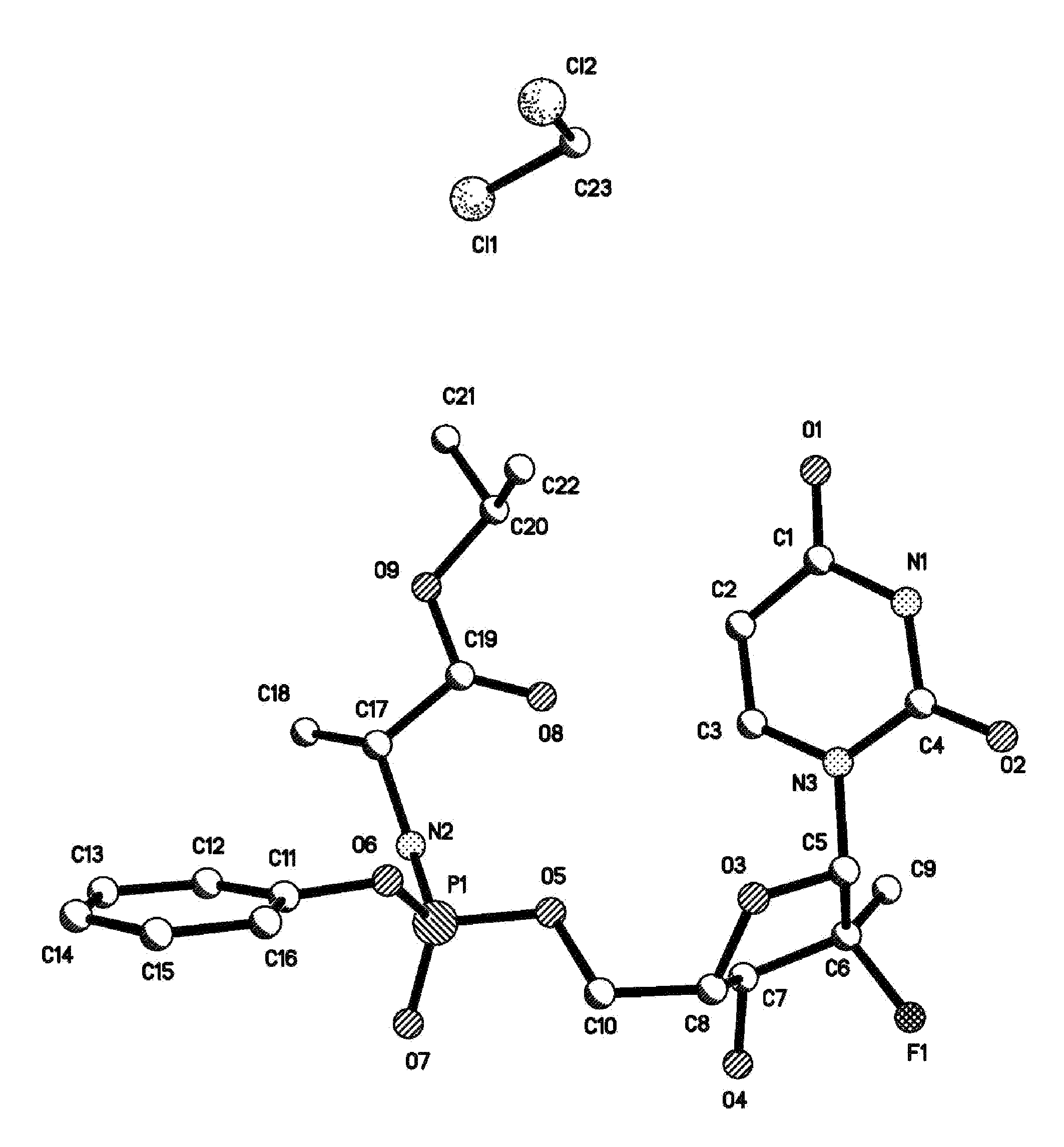
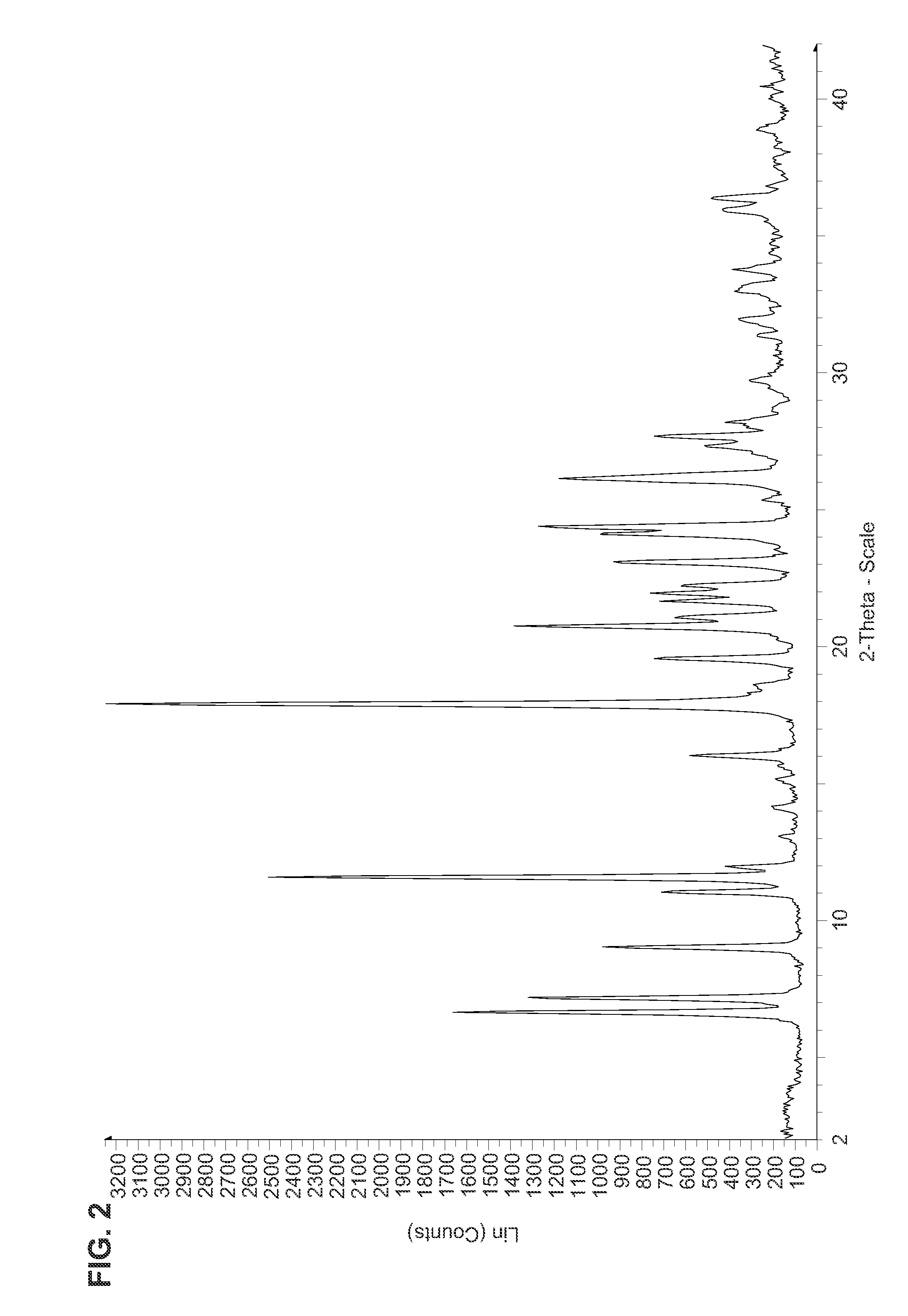
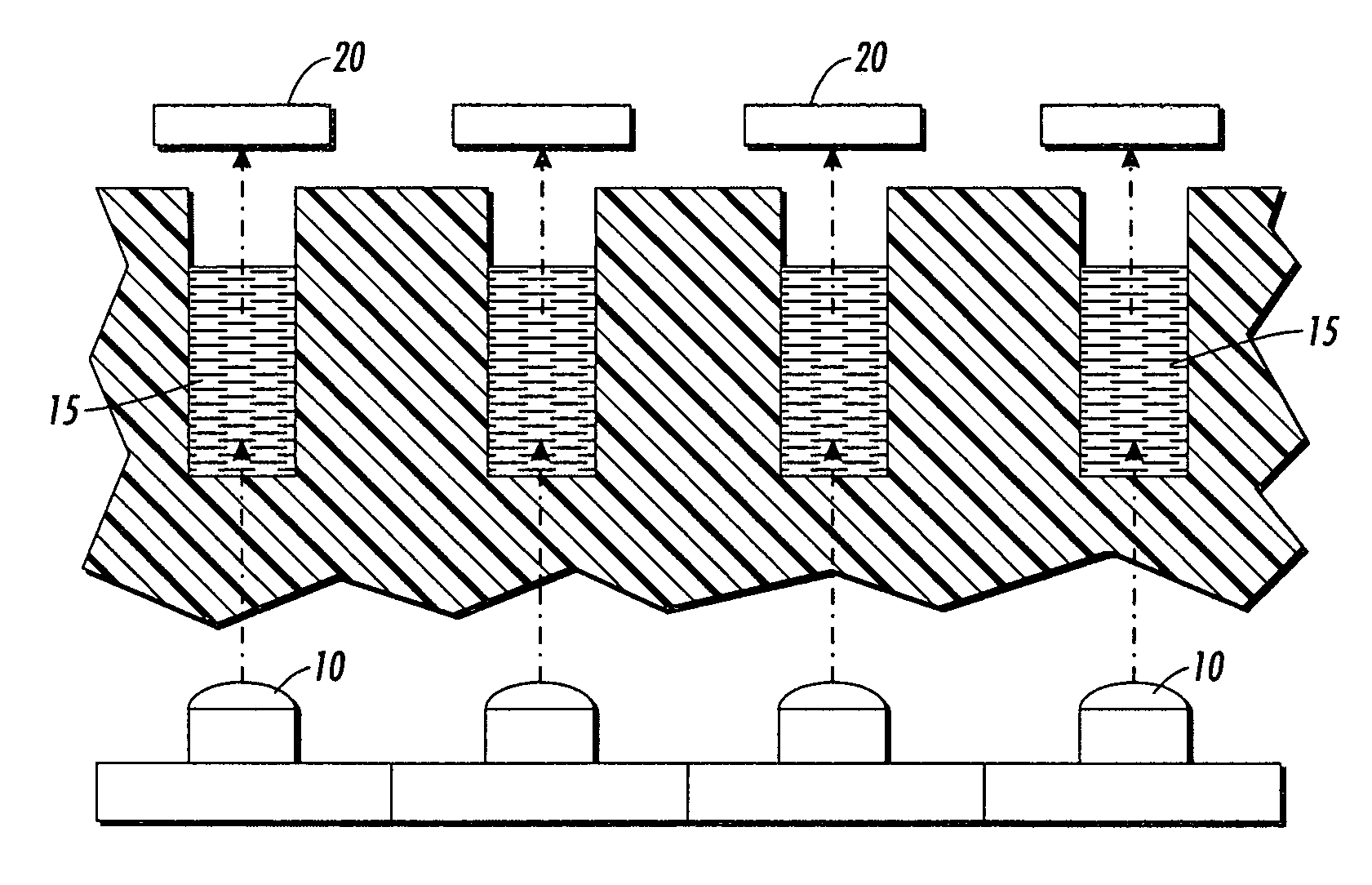
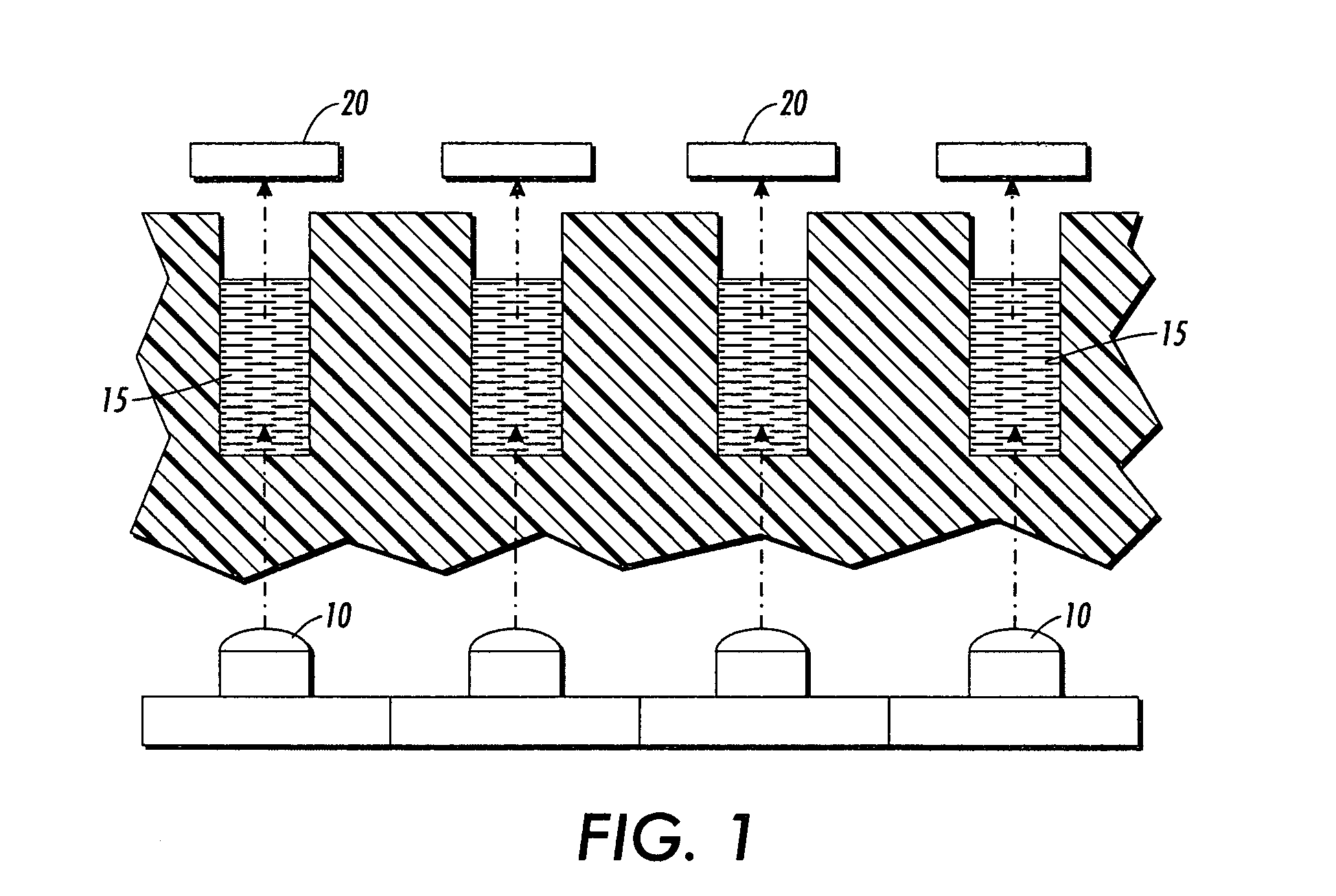
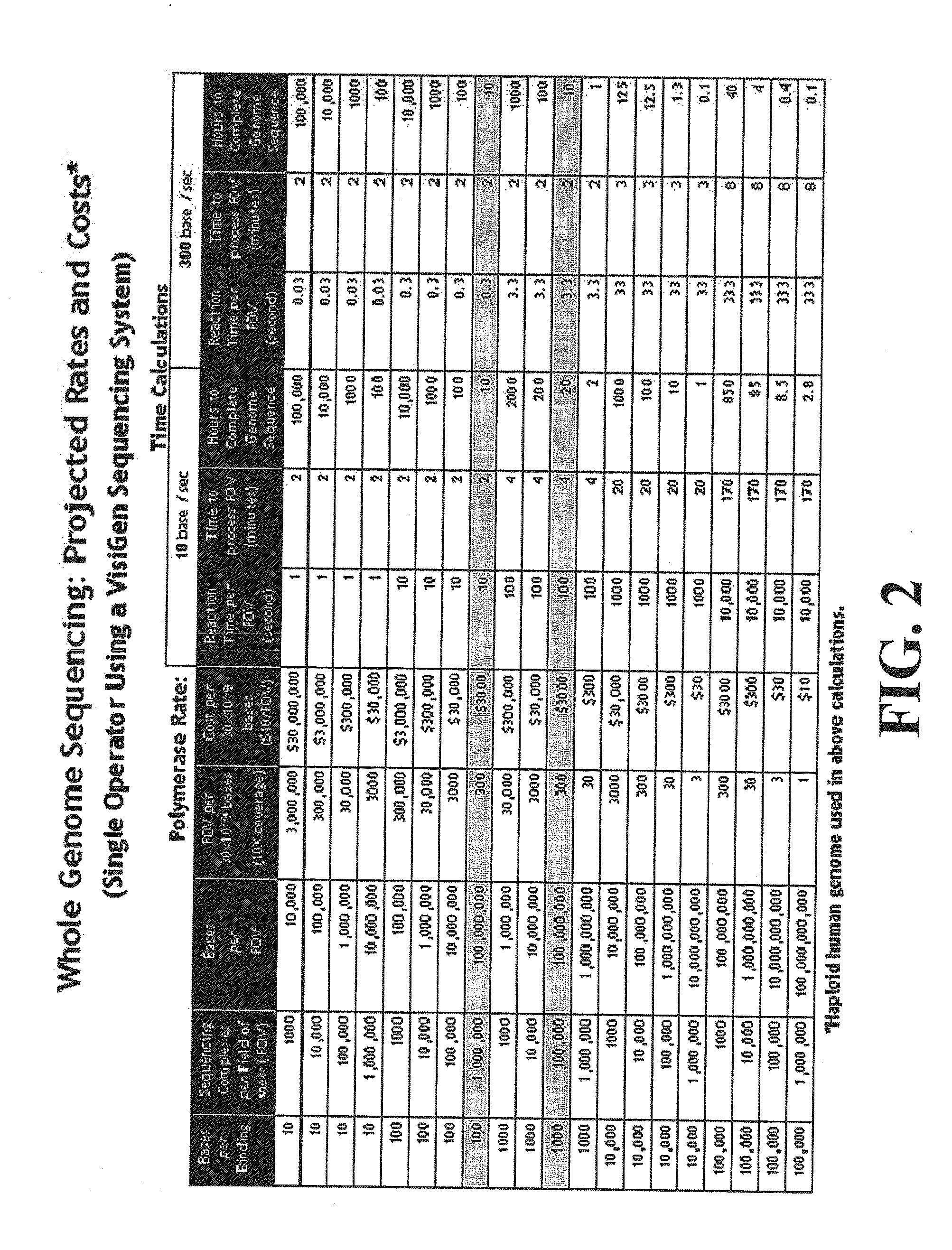
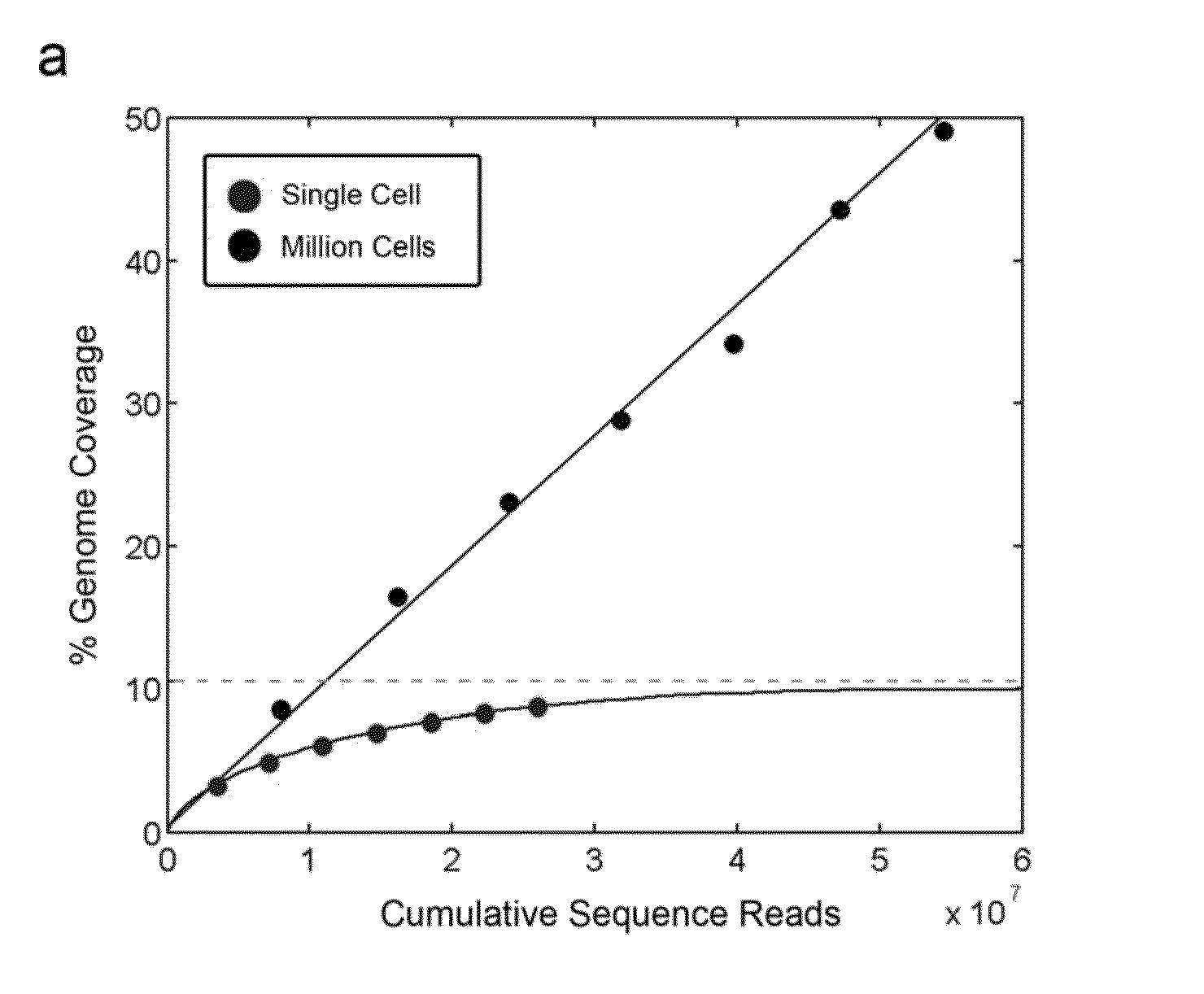
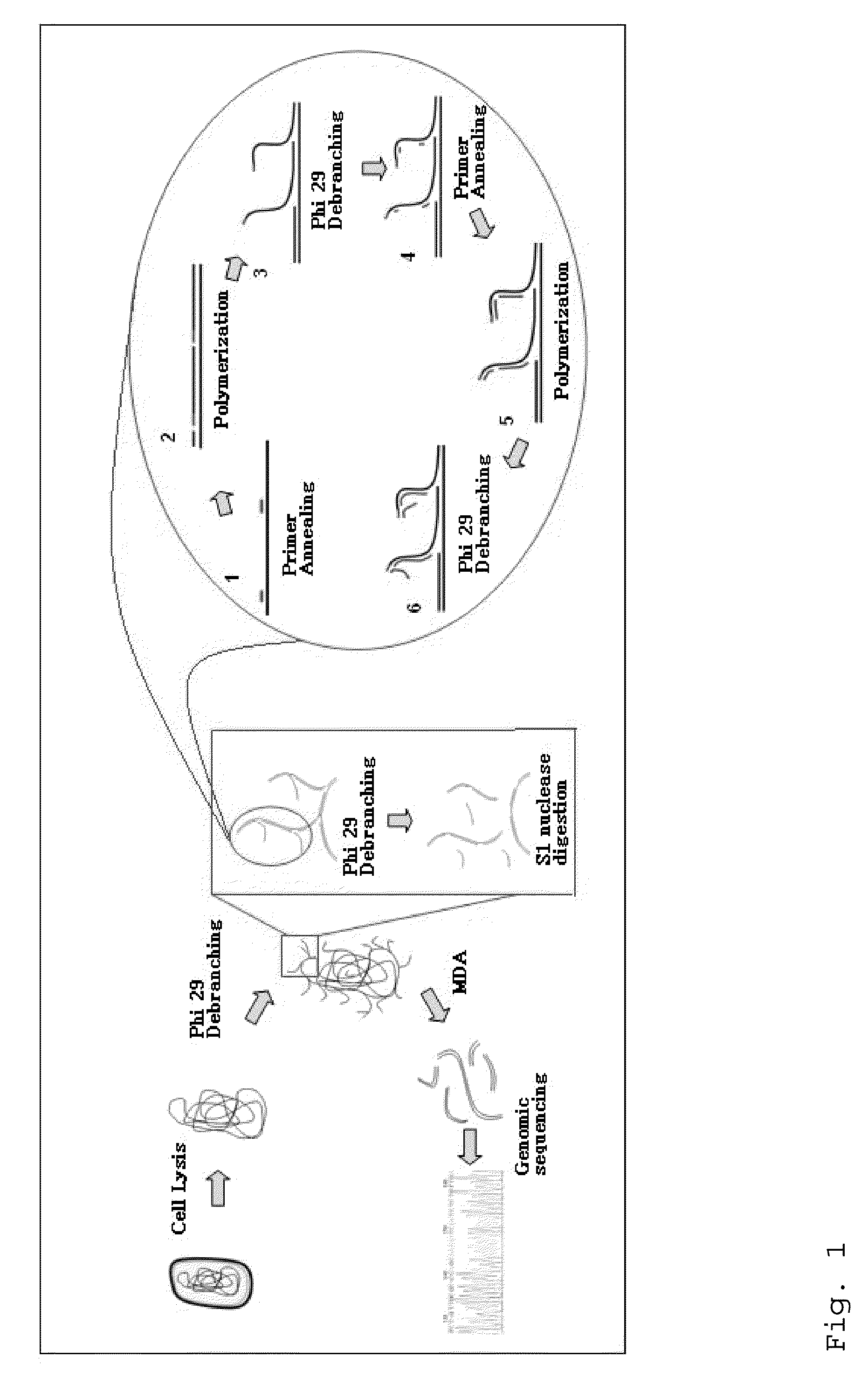
