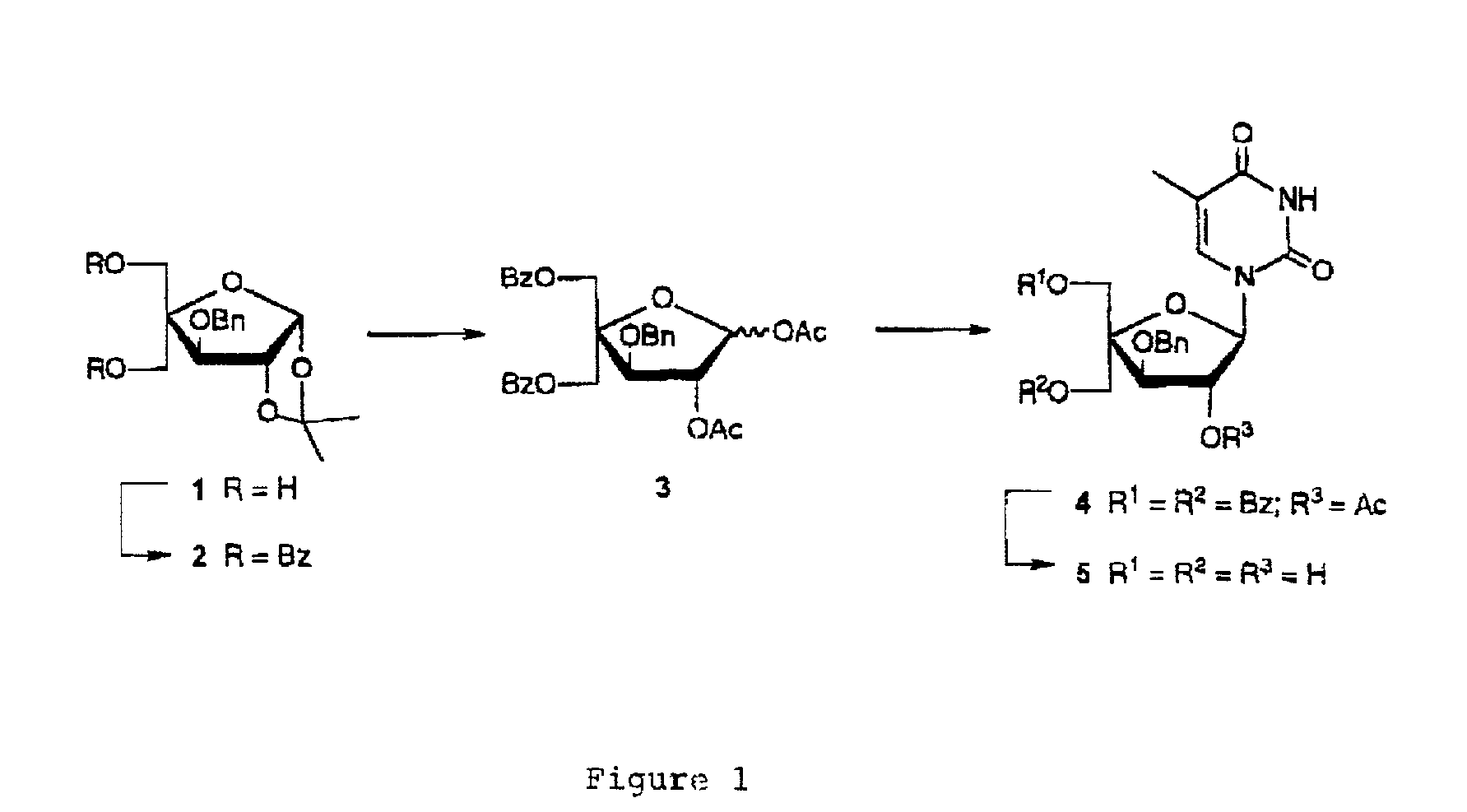
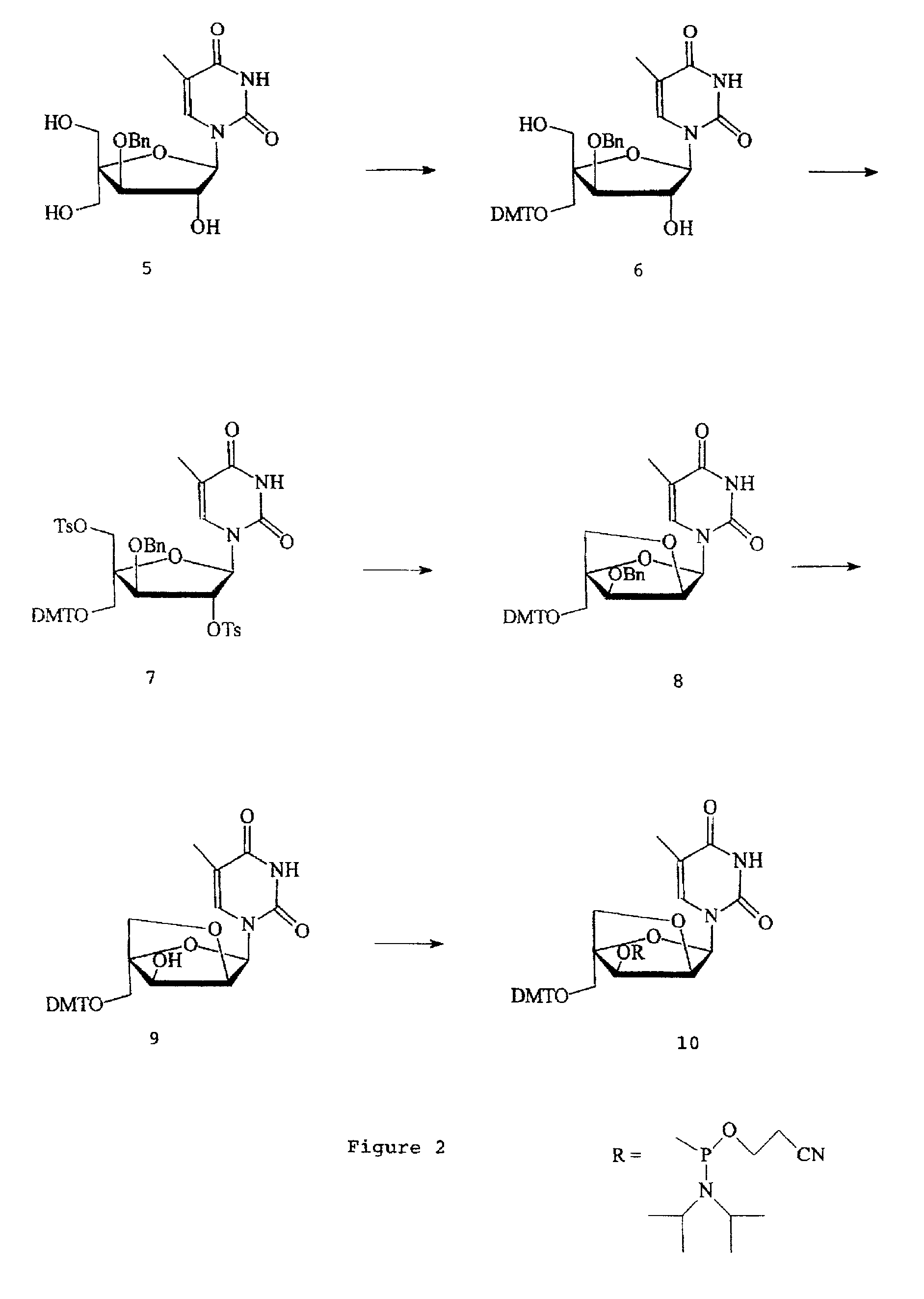
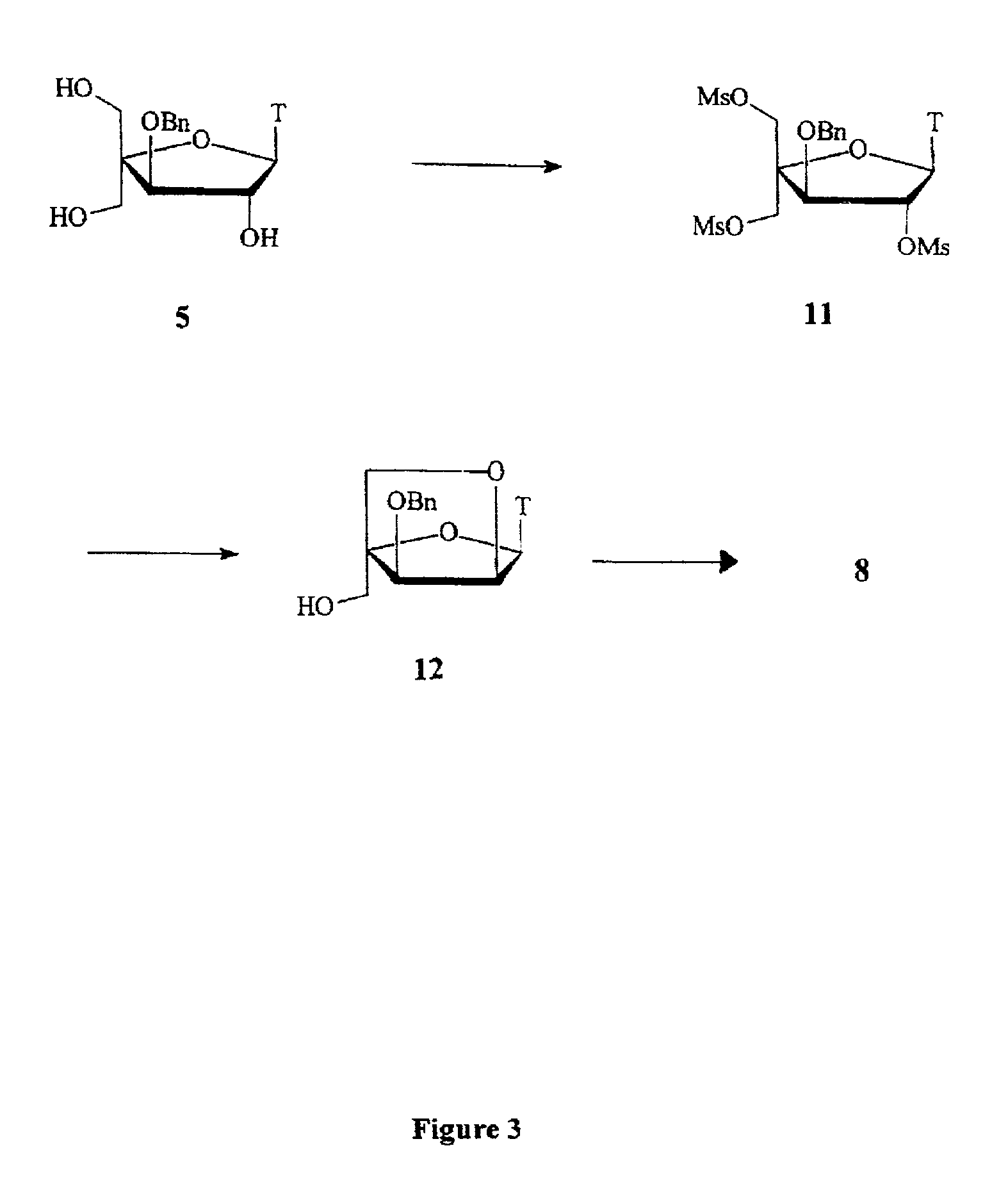
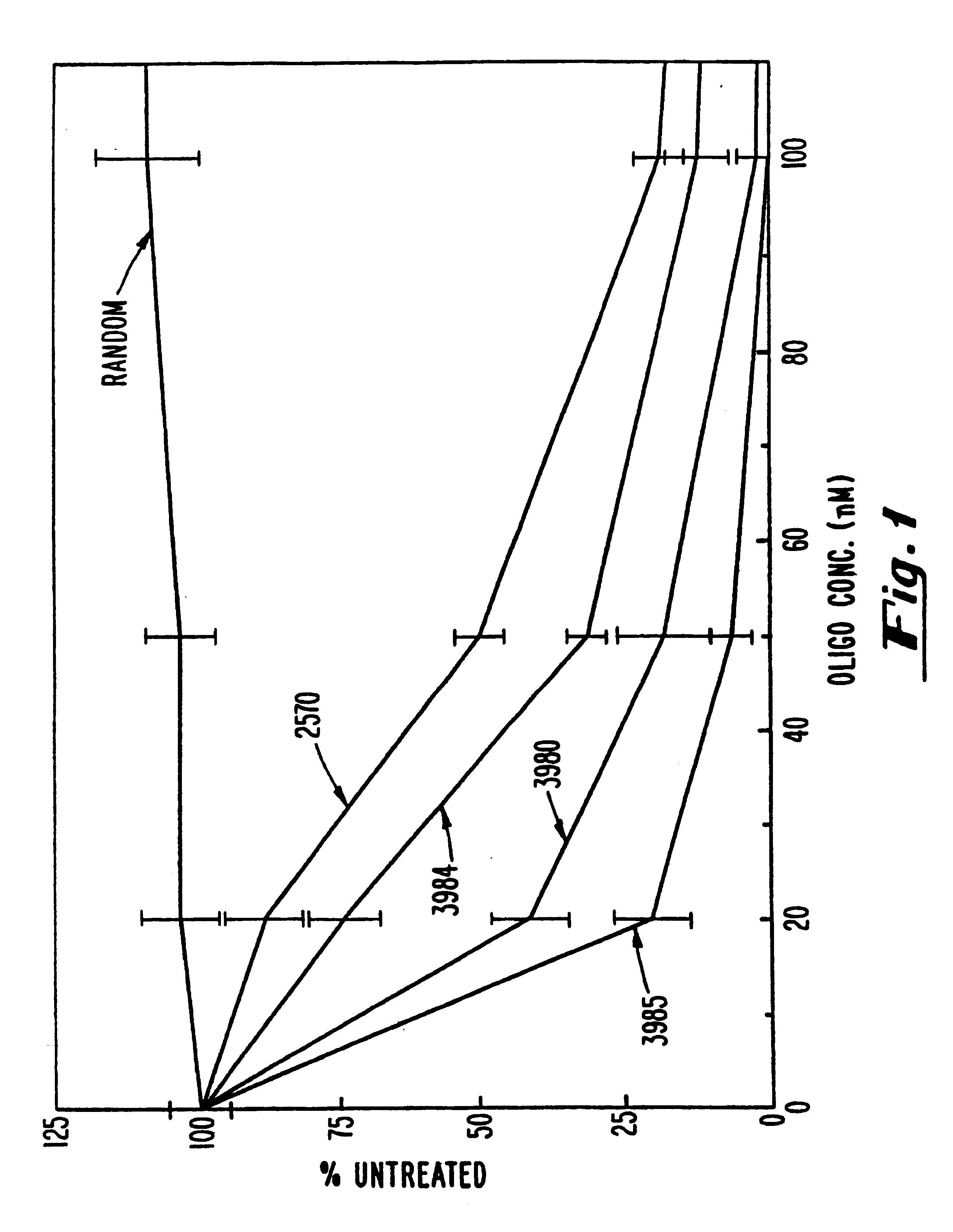
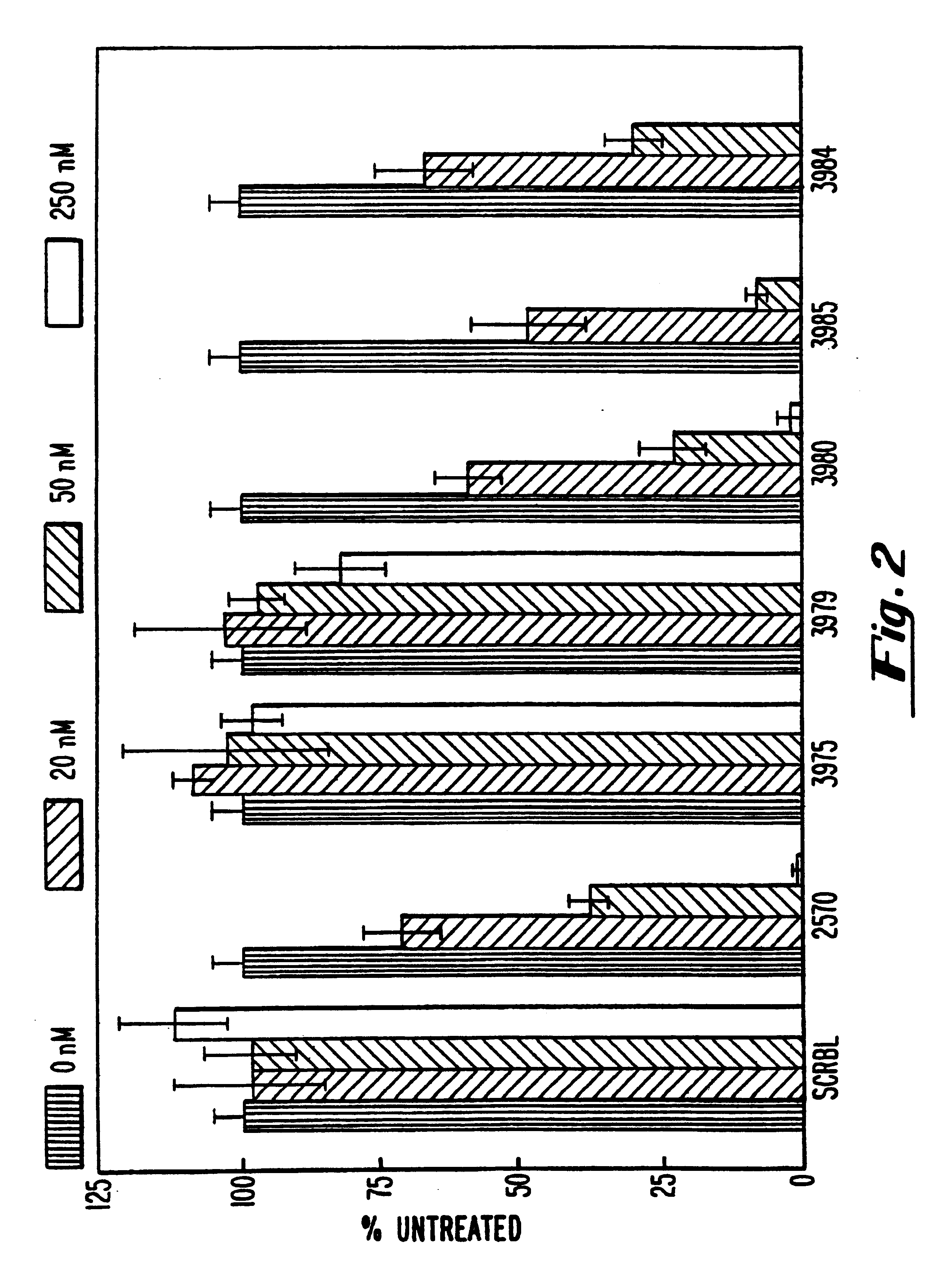
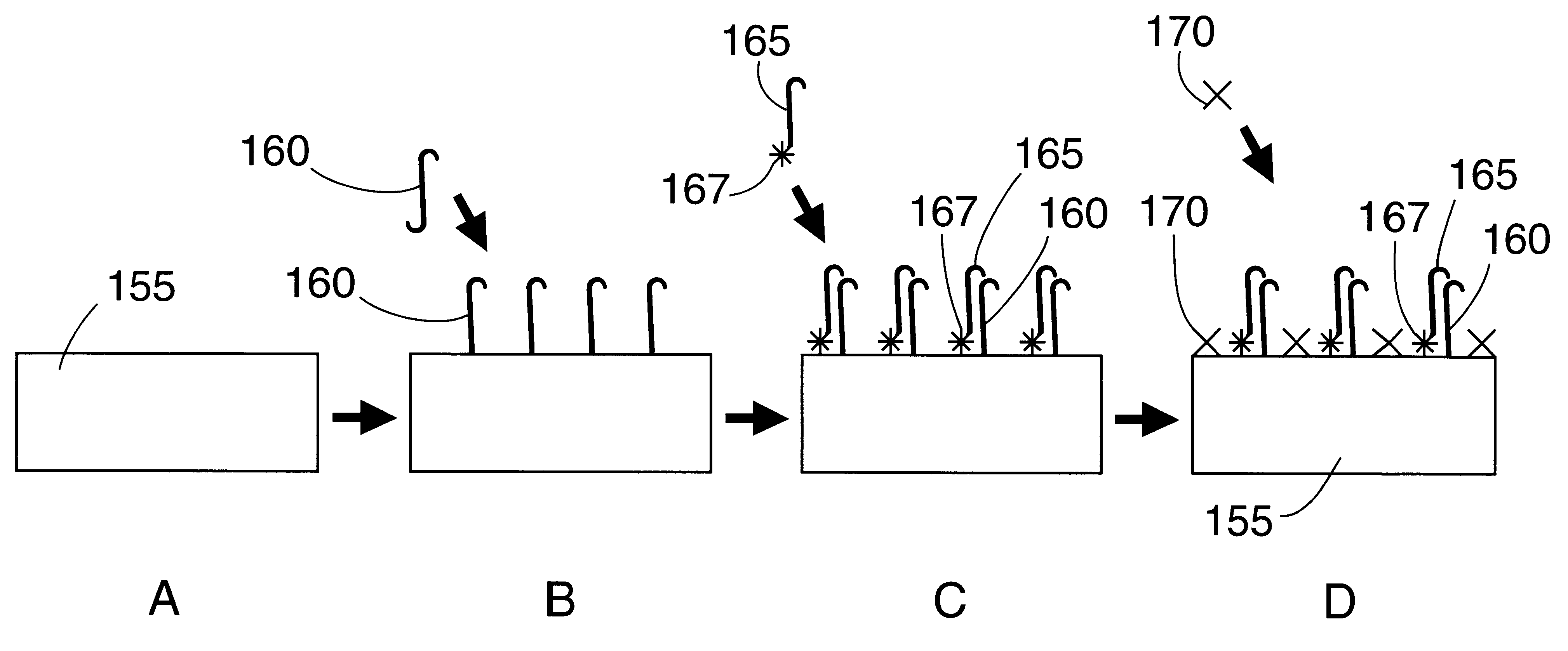
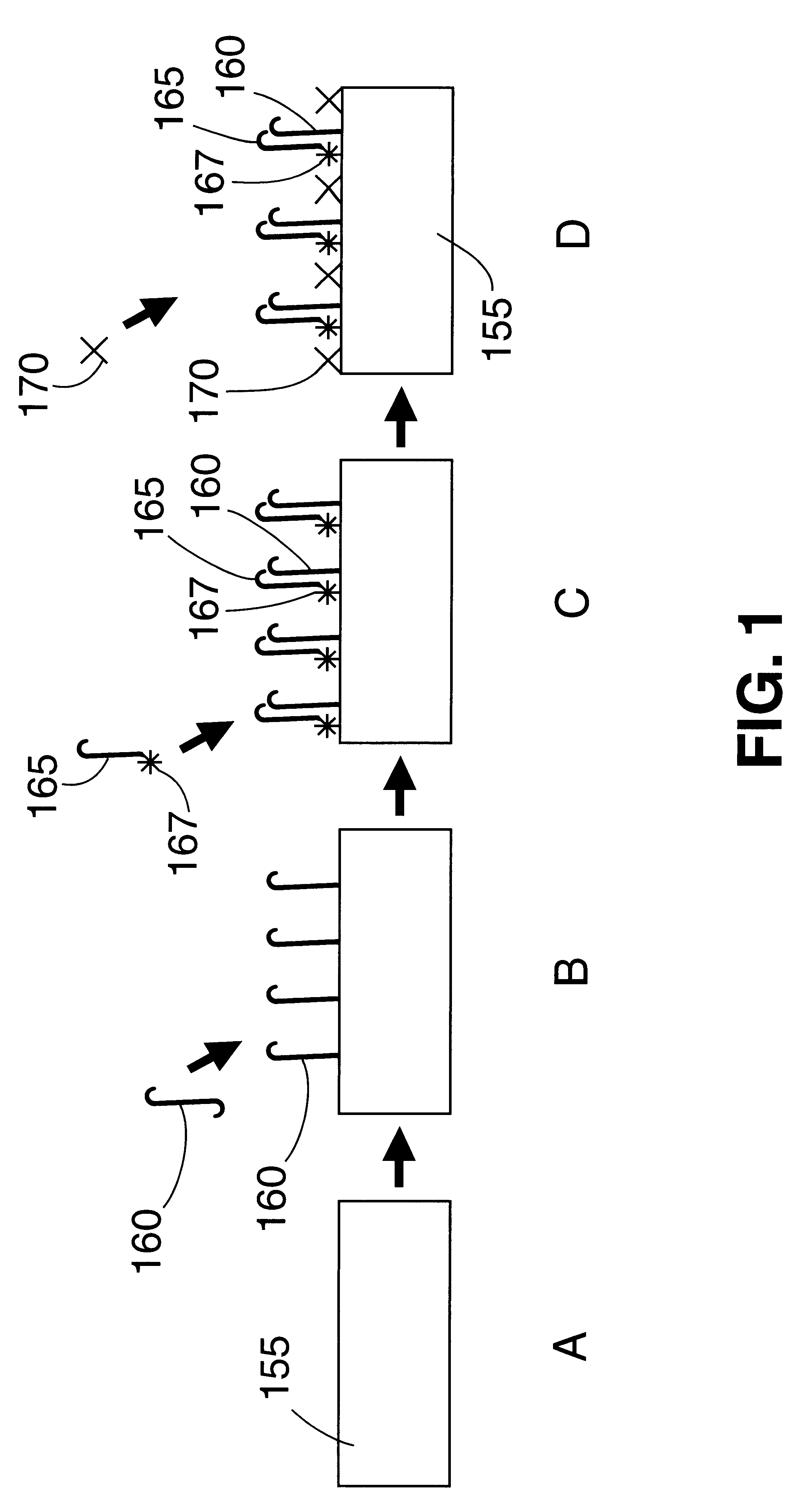
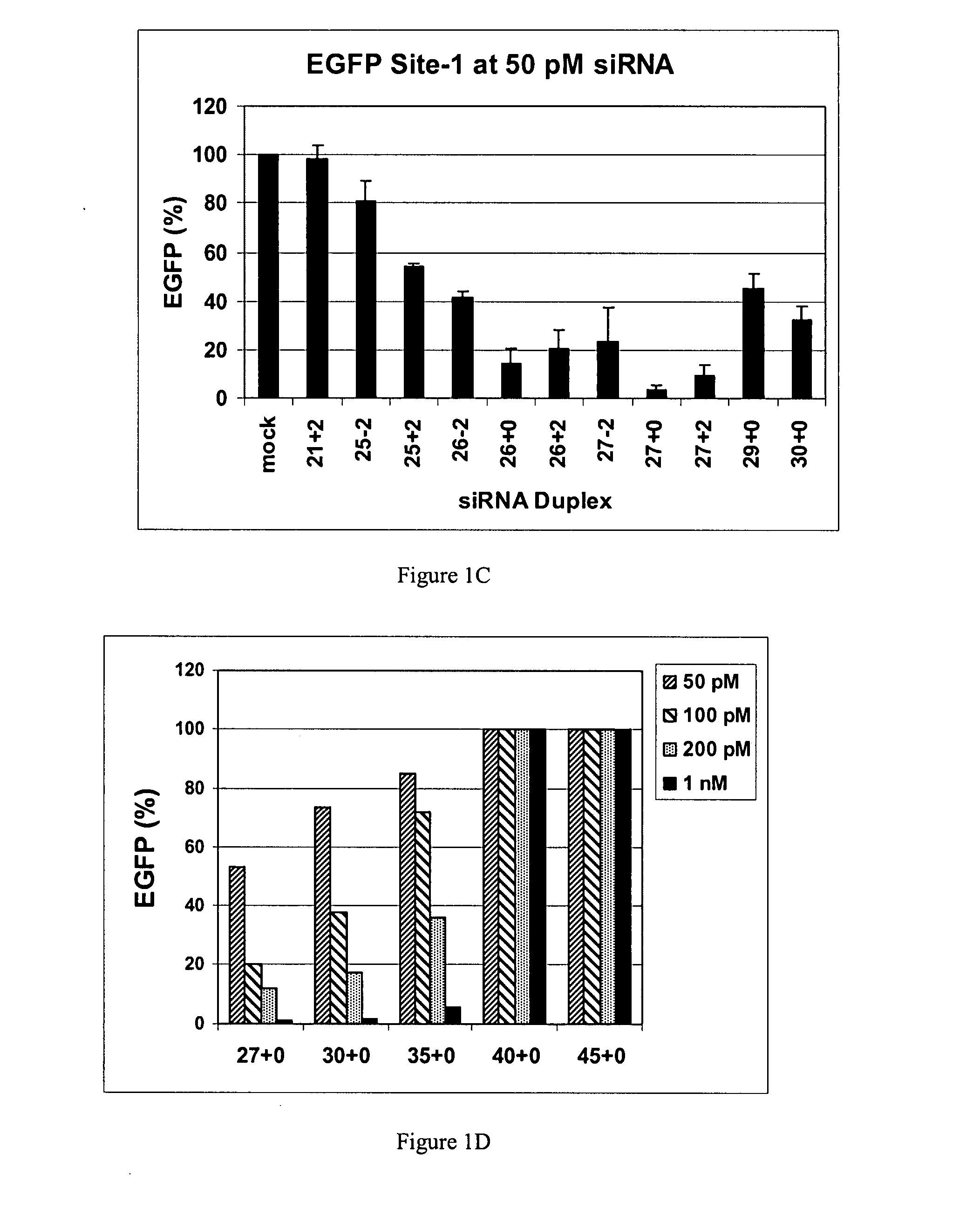
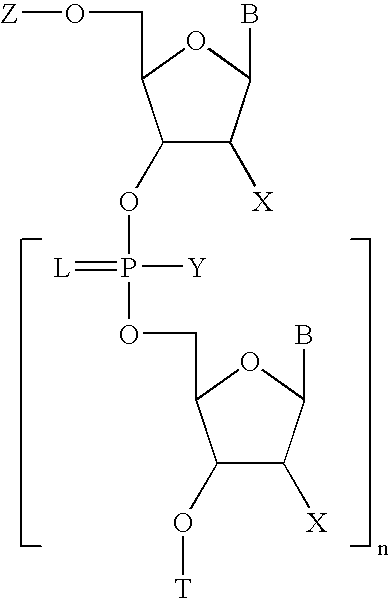
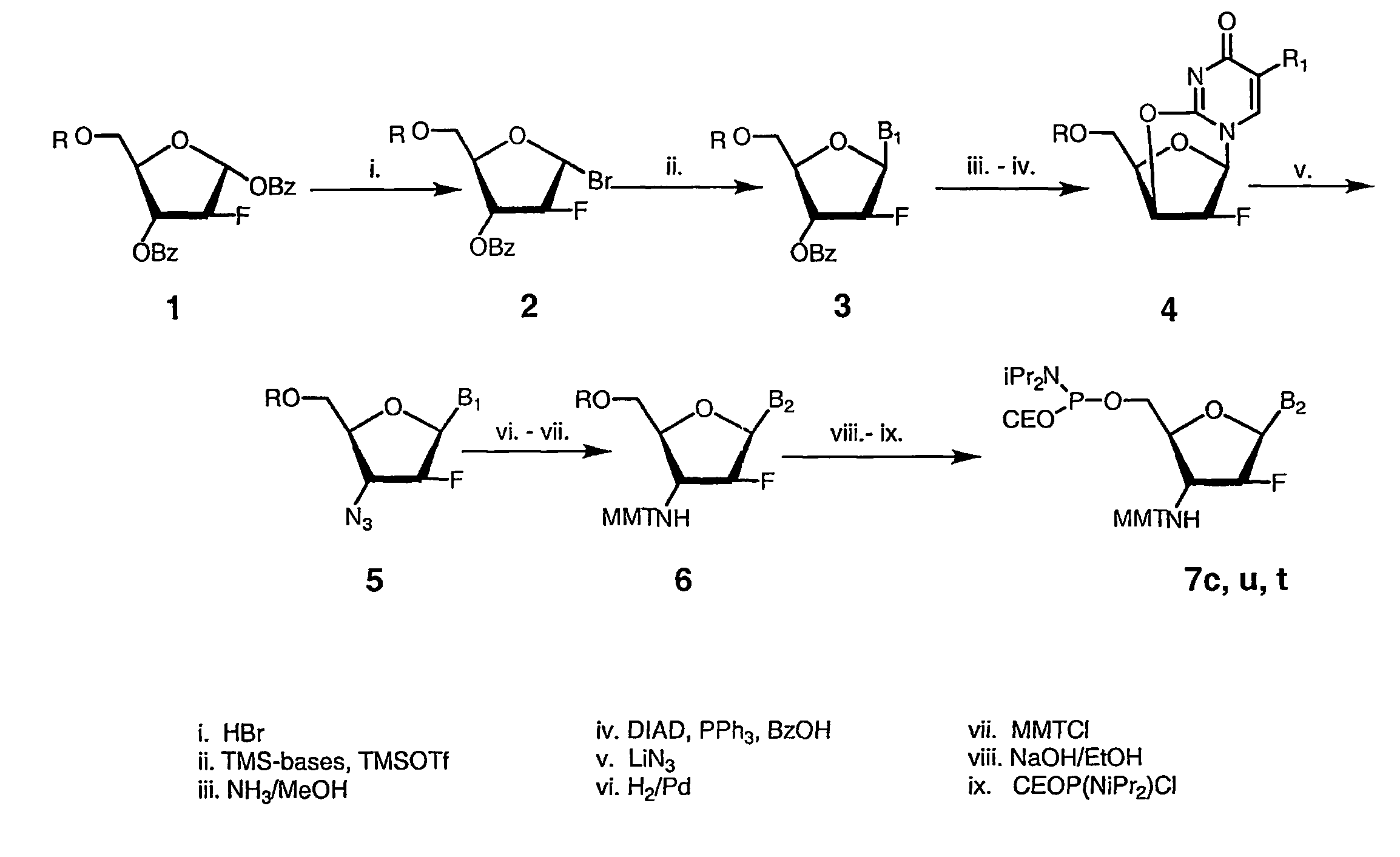
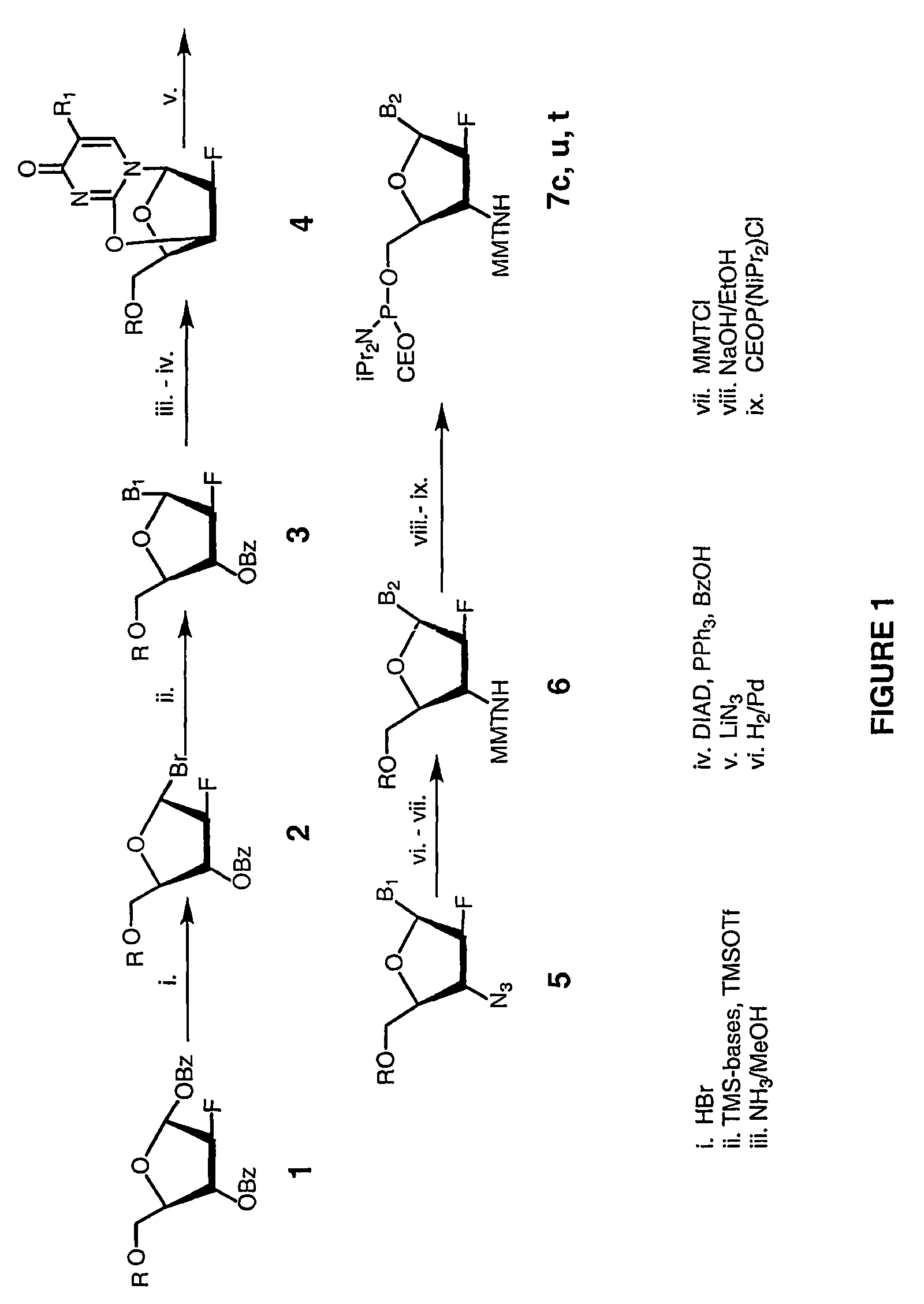
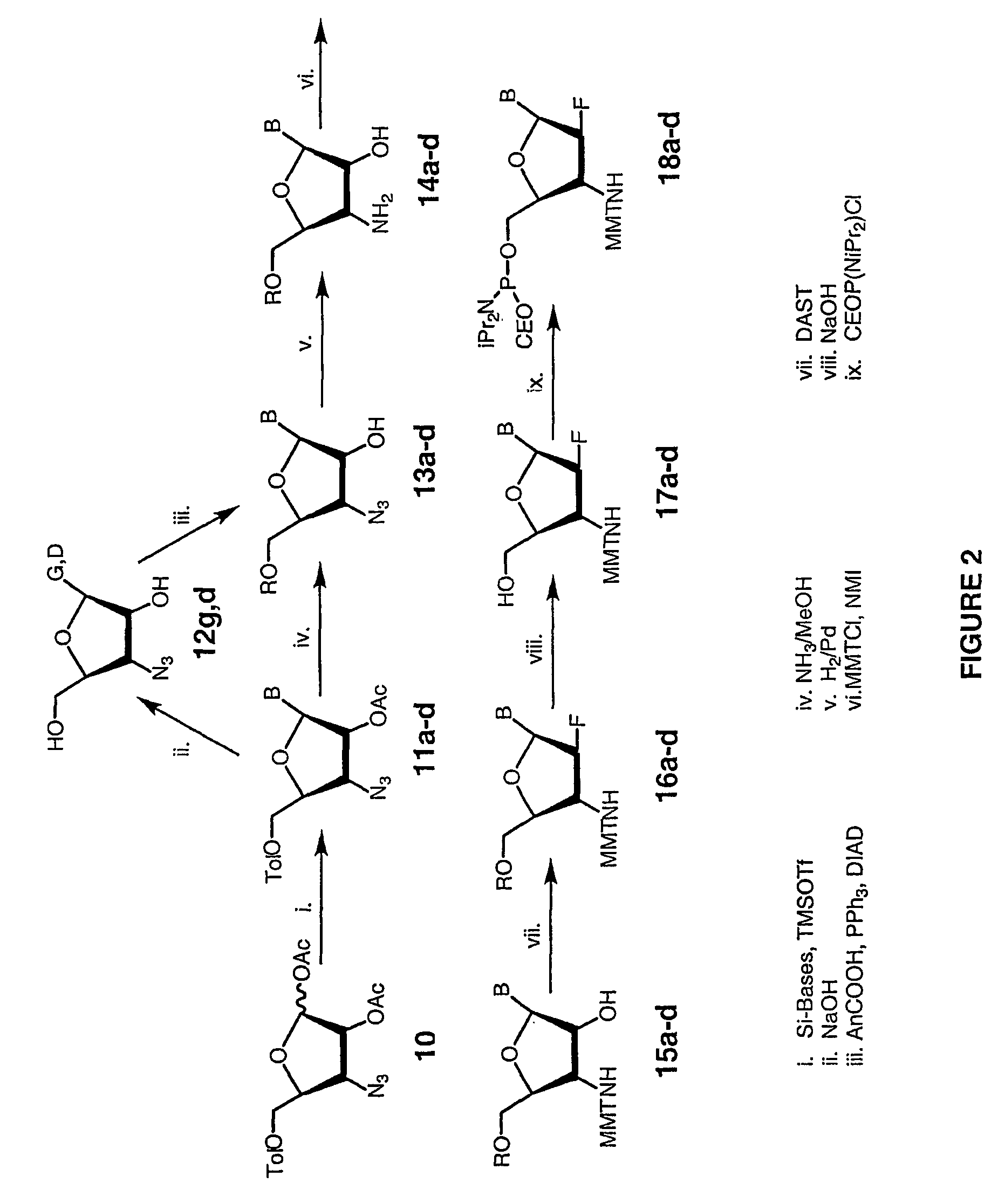
