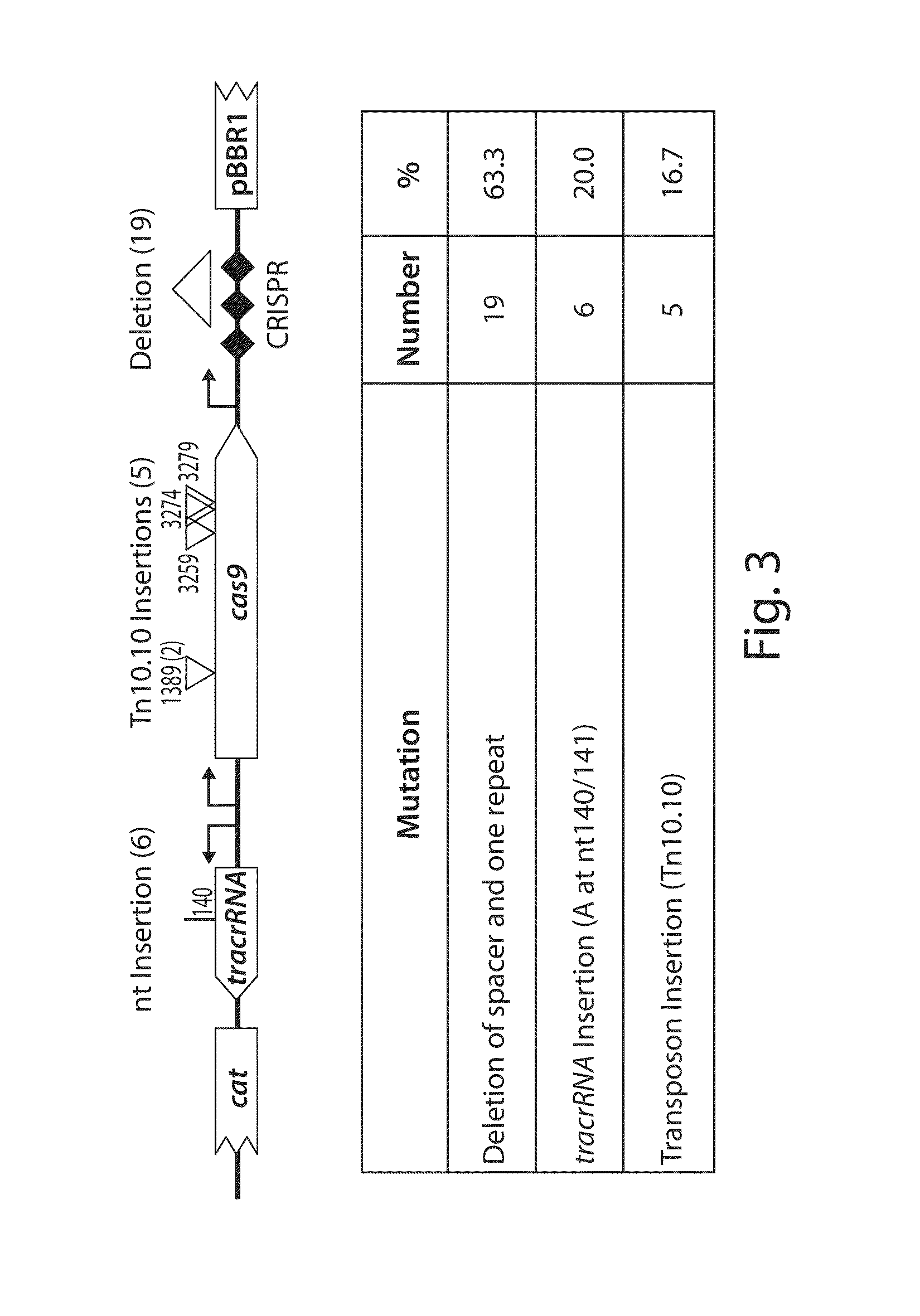
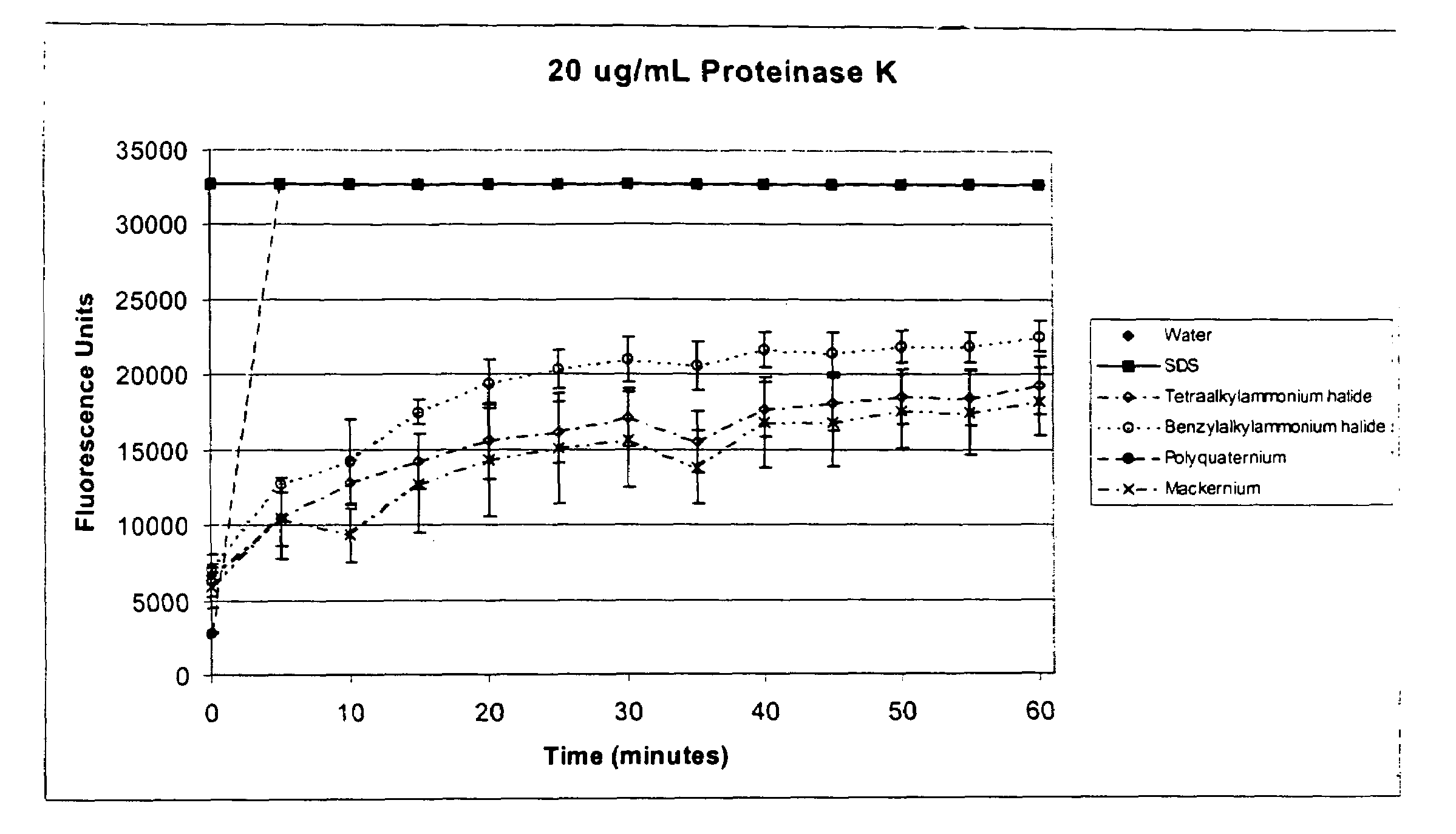
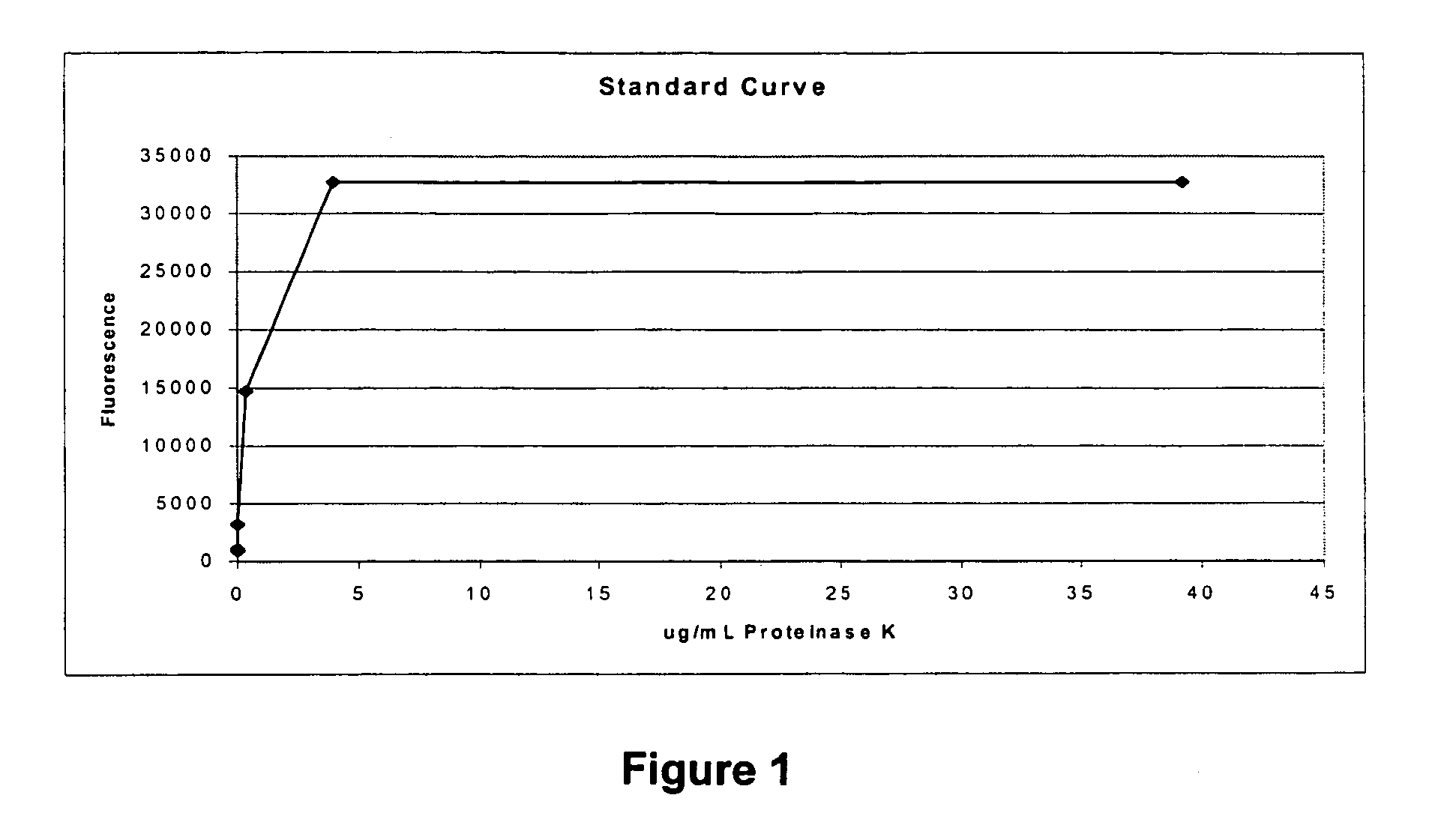
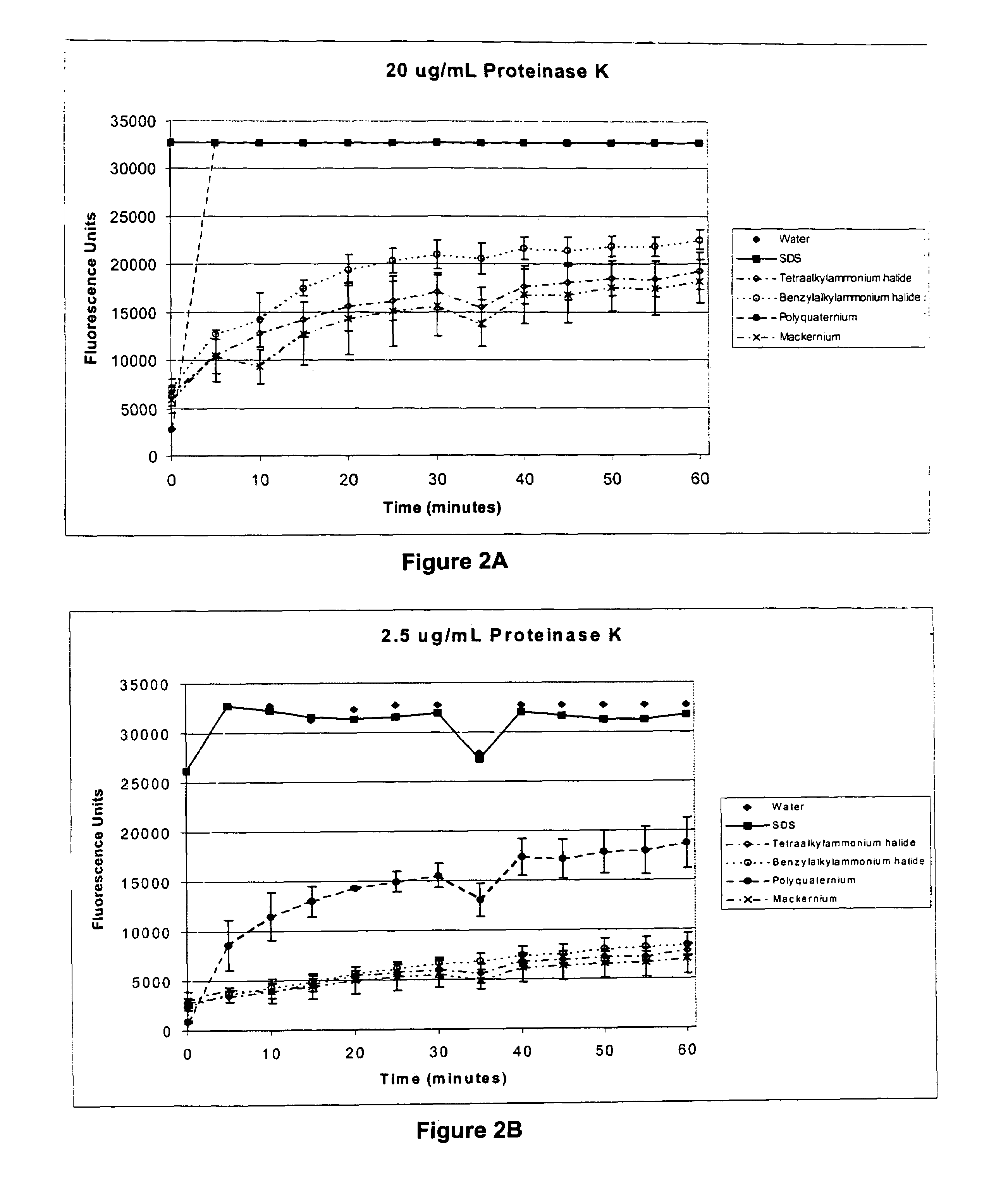
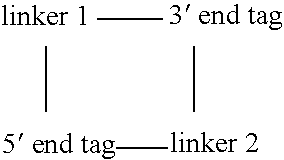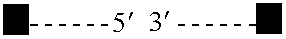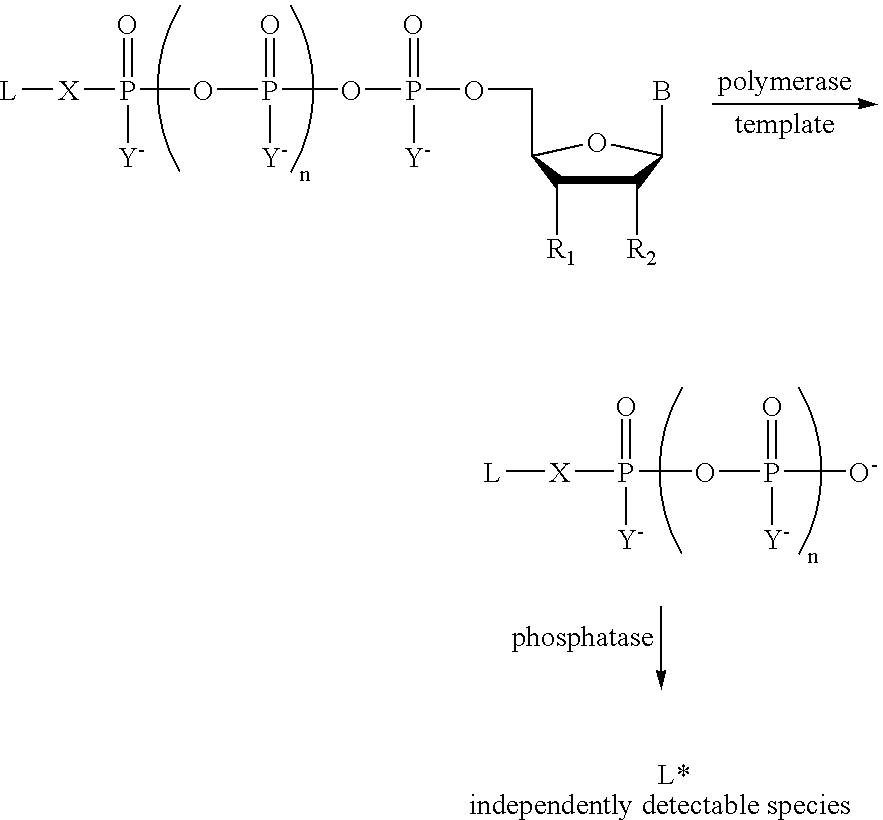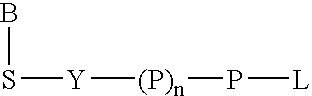Patents
Literature
1817 results about "Nuclease" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.
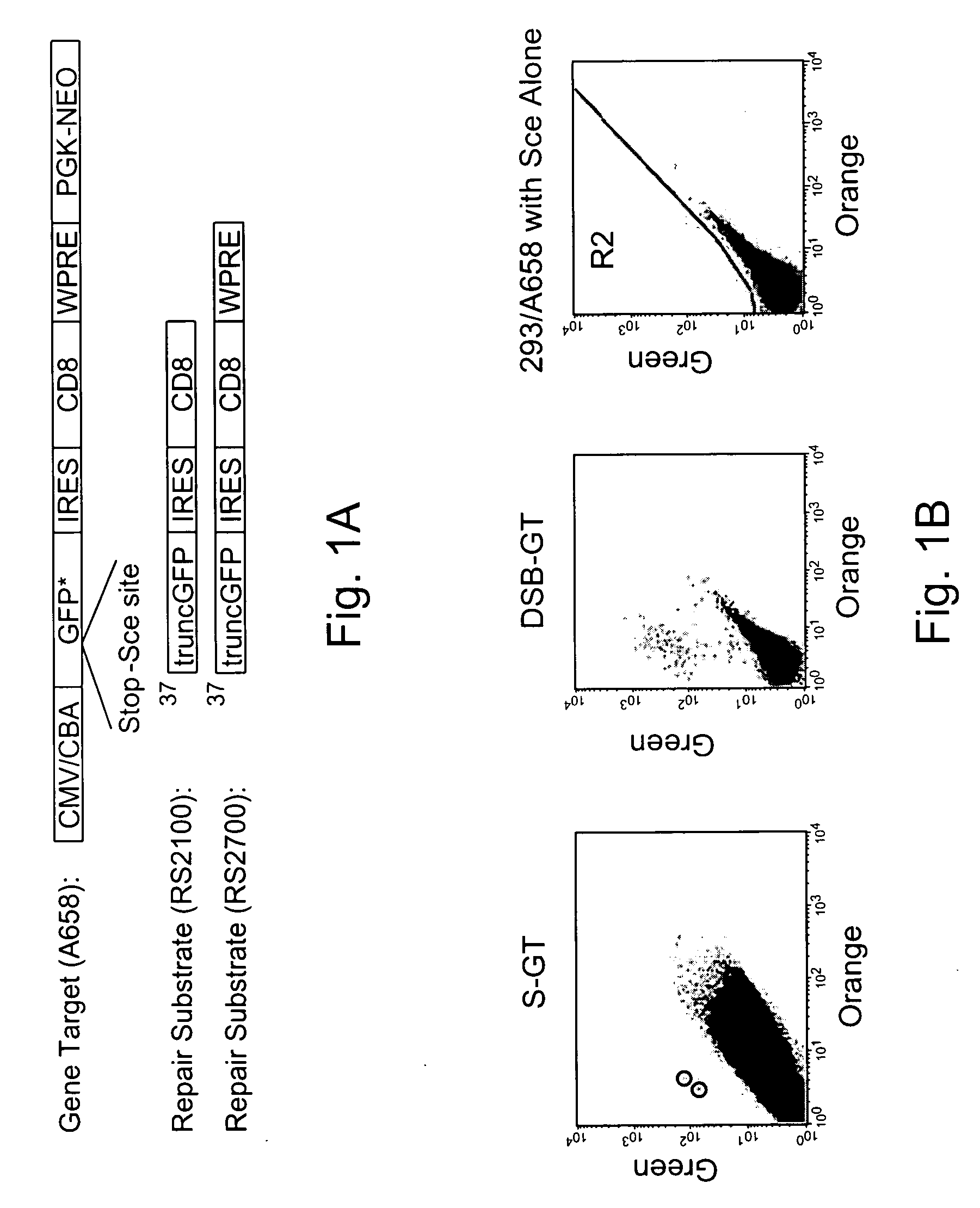
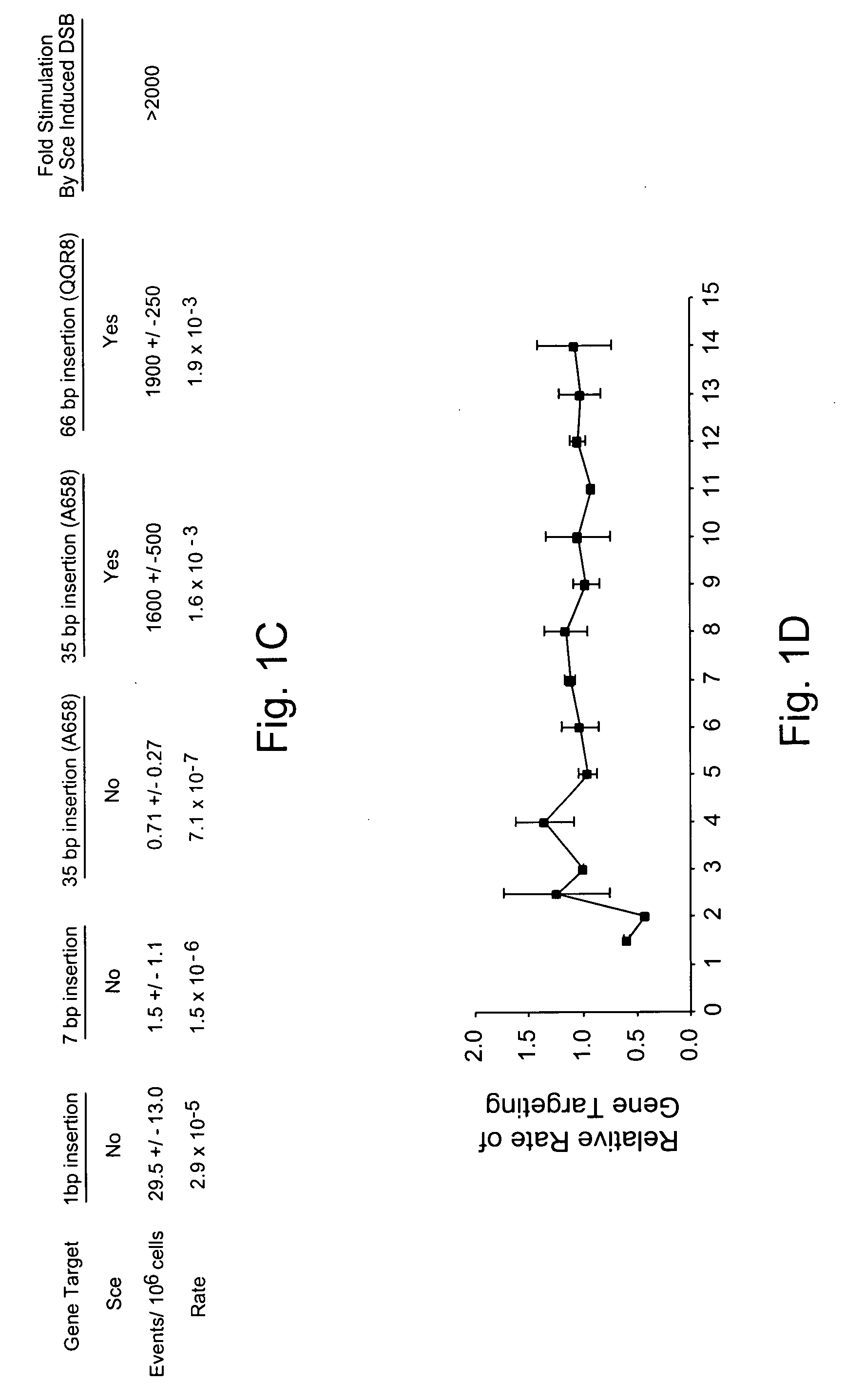
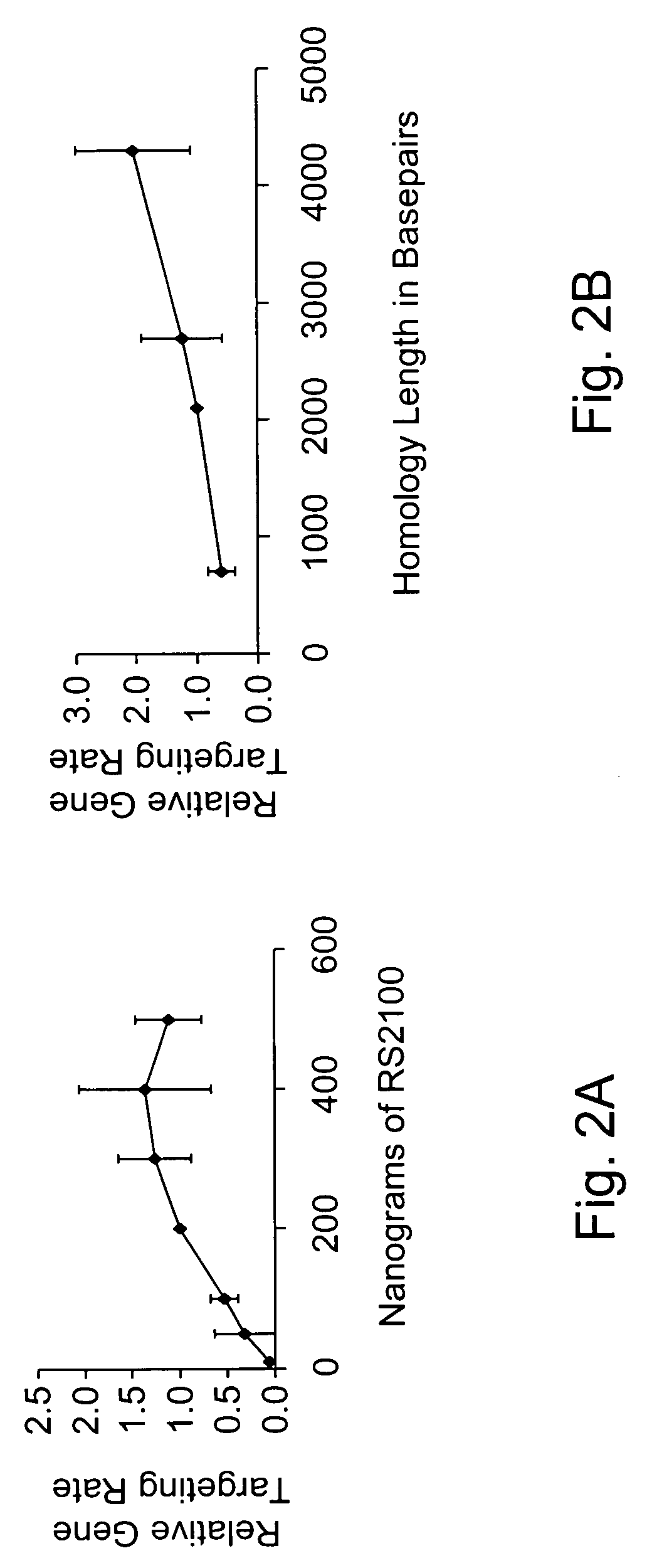
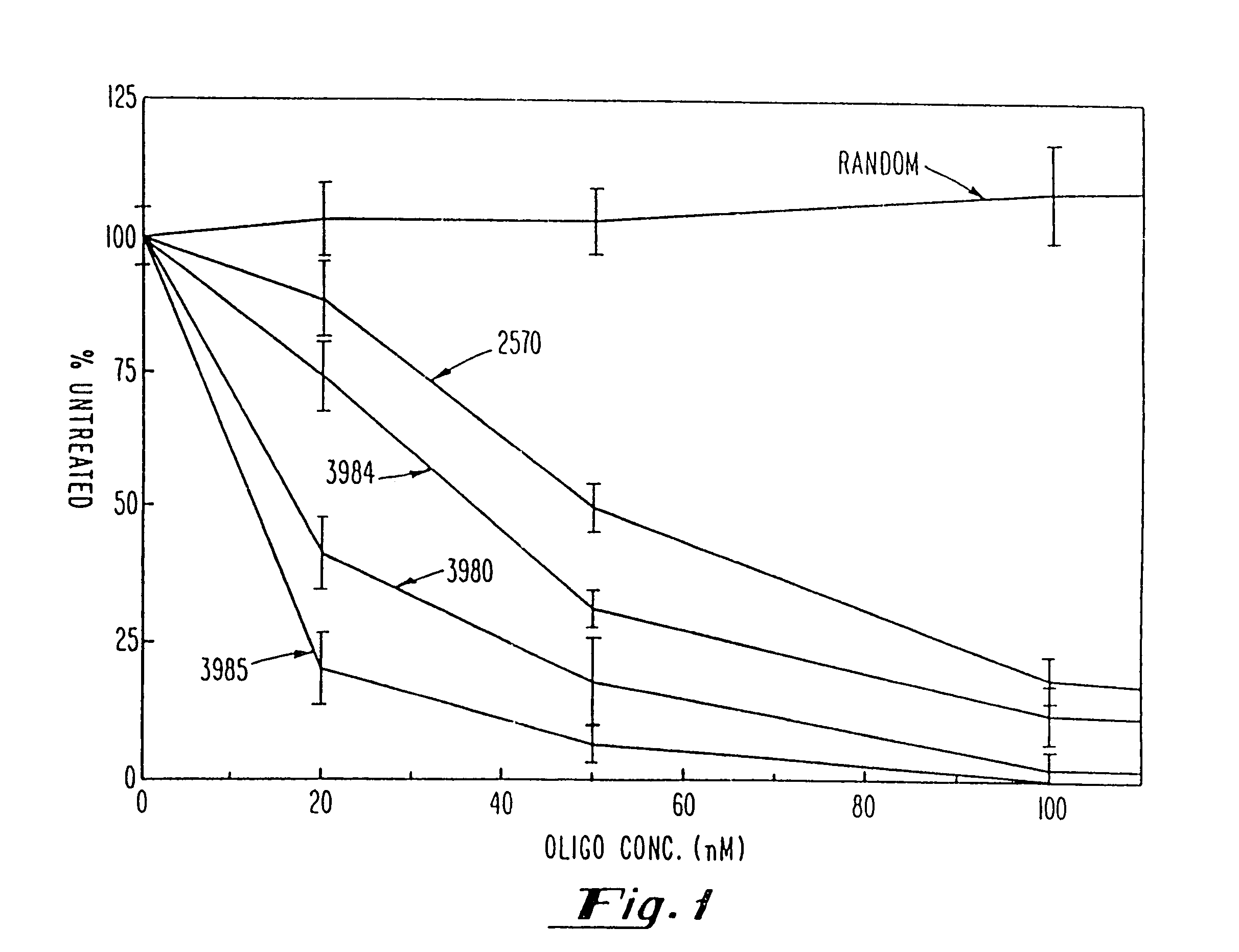
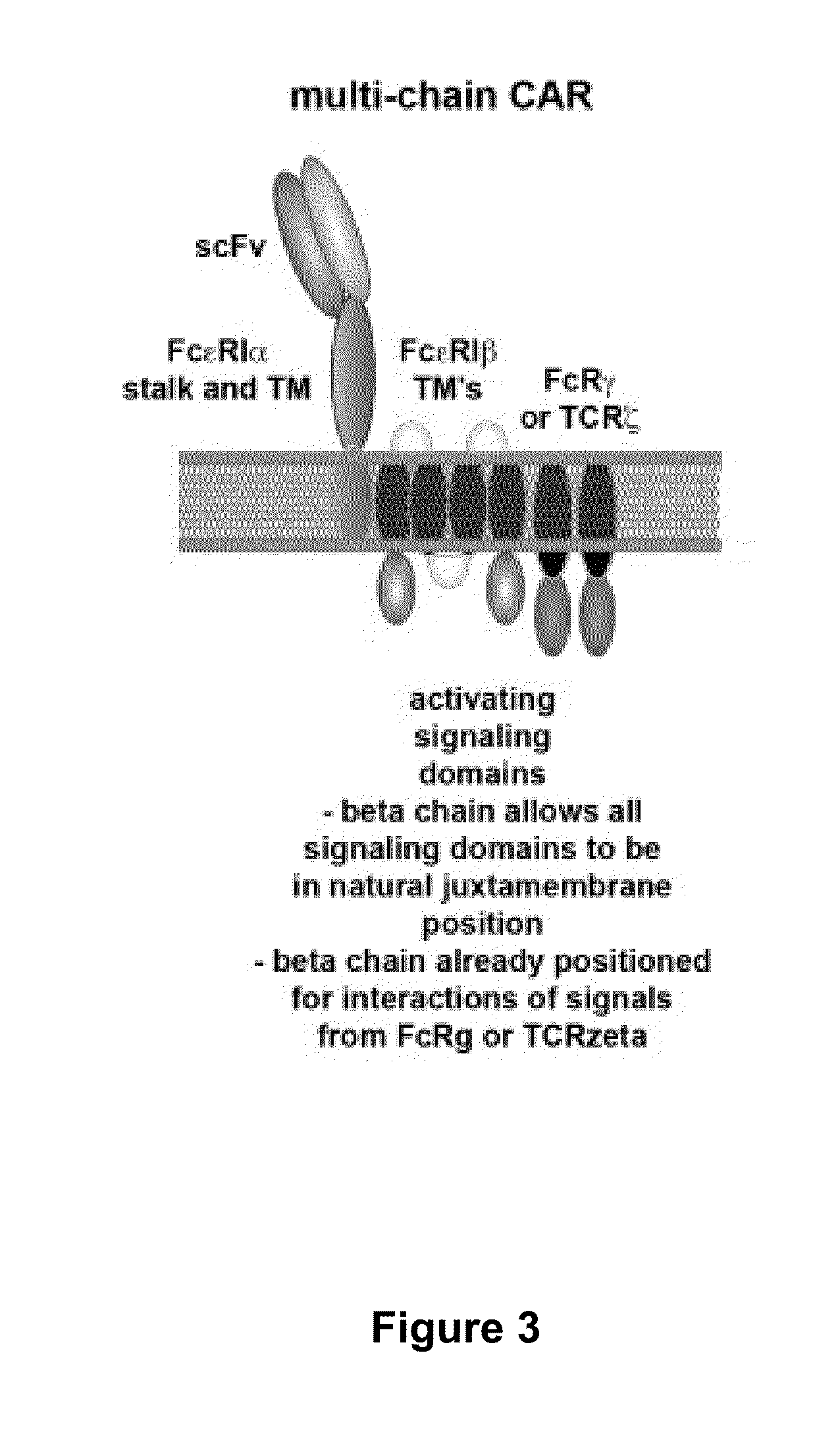
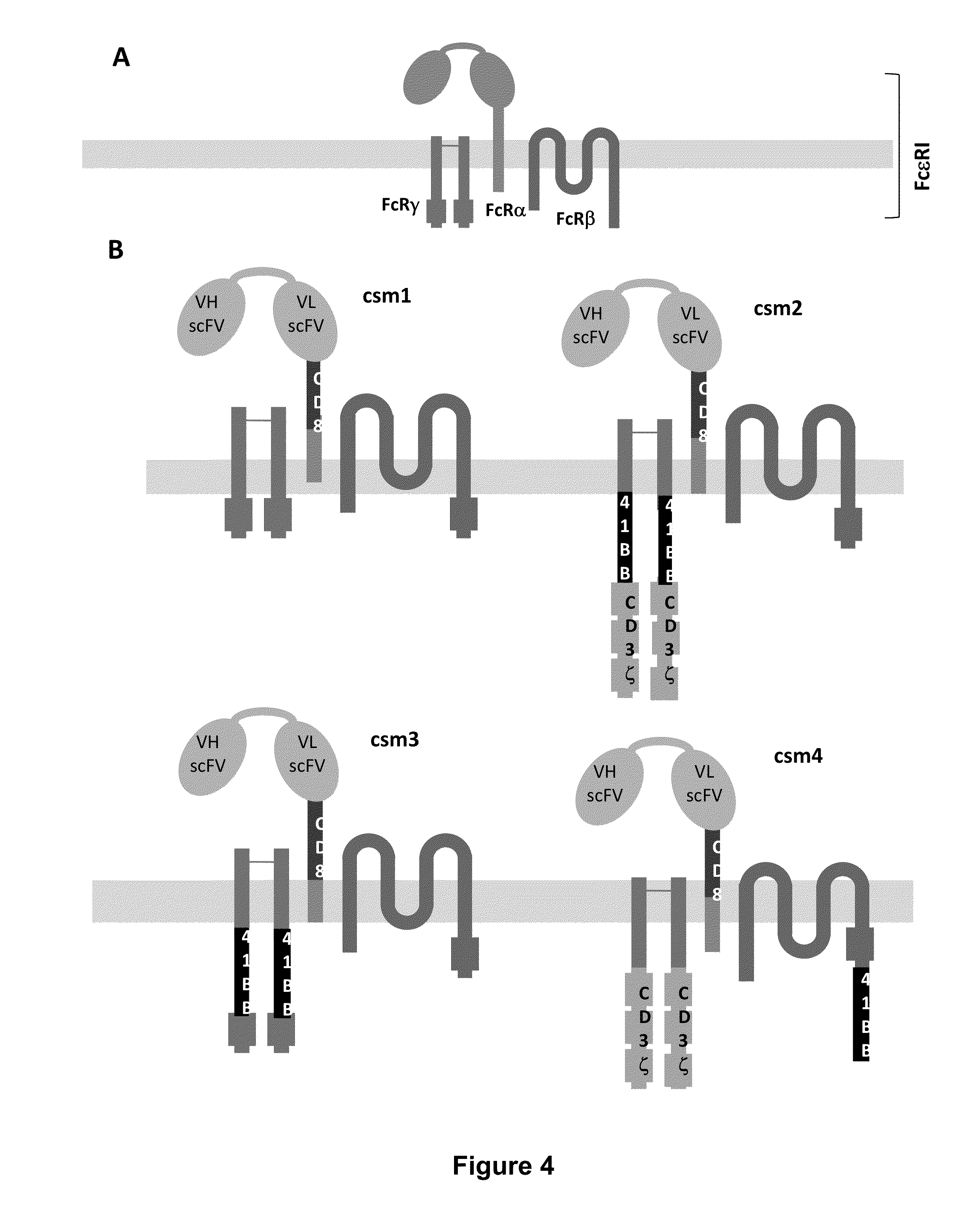
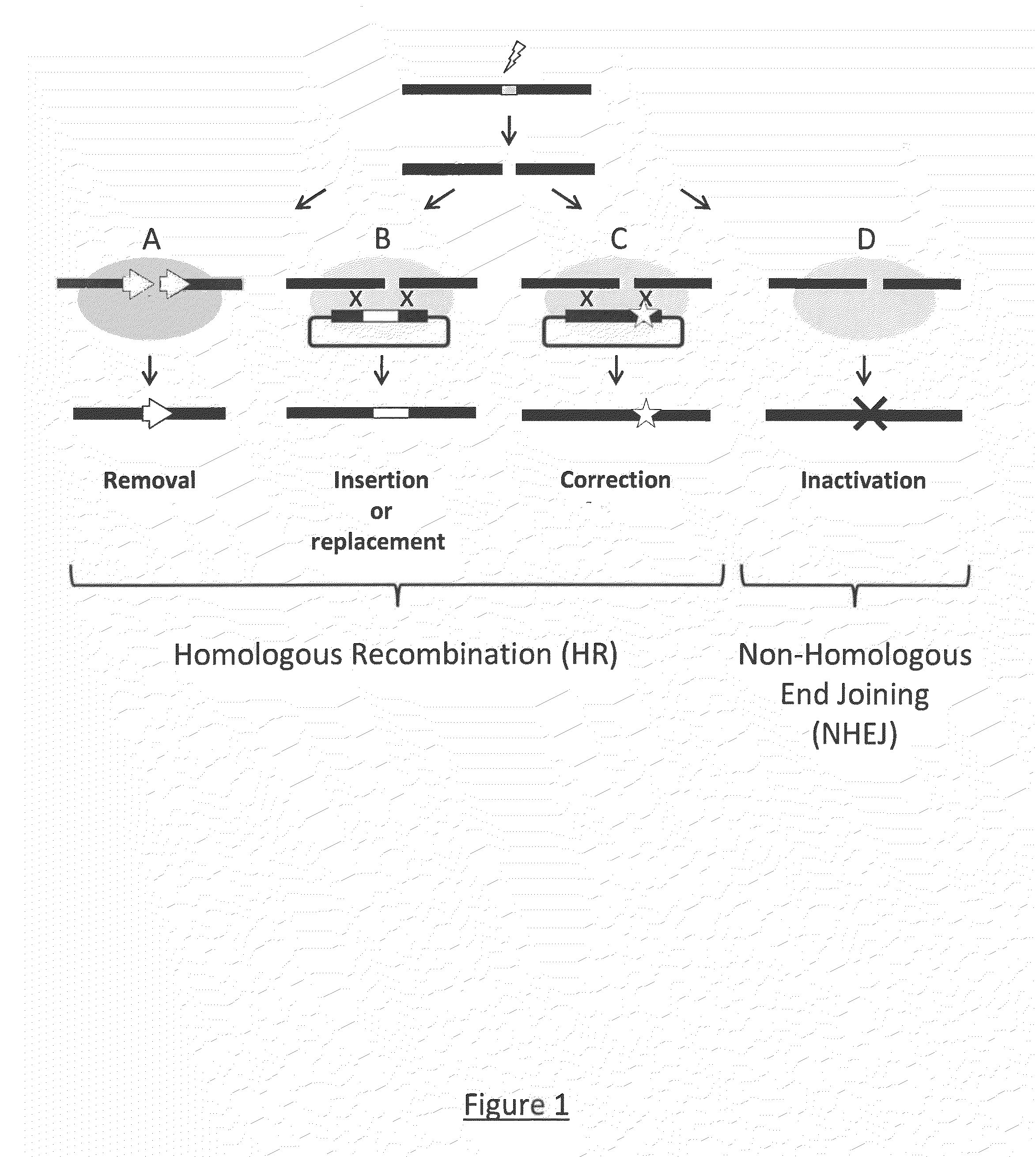
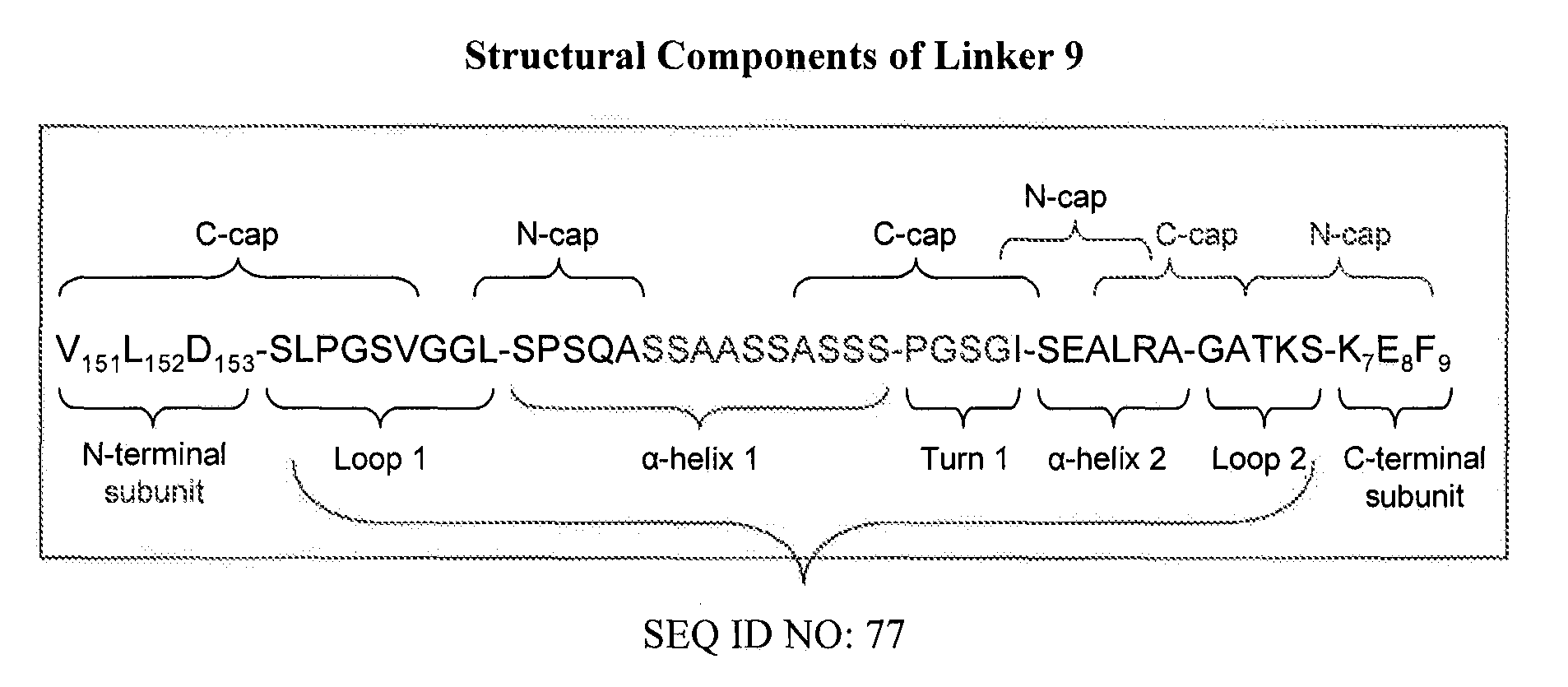
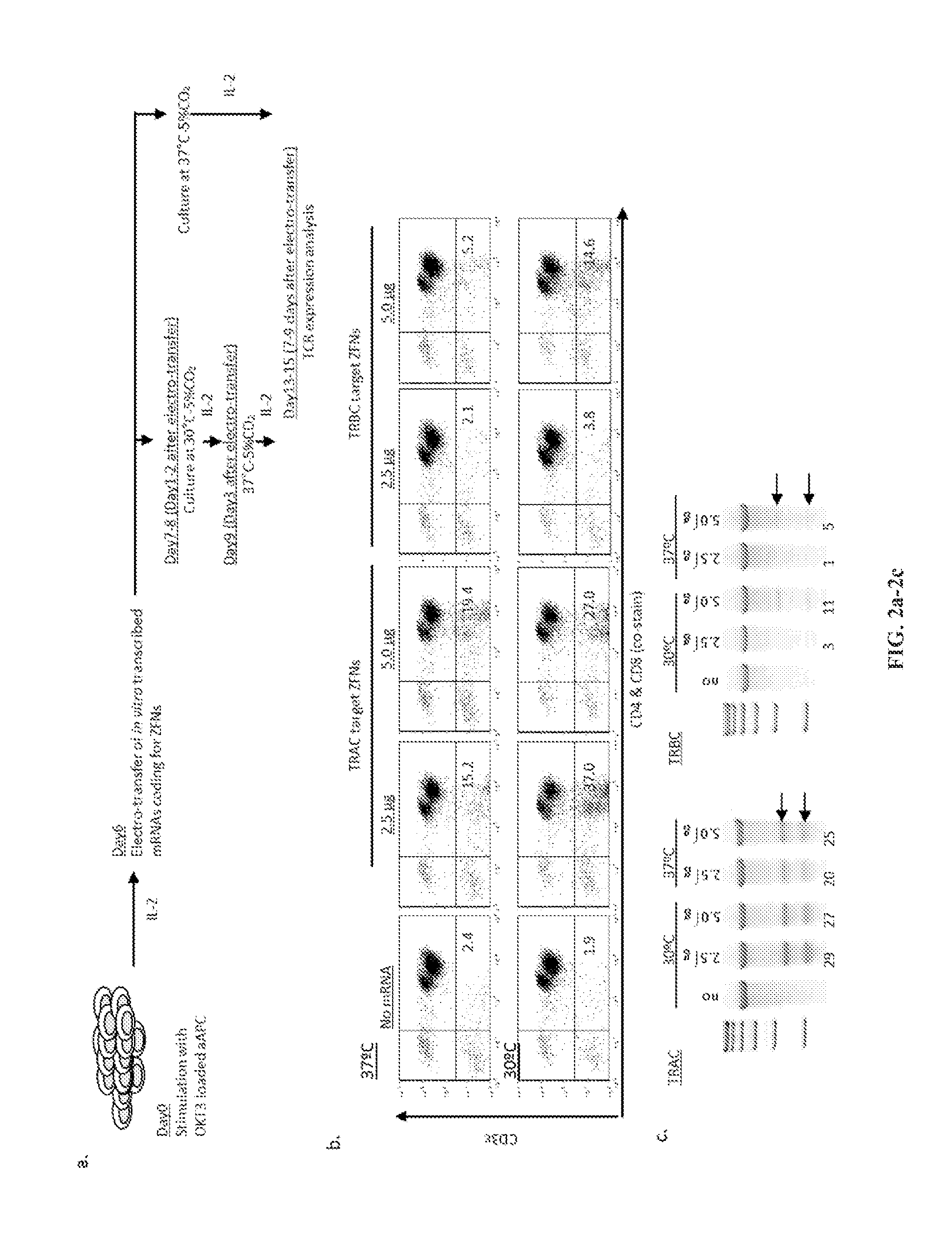