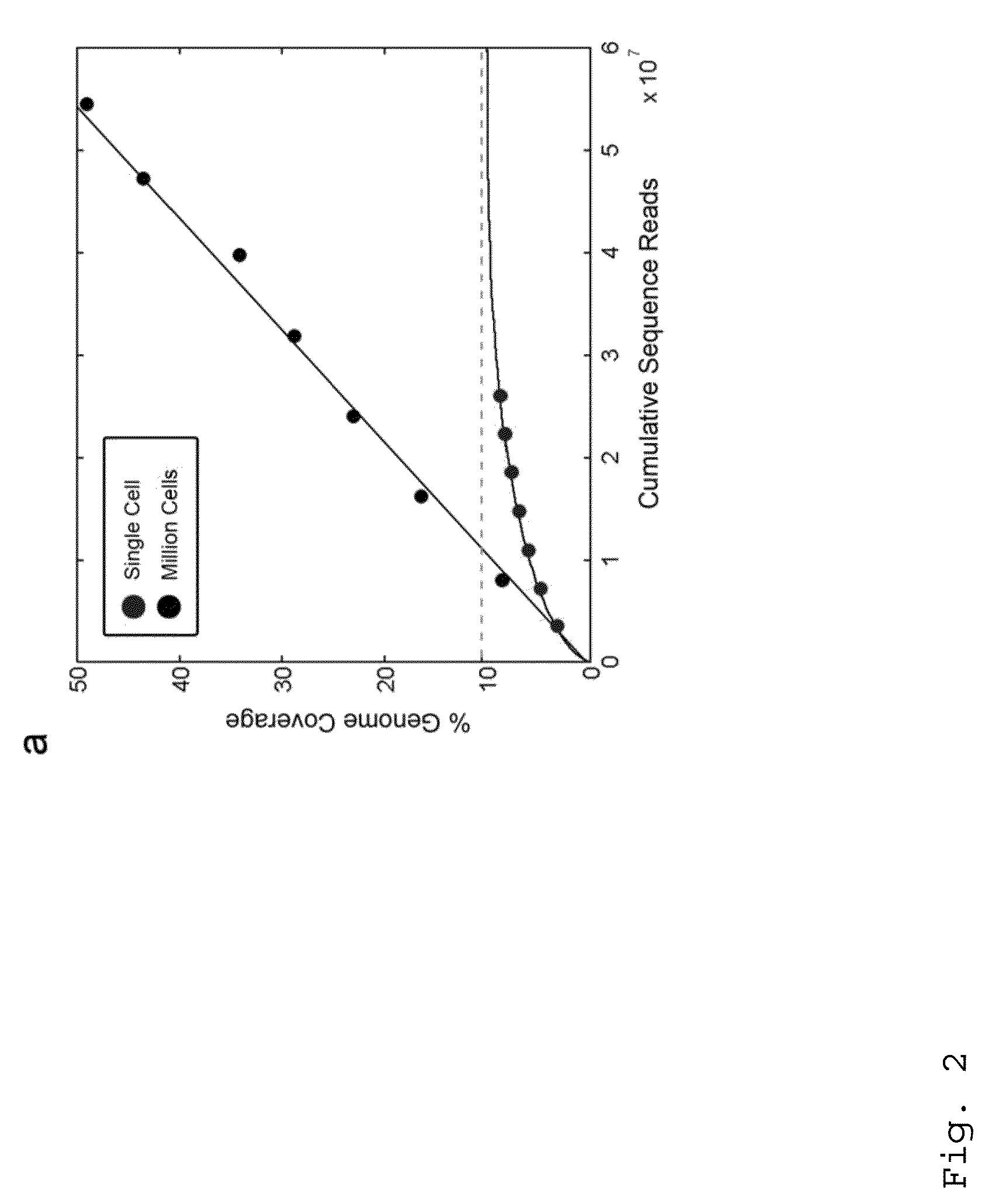
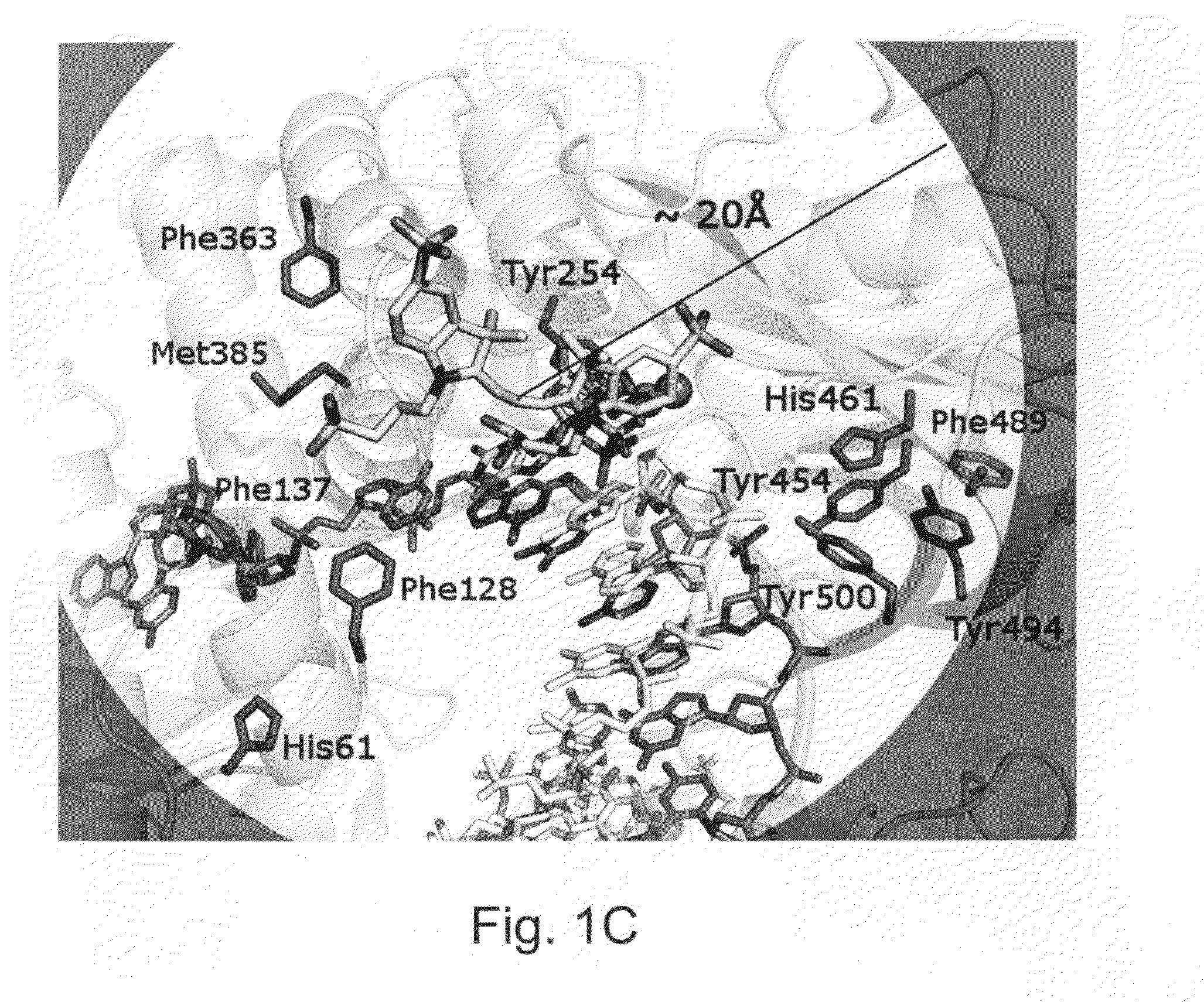
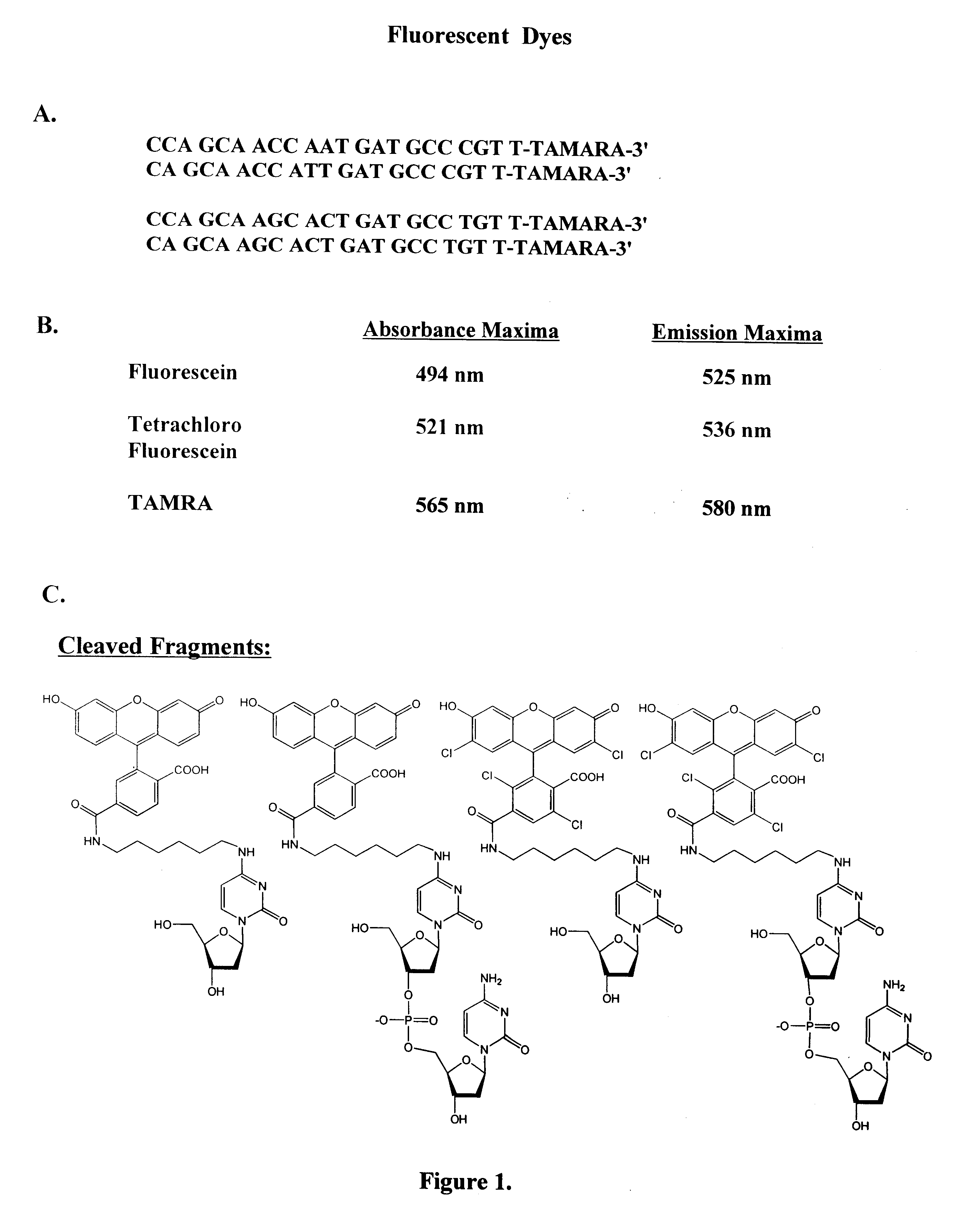
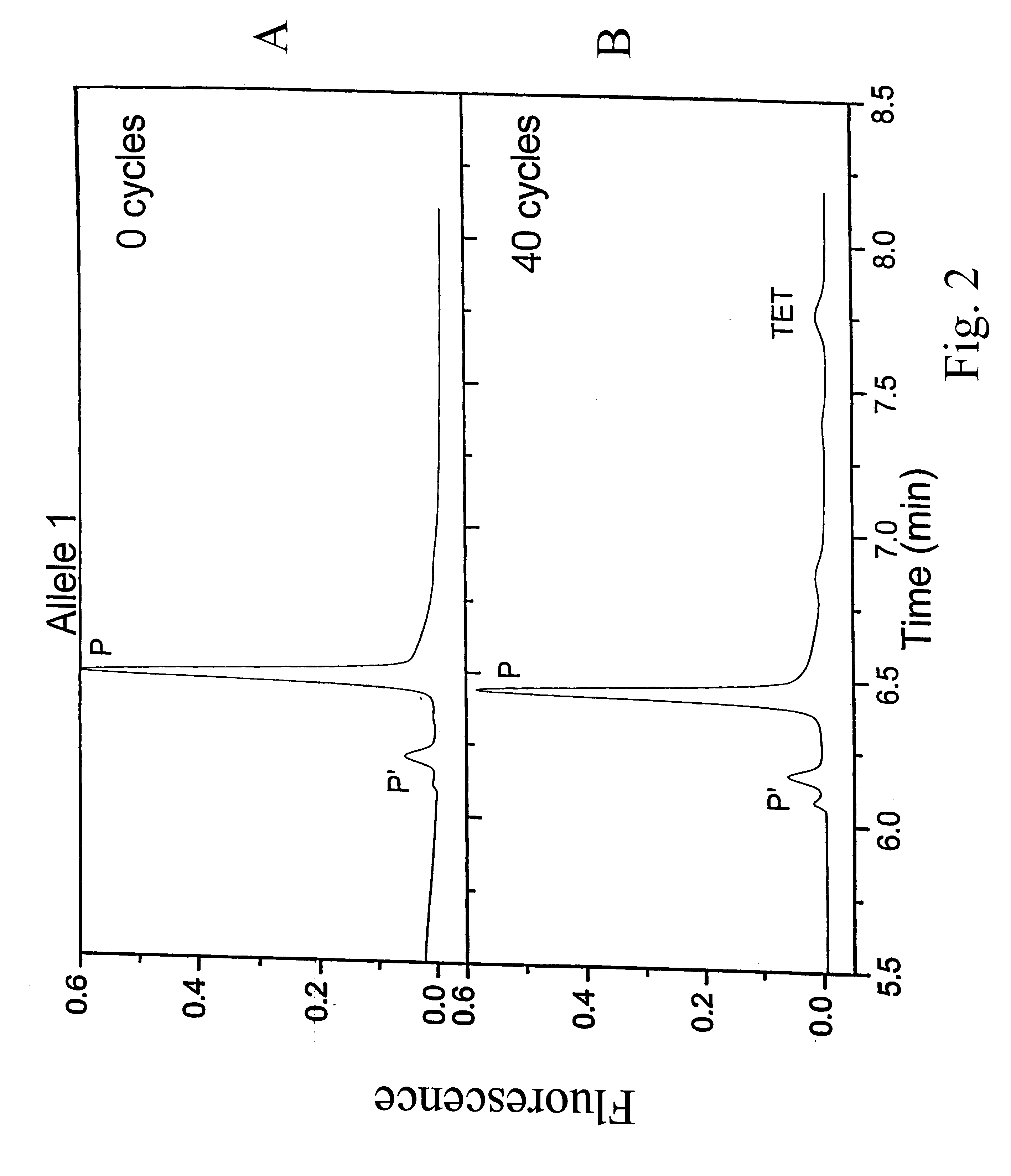
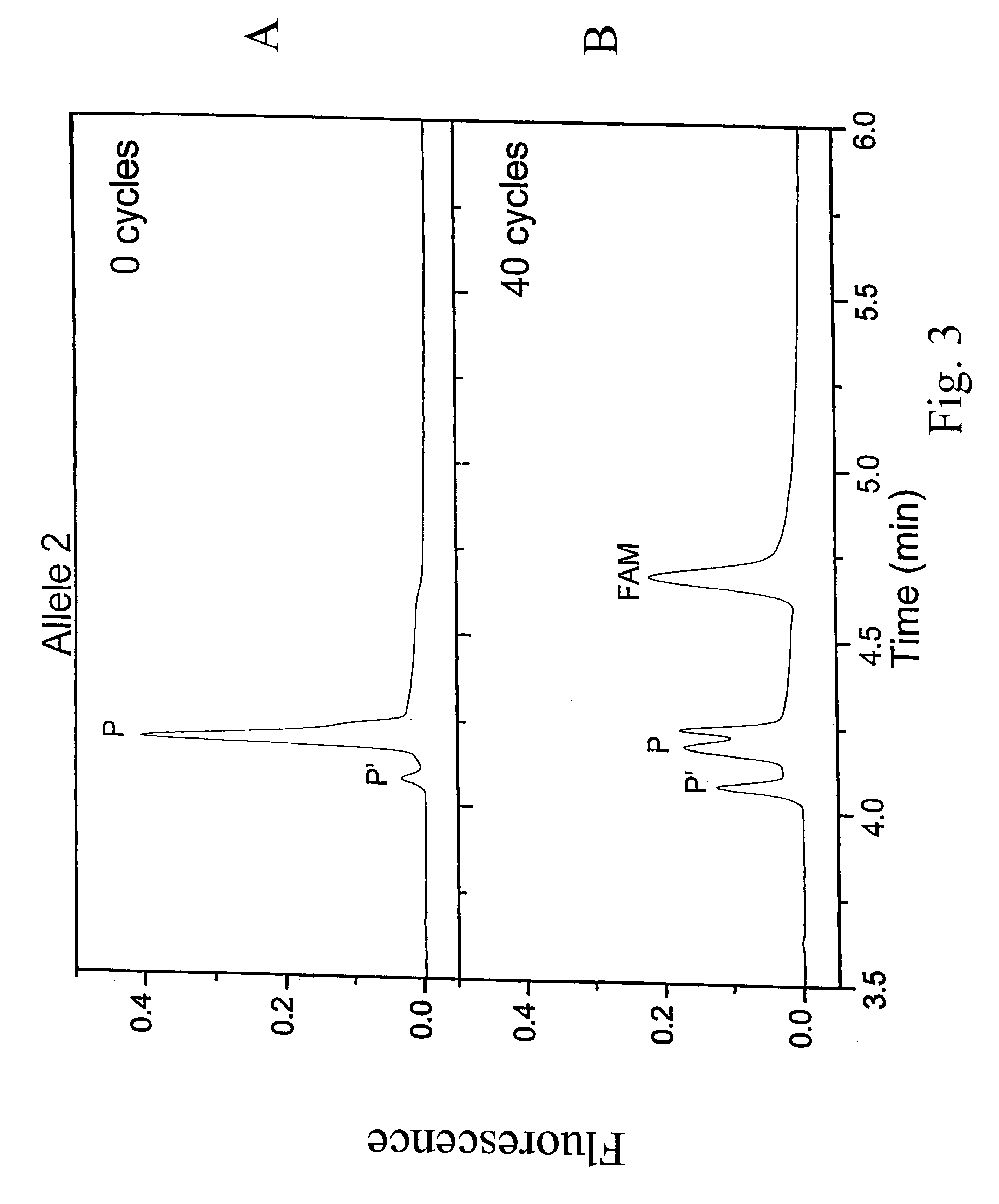
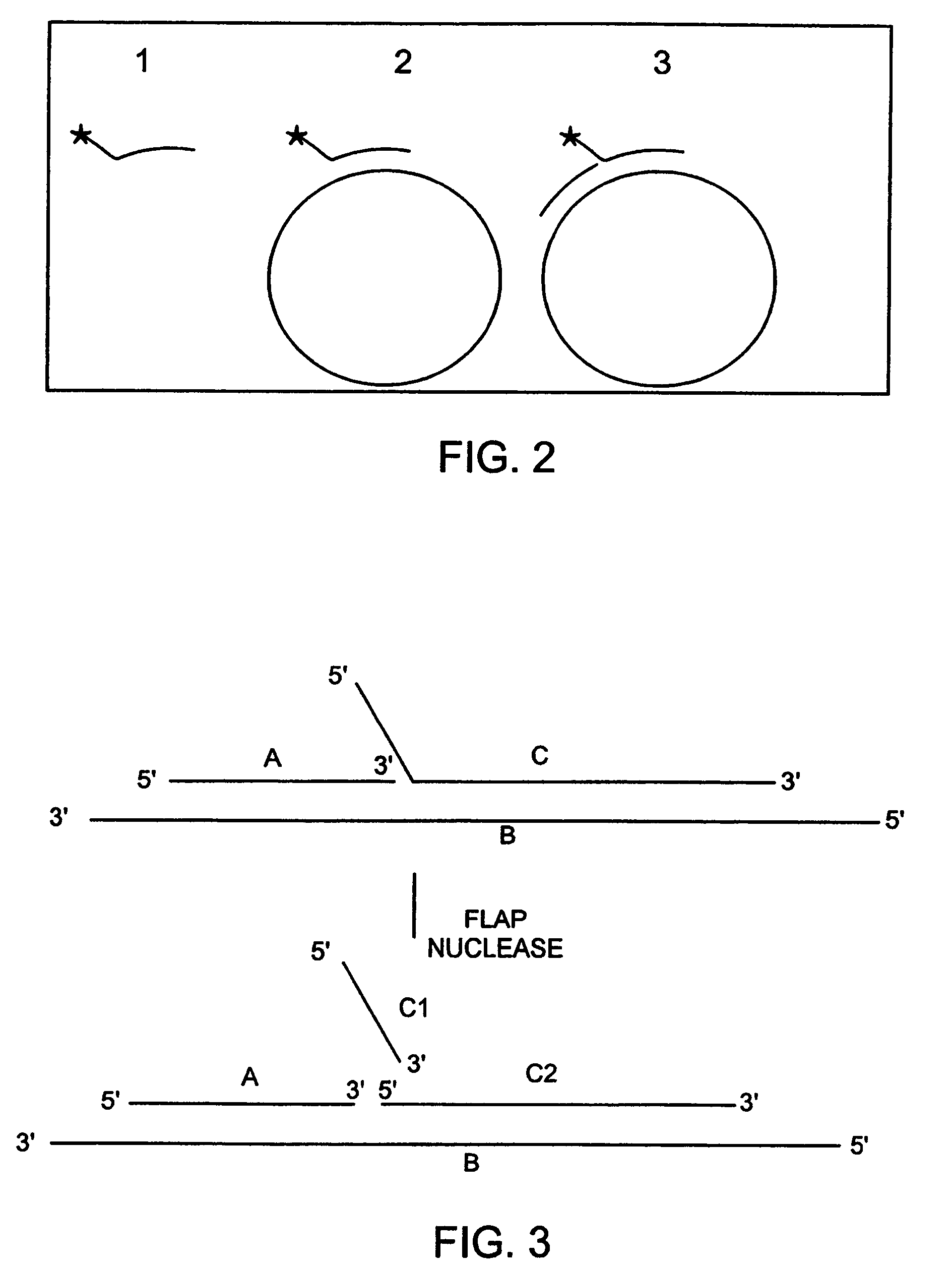
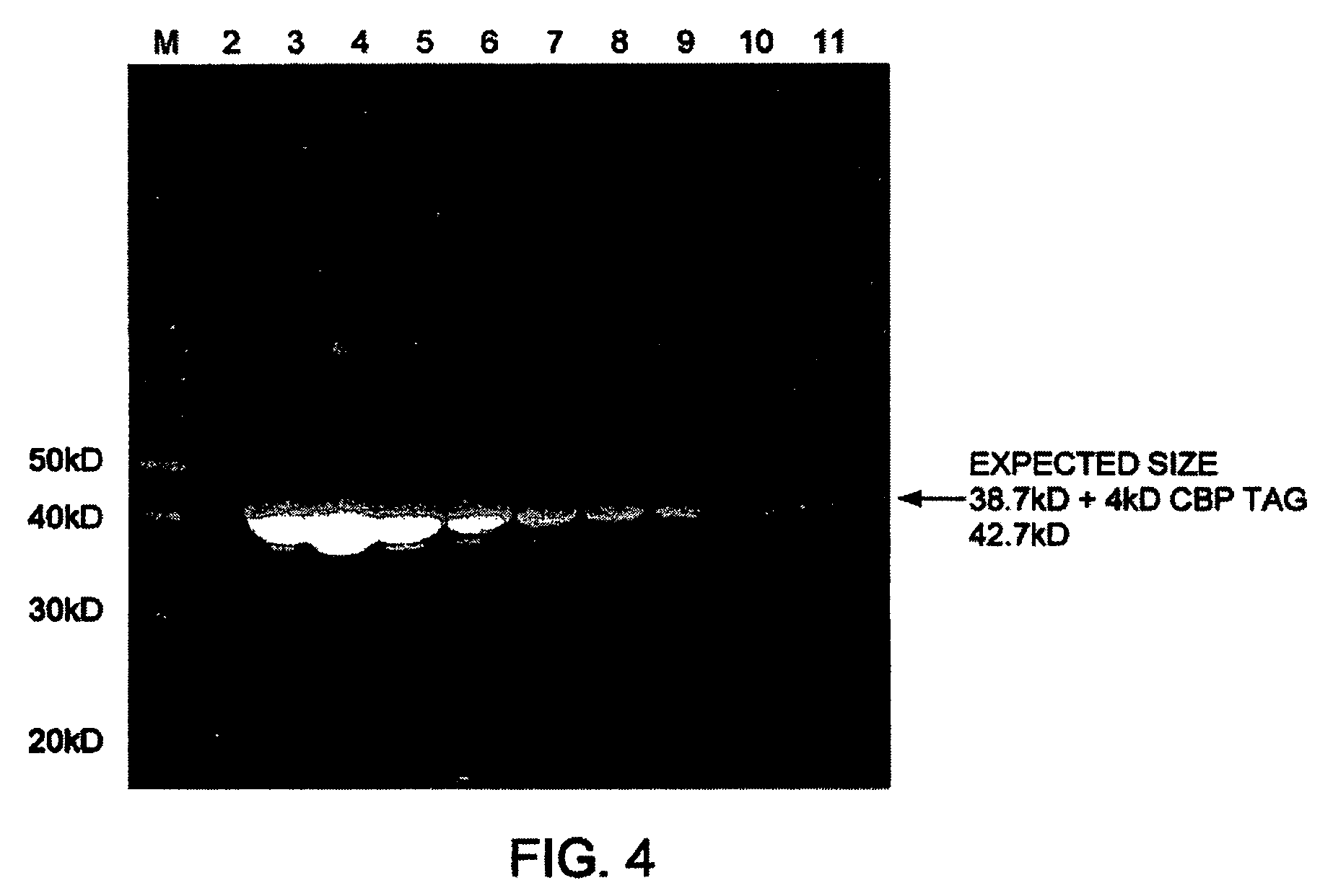
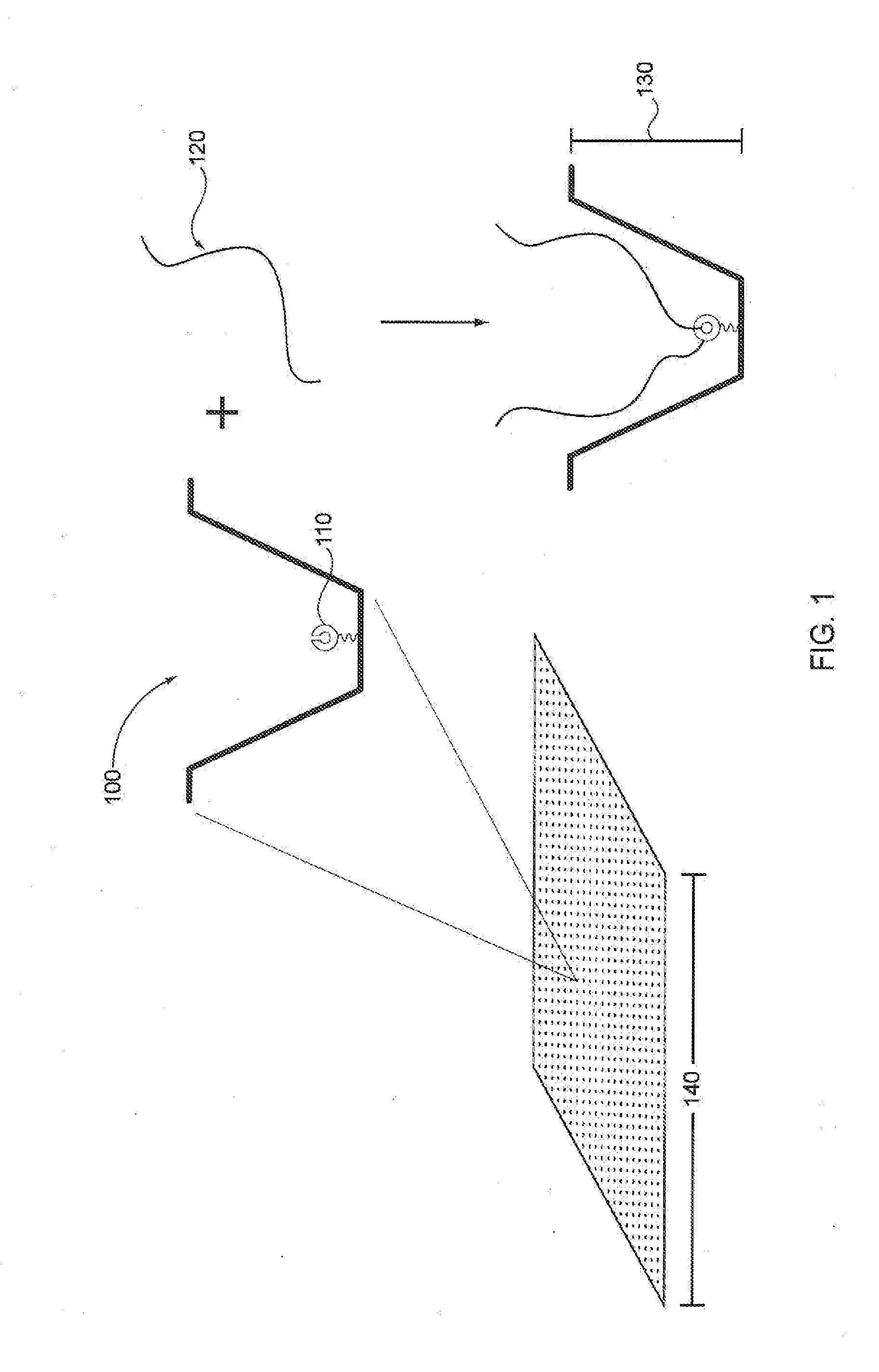
