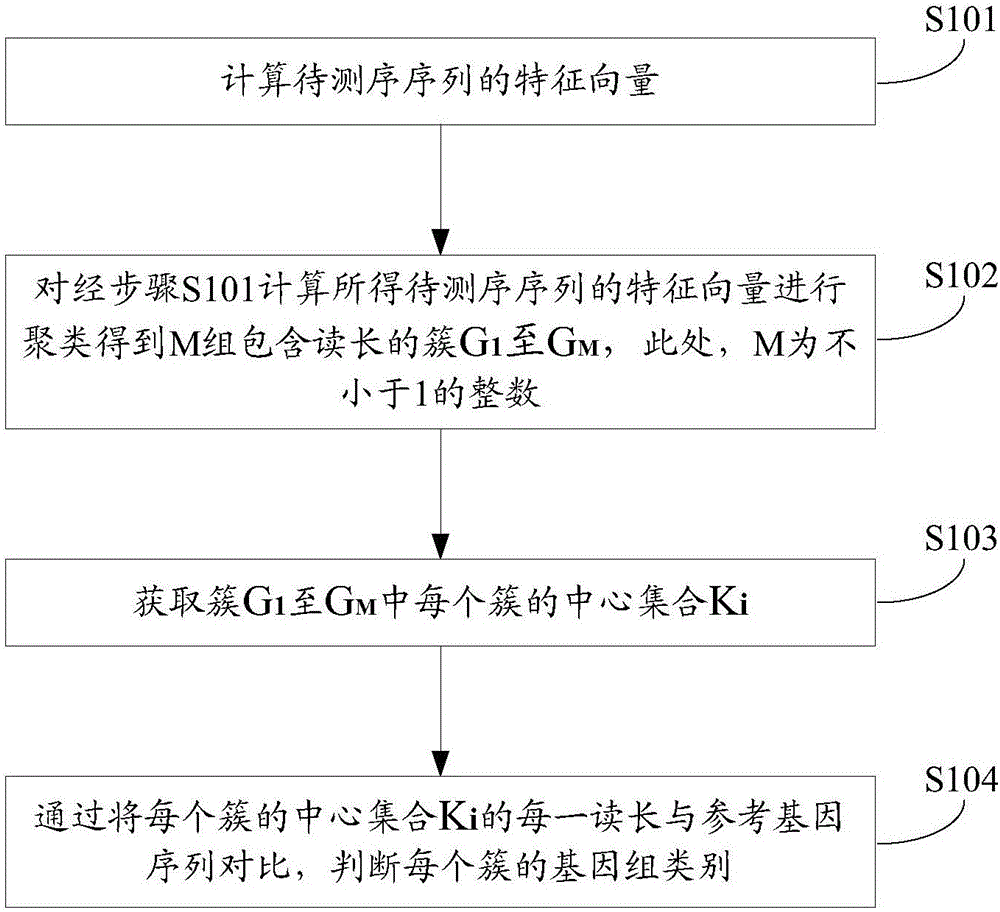
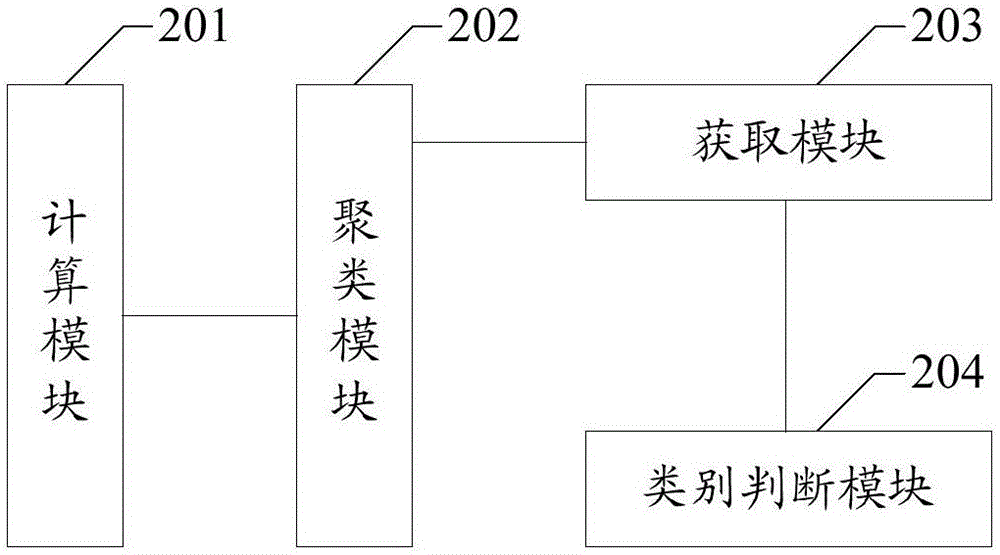
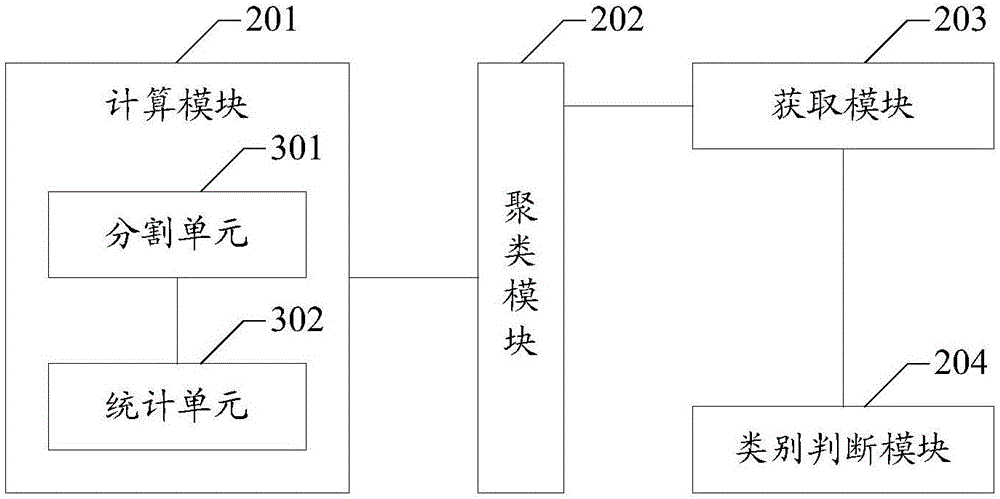
ActiveCN106682454AReduce time overheadFast operationCharacter and pattern recognitionProteomicsFeature vectorReference genes
The invention belongs to the field of gene data processing and provides a method and device for data classification of metagenomes. The genomic classification precision is improved at a small time cost. The method comprises the steps of calculating feature vectors of a sequencing sequence; clustering the feature vectors to obtain M groups of clusters G1 to GM including read length, and M is an integer not smaller than 1; obtaining a center set Ki of each cluster from the clusters G1 to GM; and through the comparison of each read length of the center set Ki of each cluster and the reference gene sequence, judging the genome category of each cluster. Compared with the prior art, according to the technical scheme, the time cost used for classification is reduced, the operating speed is improved, and the classification precision of the genome category which the sequencing sequence belongs to is remarkably improved.