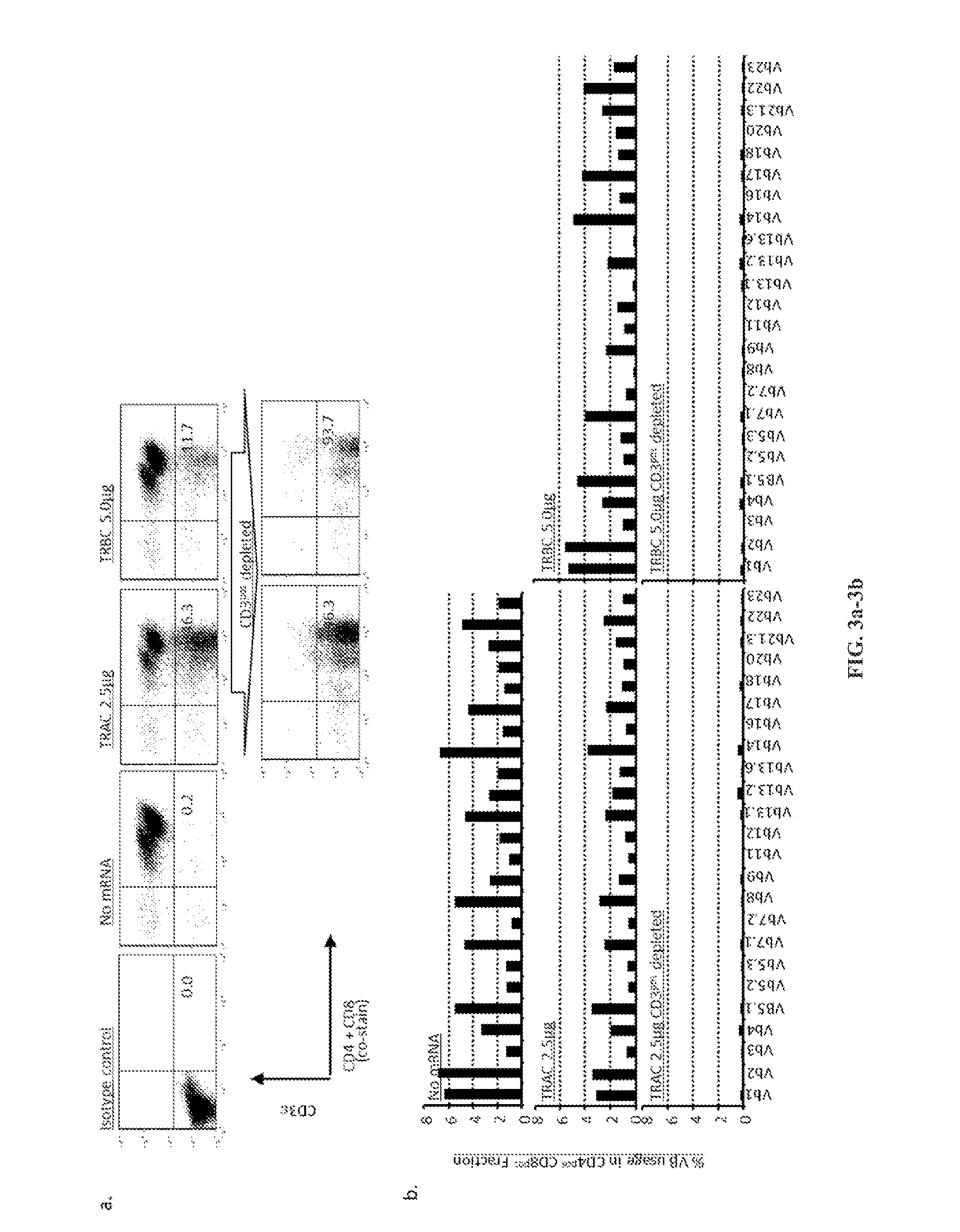
Use of chimeric nucleases to stimulate gene targeting
ActiveUS20050026157A1Ameliorate genetic disorderIncrease productionAntibacterial agentsFusion with DNA-binding domainGene targetsGenetic Change
Gene targeting is a technique to introduce genetic change into one or more specific locations in the genome of a cell. For example, gene targeting can introduce genetic change by modifying, repairing, attenuating or inactivating a target gene or other chromosomal DNA. In one aspect, this disclosure relates to methods and compositions for gene targeting with high efficiency in a cell. This disclosure also relates to methods of treating or preventing a genetic disease in an individual in need thereof. Further disclosed are chimeric nucleases and vectors encoding chimeric nucleases.
