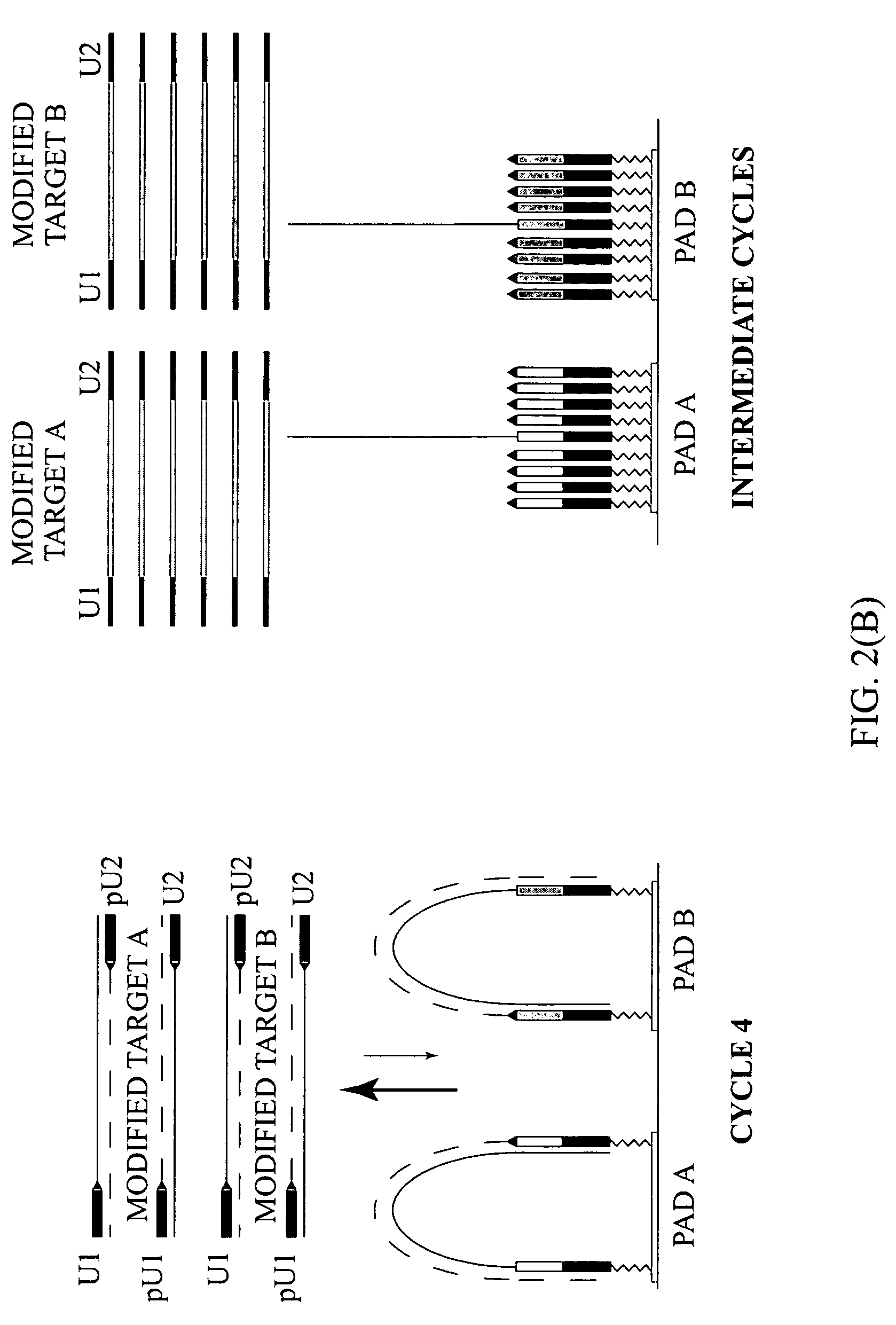
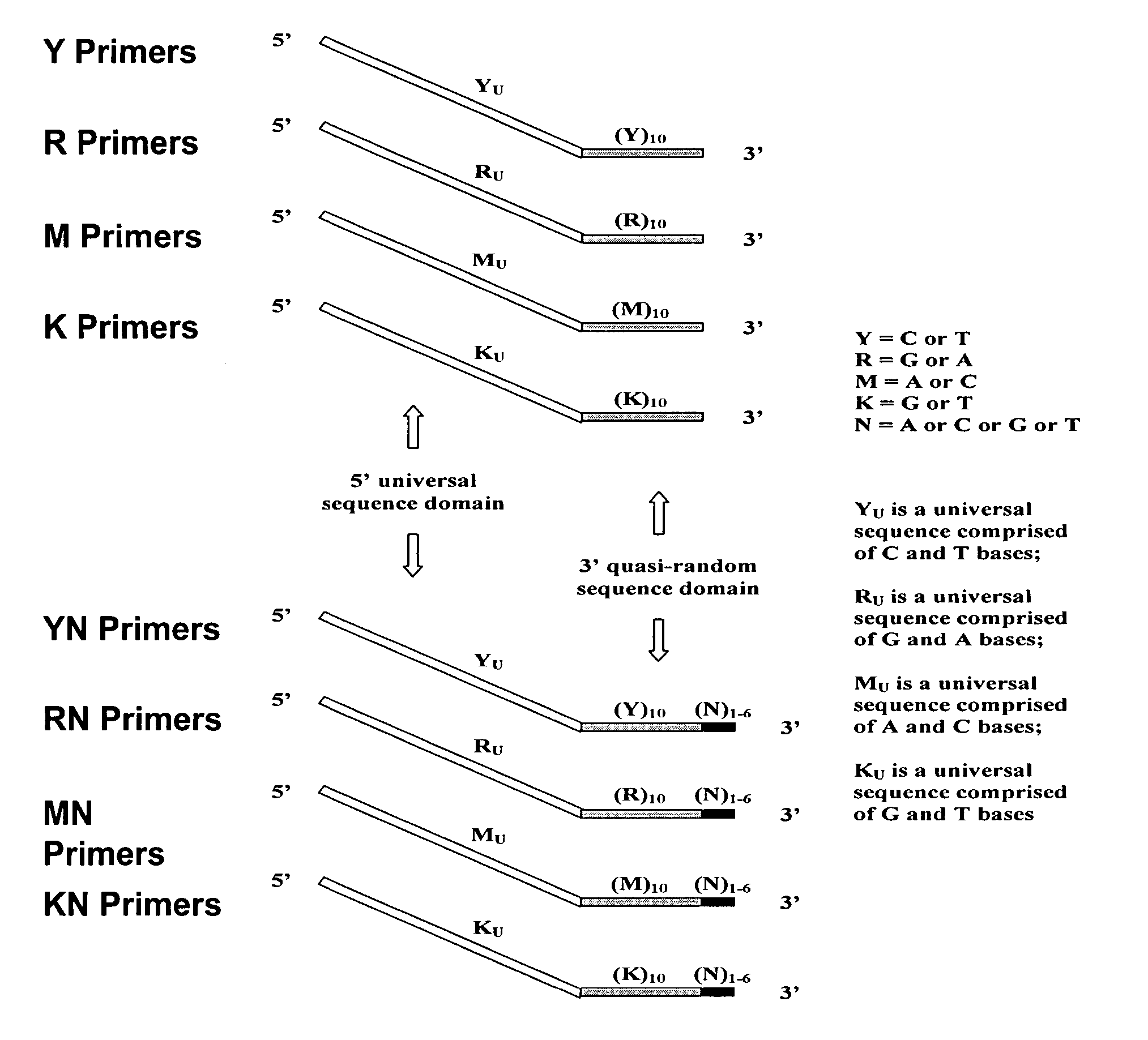
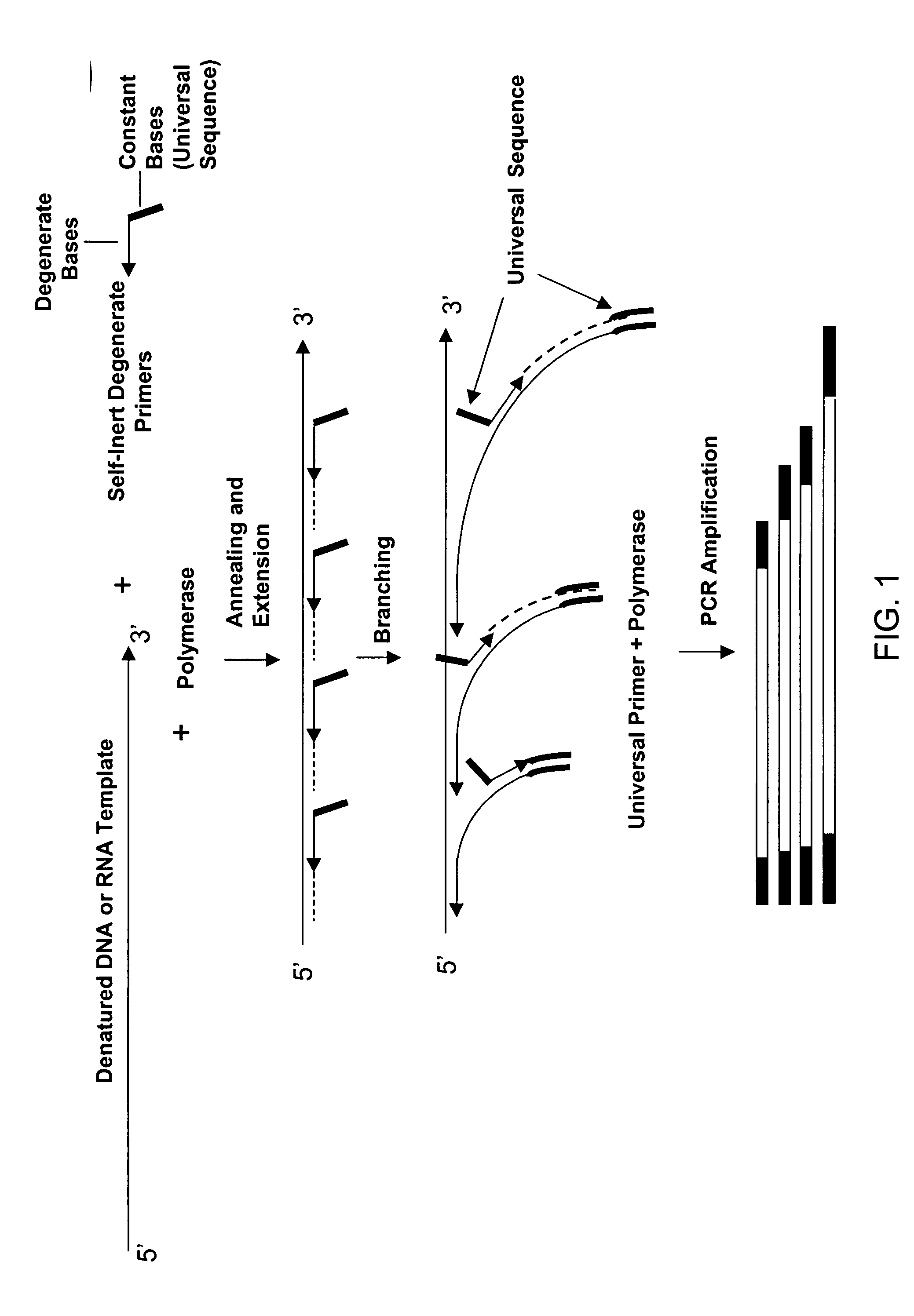
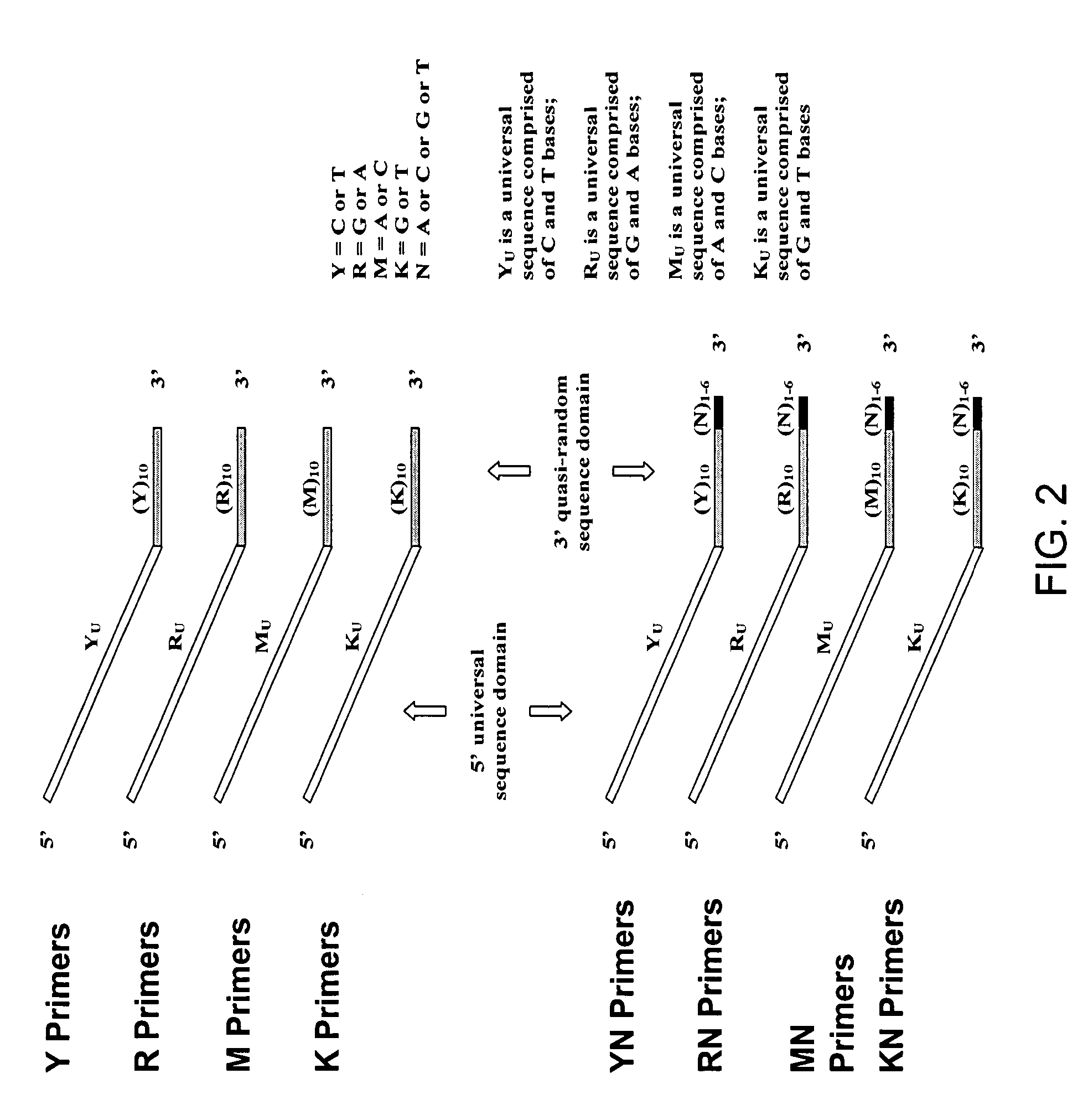
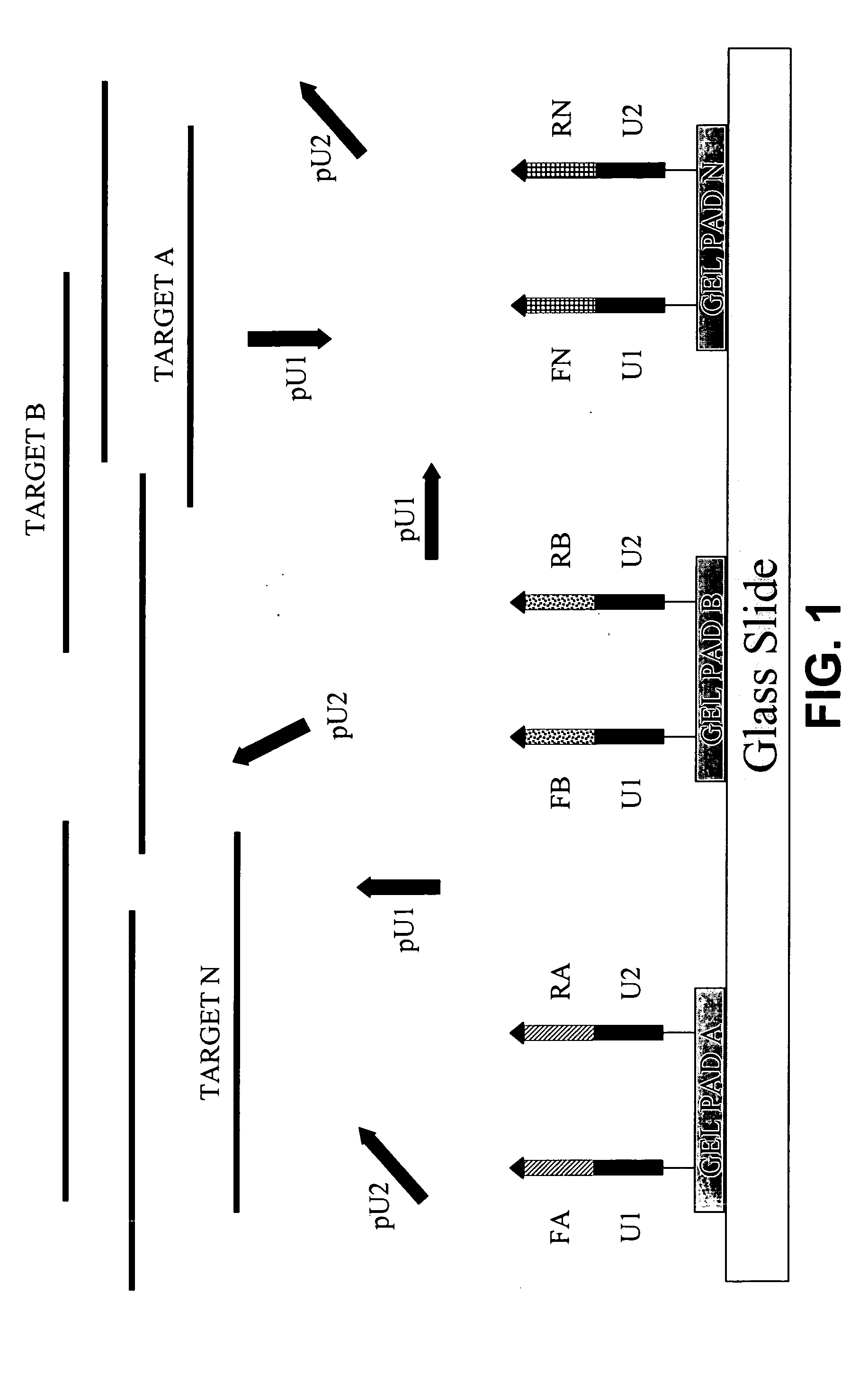
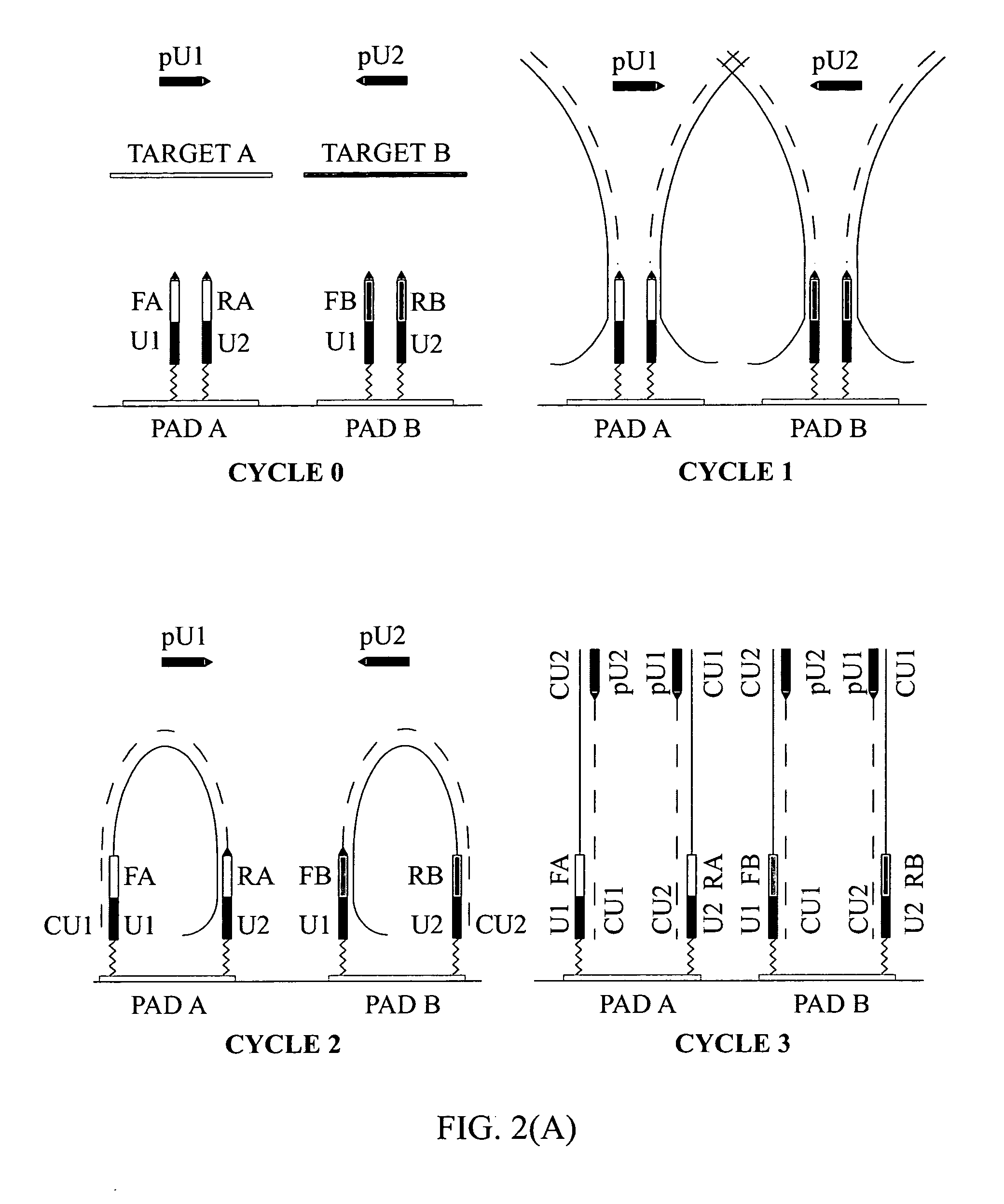
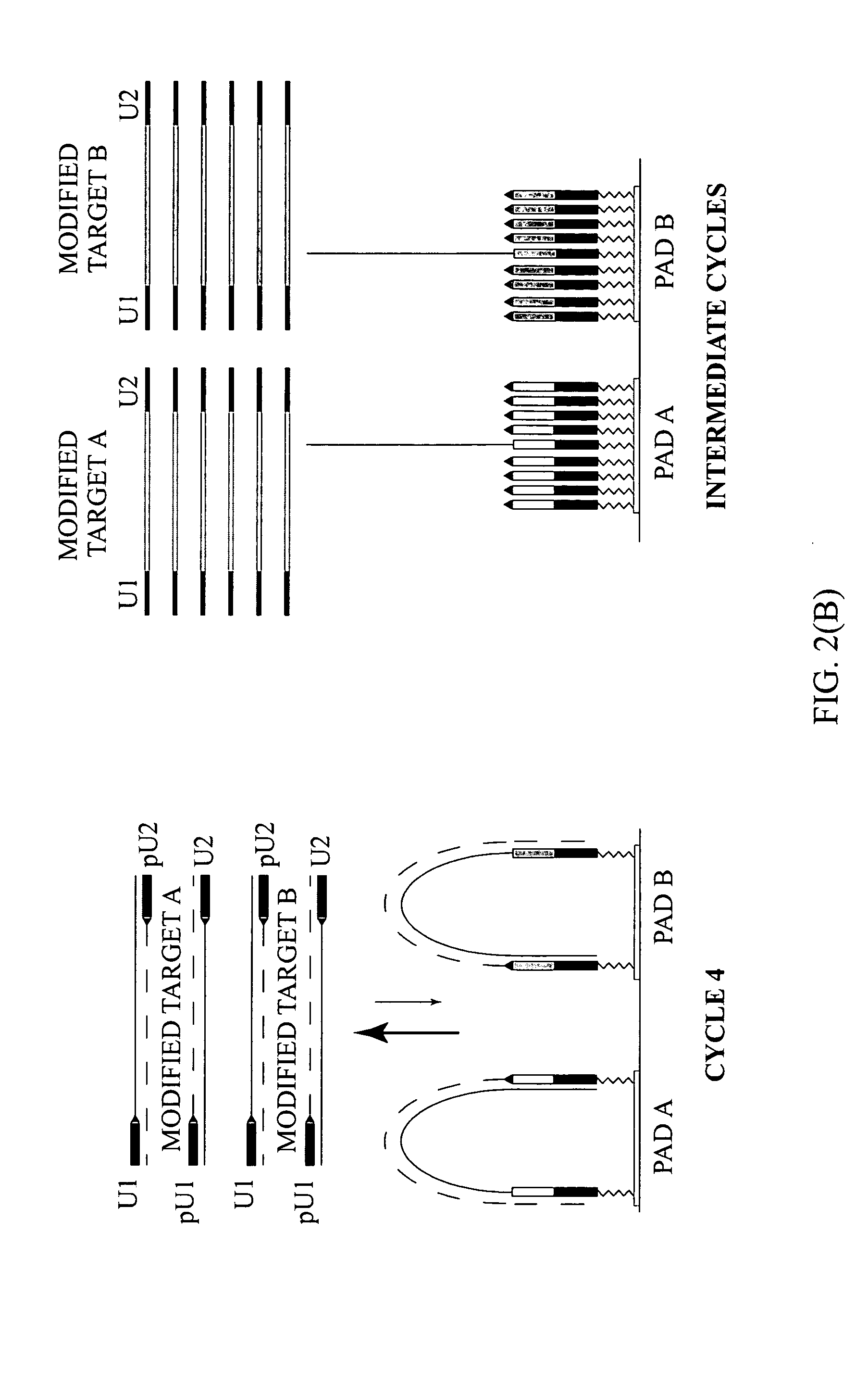
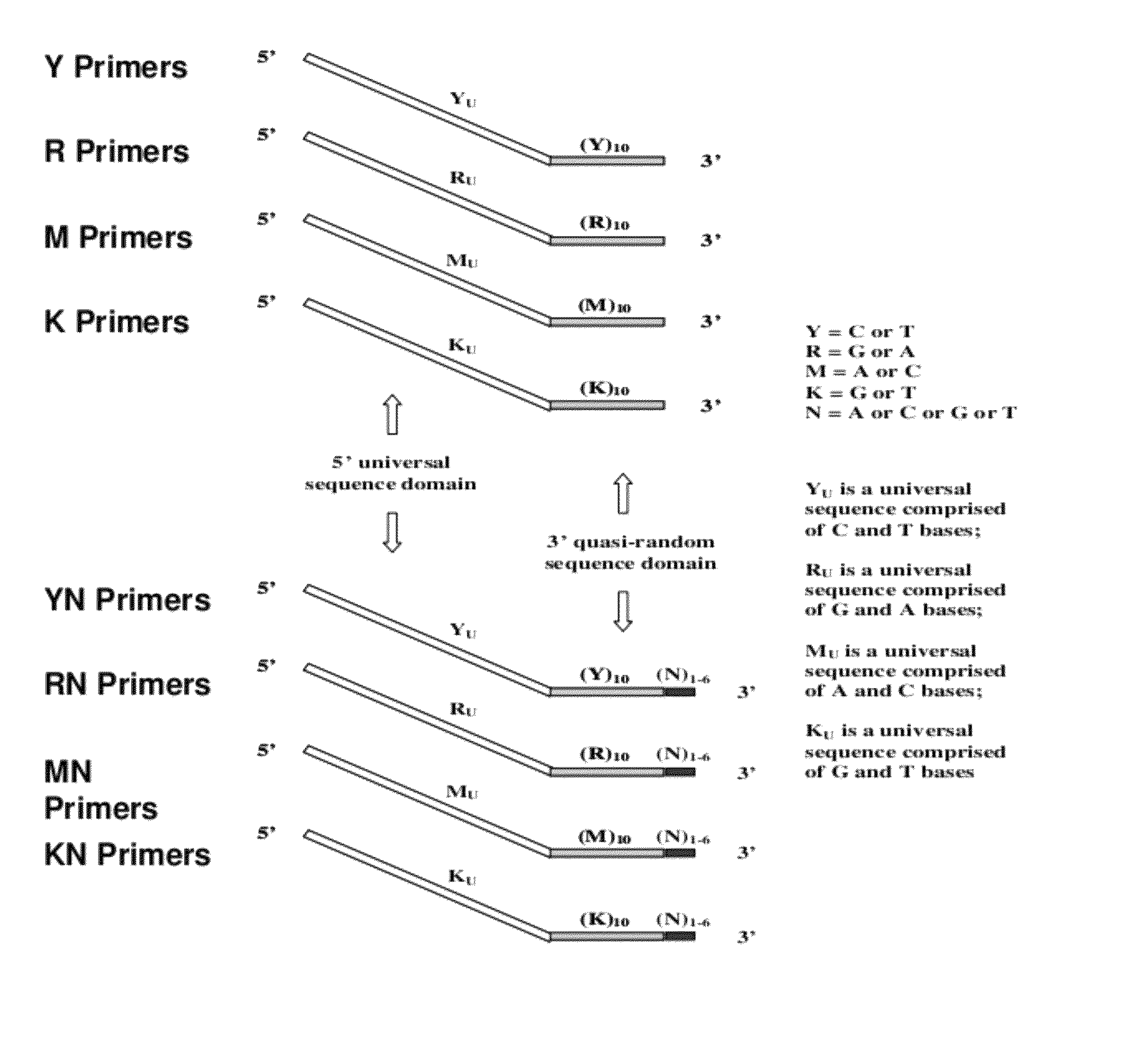
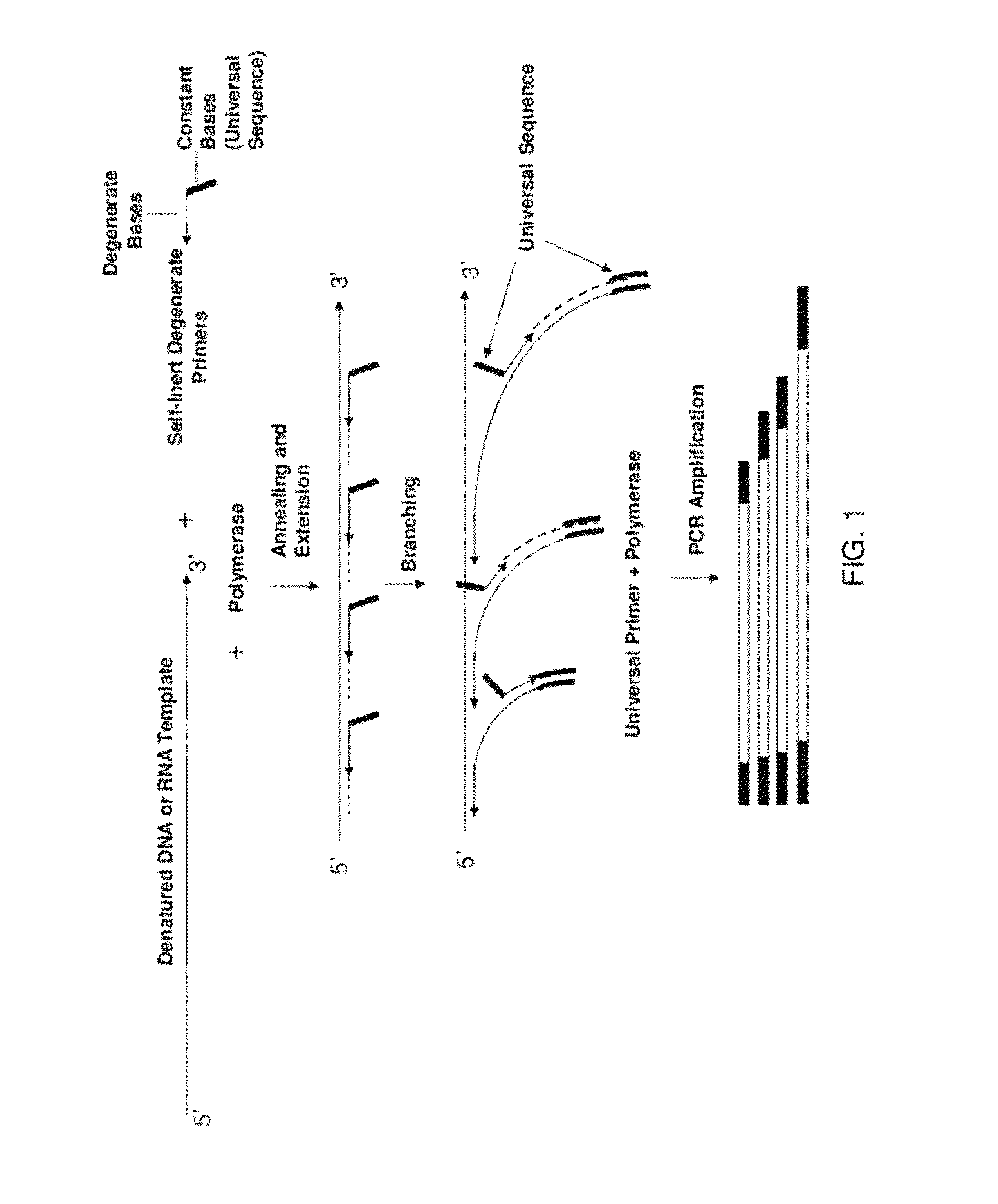
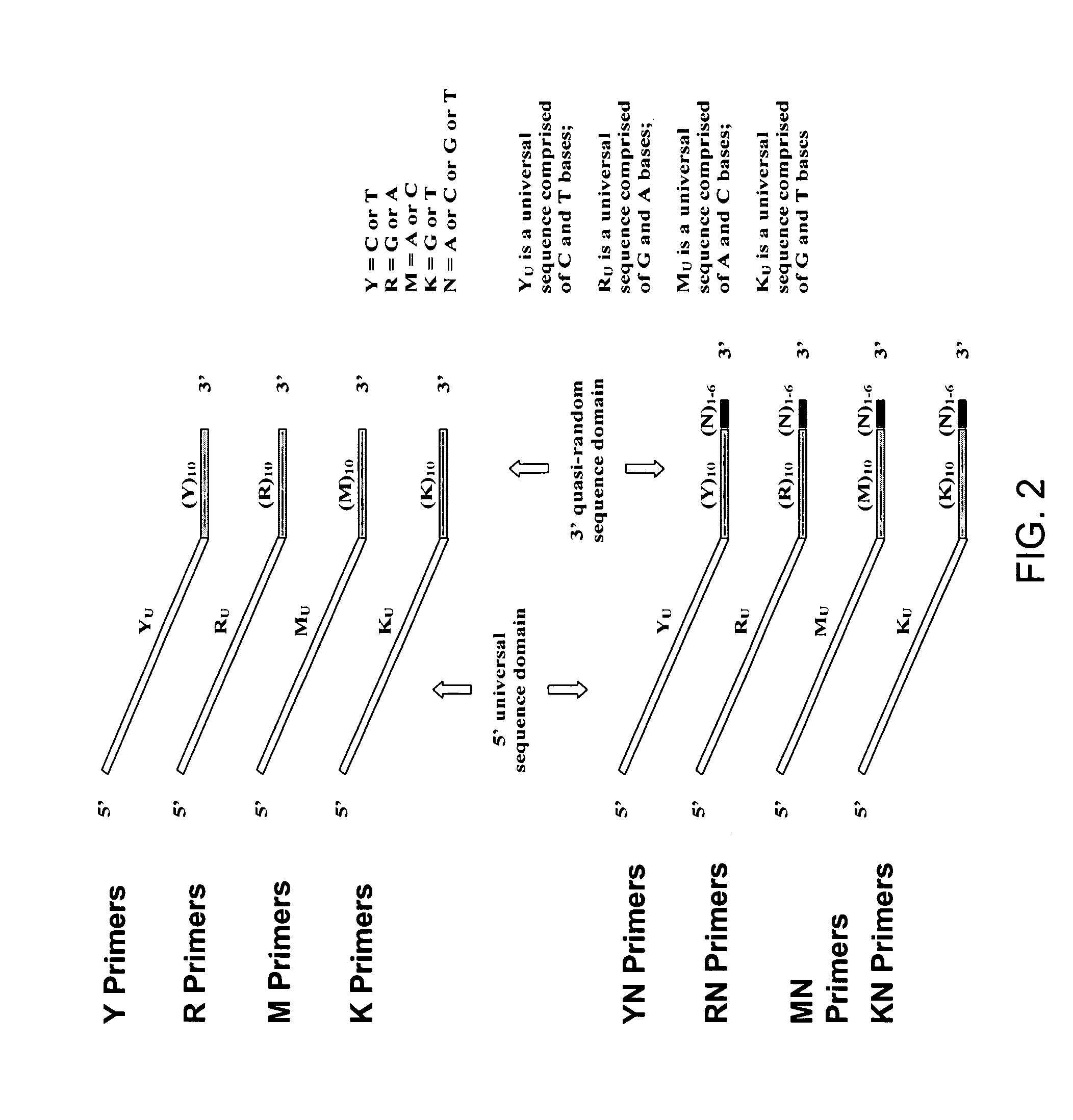
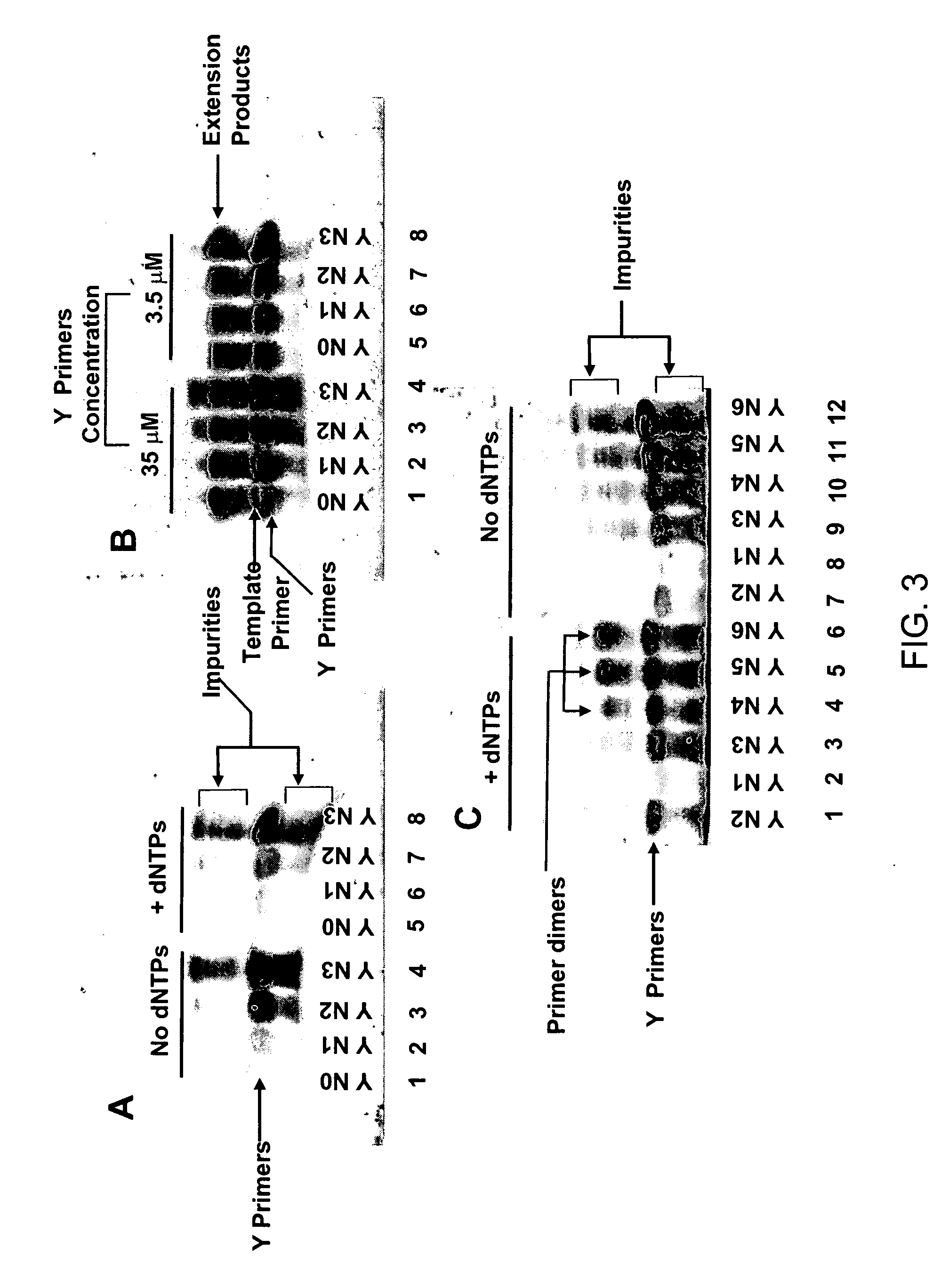
Patents
Literature
7636 results about "Specific primers" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
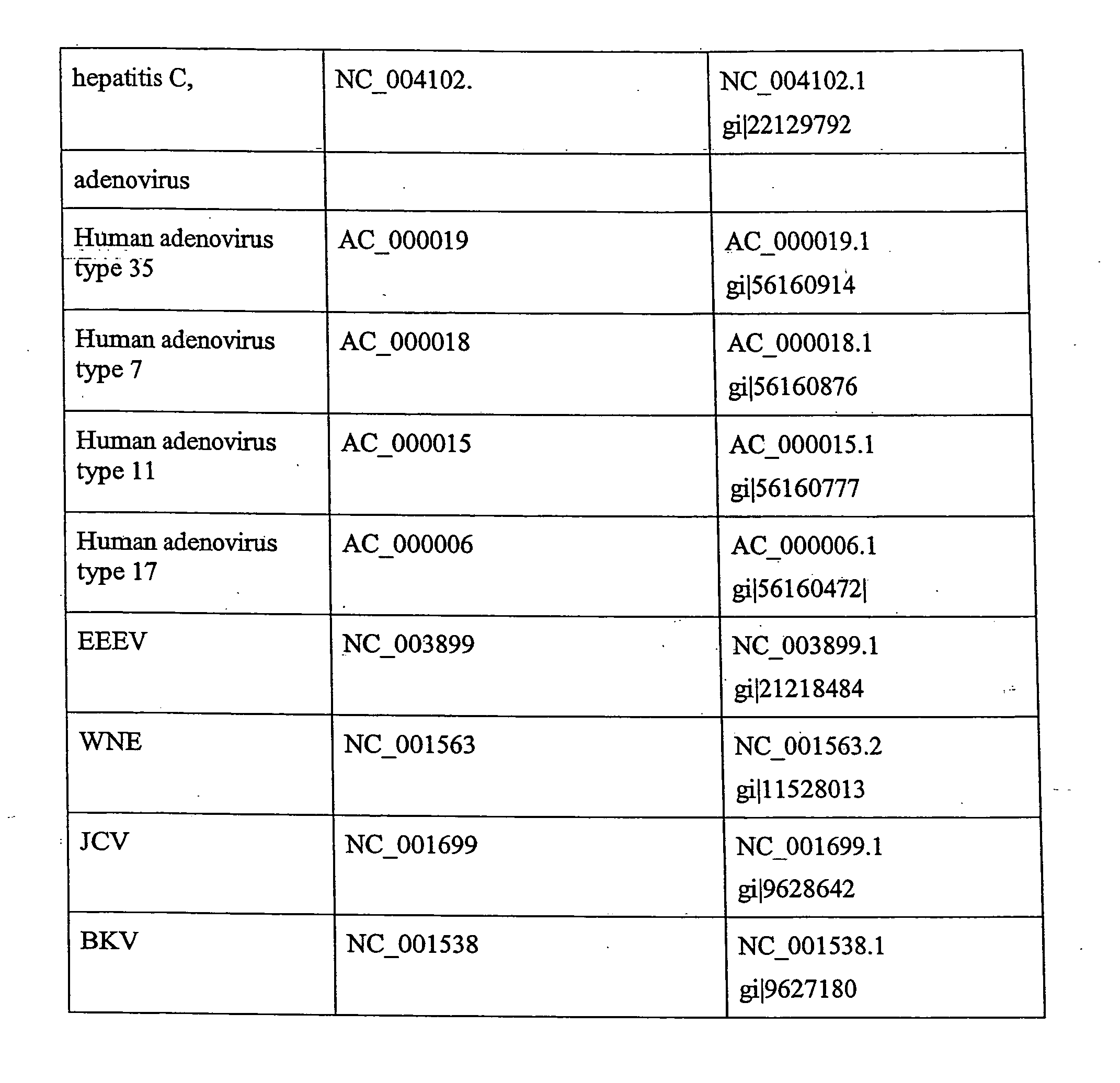
Specific primers : are used to test the presence of a gene sequence whose you already know the sequence , so you can design a specific primers to amplify that region and see if you get the amplification product.
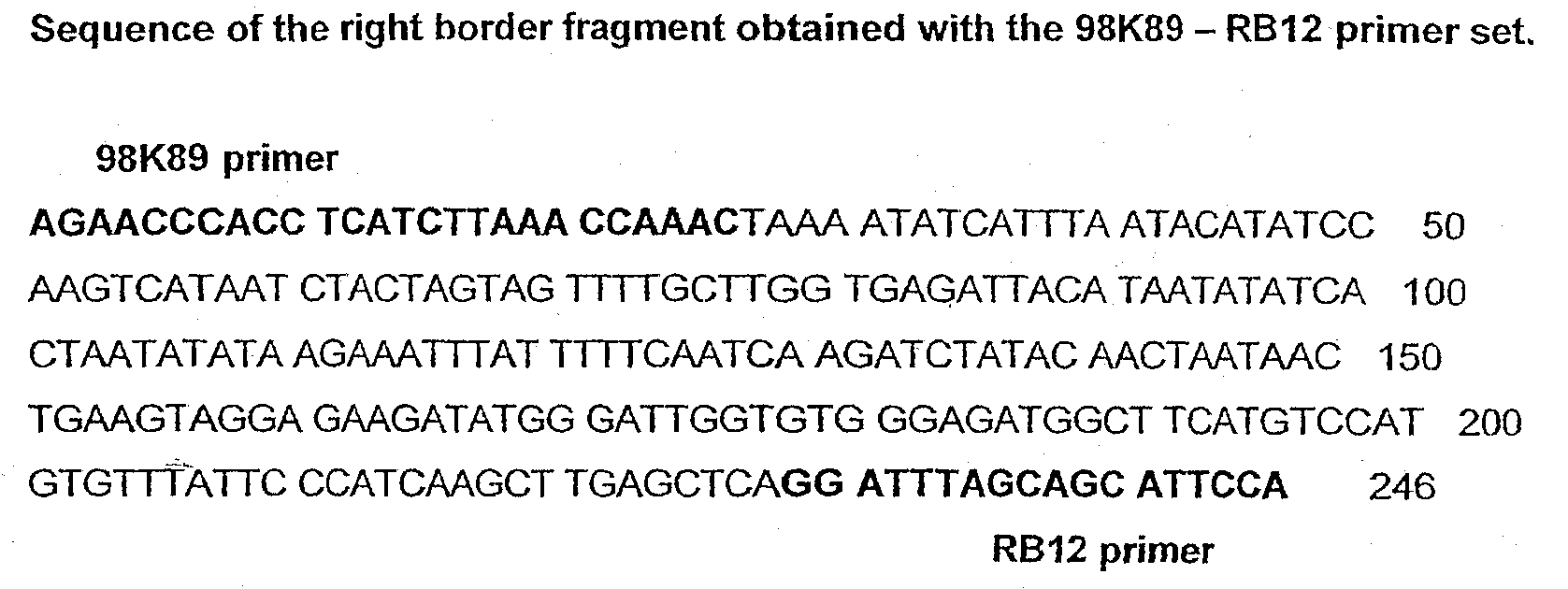