Owner:SHENZHEN INST OF ADVANCED TECH CHINESE ACAD OF SCI
Gene methylation detection method
InactiveCN104131070AImprove accuracyGood repeatabilityMicrobiological testing/measurementReference genesDuplex pcr
The invention discloses a gene methylation detection method. The method comprises the steps of: subjecting a to-be-detected DNA sample, a negative control, a positive control and a no template control to PCR amplification reaction, with the DNA sample in the PCR amplification reaction being bisulfite modified DNA, carrying out duplex PCR amplification reaction on a detection gene and a reference gene, and interpreting a parameter range to judge the methylation condition of the detection gene of the to-be-detected DNA sample. By the mode, the gene methylation detection method provided by the invention has the advantages of strong specificity, duplex reaction, high sensitivity, small pollution, simple and quick operation, and safety, etc., and the detection result has good accuracy and repeatability. Thus, the method has certain auxiliary diagnosis effect on early detection of colorectal cancer and other cancers.
Owner:JIANGSU MICRODIAG BIOMEDICINE TECH CO LTD
TaqMan probe real-time fluorescence PCR (Polymerase Chain Reaction) method for detecting HLA (Human Leukocyte Antigen)-B*5801 alleles
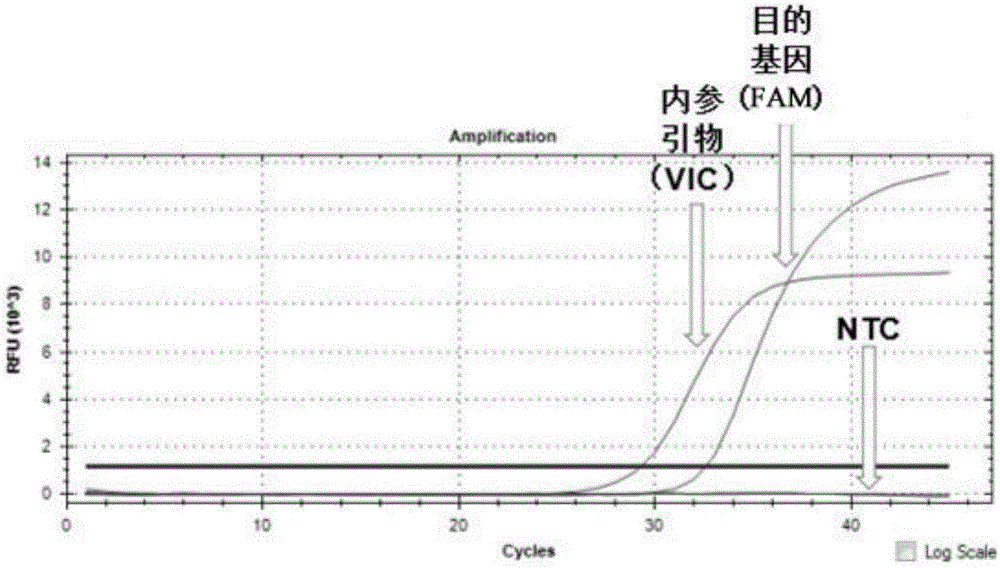
ActiveCN104232781ASave consumablesShorten the timeMicrobiological testing/measurementDNA/RNA fragmentationReference genesFluorescence
The invention discloses a primer probe assembly for high-specificity amplification HLA (Human Leukocyte Antigen)-B*5801 alleles on the basis of a TaqMan probe detection method. The primer probe assembly comprises an upstream primer Fp: 5'-AGGGGCCGGAGTATTGGGATG-3', a downstream primer Rp: 5'-TTGGCCTCAACTGAAAATGAAAC-3' and a probe: 5'-HEX-TCAGGGAGGCGGATCTCGGAC-BHQ2-3'. Primers and probes of reference genes ACTB are used simultaneously, target genes and the reference genes are added into a tube for dual-channel fluorescent quantitative PCR reaction, and a result is analyzed through an amplification curve. The method has the characteristics of simplicity, convenience, flexibility, quickness, high specificity, high flux, zero pollution, high resolution and the like, and is applicable to detection of the HLA-B*5801 alleles of a whole-genome DNA (Deoxyribonucleic Acid) sample in the peripheral blood and the saliva of a human body.
Owner:SHANXI LIFEGEN
Methods for Detecting and Treating Rhinovirus Infection
The invention provides a method for evaluating the activity of an agent for treating rhinovirus infection or a symptom thereof, a method of detecting or monitoring rhinovirus infection, and a method of treating rhinovirus infection or a symptom thereof. Various embodiments comprise measuring expression of (i) one or more genes selected from the group consisting of CRY2, B3GAT3, C10ORF95, and BATF3, and (ii) one or more genes selected from the group consisting of RNFT2, BTG4, PSD3, CAPN9, SULT1E1, HEY1, LRRC36, RAB3B, ALDH3B1, FAM134B, FAS, PLSCR1, CLEC2B, HAS2, MX1, SP110, GBP1, IFIT3, IFIT1, CXCL9, CXCL10, and CXCL11, from at least one biological sample to produce a gene expression profile, and comparing the gene expression profile to a reference gene expression profile. Systems, computer readable media, compositions, and methods for maintaining or improving respiratory health also are provided.
Owner:THE PROCTER & GAMBLE COMPANY
Screening and application of fluorescent quantitative reference genes of pear fruits at different development stages
InactiveCN107190062ATo overcome the shortcomingsOvercome deficienciesMicrobiological testing/measurementDNA/RNA fragmentationReference genesPEAR
The invention discloses screening and application of fluorescent quantitative reference genes of pear fruits at different development stages, and relates to the field of pear molecular biology researches. Reference genes, which are more stable than common housekeeping genes at different development stages of Cuiguan, Fengshui and Xueqing pear fruits, are pertinently obtained according to dynamic RNA (Ribonucleic Acid)-Seq sequencing results, post-period experience validation and data analysis of pear fruits of various varieties; the difference between samples and the difference in the samples, which are caused by the selection of the reference genes, are reduced to the greatest extent; the correction and standardization of a target gene expression level can be realized, and accurate quantification of functional gene expression of the pear fruits is realized.