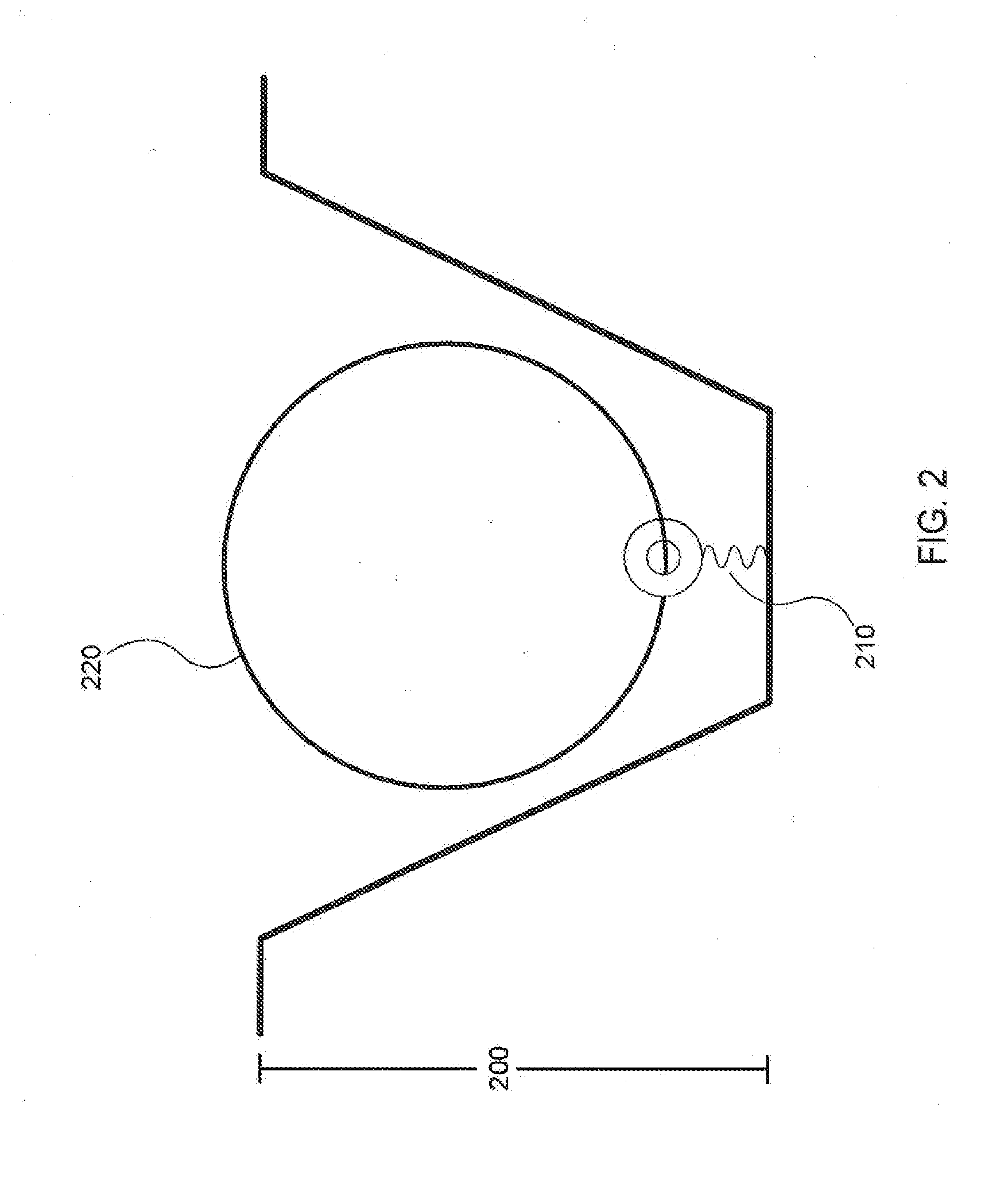
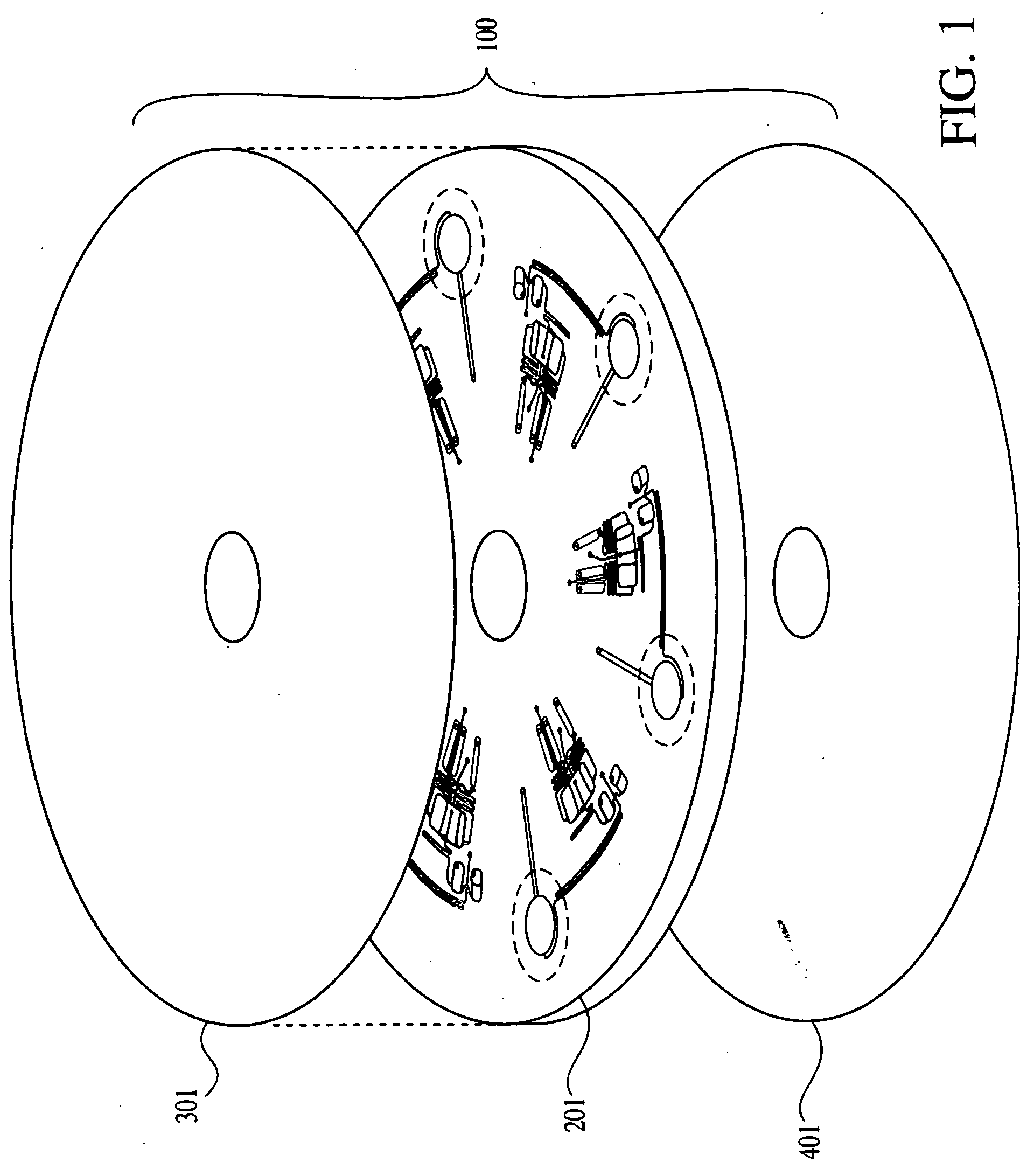
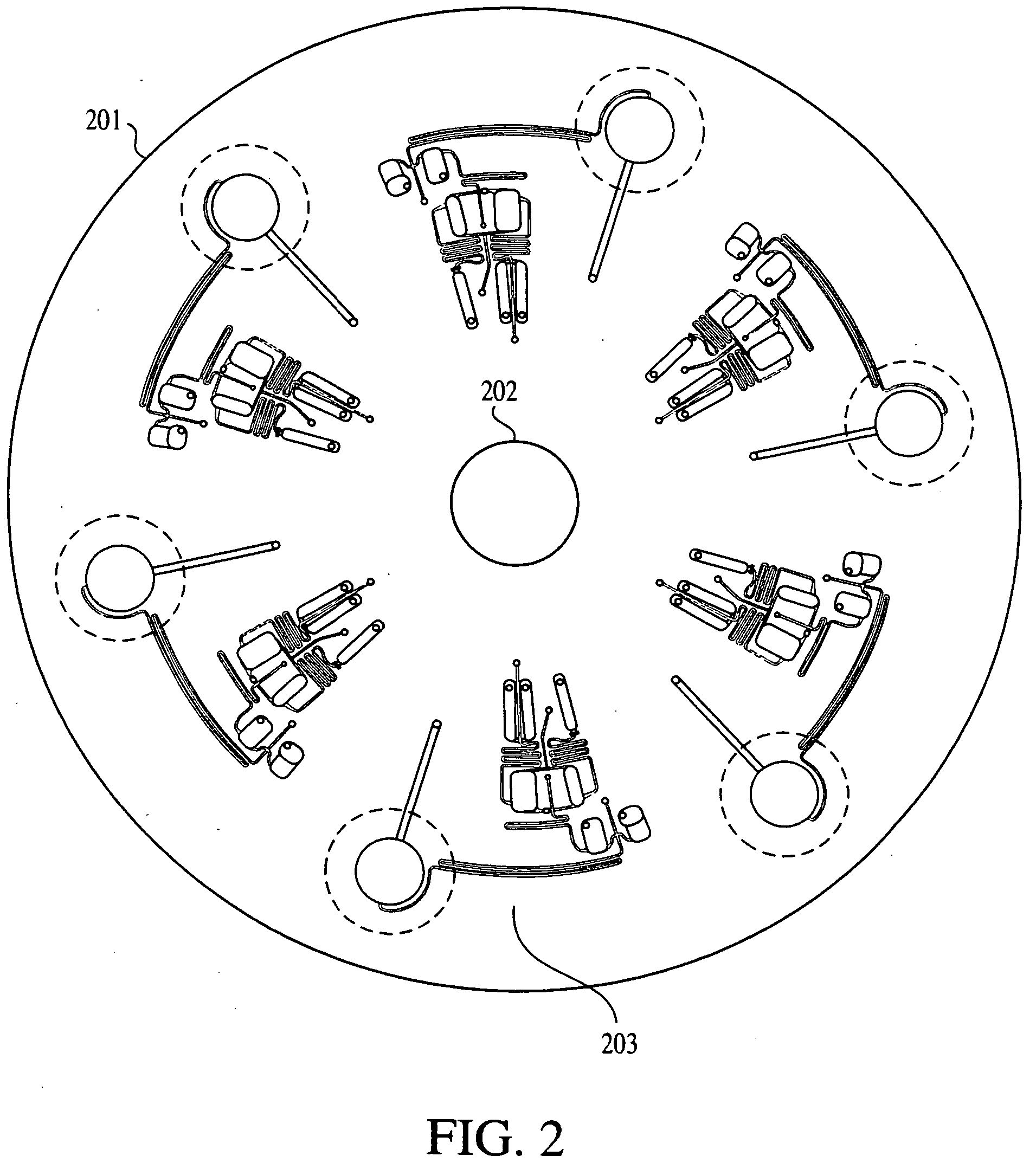
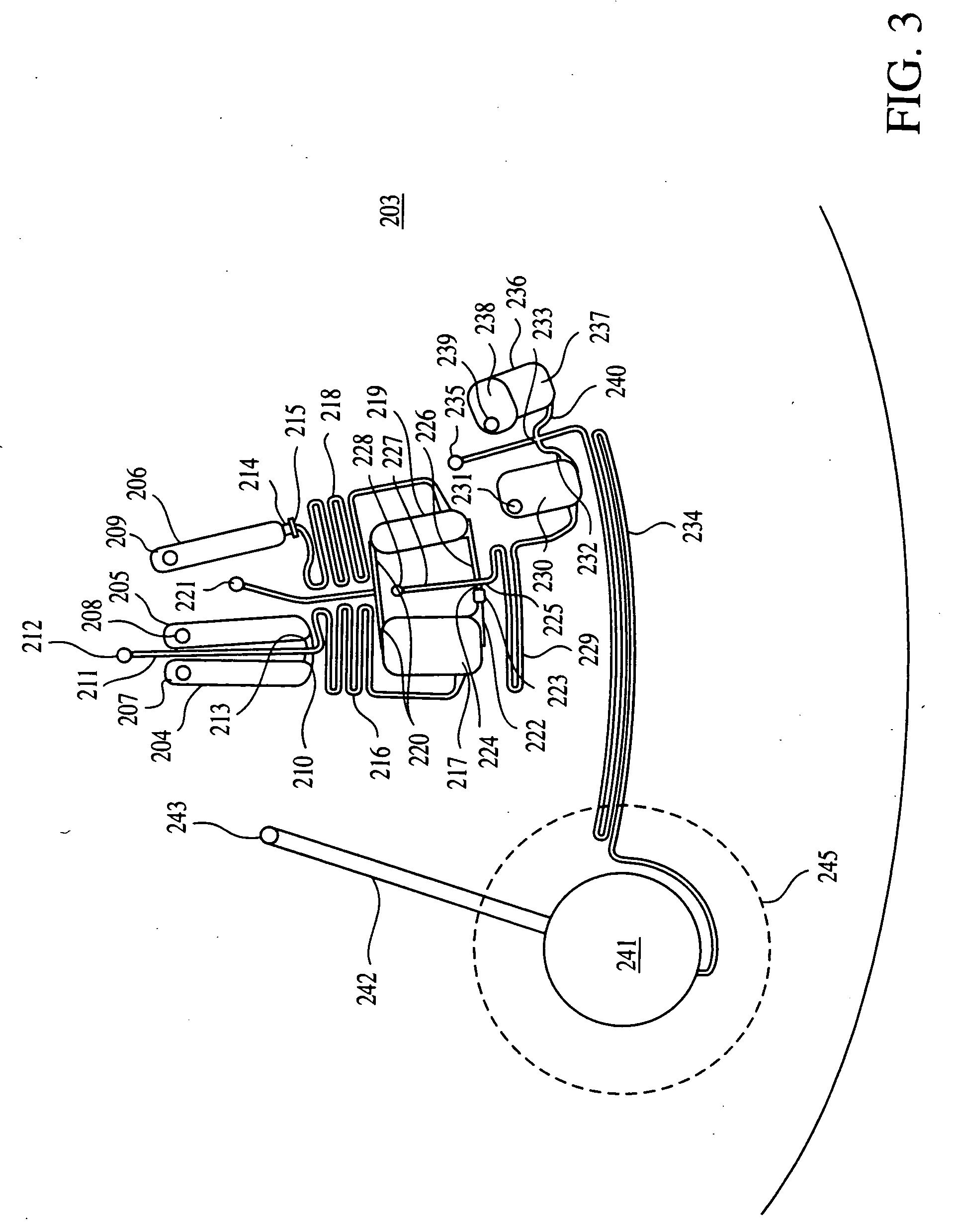
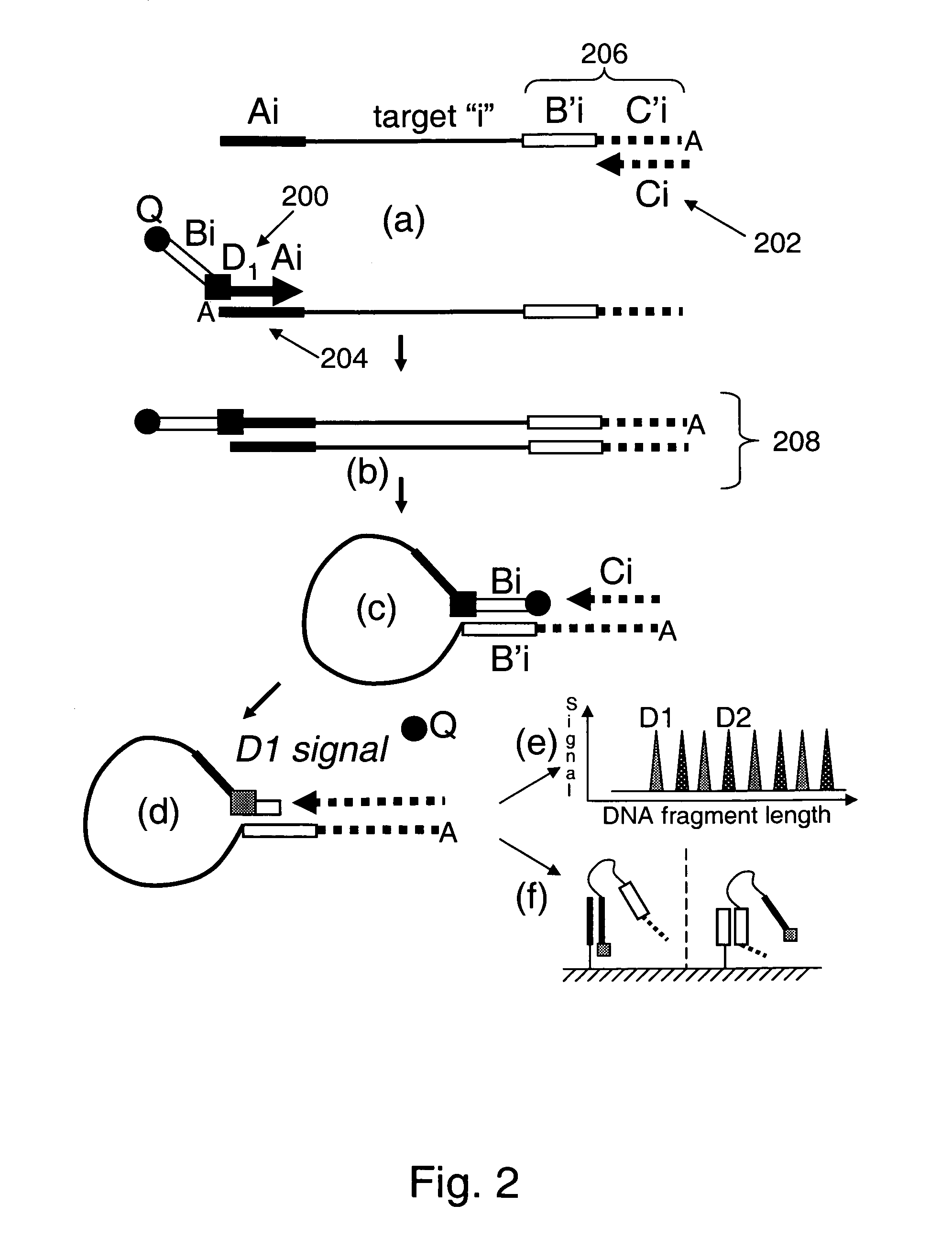
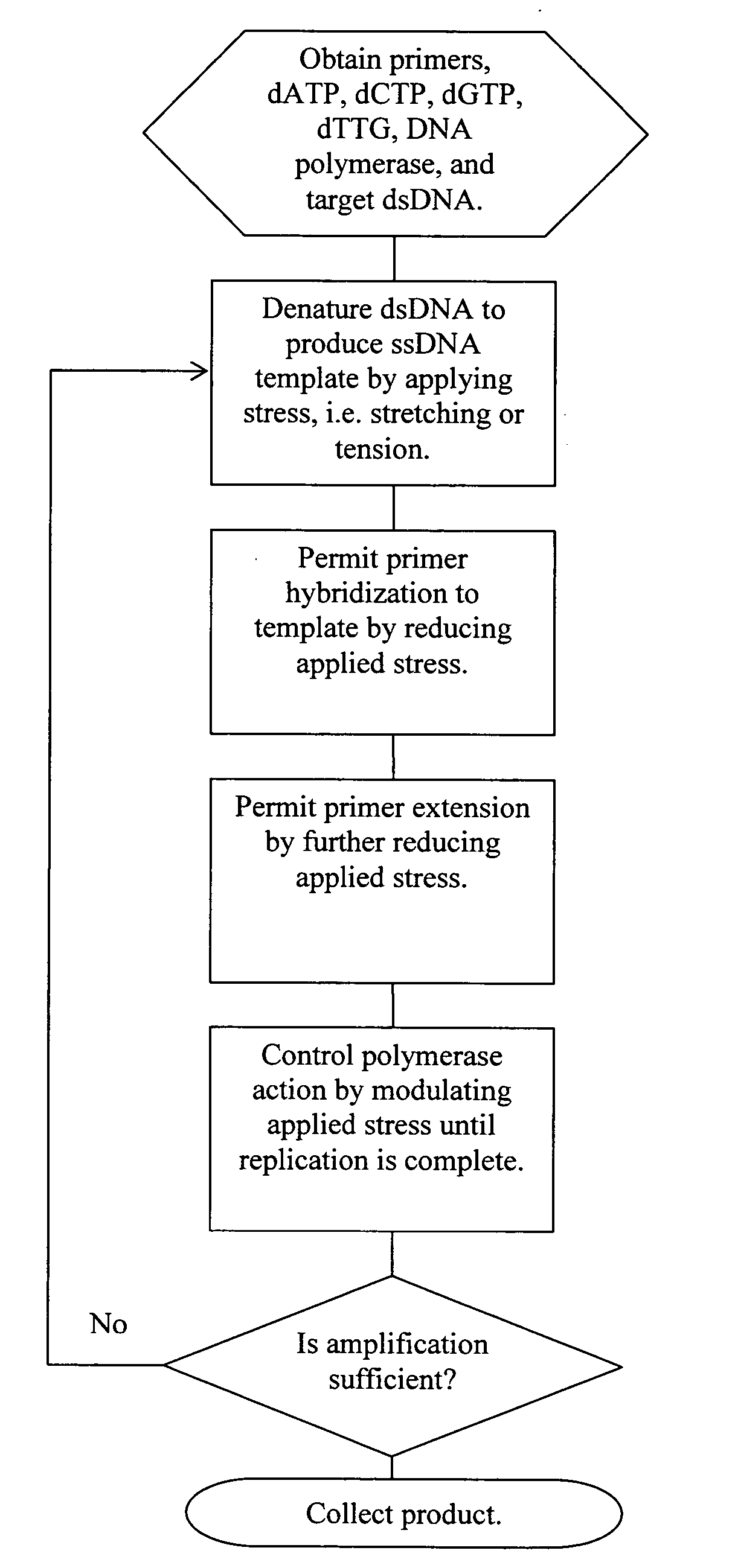
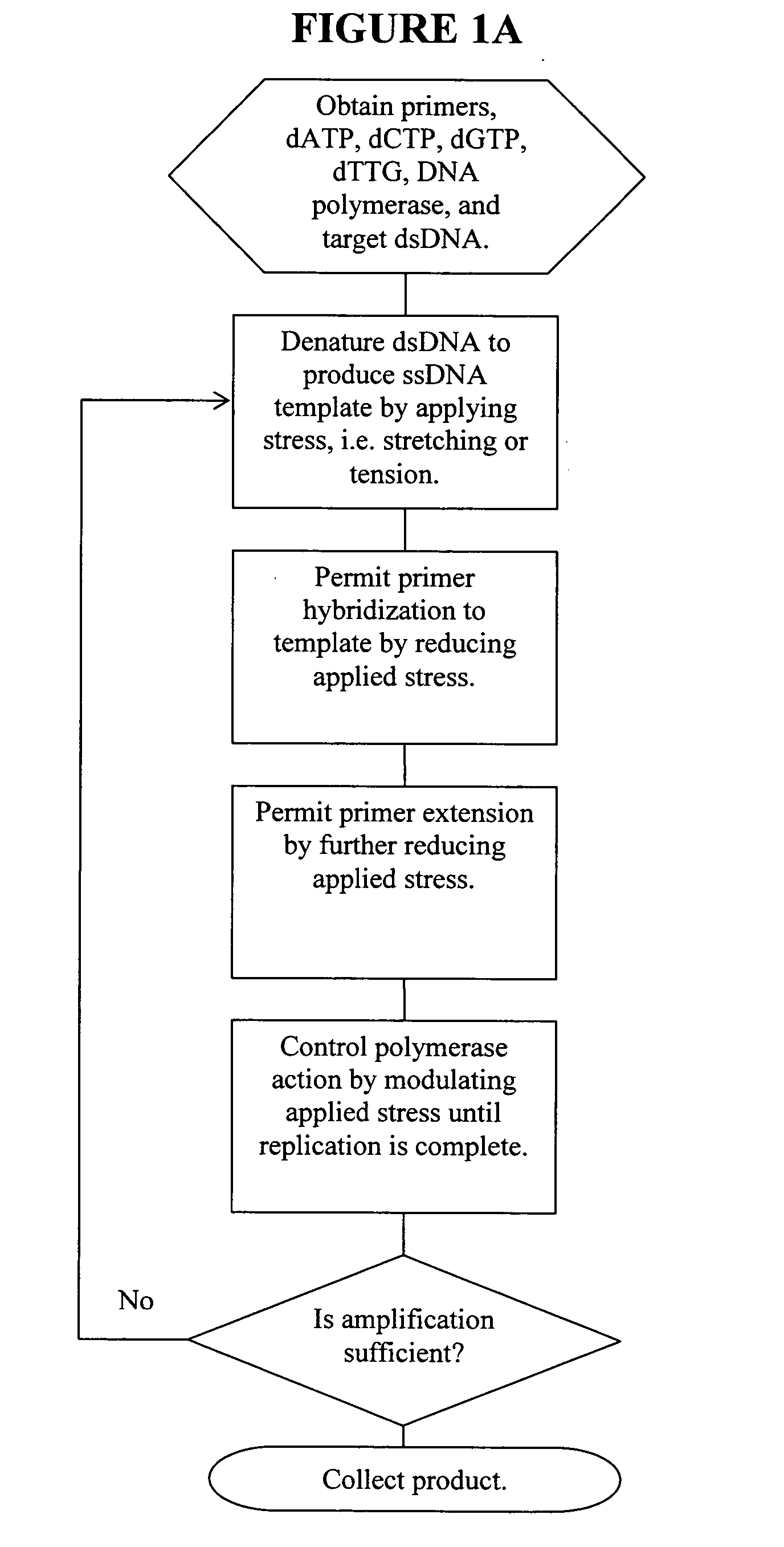