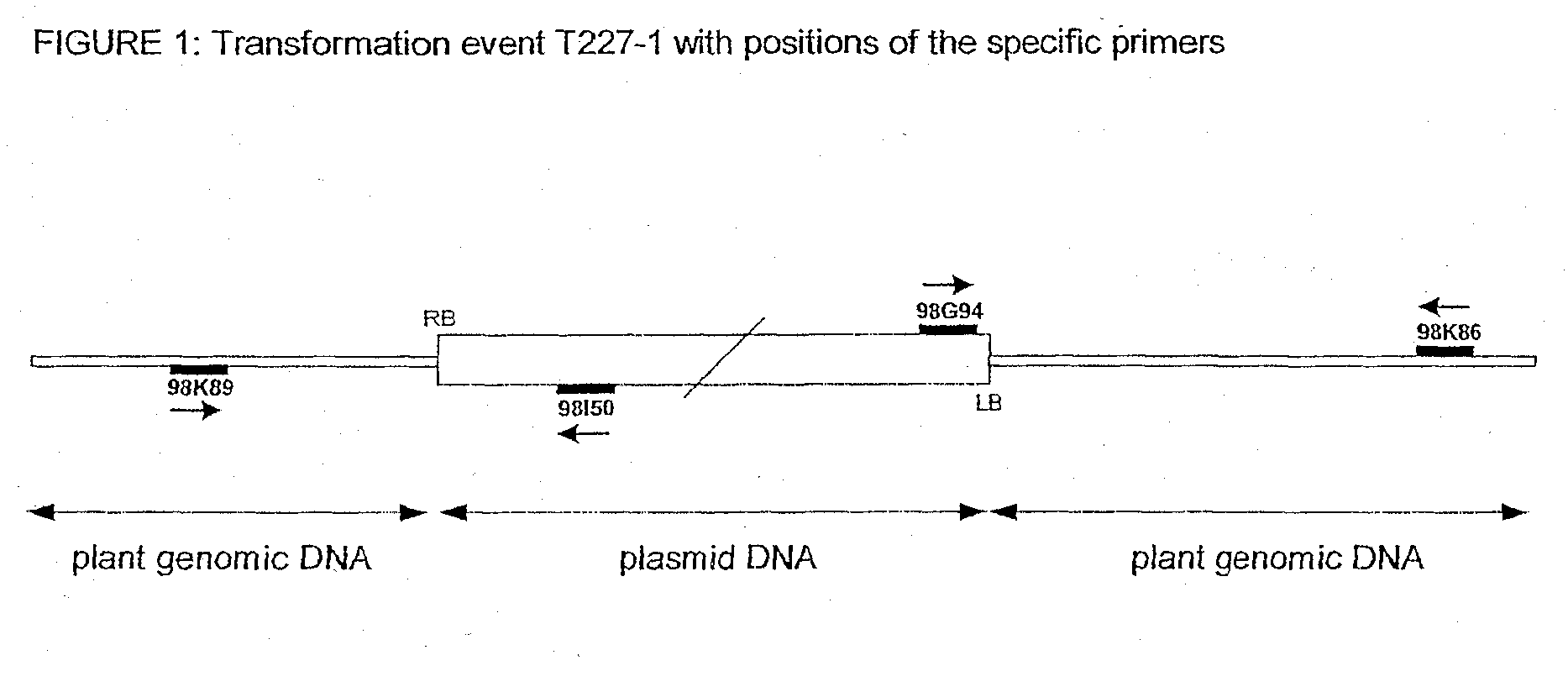
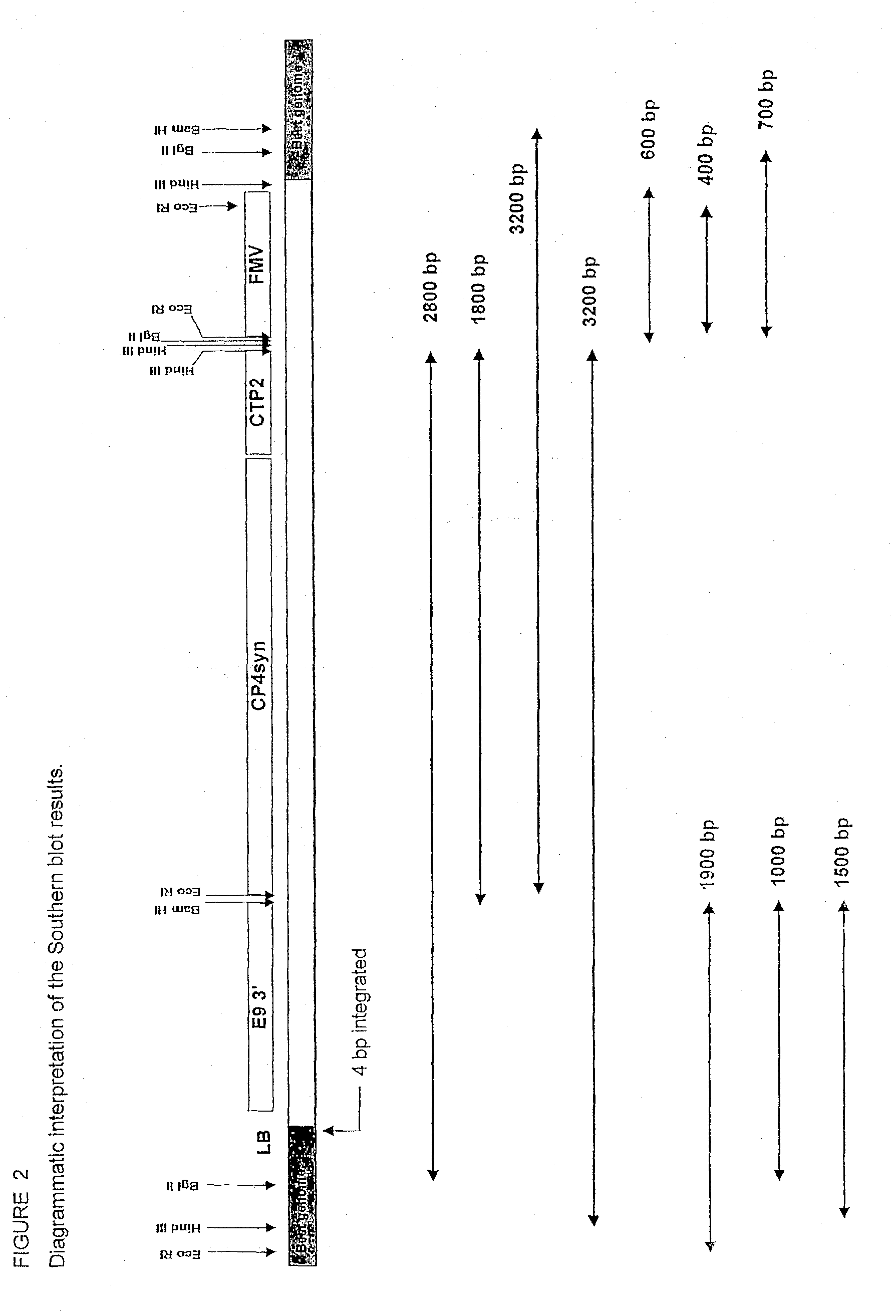
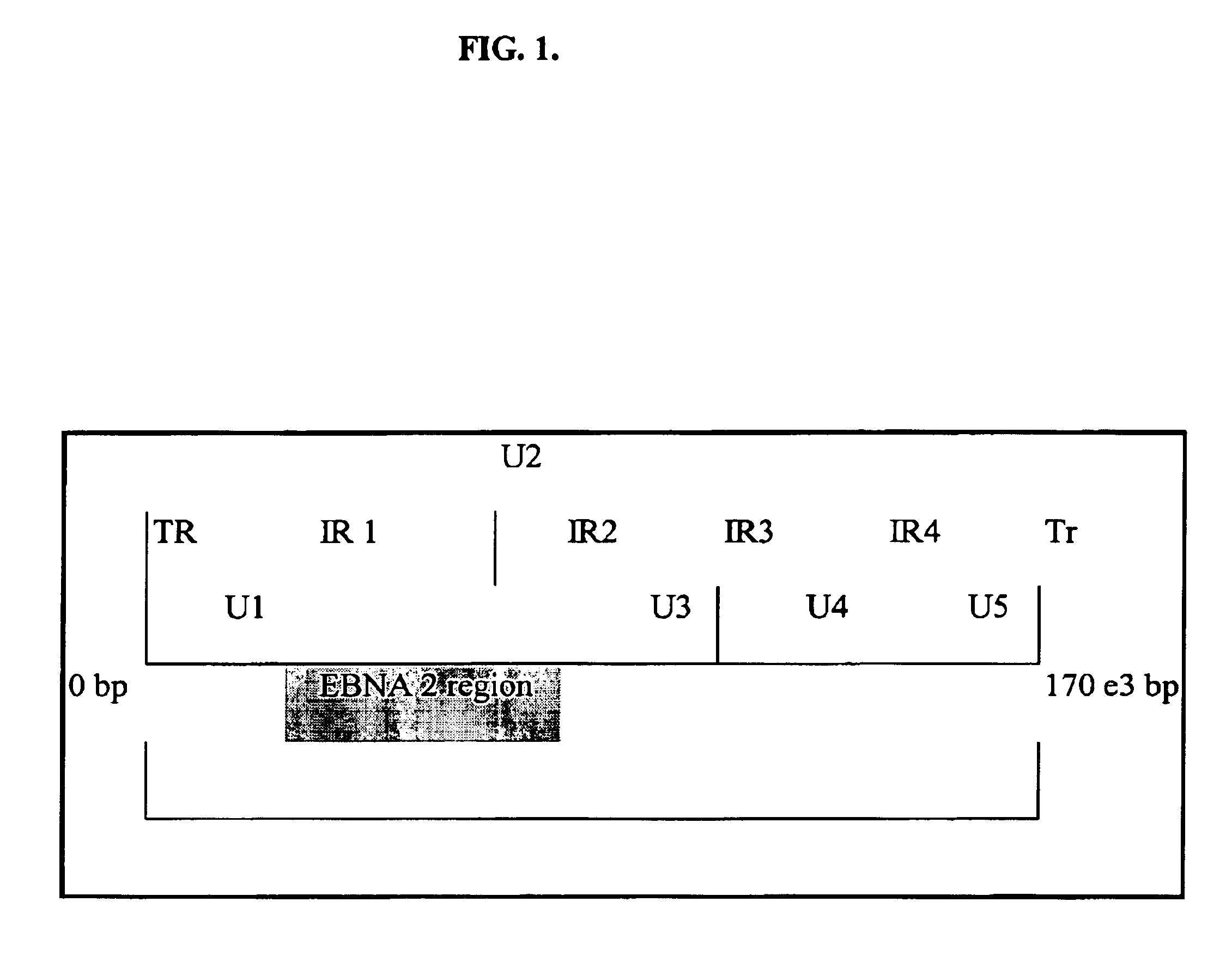
T227-1 flanking sequence
A herbicide resistant transformed sugar beet that is detectable by the specific primers developed to match the DNA sequences that flank the left and / or right border region of the inserted transgenic DNA and the method of identifying primer pairs containing plant genomic DNA / plasmid DNA. More specifically, the present invention covers a specific glyphosate resistant sugar beet plant having an insertion of the transgenic material identified as the T227-1 event. The present invention additionally covers primer pairs: plant genomic DNA / Plasmid DNA that are herein identified. Additionally, these primer pairs for either the left or the right flanking regions make an event specific test for the T227-1 insert of transgenic material.