Owner:NANJING AGRICULTURAL UNIVERSITY
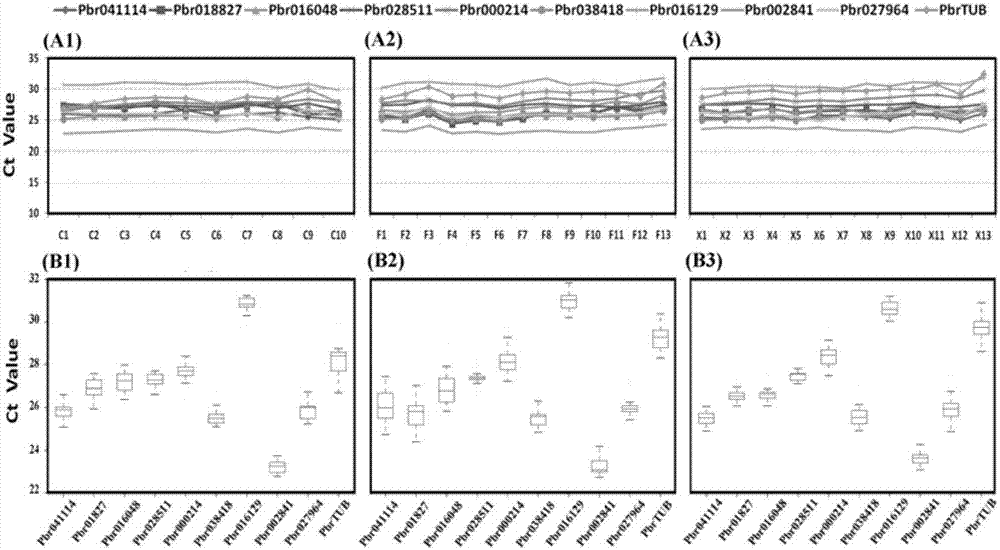
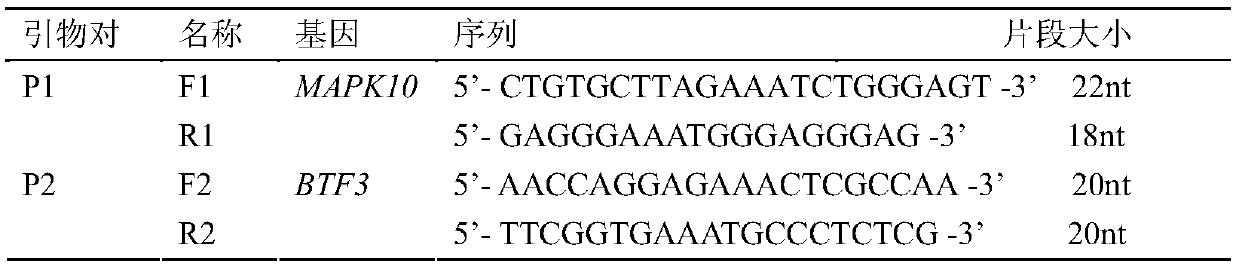
Method for detecting CNV (copy number variation) mark of MAPK10 gene of Nanyang cattle and application thereof
ActiveCN105506111AExcellent genetic resourcesFacilitates marker-assisted selectionMicrobiological testing/measurementReference genesMarker-assisted selection
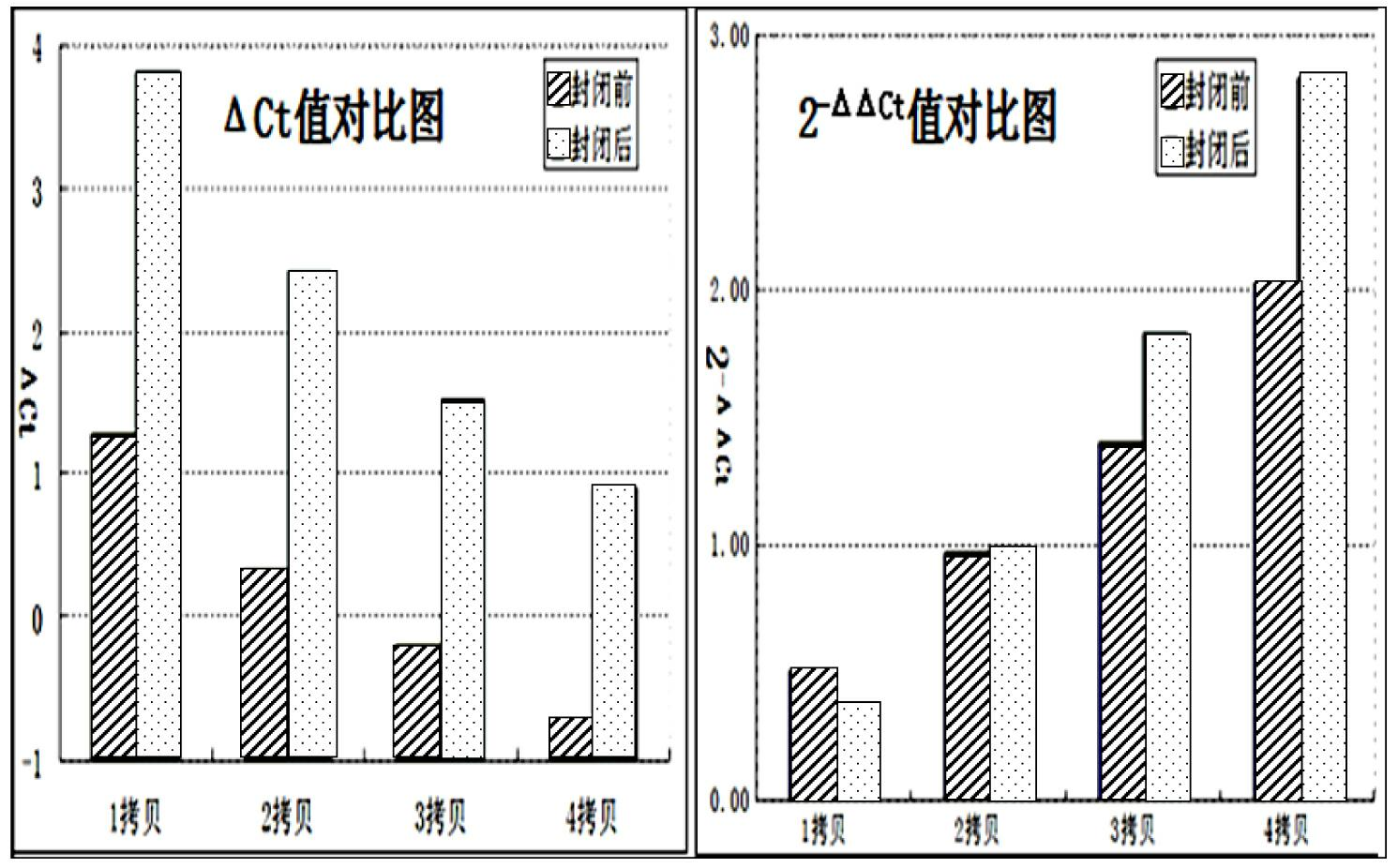
The invention discloses a method for detecting a CNV (copy number variation) mark of a MAPK10 gene of Nanyang cattle and an application thereof. The method comprises the following steps: respectively amplifying the CNV region and reference gene BTF3 of the MAPK10 gene of Nanyang cattle through a real-time fluorescence quantification PCR method by taking blood genome DNA of Nanyang cattle as a template; dividing the result into insert type, deficient type and normal type according to log22-delta delta Ct; and identifying CNV of the MAPK10 gene of Nanyang cattle. Analysis correlated to Nanyang cattle production data indicates that the growth and development of Nanyang cattle is remarkably influenced by different copy numbers of MAPK10, wherein the birth weight, 18-month-aged body height and 24-month-aged body weight and daily gain of a normal type individual body are remarkably higher than those of the insert type and deficient individual bodies. The CNV mark closely related to the production traits of Nanyang cattle is detected on DNA level, and can be used as an important candidate molecular mark for marker assisted selection of the growth traits of Nanyang cattle of China.