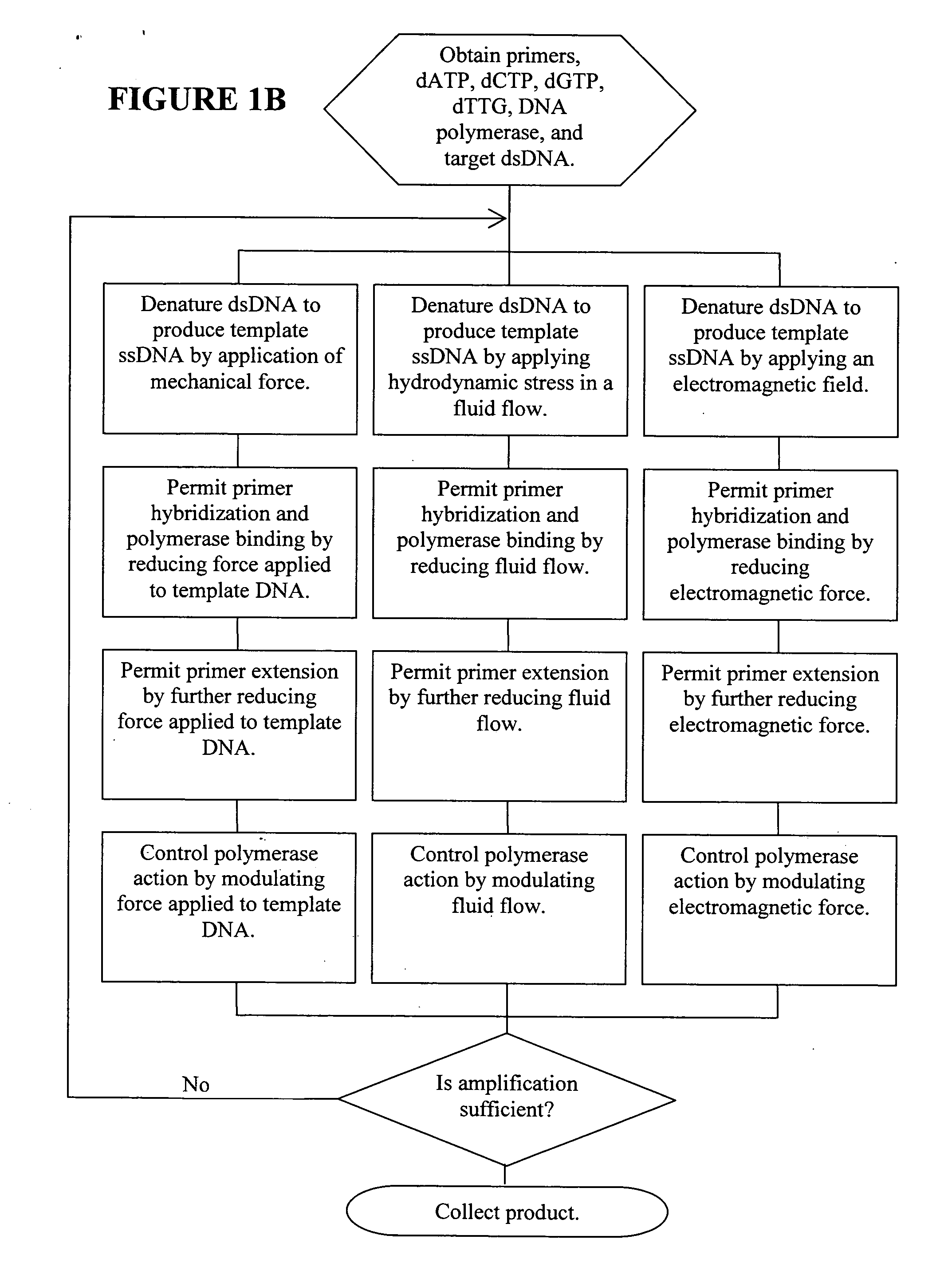
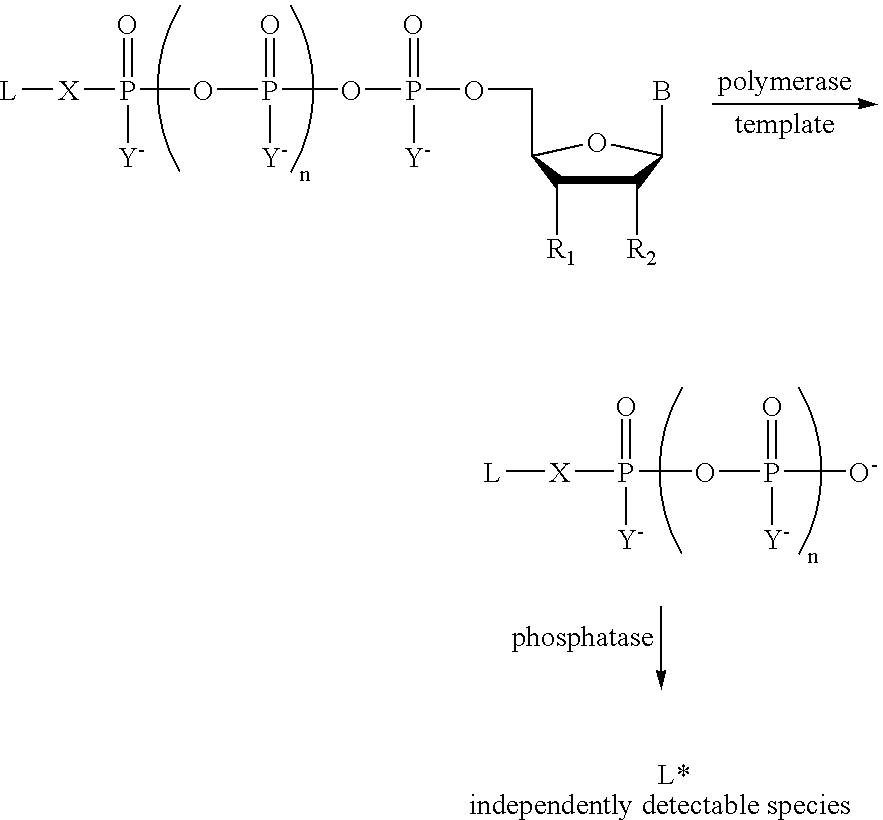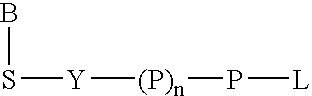Patents
Literature
3374 results about "Polymerase" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A polymerase is an enzyme (EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication.