Owner:SES EURO N V
Methods for genotyping selected polymorphism
InactiveUS20050142577A1Sugar derivativesMicrobiological testing/measurementDegenerate oligonucleotideGene
Methods for genotyping polymorphisms using a locus specific primer that is complementary to a region near a selected polymorphism are described. Methods for synthesizing pools of locus specific primers that incorporate some degenerate positions are also disclosed. A plurality of different sequence capture probes are synthesized simultaneously using degenerate oligonucleotide synthesis. The sequence of the locus specific regions of the capture probes are related in that they have some bases that are identical in each sequence in the plurality of sequences and positions that vary from one locus specific region to another. The sequences are selected based on proximity to a polymorphism of interest and because they conform to a similar sequence pattern.
Owner:AFFYMETRIX INC
Selective tagging of short nucleic acid fragments and selective protection of target sequences from degradation
InactiveUS20100285537A1Microbiological testing/measurementFermentationNucleotide sequencingBioinformatics
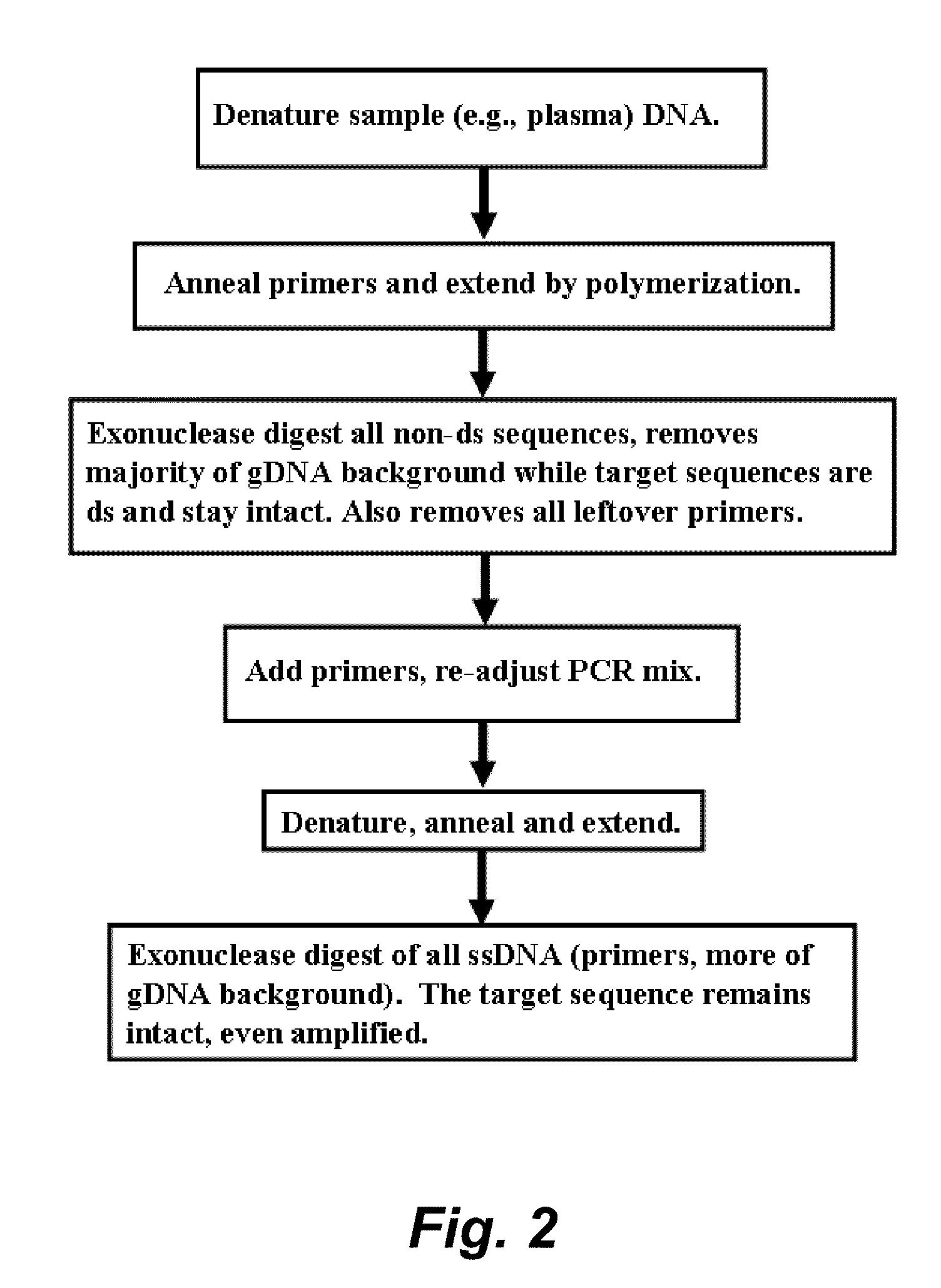
Methods are provided for selective tagging of short nucleic acids comprising a short target nucleotide sequence over longer nucleic acids comprising the same target nucleotide sequence. The methods can involve performing one or two cycles of amplification of a sample comprising long nucleic acids and short nucleic acids, each comprising the same target nucleotide sequence with at least two target-specific primers or primer pairs under suitable annealing conditions, wherein the primer pairs comprise: an inner primer or primer pair that can amplify the target nucleotide sequence on long and short nucleic acids (wherein each inner primer comprises a 5′ nucleotide tag; and an outer primer or primer pair that amplifies the target nucleotide sequence on long nucleic acids, but not on short nucleic acids); whereby the amplification after a second cycle produces at least one tagged target nucleotide sequence that comprises two nucleotide tags, one from each inner primer, with the target nucleotide sequence located between the nucleotide tags.
Owner:FLUIDIGM CORP
Methods for genotyping selected polymorphism
InactiveUS7459273B2Sugar derivativesMicrobiological testing/measurementGeneticsDegenerate oligonucleotide
Methods for genotyping polymorphisms using a locus specific primer that is complementary to a region near a selected polymorphism are described. Methods for synthesizing pools of locus specific primers that incorporate some degenerate positions are also disclosed. A plurality of different sequence capture probes are synthesized simultaneously using degenerate oligonucleotide synthesis. The sequence of the locus specific regions of the capture probes are related in that they have some bases that are identical in each sequence in the plurality of sequences and positions that vary from one locus specific region to another. The sequences are selected based on proximity to a polymorphism of interest and because they conform to a similar sequence pattern.
Owner:AFFYMETRIX INC
Solid support for high-throughput nucleic acid analysis
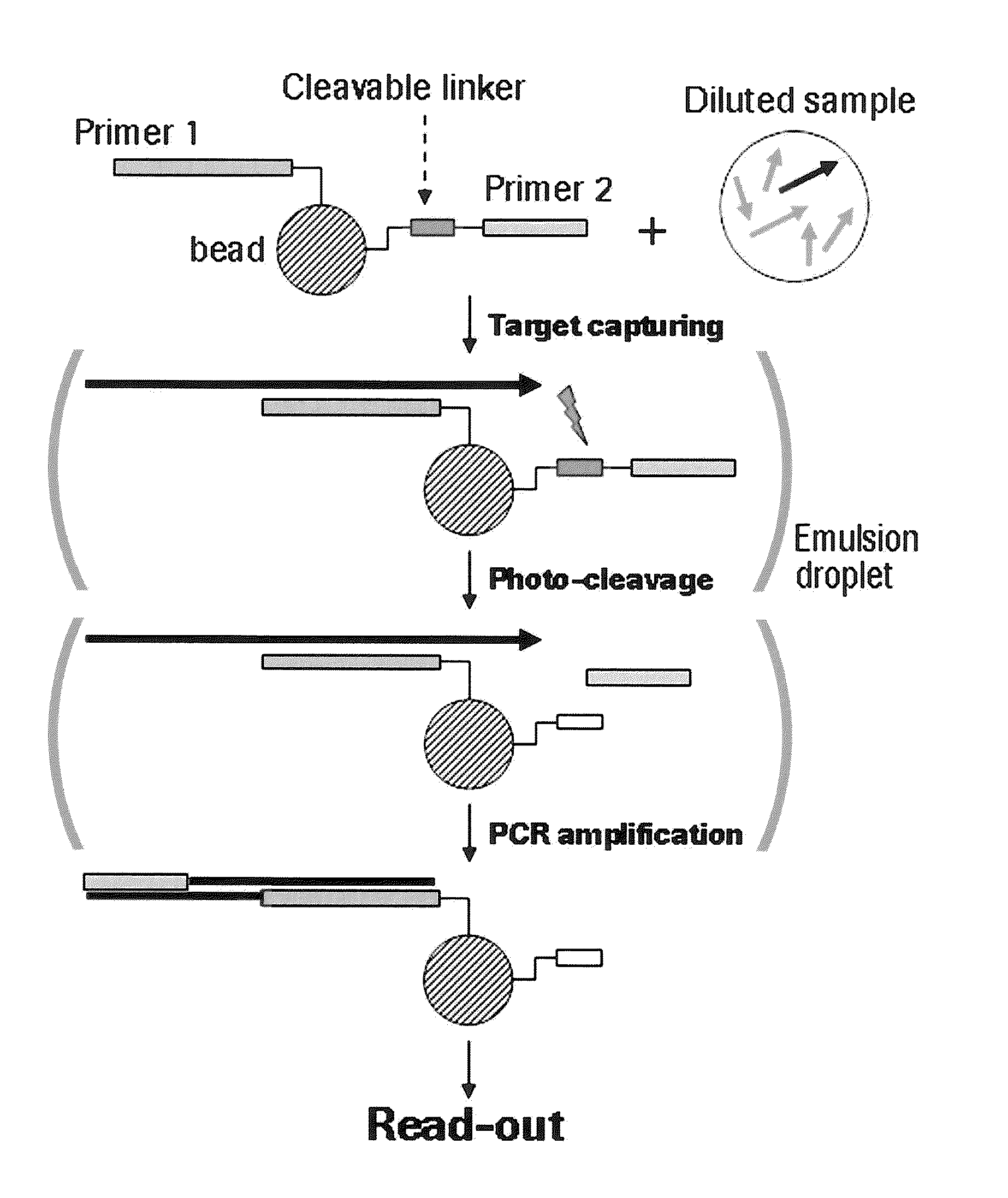
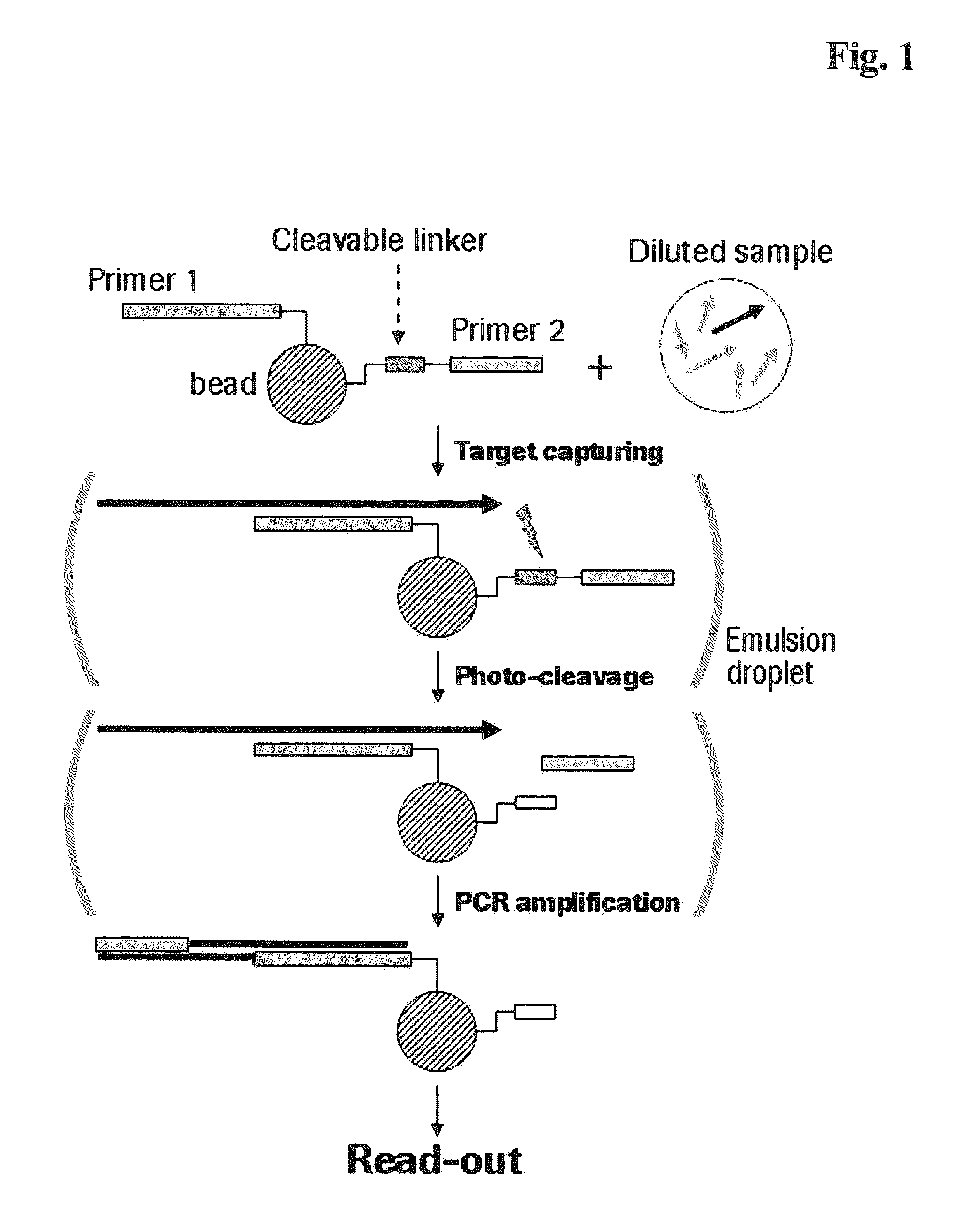
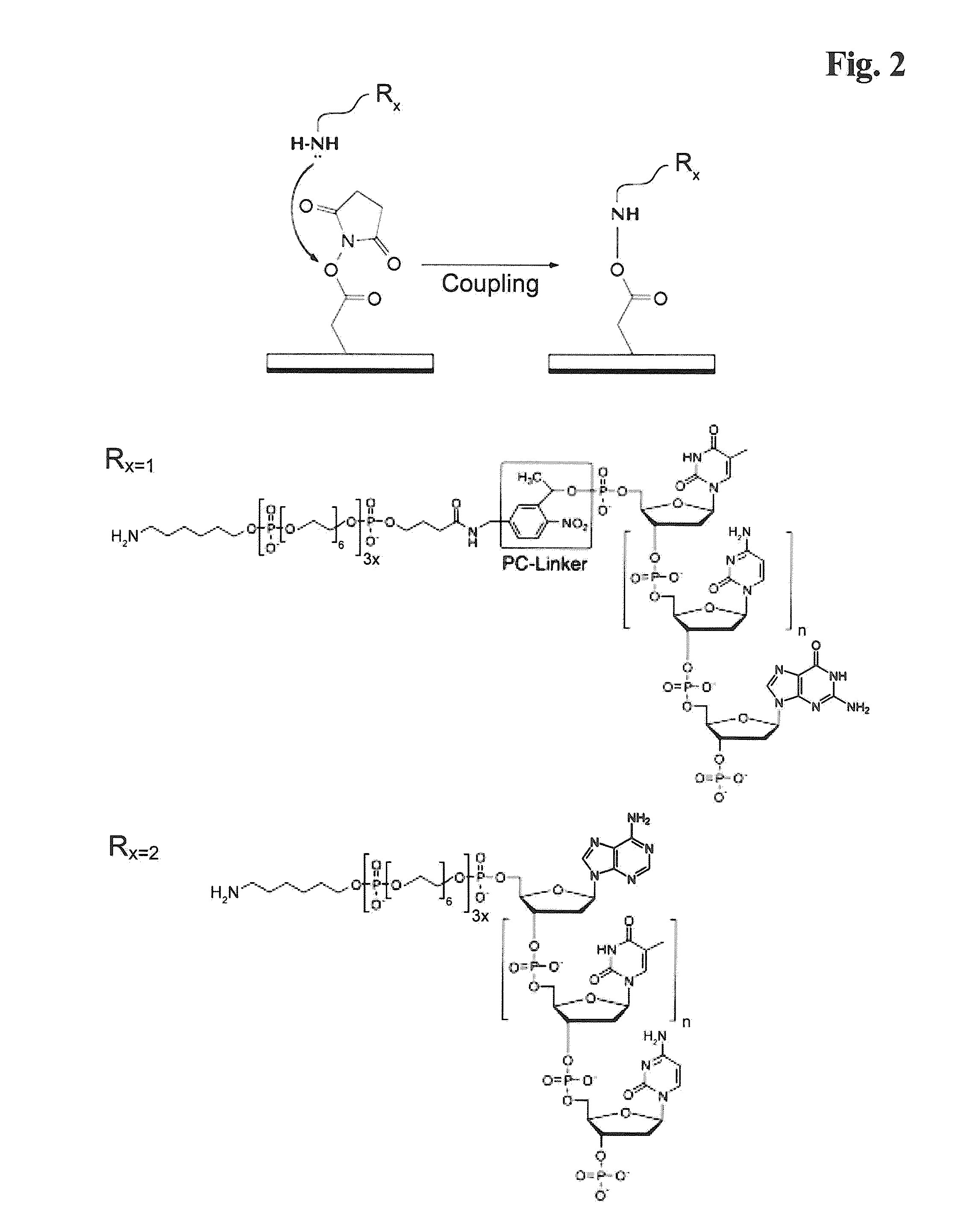
The present invention provides a solid support which is preferably a bead comprising at least two sequence specific amplification primers wherein at least one primer is bound to the support with an inducible cleavable linker. The present invention also provides various method for preparing a solid support comprising at least two sequence specific primers, further characterized in that at least one of the primers is cleavable.
Owner:ROCHE MOLECULAR SYST INC
Multiplexed quantitative detection of pathogens
InactiveUS20070134652A1Easy to adaptQuantitative precisionMicrobiological testing/measurementSingle samplePolynucleotide
The invention allows for the quantitative detection of a plurality of pathogens in a single sample. The method includes the amplification of a sample with a plurality of pathogen-specific primer pairs to generate amplicons of distinct sizes from each of the pathogen specific primer pairs. The method further includes the use of a plurality of competitor polynucleotide targets that correspond to each of the pathogen-specific primer pairs. The competitor polynucleotides are added to the reaction mixture at a known concentration to allow for the quantitation of the amount of pathogen in the sample. The method can be used for monitoring pathogen infection in an individual, preferably an immunocompromised individual.