Owner:NORTHWEST A & F UNIV
Quantitative detection method for gene copy number
InactiveCN102618646ASync detectionHigh sensitivityMicrobiological testing/measurementReference genesReference sample
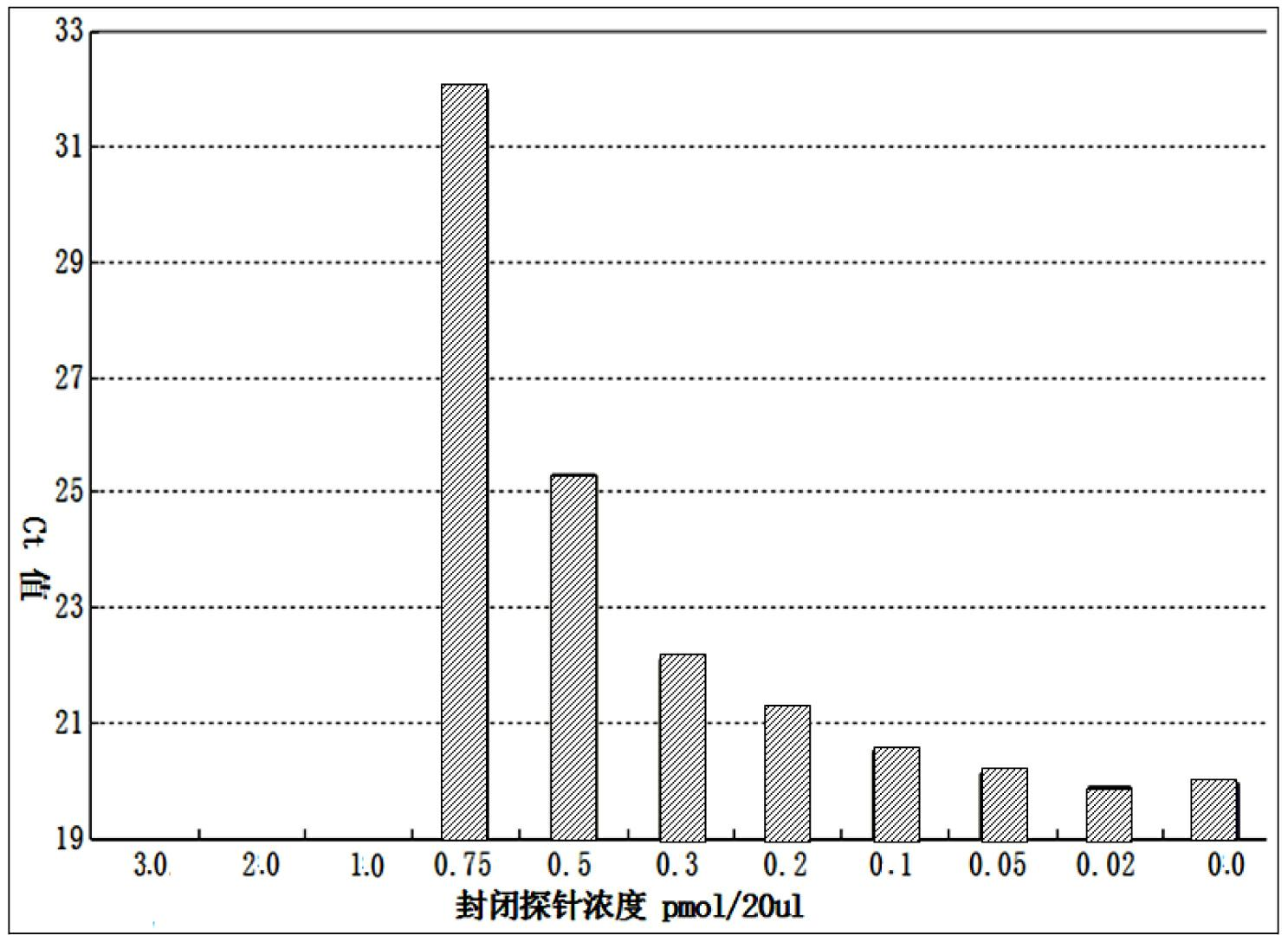
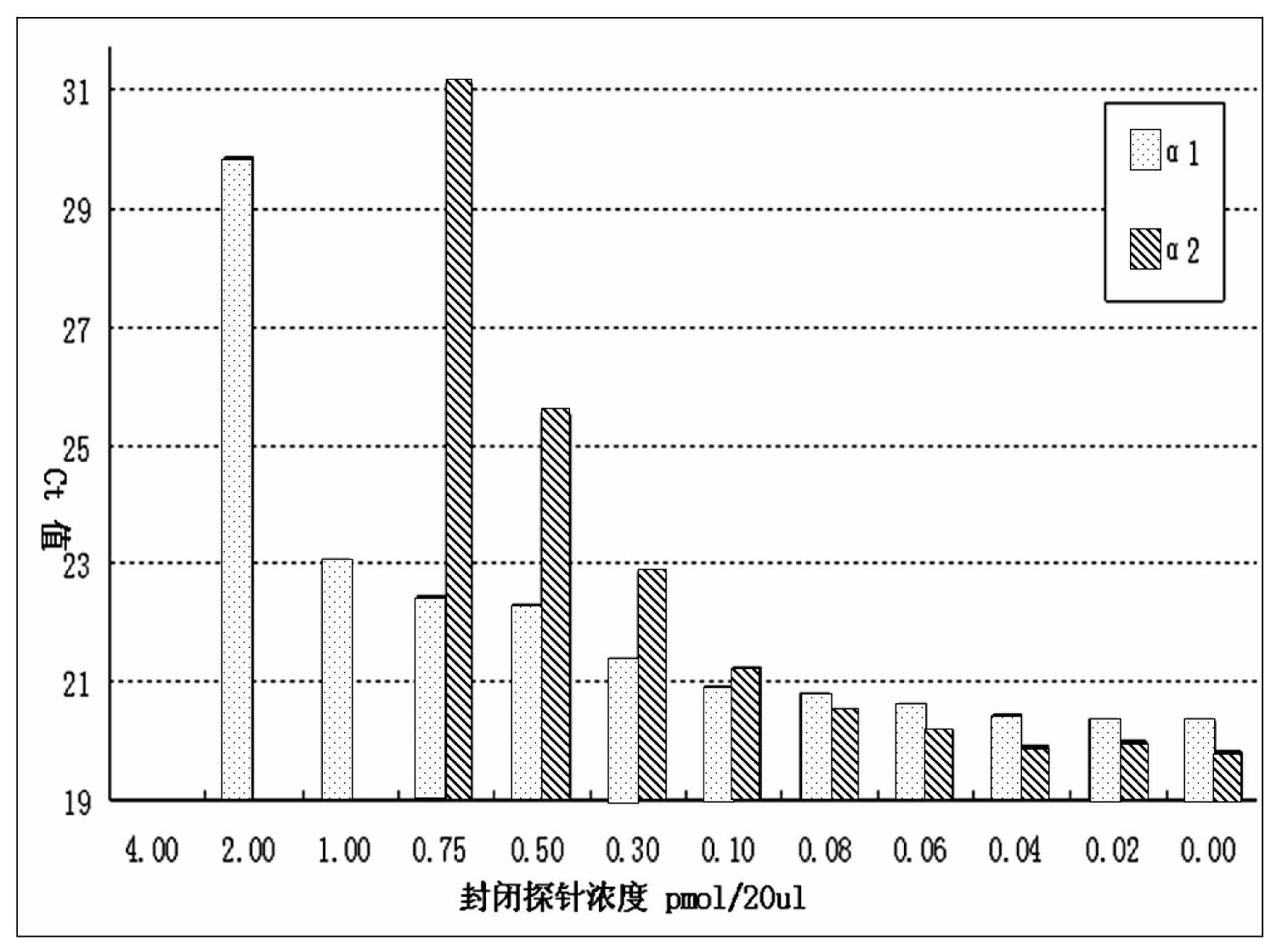
The invention discloses a quantitative detection method for a gene copy number. The method comprises the following steps of: respectively designing primers and fluorescence probes for target gene and reference gene amplification sequences, and designing a closed probe for the target gene amplification sequence; detecting a sample: performing multiple TaqMan fluorescent quantitative polymerase chain reaction (PCR) on target genes and reference genes by using the DNA sample to be detected as a template, and respectively recording Ct values by using a DNA sample with known target gene copy number as a reference sample; and analyzing the result. By the method, the copy number of the functional genes can be directly and quantitatively detected, and deletion, deletion number and multiple copies (CNVs) of