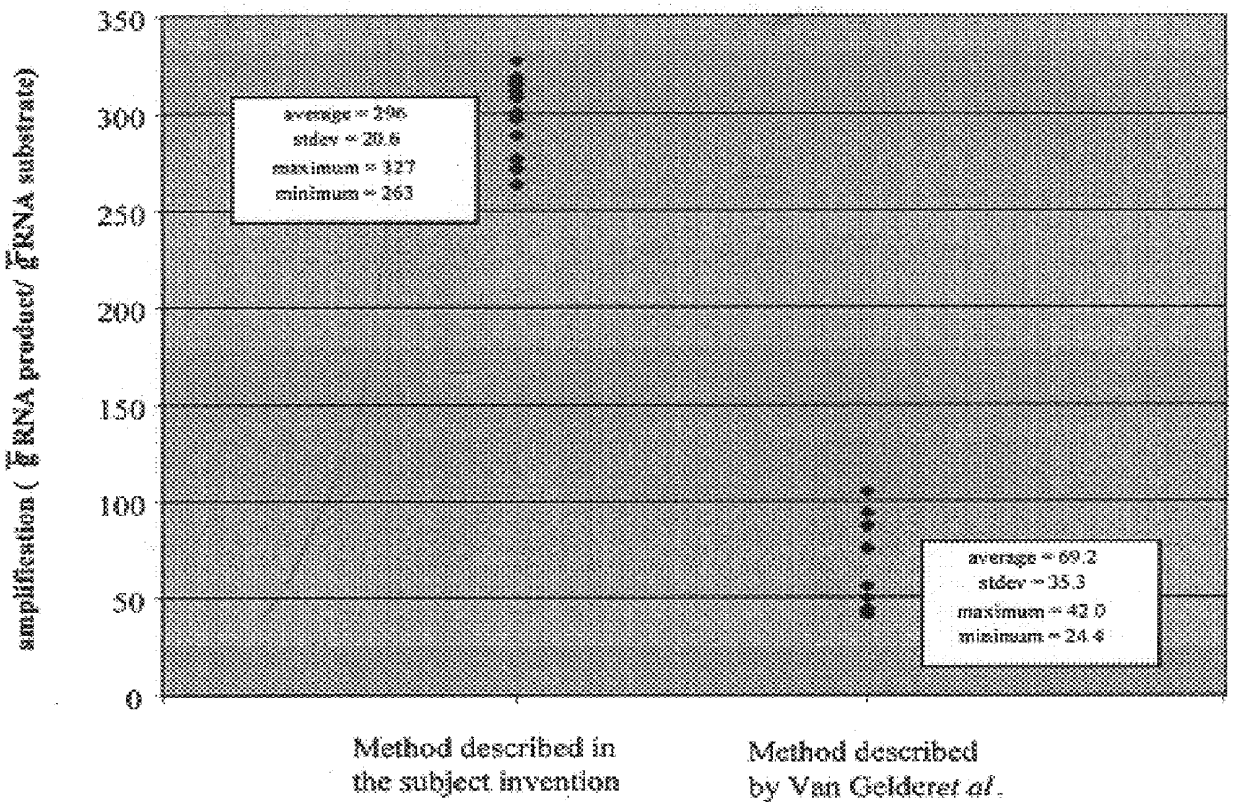
Method for linear mRNA amplification
InactiveUS6132997ASugar derivativesMicrobiological testing/measurementAntisense RNAReverse transcriptase
Methods for linearly amplifying mRNA to produce antisense RNA are provided. In the subject methods, mRNA is converted to double-stranded cDNA using a promoter-primer having a poly-dT primer site linked to a promoter sequence so that the resulting double-stranded cDNA is recognized by an RNA polymerase. The resultant double-stranded cDNA is then transcribed into antisense RNA in the presence of a reverse transcriptase that is rendered incapable of RNA-dependent DNA polymerase activity during this transcription step. The subject methods find use a variety of different applications in which the preparation of linearly amplified amounts of antisense RNA is desired. Also provided are kits for practicing the subject methods.