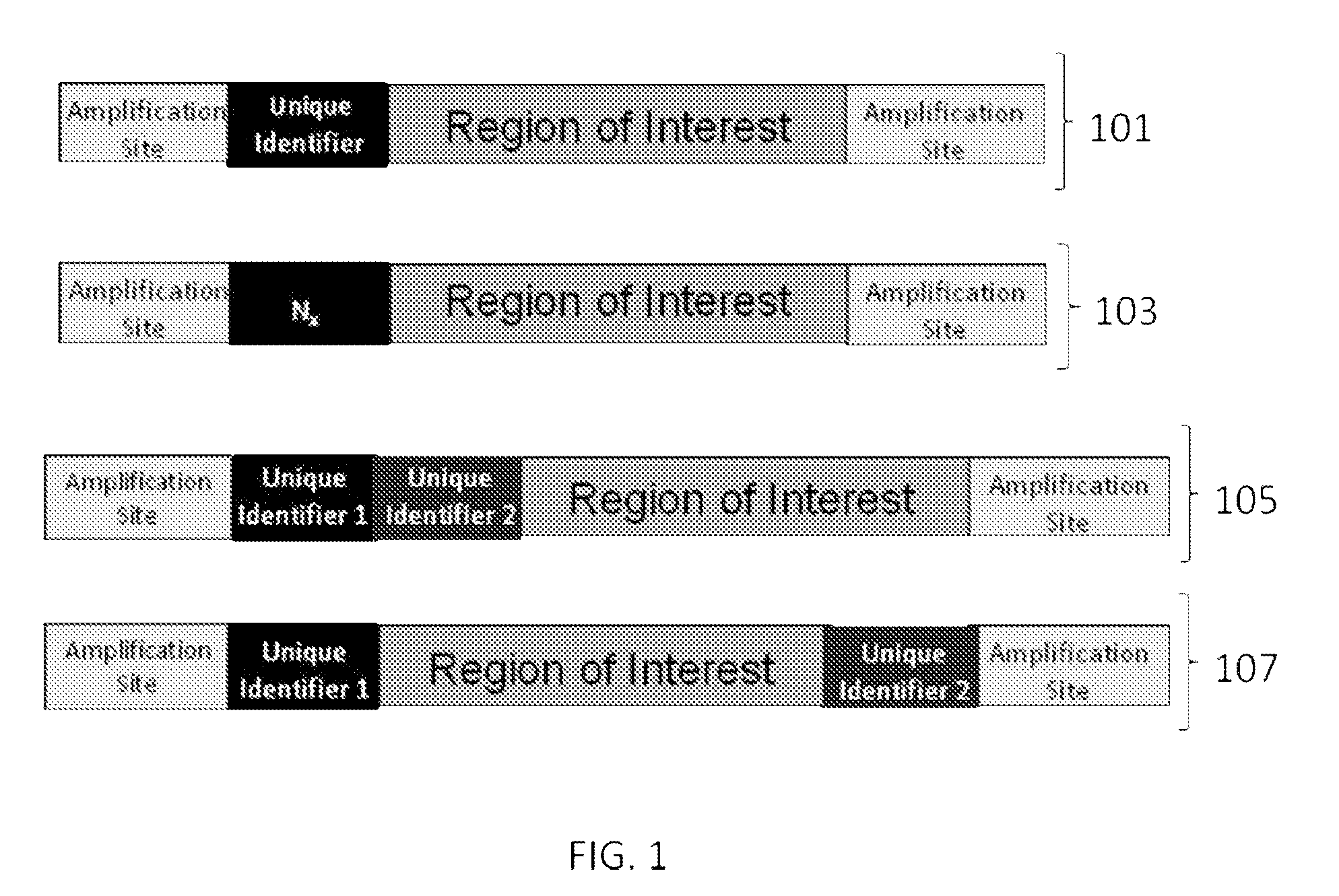
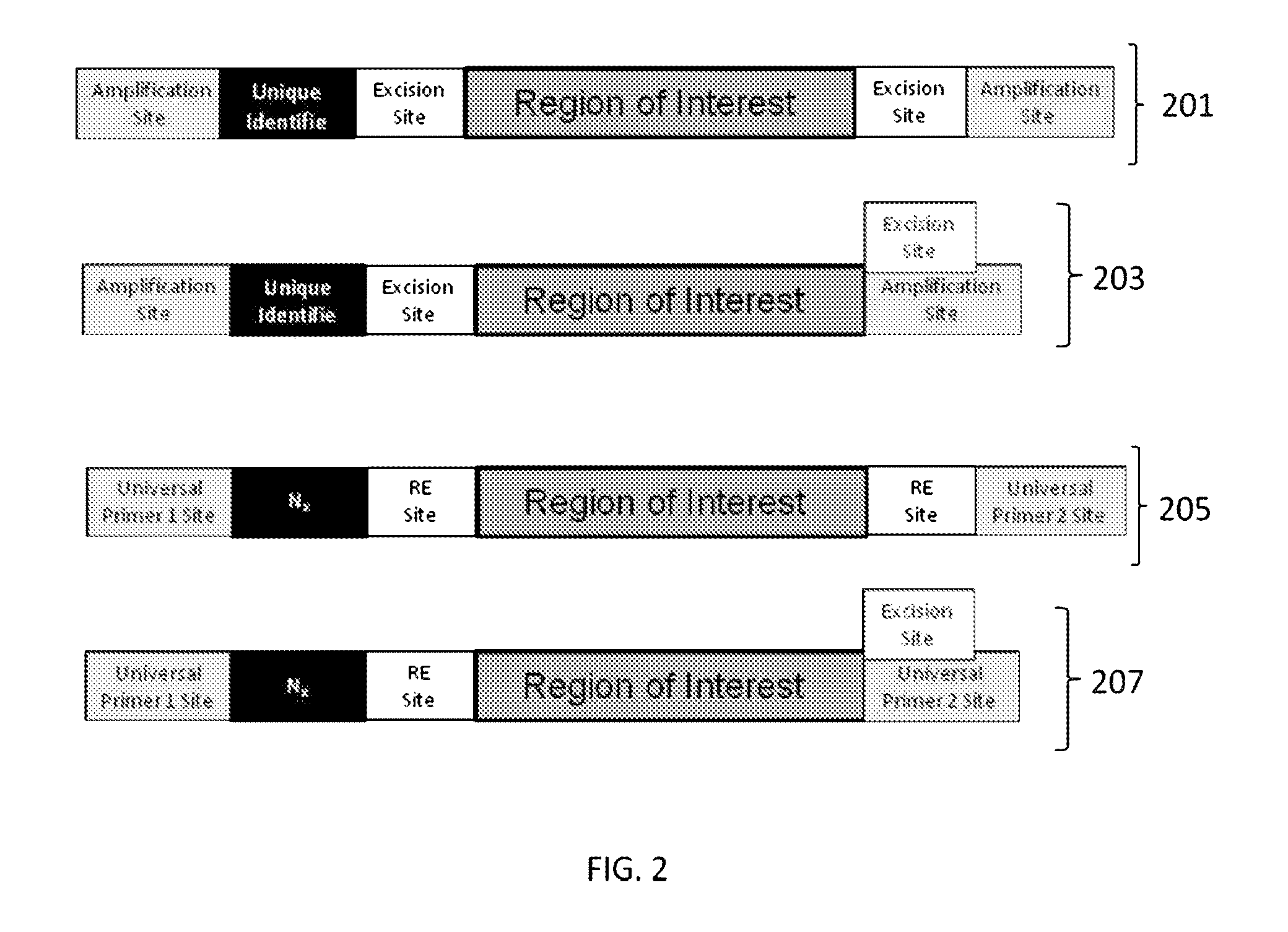
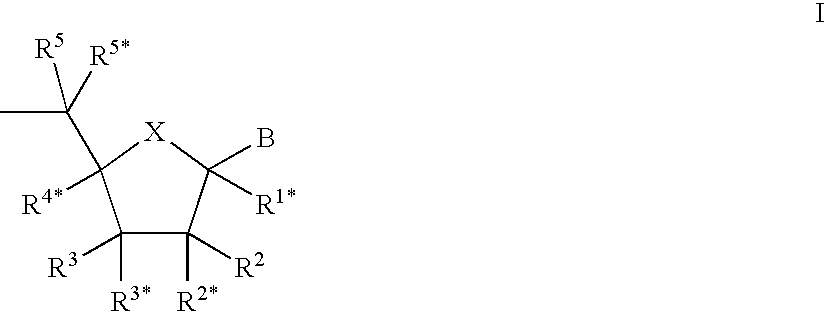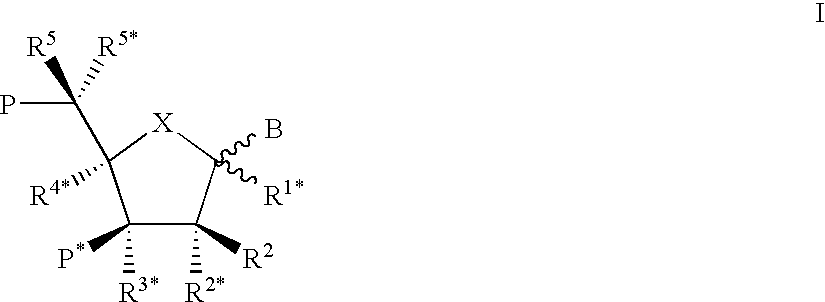Patents
Literature
6054 results about "Oligonucleotide" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
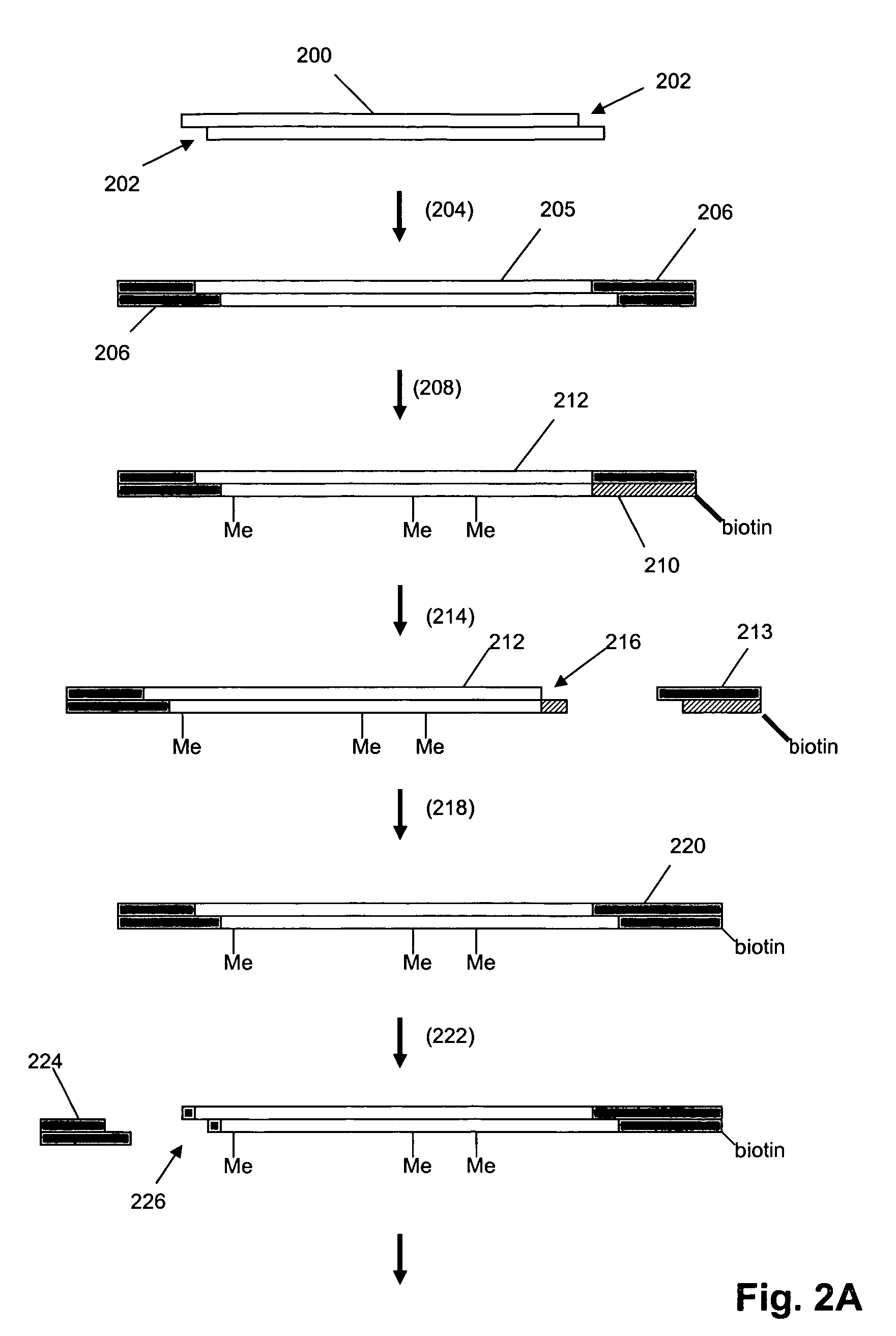
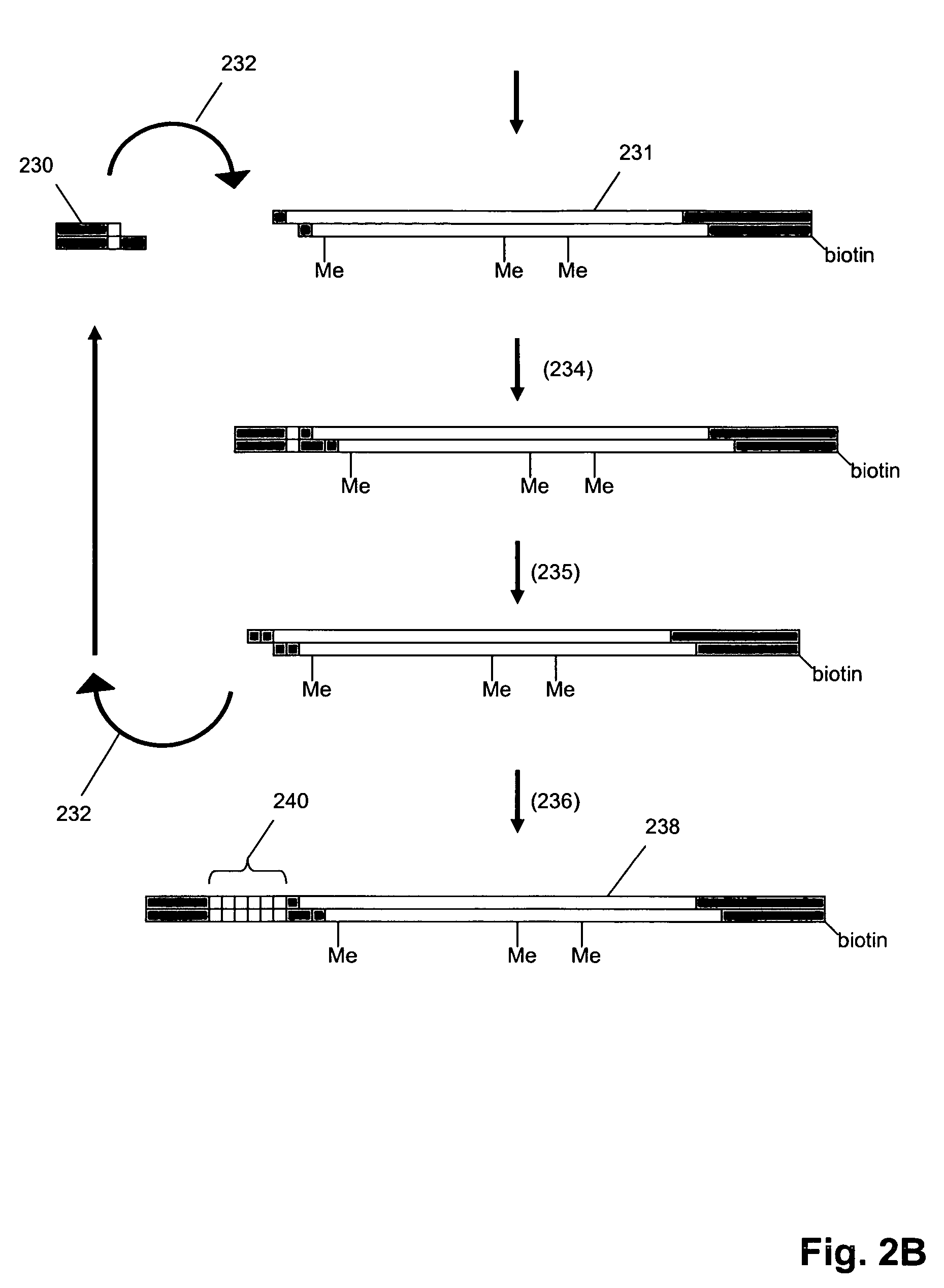
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, library construction and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.