Owner:CALIFORNIA INST OF TECH
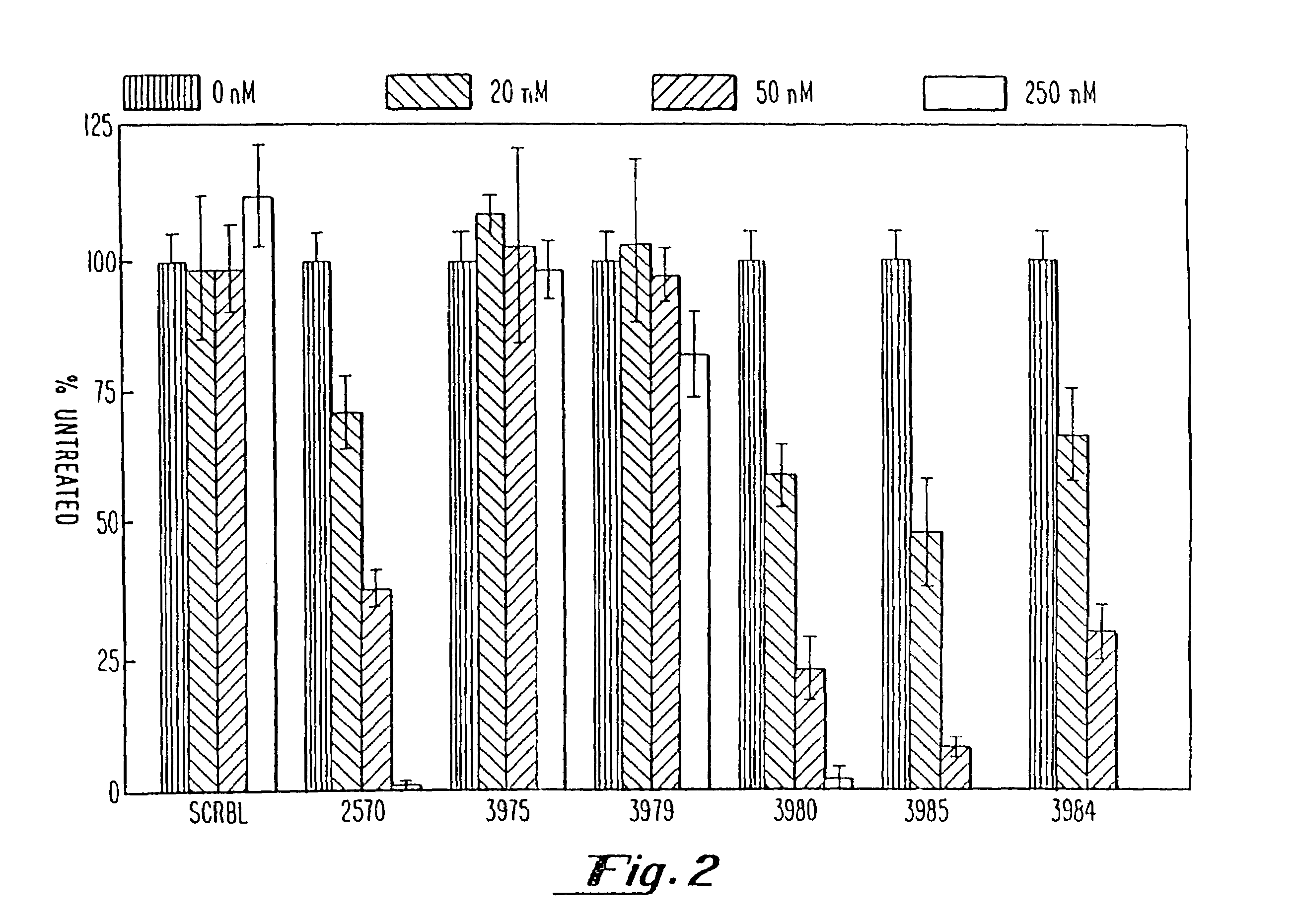
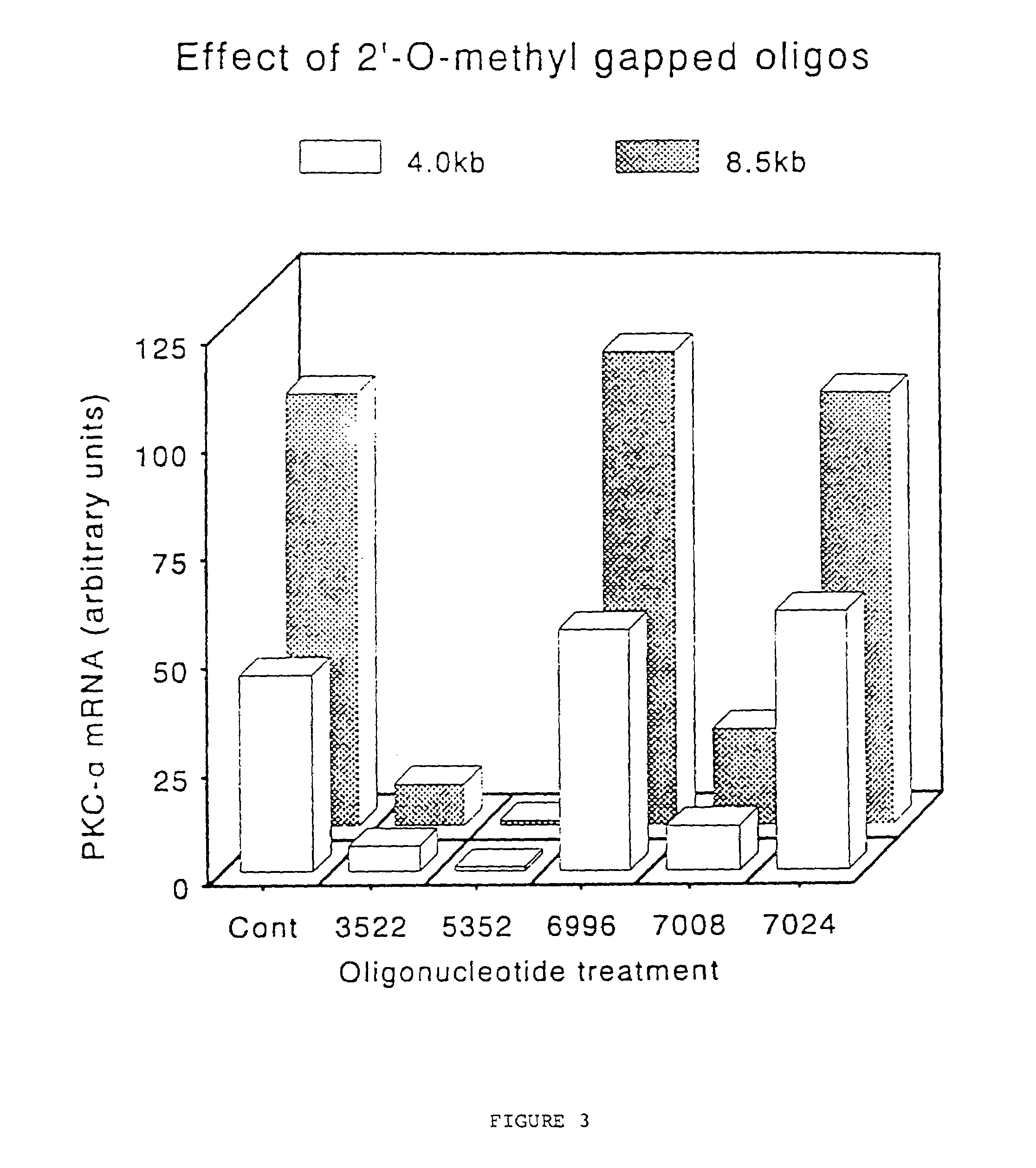
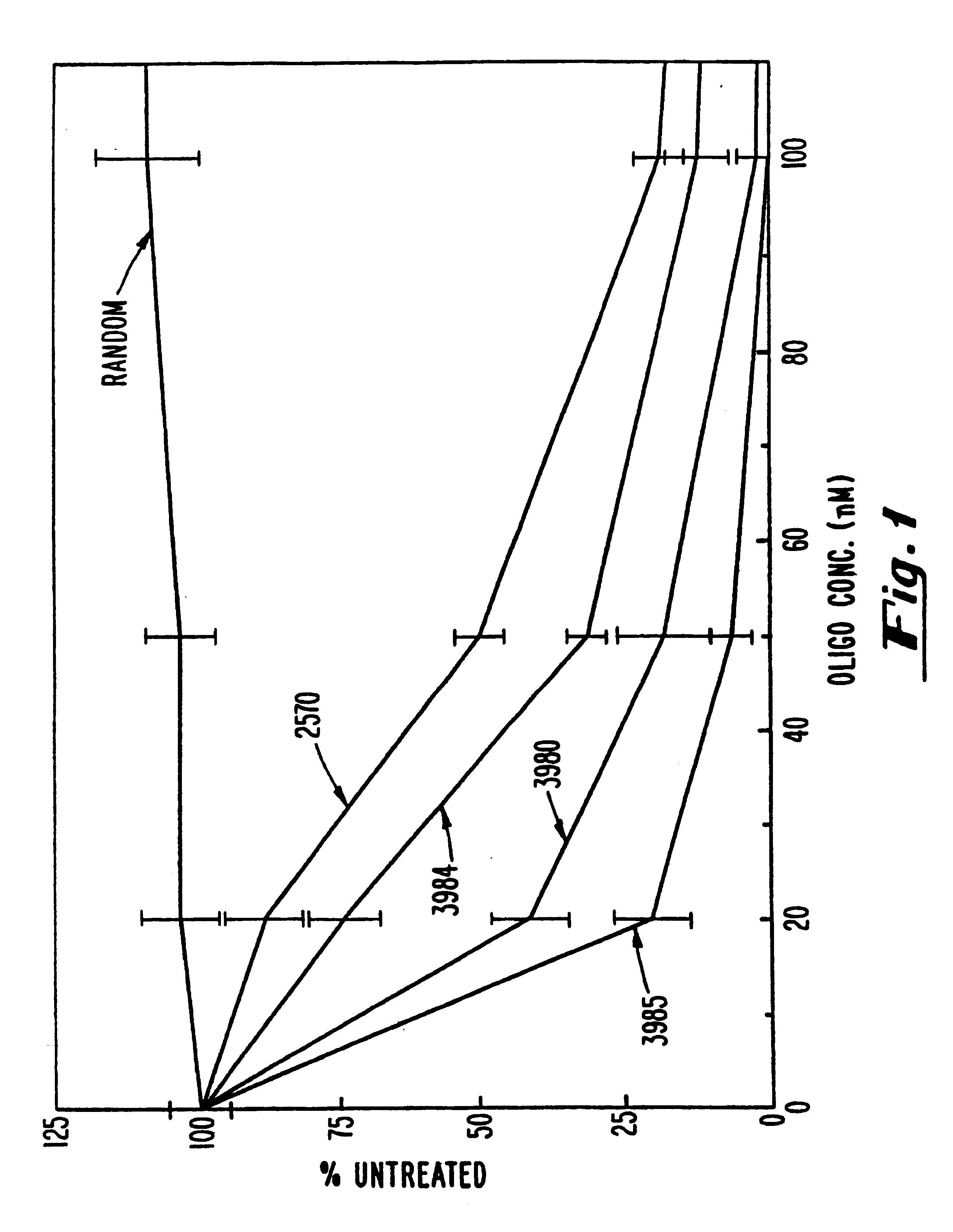
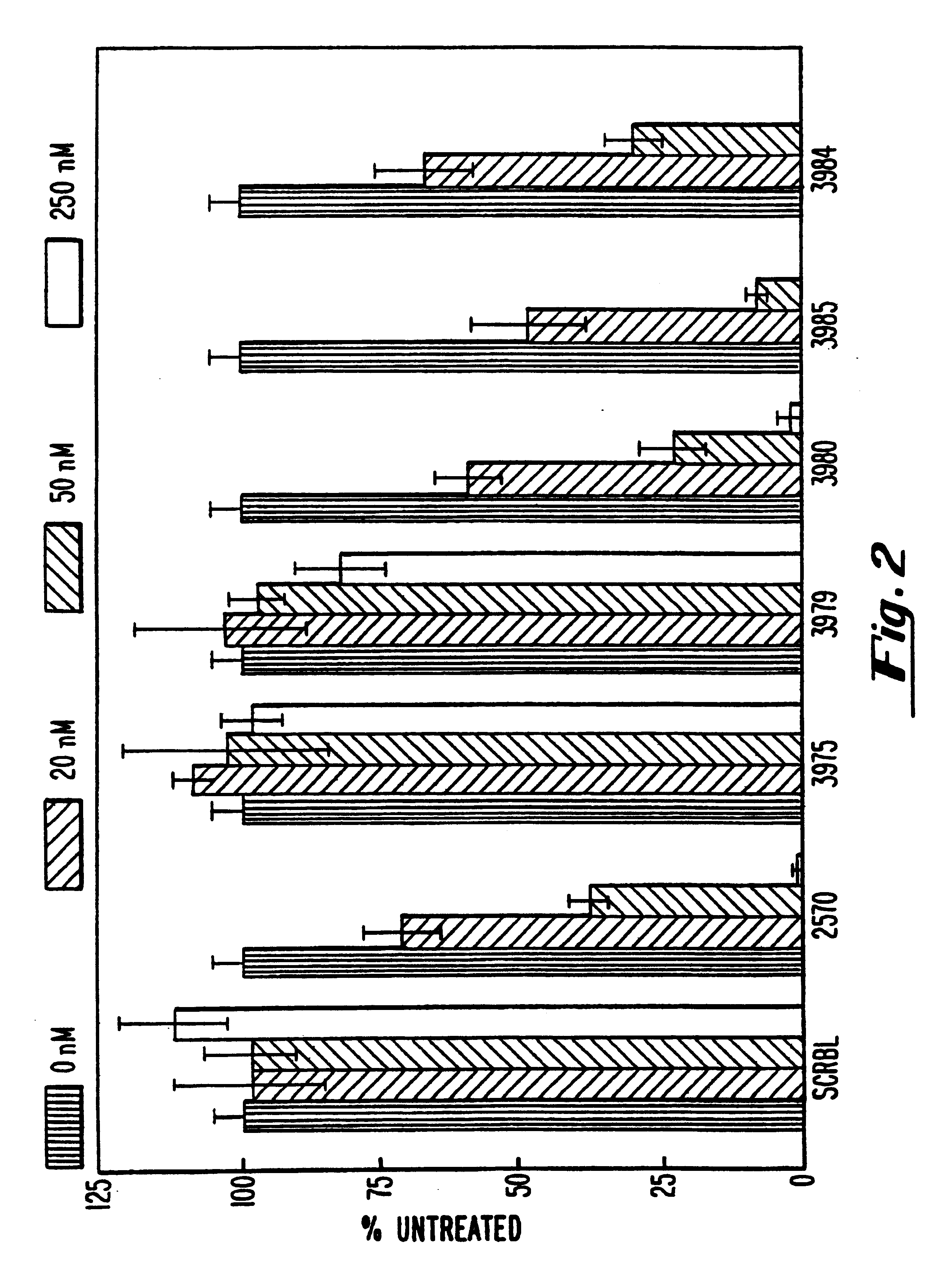
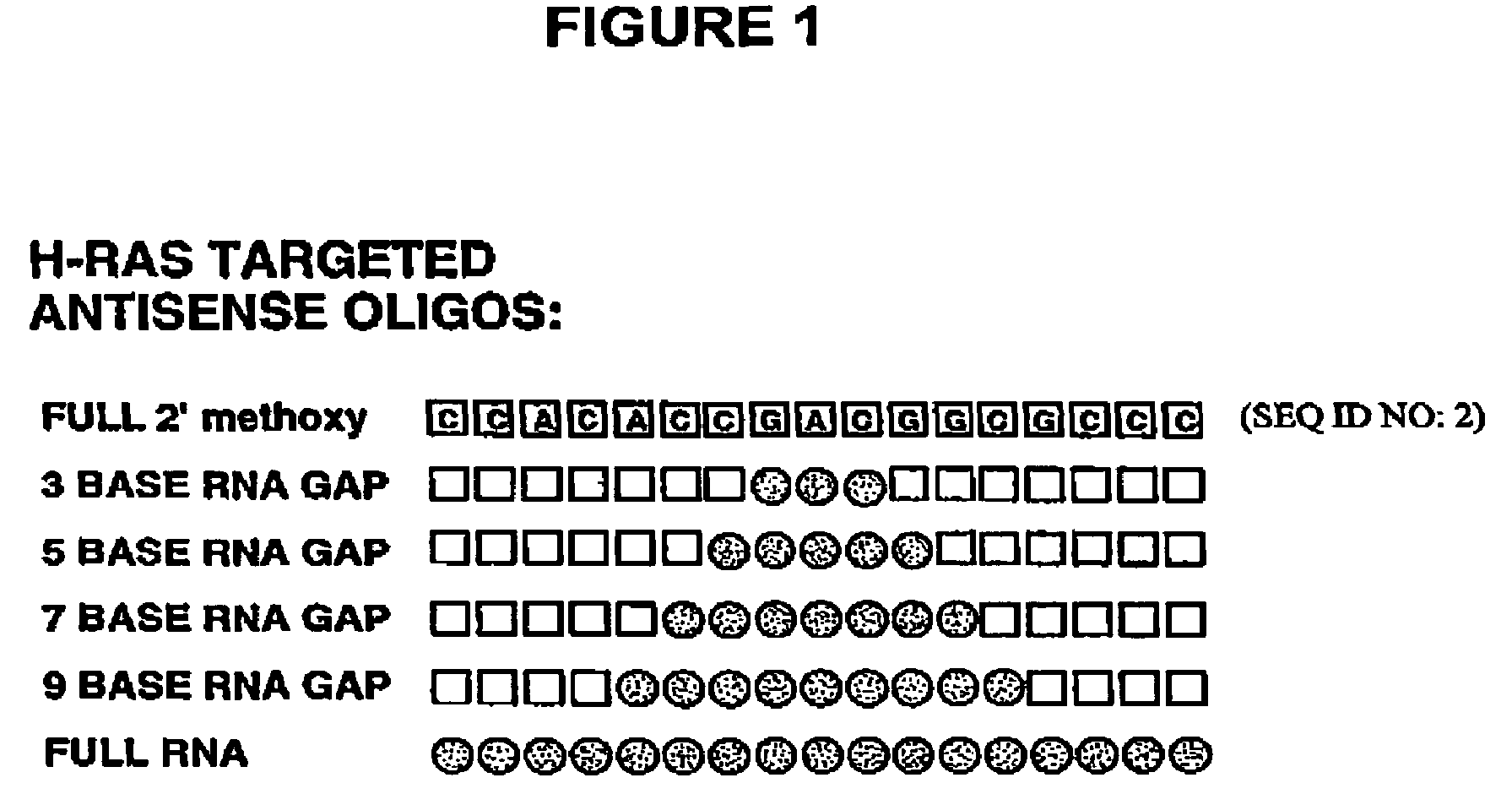
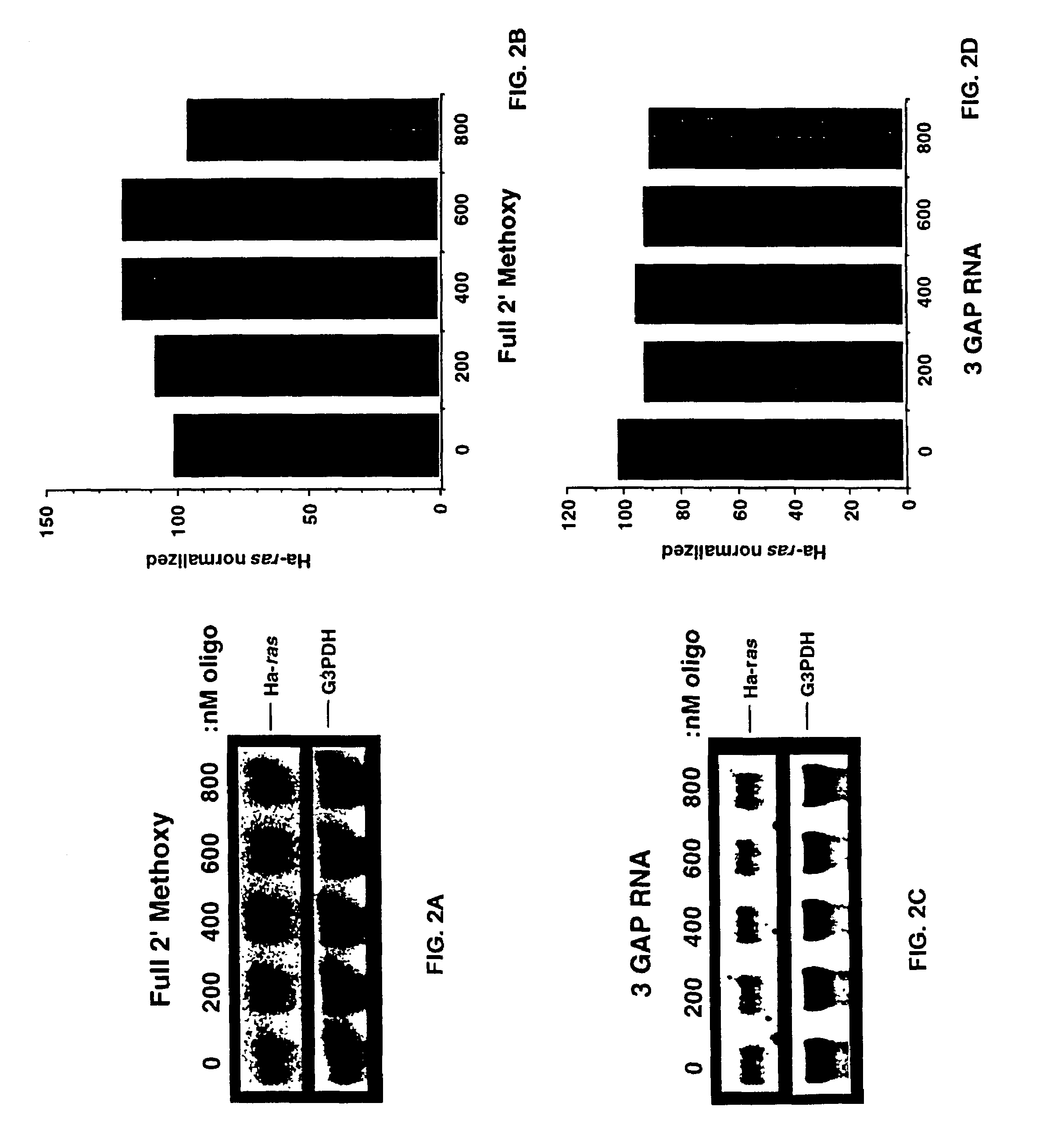
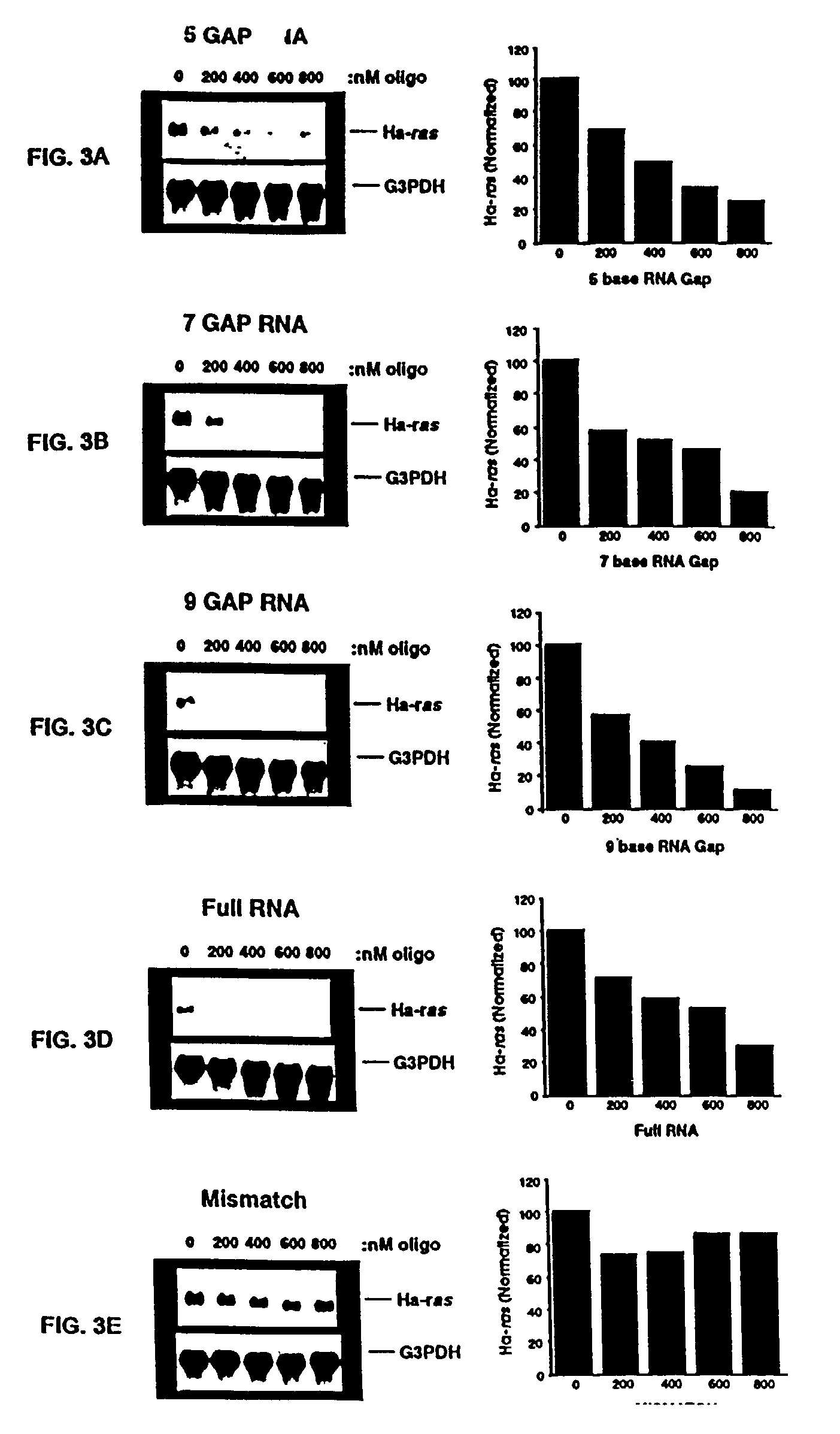
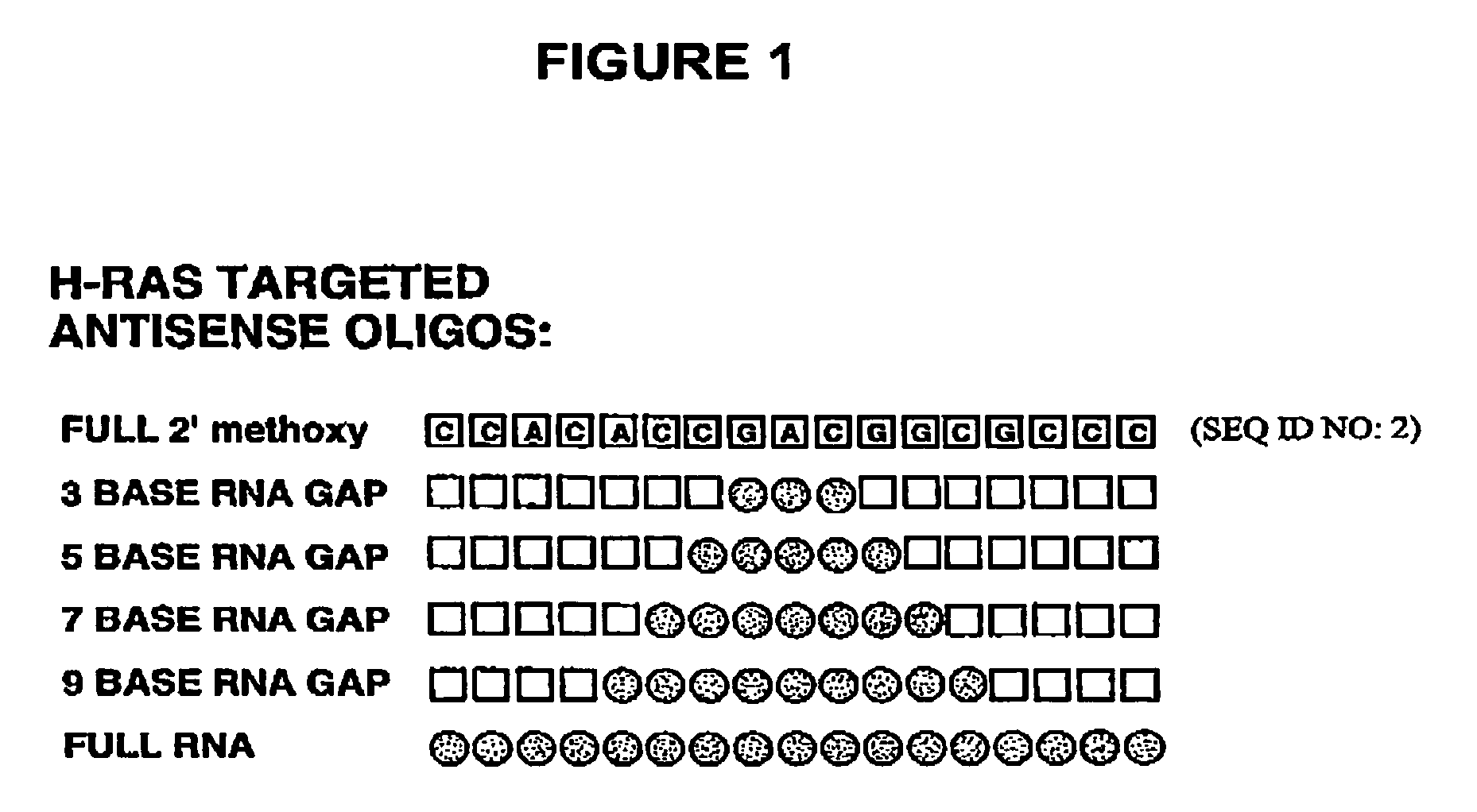
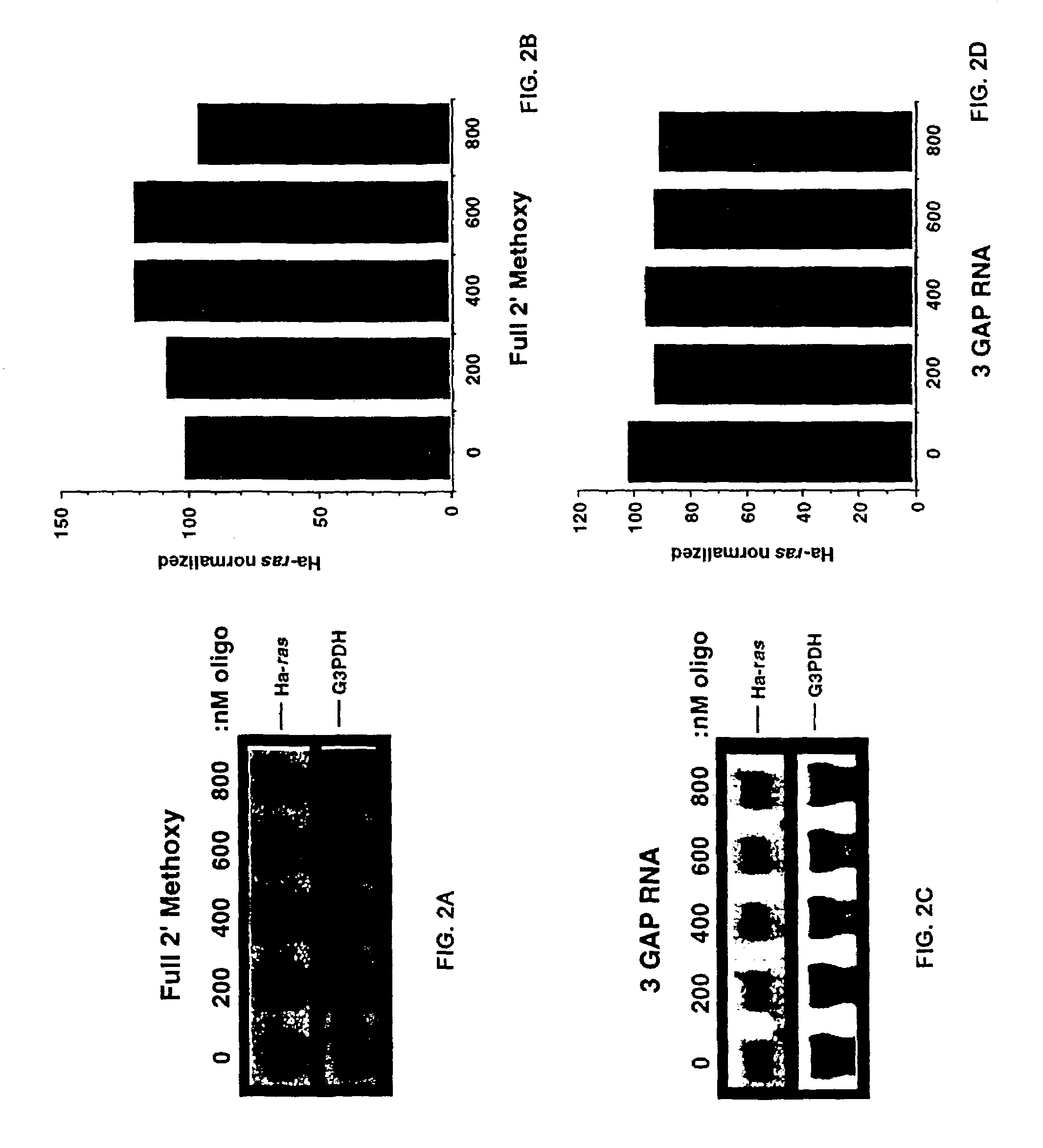
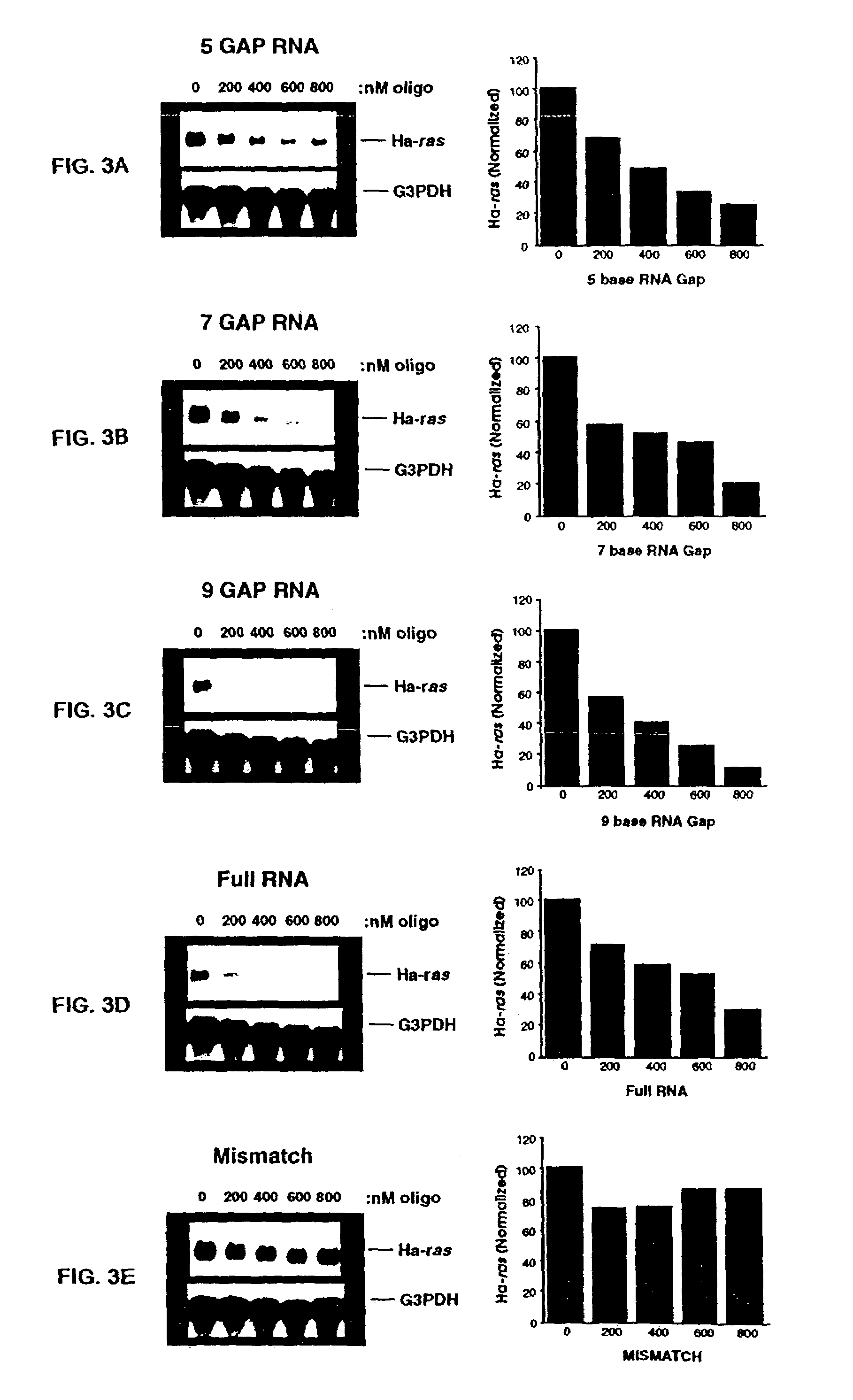
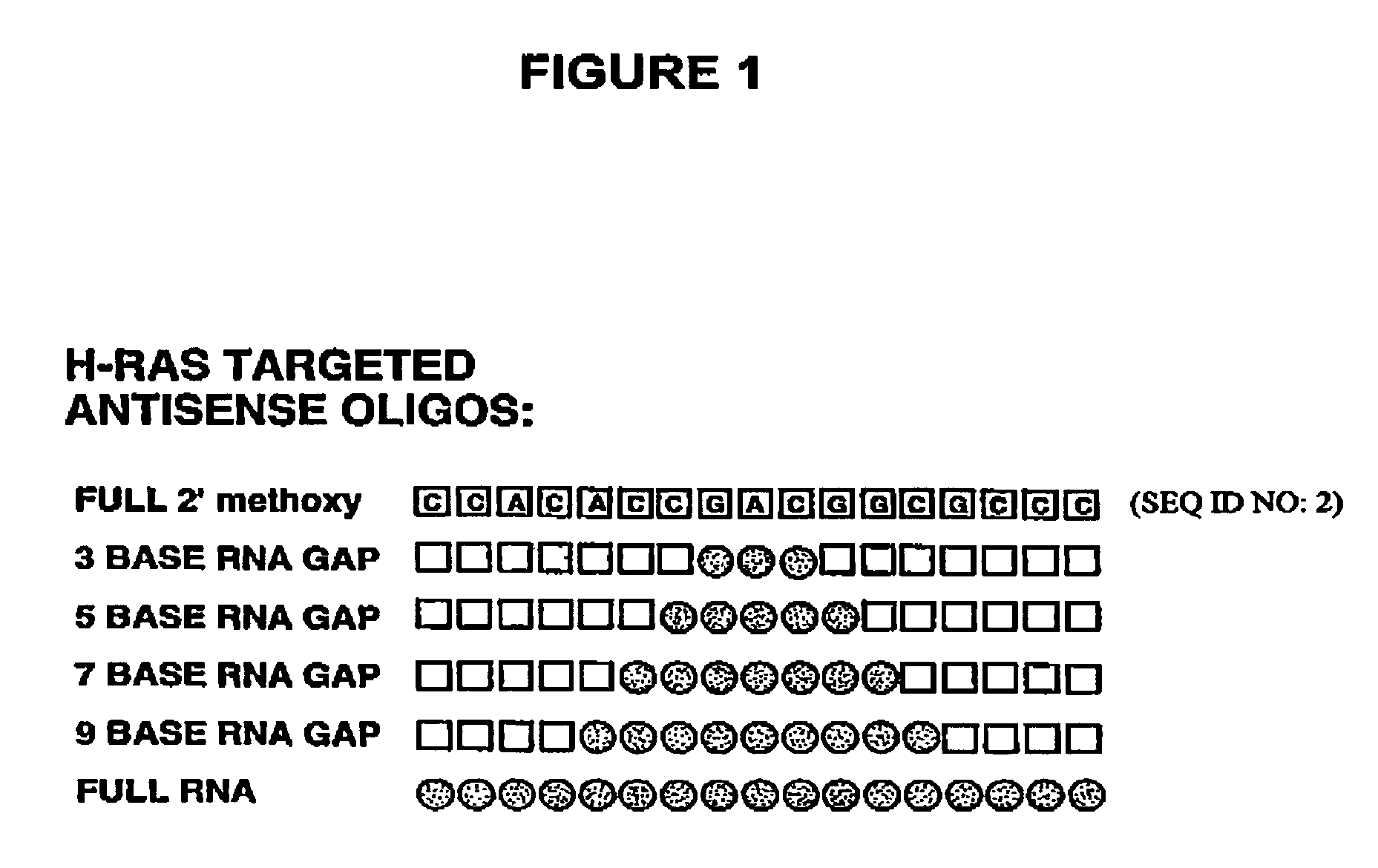
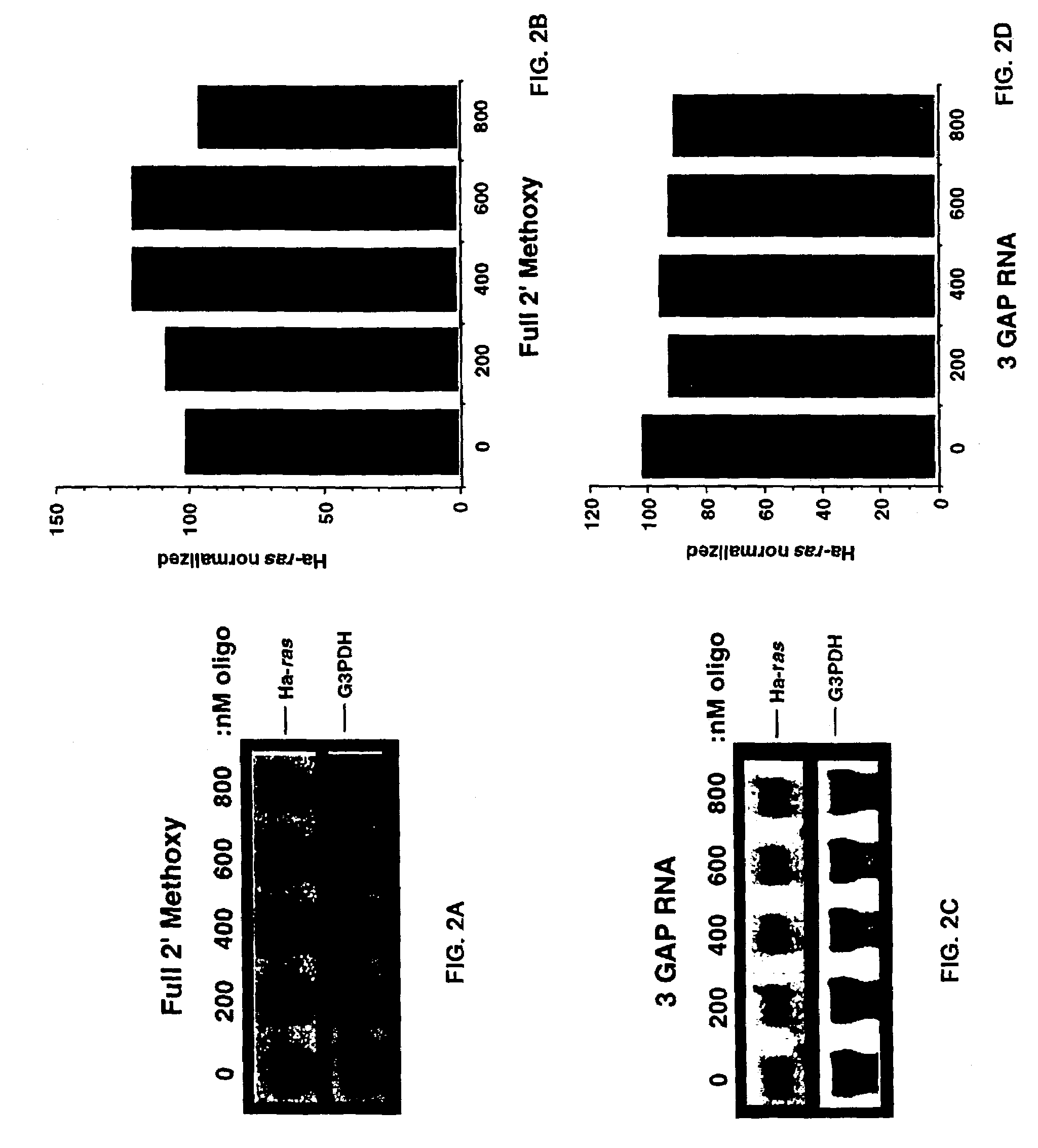
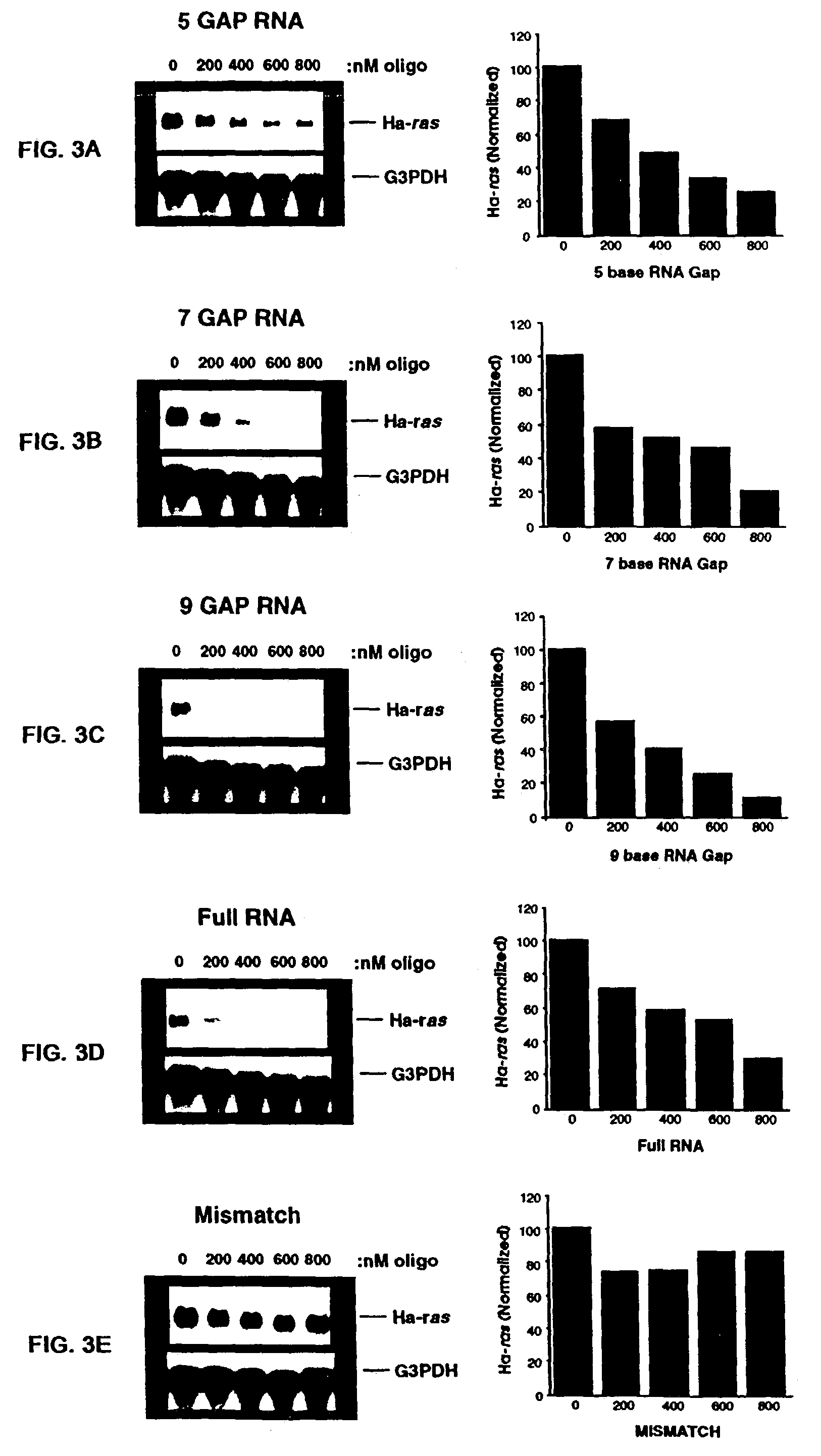
Gapped oligonucleotides
InactiveUS7015315B1Increased nuclease resistanceHigh binding affinityPeptide/protein ingredientsGenetic material ingredientsADAMTS ProteinsNuclease
Oligonucleotides and other macromolecules are provided which have increased nuclease resistance, substituent groups for increasing binding affinity to complementary strand, and subsequences of 2′-deoxy-erythro-pentofuranosyl nucleotides that activate RNase H. Such oligonucleotides and macromolecules are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics.
Owner:IONIS PHARMA INC
Gapped 2' modified oligonucleotides
InactiveUS6326199B1High affinityAvoid degradationHydrolasesPeptide/protein ingredientsNucleotideNuclease
Oligonucleotides and other macromolecules are provided that have increased nuclease resistance, substituent groups for increasing binding affinity to complementary strand, and sub-sequences of 2'-deoxy-erythro-pentofuranosyl nucleotides that activate RNase H enzyme. Such oligonucleotides and macromolecules are useful for diagnostics and other research purposes, for modulating protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to antisense therapeutics.