the detected genes are directly reflected, so that synchronous quick detection of gene deletion and multiple gene copies is realized; and the method is high in sensitivity, the synchronous quick detection of the gene deletion and the multiple gene copies can be finished only by a common fluorescent quantitative PCR instrument, and experiments prove that the method is accurate and reliable.
Owner:SOUTHERN MEDICAL UNIVERSITY
Determination of gene expression levels of a cell type
ActiveUS20130190194A1Nucleotide librariesMicrobiological testing/measurementReference genesCell type specific
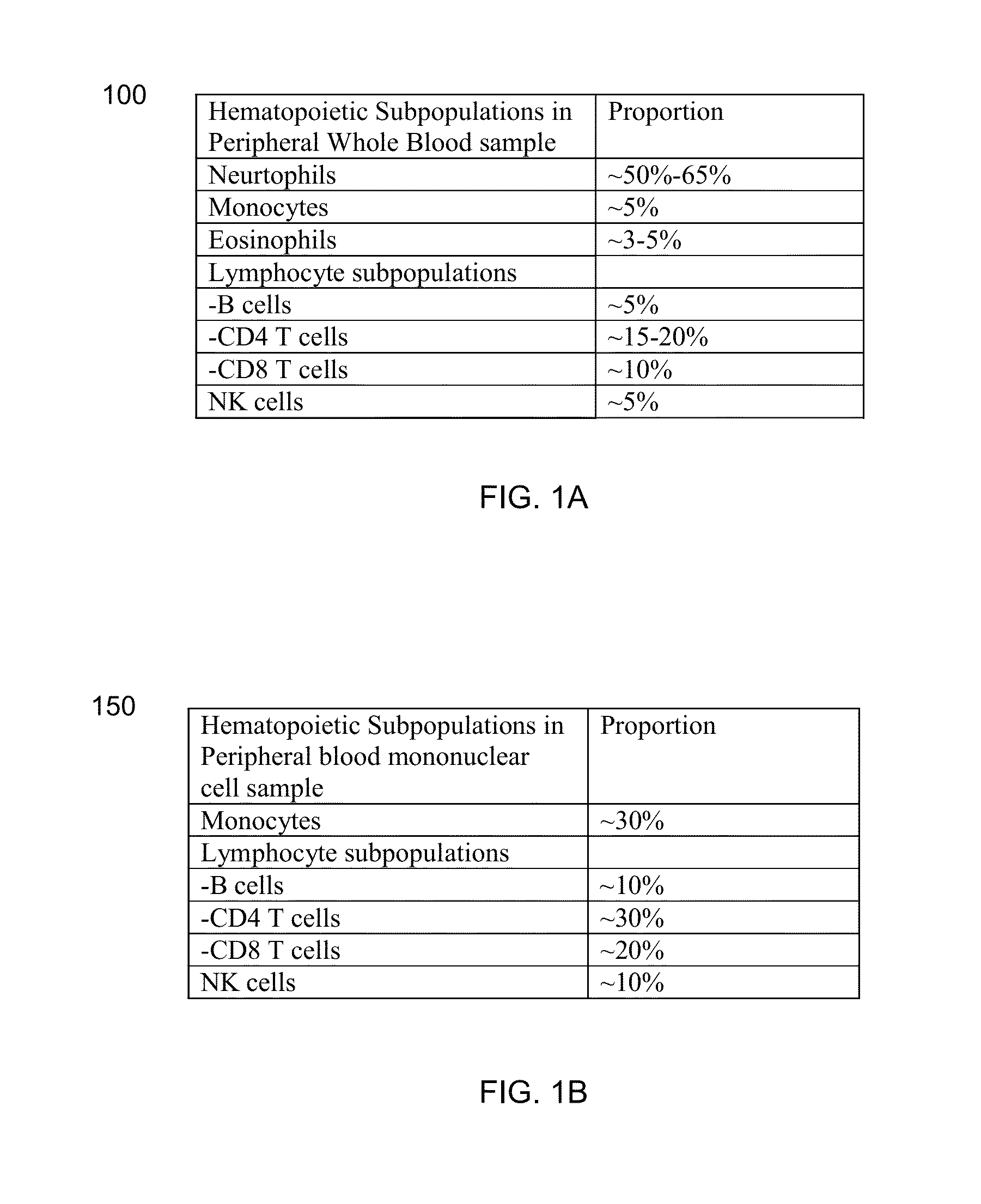
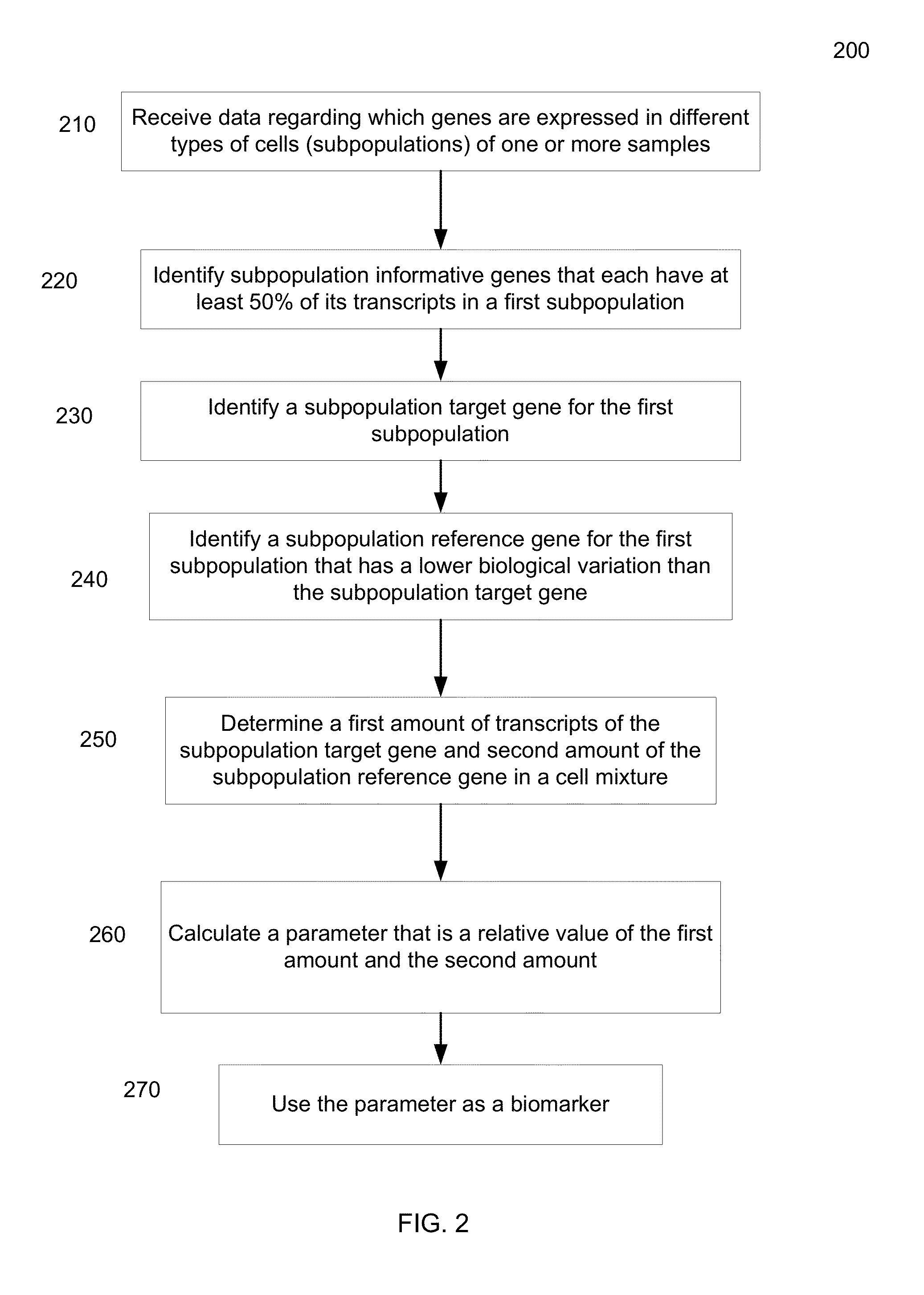
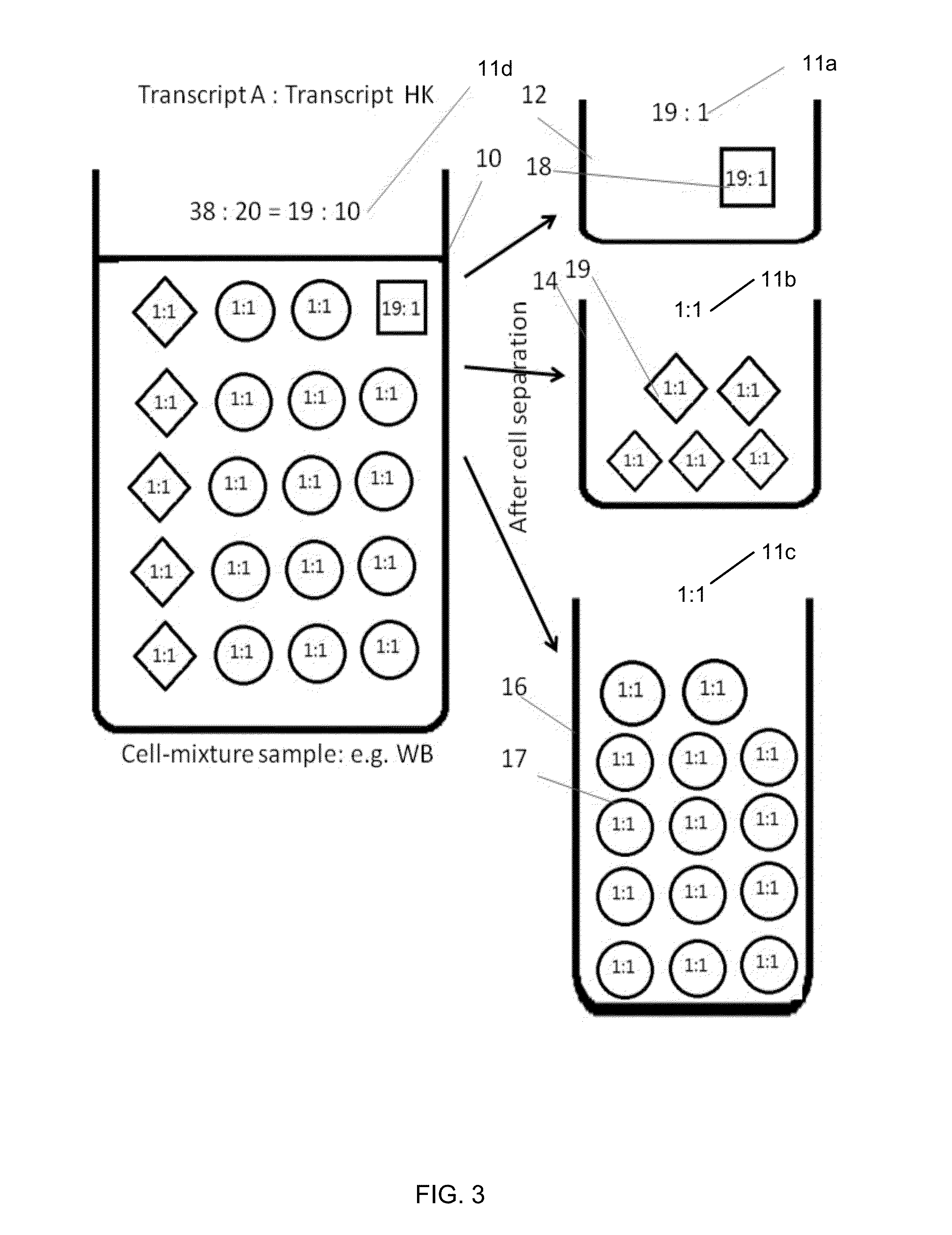
Methods, systems, and compositions can determine gene expression level of a specified cell-type subpopulation by direct analysis of a cell mixture sample composed of multiple subpopulations of various cell-types without the need of prior separation of the component cell-type subpopulations. A target gene and a reference gene can be identified as being informative for a