Owner:PRIMERADX
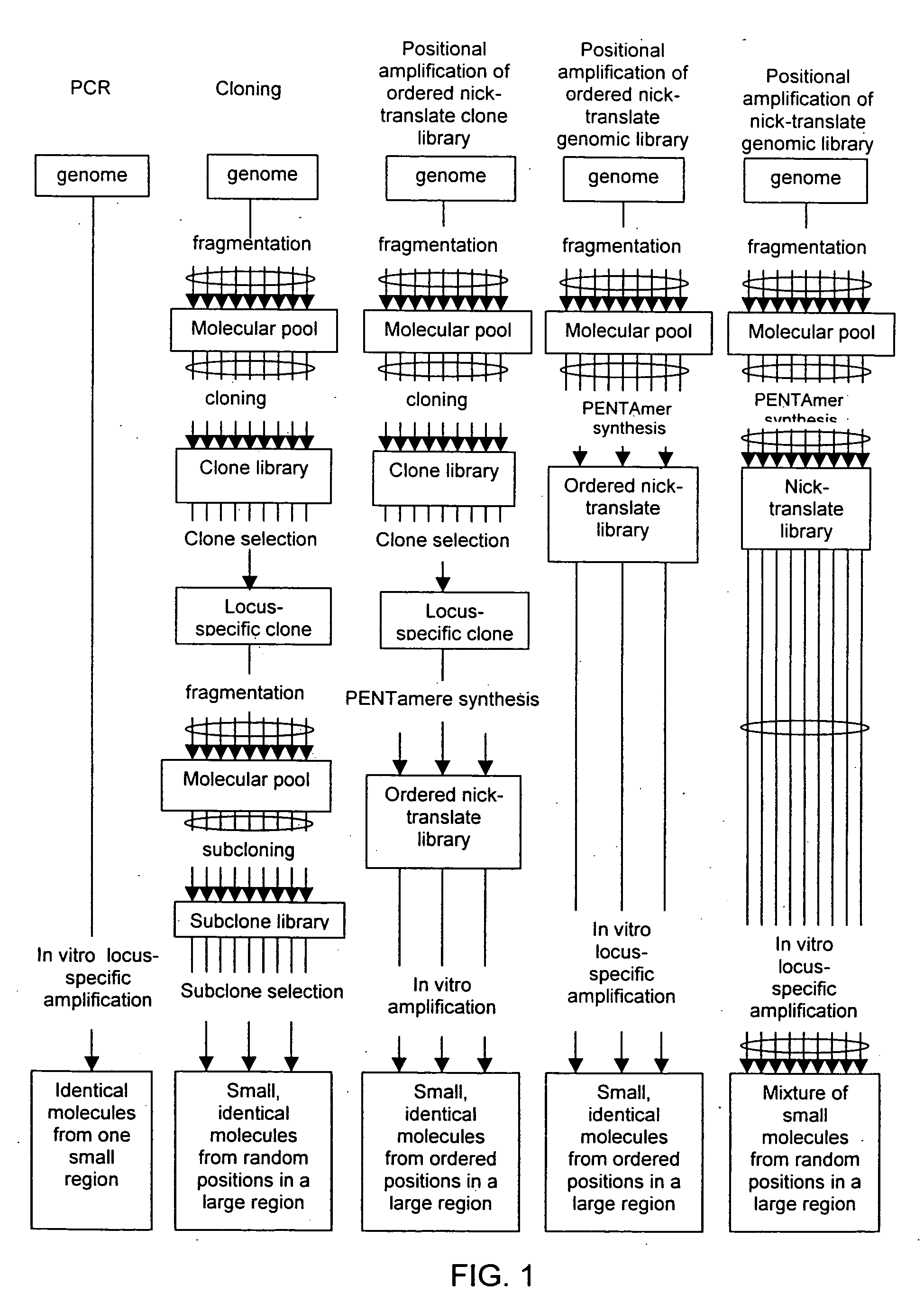
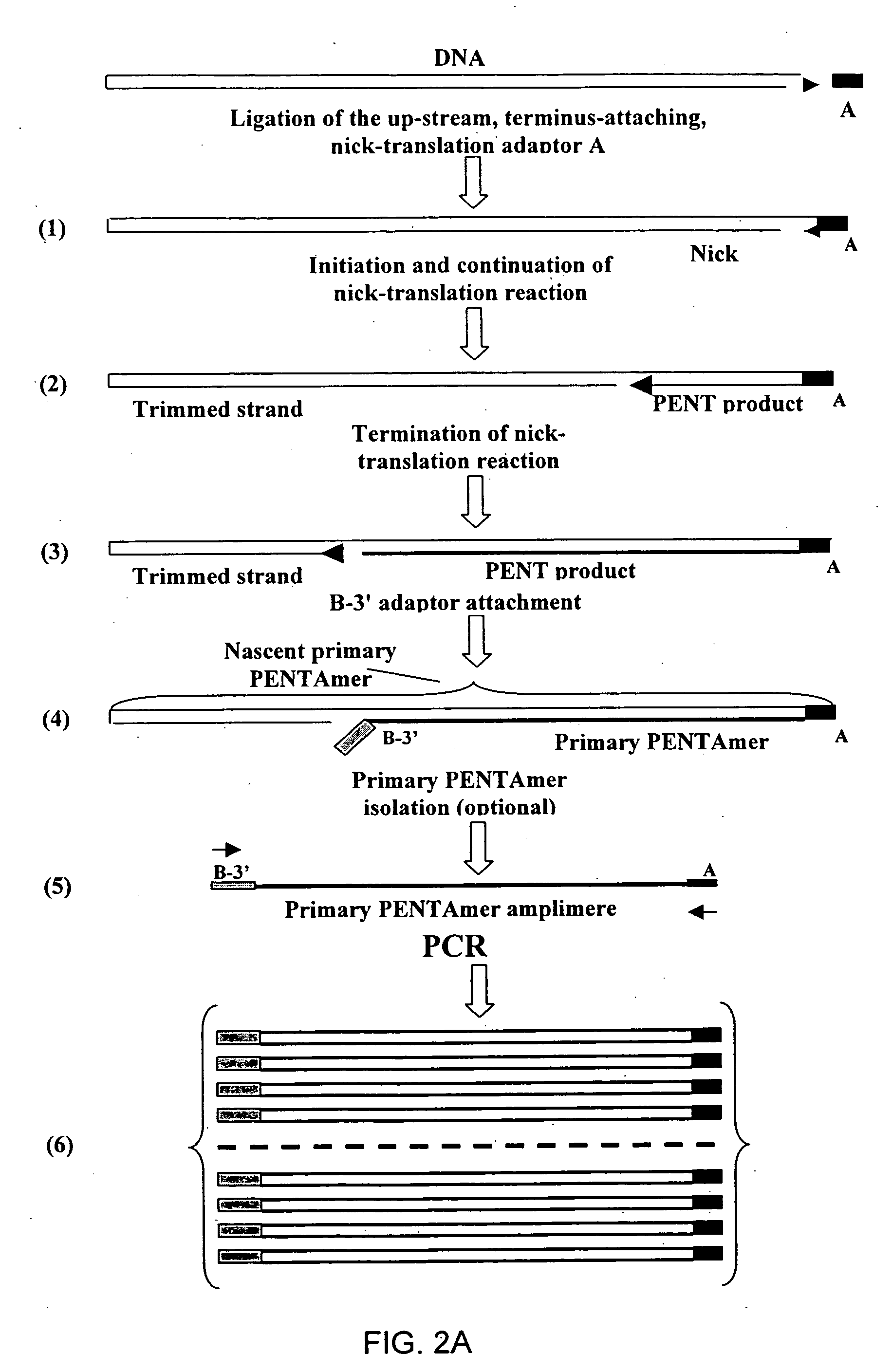
Method of producing a DNA library using positional amplification
InactiveUS20060068394A1Microbiological testing/measurementOrganic chemistry methodsCDNA libraryPentamer
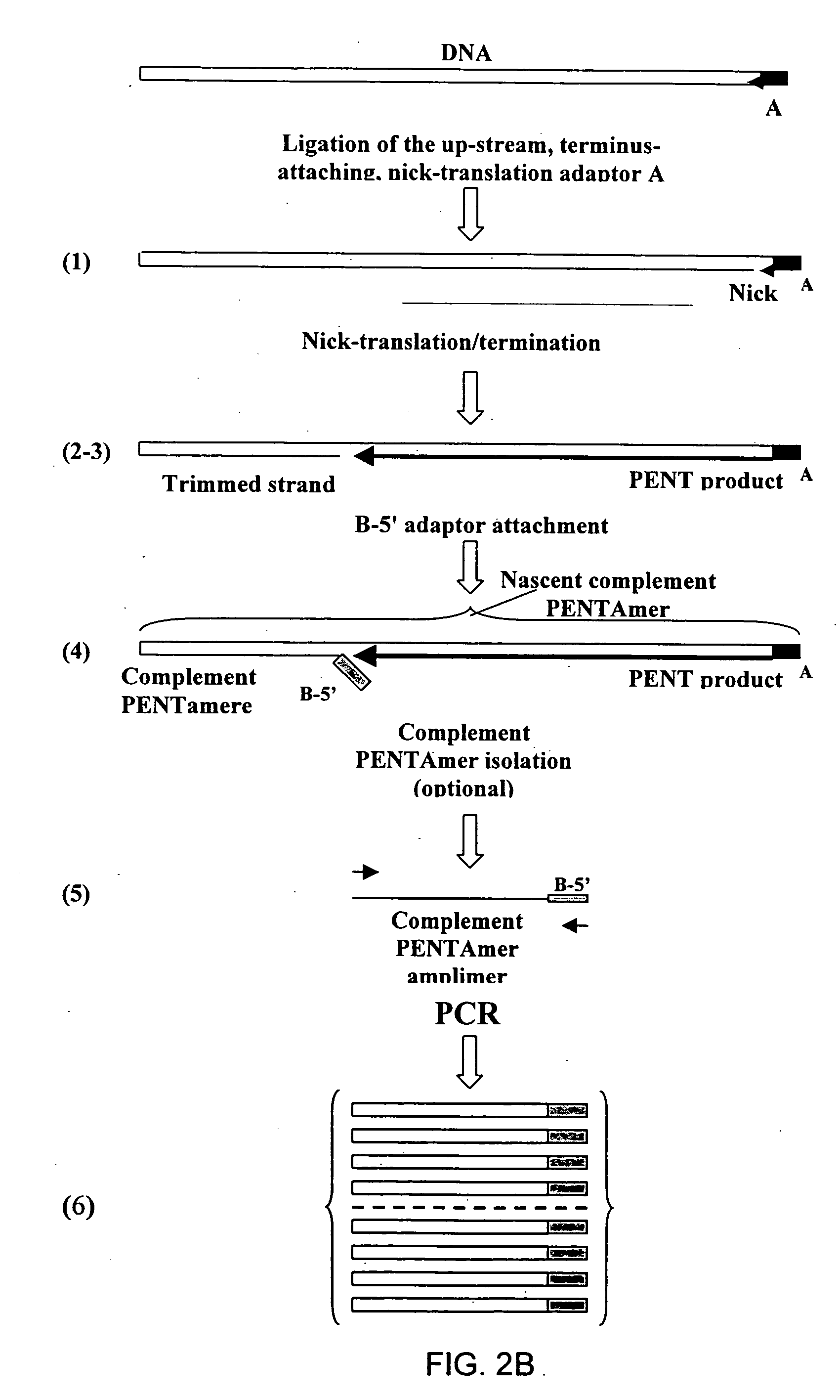
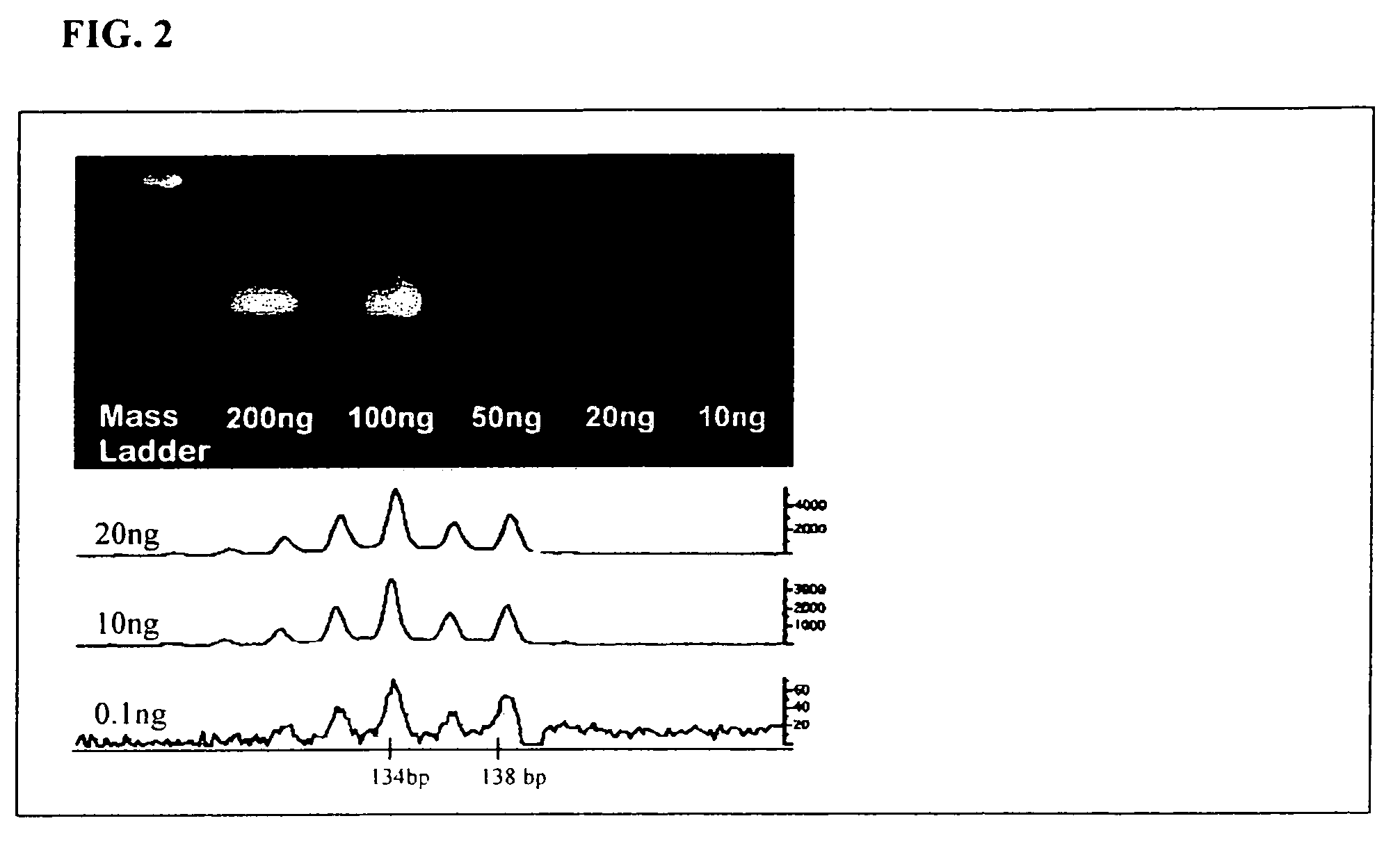
The disclosed invention relates to general and specific methods to use the Primer Extension / Nick Translation (PENT) reaction to create an amplifiable DNA strand, called a PENTAmer. A PENTAmers can be made for the purpose of amplifying a controlled length of DNA located at a controlled position within a DNA molecule, a process referred to as Positional Amplification by Nick Translation (PANT). In contrast to PCR, which amplifies DNA between two specific sequences, PANT can amplify DNA between two specific positions. PENTAmers can be created to amplify-very large regions of DNA (up to 500,000 bp) as random mixtures (unordered positional libraries), or as molecules sorted according to position (ordered positional libraries). PANT is fast and economical, because PENTAmer preparation can be multiplexed. A single PENTAmer preparation can include very complex mixtures of DNA such as hundreds of large-insert clones, complete genomes, or cDNA libraries. Subsequent PCR amplification of the preparation using a single specific primer can positionally amplify contiguous regions along a specific clone, along a specific genomic region, or along a specific expressed sequence.
Owner:LANGMORE JOHN +1
Methods for detecting circulating mutant BRAF DNA
ActiveUS7442507B2High sensitivityMicrobiological testing/measurementFermentationMelanomaNucleic acid sequencing
The present invention relates to a method for detecting the presence of circulating mutant BRAF DNA, which may be present in circulating melanoma cells or as DNA shed from tumor cells. Methods, compositions and kits which employ one or more sets of BRAF mutant specific primer pairs for detection of circulating mutant BRAF DNA are presented. Also provided are methods for diagnosing and / or determining stage / progression of a melanoma in a mammal based on detection of a BRAF mutant nucleic acid sequence. Such methods are also well suited to monitoring disease activity in patients with active disease or those in remission.
Owner:NEW YORK UNIV +1
Dual phase multiplex polymerase chain reaction
ActiveUS7432055B2Further amplificationMicrobiological testing/measurementLibrary member identificationGenomic DNASingle pair
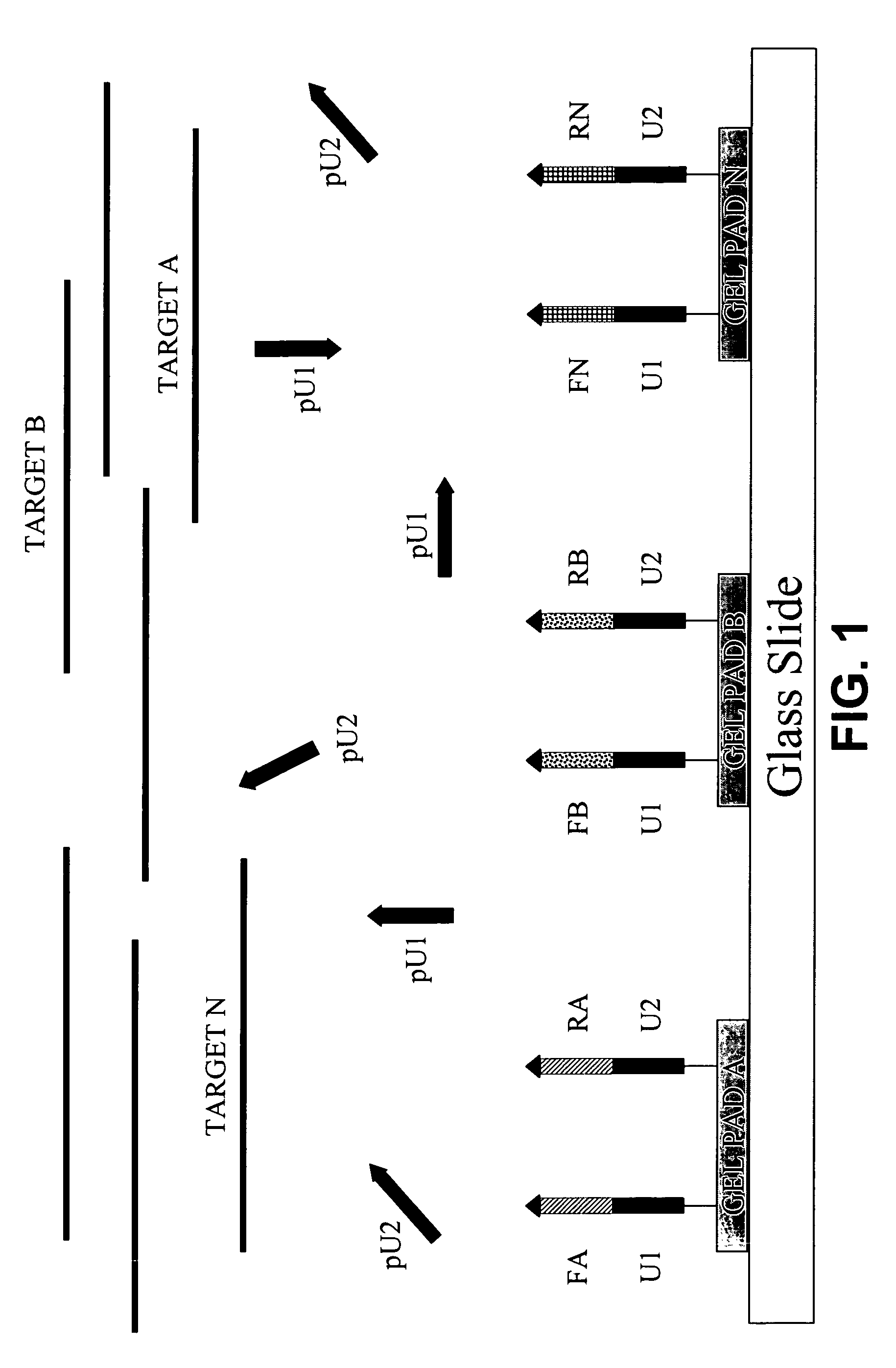
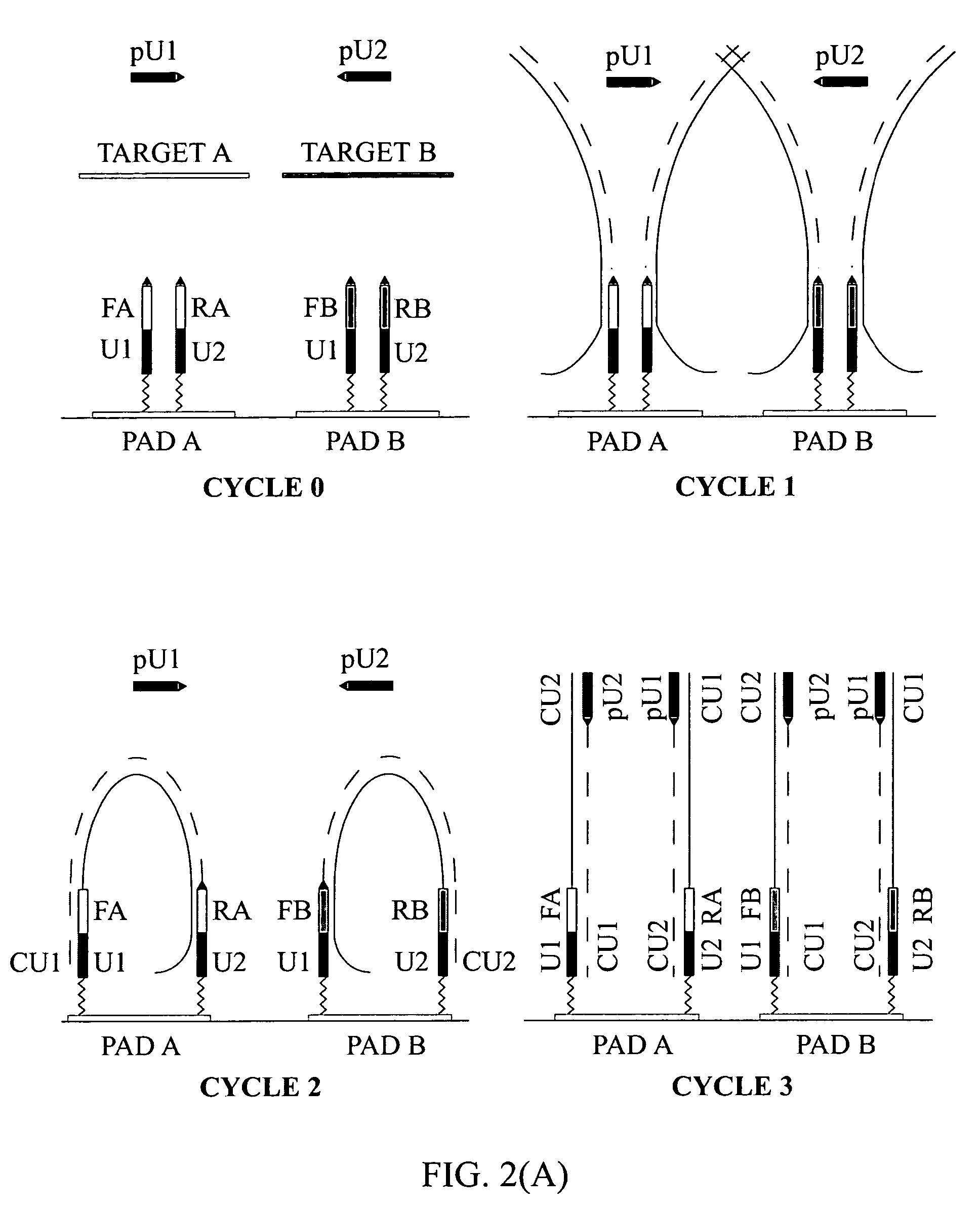
Highly specific and sensitive methods were developed for multiplex amplification of nucleic acids on supports such as microarrays. Based on a specific primer design, methods include five types of amplification that proceed in a reaction chamber simultaneously. These relate to four types of multiplex amplification of a target DNA on a solid support, directed by forward and reverse complex primers immobilized to the support and a fifth type—pseudo-monoplex polymerase chain reaction (PCR) of multiple targets in solution, directed by a single pair of unbound universal primers. The addition of the universal primers in the reaction mixture increases the yield over the traditional “bridge” amplification on a solid support by approximately ten times. Methods that provide multitarget amplification and detection of as little as 0.45-4.5×10−12 g (equivalent to 102-103 genomes) of a bacterial genomic DNA are disclosed.