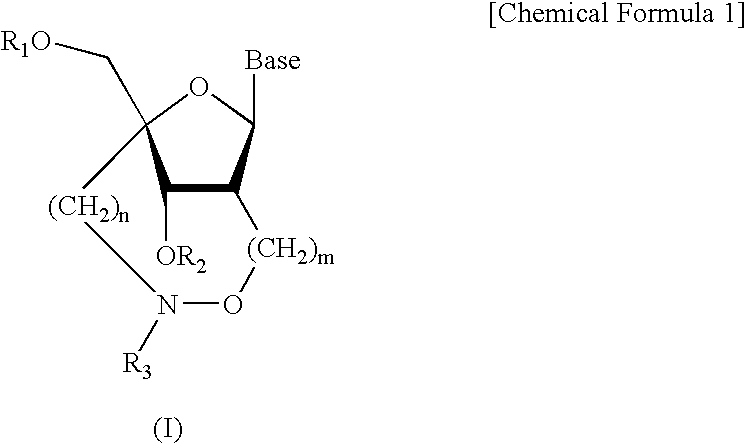
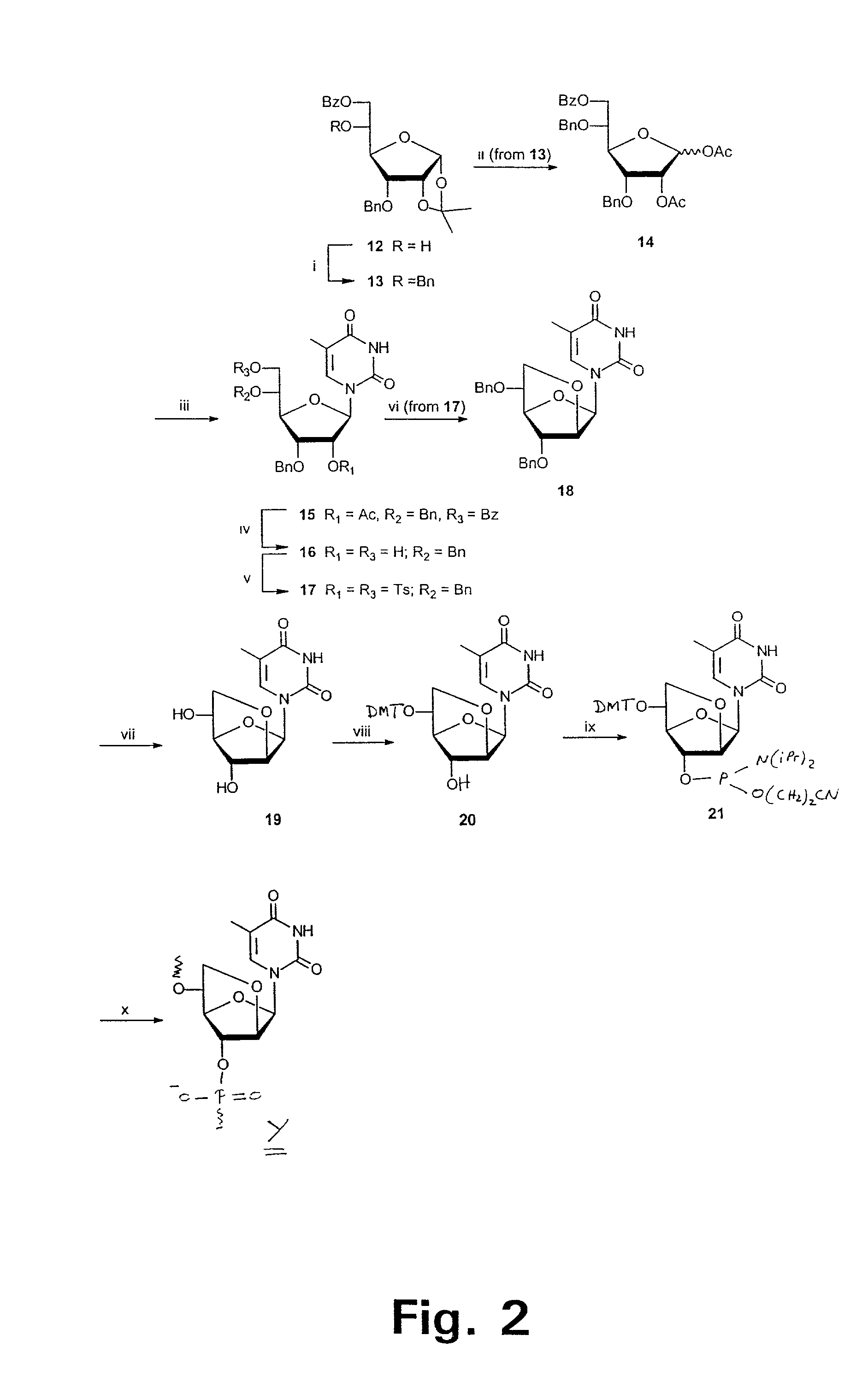
L-ribo-LNA analogues
Provided are L-ribo bicyclic nucleotide compounds as well as syntheses of such compounds. The nucleoside compounds of the invention are useful in forming oligonucleotides that can produce nucleobase specific duplexes with complementary single stranded and double stranded nucleic acids.
Owner:SANTARIS PHARMA AS
Oligonucleotide analogues
Novel oligomers, and synthesis thereof, comprising one or more bi-, tri, or polycyclic nucleoside analogues are disclosed herein. The nucleoside analogues have a “locked” structure, termed Locked Nucleoside Analogues (LNA). LNA's exhibit highly desirable and useful properties. LNA's are capable of forming nucleobase specific duplexes and triplexes with single and double stranded nucleic acids. These complexes exhibit higher thermostability than the corresponding complexes formed with normal nucleic acids. The properties of LNA's allow for a wide range of uses such as diagnostic agents and therapeutic agents in a mammal suffering from or susceptible to, various diseases.
Owner:EXIQON AS
Artificial nucleic acids of n-o bond crosslinkage type
ActiveUS7427672B2High sensitivity analysisConfirmed its usefulnessBiocideSugar derivativesSingle strandDouble strand
Owner:RIKEN GENESIS
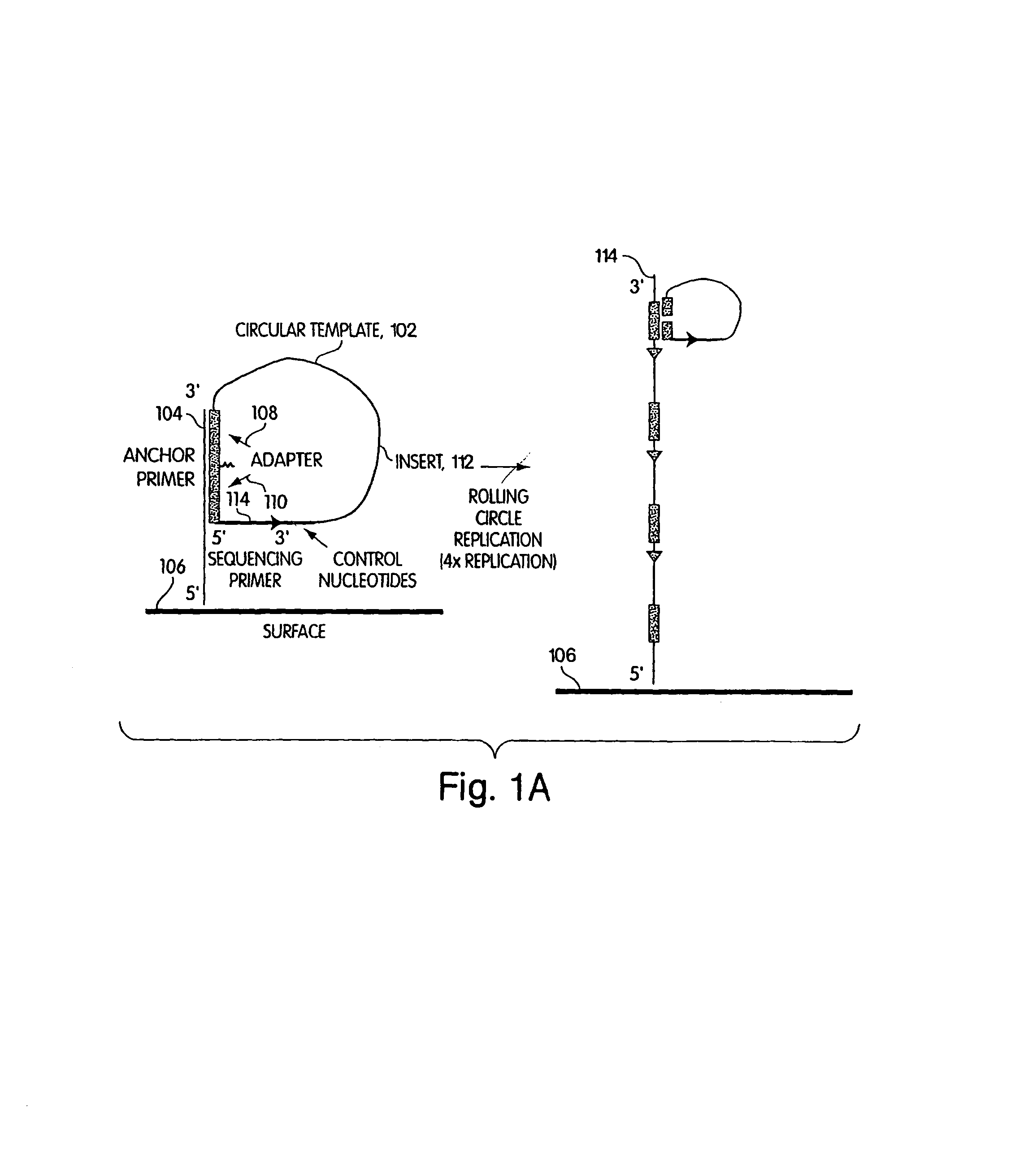
Method of sequencing a nucleic acid
InactiveUS7244559B2Bioreactor/fermenter combinationsMaterial nanotechnologyNucleic acid detectionOligonucleotide
Owner:454 LIFE SCIENCES CORP
Analying polynucleotide sequences
InactiveUS6054270AStable duplexReduce impactSequential/parallel process reactionsSugar derivativesHybridization reactionSequence determination
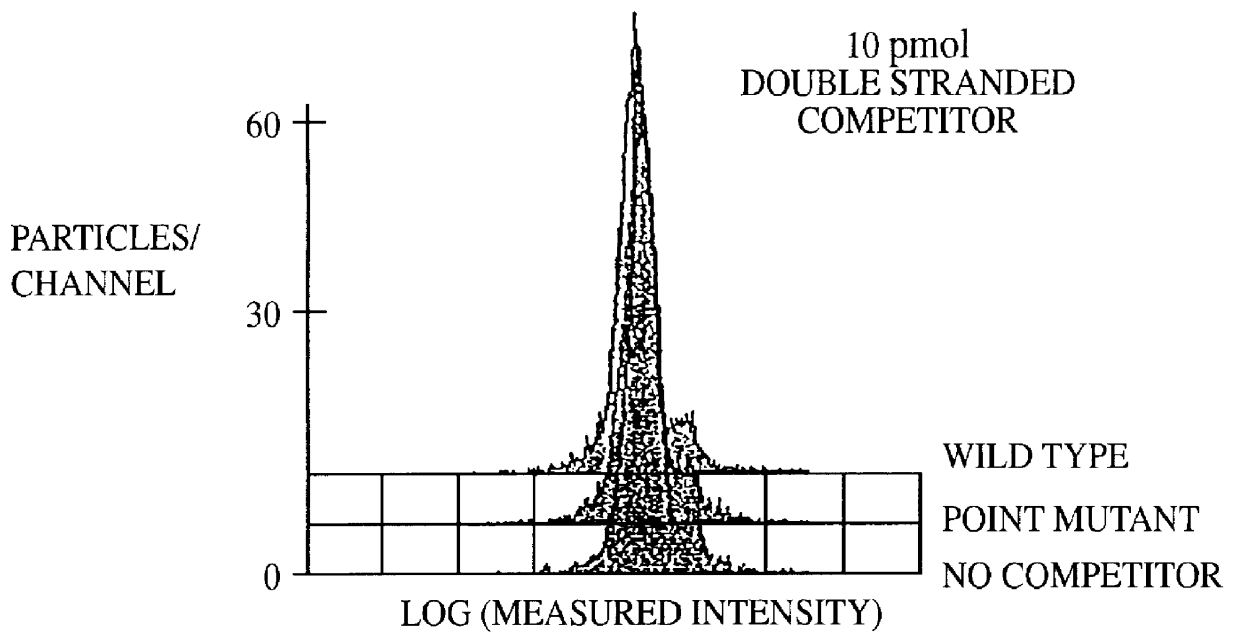
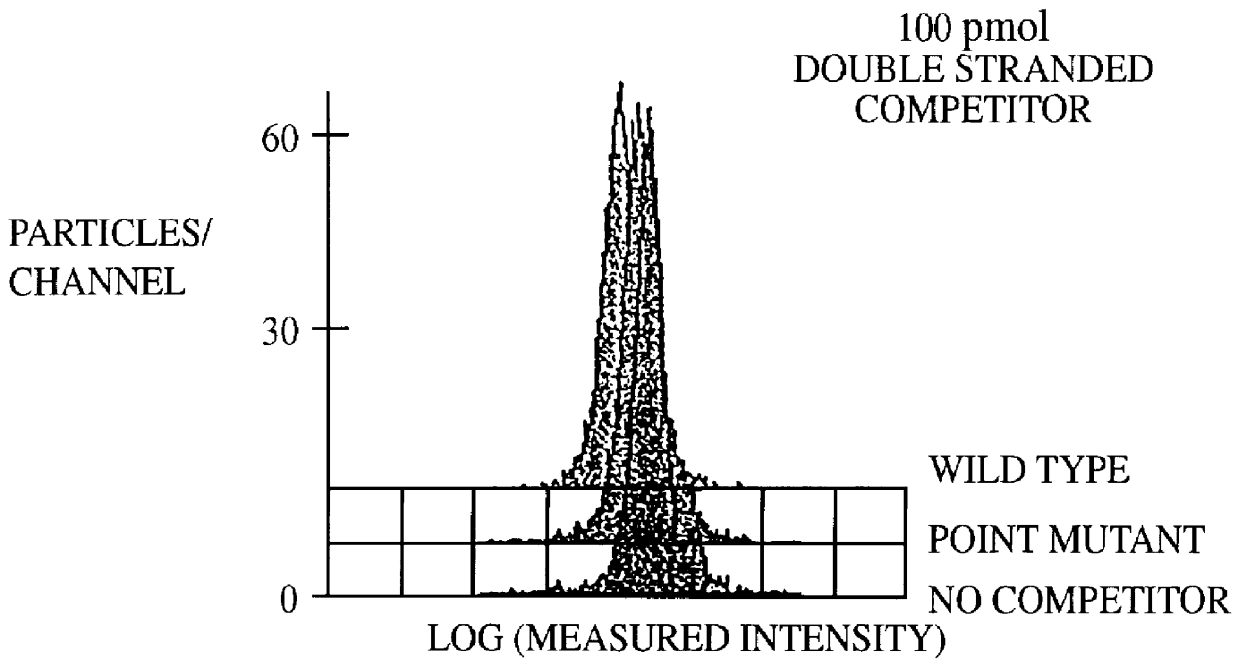
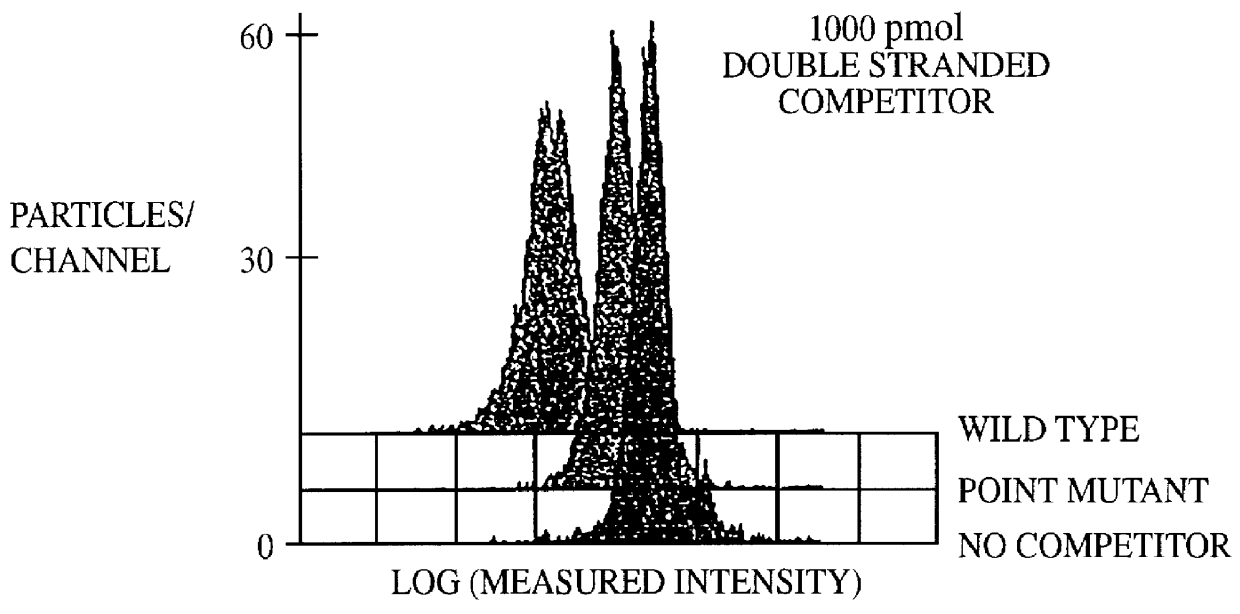
This invention provides an apparatus and method for analyzing a polynucleotide sequence; either an unknown sequence or a known sequence. A support, e.g. a glass plate, carries an array of the whole or a chosen part of a complete set of oligonucleotides which are capable of taking part in hybridization reactions. The array may comprise one or more pair of oligonucleotides of chosen lengths. The polynucleotide sequence, or fragments thereof, are labelled and applied to the array under hybridizing conditions. Applications include analyses of known point mutations, genomic fingerprinting, linkage analysis, characterization of mRNAs, mRNA populations, and sequence determination.