specific cell-type subpopulation when at least 50% of the gene's transcripts in the cell mixture are from the subpopulation. This relative expression level in the cell mixture of the informative target and reference genes can correlate to the relative expression when measured in the isolated subpopulation. Thus, a similar biomarker can be obtained without the difficult step of isolating the cells of the subpopulation.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Method for screening real-time fluorescence quantification PCR internal reference molecules of syntrichia caninervis in desert
InactiveCN103866007AEasy to correctImprove standardizationMicrobiological testing/measurementReference genesFluorescence
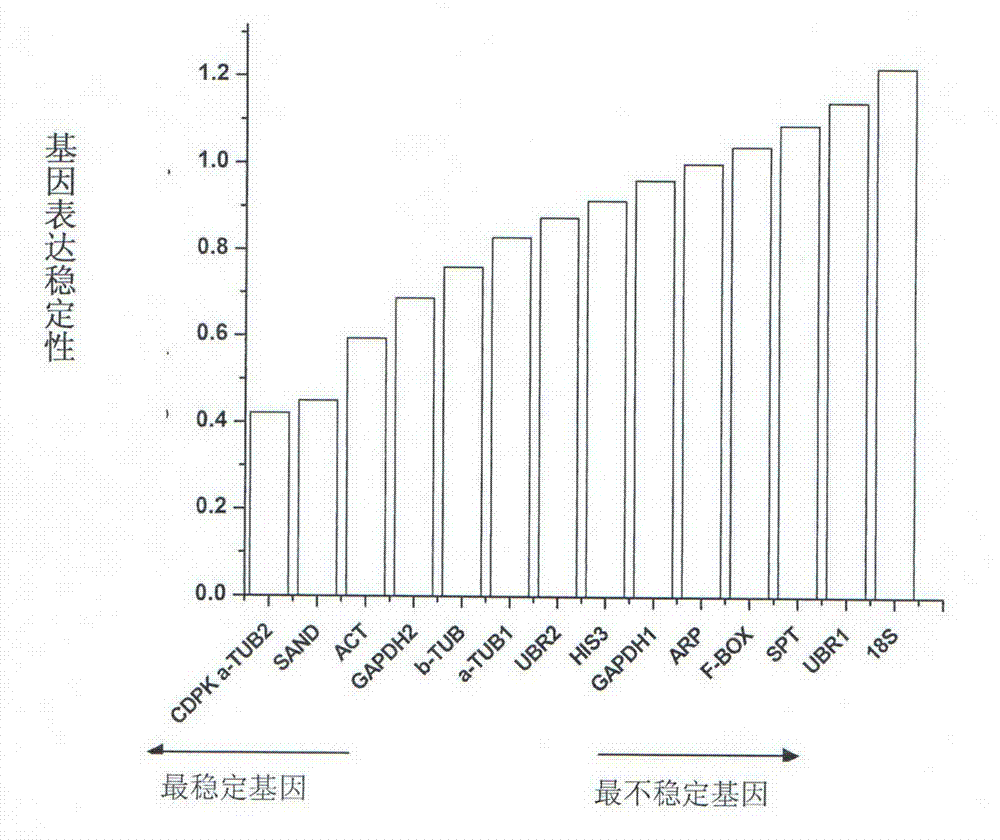
The invention relates to a method for screening real-time fluorescence quantification PCR internal reference molecules of syntrichia caninervis in a desert. The method comprises the following steps: selecting 15 internal reference candidate genes by utilizing a syntrichia caninervis transcriptome database, designing an internal reference gene specific primer of real-time fluorescence quantification PCR by taking 15 internal reference genes as templates; carrying out a fluorescence quantification PCR experiment by selecting syntrichia caninervis gametophytes stressed by 10 non-living things and free of stress (in contrast) as experimental materials; carrying out fluorescence quantification data analysis by using geNorm, NormFinder and Refinder software, so as to screen out the best fit and the most stable internal reference molecules CDPK and alpha-TUB2 of the syntrichia caninervis for developing fluorescence quantification research under various abiotic stresses. By adopting the method disclosed by the invention, the errors of the syntrichia caninervis transcriptome material under different abiotic stresses in ribonucleic acid (RNA) quality, yield, reverse transcription efficiency and the like can be avoided, the real-time fluorescence quantification detection data can be better corrected and standardized, and the accuracy and the reliability of the gene quantification research are improved.