Owner:UCHICAGO ARGONNE LLC +1
Methods, Compositions, and Kits for Detection of microRNA
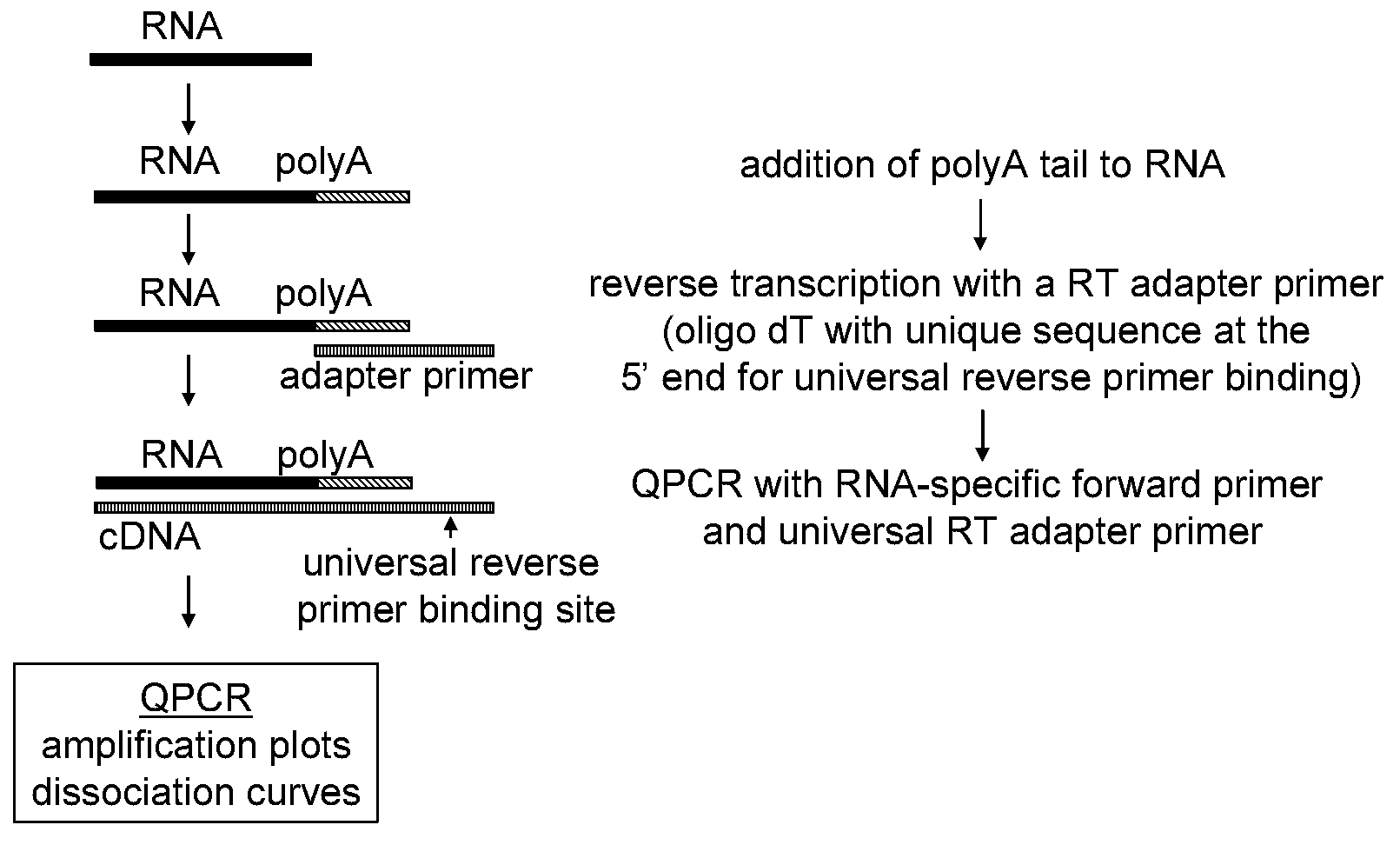
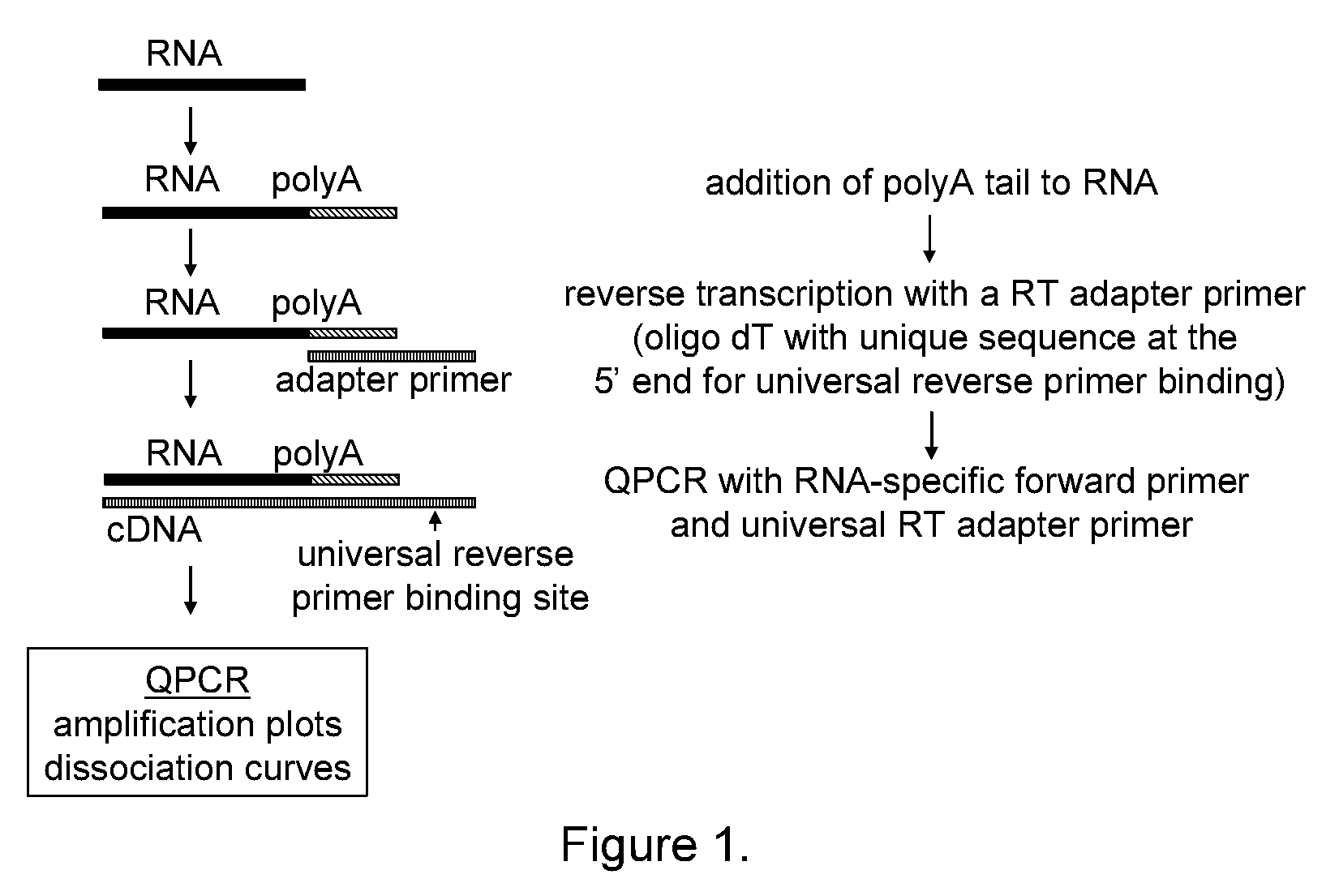
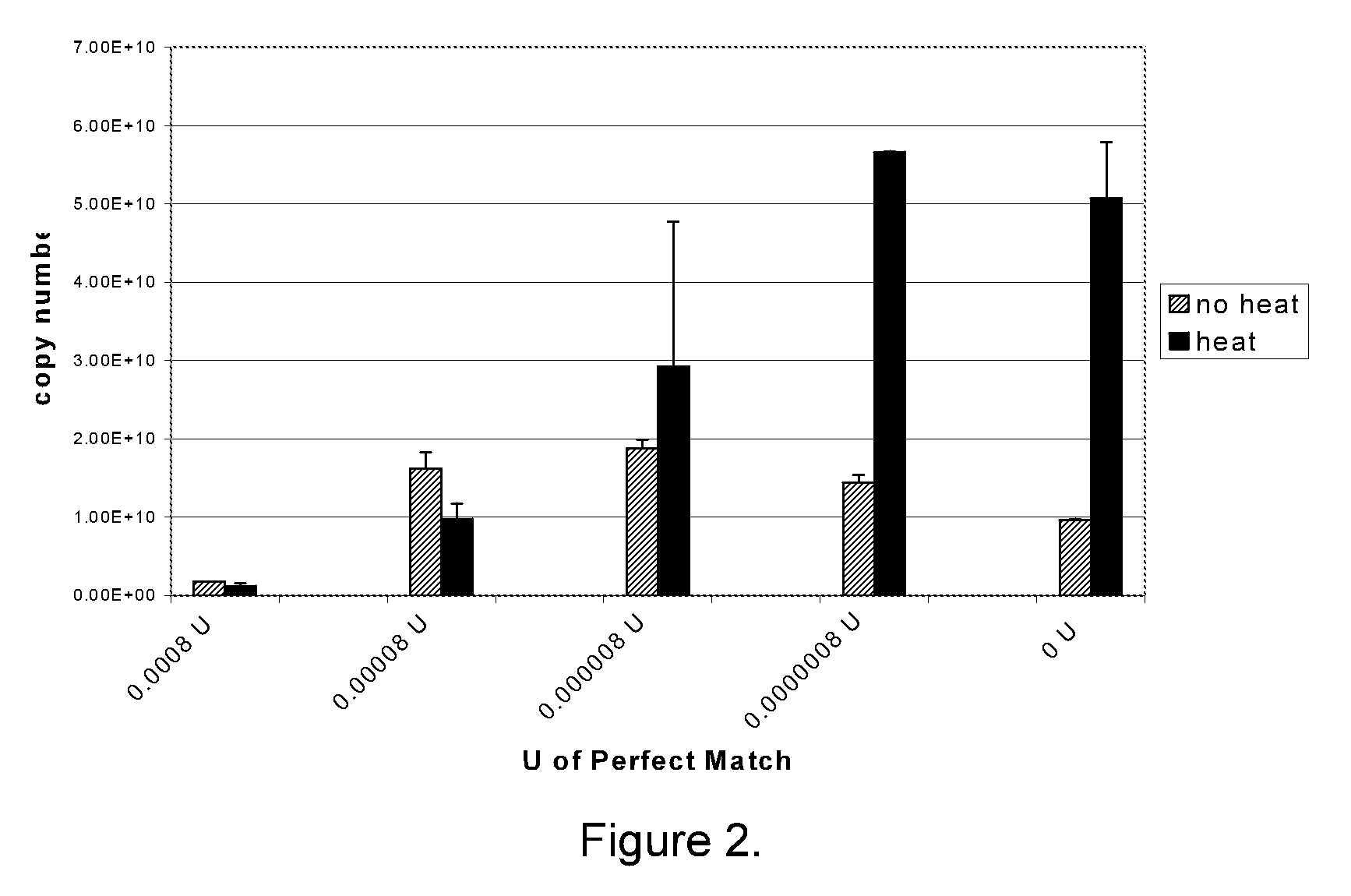
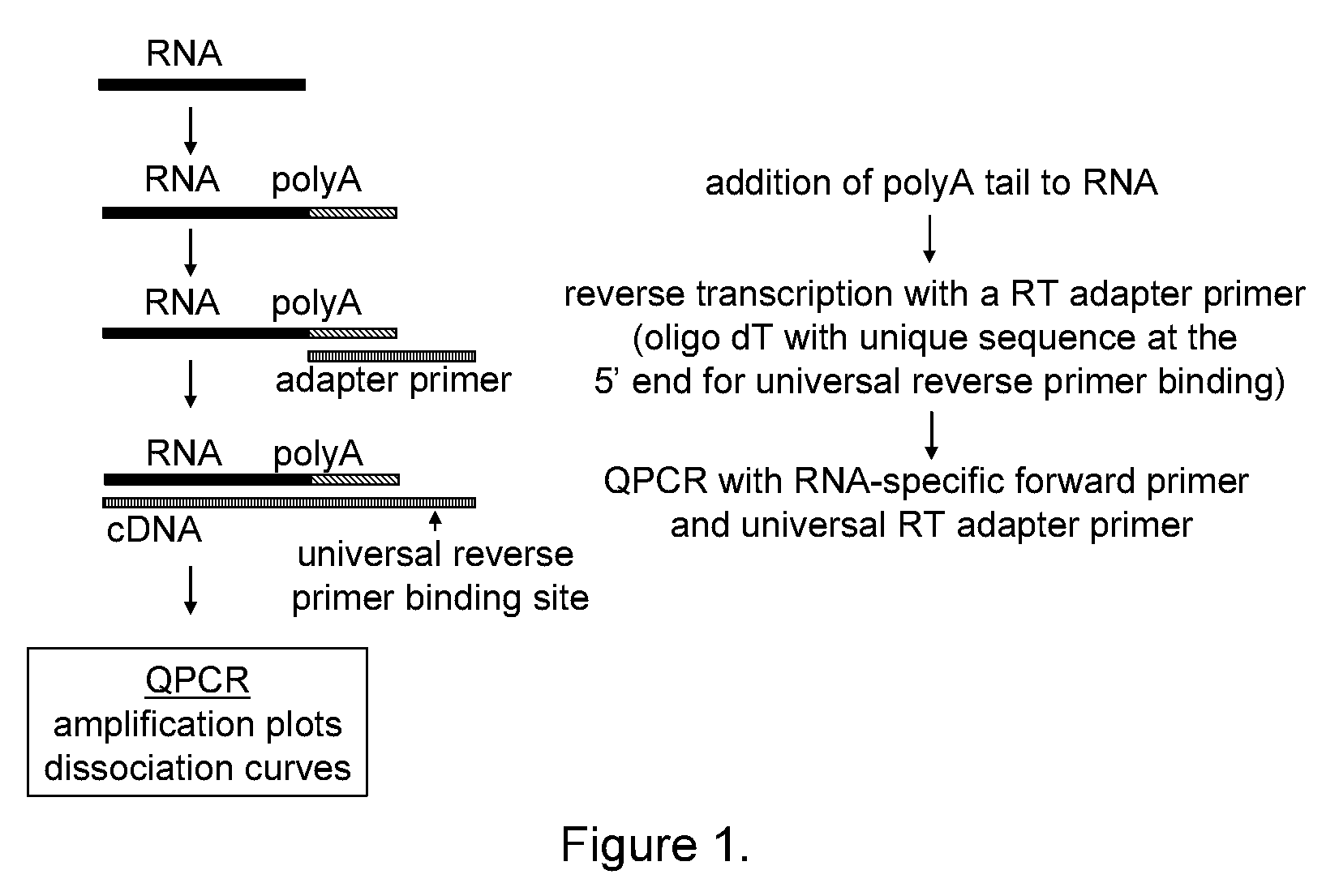
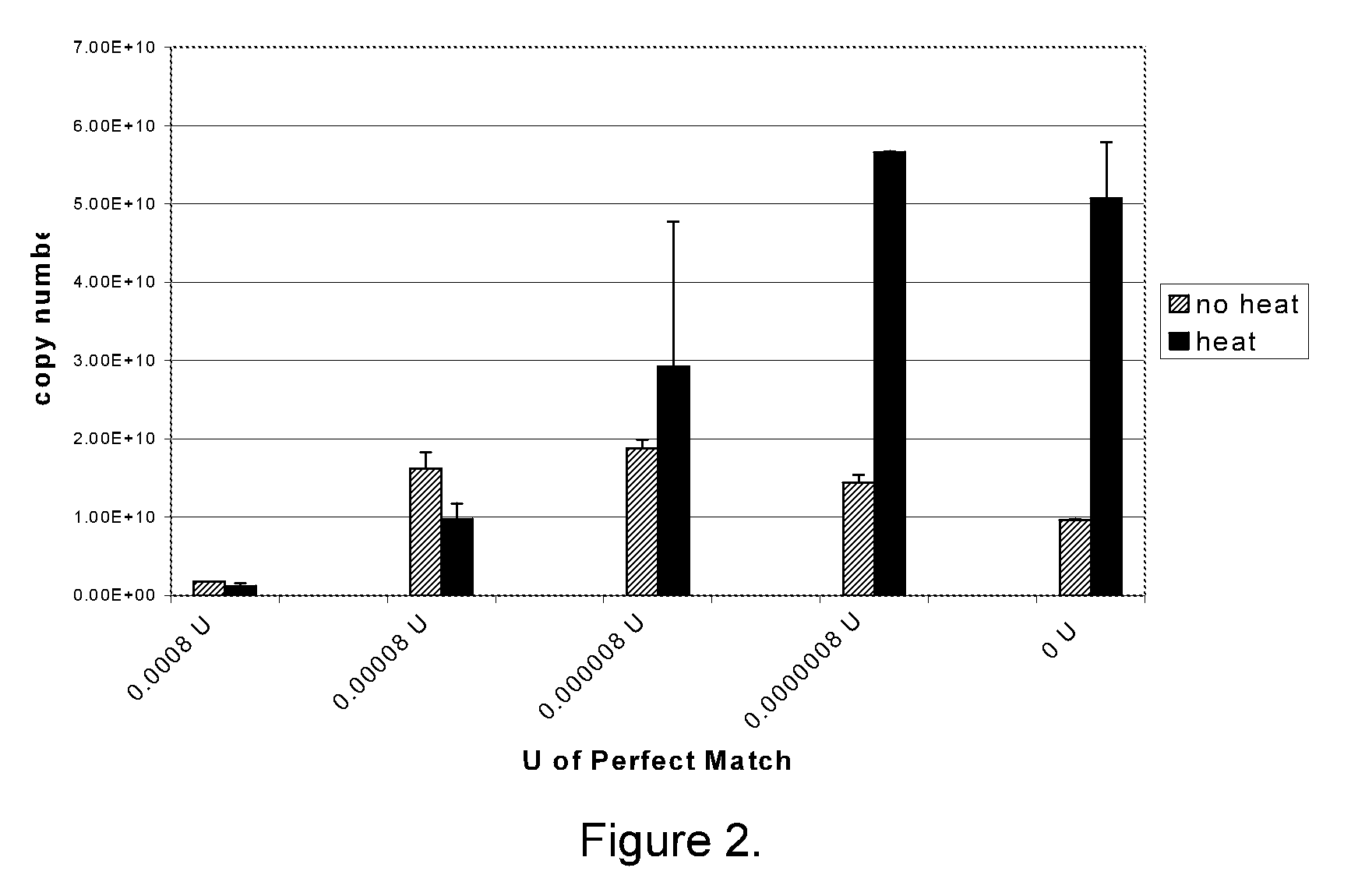
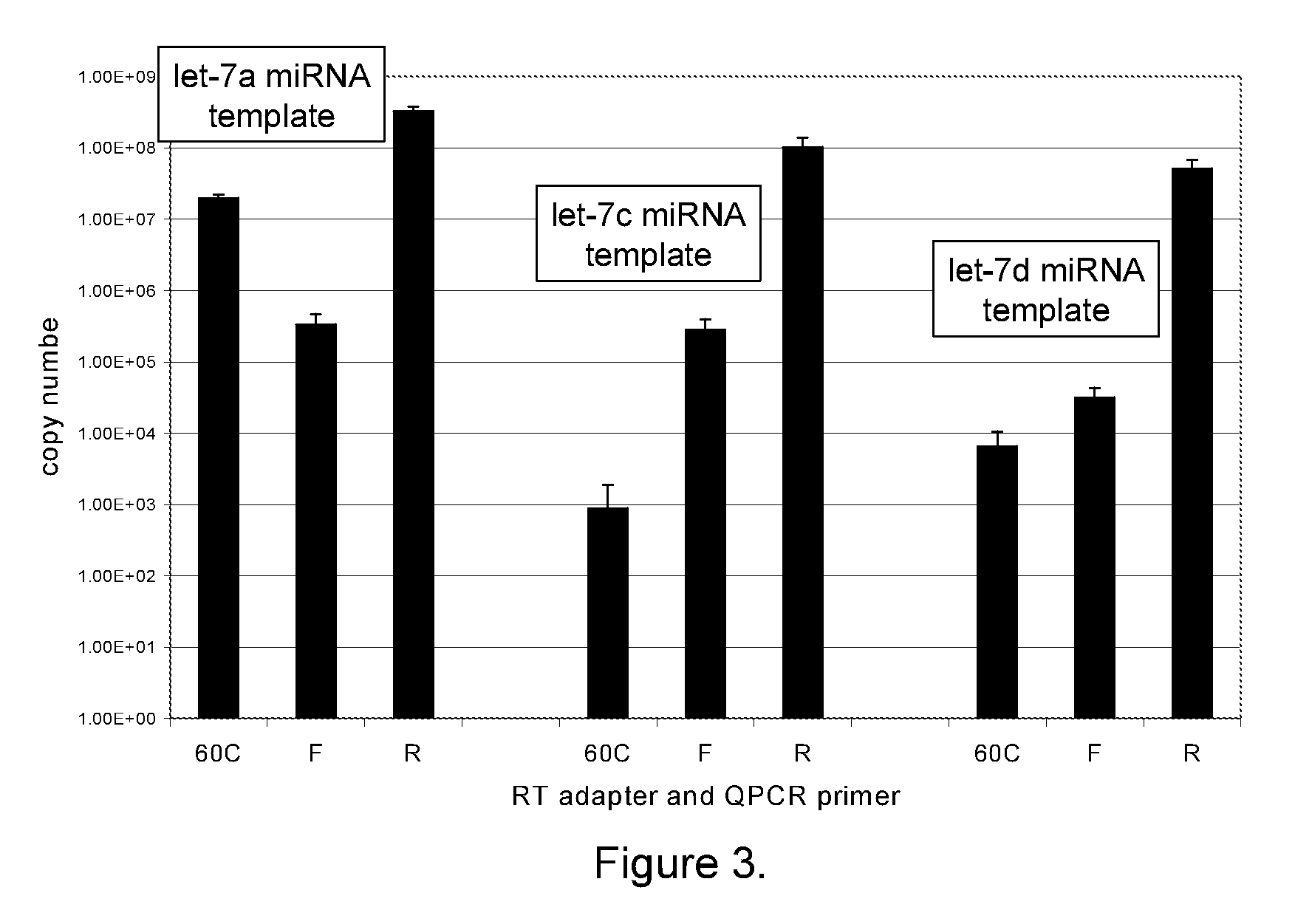
The present invention provides methods, nucleic acids, compositions, and kits for detecting microRNA (miRNA) in samples. The methods comprise designing mRNA-specific primers, adding a polyA tail to the miRNA, and using reverse transcription and amplification to detect the miRNA. The nucleic acids, compositions, and kits typically comprise some or all of the components necessary to practice the methods of the invention.
Owner:AGILENT TECH INC
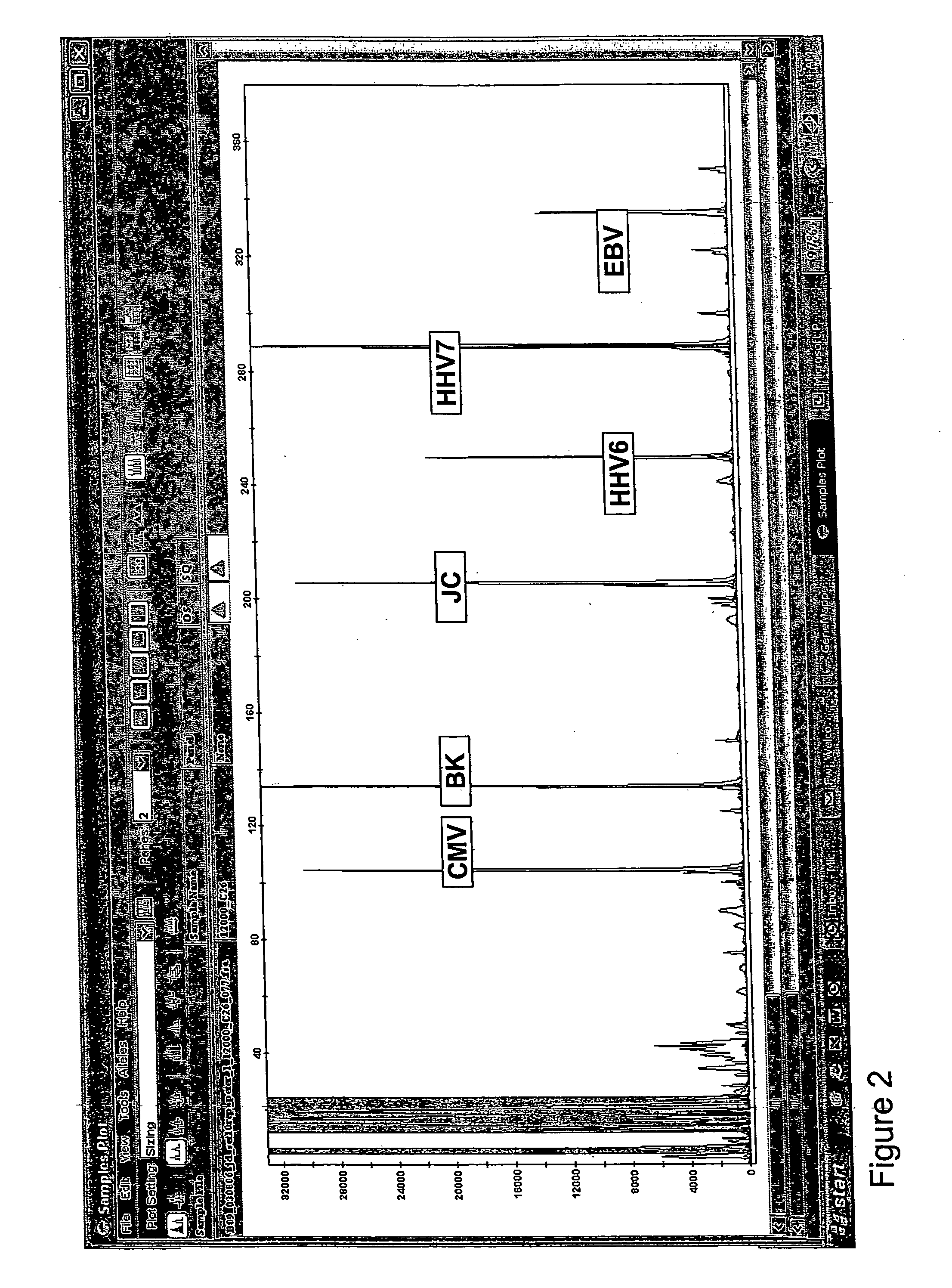
Quantitative epstein barr virus PCR rapid assay
InactiveUS6790952B2Sensitive and accurateSugar derivativesMicrobiological testing/measurementEpstein bar virusNucleotide
The present invention provides novel compositions comprising Epstein-Barr virus-specific oligonucleotides that are useful as primers to amplify particular regions of the genome during enzymatic nucleic acid amplification. The invention also provides a rapid, sensitive and specific method for the detection and quantitation of the virus which may be present in a clinical specimen, using the virus-specific primers and enzymatic nucleic acid amplification; hybridization of amplified target sequences, if present, with one or more Epstein-Barr virus-specific oligonucleotide probes which are labeled with a detectable moiety; and detection of the detectable moiety of labeled oligonucleotide probe hybridized to amplified target sequences of Epstein-Barr virus DNA.
Owner:CHILDRENS HOSPITAL RES FOUND
Methods and compositions for optimizing multiplex pcr primers
This invention relates generally to the field of PCR. In particular, the invention provides methods, compositions and kits for optimizing multiplex PCR primers using, inter alia, a plurality of 5′ and 3′ specific primers and a 5′ and a 3′ universal primer at various ratios.