Owner:OXFORD GENE TECH
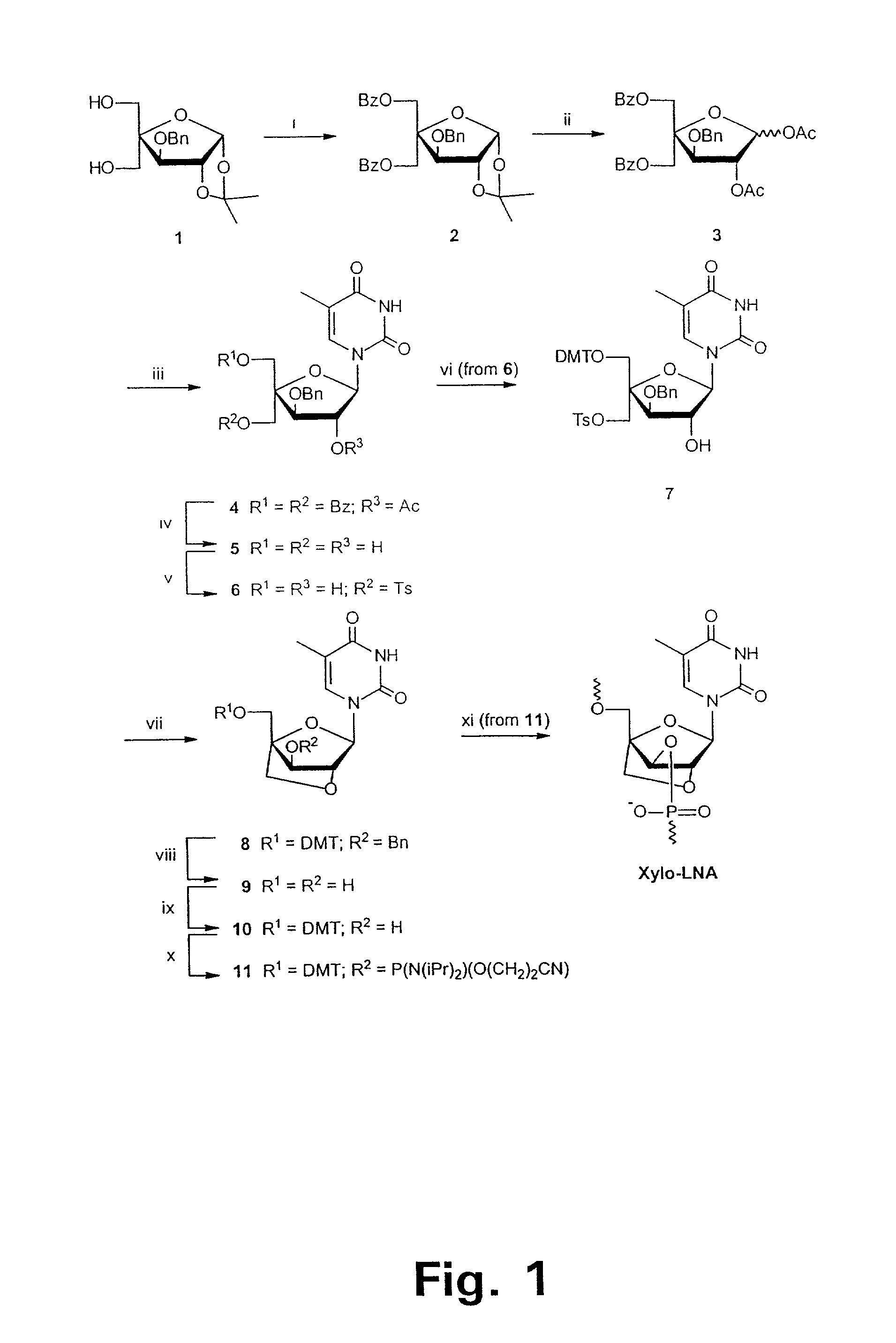
Xylo-LNA analogues
Based on the above and on the remarkable properties of the 2′-O,4′-C-methylene bridged LNA monomers it was decided to synthesise oligonucleotides comprising one or more 2′-O,4′-C-methylene-β-D-xylofuranosyl nucleotide monomer(s) as the first stereoisomer of LNA modified oligonucleotides. Modelling clearly indicated the xylo-LNA monomers to be locked in an N-type furanose conformation. Whereas the parent 2′-deoxy-β-D-xylofuranosyl nucleosides were shown to adopt mainly an N-type furanose conformation, the furanose ring of the 2′-deoxy-β-D-xylofuranosyl monomers present in xylo-DNA were shown by conformational analysis and computer modelling to prefer an S-type conformation thereby minimising steric repulsion between the nucleobase and the 3′-O-phopshate group (Seela, F.; Wömer, Rosemeyer, H. Helv. Chem. Acta 1994, 77, 883). As no report on the hybridisation properties and binding mode of xylo-configurated oligonucleotides in an RNA context was believed to exist, it was the aim to synthesise 2′-O,4′-C-methylene-β-D-xylofuranosyl nucleotide monomer and to study the thermal stability of oligonucleotides comprising this monomer. The results showed that fully modified or almost fully modified Xylo-LNA is useful for high-affinity targeting of complementary nucleic acids. When taking into consideration the inverted stereochemistry at C-3′ this is a surprising fact. It is likely that Xylo-LNA monomers, in a sequence context of Xylo-DNA monomers, should have an affinity-increasing effect.
Owner:QIAGEN GMBH
Nucleic acid amplification oligonucleotides with molecular energy transfer labels and methods based thereon
InactiveUS6117635AReduce the possibilityRapid and sensitive and reliableSugar derivativesMicrobiological testing/measurementEnergy transferFluorophore
The present invention provides labeled nucleic acid amplification oligonucleotides, which can be linear or hairpin primers or blocking oligonucleotides. The oligonucleotides of the invention are labeled with donor and / or acceptor moieties of molecular energy transfer pairs. The moieties can be fluorophores, such that fluorescent energy emitted by the donor is absorbed by the acceptor. The acceptor may be a fluorophore that fluoresces at a wavelength different from the donor moiety, or it may be a quencher. The oligonucleotides of the invention are configured so that a donor moiety and an acceptor moiety are incorporated into the amplification product. The invention also provides methods and kits for directly detecting amplification products employing the nucleic acid amplification primers. When labeled linear primers are used, treatment with exonuclease or by using specific temperature eliminates the need for separation of unincorporated primers. This "closed-tube" format greatly reduces the possibility of carryover contamination with amplification products, provides for high throughput of samples, and may be totally automated.
Owner:MILLIPORE CORP
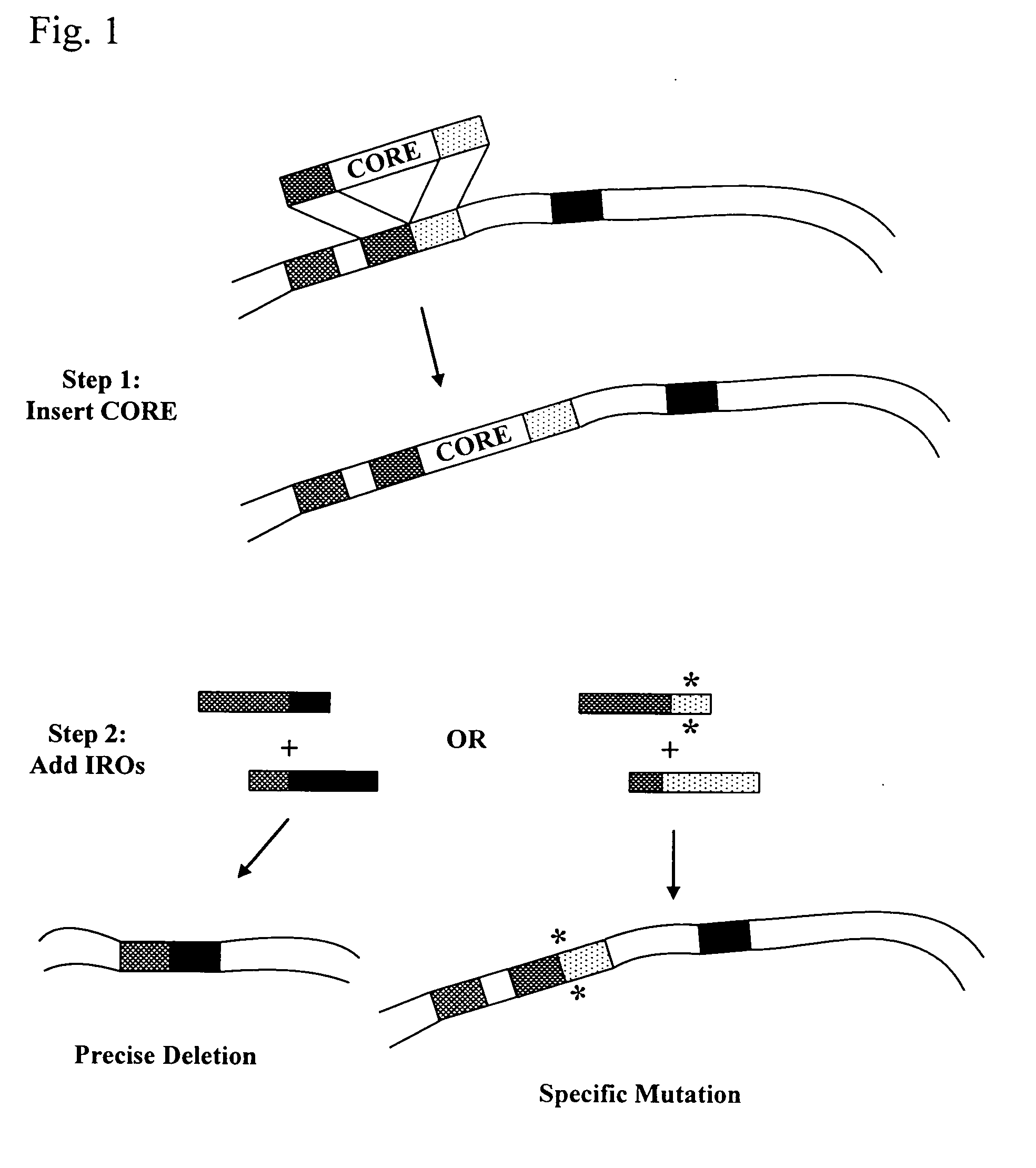
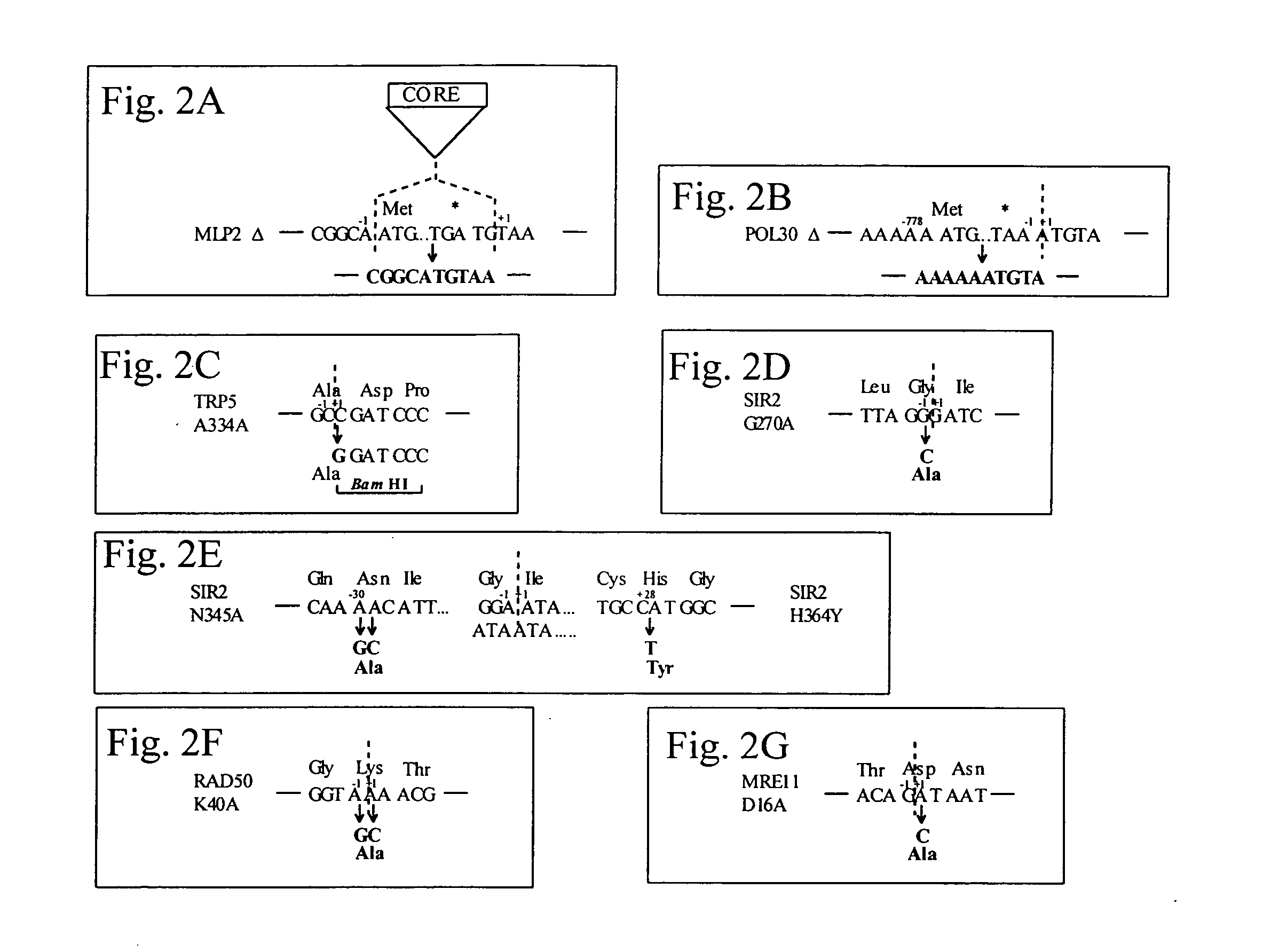
Systems for in vivo site-directed mutagenesis using oligonucleotides
InactiveUS20040171154A1Large deletionImprove applicabilitySugar derivativesMicrobiological testing/measurementHeterologousSite-directed mutagenesis
This disclosure provides several methods to generate nucleic acid mutations in vivo, for instance in such a way that no heterologous sequence is retained after the mutagenesis is complete. The methods employ integrative recombinant oligonucleotides (IROs). Specific examples of the described mutagenesis methods enable site-specific point mutations, deletions, and insertions. Also provided are methods that enable multiple rounds of mutation and random mutagenesis in a localized region. The described methods are applicable to any organism that has a homologous recombination system.
Owner:HEALTH & HUMAN SERVICES DEPT OF THE GOVERNMENT OF THE US SEC THE
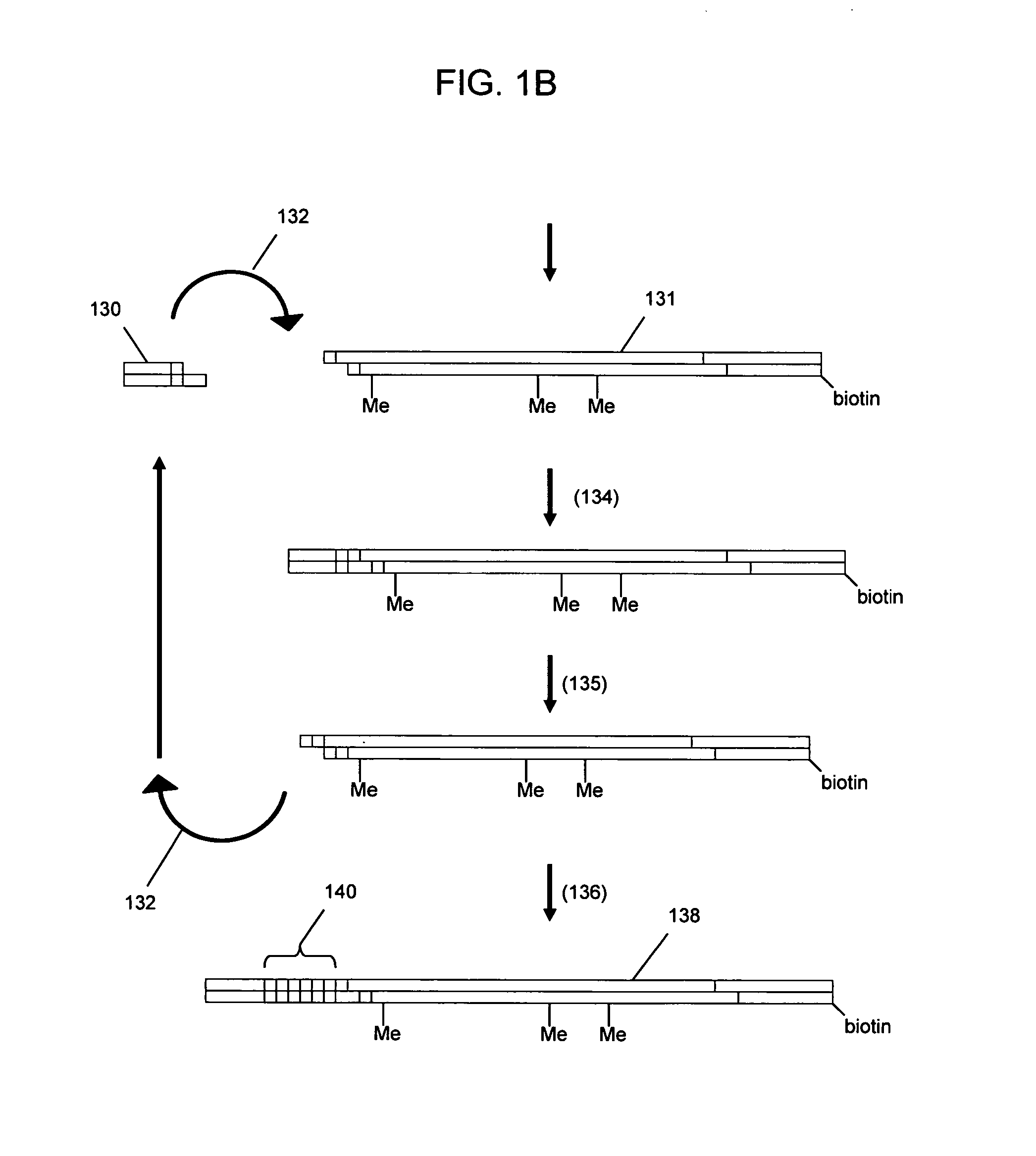
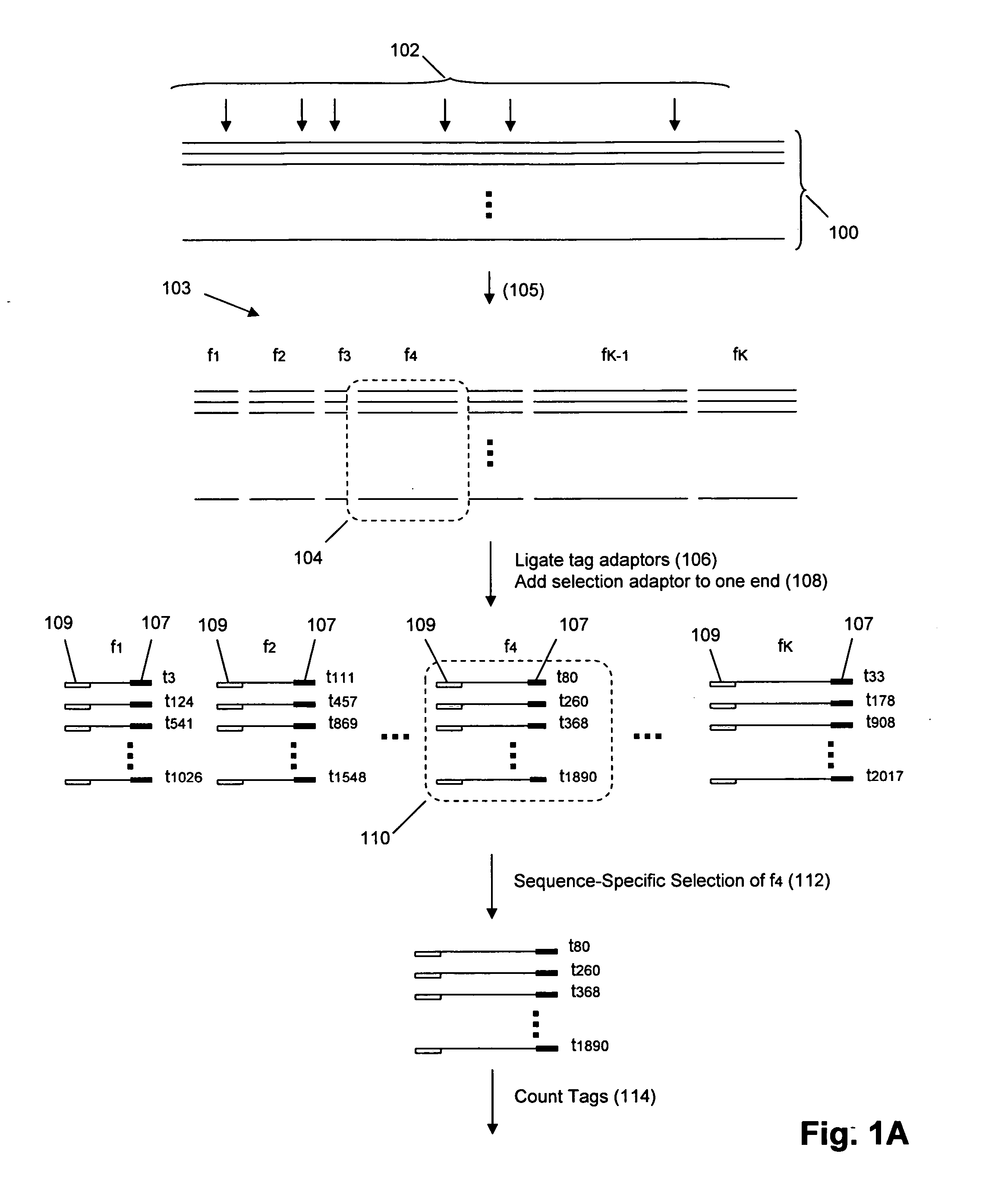
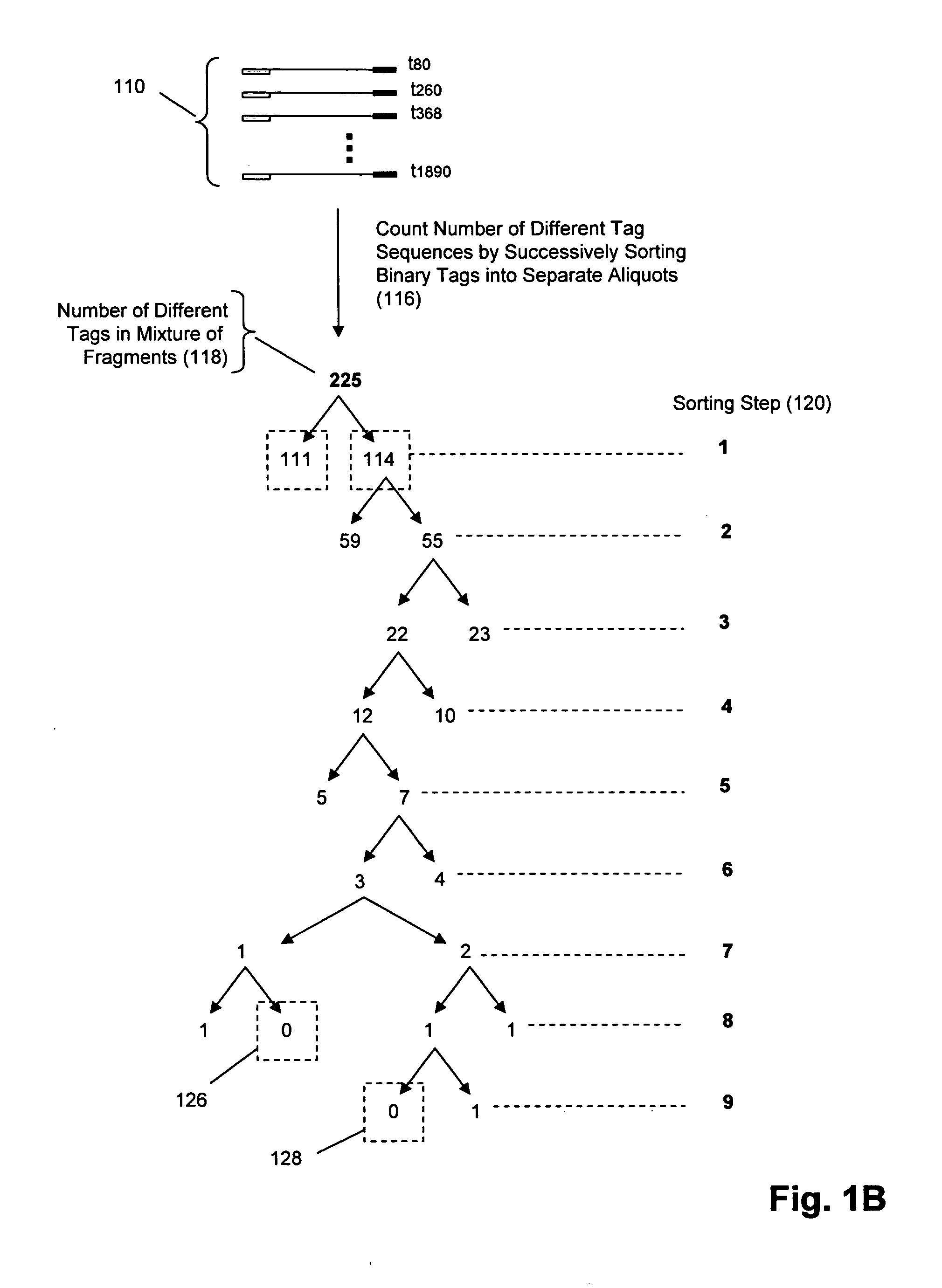
Molecular counting
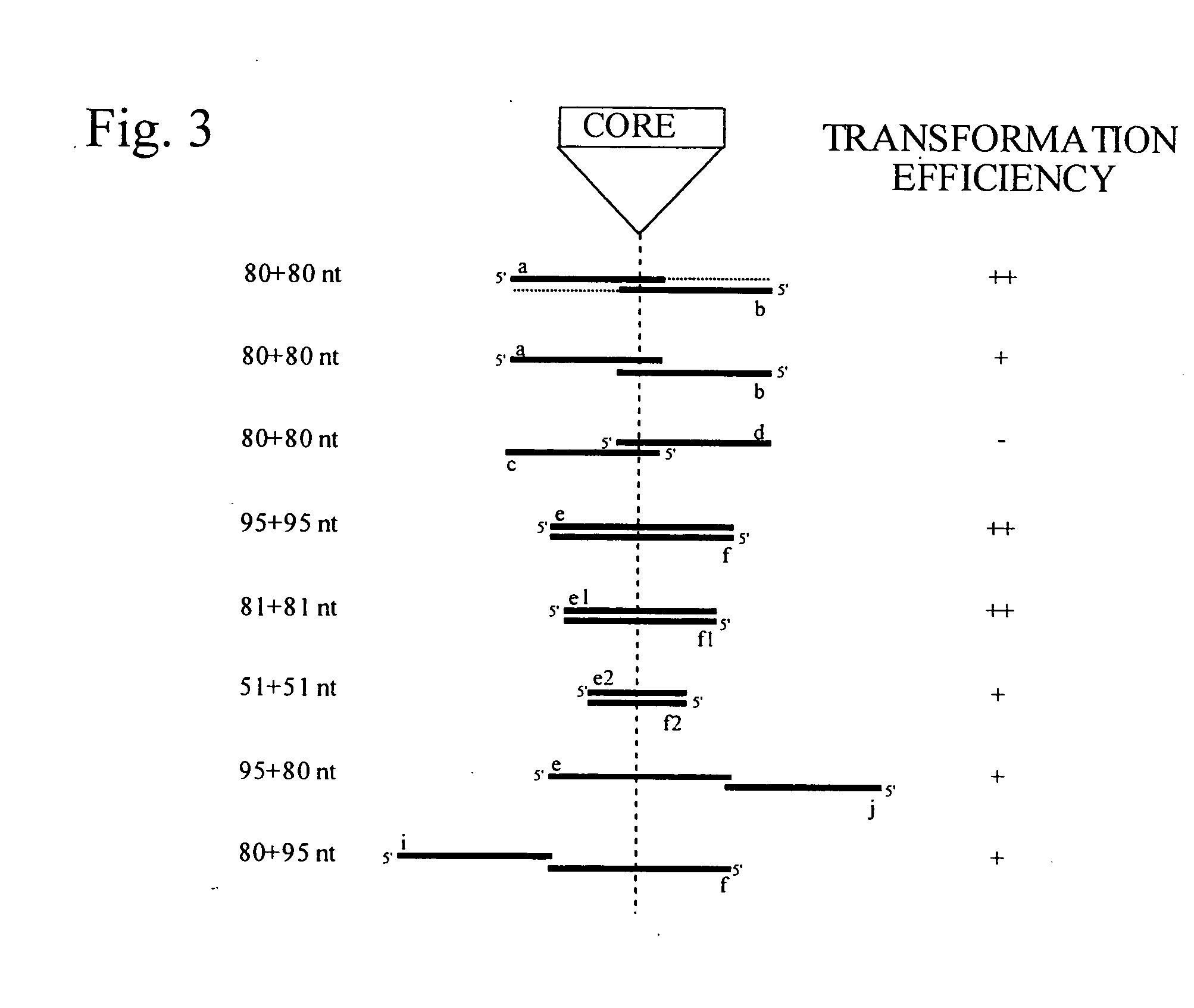
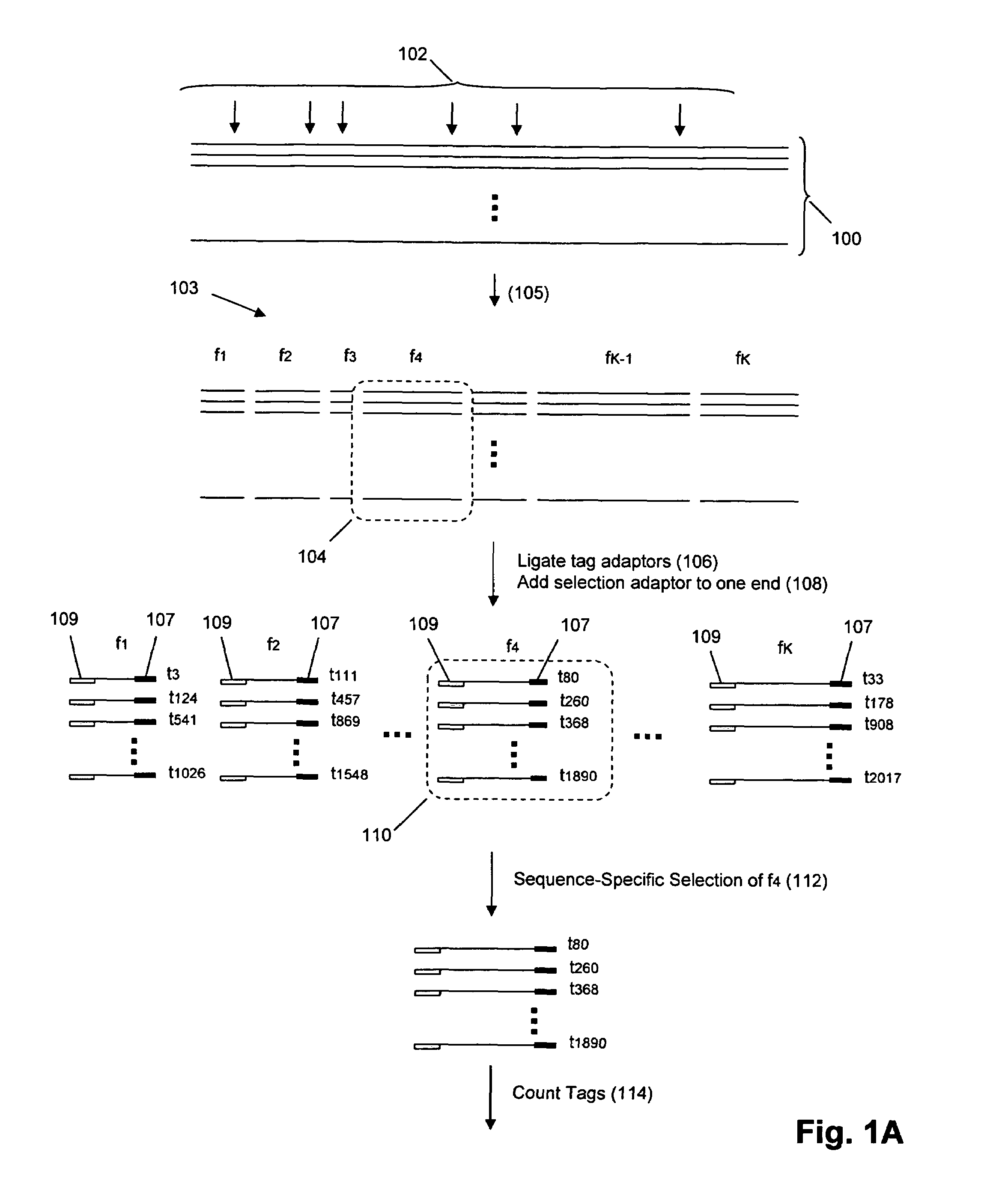
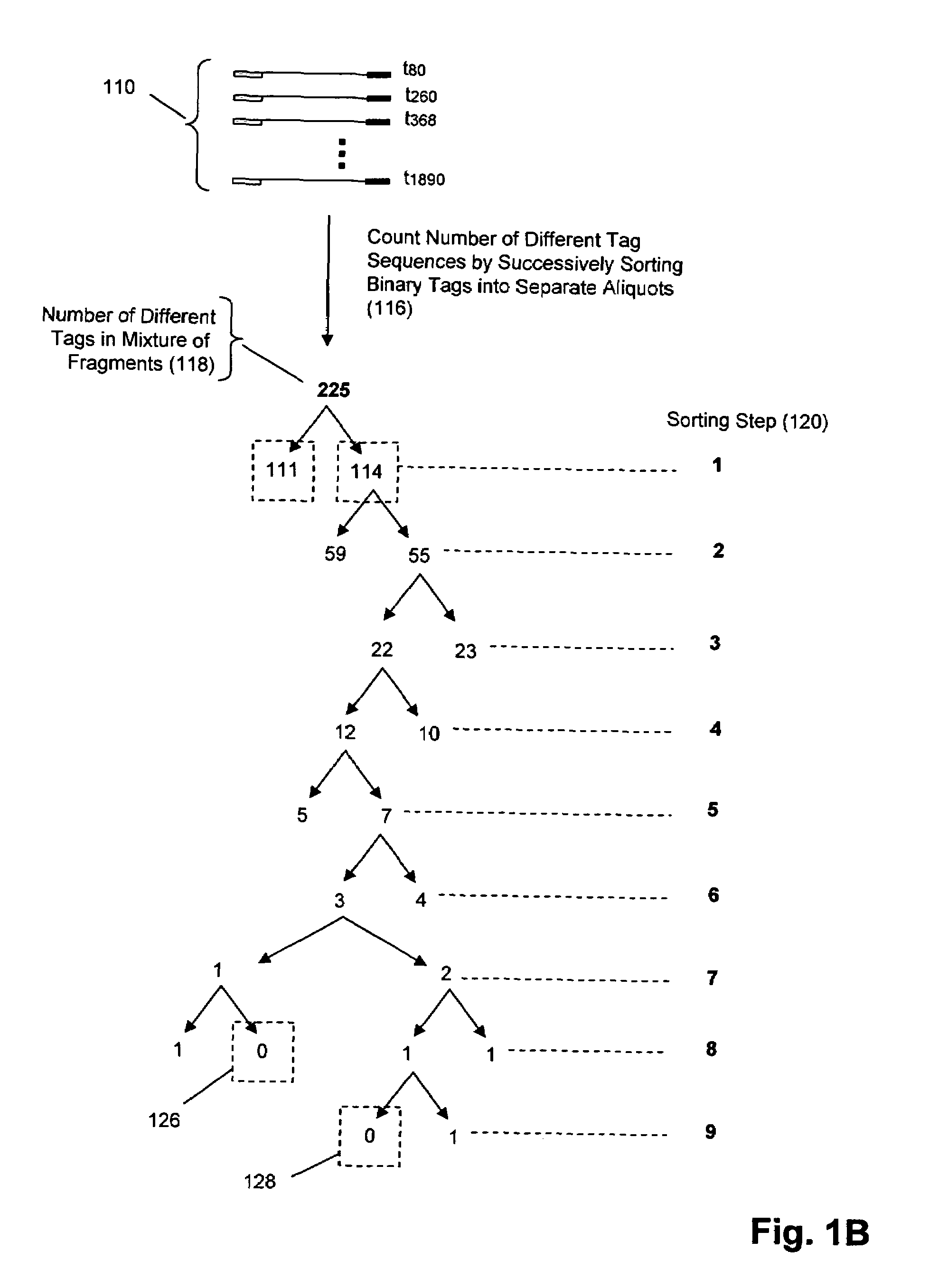
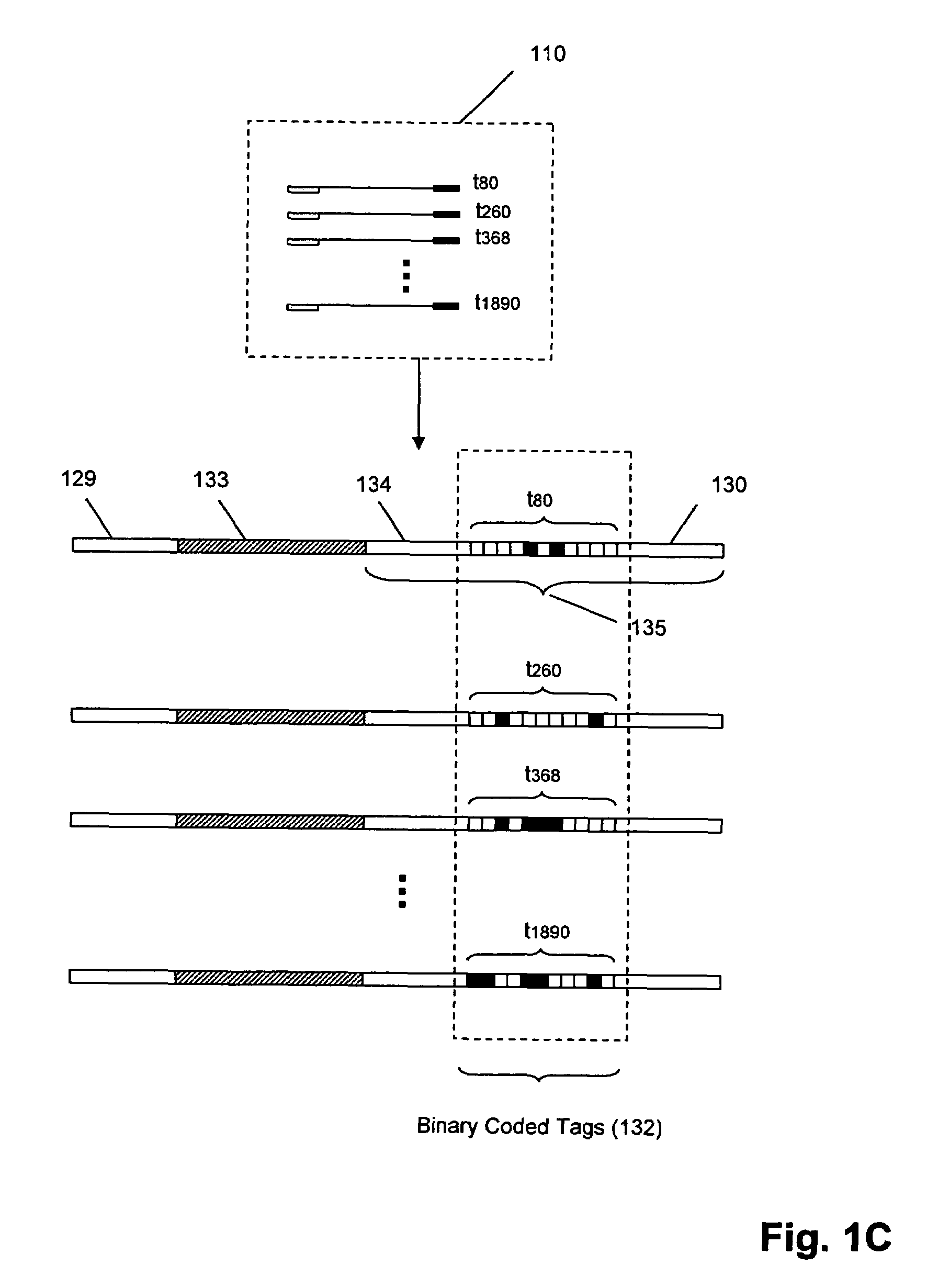