Owner:IONIS PHARMA INC
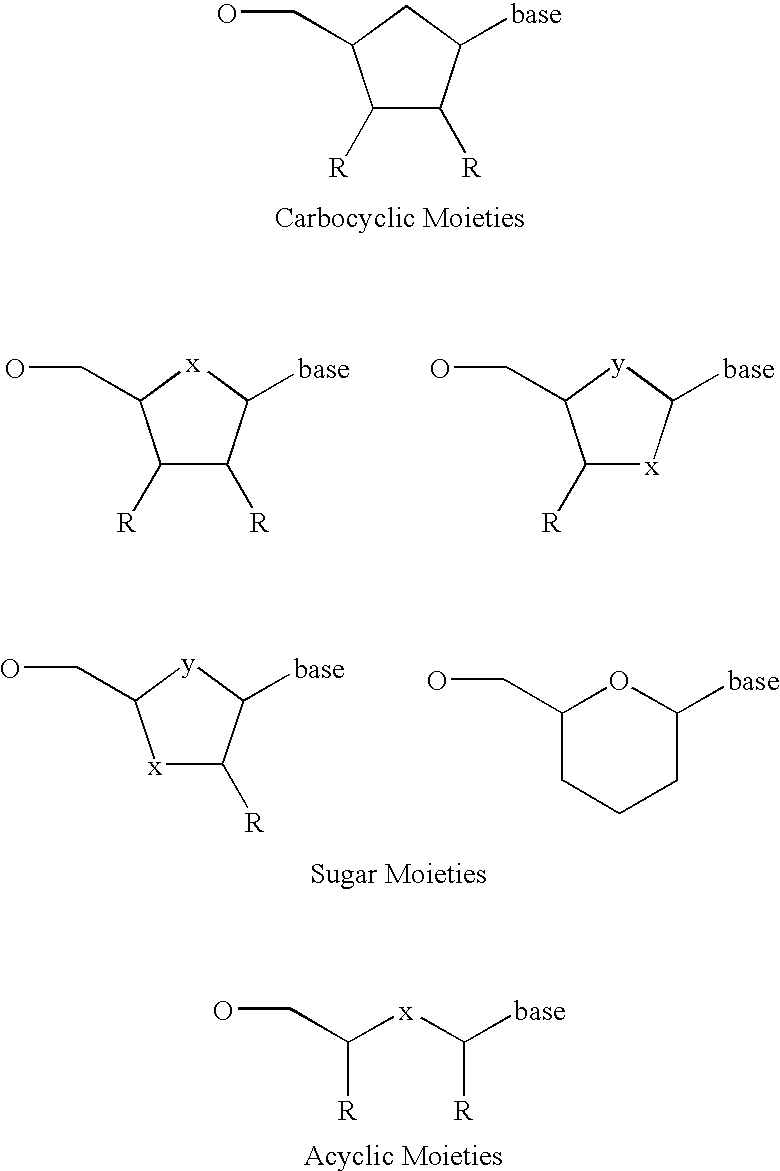
Carbocyclic bicyclic nucleic acid analogs
Provided herein are saturated and unsaturated carbocyclic bicyclic nucleosides, oligomeric compounds prepared therefrom and methods of using these oligomeric compounds. The saturated and unsaturated carbocyclic bicyclic nucleosides are useful for enhancing properties of oligomeric compounds including nuclease resistance.
Owner:IONIS PHARMA INC
Methods for producing a paired tag from a nucleic acid sequence and methods of use thereof
InactiveUS20060024681A1Microbiological testing/measurementFermentationSmaI restriction endonucleaseEcoRI
Methods for producing a paired tag from a nucleic acid sequence are provided in which the paired tag comprises the 5′ end tag and 3′ end tag of the nucleic acid sequence. In one embodiment, the nucleic acid sequence comprises two restriction endonuclease recognition sites specific for a restriction endonuclease that cleaves the nucleic acid sequence distally to the restriction endonuclease recognition sites. In another embodiment, the nucleic acid sequence further comprises restriction endonuclease recognition sites specific for a rare cutting restriction endonuclease. Methods of using paired tags are also provided. In one embodiment, paired tags are used to characterize a nucleic acid sequence. In a particular embodiment, the nucleic acid sequence is a genome. In one embodiment, the characterization of a nucleic acid sequence is karyotyping. Alternatively, in another embodiment, the characterization of a nucleic acid sequence is mapping of the sequence. In a further embodiment, a method is provided for identifying nucleic acid sequences that encode at least two interacting proteins.
Owner:APPL BIOSYSTEMS INC
Modified nucleotide compounds
ActiveUS7495088B1Increased nuclease resistanceSugar derivativesMicrobiological testing/measurementBiologyModified nucleosides
Disclosed is a nuclease resistant nucleotide compound capable of hybridizing with a complementary RNA in a manner which inhibits the function thereof, which modified nucleotide compound includes at least one component selected from the group consisting of MN3M, B(N)xM and M(N)xB wherein N is a phosphodiester-linked modified 2′-deoxynucleoside moiety; M is a moiety that confers endonuclease resistance on said component and that contains at least one modified or unmodified nucleic acid base; B is a moiety that confers exonuclease resistance to the terminus to which it is attached; and x is an integer of at least 2.
Owner:ENZO BIOCHEM
6-disubstituted or unsaturated bicyclic nucleic acid analogs
The present disclosure describes 6-disubstituted bicyclic nucleosides, oligomeric compounds prepared therefrom and methods of using the oligomeric compounds. More particularly, the 6-disubstituted bicyclic nucleosides each comprise a 2′-O—C(Ri)(R2)-4′ or 2′-O—C=(R3)(R.4)-4′ bridge wherein each R is, independently a substituent group and Ri and R2 include H. The 6-disubstituted bicyclic nucleosides are useful for enhancing properties of oligomeric compounds including nuclease resistance. In certain embodiments, the oligomeric compounds provided herein hybridize to a portion of a target RNA resulting in loss of normal function of the target RNA.
Owner:IONIS PHARMA INC
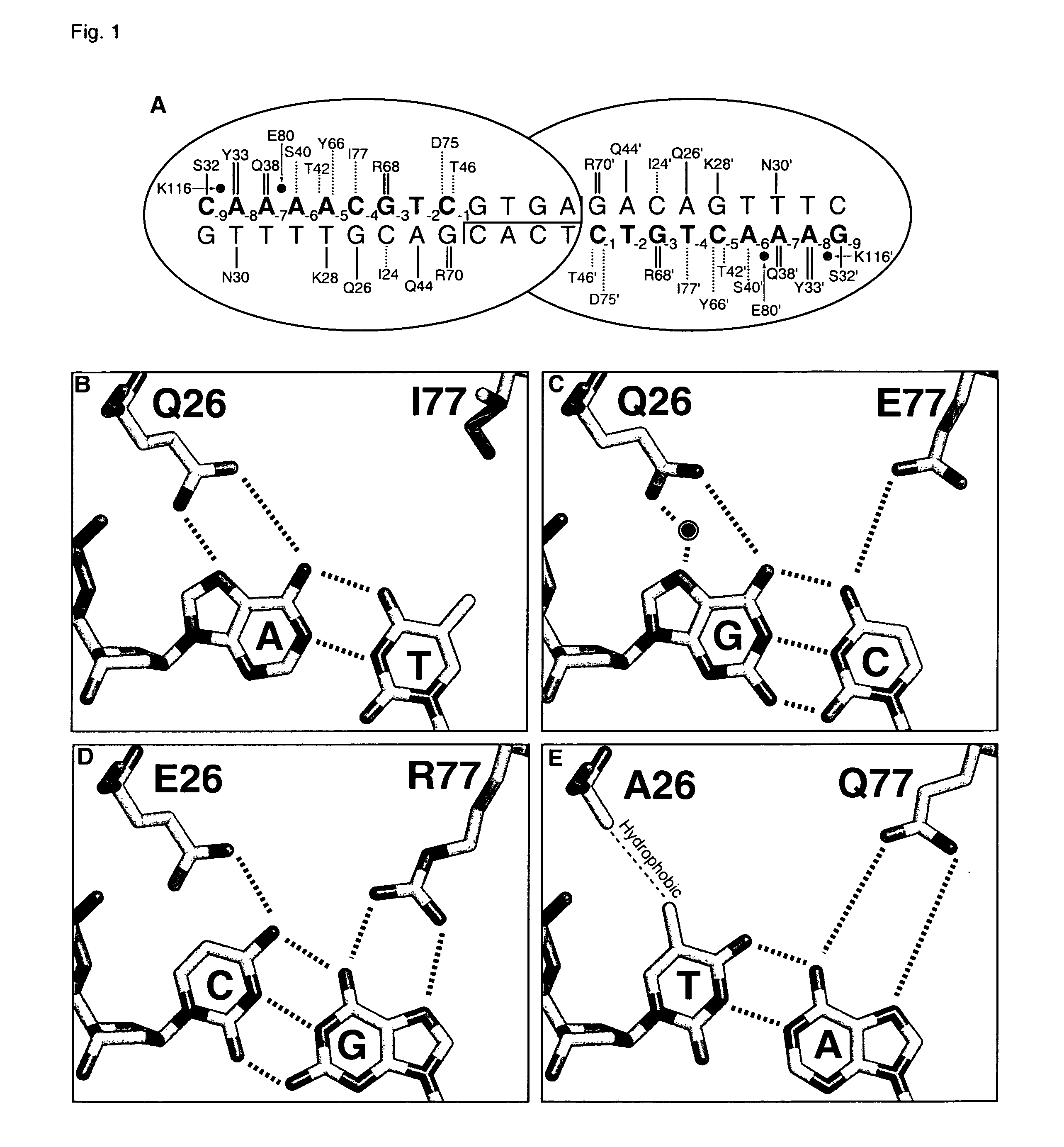
Rationally-designed meganucleases with altered sequence specificity and DNA-binding affinity
ActiveUS8021867B2Affect specificityAffect activityPeptide/protein ingredientsHydrolasesBiological bodyNuclease
Rationally-designed LAGLIDADG (SEQ ID NO: 37) meganucleases and methods of making such meganucleases are provided. In addition, methods are provided for using the meganucleases to generate recombinant cells and organisms having a desired DNA sequence inserted into a limited number of loci within the genome, as well as methods of gene therapy, for treatment of pathogenic infections, and for in vitro applications in diagnostics and research.
Owner:DUKE UNIV
Method for producing complex DNA methylation fingerprints
InactiveUS6214556B1Accurately determineSugar derivativesMicrobiological testing/measurementFingerprintGenomic DNA
Method for characterizing, classifying and differentiating tissues and cell types, for predicting the behavior of tissues and groups of cells, and for identifying genes with changed expression. The method involves obtaining genomic DNA from a tissue sample, the genomic DNA subsequently being subjected to shearing, cleaved by means of a restriction endonuclease or not treated by either one of these methods. The base cytosine, but not 5-methylcytosine, from the thus-obtained genomic DNA is then converted into uracil by treatment with a bisulfite solution. Fractions of the thus-treated genomic DNA are then amplified using either very short or degenerated oligonucleotides or oligonuclcotides which are complementary to adaptor oligonucleotides that have been ligated to the ends of the cleaved DNA. The quantity of the remaining cytosines on the guanine-rich DNA strand and / or the quantity of guanines on the cytosine-rich DNA strand from the amplified fractions are then detected by hybridization or polymerase reaction, which quantities are such that the data generated thereby and automatically applied to a processing algorithm allow the drawing of conclusions concerning the phenotype of the sample material.
Owner:EPIGENOMICS AG
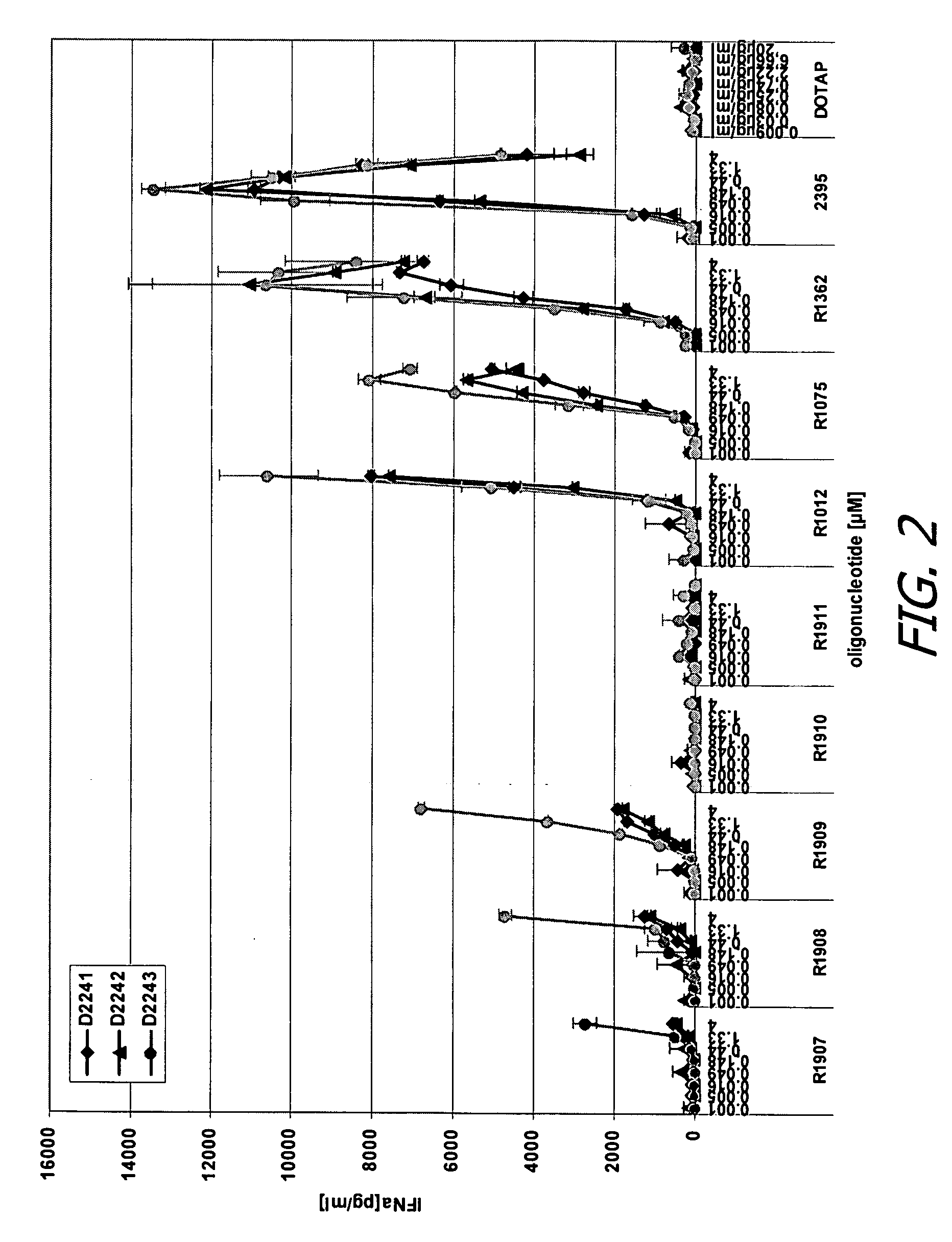
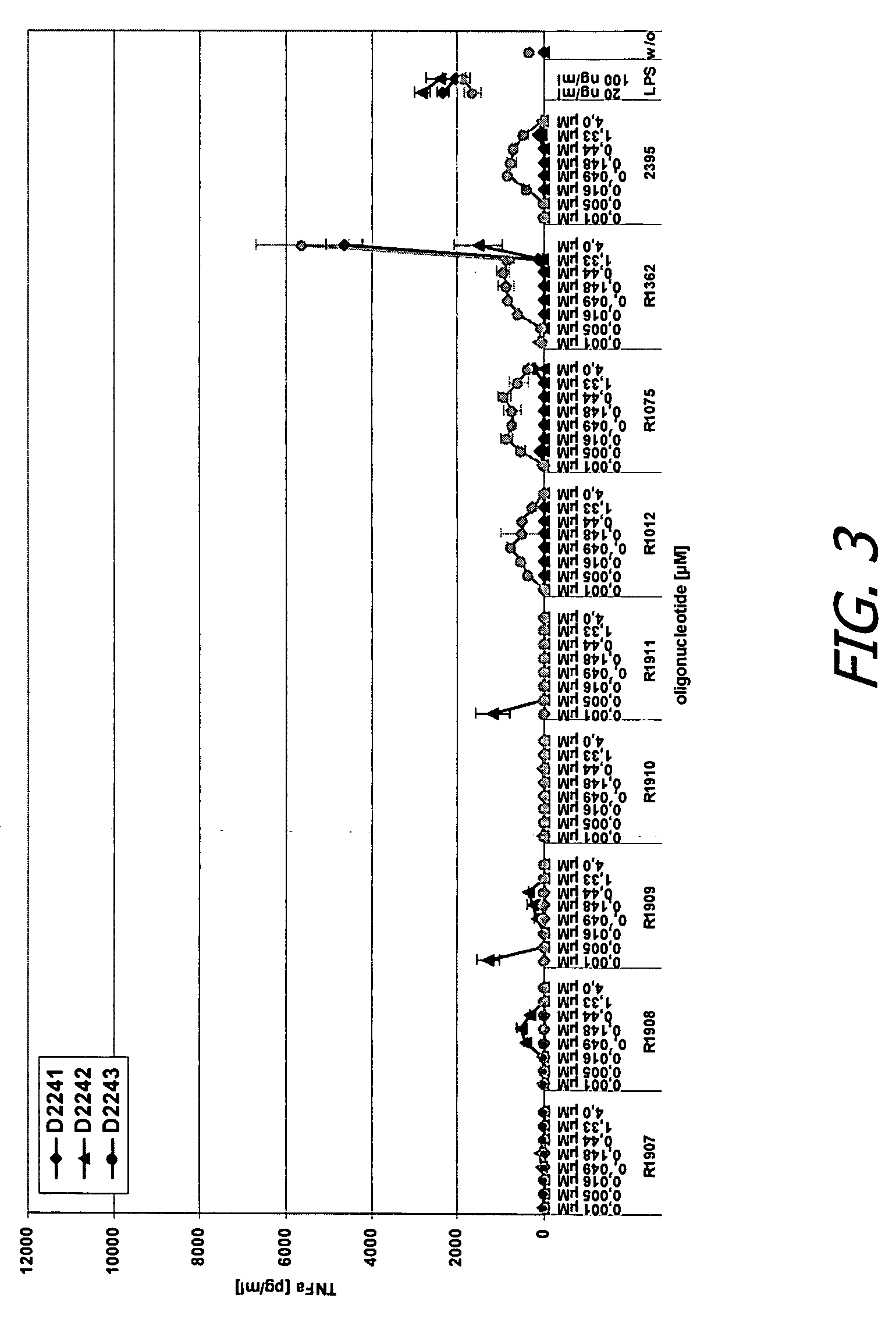
Modified oligoribonucleotide analogs with enhanced immunostimulatory activity
The invention provides immunostimulatory compositions and methods for their use. In particular, the immunostimulatory compositions of the invention include RNA-like polymers that incorporate an immunostimulatory sequence motif and at least one chemical modification to confer improved stability against nuclease degradation and improved activity. Specific modifications involving phosphate linkages, nucleotide analogs, and combinations thereof are provided. Compositions of the invention optionally include an antigen and can be used to stimulate an immune response. Also provided are compositions and methods useful for treating a subject having an infection, a cancer, an allergic condition, or asthma. Modified oligoribonucleotide analogs of the invention are believed to stimulate Toll-like receptors TLR7 and TLR8.
Owner:COLEY PHARMA GRP INC +1
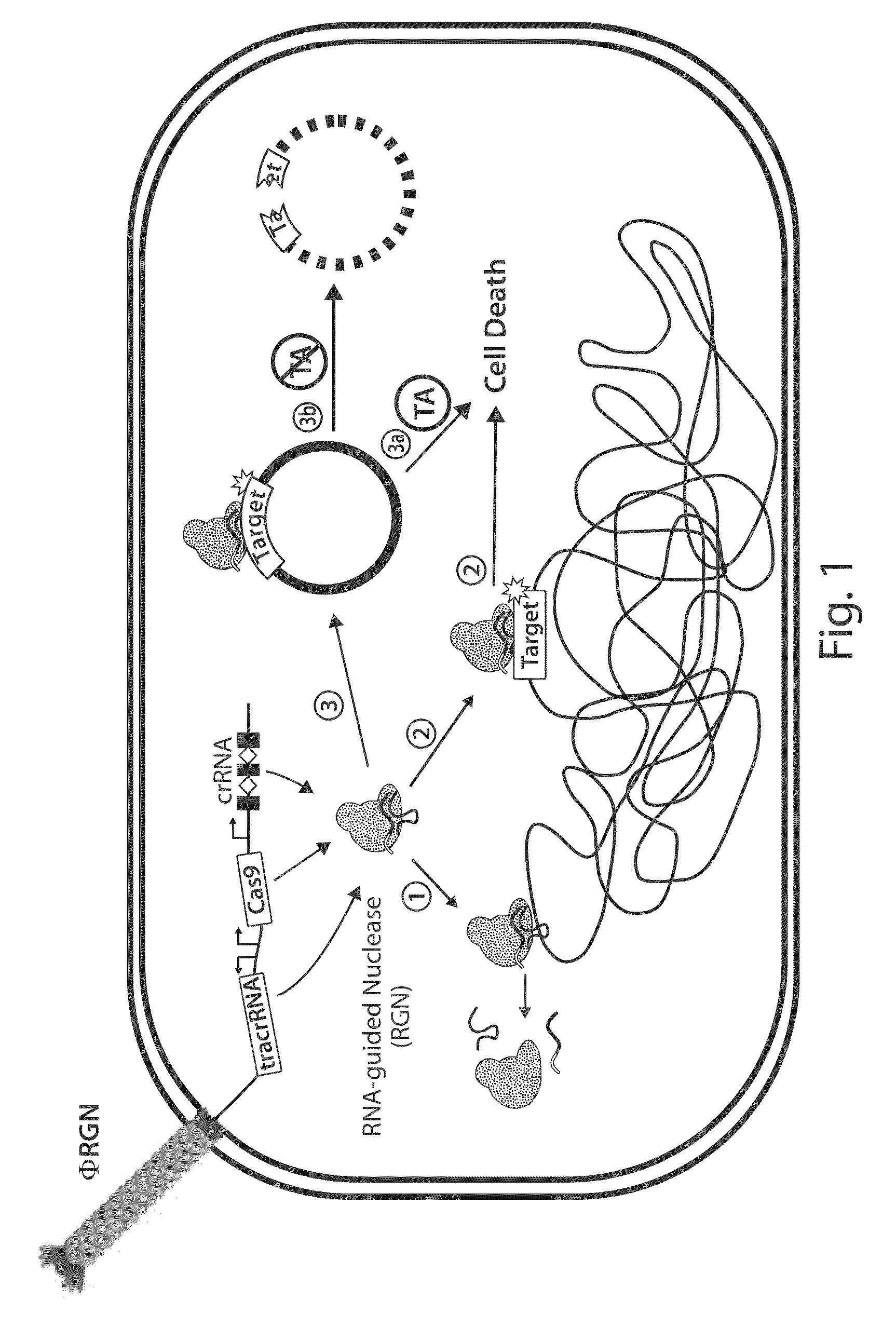
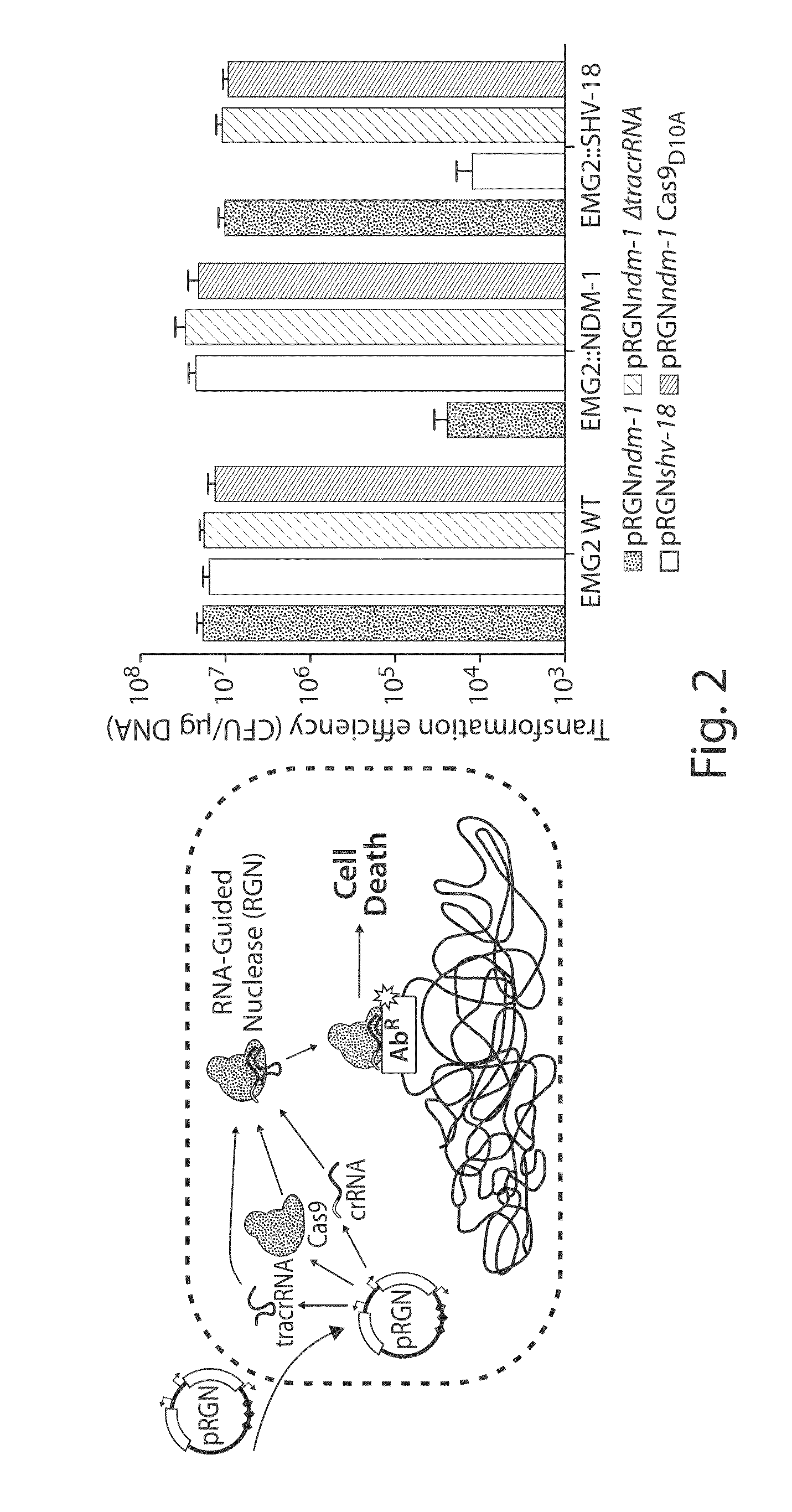
Tuning microbial populations with programmable nucleases
ActiveUS20150064138A1Efficiently and rapidly encoded into synthetic constructBroad deliveryBiocideSugar derivativesMicroorganismVirulent characteristics
Various aspects and embodiments of the invention are directed to methods and compositions for reversing antibiotic resistance or virulence in and / or destroying pathogenic microbial cells such as, for example, pathogenic bacterial cells. The methods include exposing microbial cells to a delivery vehicle with at least one nucleic acid encoding an engineered autonomously distributed circuit that contains a programmable nuclease targeted to one or multiple genes of interest.