Owner:XINJIANG INST OF ECOLOGY & GEOGRAPHY CHINESE ACAD OF SCI
Composition for noninvasive screening of early lung cancer and application of composition
ActiveCN107034296AImprove the detection rateHigh sensitivityMicrobiological testing/measurementReference genesProstaglandin E Receptor
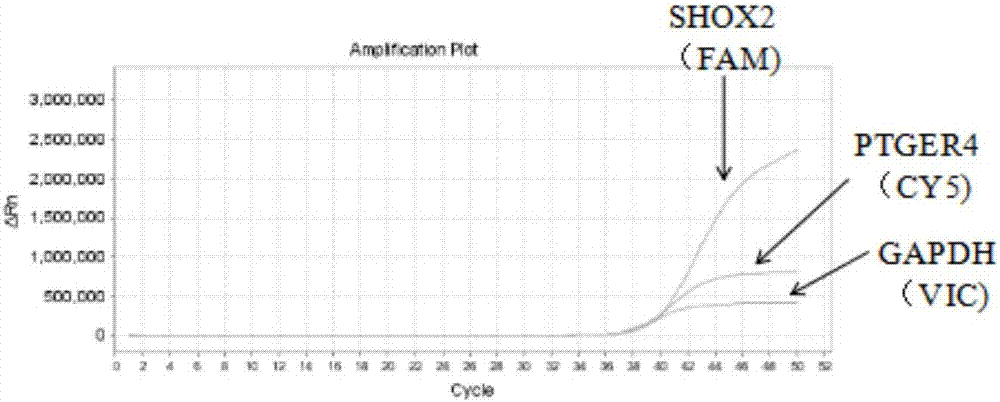
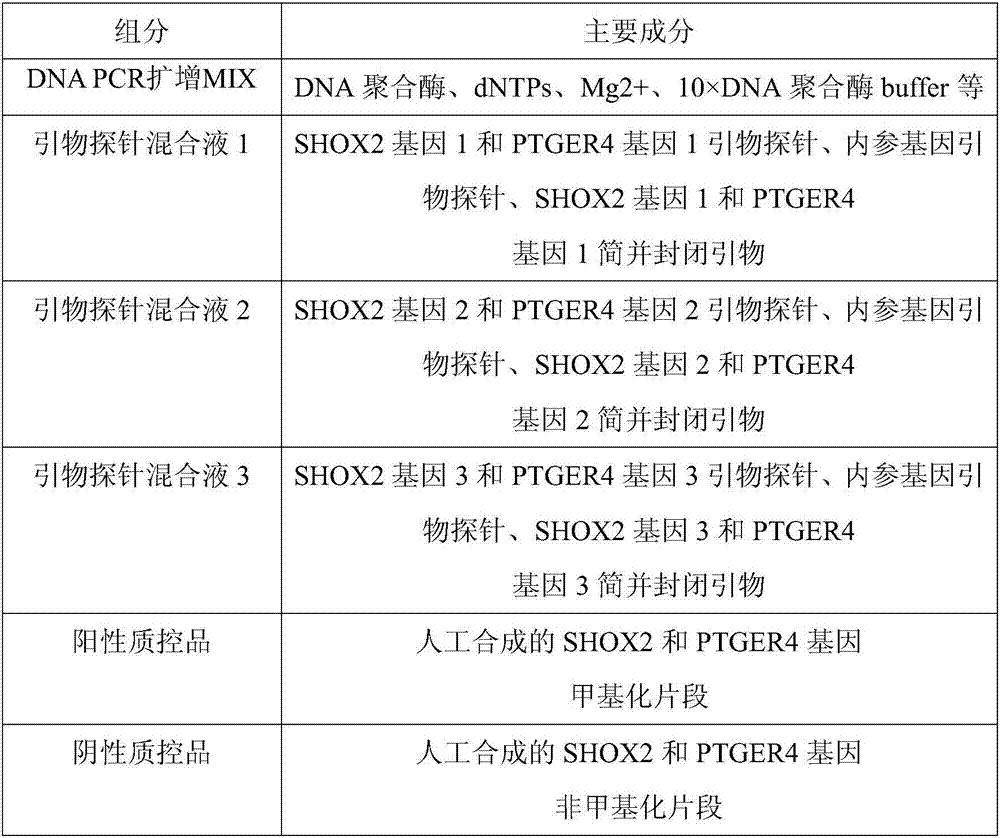
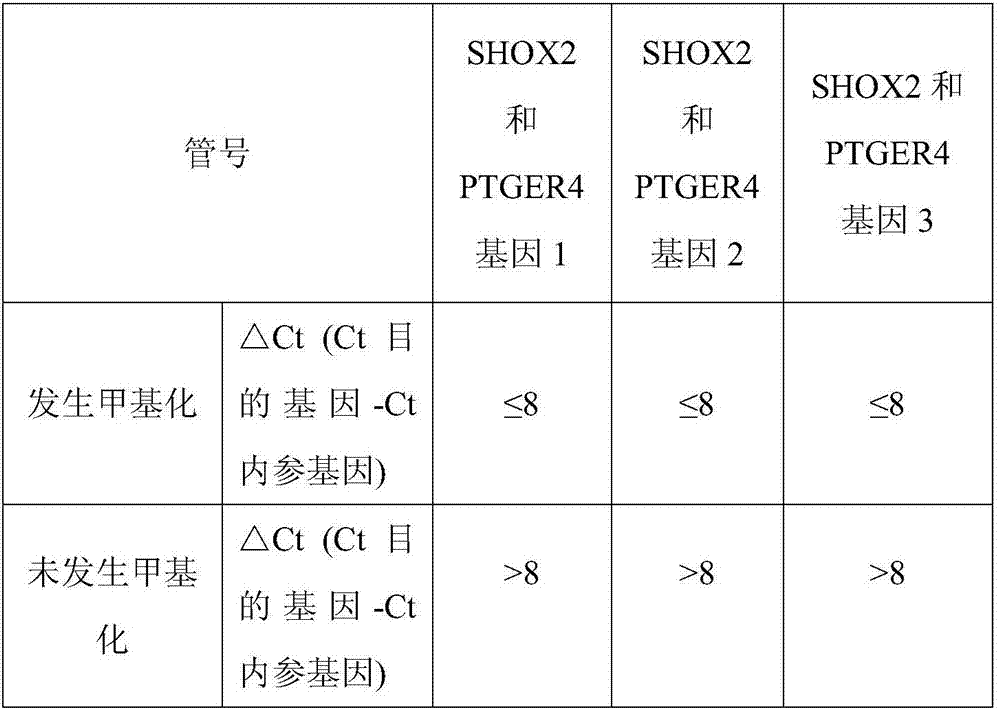
The invention discloses a composition for noninvasive screening of an early lung cancer and application of the composition. The composition comprises three pairs of methylation-specific primer probes of SHOX2 (Short Stature Homebox2) genes, three pairs of methylation-specific primers of PTGER4 (Prostaglandin E Receptor 4) genes and one pair of reference gene primer probes and further comprises three MGB probes corresponding to non-methylation target genes in the SHOX2 genes and three MGB probes corresponding to the non-methylation target genes in the PTGER4 genes. A methylation detection kit for the noninvasive screening of the early lung cancer comprises the composition. Patients suffering from the early lung cancer are screened by detecting promoters of the SHOX2 genes and the PTGER4 genes of free DNA in blood of high-risk lung cancer individuals and methylation conditions of three target sites in gene coding and non-coding areas, and early diagnosis of the lung cancer is aided. The methylation-specific PCR / MSP detection method is simple and convenient in operation and low in time consumption, and the results are easily analyzed.
Owner:北京鑫诺美迪基因检测技术有限公司
Multi-purpose real-time fluorescent quantitative PCR kit, method and primer probe composition for detecting 2019 novel coronavirus
InactiveCN112725537AShort detection cycleStrong specificityMicrobiological testing/measurementAgainst vector-borne diseasesReference genesBlood specimen
The invention relates to a multi-purpose reverse transcription real-time fluorescent quantitative PCR detection kit for synchronously detecting a 2019 novel coronavirus ORF1ab gene, an N gene, an S gene and a human B2M gene as well as a special primer and probe combination thereof, and the kit adopts a single tube and four fluorescent channels to simultaneously detect three 2019 novel coronavirus genes and a human reference gene, and can detect the existence of the 2019 novel coronavirus RNA in respiratory tracts and blood samples. The kit has the advantages of short detection period, strong specificity, high sensitivity, low omission ratio and good repeatability, can perform quality monitoring on the extraction and amplification process of the sample according to the detection result of the human reference gene, and reduces the occurrence of false positive results caused by aerosol pollution through a dUTP-UNG enzyme anti-pollution system.
Owner:NINGBO HEALTH GENE TECHNOLOGIES CO LTD +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com