The invention provides methods and compositions for counting molecules in a sample, wherein each molecule is labeled with a unique oligonucleotide tag. Such tags are amplified and identified rather than the molecules themselves; that is, the problem of counting molecules is converted into the problem of counting tags. In one aspect of the invention, molecules to be counted are labeled by sampling. That is, conjugates are formed between the molecules to be counted and oligonucleotide tags of a very large set, or repertoire.After conjugation, a sample of conjugates is taken that is sufficiently small so that substantially every molecule has a unique oligonucleotide tag. Counting of different tags may be accomplished in a variety of ways. In one aspect, different tags may be counted by carrying out a series of sorting steps to generate successively less complex mixtures in which tags are enumerated using length-encoded “metric” tags. In another aspect, different tags may be counted by directly sequencing a sample of tags using any one of several different sequencing methodologies.
Owner:AGILENT TECH INC
Methods and compositions for flow cytometric determination of DNA sequences
InactiveUS6057107AReduce fluorescenceReduce intensityMicrobiological testing/measurementIndividual particle analysisAnalysis dnaTest sample
A method for the analysis of DNA sequences and PCR products comprises the steps of constructing an oligonucleotide-labeled beadset, and labeled complementary probe, and exposing the beadset and probe to a DNA fragment or PCR product under hybridizing conditions and analyzing the combined sample / beadset by flow cytometry. Flow cytometric measurements are used to classify beads within an exposed beadset to determine the presence of identical or nonidentical sequences within the test sample. The inventive technology enables the rapid analysis of DNA sequences and detection of point mutations, deletions and / or inversions while also reducing the cost and time for performing genetic assays.
Owner:LUMINEX
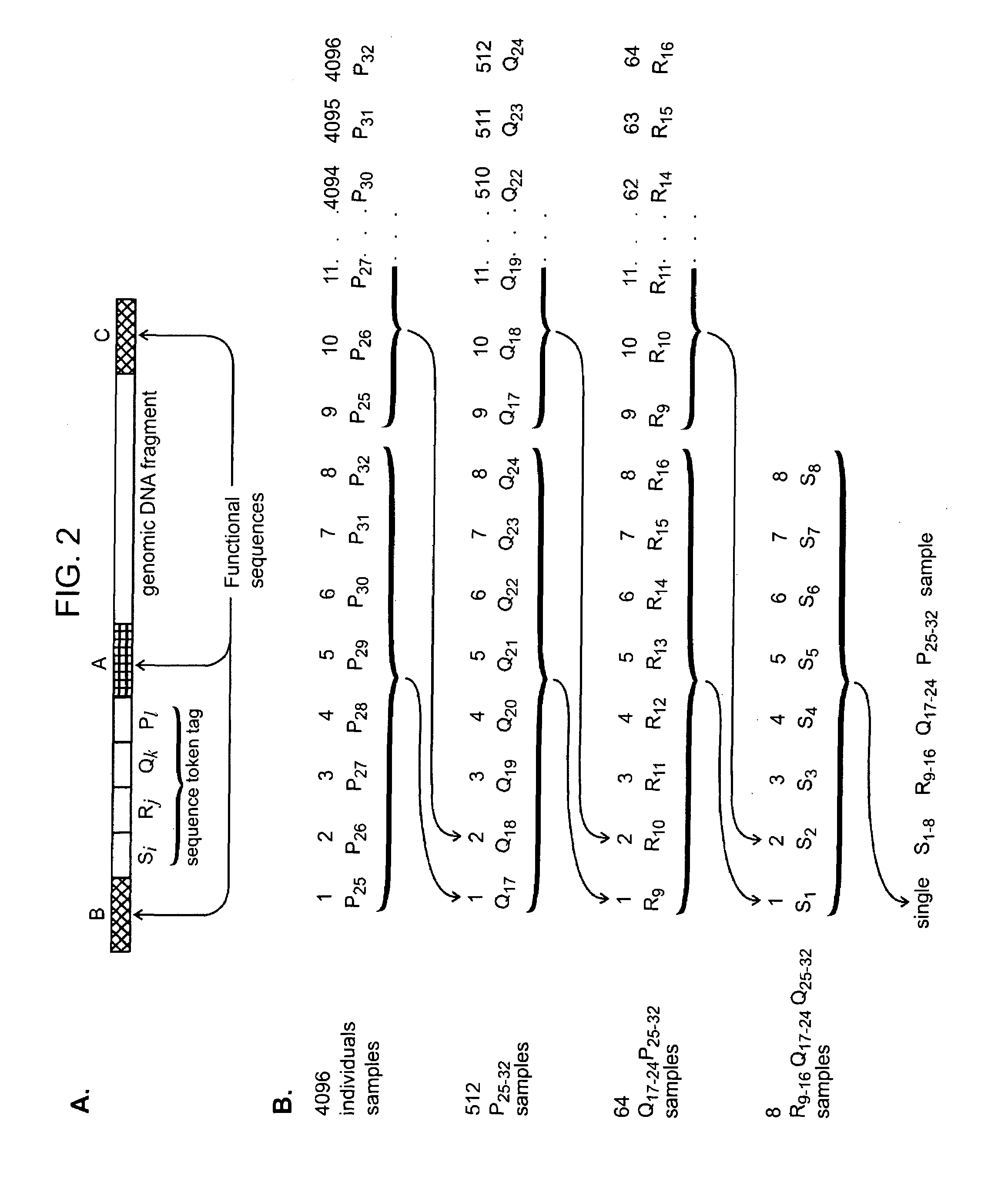
Nucleic acid analysis using sequence tokens
ActiveUS7544473B2Efficiently determine variations in nucleotide sequences in the associated nucleic acid sequence fragmentsBioreactor/fermenter combinationsBiological substance pretreatmentsDon't repeat yourselfNucleic acid sequencing
The present invention provides methods and compositions for tagging nucleic acid sequence fragments, e.g., a set of nucleic acid sequence fragments from a single genome, with one or more unique members of a collection of oligonucleotide tags, or sequence tokens, which, in turn, can be identified using a variety of readout platforms. As a general rule, a given sequence token is used once and only once in any tag sequence. In addition, the present invention also provides methods for using the sequence tokens to efficiently determine variations in nucleotide sequences in the associated nucleic acid sequence fragments.
Owner:PERSONAL GENOME DIAGNOSTICS INC
Molecular counting
The invention provides methods and compositions for counting molecules in a sample, wherein each molecule is labeled with a unique oligonucleotide tag. Such tags are amplified and identified rather than the molecules themselves; that is, the problem of counting molecules is converted into the problem of counting tags. In one aspect of the invention, molecules to be counted are labeled by sampling. That is, conjugates are formed between the molecules to be counted and oligonucleotide tags of a very large set, or repertoire. After conjugation, a sample of conjugates is taken that is sufficiently small so that substantially every molecule has a unique oligonucleotide tag. Counting of different tags may be accomplished in a variety of ways. In one aspect, different tags may be counted by carrying out a series of sorting steps to generate successively less complex mixtures in which tags are enumerated using length-encoded “metric” tags. In another aspect, different tags may be counted by directly sequencing a sample of tags using any one of several different sequencing methodologies.
Owner:AGILENT TECH INC
Oligonucleotides having chiral phosphorus linkages
InactiveUS6239265B1Improved pharmacokinetic propertiesImprove propertiesPeptide/protein ingredientsGenetic material ingredientsSugar moietyPhosphoramidate
Sequence-specific oligonucleotides are provided having substantially pure chiral Sp phosphorothioate, chiral Rp phosphorothioate, chiral Sp alkylphosphonate, chiral Rp alkylphosphonate, chiral Sp phosphoamidate, chiral Rp phosphoamidate, chiral Sp phosphotriester, and chiral Rp phosphotriester linkages. The novel oligonucleotides are prepared via a stereospecific SN2 nucleophilic attack of a phosphodiester, phosphorothioate, phosphoramidate, phosphotriester or alkylphosphonate anion on the 3' position of a xylonucleotide. The reaction proceeds via inversion at the 3' position of the xylo reactant species, resulting in the incorporation of phosphodiester, phosphorothioate, phosphoramidate, phosphotriester or alkylphosphonate linked ribofuranosyl sugar moieties into the oligonucleotide.