Owner:MASSACHUSETTS INST OF TECH
Compositions, methods, and kits for isolating nucleic acids using surfactants and proteases
The invention provides compositions and methods for releasing and for isolating nucleic acids from biological samples, preferably from whole tissue, using cationic surfactants and proteases. The surfactant-protease combinations, when used with whole tissue, macerate the tissue, lyse individual cells, release nucleic acids, and inactivate nucleases. Kits for isolating nucleic acids from biological samples, particularly from whole tissue, are also provided.
Owner:APPL BIOSYSTEMS INC
Oligoribonucleotides and ribonucleases for cleaving RNA
InactiveUS7432250B2High affinityStrong specificityHydrolasesPeptide/protein ingredientsOrganismResearch purpose
Oligomeric compounds including oligoribonucleotides and oligoribonucleosides are provided that have subsequences of 2′-pentoribofuranosyl nucleosides that activate dsRNase. The oligoribonucleotides and oligoribonucleosides can include substituent groups for increasing binding affinity to complementary nucleic acid strand as well as substituent groups for increasing nuclease resistance. The oligomeric compounds are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics. Also included in the invention are mammalian ribonucleases, i.e., enzymes that degrade RNA, and substrates for such ribonucleases. Such a ribonuclease is referred to herein as a dsRNase, wherein “ds” indicates the RNase's specificity for certain double-stranded RNA substrates. The artificial substrates for the dsRNases described herein are useful in preparing affinity matrices for purifying mammalian ribonuclease as well as non-degradative RNA-binding proteins.
Owner:IONIS PHARMA INC
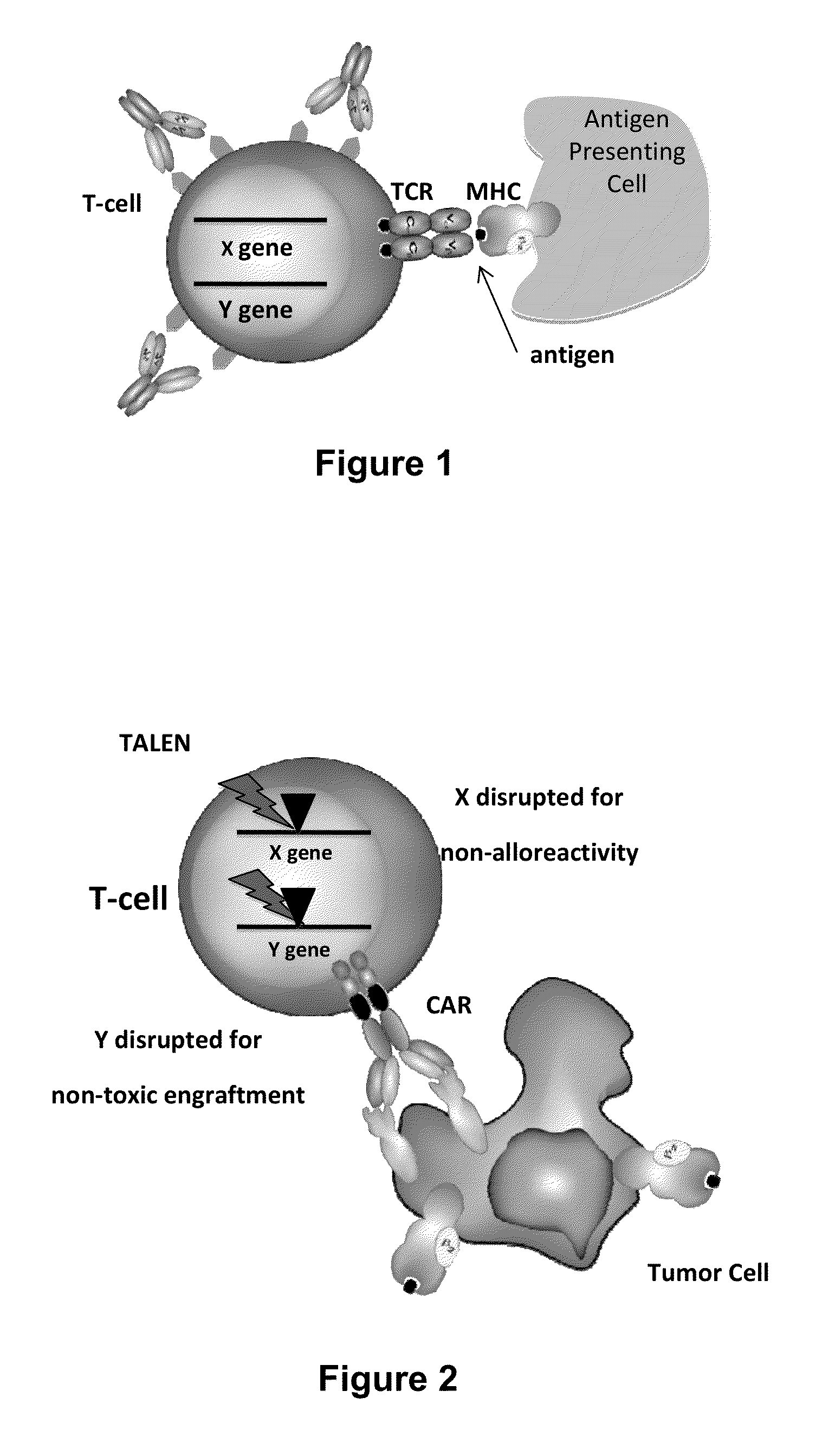
Methods for engineering allogeneic and immunosuppressive resistant t cell for immunotherapy
ActiveUS20130315884A1Precise positioningPeptide/protein ingredientsAntibody mimetics/scaffoldsImmunosuppressive drugPrimary cell
Methods for developing engineered T-cells for immunotherapy that are both non-alloreactive and resistant to immunosuppressive drugs. The present invention relates to methods for modifying T-cells by inactivating both genes encoding target for an immunosuppressive agent and T-cell receptor, in particular genes encoding CD52 and TCR. This method involves the use of specific rare cutting endonucleases, in particular TALE-nucleases (TAL effector endonuclease) and polynucleotides encoding such polypeptides, to precisely target a selection of key genes in T-cells, which are available from donors or from culture of primary cells. The invention opens the way to standard and affordable adoptive immunotherapy strategies for treating cancer and viral infections.
Owner:CELLECTIS SA
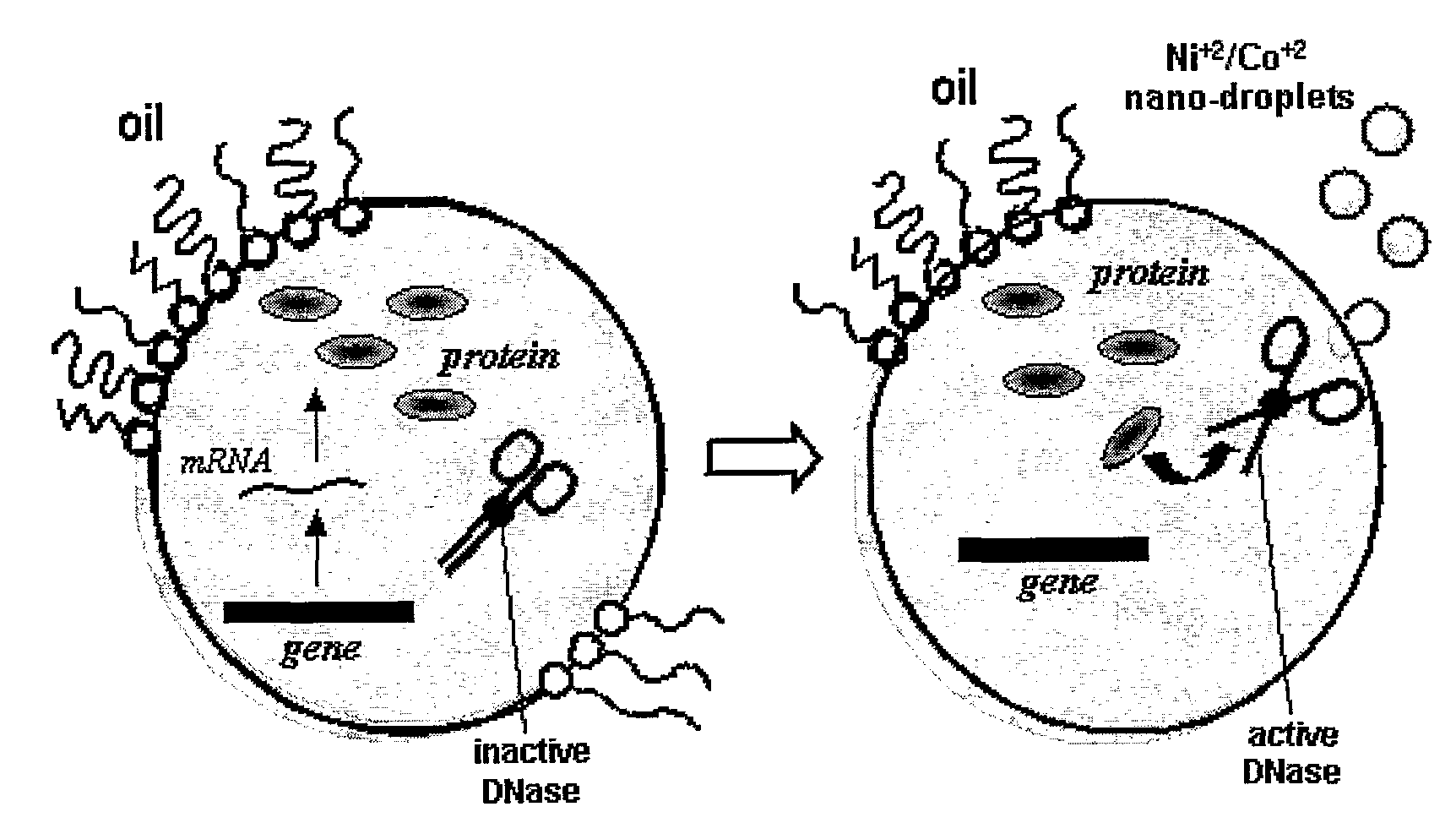
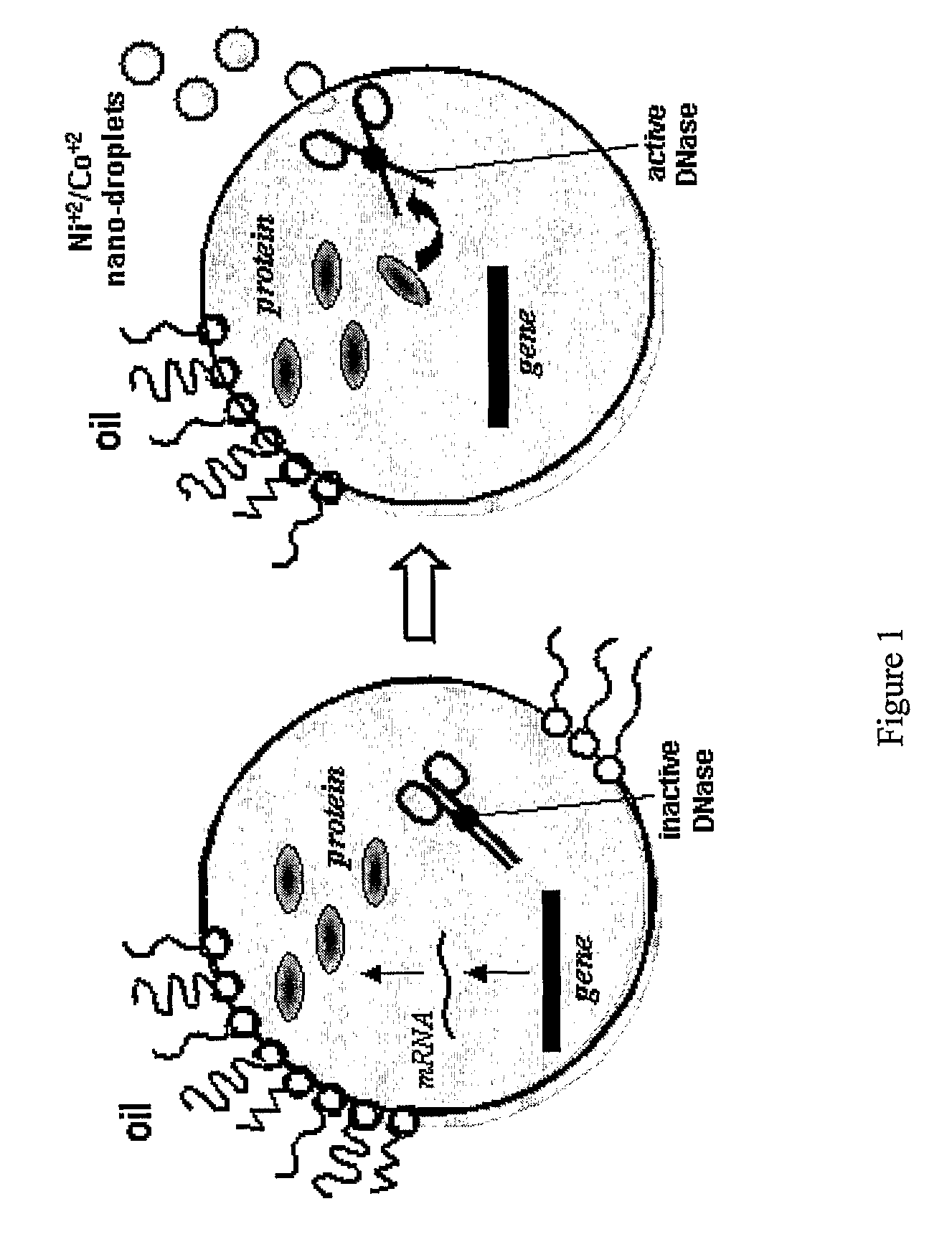
Directed Evolution and Selection Using in Vitro Compartmentalization
InactiveUS20080004436A1Easy to adjustImprove concentrationSugar derivativesDNA preparationWater in oil emulsionGenetic element
The present invention is related to the field of compartmentalized libraries of genetic elements and the selection of biologically active molecules and the genes encoding same from said libraries. The selection assay of the invention utilizes water-in-oil emulsions and is particularly advantageous in applications in the field of directed-evolution, as exemplified herein for selection of protein inhibitors of DNA nucleases.
Owner:YISSUM RES DEV CO OF THE HEBREWUNIVERSITY OF JERUSALEM LTD +1
Bicyclic cyclohexose nucleic acid analogs
The present invention provides bicyclic cyclohexose nucleoside analogs and oligomeric compounds comprising these nucleoside analogs. These bicyclic nucleoside analogs are useful for enhancing properties of oligomeric compounds including nuclease resistance.
Owner:IONIS PHARMA INC
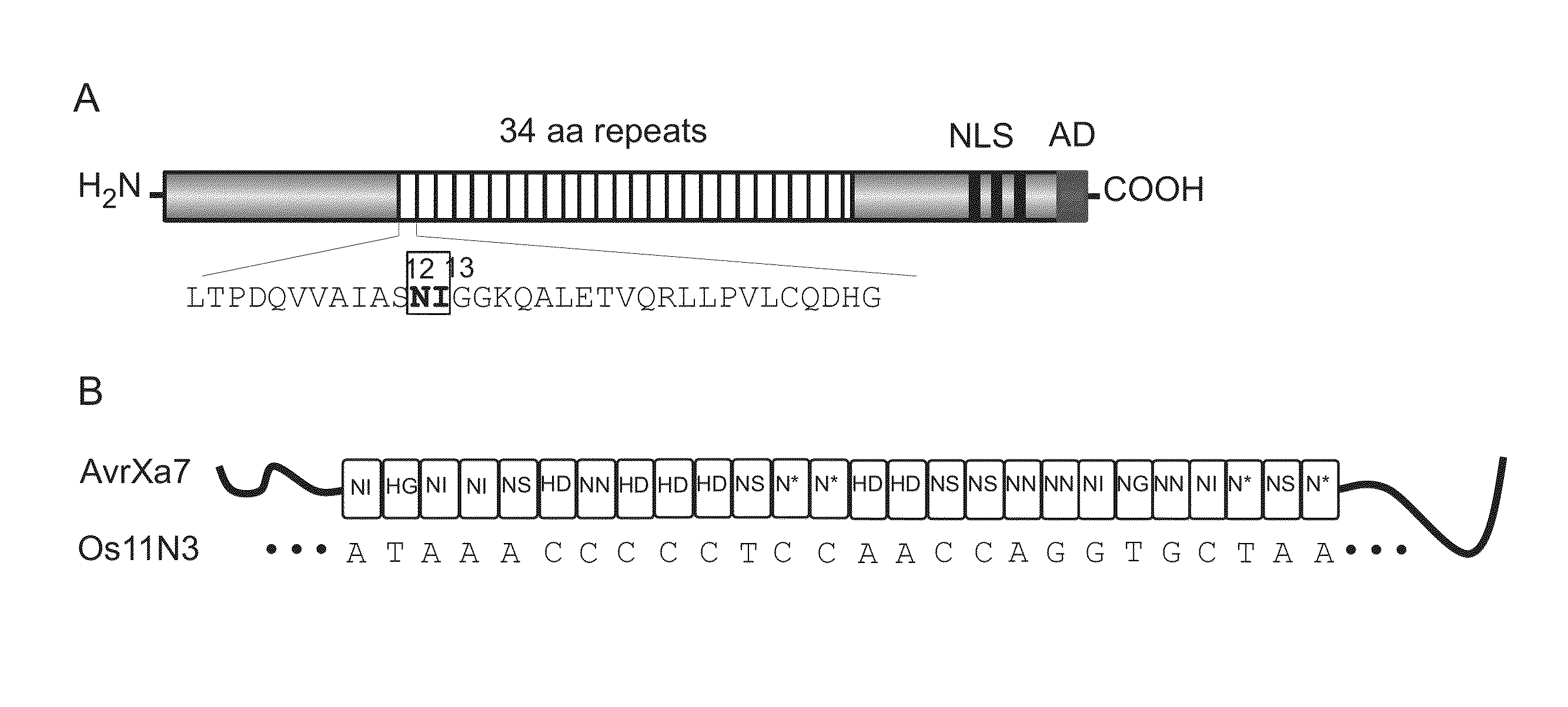
Method for the generation of compact tale-nucleases and uses thereof
ActiveUS20130117869A1Simple processSimple and efficient vectorizationFusion with DNA-binding domainHydrolasesDNA-binding domainNuclease
The present invention relates to a method for the generation of compact Transcription Activator-Like Effector Nucleases (TALENs) that can efficiently target and process double-stranded DNA. More specifically, the present invention concerns a method for the creation of TALENs that consist of a single TALE DNA binding domain fused to at least one catalytic domain such that the active entity is composed of a single polypeptide chain for simple and efficient vectorization and does not require dimerization to target a specific single double-stranded DNA target sequence of interest and process DNA nearby said DNA target sequence. The present invention also relates to compact TALENs, vectors, compositions and kits used to implement the method.
Owner:CELLECTIS SA
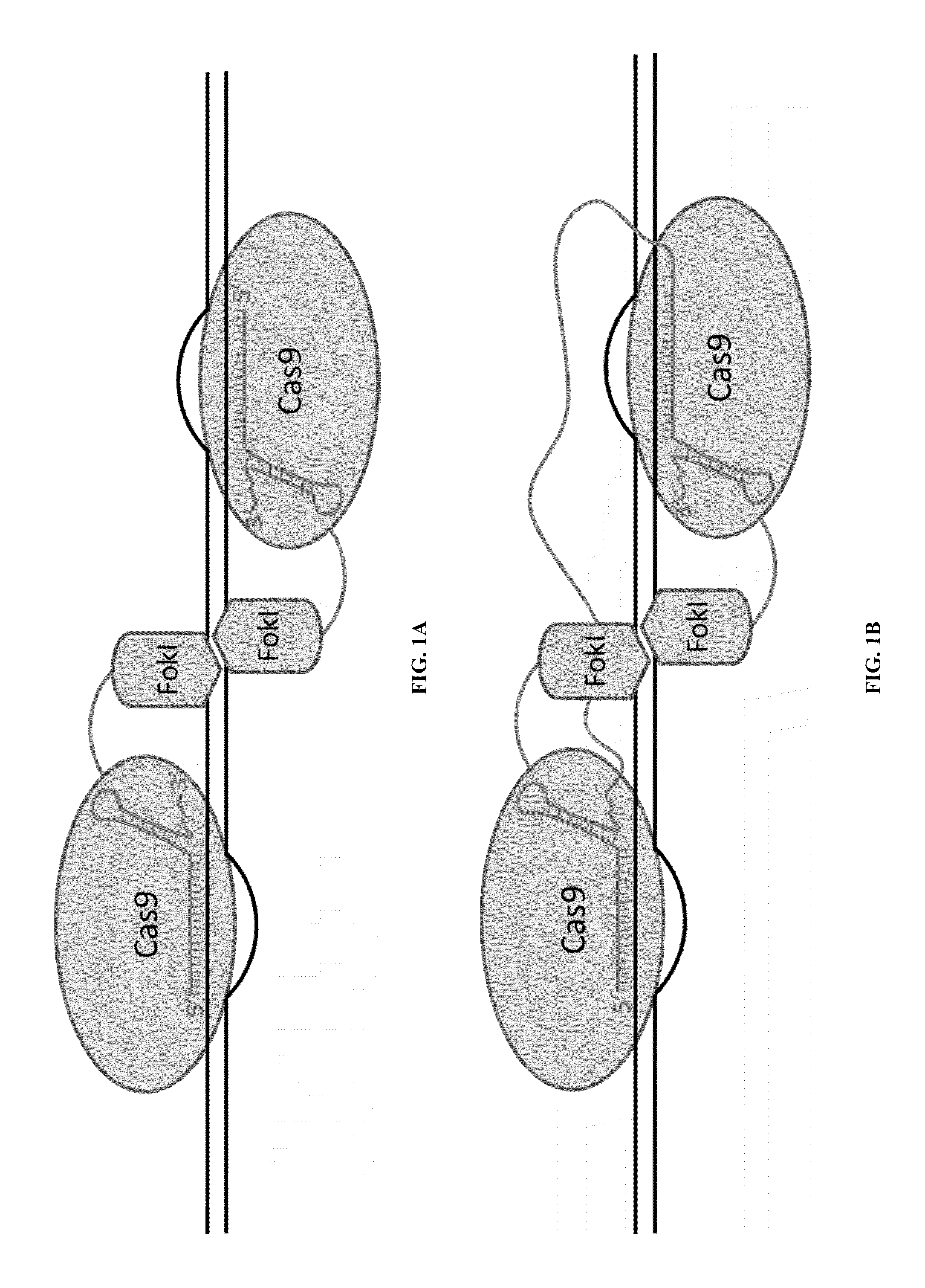
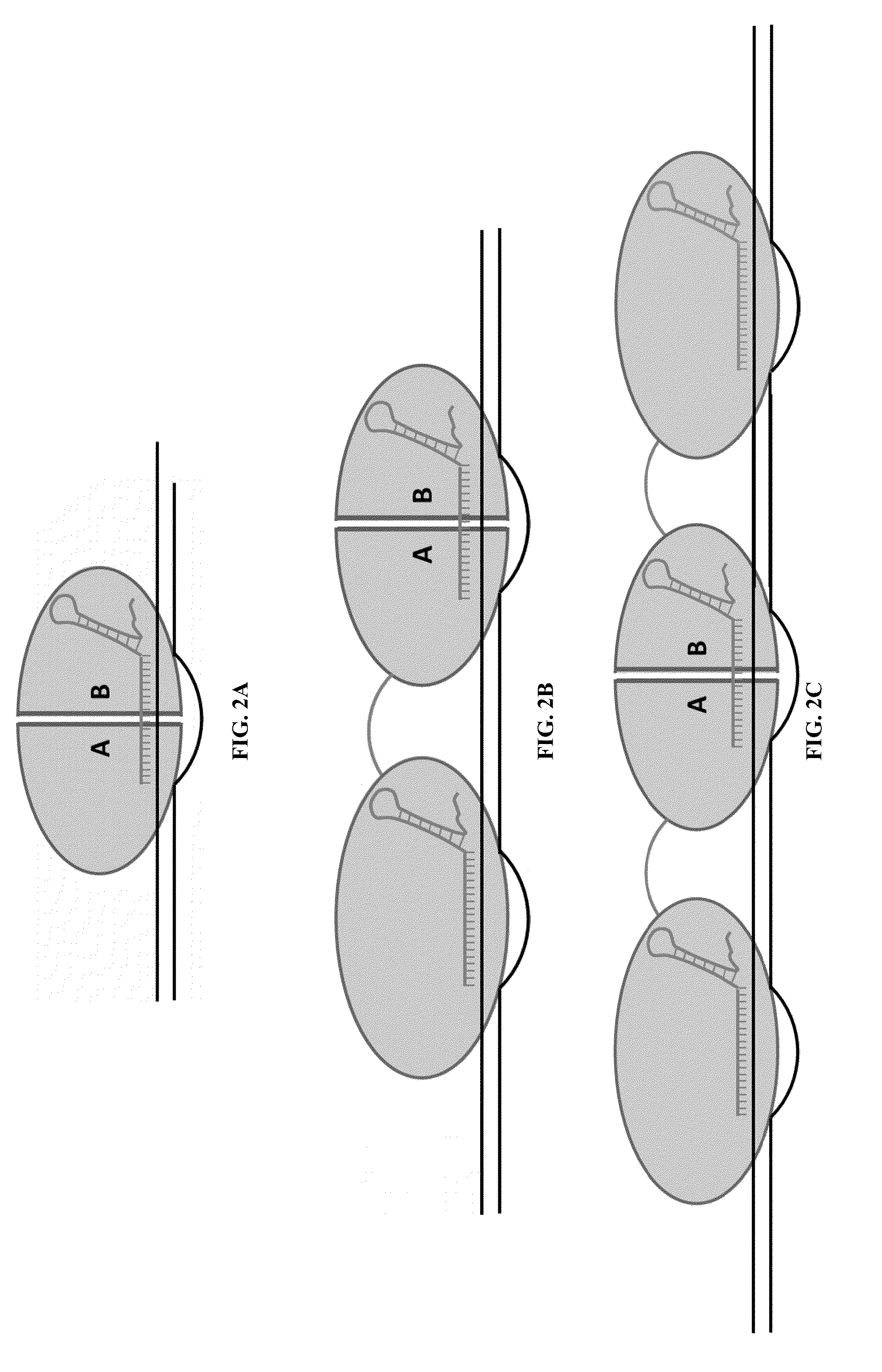
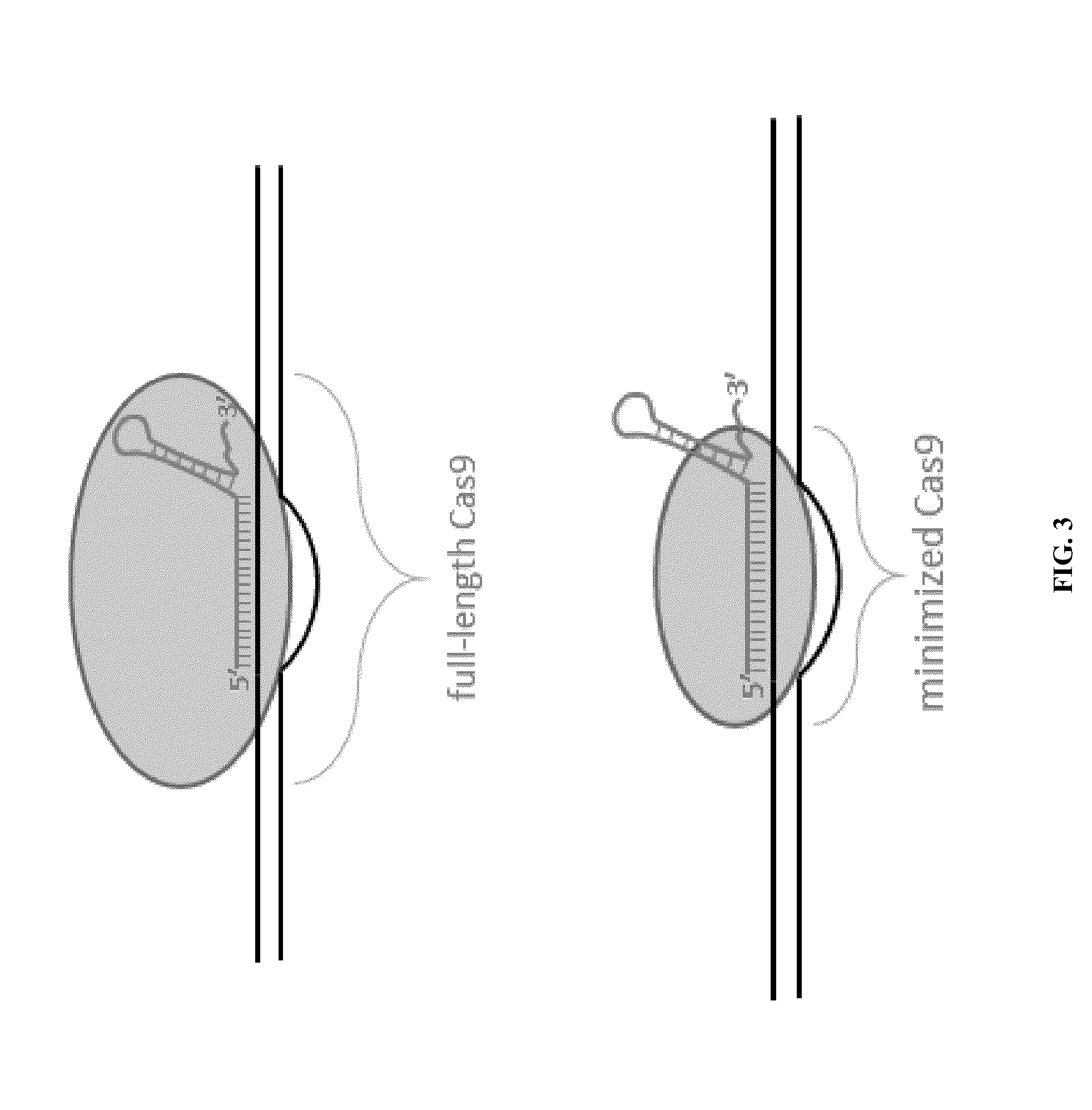
Cas9-recombinase fusion proteins and uses thereof
ActiveUS20150071898A1Strong specificityReduce the possibilityFusion with DNA-binding domainBacteriaSite-specific recombinationResearch setting
Some aspects of this disclosure provide compositions, methods, and kits for improving the specificity of RNA-programmable endonucleases, such as Cas9. Also provided are variants of Cas9, e.g., Cas9 dimers and fusion proteins, engineered to have improved specificity for cleaving nucleic acid targets. Also provided are compositions, methods, and kits for site-specific recombination, using Cas9 fusion proteins (e.g., nuclease-inactivated Cas9 fused to a recombinase catalytic domain). Such Cas9 variants are useful in clinical and research settings involving site-specific modification of DNA, for example, genomic modifications.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Analyte detection
A method of characterizing an analyte sample is provided that includes the steps of: (a) anchoring the analyte to a nucleic acid template of known sequence; (b) conducting a DNA polymerase reaction that includes the reaction of a template, a non-hydrolyzable primer, at least one terminal phosphate-labeled nucleotide, DNA polymerase, and an enzyme having 3'->5' exonuclease activity which reaction results in the production of labeled polyphosphate; (c) permitting the labeled polyphosphate to react with a phosphatase to produce a detectable species characteristic of the sample; (d) detecting the detectable species. The method may include the step of characterizing the nucleic acid sample based on the detection. Also provided are methods of analyzing multiple analytes in a sample, and kits for characterizing analyte samples.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
Rationally-designed single-chain meganucleases with non-palindromic recognition sequences
Disclosed are rationally-designed, non-naturally-occurring meganucleases in which a pair of enzyme subunits having specificity for different recognition sequence half-sites are joined into a single polypeptide to form a functional heterodimer with a non-palindromic recognition sequence. The invention also relates to methods of producing such meganucleases, and methods of producing recombinant nucleic acids and organisms using such meganucleases.