Owner:CAPITALBIO CORP +1
Amplification and analysis of whole genome and whole transcriptome libraries generated by a DNA polymerization process
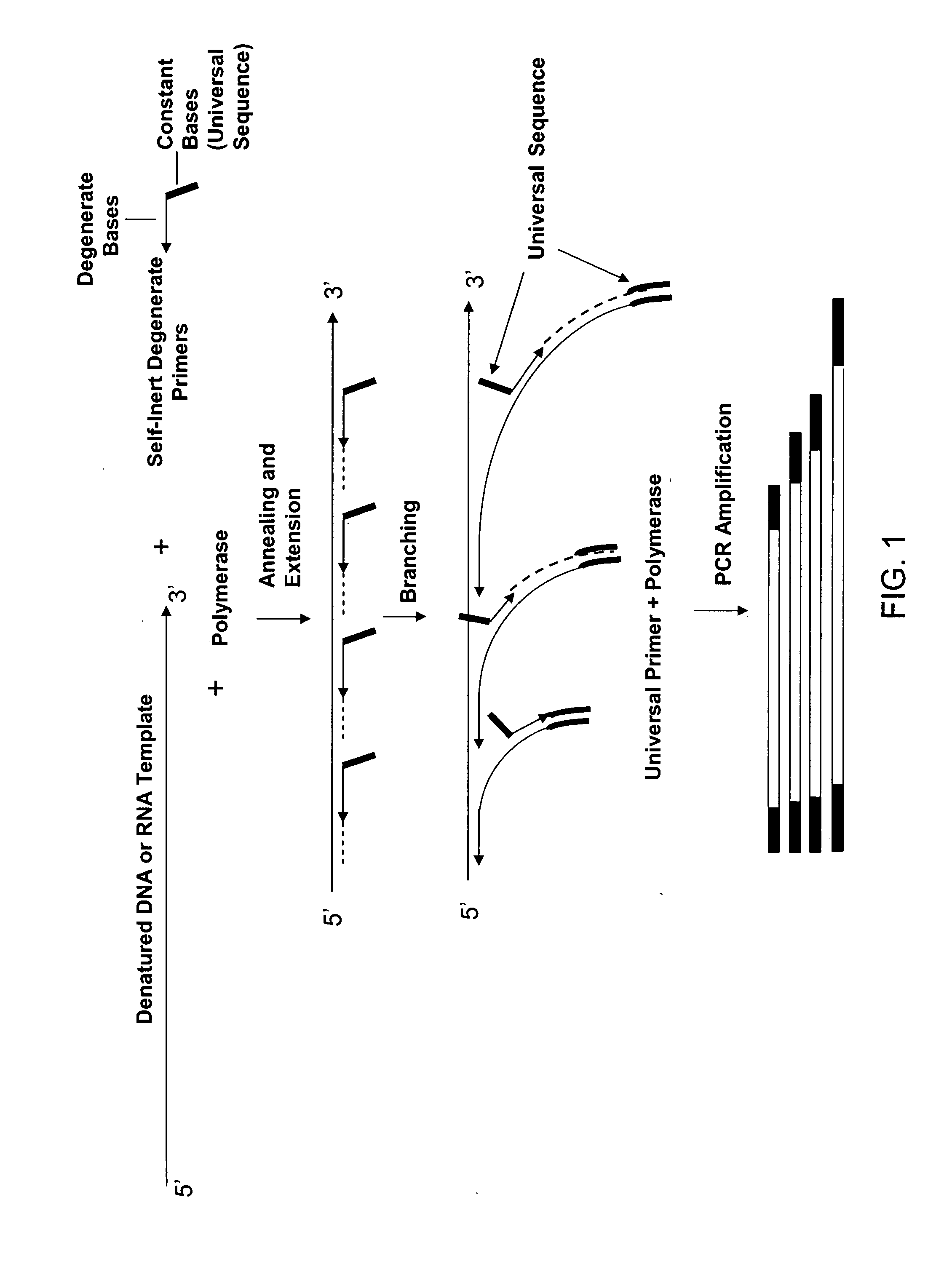
The present invention regards a variety of methods and compositions for whole genome amplification and whole transcriptome amplification. In a particular aspect of the present invention, there is a method of amplifying a genome comprising a library generation step followed by a library amplification step. In specific embodiments, the library generating step utilizes specific primer mixtures and a DNA polymerase, wherein the specific primer mixtures are designed to eliminate ability to self-hybridize and / or hybridize to other primers within a mixture but efficiently and frequently prime nucleic acid templates.
Owner:TAKARA BIO USA INC
Dual phase multiplex polymerase chain reaction
ActiveUS20050196760A1Increase volumeFurther amplificationMicrobiological testing/measurementLibrary member identificationGenomic DNASingle pair
Highly specific and sensitive methods were developed for multiplex amplification of nucleic acids on supports such as microarrays. Based on a specific primer design, methods include five types of amplification that proceed in a reaction chamber simultaneously. These relate to four types of multiplex amplification of a target DNA on a solid support, directed by forward and reverse complex primers immobilized to the support and a fifth type—pseudo-monoplex polymerase chain reaction (PCR) of multiple targets in solution, directed by a single pair of unbound universal primers. The addition of the universal primers in the reaction mixture increases the yield over the traditional “bridge” amplification on a solid support by approximately ten times. Methods that provide multitarget amplification and detection of as little as 0.45-4.5×10−12 g (equivalent to 102-103 genomes) of a bacterial genomic DNA are disclosed.
Owner:UCHICAGO ARGONNE LLC +1
Amplification and analysis of whole genome and whole transcriptome libraries generated by a DNA polymerization process
ActiveUS8206913B1Increase success rateEnhance single-copy sensitivityMicrobiological testing/measurementFermentationPolymerase LA-DNA
The present invention regards a variety of methods and compositions for whole genome amplification and whole transcriptome amplification. In a particular aspect of the present invention, there is a method of amplifying a genome comprising a library generation step followed by a library amplification step. In specific embodiments, the library generating step utilizes specific primer mixtures and a DNA polymerase, wherein the specific primer mixtures are designed to eliminate ability to self-hybridize and / or hybridize to other primers within a mixture but efficiently and frequently prime nucleic acid templates.
Owner:TAKARA BIO USA INC
Methods, compositions, and kits for detection of microRNA
The present invention provides methods, nucleic acids, compositions, and kits for detecting microRNA (miRNA) in samples. The methods comprise designing mRNA-specific primers, adding a polyA tail to the miRNA, and using reverse transcription and amplification to detect the miRNA. The nucleic acids, compositions, and kits typically comprise some or all of the components necessary to practice the methods of the invention.
Owner:AGILENT TECH INC
Amplification and analysis of whole genome and whole transcriptome libraries generated by a DNA polymerization process
InactiveUS20070054311A1High throughput formatNucleotide librariesMicrobiological testing/measurementDna polymerasenPolymerase L
The present invention regards a variety of methods and compositions for whole genome amplification and whole transcriptome amplification. In a particular aspect of the present invention, there is a method of amplifying a genome comprising a library generation step followed by a library amplification step. In specific embodiments, the library generating step utilizes specific primer mixtures and a DNA polymerase, wherein the specific primer mixtures are designed to eliminate ability to self-hybridize and / or hybridize to other primers within a mixture but efficiently and frequently prime nucleic acid templates.
Owner:KAMBEROV EMMANUEL +6
Rapid fluorescence PCR detection kit for ASFV (African swine fever virus)
PendingCN109593893ADetection fitReduce manual operationsMicrobiological testing/measurementDNA/RNA fragmentationSerum igeFluorescence
The invention discloses a rapid fluorescence PCR detection kit for ASFV (African swine fever virus). The kit comprises specific primers ASFV-F and ASFV-R as well as a TaqMan probe ASFV-P, the 5' end of the probe labels fluorescence dye as FAM and the 3' end labels a fluorescence quenching group as BHQ-1. The kit can be used for detecting the ASFV in nasal swabs, blood, serum, plasma and tissue ofswine to realize rapid detection of the ASFV. In the whole ASFV detection process, only 40 min is taken from DNA extraction to obtaining of detection results, so that the detection time is greatly shortened, and the detection efficiency is improved.
Owner:ZHENGZHOU ZHONGDAO BIOTECHNOLOGY CO LTD +2
Vibrio parahemolyticus detection kit and detection method thereof
InactiveCN102586452AEfficient analysisHigh throughput processingMicrobiological testing/measurementVibrio parahemolyticusMicrobiology
The invention relates to a vibrio parahemolyticus detection kit. The vibrio parahemolyticus detection kit is characterized by comprising the following components: (1) an immune enrichment reaction system component, (2) a loop-mediated isothermal amplification (LAMP) system component and (3) a set of specific primers based on an LAMP technology of vibrio parahemolyticus tlh genes, wherein the specific primers comprise two outer primers F3 and B3, two inner primers FIP and BIP and two ring primers LF and LB. The detection method for the vibrio parahemolyticus detection kit comprises the following steps of: detecting the vibrio parahemolyticus by adopting a method of combining immune enrichment and the LAMP technology; preparing an immunomagnetic bead by adopting a vibrio parahemolyticus polyclonal antibody; preliminarily screening the vibrio parahemolyticus in an actual sample by adopting an immunology method; and designing an LAMP specific primer according to the species specificity gene of the vibrio parahemolyticus. According to the invention, molecular detection is carried out on a nucleic acid level, therefore, the condition of false positive or undetection of a simple immunology method or a molecular method is effectively avoided, and the invention is a new development direction of the quick detection of the vibrio parahemolyticus.
Owner:SHANGHAI OCEAN UNIV
Method for preparing II-type pig's ring-virus nucleic vaccine and the use thereof
InactiveCN1579553AShorten the production cycleAvoid pollutionViral antigen ingredientsGenetic material ingredientsPeripheral blood mononuclear cellBiology
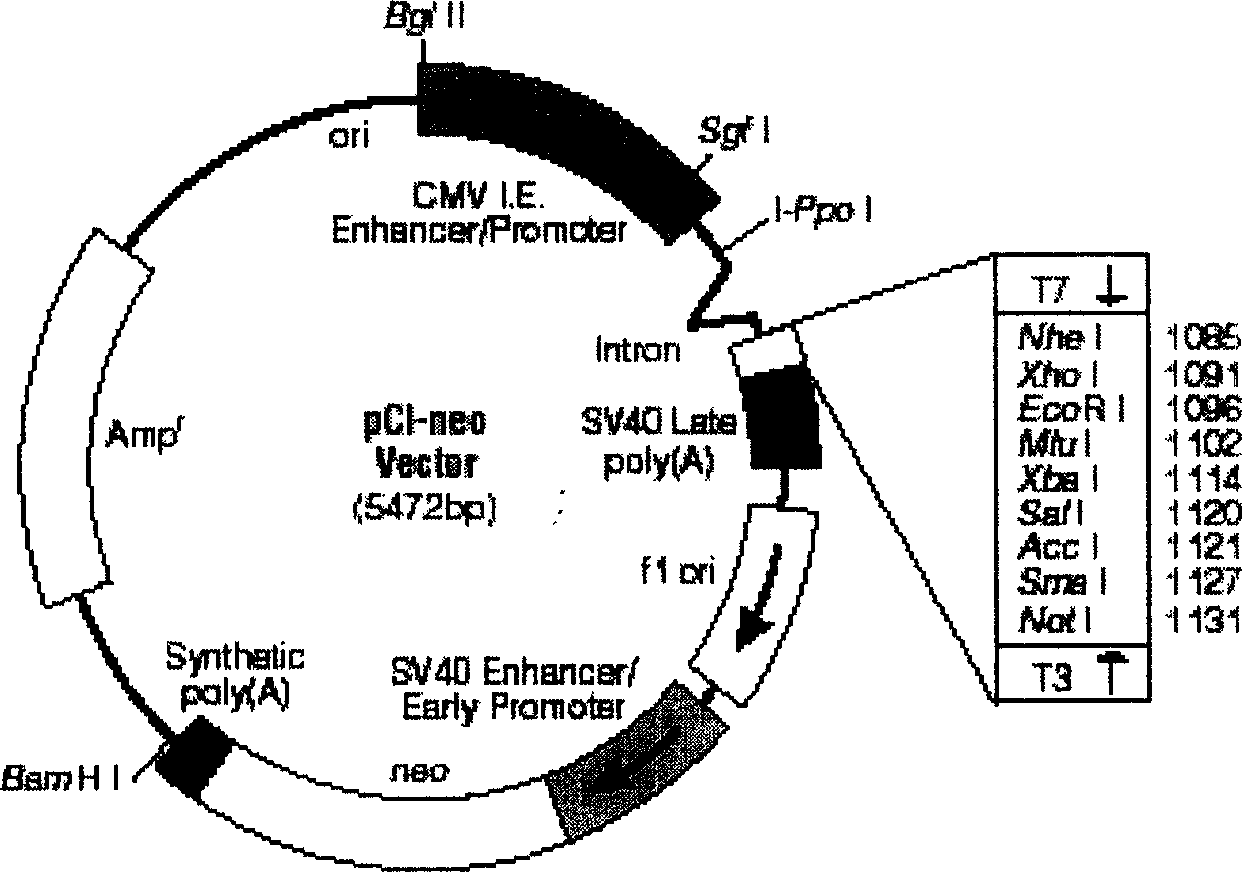
The invention discloses a manufacturing method and application of II type pig ring virus nucleic acid vaccine. The method is: 1) designs the specific primer, uses PCV2 Hangzhou (HZ201) gene group as template, closes ORF1, ORF2, ORF3 and ORF4 genes of PCV-2 with PCR method, and they are constructed into eucaryon expressing carrier with pCI-neo; 2) separates the pig external blood single nucleus cell, clones the genes IFN, IL-2 and IL-4 with RT-PCR method, and they are constructed into eucaryon expressing carrier with pCI-neo; 3) based on above mentioned reconstructed carrier, constructs the fused expressing carrier of the other genes of PCV2, ORF2 and PCV2 or pig cell factor gene; the merits of the invention lie in: (1) it needs not to culture virus, the producing period is short; (2) it needs not cell culture, thus can prevent the contamination from other pig source virus; (3) it does not express the pathogenesis protein which is harmful to body, the safety is high. (4) it can activates body secretion and cell immunity replay at the same time.
Owner:ZHEJIANG UNIV
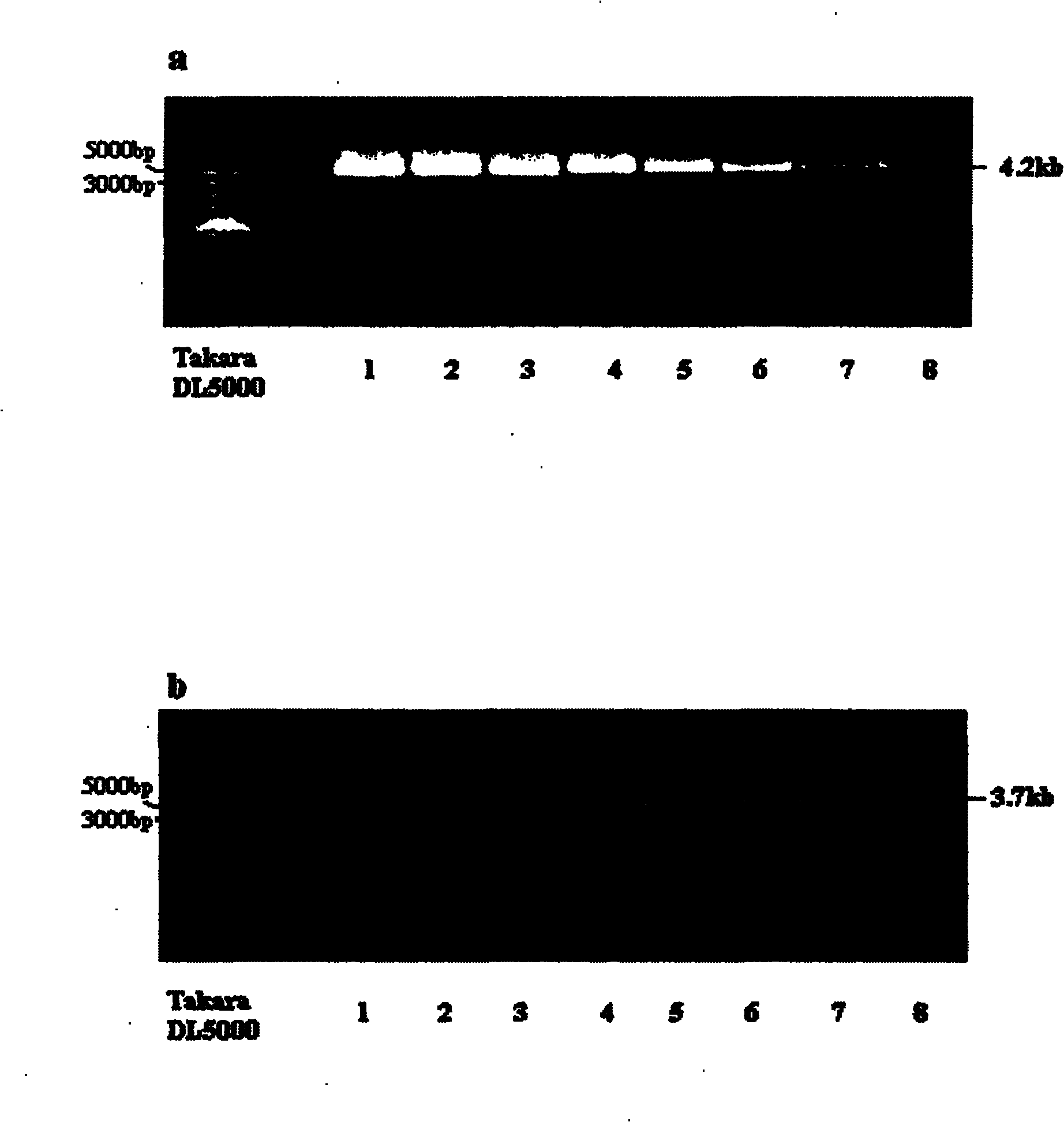
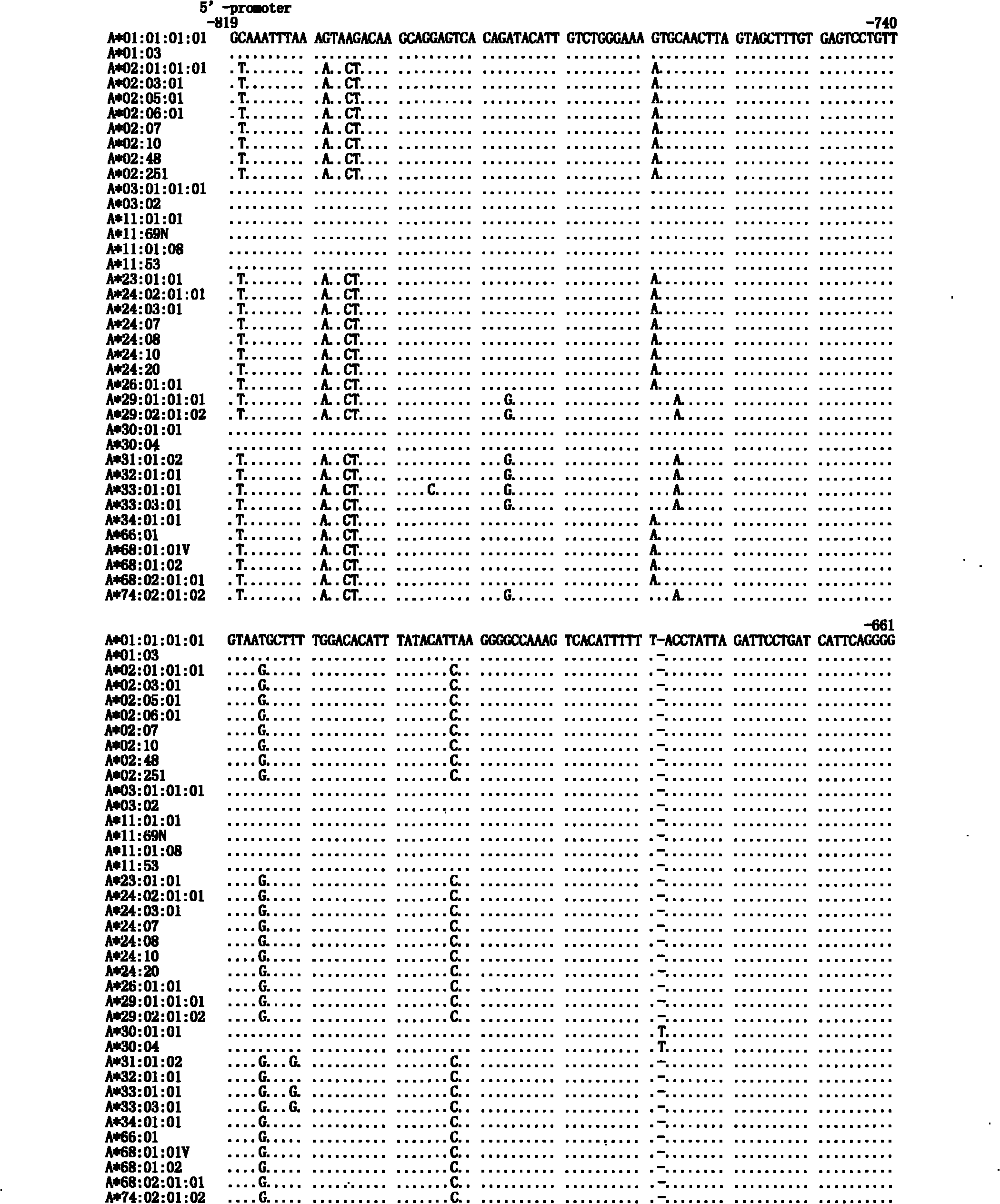
Human leukocyte antigen HLA-A and HLA-B gene full-length sequencing method and HLA gene sequencing and typing method
InactiveCN101962676AWide detection coverageResolving Ambiguous Results IssuesMicrobiological testing/measurementDNA/RNA fragmentationTyping methodsHLA-B gene
The invention discloses a human leukocyte antigen HLA-A and HLA-B gene full-length sequencing method and an HLA gene sequencing and typing method. The HLA-A and HLA-B gene full-length sequencing method comprises the following steps of: a, performing PCR amplification on about 4kb full-length sequences of HLA-A and HLA-B genes by using a pair of primers respectively; and b, cloning the amplification products to a pGEM-Tea sy vector, sequencing the full-length sequences by using ten walking sequencing primers in positive and negative directions respectively, and totally obtaining 38 allele 4.3kb full-length sequences of the HLA-A and 30 allele 3.7kb full-length sequences of the HLA-B. The HLA-A and HLA-B sequencing and typing method comprises the following steps of: performing PCR amplification on typing target areas of the HLA-A and HLA-B by using two pairs of primers respectively; and performing two directional sequencing on products by using fourteen sequencing primers respectively, wherein the HLA-DRB1 and HLA-DQB1 sequencing and typing method comprises the following steps of: amplifying sequences of second and third exons of DRB1 and DQB1 by adopting group specificity primers respectively; performing two directional sequencing on the second and third exons of the DRB1 by adopting eight group specificity primers and three sequencing primers; and performing two directional sequencing on the second and third exons of the DQB1 by adopting four sequencing primers respectively.