Owner:IONIS PHARMA INC
Molecular encoding of nucleic acid templates for PCR and other forms of sequence analysis
InactiveUS20070020640A1Microbiological testing/measurementBiological testingOligonucleotideDouble stranded
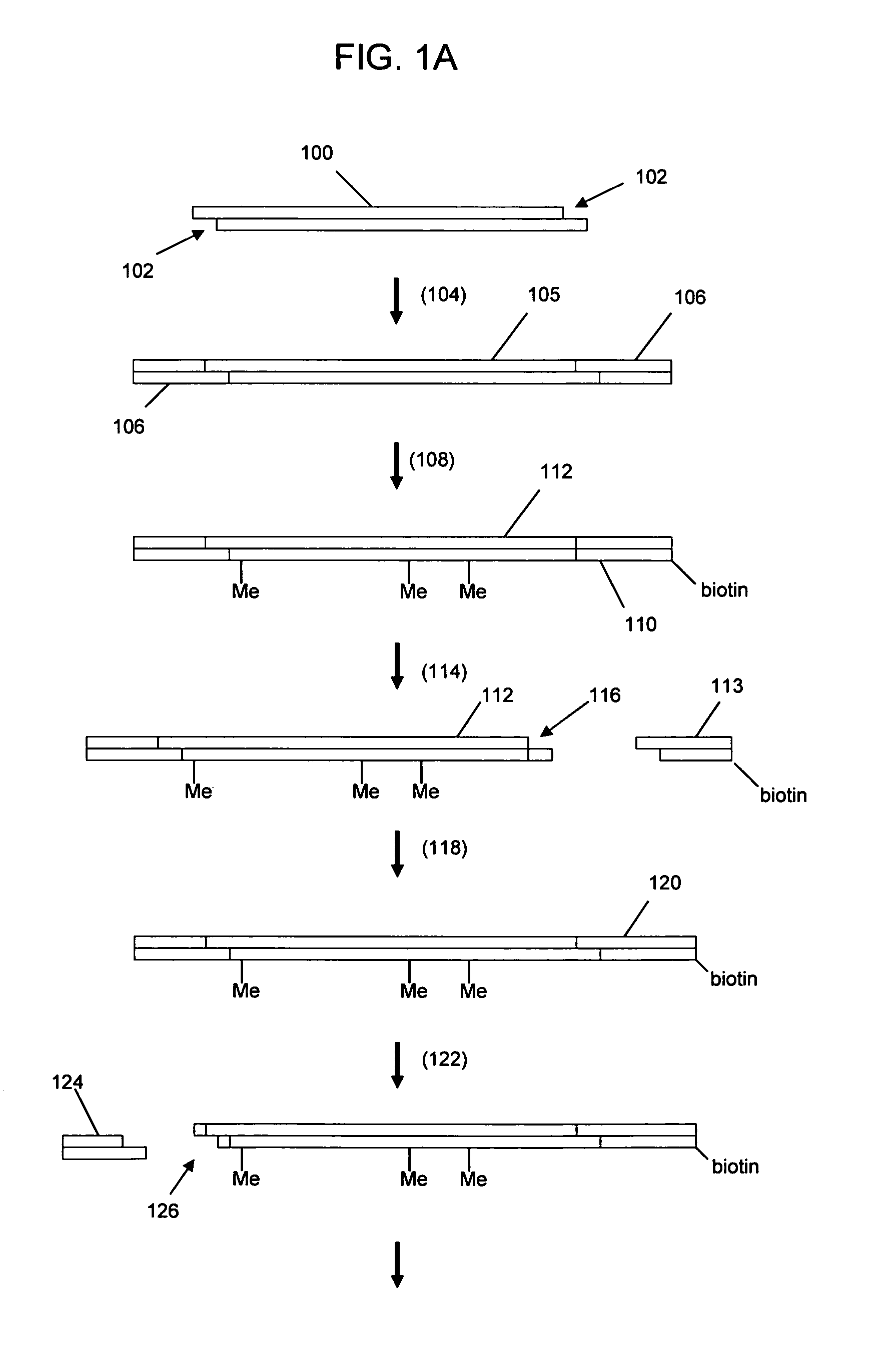
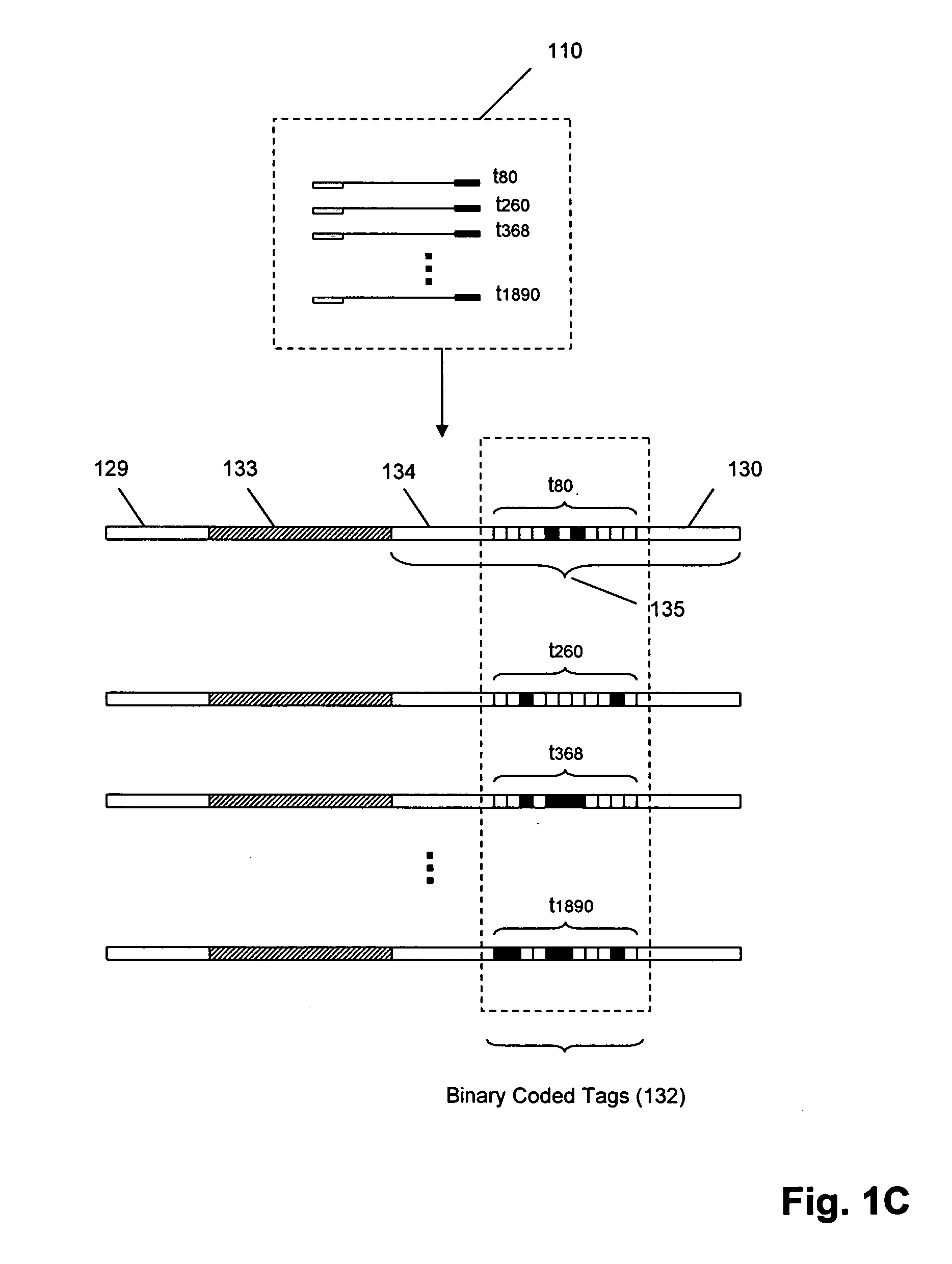
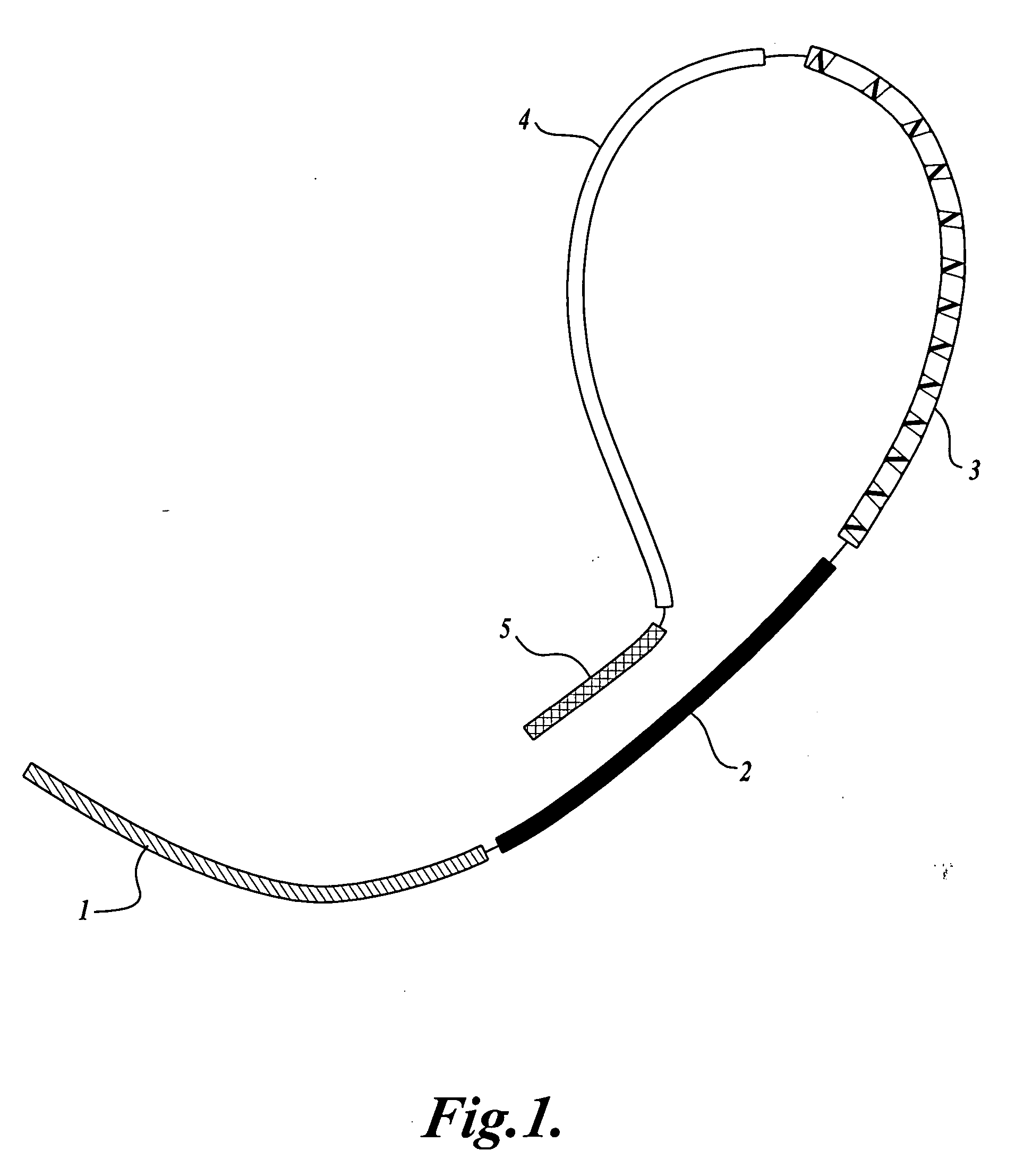
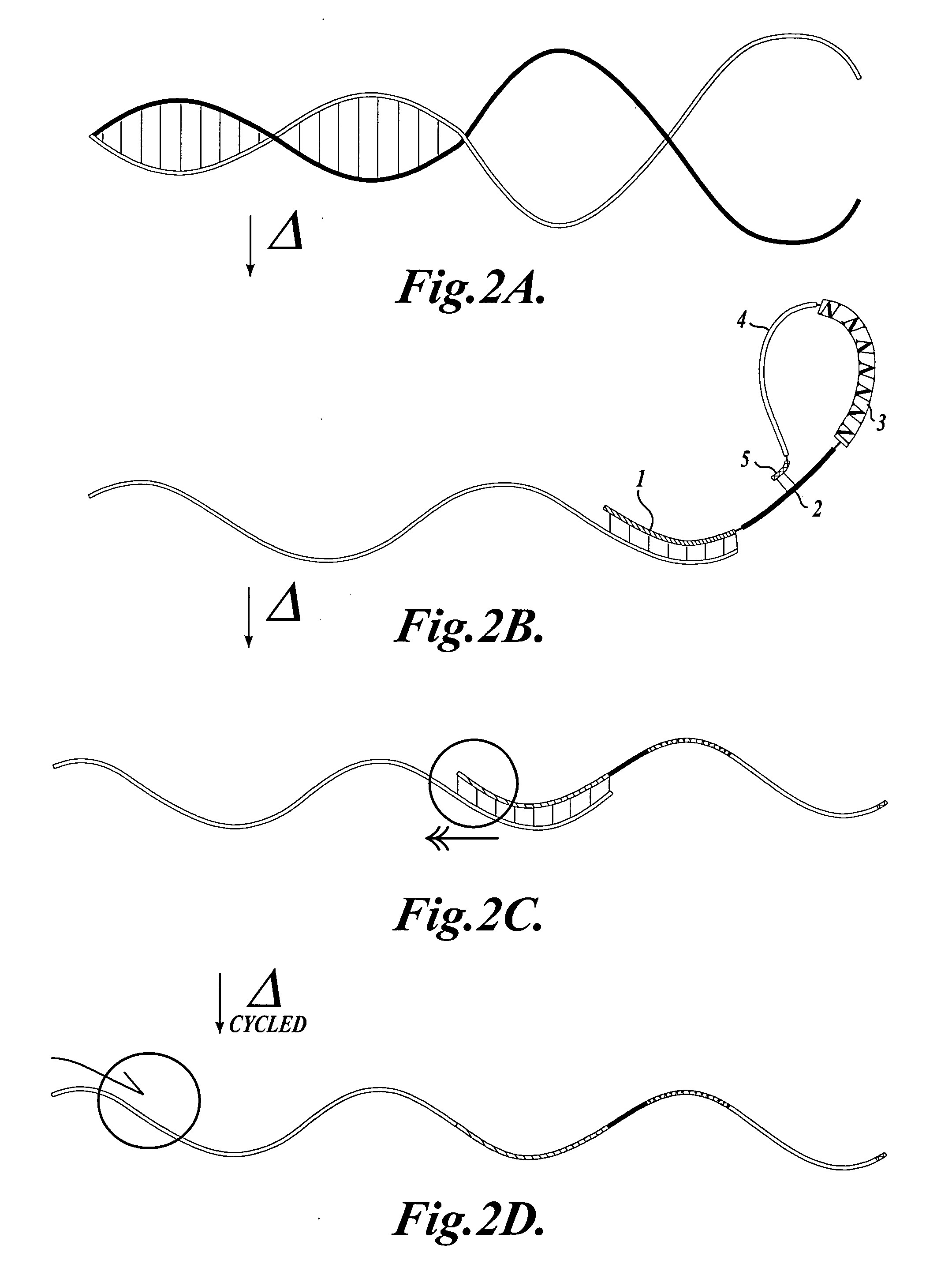
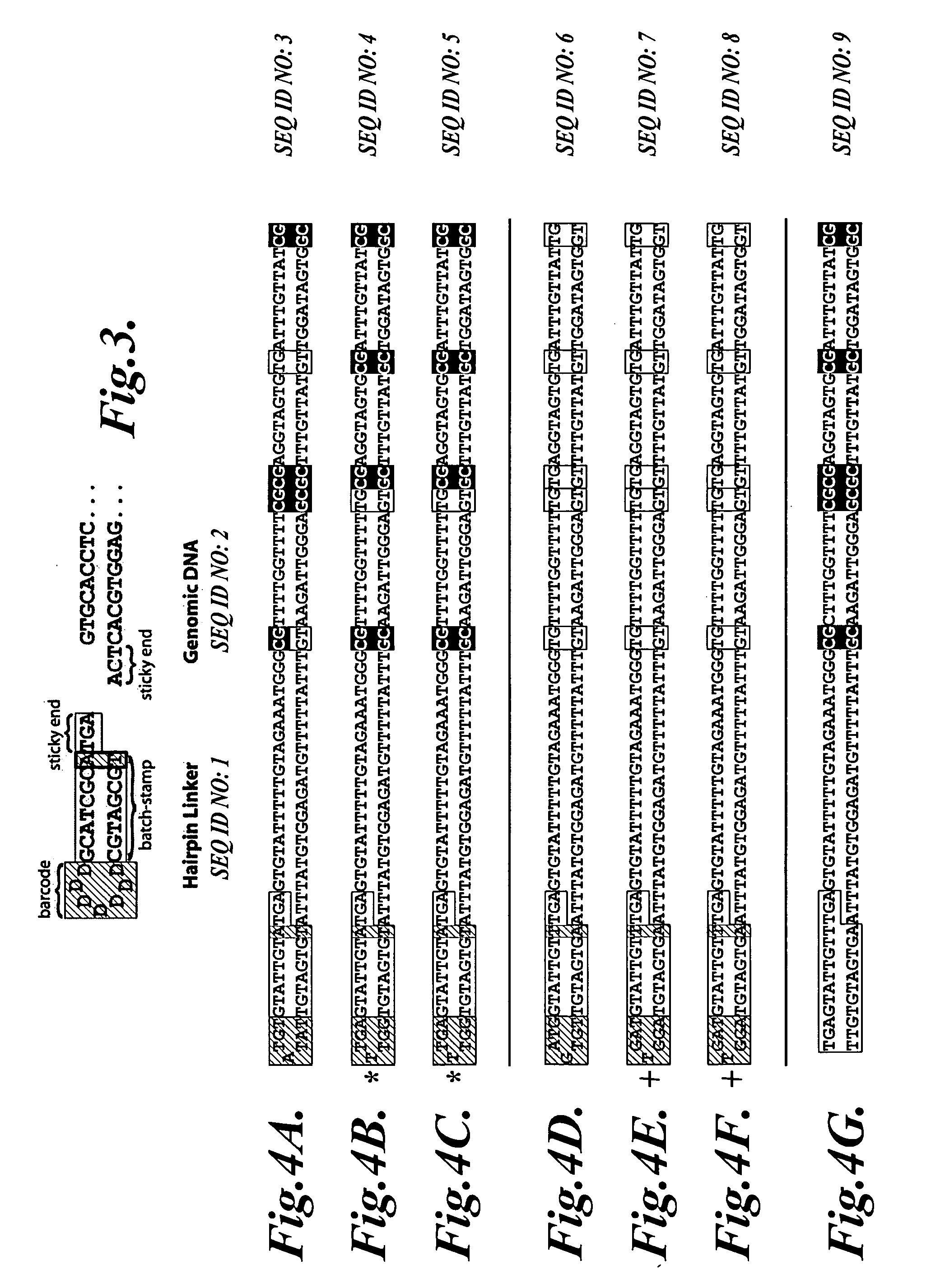
In a first aspect, the present invention provides methods for authenticating a nucleic acid molecule and its sequence with a molecular barcode and batch-stamp. In another aspect, the present invention provides methods for authenticating a nucleic acid amplification product. In a further aspect, the present invention provides compositions for encoding both single-stranded and double-stranded target nucleic acids with coded oligonucleotides. The compositions are useful in the practice of the methods of the invention.
Owner:UNIV OF WASHINGTON
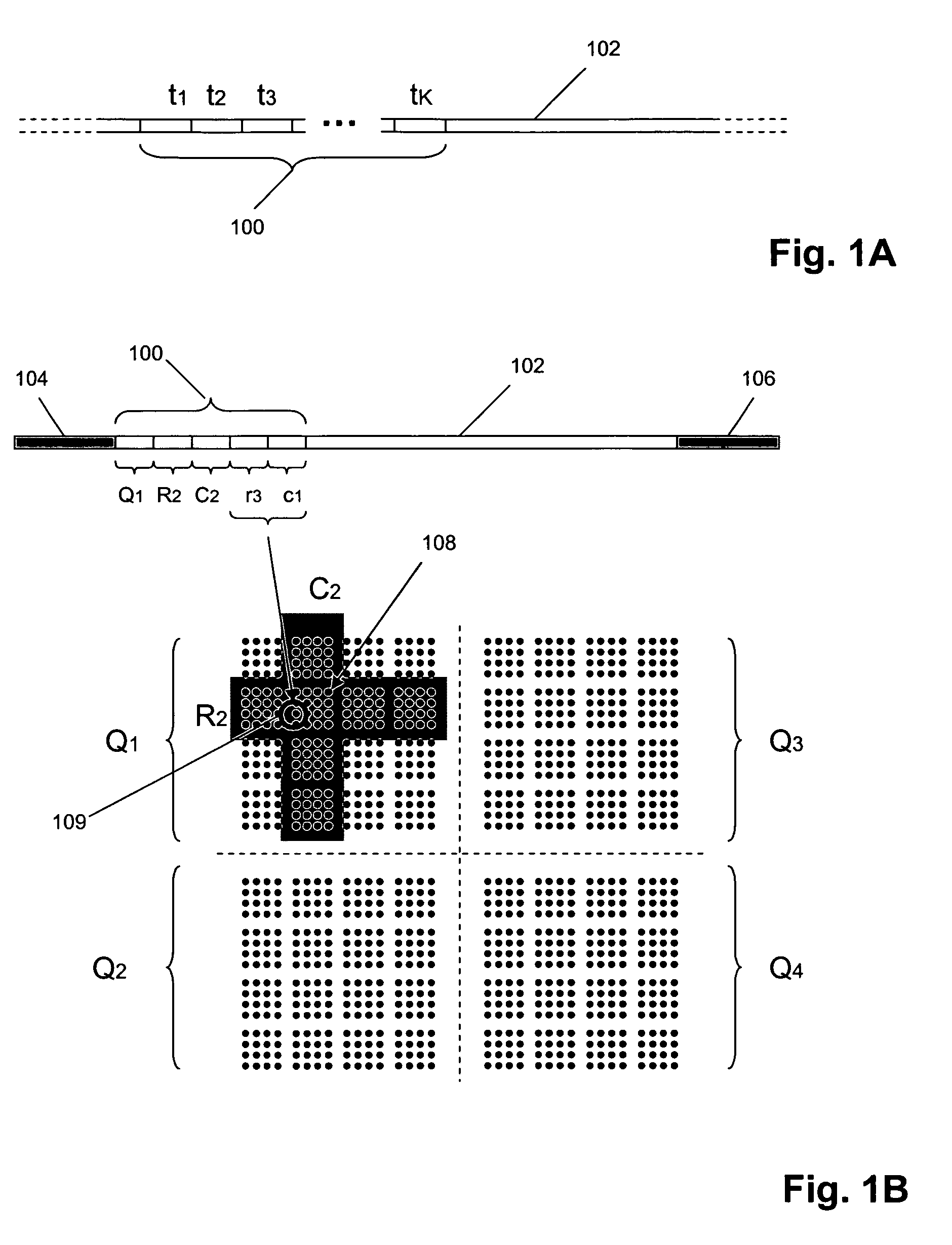
Methods and compositions for tagging and identifying polynucleotides
The invention provides methods and compositions for attaching oligonucleotide tags to polynucleotides for the purpose of carrying out analytical assays in parallel and for decoding the oligonucleotide tags of polynucleotides selected in such assays. Words, or subunits, of oligonucleotide tags index submixtures in successively more complex sets of submixtures (referred to herein as “tiers” of submixtures) that a polynucleotide goes through while successive words are added to a growing tag. By identifying each word of an oligonucleotide tag, a series of submixtures is identified including the first submixture that contains only a single polynucleotide, thereby providing the identity of the selected polynucleotide. The analysis of the words of an oligonucleotide tag can be carried out in parallel, e.g. by specific hybridization of the oligonucleotide tag to its tag complement on an addressable array; or such analysis can be carried out serially by successive specific hybridizations of labeled word complements, or the like.
Owner:AGILENT TECH INC
Methods for detecting and identifying single molecules
InactiveUS6287765B1Highly specific controlImprove securityNanotechSugar derivativesSynthetic nucleotideMolecular adsorption
Multimolecular devices and drug delivery systems prepared from synthetic heteropolymers, heteropolymeric discrete structures, multivalent heteropolymeric hybrid structures, aptameric multimolecular devices, multivalent imprints, tethered specific recognition devices, paired specific recognition devices, nonaptameric multimolecular devices and immobilized multimolecular structures are provided, including molecular adsorbents and multimolecular adherents, adhesives, transducers, switches, sensors and delivery systems. Methods for selecting single synthetic nucleotides, shape-specific probes and specifically attractive surfaces for use in these multimolecular devices are also provided. In addition, paired nucleotide-nonnucleotide mapping libraries for transposition of selected populations of selected nonoligonucleotide molecules into selected populations of replicatable nucleotide sequences are described.
Owner:MOLECULAR MACHINES
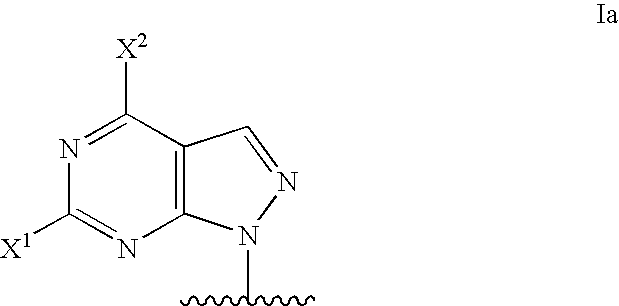
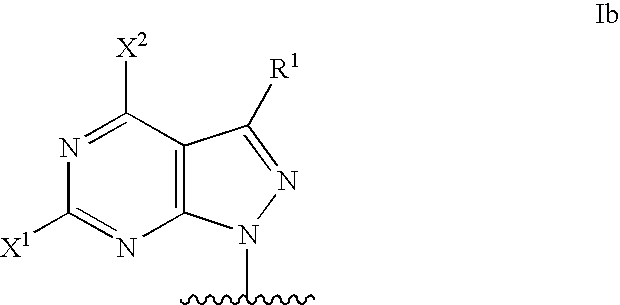
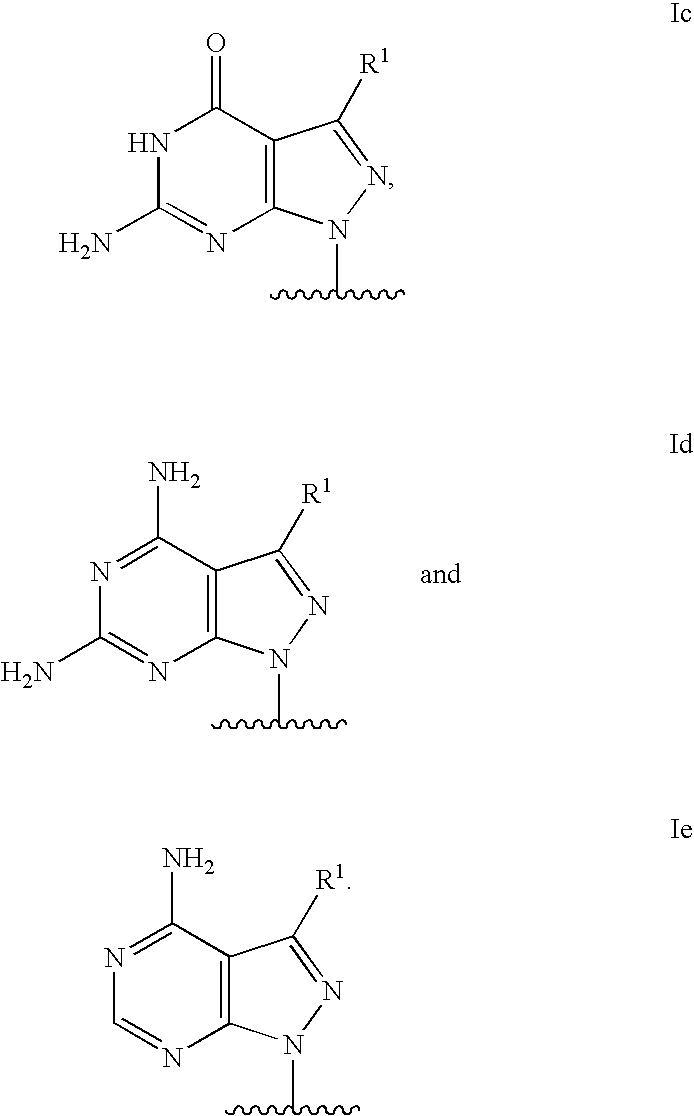
Modified oligonucleotides for mismatch discrimination
Modified oligonucleotides are provided containing bases selected from unsubstituted and 3-substituted pyrazolo[3,4-d]pyrimidines and 5-substituted pyrimidines, and optionally have attached minor groove binders and reporter groups.