Owner:PRECISION BIOSCI
Oligoribonucleotides and ribonucleases for cleaving RNA
InactiveUS7432249B2High affinityStrong specificityPeptide/protein ingredientsHydrolasesADAMTS ProteinsOrganism
Oligomeric compounds including oligoribonucleotides and oligoribonucleosides are provided that have subsequences of 2′-pentoribofuranosyl nucleosides that activate dsRNase. The oligoribonucleotides and oligoribonucleosides can include substituent groups for increasing binding affinity to complementary nucleic acid strand as well as substituent groups for increasing nuclease resistance. The oligomeric compounds are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics. Also included in the invention are mammalian ribonucleases, i.e., enzymes that degrade RNA, and substrates for such ribonucleases. Such a ribonuclease is referred to herein as a dsRNase, wherein “ds” indicates the RNase's specificity for certain double-stranded RNA substrates. The artificial substrates for the dsRNases described herein are useful in preparing affinity matrices for purifying mammalian ribonuclease as well as non-degradative RNA-binding proteins.
Owner:IONIS PHARMA INC
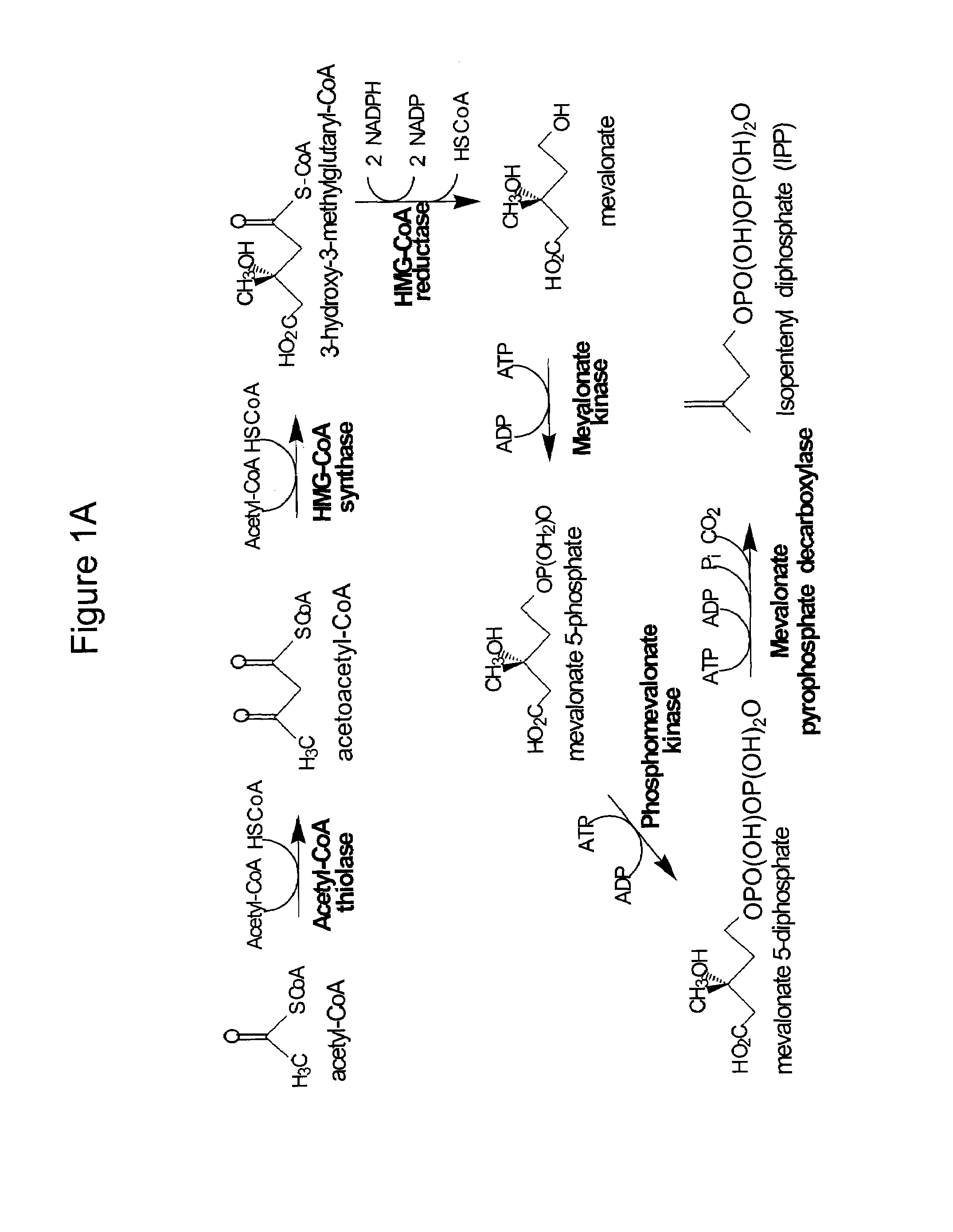
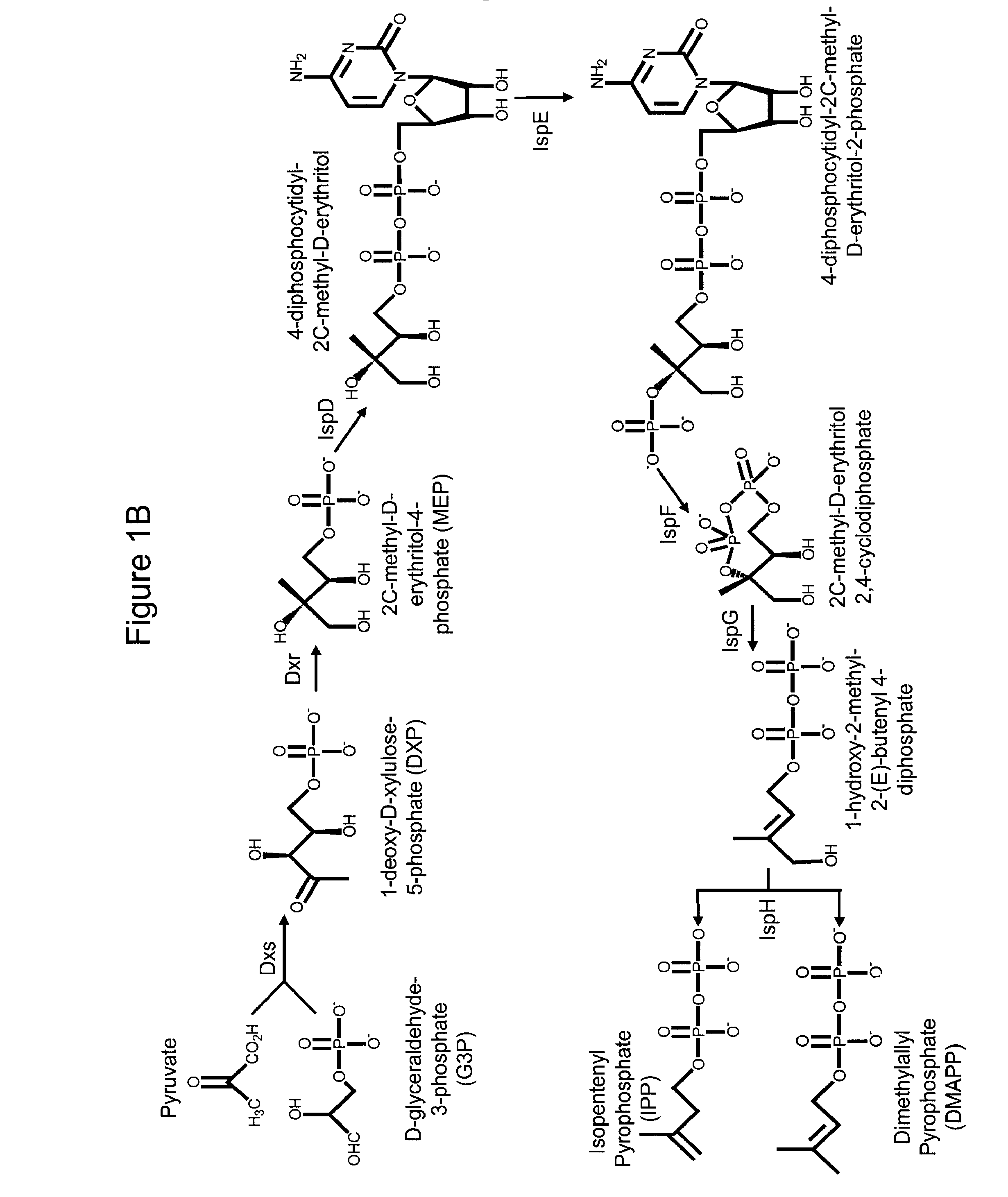
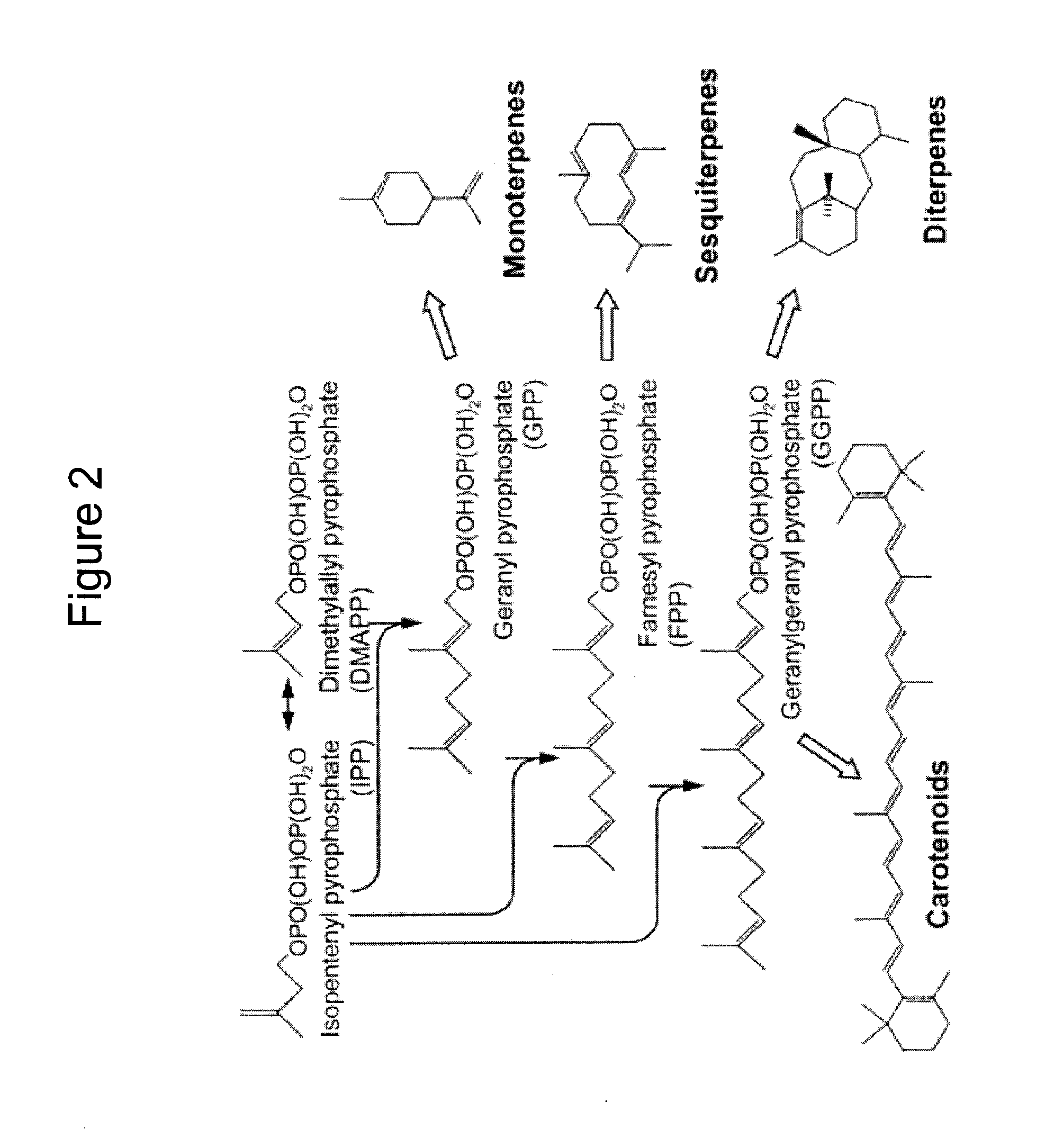
Production of isoprenoids
The present invention provides methods for a robust production of isoprenoids via one or more biosynthetic pathways. The invention also provides nucleic acids, enzymes, expression vectors, and genetically modified host cells for carrying out the subject methods. The invention also provides fermentation methods for high productivity of isoprenoids from genetically modified host cells.
Owner:AMYRIS INC
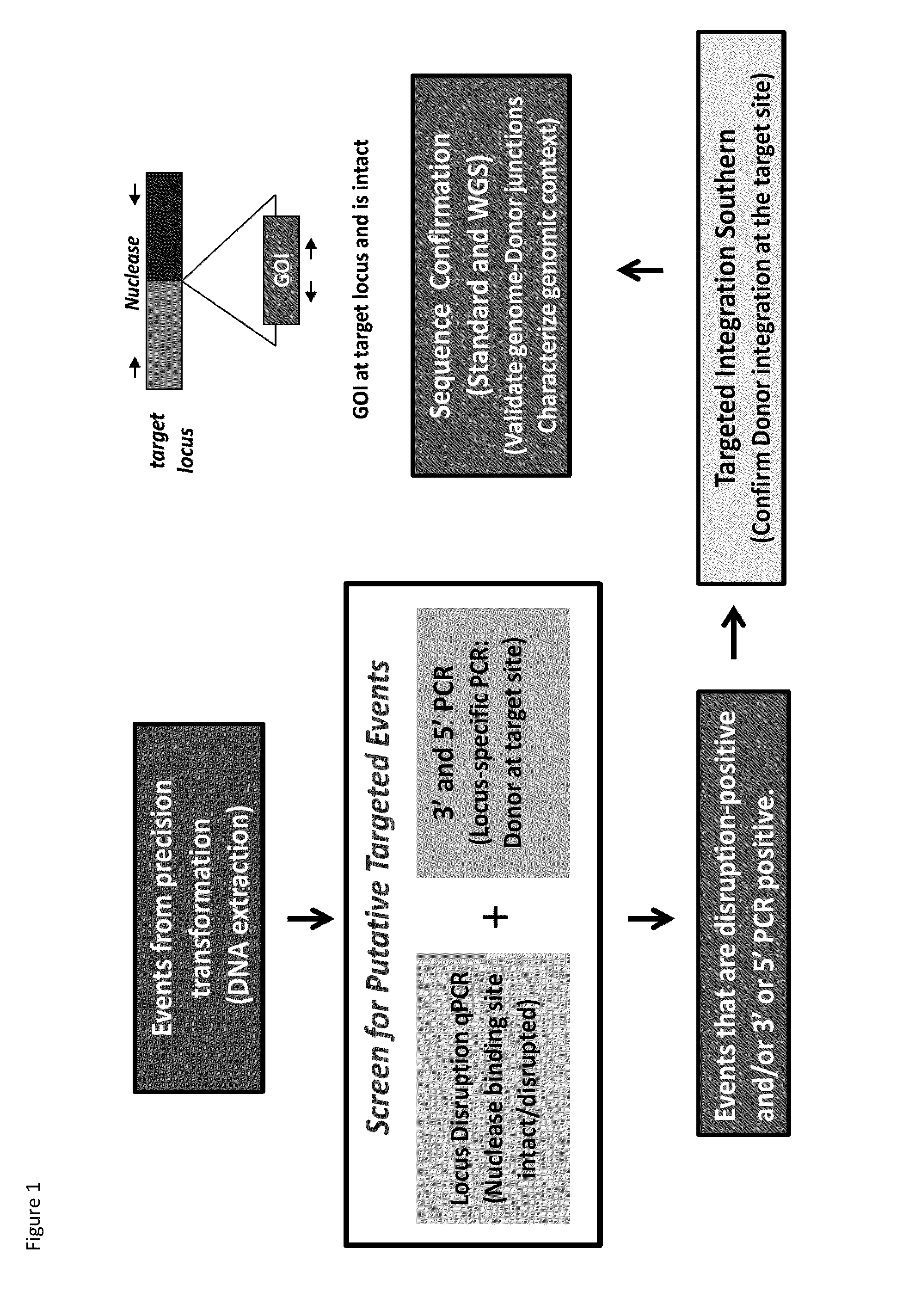
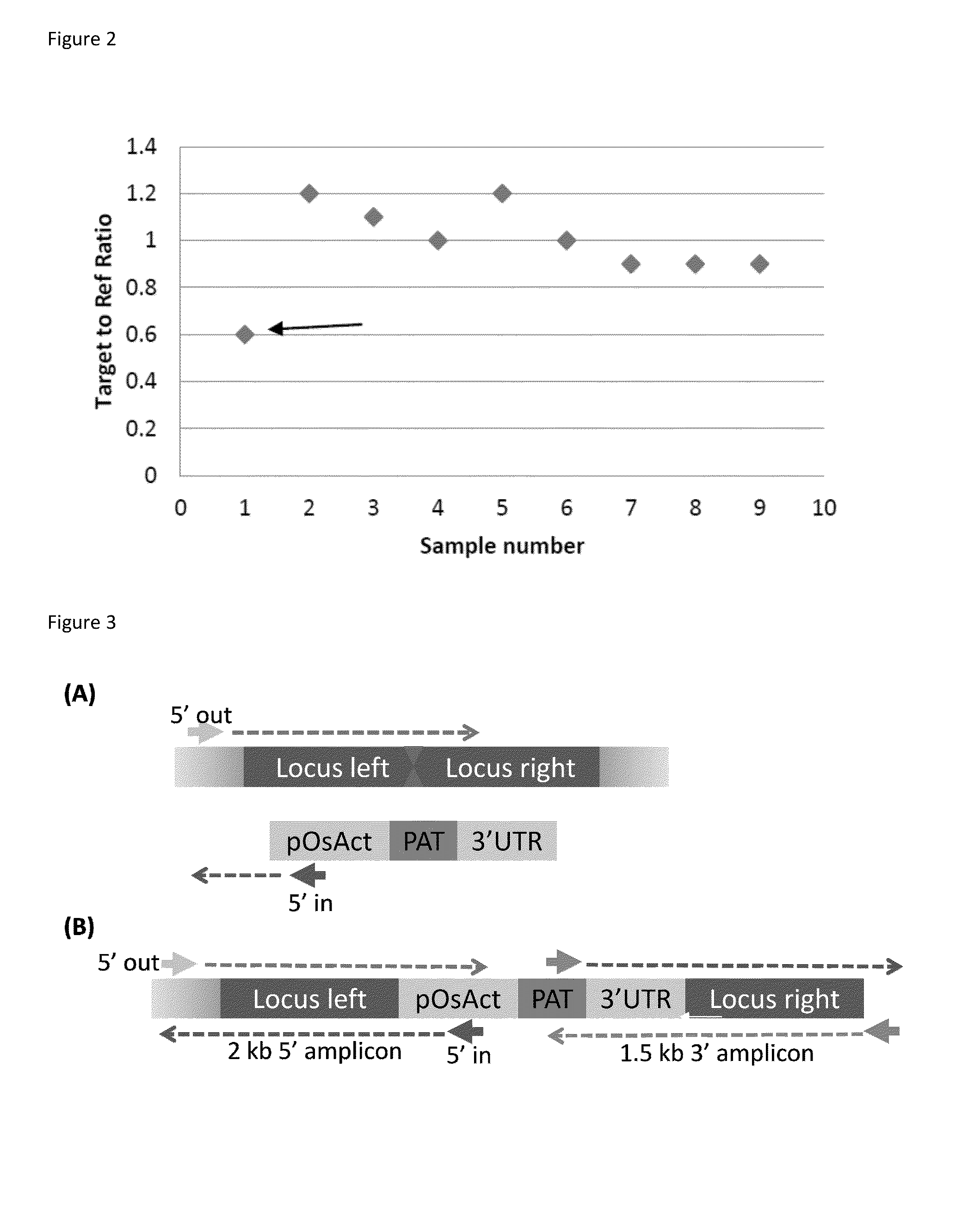
DNA detection methods for site specific nuclease activity
The present disclosure provides methods for detecting and identifying plant events that contain precision targeted genomic loci, and plants and plant cells comprising such targeted genomic loci. The method can be deployed as a high throughput process utilized for screening the intactness or disruption of a targeted genomic loci and optionally for detecting a donor DNA polynucleotide insertion at the targeted genomic loci. The methods are readily applicable for the identification of plant events produced via a targeting method which results from the use of a site specific nuclease.
Owner:CORTEVA AGRISCIENCE LLC
Genome editing of genes associated with trinucleotide repeat expansion disorders in animals
InactiveUS20110016540A1Vertebrate cellsArtificial cell constructsZinc finger nucleaseChemical toxicity
The present invention provides genetically modified animals and cells comprising edited chromosomal sequences encoding proteins that are associated with trinucleotide repeat expansion disorders. In particular, the animals or cells are generated using a zinc finger nuclease-mediated editing process. Also provided are methods of using the genetically modified animals or cells disclosed herein to screen agents for toxicity and other effects.