Owner:SHENZHEN BLOOD CENT
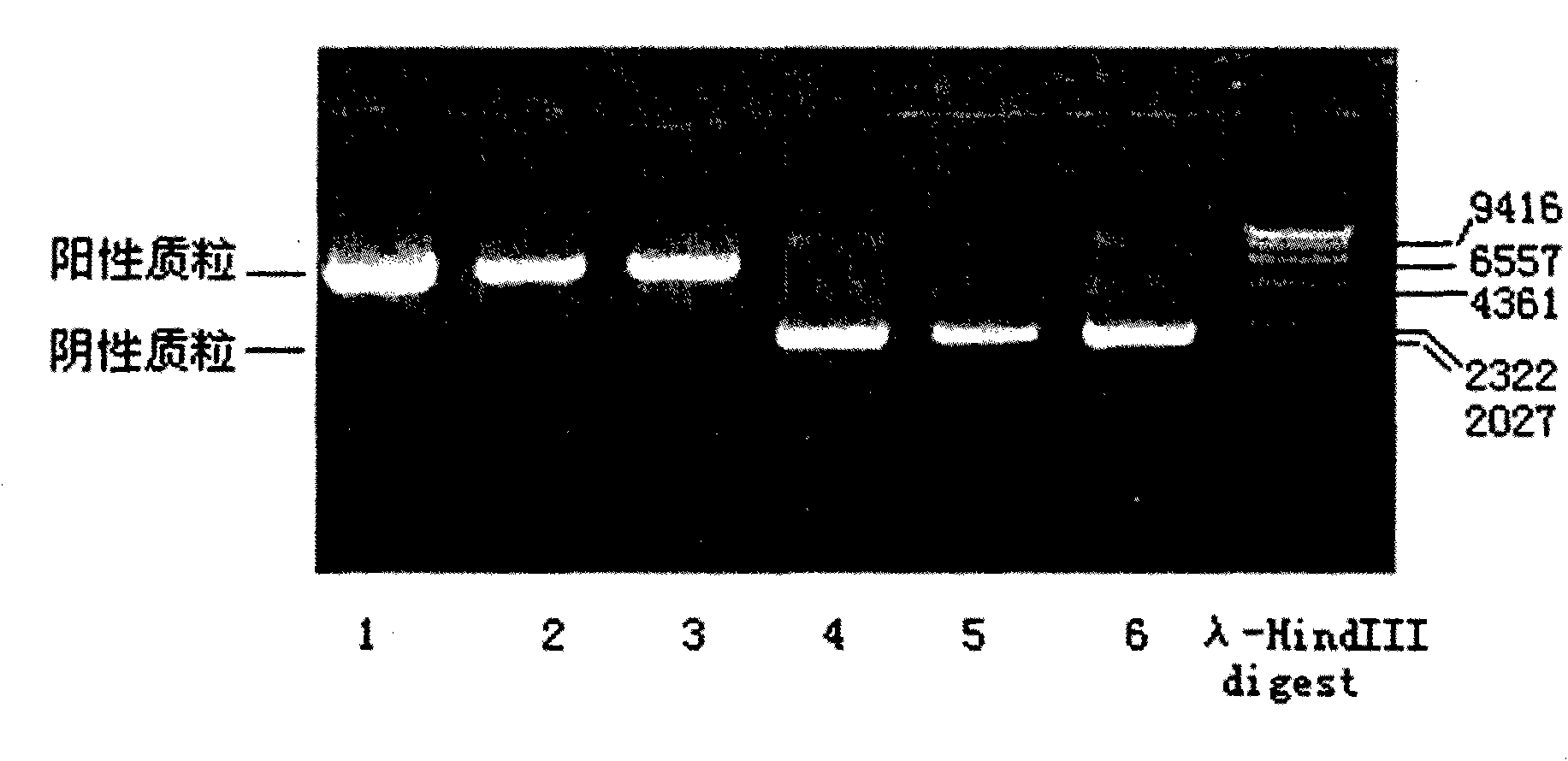
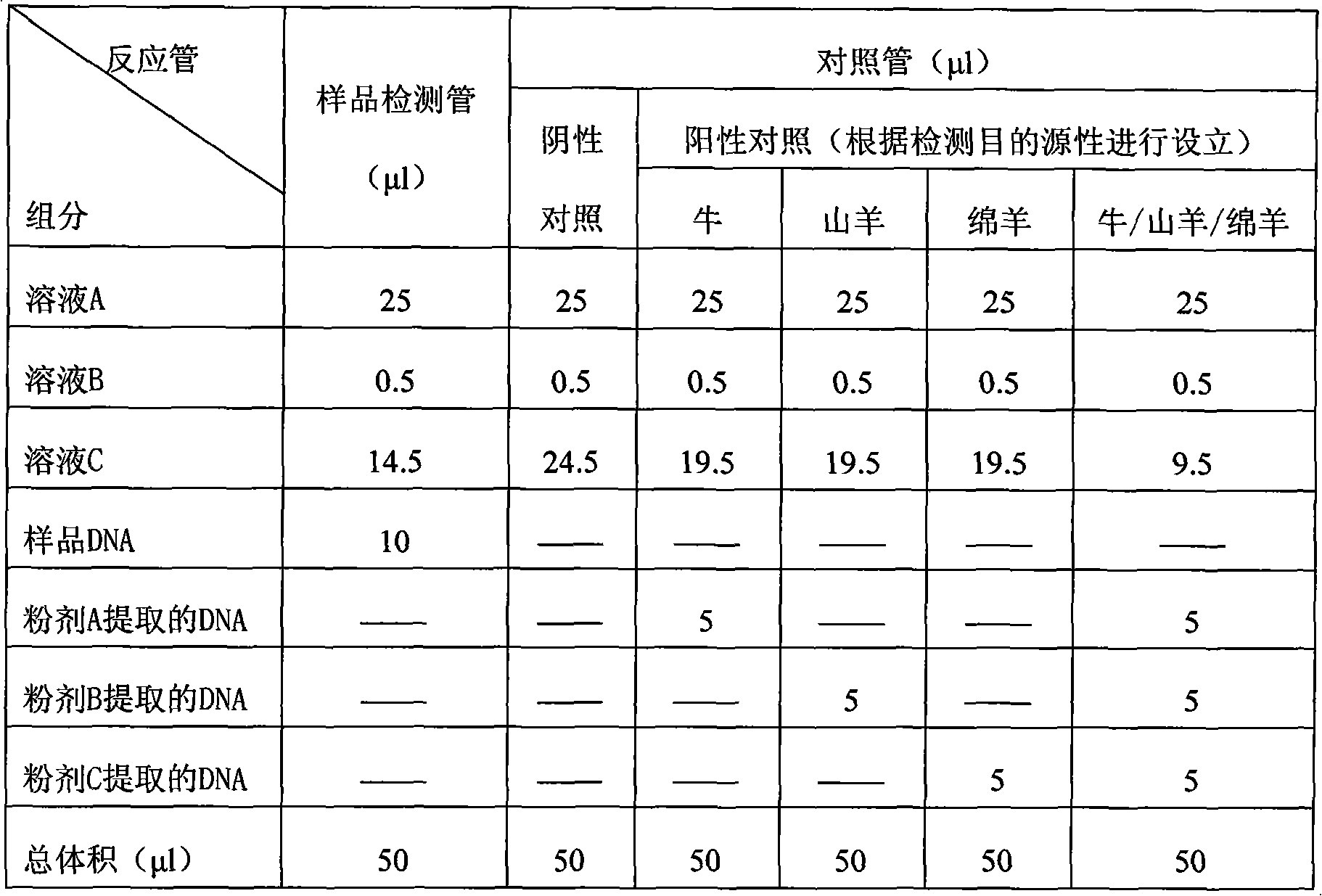
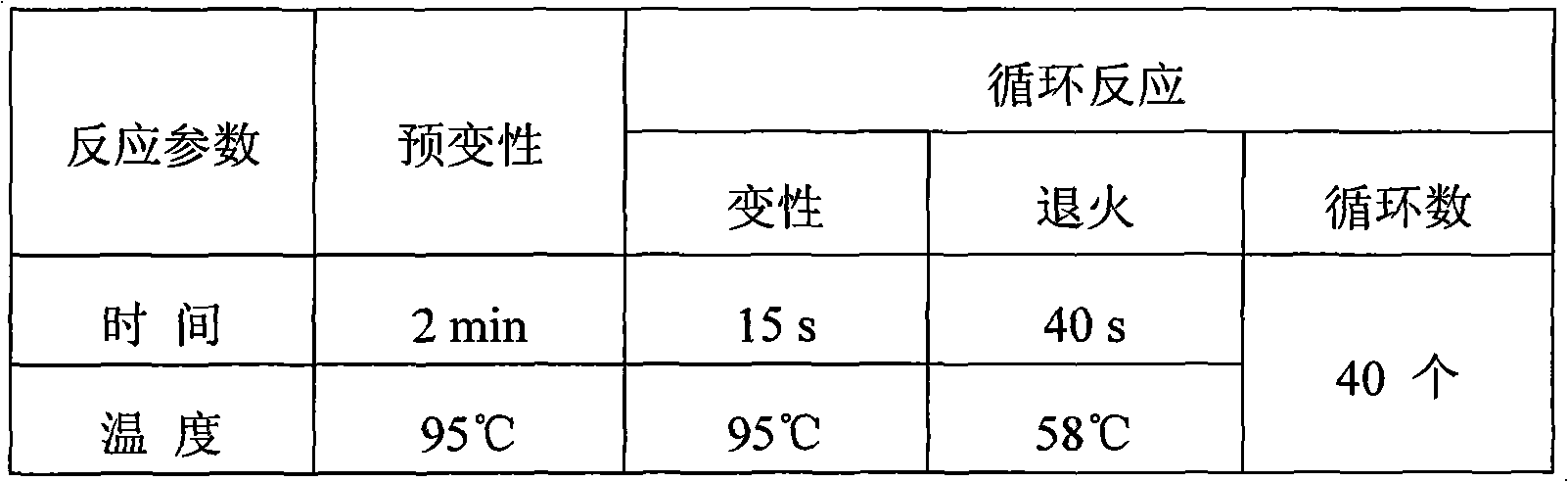
Fluorescence PCR detection reagent capable of discriminating source components of ruminant animal, preparation method and application thereof
InactiveCN101659996AStrong specificityHigh sensitivityMicrobiological testing/measurementFluorescence/phosphorescenceRuminant animalConserved sequence
The invention relates to a biological detection reagent capable of discriminating three types of ruminant animal varieties simultaneously, a preparation method and an application thereof. The invention selects the specificity of the cow, goat and sheep, and the conservative sequence segment of conservative mitochondrial gene as target, applies Primer Eexpress 3.0 software and Primer Select software in DNAStar, and designs and synthesizes a plurality of groups of primers and probes. After being synthesized and marked, the plurality of pairs of designed primers and the probes are carried out best pairing screening experiment so as to obtain the fittest combination of primers and probes. The reagent contains two pairs of specificity primers, and three Taq Man probes, wherein one pair of theprimer aims at cow source components, the other pair of primer aims at common primers of goats and sheep source component detection; the three probes are respective source components to the cows, thegoats and the sheep; and the amplified target fragment length to the source component detection of the cows, the goats and the sheep is respectively 92bp, 136bp and 136bp. The triple Taq Man fluorescence PCR detection method can simultaneously carry out accurate quantitative detection to the source components of the cows, the goats and the sheep, has simple and convenient operation, intuitive result, strong specificity, high sensitivity and good repeatability and can realize high throughput detection of various source components.
Owner:花群义
Universal RT-coupled PCR method for the specific amplification of mRNA
InactiveUS7141372B2Genomic DNA amplification is avoidedSugar derivativesMicrobiological testing/measurementForward primerSpecific detection
The invention relates to a novel Universal RT-coupled PCR strategy for the specific detection and accurate quantitation of mRNA. Claimed and disclosed are novel Universal reverse transcription (RT) primers, a specific primer mix containing the Universal RT-primers, a transcript specific forward primer and a reverse PCR primer identical to a unique tag sequence, and methods and kits thereof for avoiding the amplification of genomic DNA and / or pseudogenes.
Owner:HEALTH RES INC
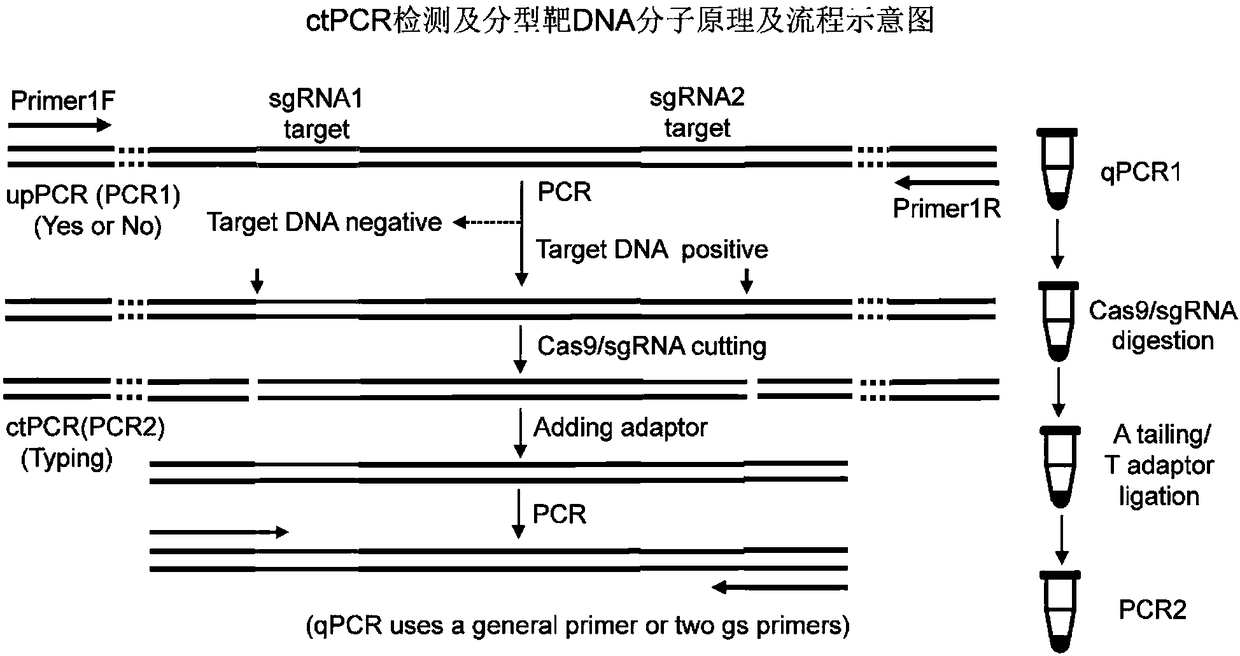
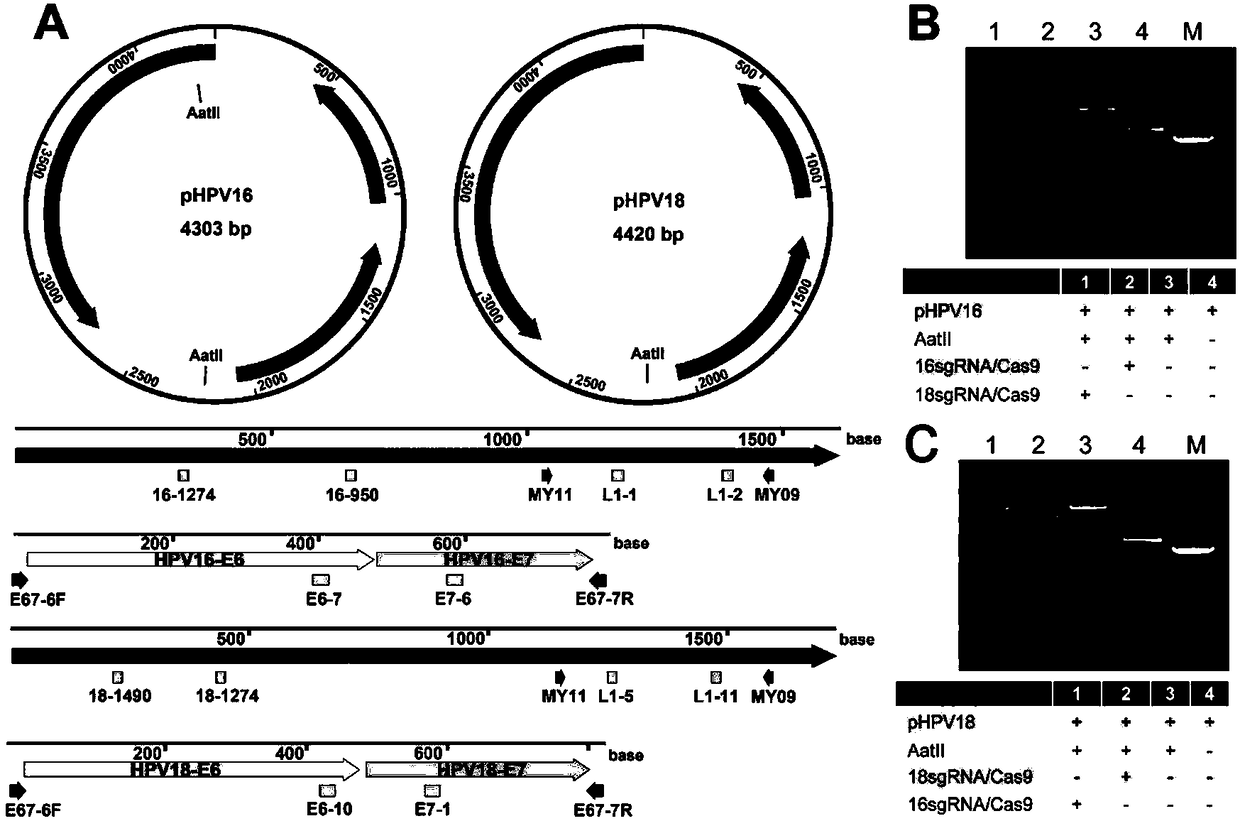
DNA detection and analysis method based on Cas9 nuclease and application thereof
ActiveCN108192956AImprove featuresHigh sensitivityMicrobiological testing/measurementNucleic acid detectionTyping
The invention provides a DNA detection and analysis method based on Cas9 nuclease and application thereof. The method comprises the following three steps: (1) carrying out PCR amplification on targetDNAs; (2) treating a PCR amplification product by using a CAT method; and (3) carrying out PCR amplification with DNAs treated by using the CAT method as a template. The method can successfully detectL1 and E6 / E7 genes of HPV16 and HPV18 in human cervical carcinoma cells. According to the invention, the method successfully avoids current key bottleneck problems in nucleic acid hybridization, designing of specific PCR primers and the like in the fields of nucleic acid detection and typing.
Owner:SOUTHEAST UNIV
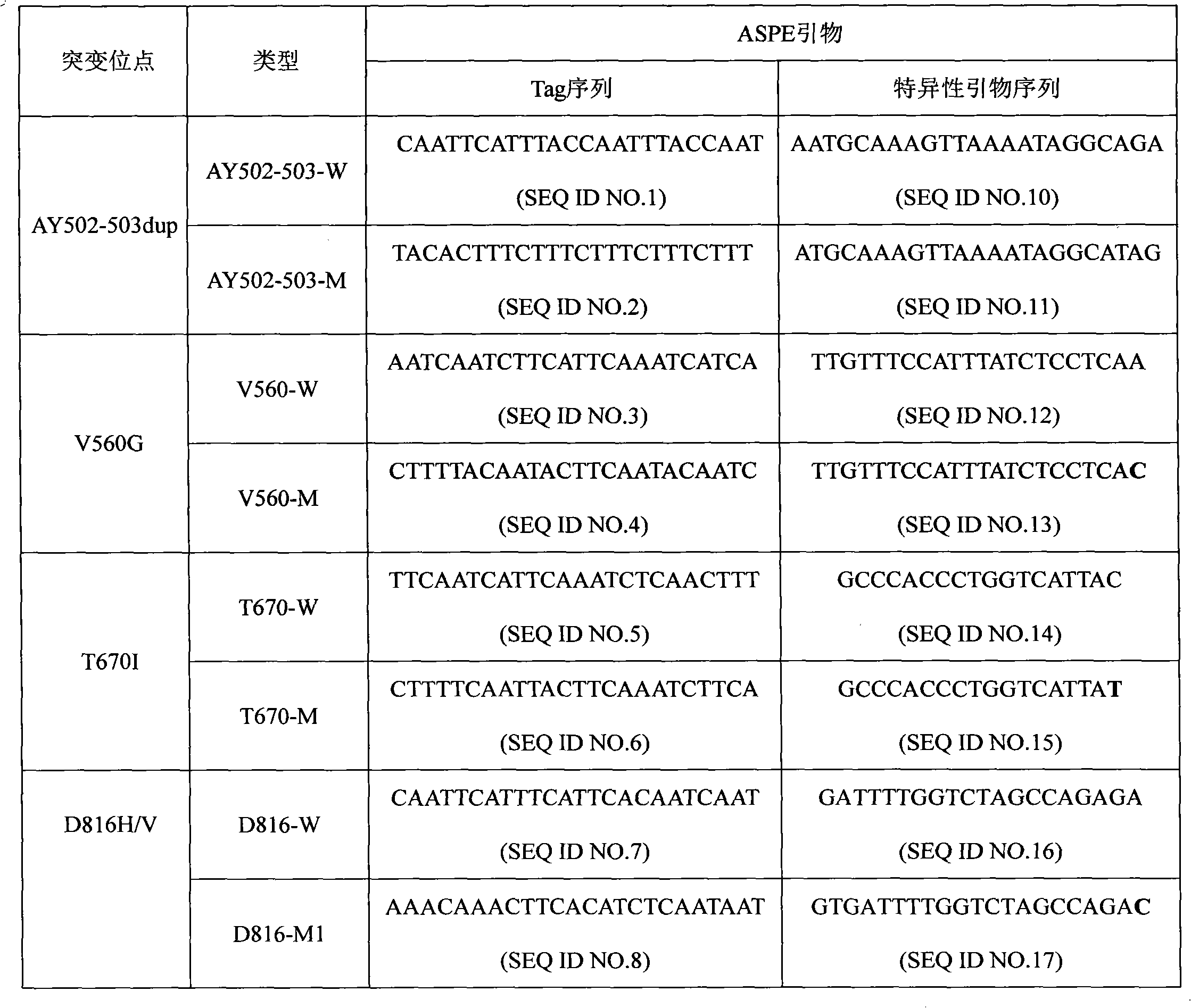
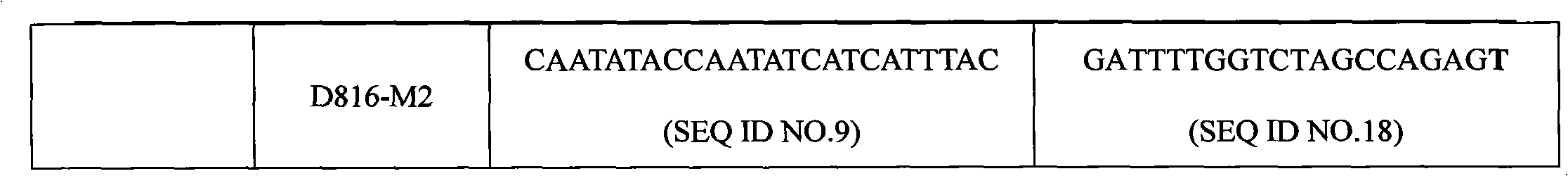
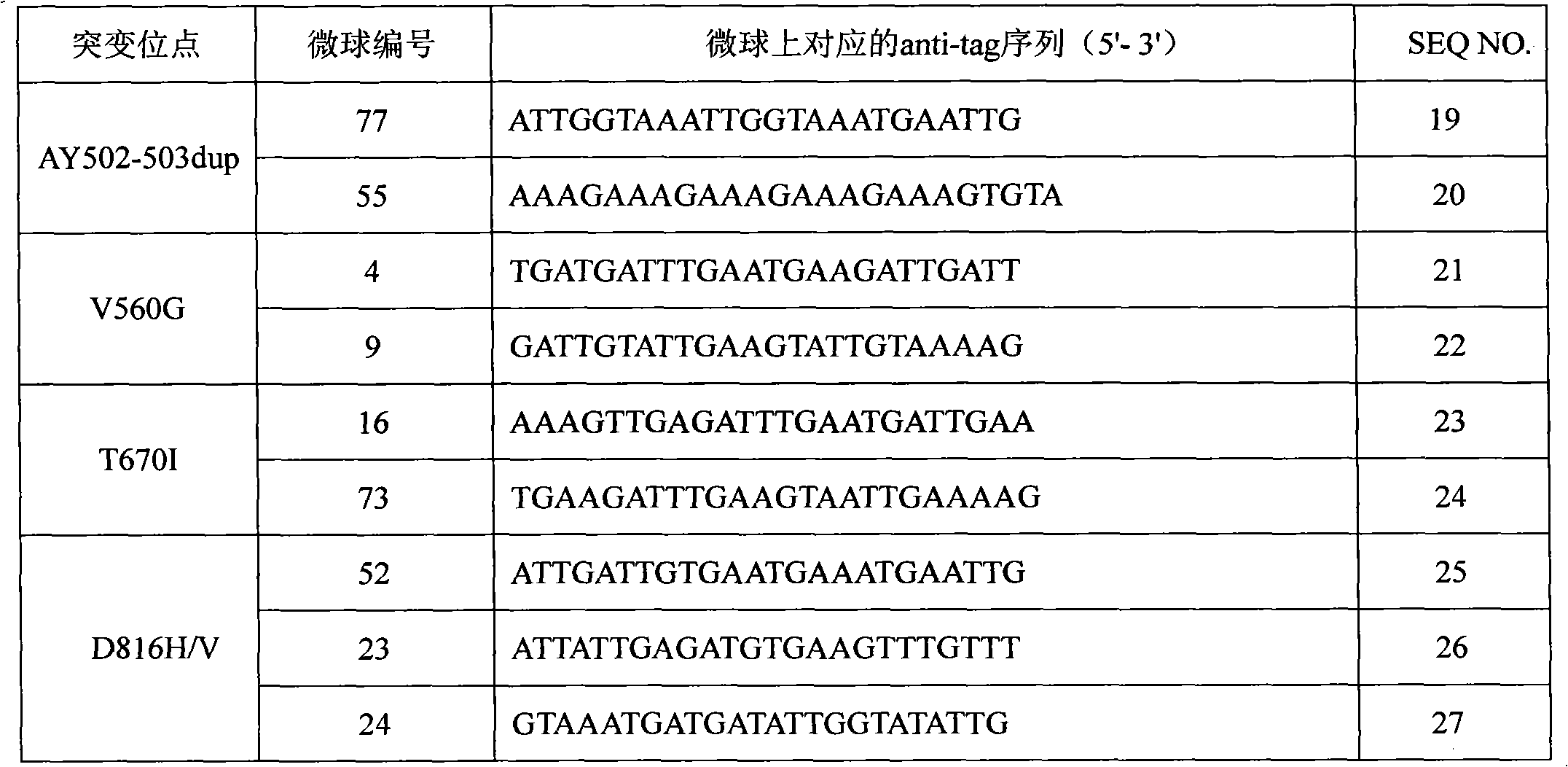
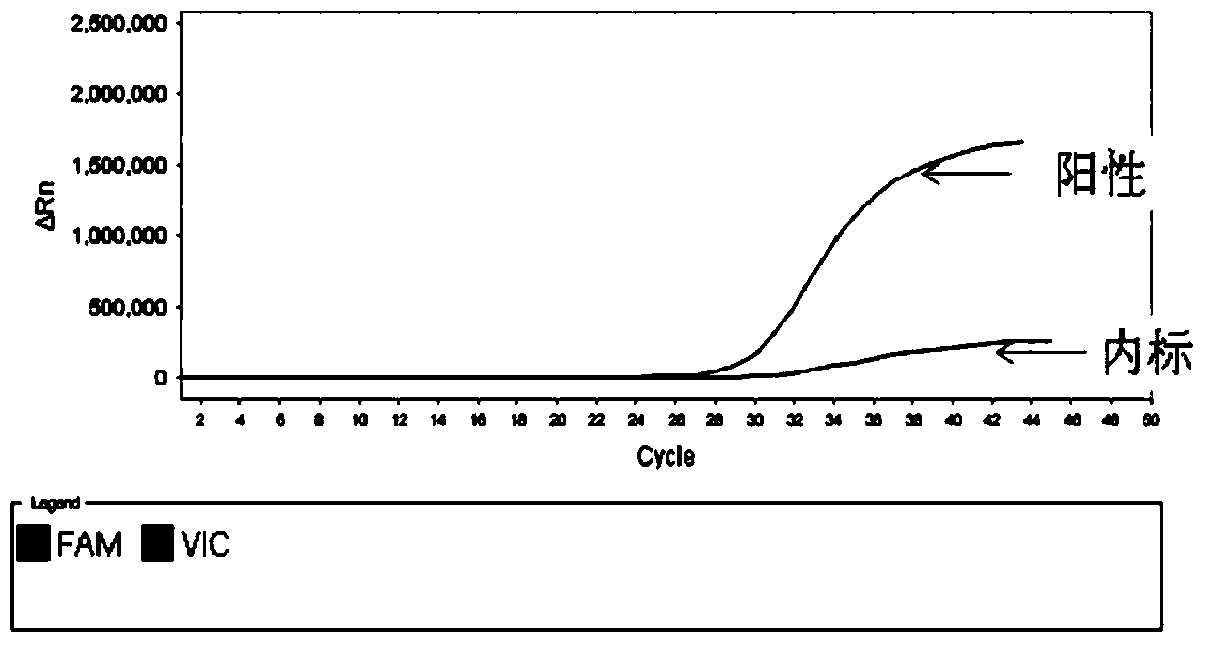
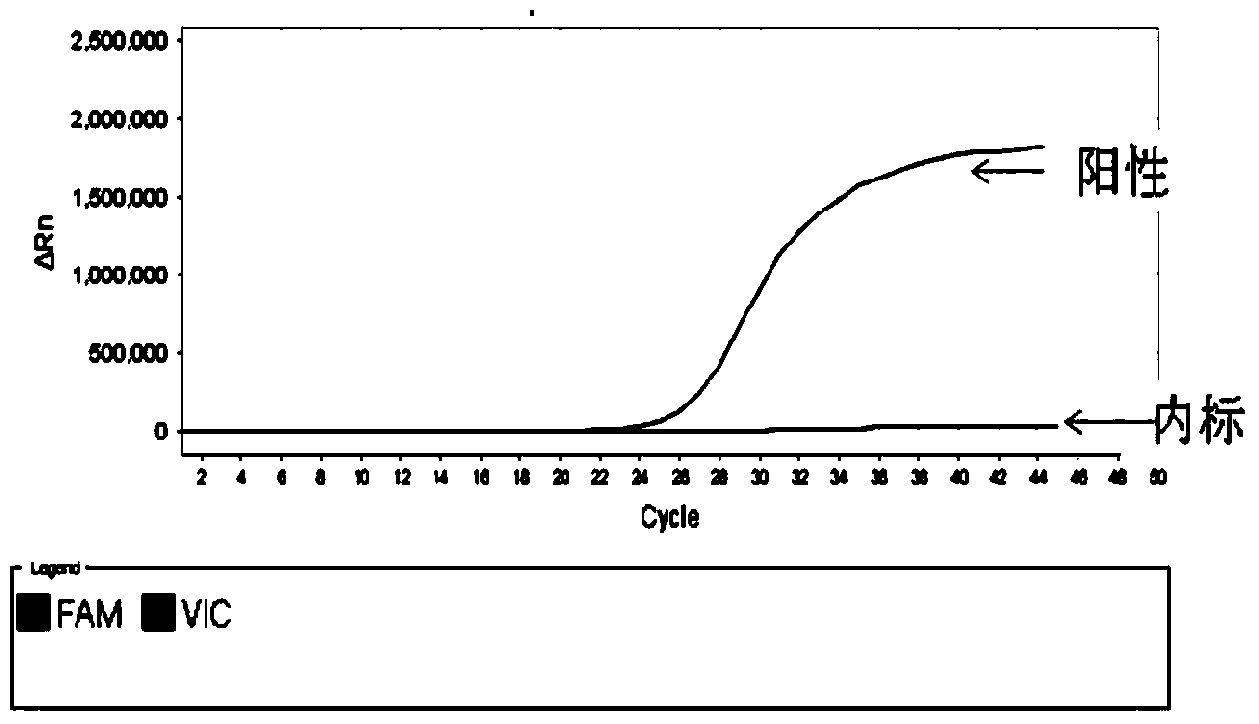
c-KIT gene mutation detection liquid-phase chip
ActiveCN101984070AImprove signal-to-noise ratioImplement parallel detectionMicrobiological testing/measurementSignal-to-noise ratio (imaging)Mutation detection
The invention discloses a c-KIT gene mutation detection liquid-phase chip, which comprises tag sequence at 5' terminal and ASPE (Allele Specific Primer Extension) primers consisting of specific primers specific to mutational sites at 3' terminal. The specific primers consist of sequences shown in SEQ ID No.10 and SEQ ID No.11 specific to AY502-503dup mutational sites, sequences shown in SEQ ID No.12 and SEQ ID No.13 specific to V560G mutational site, sequences shown in SEQ ID No.14 and SEQ ID No.15 specific to T670I mutational site and / or sequences shown in SEQ ID No.16-SEQ ID No.18specific to D816H / V mutational site, the tag sequence which is selected from sequences shown in SEQ ID No.1-SEQ ID No.9, microballoons which have different color codings and are respectively enveloped with a specific anti-tag sequence and amplification primers. The coincidence ratio of the result of sequencing method with that of the c-KIT gene mutation detection liquid-phase chip reaches up to 100 percent. The c-KIT gene mutation detection liquid-phase chip can simultaneously detect a plurality of mutational sites with excellent signal to noise ratio.