Owner:ELITECHGRP +1
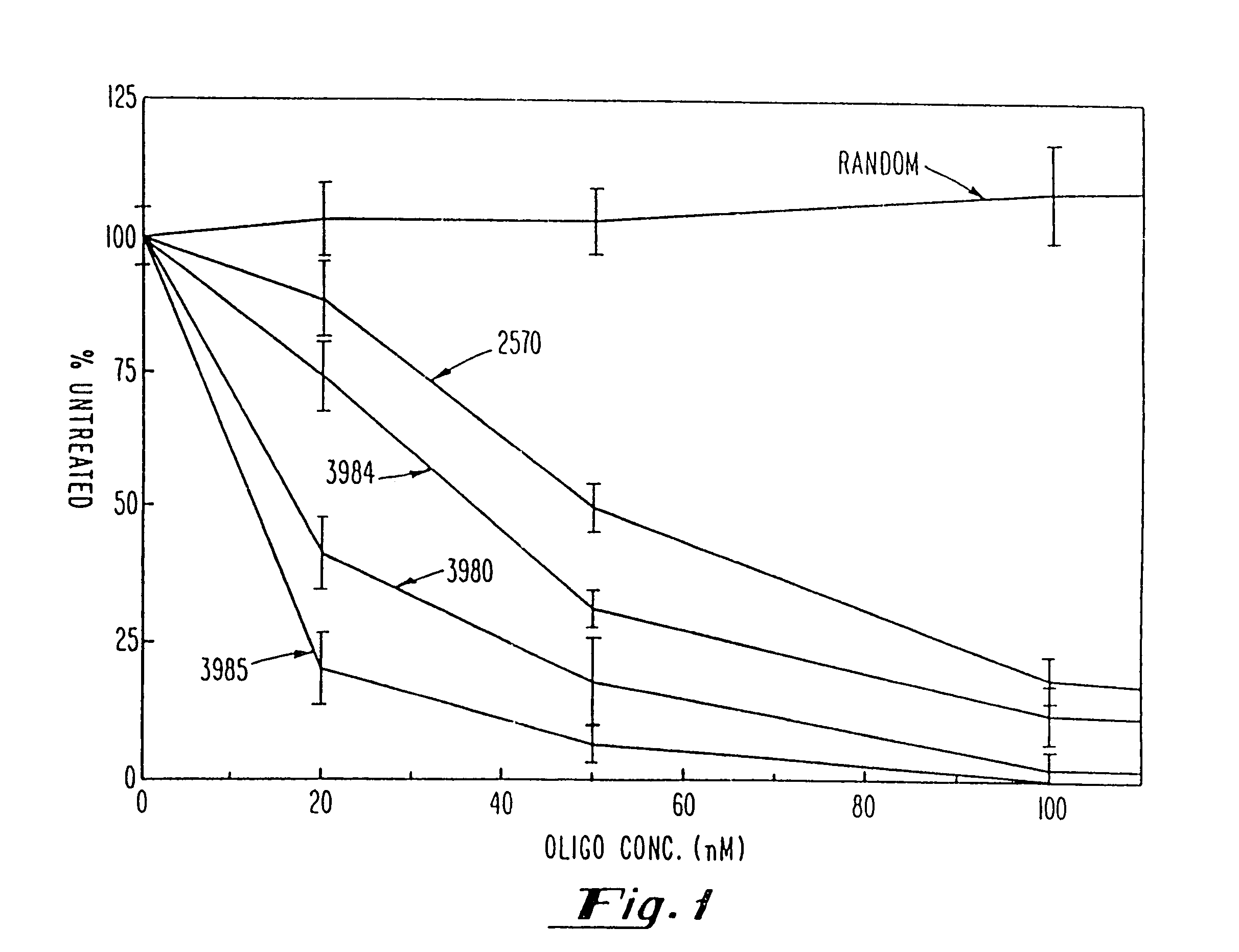
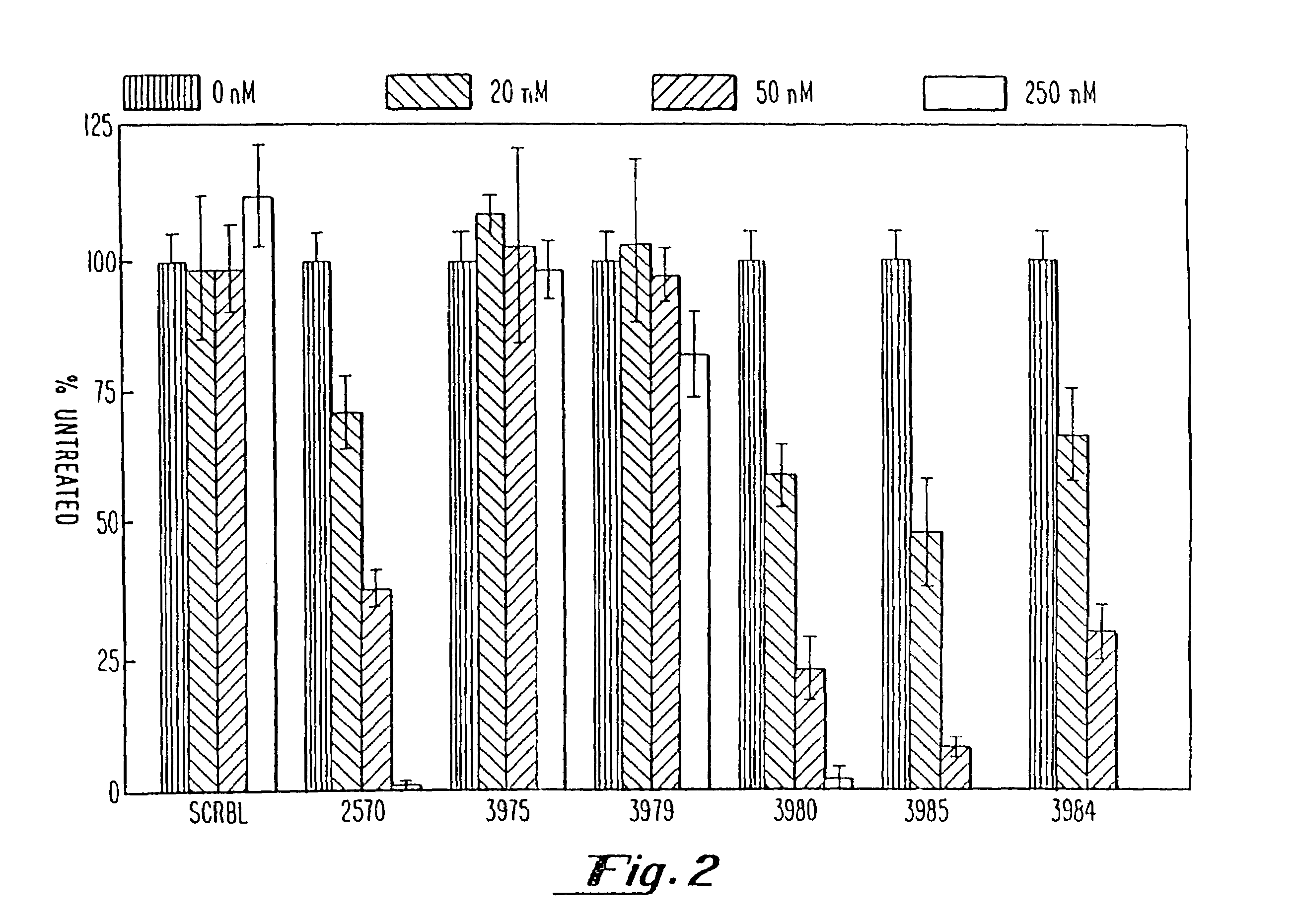
Gapped oligonucleotides
InactiveUS7015315B1Increased nuclease resistanceHigh binding affinityPeptide/protein ingredientsGenetic material ingredientsADAMTS ProteinsNuclease
Oligonucleotides and other macromolecules are provided which have increased nuclease resistance, substituent groups for increasing binding affinity to complementary strand, and subsequences of 2′-deoxy-erythro-pentofuranosyl nucleotides that activate RNase H. Such oligonucleotides and macromolecules are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics.
Owner:IONIS PHARMA INC
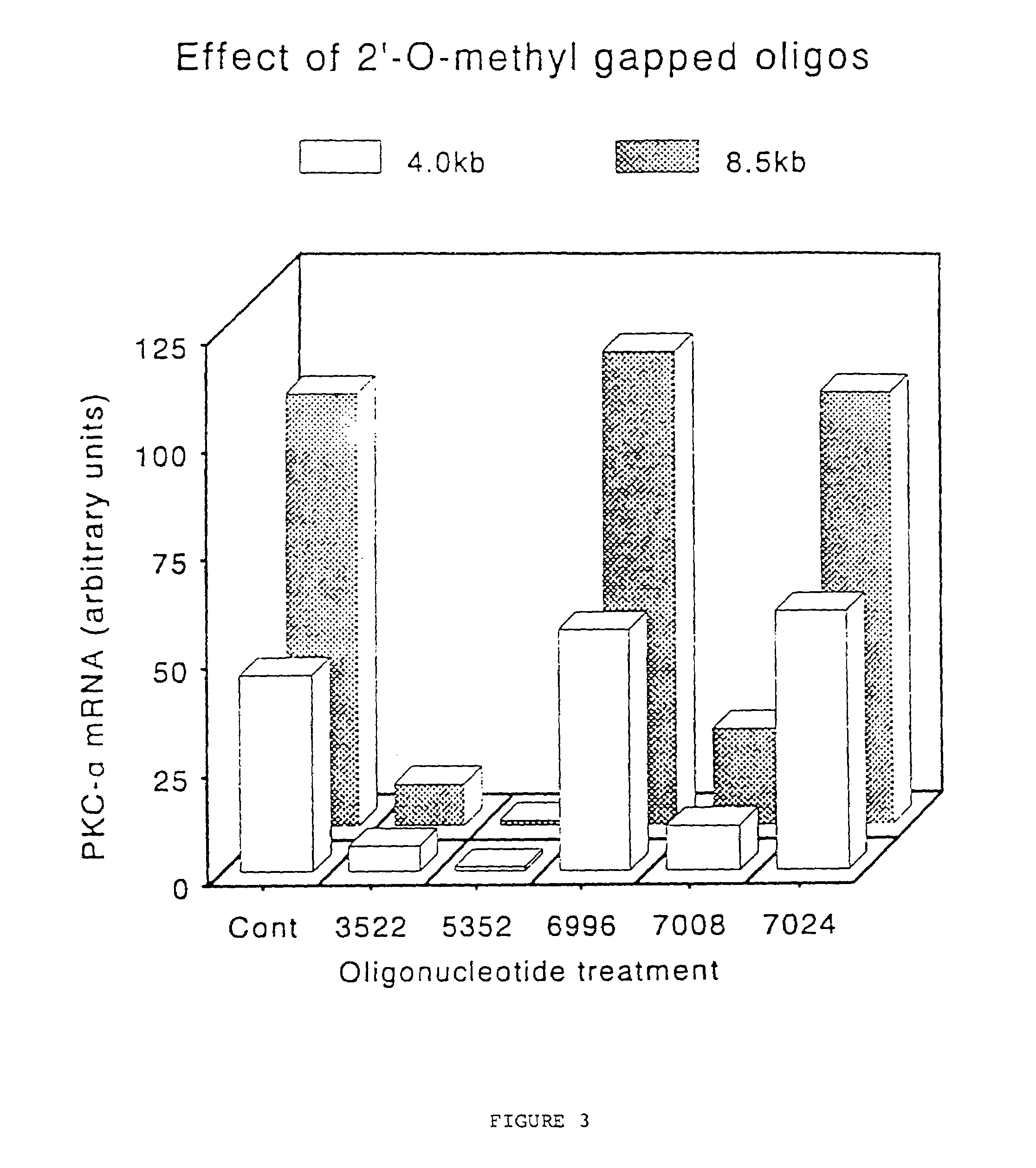
Gapped 2' modified oligonucleotides
InactiveUS6326199B1High affinityAvoid degradationHydrolasesPeptide/protein ingredientsNucleotideNuclease
Oligonucleotides and other macromolecules are provided that have increased nuclease resistance, substituent groups for increasing binding affinity to complementary strand, and sub-sequences of 2'-deoxy-erythro-pentofuranosyl nucleotides that activate RNase H enzyme. Such oligonucleotides and macromolecules are useful for diagnostics and other research purposes, for modulating protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to antisense therapeutics.
Owner:IONIS PHARMA INC
Advanced surface-enhanced Raman gene probe systems and methods thereof
InactiveUS6174677B1Produce roughnessEffective supportBioreactor/fermenter combinationsRadiation pyrometryNucleotideGene probe
The subject invention is a series of methods and systems for using the Surface-Enhanced Raman (SER)-labeled Gene Probe for hybridization, detection and identification of SER-labeled hybridized target oligonucleotide material comprising the steps of immobilizing SER-labeled hybridized target oligonucleotide material on a support means, wherein the SER-labeled hybridized target oligonucleotide material comprise a SER label attached either to a target oligonucleotide of unknown sequence or to a gene probe of known sequence complementary to the target oligonucleotide sequence, the SER label is unique for the target oligonucleotide strands of a particular sequence wherein the SER-labeled oligonucleotide is hybridized to its complementary oligonucleotide strand, then the support means having the SER-labeled hybridized target oligonucleotide material adsorbed thereon is SERS activated with a SERS activating means, then the support means is analyzed.
Owner:LOCKHEED MARTIN ENERGY SYST INC
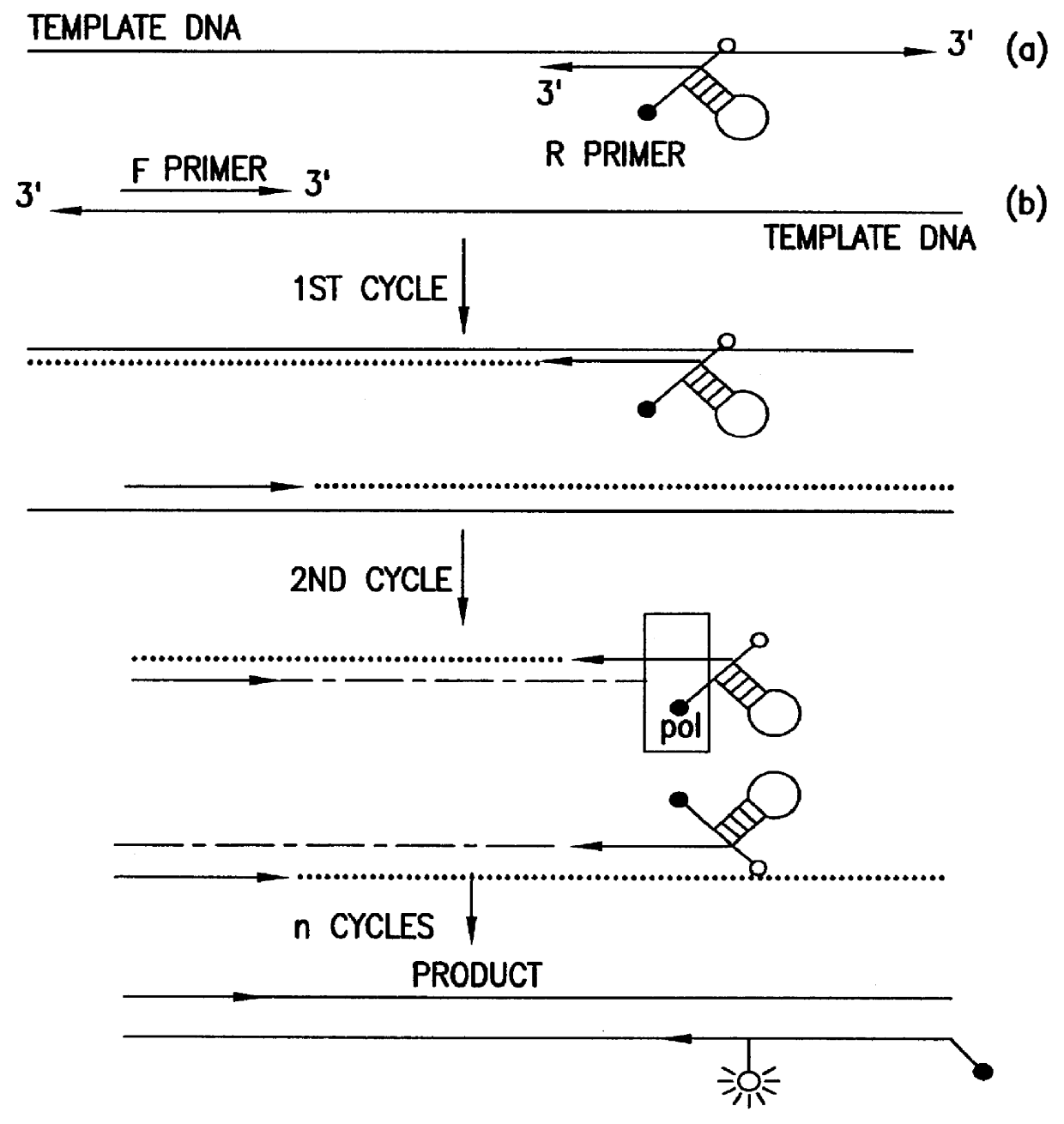
Method of determining the nucleotide sequence of oligonucleotides and DNA molecules
InactiveUS7037687B2Eliminate needBioreactor/fermenter combinationsBiological substance pretreatmentsOligonucleotide primersThermopile
The present invention relates to a novel method for analyzing nucleic acid sequences based on real-time detection of DNA polymerase-catalyzed incorporation of each of the four nucleotide bases, supplied individually and serially in a microfluidic system, to a reaction cell containing a template system comprising a DNA fragment of unknown sequence and an oligonucleotide primer. Incorporation of a nucleotide base into the template system can be detected by any of a variety of methods including but not limited to fluorescence and chemiluminescence detection. Alternatively, microcalorimetic detection of the heat generated by the incorporation of a nucleotide into the extending template system using thermopile, thermistor and refractive index measurements can be used to detect extension reactions.
Owner:ALBERTA UNIV OF +1
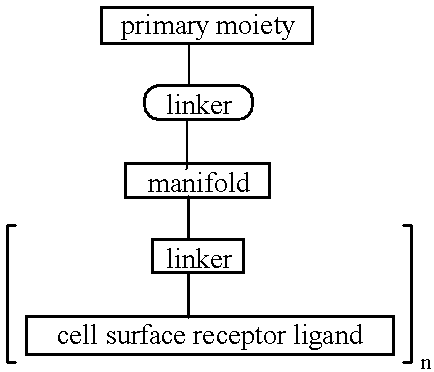
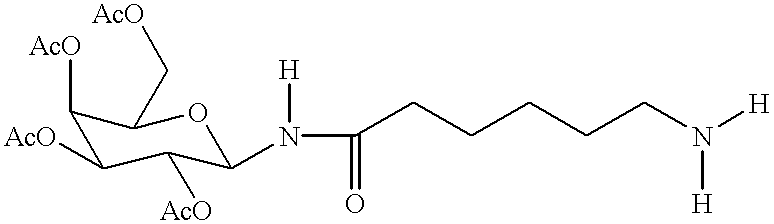
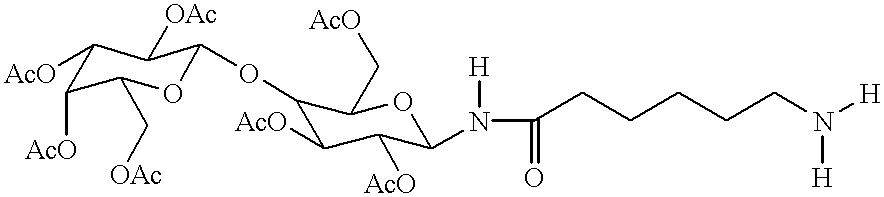
Targeted oligonucleotide conjugates
InactiveUS6300319B1Simple methodReduce interferenceBiocideOrganic active ingredientsReceptor degradationOligonucleotide
The present invention provides improved ingress of therapeutic and other moieties into cellular targets. In accordance with preferred embodiments, complexes are provided which carry primary moieties, chiefly therapeutic moieties, to such target cells. Such complexes preferably feature cell surface receptor ligands to provide specificity. Such ligands are preferably bound to primary moieties through polyfunctional manifold compounds.
Owner:IONIS PHARMA INC
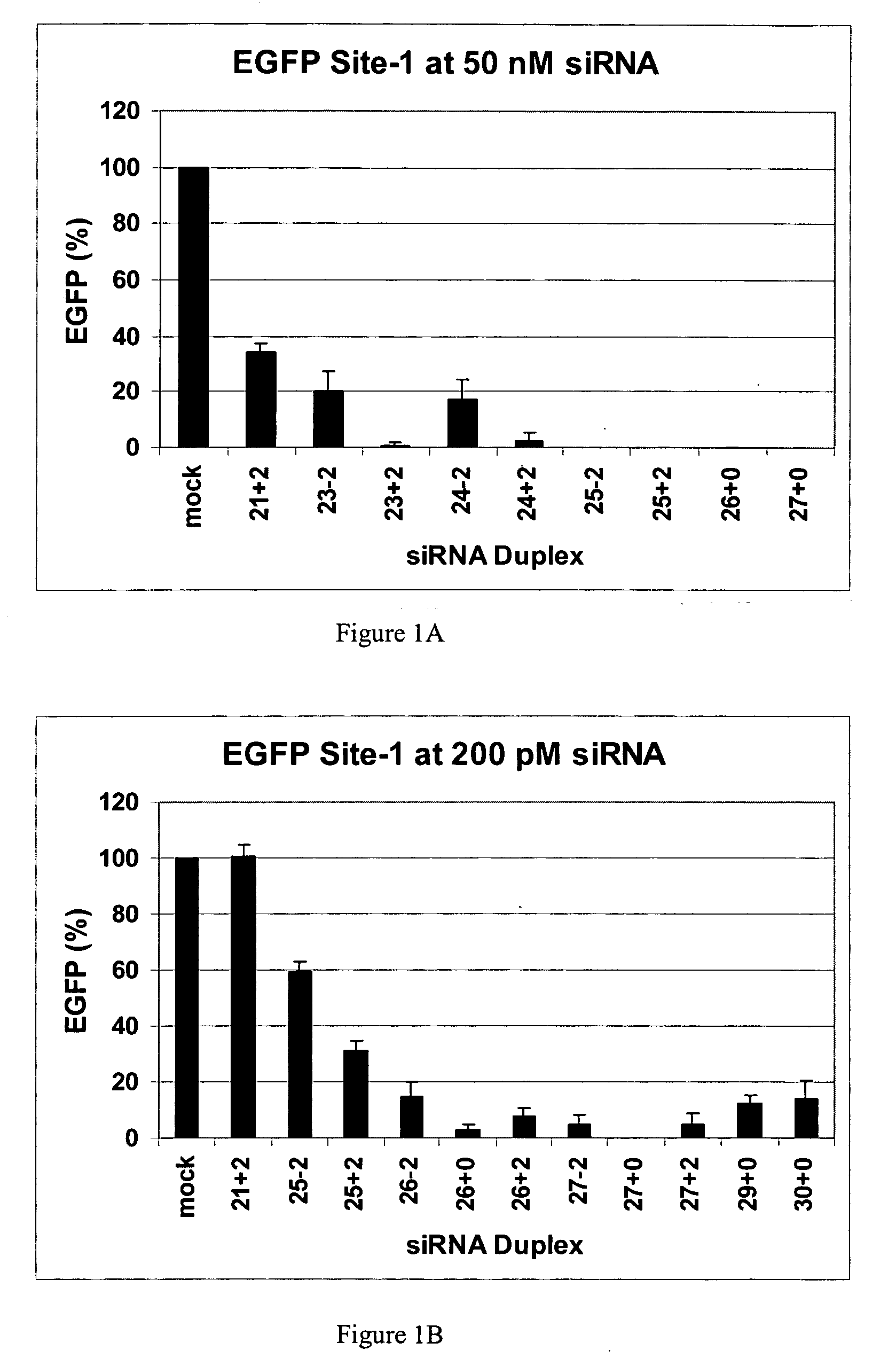
Methods and compositions for the specific inhibition of gene expression by double-stranded RNA
The invention is directed to compositions and methods for selectively reducing the expression of a gene product from a desired target gene in a cell, as well as for treating diseases caused by the expression of the gene. More particularly, the invention is directed to compositions that contain double stranded RNA (“dsRNA”), and methods for preparing them, that are capable of reducing the expression of target genes in eukaryotic cells. The dsRNA has a first oligonucleotide sequence that is between 25 and about 30 nucleotides in length and a second oligonucleotide sequence that anneals to the first sequence under biological conditions. In addition, a region of one of the sequences of the dsRNA having a sequence length of at least 19 nucleotides is sufficiently complementary to a nucleotide sequence of the RNA produced from the target gene to trigger the destruction of the target RNA by the RNAi machinery.