Owner:SIGMA ALDRICH CO LLC
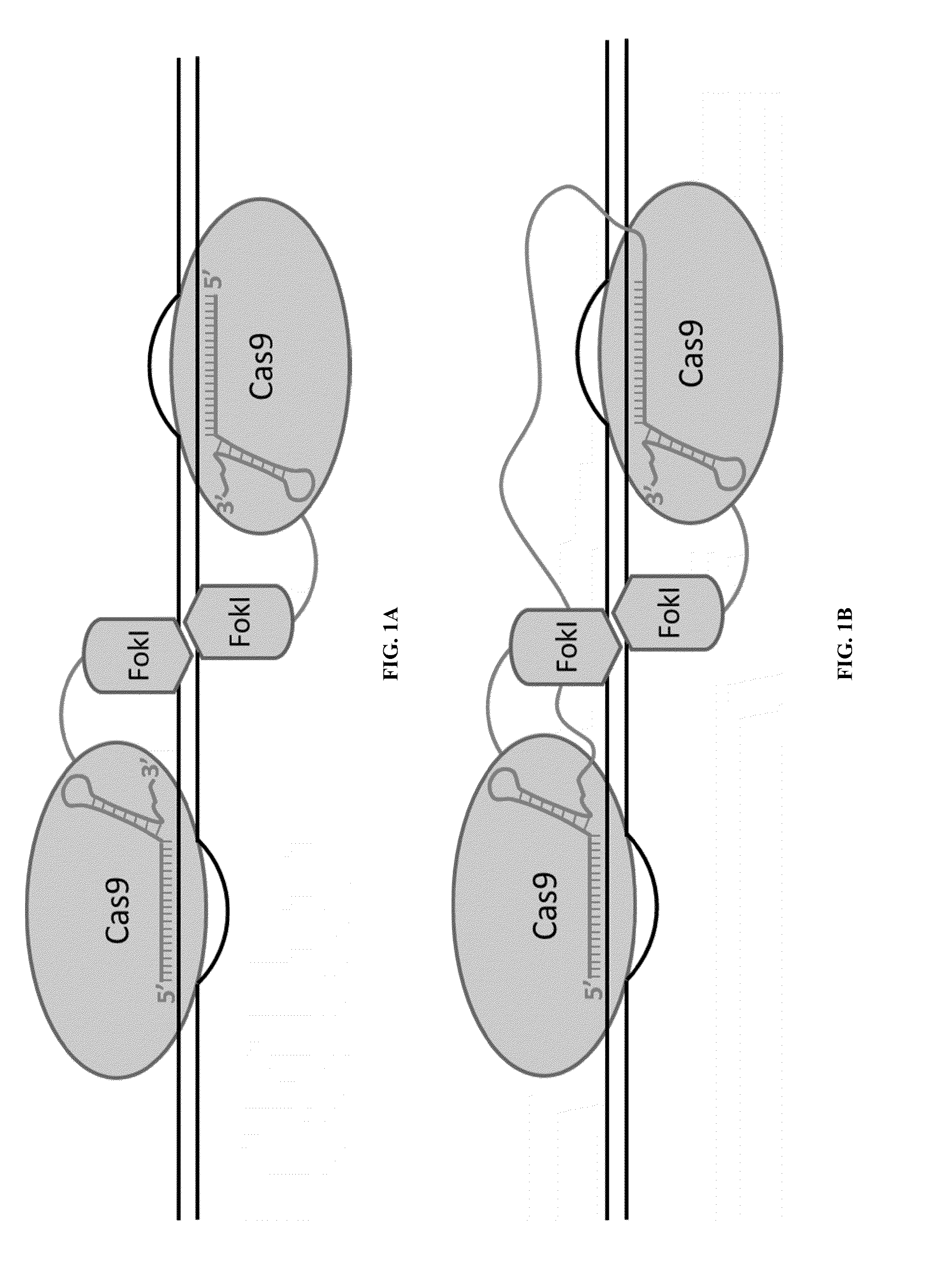
Cas9-foki fusion proteins and uses thereof
ActiveUS20150071899A1Strong specificityReduce the possibilityFusion with DNA-binding domainPeptide/protein ingredientsResearch settingNuclease
Some aspects of this disclosure provide compositions, methods, and kits for improving the specificity of RNA-programmable endonucleases, such as Cas9. Also provided are variants of Cas9, e.g., Cas9 dimers and fusion proteins, engineered to have improved specificity for cleaving nucleic acid targets. Also provided are compositions, methods, and kits for site-specific nucleic acid modification using Cas9 fusion proteins (e.g., nuclease-inactivated Cas9 fused to a nuclease catalytic domain). Such Cas9 variants are useful in clinical and research settings involving site-specific modification of DNA, for example, genomic modifications.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
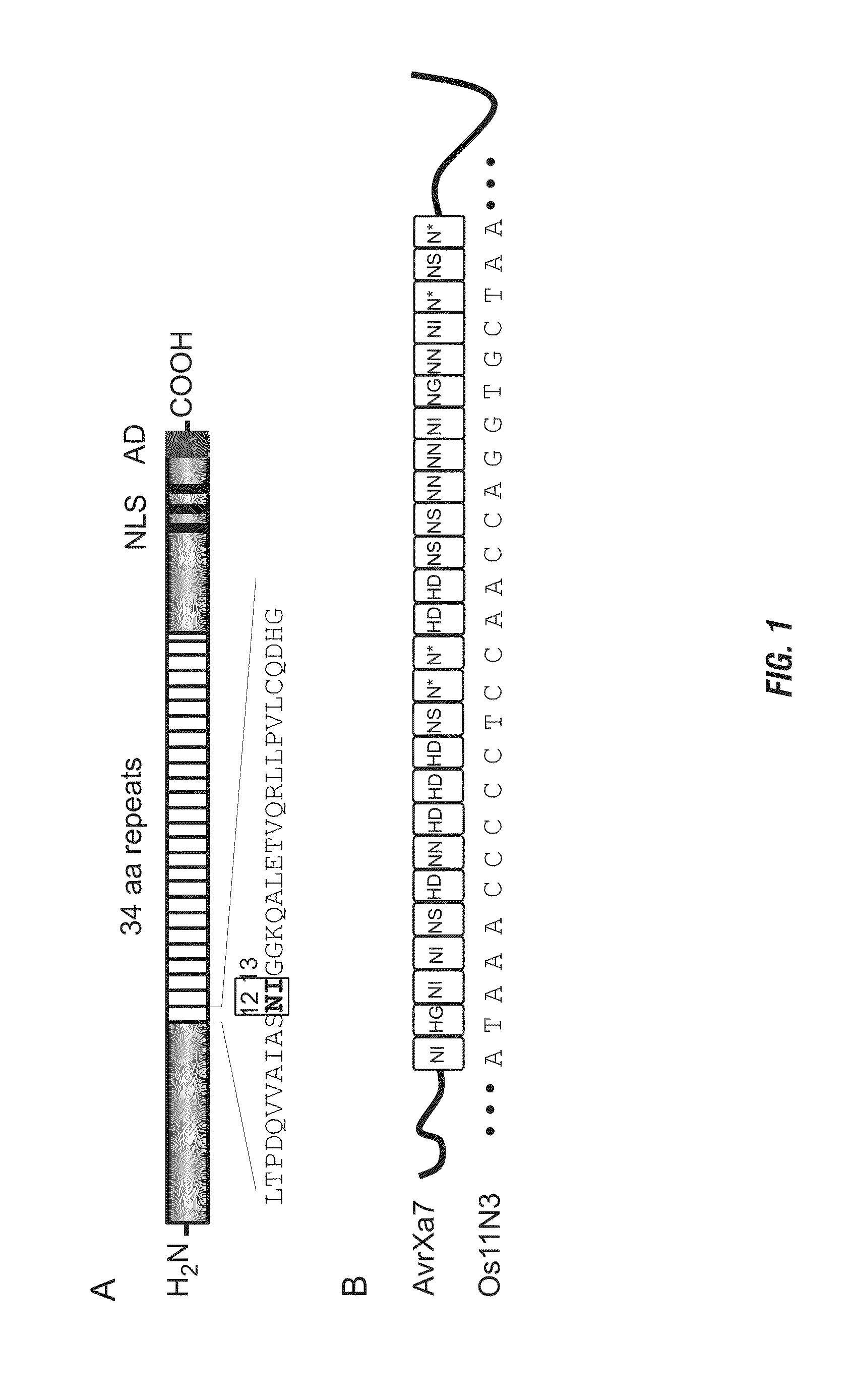
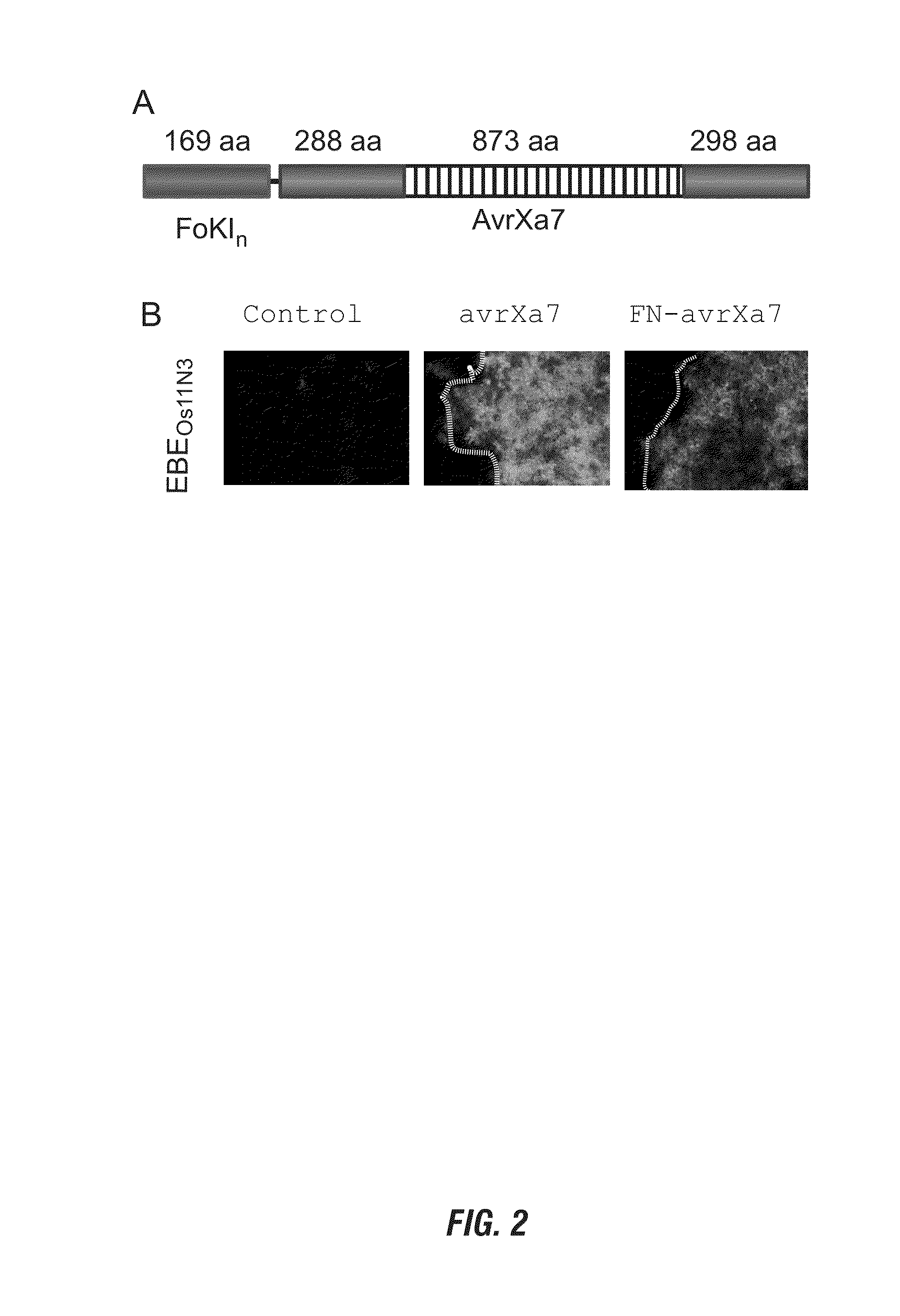
Nuclease activity of tal effector and foki fusion protein
The present invention provides compositions and methods for targeted cleavage of cellular chromatin in a region of interest and / or homologous recombination at a predetermined site in cells. Compositions include fusion polypeptides comprising a TAL effector binding domain and a cleavage domain. The cleavage domain can be from any endonuclease. In certain embodiments, the endonuclease is a Type IIS restriction endonuclease. In further embodiments, the Type IIS restriction endonuclease is FokI.
Owner:IOWA STATE UNIV RES FOUND
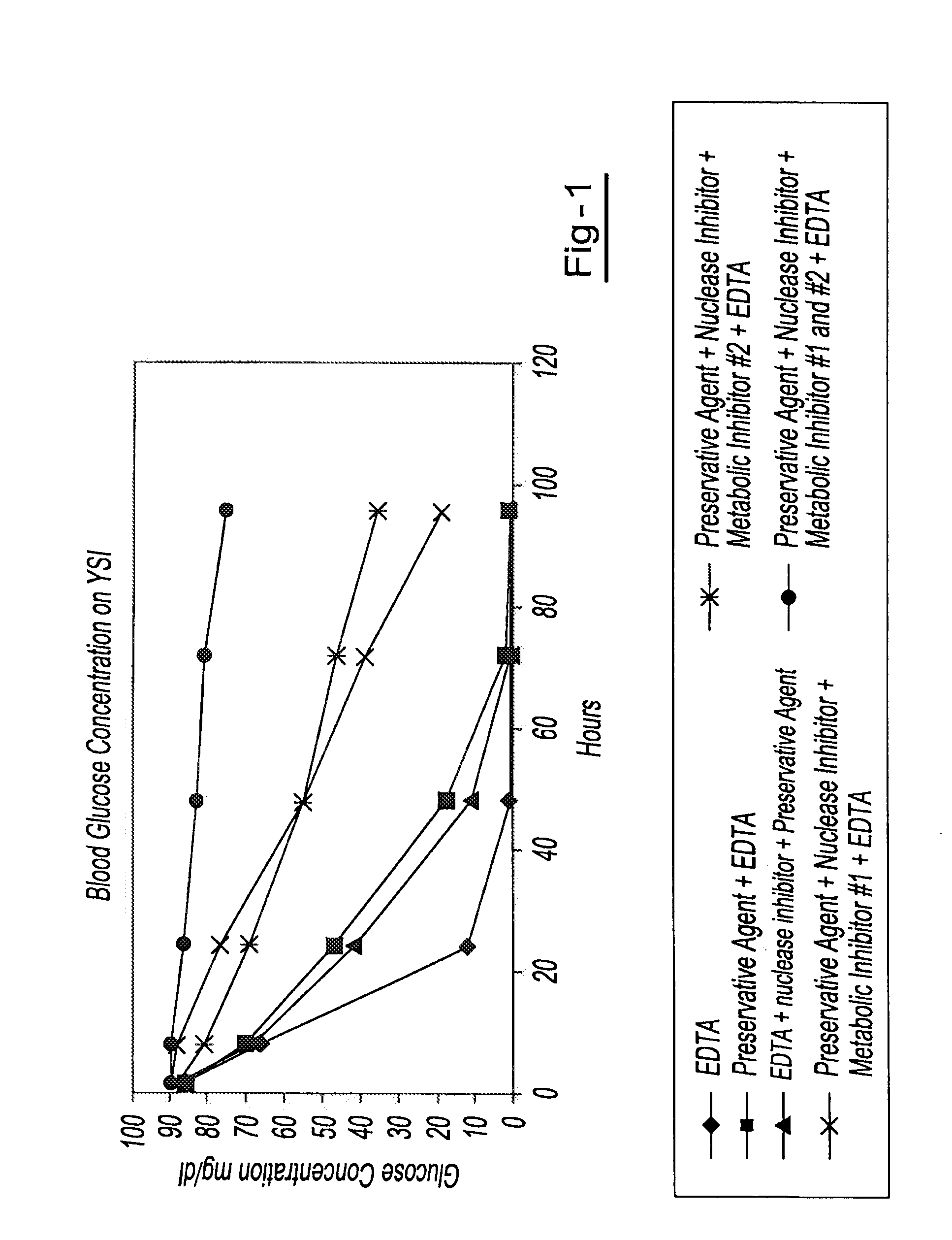
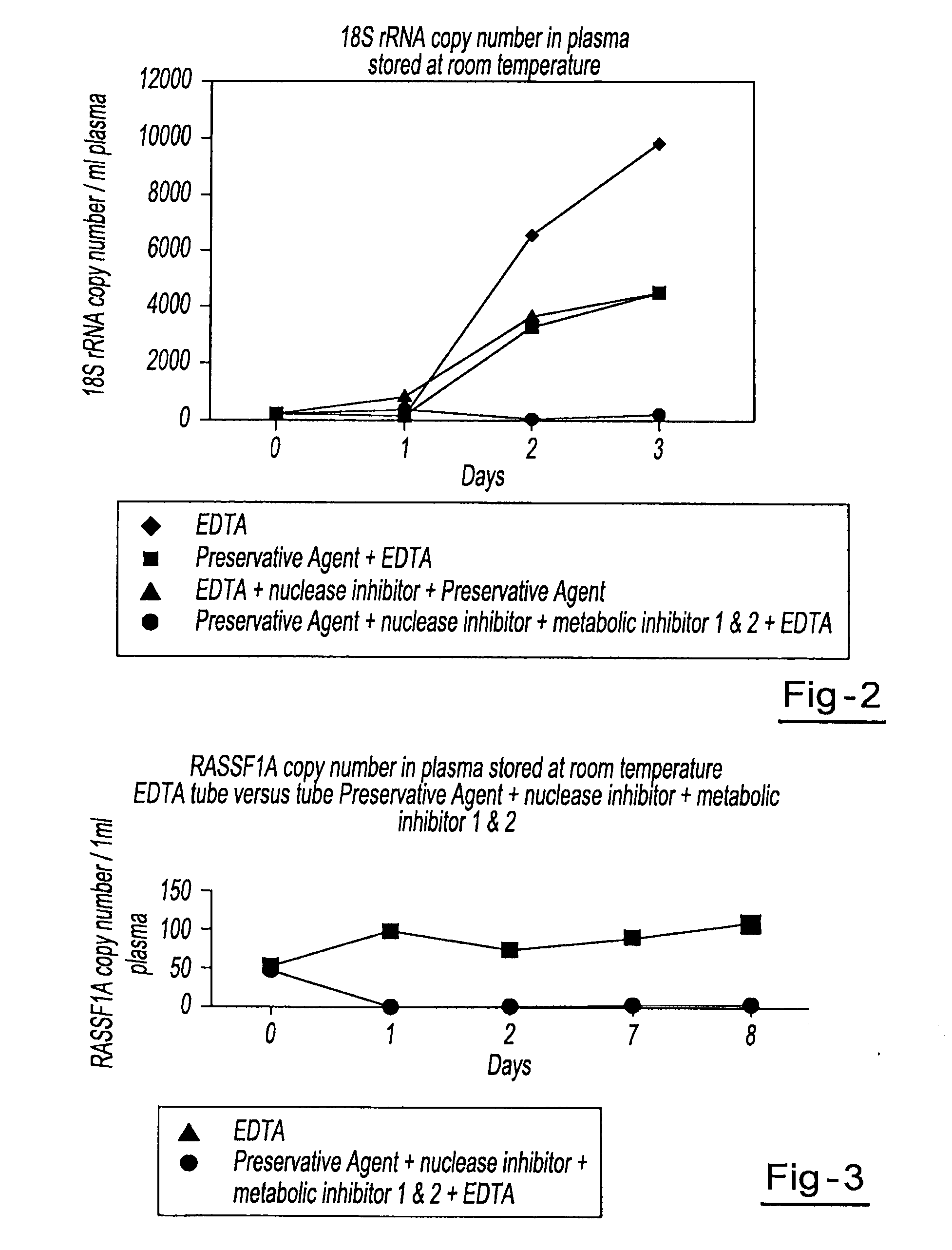
Preservation of cell-free nucleic acids
ActiveUS20100209930A1Prevent leakageInhibit synthesisMicrobiological testing/measurementChemical inhibitorsLysisNuclease
A method for preserving and processing cell-free nucleic acids located within a blood sample is disclosed, wherein a blood sample containing cell-free nucleic acids is treated to reduce both blood cell lysis and nuclease activity within the blood sample. The treatment of the sample aids in increasing the amount of cell-free nucleic acids that can be identified and tested while maintaining the structure and integrity of the nucleic acids.
Owner:STRECK LLC
Oligoribonucleotides and ribonucleases for cleaving RNA
InactiveUS7629321B2High affinityStrong specificityHydrolasesPeptide/protein ingredientsOrganismResearch purpose
Oligomeric compounds including oligoribonucleotides and oligoribonucleosides are provided that have subsequences of 2′-pentoribofuranosyl nucleosides that activate dsRNase. The oligoribonucleotides and oligoribonucleosides can include substituent groups for increasing binding affinity to complementary nucleic acid strand as well as substituent groups for increasing nuclease resistance. The oligomeric compounds are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics. Also included in the invention are mammalian ribonucleases, i.e., enzymes that degrade RNA, and substrates for such ribonucleases. Such a ribonuclease is referred to herein as a dsRNase, wherein “ds” indicates the RNase's specificity for certain double-stranded RNA substrates. The artificial substrates for the dsRNases described herein are useful in preparing affinity matrices for purifying mammalian ribonuclease as well as non-degradative RNA-binding proteins.
Owner:IONIS PHARMA INC
Car+ t cells genetically modified to eliminate expression of t-cell receptor and/or HLA
ActiveUS20140349402A1Suitable for storageReduce/eliminate T cellsGenetically modified cellsMammal material medical ingredientsAntigen receptorsAutoimmunity
The present invention concerns methods and compositions for immunotherapy employing a modified T cell comprising disrupted T cell receptor and / or HLA and comprising a chimeric antigen receptor. In certain embodiments, the compositions are employed allogeneically as universal reagents for “off-the-shelf treatment of medical conditions such as cancer, autoimmunity, and infection. In particular embodiments, the T cell receptor-negative and / or HLA-negative T cells are generated using zinc finger nucleases, for example.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Single nucleotide detection using degradation of a fluorescent sequence
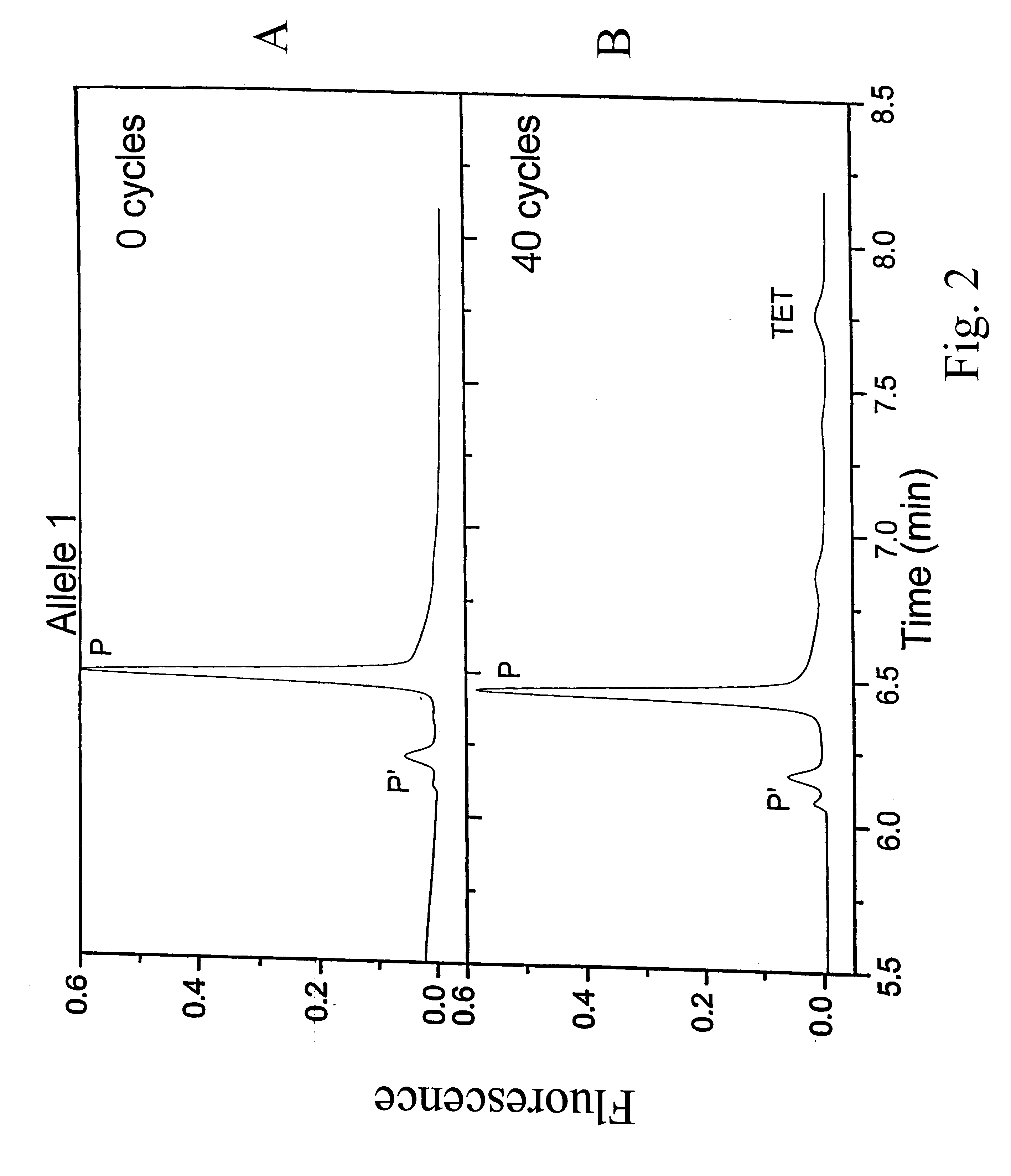
Methods and compositions are provided for detecting single nucleotide polymorphisms using a pair of oligonucleotides, a primer and a snp detection sequence, where the snp detection sequence hybridizes to the target DNA downstream from the primer and in the direction of primer extension. The snp detection sequence is characterized by having a nucleotide complementary to the snp and adjacent nucleotide complementary to adjacent nucleotides in the target and an electophoretic tag bonded to the 5'-nucleotide. The pair of oligonucleotides is combined with the target DNA under primer extension conditions, where the polymerase has 5'-3' exonuclease activity. When the snp is present, the electophoretic tag is released from the snp detection sequence, and can be detected by electrophoresis as indicative of the presence of the snp in the target DNA.
Owner:MONOGRAM BIOSCIENCES
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com