Owner:SUREXAM BIO TECH
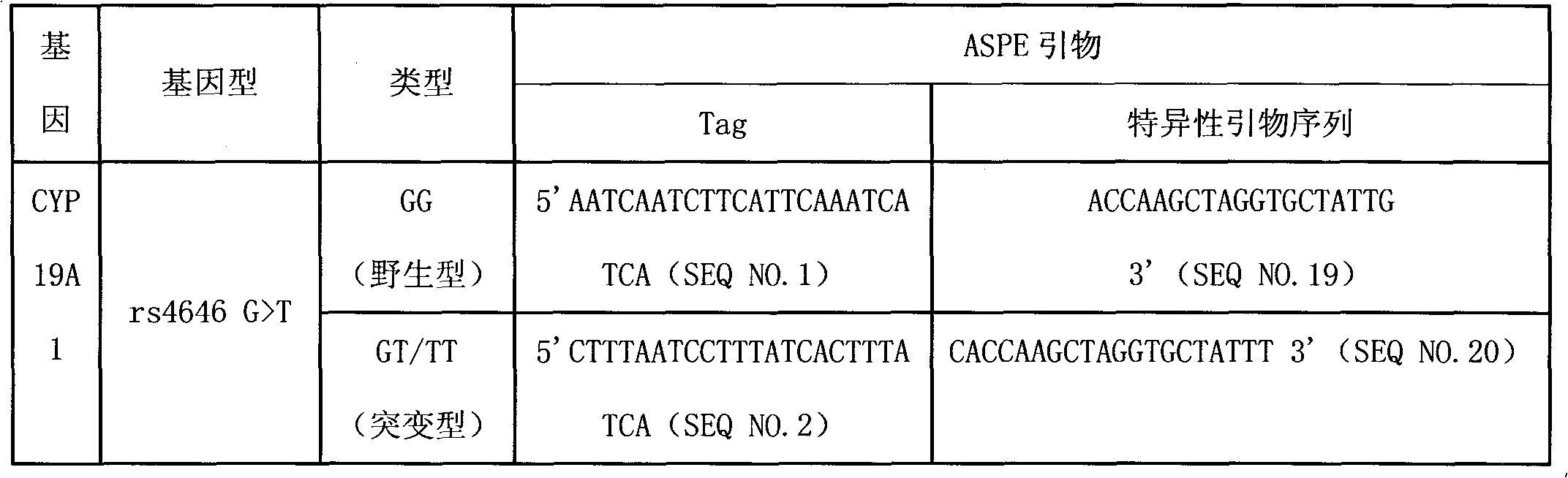
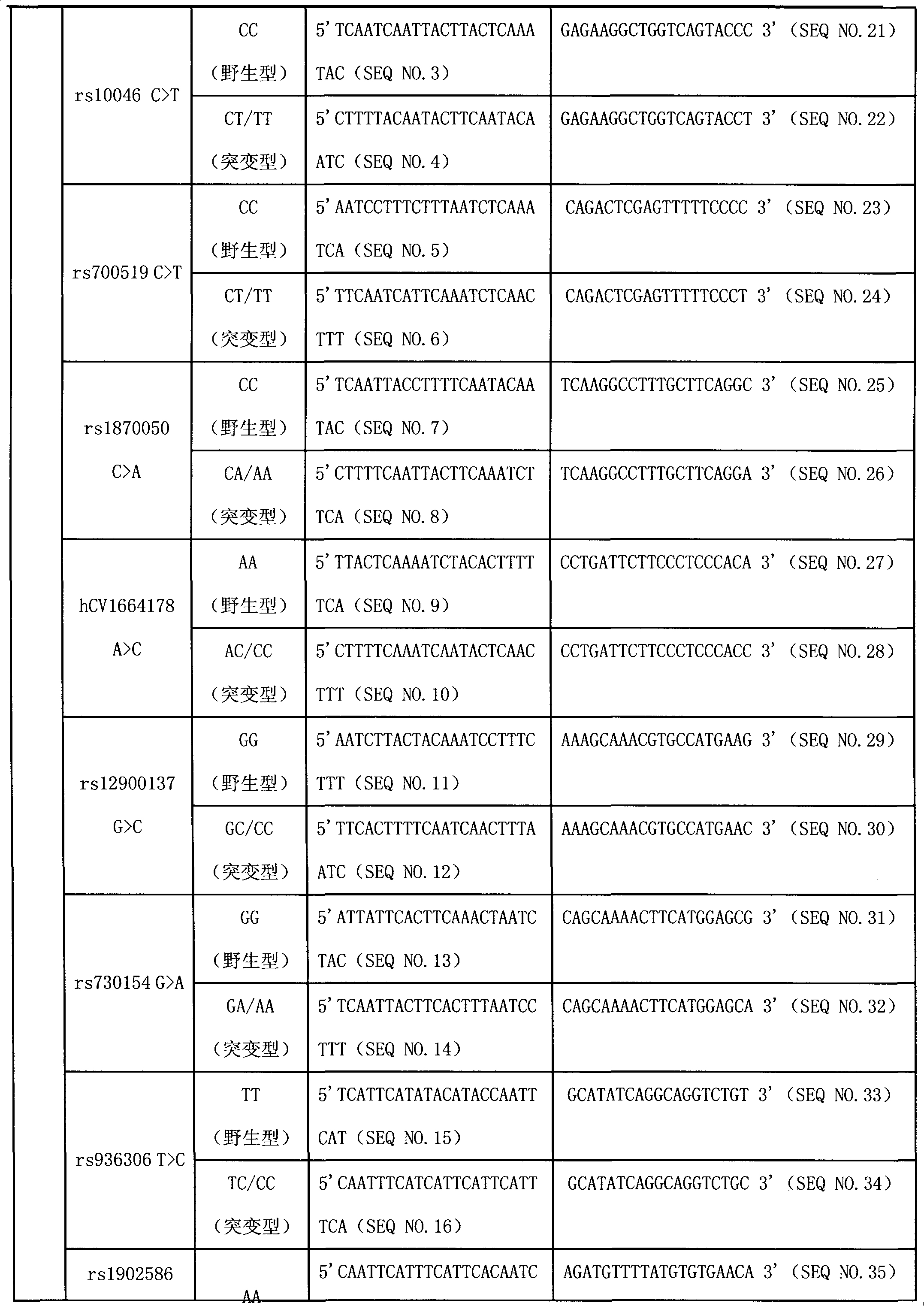
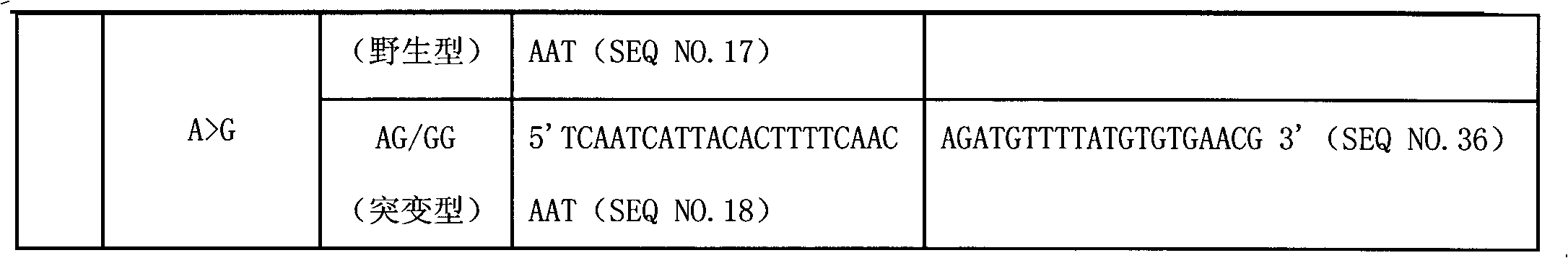
Liquid phase chip for CYP19A1 gene SNP (Single Nucleotide Polymorphism) detection and detection method thereof
ActiveCN101781684AImprove signal-to-noise ratioImplement parallel detectionMicrobiological testing/measurementType specificWild type
The invention discloses a liquid phase chip for CYP19A1 gene SNP (Single Nucleotide Polymorphism) detection, which comprises wild type and mutant type specific ASPE primers designed targeting CYP19A1 gene SNPs of rs4646, rs10046C>T, rs700519C>T, rs1870050C>A, hCV1664178A>C, rs12900137G>C, rs730154G>A, rs936306T>C and rs1902586A>G, microballoon spheres respectively coated with specific anti-tag sequences and primers used for amplifying CYP19A1 gene SNPs with target sequences to be detected. Each ASPE primer comprises a tag sequence at 5' end and a specific primer sequence at 3' end, wherein the specific primers are selected from SEQ ID NO. 19-36 and the tag sequences are selected from SEQ ID NO.1-18; and the anti-tag sequences can be correspondingly in complementary pairing with the tag sequences. The invention also discloses a CYP19A1 gene SNP detection method. The coincidence rate of the detection method provided by the invention and a sequencing method is as high as 100%; and the prepared liquid phase chip for CYP19A1 gene SNP (Single Nucleotide Polymorphism) detection has excellent signal-to-noise rate, and basically no cross reaction exists between a design probe and the anti-tag sequences.
Owner:SUREXAM BIO TECH
Fluorescent quantitative PCR (Polymerase Chain Reaction) detection reagent, kit and detection method for African swine fever virus (ASFV)
ActiveCN103757134AImprove throughputNo biological safety hazardMicrobiological testing/measurementDimethyl pyrocarbonateBiology
The invention discloses a fluorescent quantitative PCR (Polymerase Chain Reaction) detection reagent, a kit and a detection method for an African swine fever virus (ASFV). The detection reagent is targeted to a conserved segment of an ASFV VP72 gene, and comprises a pair of specific primers, a specific probe and an internal labeling probe. The kit comprises a PCR mixed solution, Taq DNA polymerase, DEPC (diethylpyrocarbonate)-H2O, an ASFV positive quality product, an ASFV negative quality product, a quantification standard substance and a pseudovirus internal labeling solution, wherein the PCR mixed solution comprises a pair of specific primers, a specific probe and an internal labeling probe. The detection method comprises the steps of extraction of total DNAs, preparation of reaction components, making of a standard curve by diluting the standard substance, amplifying and result judgment. The reagent and the kit have the advantages of high specificity, high sensitivity, no pollution and high flux, and can be used for accurately detecting infection, inapparent infection or continuous carrying of a toxic host of low-content ASFV. The detection method is quick, specific and sensitive, and can be used for quantitatively detecting a large quantity of samples at the same time.
Owner:深圳澳东检验检测科技有限公司
Kit for detecting guinea pig aeromonas by utilizing Loop-mediated isothermal amplification technique
InactiveCN101235411AAccurate identificationRapid identificationMicrobiological testing/measurementAeromonas caviaeLoop-mediated isothermal amplification
The invention relates to a reagent kit and a detecting method for using loop-mediated isothermal amplification (LAMP) technique to detect aeromonas caviae, which belongs to the pathogen diagnosis field. The main technical scheme of the invention comprises: utilizing the LAMP technique, designing four pairs of high specificity primers aiming at six zones of 16s and 23s ribosome RNA gene transcription spacer region, and achieving the purpose for detecting the aeromonas caviae with specificity. Compared with a traditional method, equipment and operational steps which are needed in the method are simple, the method has the advantages of rapidness and high specificity, and the detecting cost is lower, which is suitable for common clinics and field detections.
Owner:NANKAI UNIV
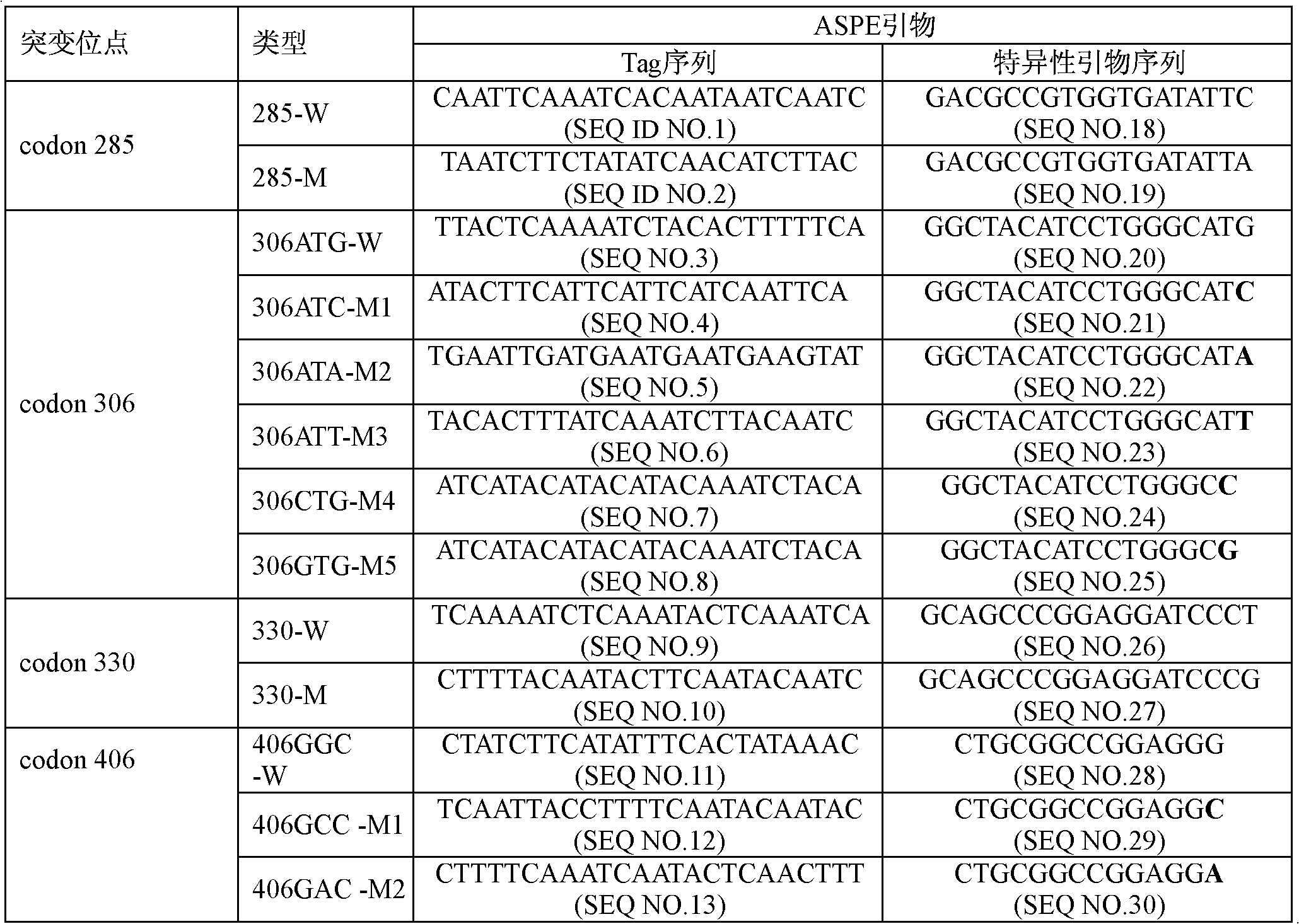
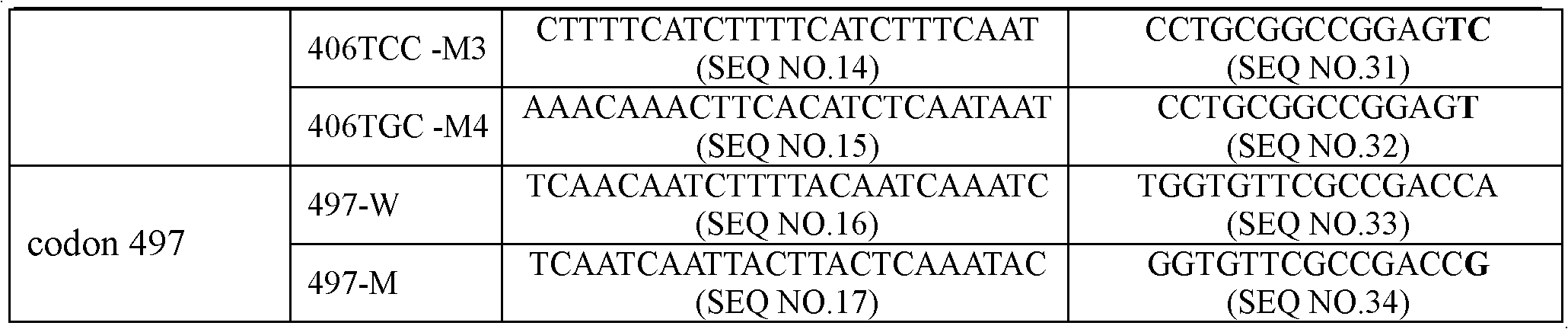
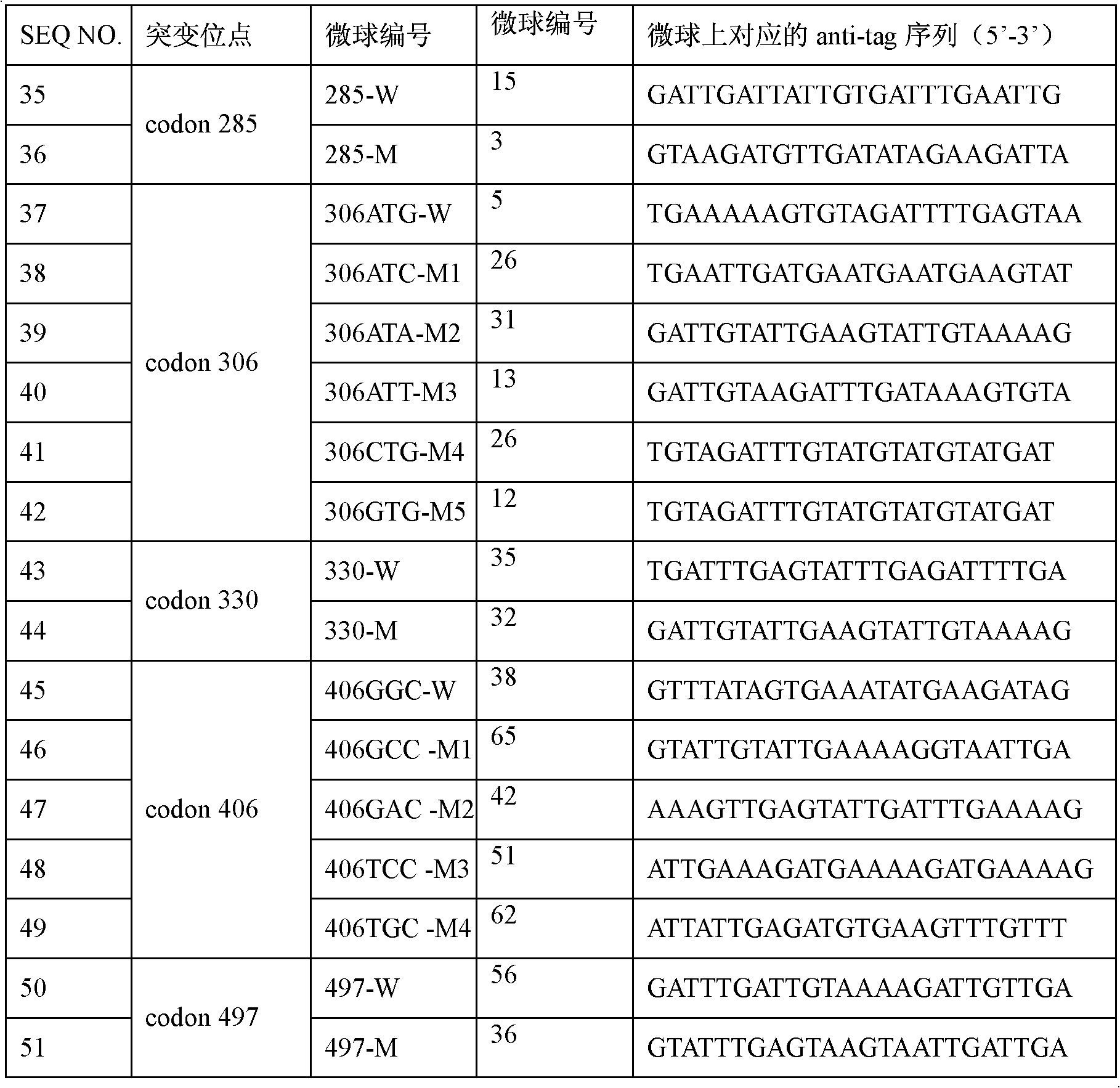
Emb B gene mutation detection specific primers and liquid phase chip
InactiveCN102010906AImprove signal-to-noise ratioImplement parallel detectionMicrobiological testing/measurementDNA/RNA fragmentationMicrosphereGenetics
The invention discloses ethambutol (emb) B gene mutation detection specific primers and a liquid phase chip. The liquid phase chip comprises ASPE primers, microspheres and amplification primers, wherein the specific primers of the ASPE primers are selected from SEQ ID No.18 and SEQ ID No.19 at the codon 285th site, SEQ ID No.20 and more than one of sequences from SEQ ID No.21-25 at the codon 306th site, SEQ ID No.26 and SEQ ID No.27 at the codon 330th site, SEQ ID No.28 and more than one of sequences from SEQ ID No.29-32 at the codon 406th site, and / or SEQ ID No.33 and SEQ ID No.34 at the codon 497th site. The detection result of the emb B gene mutation detection liquid phase chip provided by the invention is completely matched with that of a sequencing method, and the liquid phase chip has good signal-noise ratio.
Owner:SUREXAM BIO TECH
Kit and method for staphylococcus aureus CRISPR locus detection
InactiveCN106167821AEasy to operateIncreased sensitivityMicrobiological testing/measurementMicroorganism based processesStaphylococcus cohniiTrue positive rate
The invention discloses a kit and method for staphylococcus aureus CRISPR locus detection. The kit comprises three pairs of specific primers designed according to three CRISPR locus gene sequences of staphylococcus aureus. The invention further discloses the method for staphylococcus aureus CRISPR locus detection. The three pairs of specific primers are utilized to amplify corresponding CRISPR fragments from sample genome DNAs. The kit and the detection method are simple to operate, high in sensitivity and specificity and low in detection cost and have good popularization and application values.
Owner:ZHENGZHOU UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com