Owner:CITY OF HOPE +1
Method of sequencing genomes by hybridization of oligonucleotide probes
InactiveUS6018041ASugar derivativesMicrobiological testing/measurementMammalian genomeHomologous sequence
The conditions under which oligonucleotides hybridize only with entirely homologous sequences are recognized. The sequence of a given DNA fragment is read by the hybridization and assembly of positively hybridizing probes through overlapping portions. By simultaneous hybridization of DNA molecules applied as dots and bound onto a filter, representing single-stranded phage vector with the cloned insert, with about 50,000 to 100,000 groups of probes, the main type of which is (A,T,C,G)(A,T,C,G)N8(A,T,C,G), information for computer determination of a sequence of DNA having the complexity of a mammalian genome are obtained in one step. To obtain a maximally completed sequence, three libraries are cloned into the phage vector, M13, bacteriophage are used: with the 0.5 kb and 7 kbp insert consisting of two sequences, with the average distance in genomic DNA of 100 kbp. For a million bp of genomic DNA, 25,000 subclones of the 0.5 kbp are required as well as 700 subclones 7 kb long and 170 jumping subclones. Subclones of 0.5 kb are applied on a filter in groups of 20 each, so that the total number of samples is 2,120 per million bp. The process can be easily and entirely robotized for factory reading of complex genomic fragments or DNA molecules.
Owner:HYSEQ
Methods and compositions for the specific inhibition of gene expression by double-stranded RNA
InactiveUS20070265220A1Improve stabilityConvenience to mergeSenses disorderAntipyreticDouble strandOligonucleotide
The invention is directed to compositions and methods for selectively reducing the expression of a gene product from a desired target gene in a cell, as well as for treating diseases caused by the expression of the gene. More particularly, the invention is directed to compositions that contain double stranded RNA (“dsRNA”), and methods for preparing them, that are capable of reducing the expression of target genes in eukaryotic cells. The dsRNA has a first oligonucleotide sequence that is between 25 and about 30 nucleotides in length and a second oligonucleotide sequence that anneals to the first sequence under biological conditions. In addition, a region of one of the sequences of the dsRNA having a sequence length of at least 19 nucleotides is sufficiently complementary to a nucleotide sequence of the RNA produced from the target gene to trigger the destruction of the target RNA by the RNAi machinery.
Owner:CITY OF HOPE +1
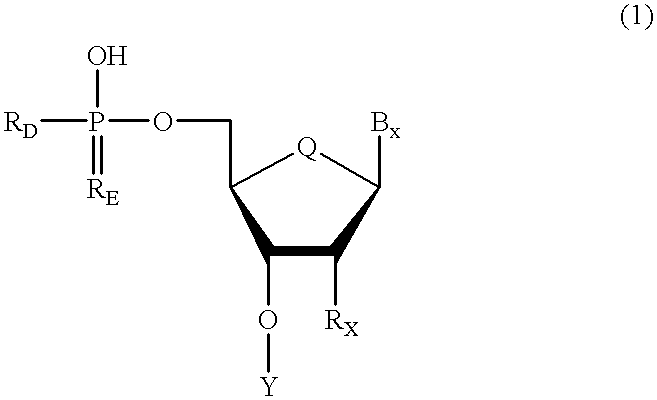
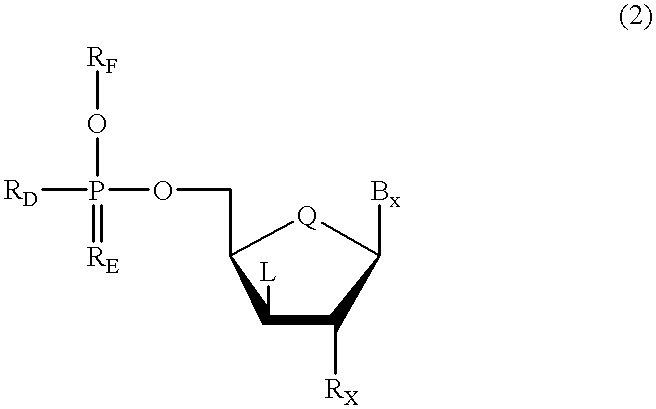
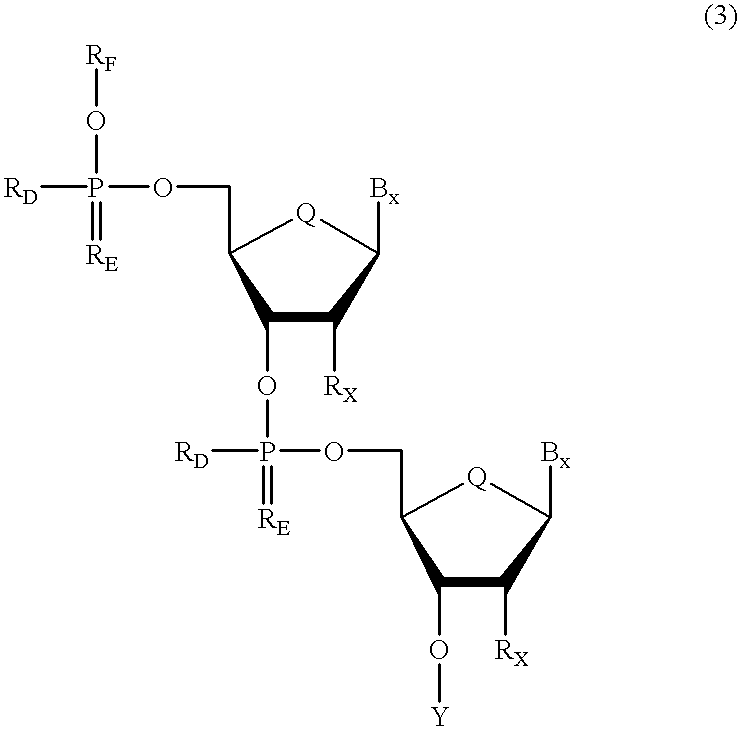
Methods for synthesis of oligonucleotides
Improved methods for synthesis of oligonucleotides and other phosphorus-linked oligomers are disclosed. The methods include the use of aromatic solvents, alkyl aromatic solvents, halogenated aromatic solvents, halogenated alkyl aromatic solvents, or aromatic ether solvents to achieve deprotection of protected hydroxyl groups.
Owner:IONIS PHARMA INC
2'-arabino-fluorooligonucleotide N3'->P5' phosphoramidates: their synthesis and use
Oligonucleotides with a novel sugar-phosphate backbone containing at least one 2′-arabino-fluoronucleoside and an internucleoside 3′-NH—P(—O)(OR)—O-5′ linkage, where R is a positively charged counter ion or hydrogen, and methods of synthesizing and using the inventive oligonucleotides are provided. The inventive phosphoramidate 2′-arabino-fluorooligonucleotides have a high RNA binding affinity to complementary nucleic acids and are base and acid stable.
Owner:GERON CORPORATION
Nucleic acid constructs and methods of use
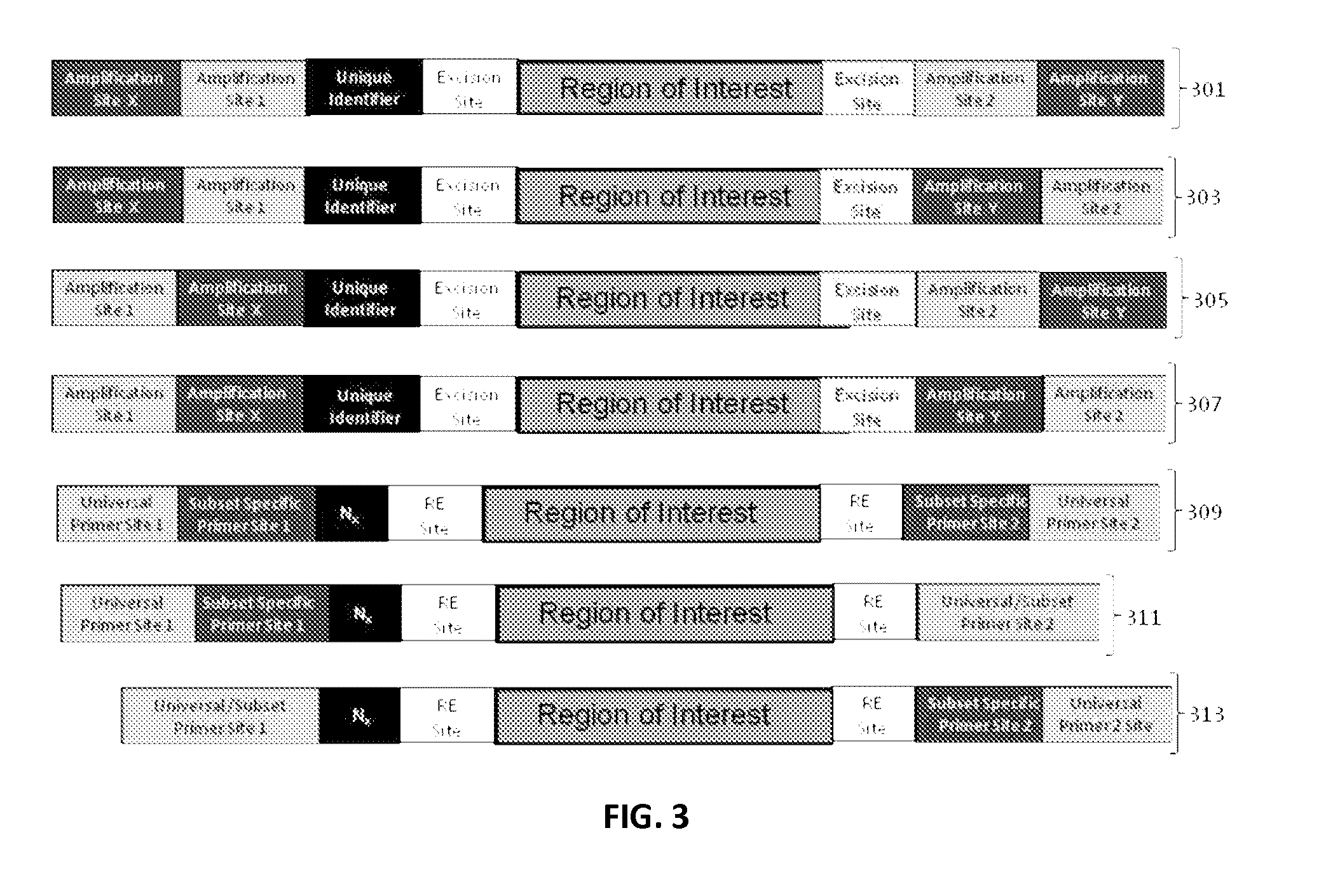
ActiveUS9085798B2Minimization requirementsSugar derivativesMicrobiological testing/measurementSynthetic biologyMutation screening
The present invention provides oligonucleotide constructs, sets of such oligonucleotide constructs, and methods of using such oligonucleotide constructs to provide validated sequences or sets of validated sequences corresponding to desired ROIs. Such validated ROIs and constructs containing these have a wide variety of uses, including in synthetic biology, quantitative nucleic acid analysis, polymorphism and / or mutation screening, and the like.
Owner:PROGNOSYS BIOSCI
Method of determining the nucleotide sequence of oligonucleotides and DNA molecules
InactiveUS6780591B2Eliminate needHigh purityMicrobiological testing/measurementRecombinant DNA-technologyFluorescenceOligonucleotide primers
The present invention relates to a novel method for analyzing nucleic acid sequences based on real-time detection of DNA polymerase-catalyzed incorporation of each of the four nucleotide bases, supplied individually and serially in a microfluidic system, to a reaction cell containing a template system comprising a DNA fragment of unknown sequence and an oligonucleotide primer. Incorporation of a nucleotide base into the template system can be detected by any of a variety of methods including but not limited to fluorescence and chemiluminescence detection. Alternatively, microcalorimetic detection of the heat generated by the incorporation of a nucleotide into the extending template system using thermopile, thermistor and refractive index measurements can be used to detect extension reactions.
Owner:LIFE TECH CORP
Microfabricated integrated DNA analysis system
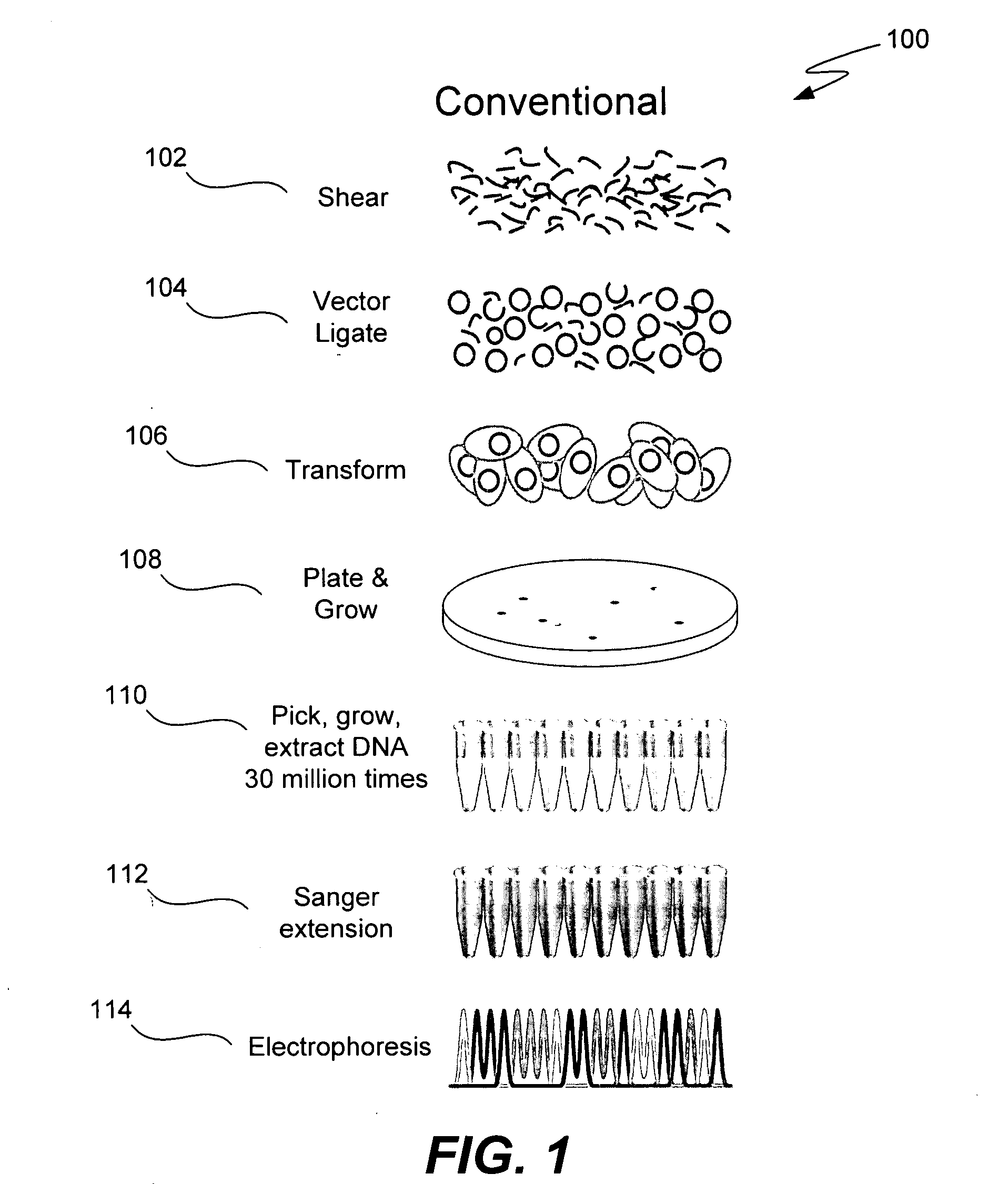
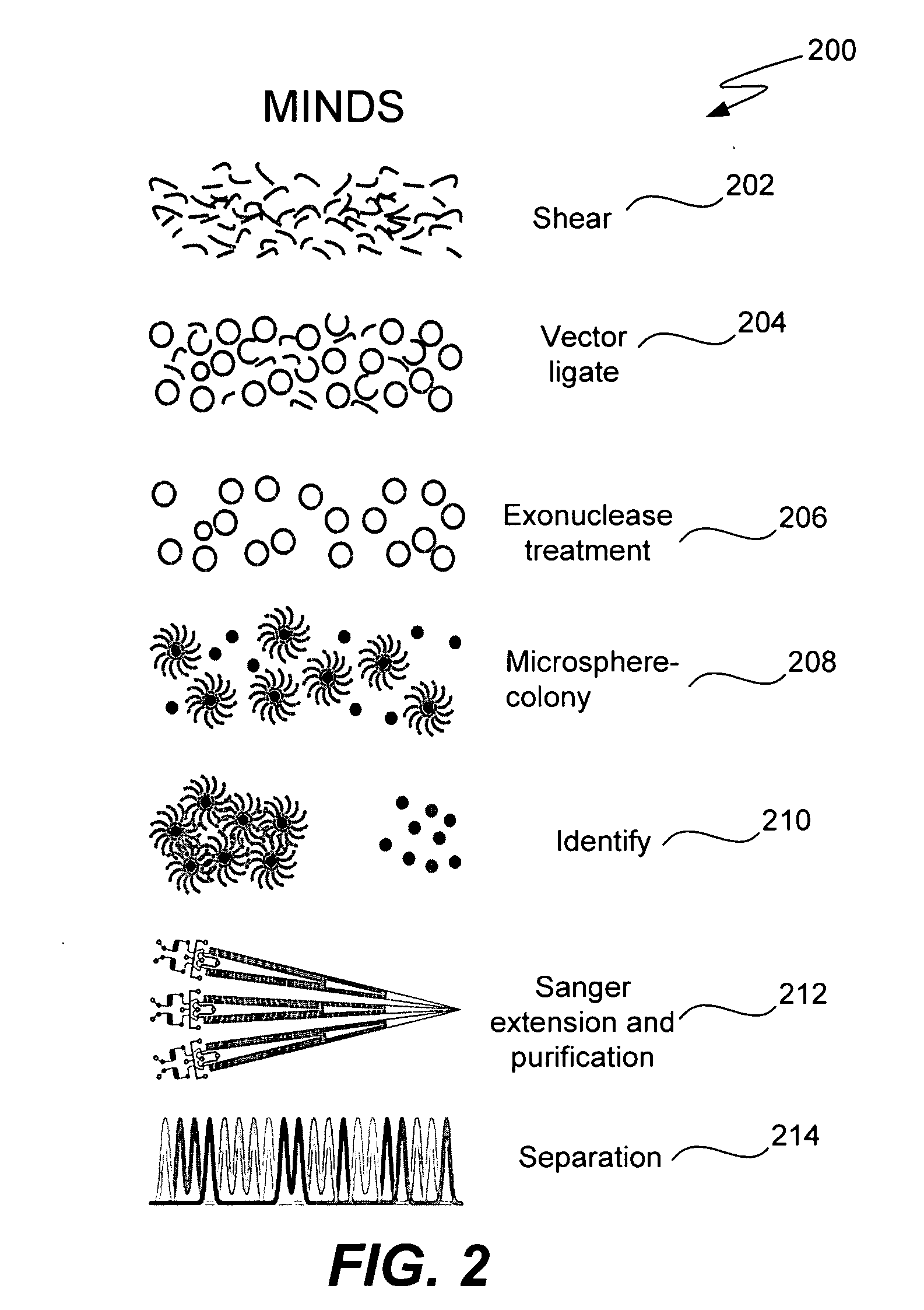
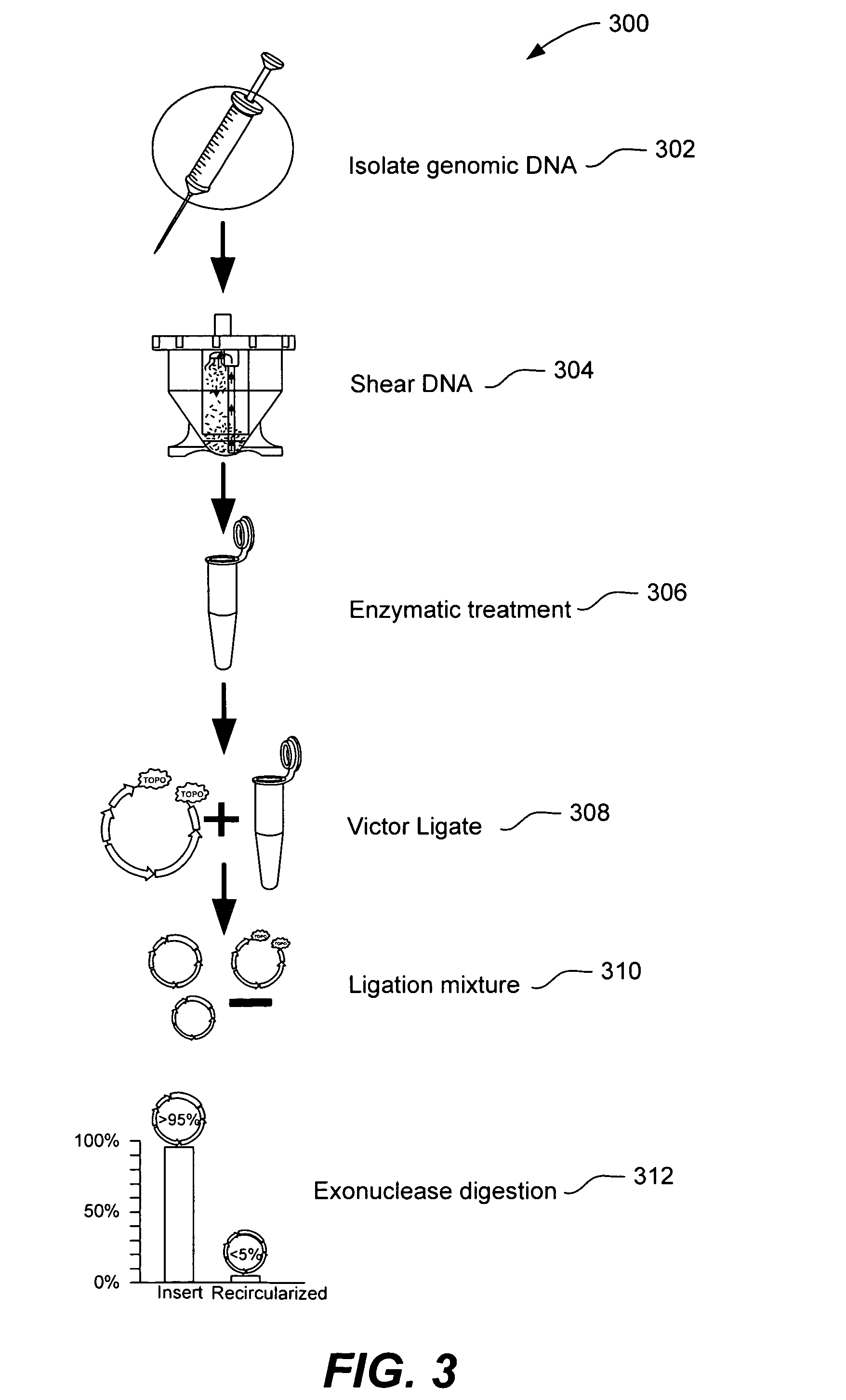
ActiveUS20050287572A1Improve automationBig spaceBioreactor/fermenter combinationsElectrolysis componentsMicroreactorMicrosphere
Methods and apparatus for genome analysis are provided. A microfabricated structure including a microfluidic distribution channel is configured to distribute microreactor elements having copies of a sequencing template into a plurality of microfabricated thermal cycling chambers. A microreactor element may include a microcarrier element carrying the multiple copies of the sequencing template. The microcarrier element may comprise a microsphere. An autovalve at an exit port of a thermal cycling chamber, an optical scanner, or a timing arrangement may be used to ensure that only one microsphere will flow into one thermal cycling chamber wherein thermal cycling extension fragments are produced. The extension products are captured, purified, and concentrated in an integrated oligonucleotide gel capture chamber. A microfabricated component separation apparatus is used to analyze the purified extension fragments. The microfabricated structure may be used in a process for performing sequencing and other genetic analysis of DNA or RNA.
Owner:RGT UNIV OF CALIFORNIA
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com