Owner:AGILENT TECH INC
Devices and methods for the performance of miniaturized in vitro amplification assays
InactiveUS6706519B1Optimize thermal cycling parametersEfficient use ofBioreactor/fermenter combinationsBiological substance pretreatmentsCentripetal forcePolymerase chain reaction
This invention relates to methods and apparatus for performing microanalytic and microsynthetic analyses and procedures. The invention provides a microsystem platform and a micromanipulation device for manipulating the platform that utilizes the centripetal force resulting from rotation of the platform to motivate fluid movement through microchannels. The invention specifically provides devices and methods for performing miniaturized in vitro amplification assays such as the polymerase chain reaction. Methods specific for the apparatus of the invention for performing PCR are provided.
Owner:TECAN TRADING AG +1
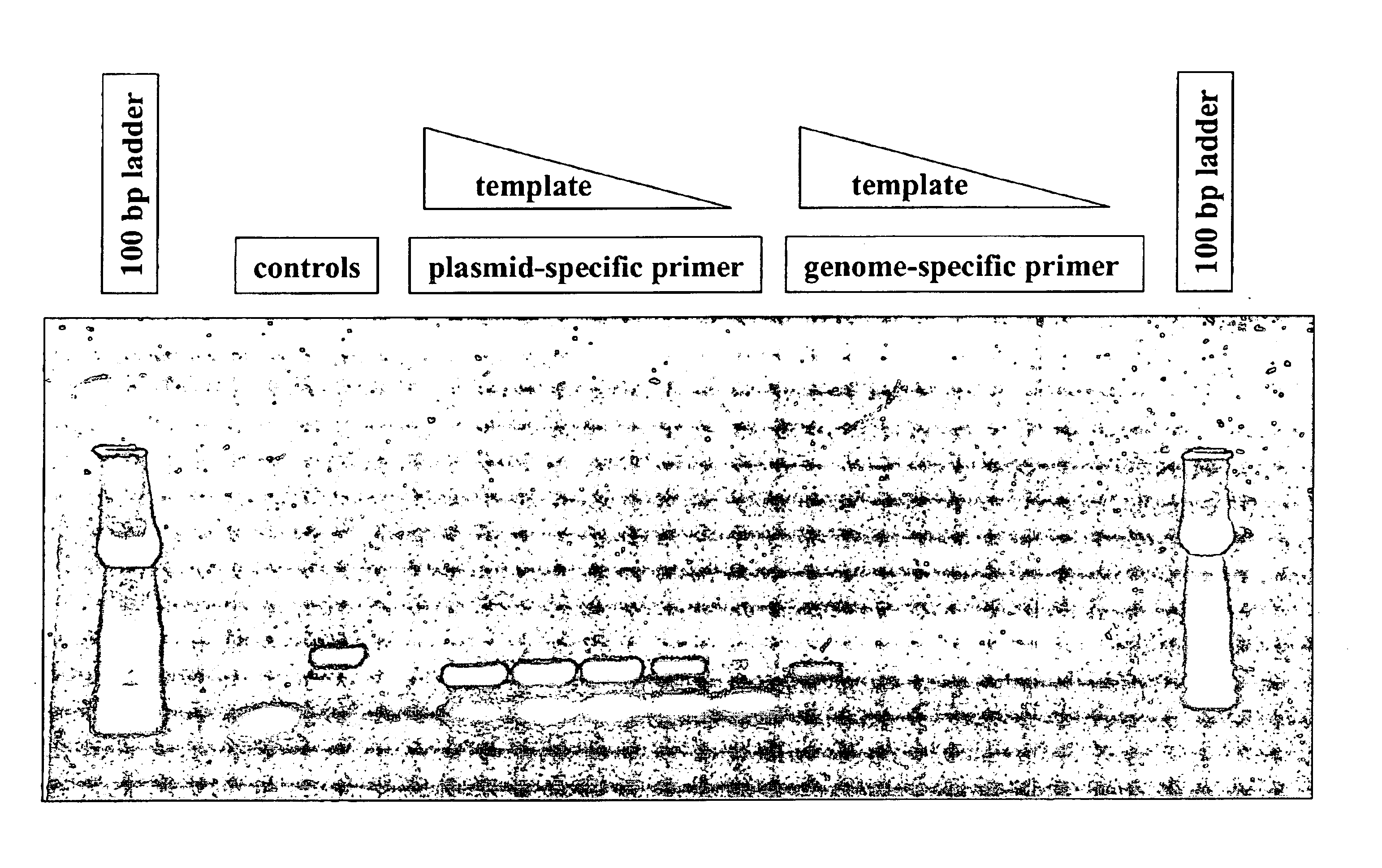
Single-primer nucleic acid amplification methods
ActiveUS7374885B2Reduce appearanceHigh levelMicrobiological testing/measurementFermentationNucleotideNucleic acid sequencing
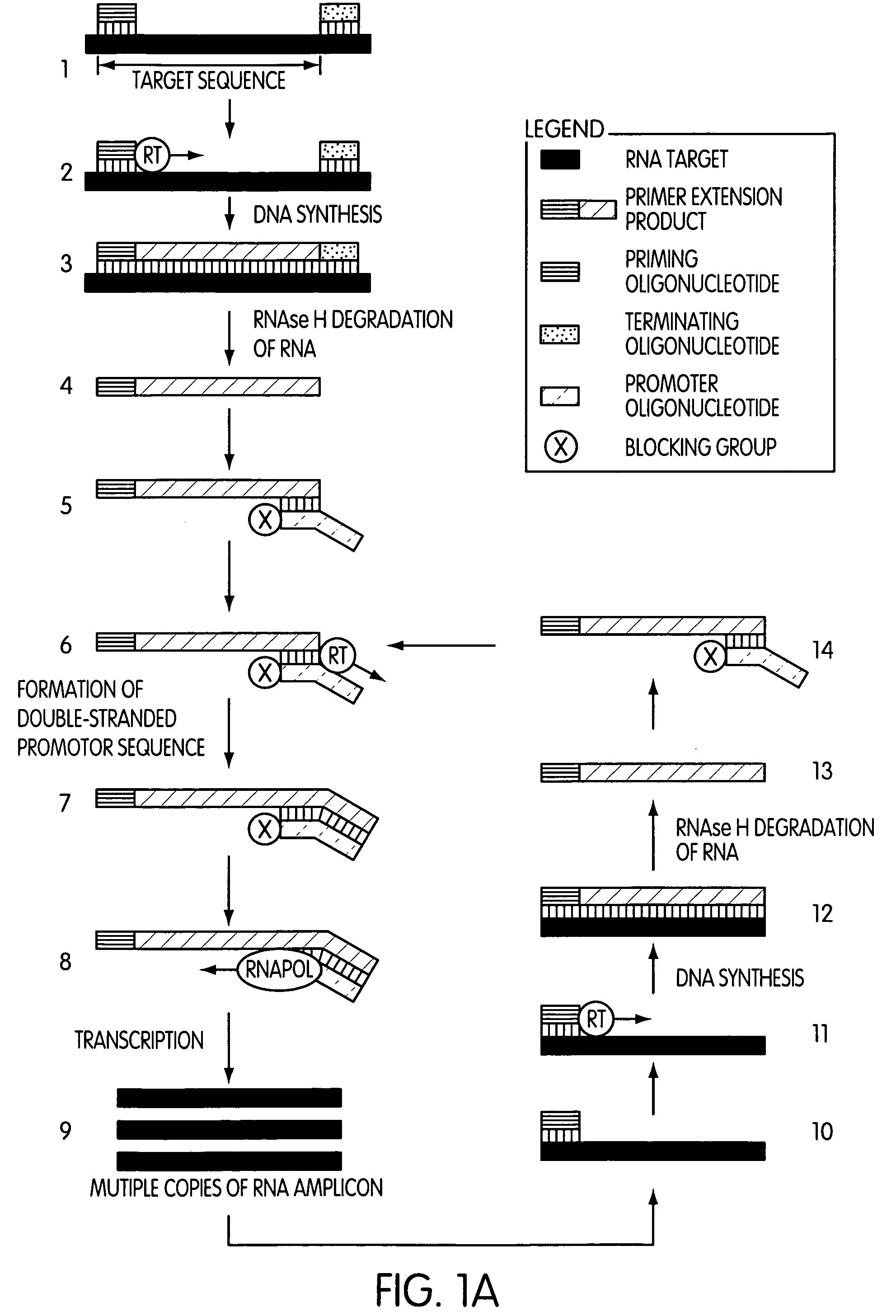
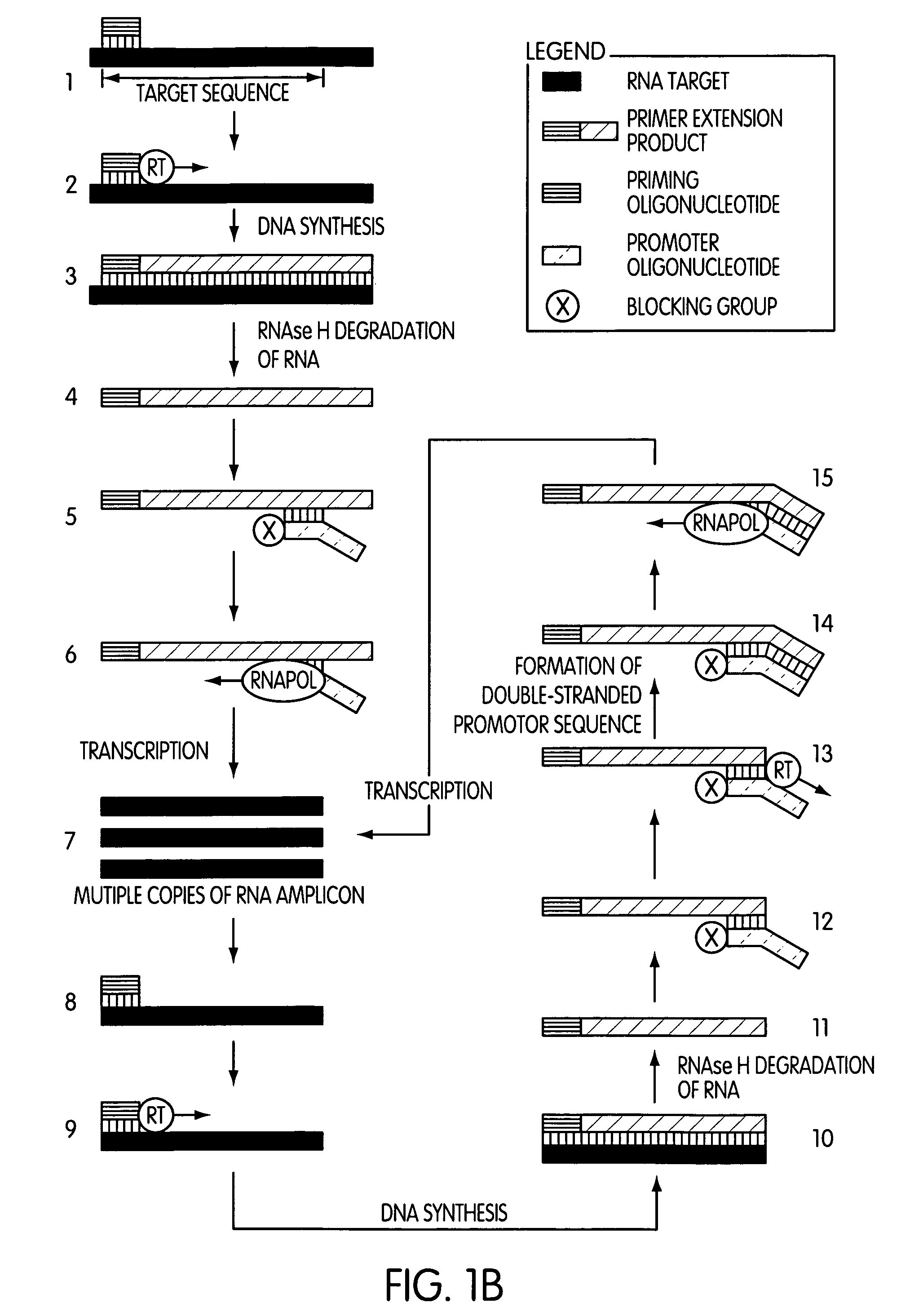
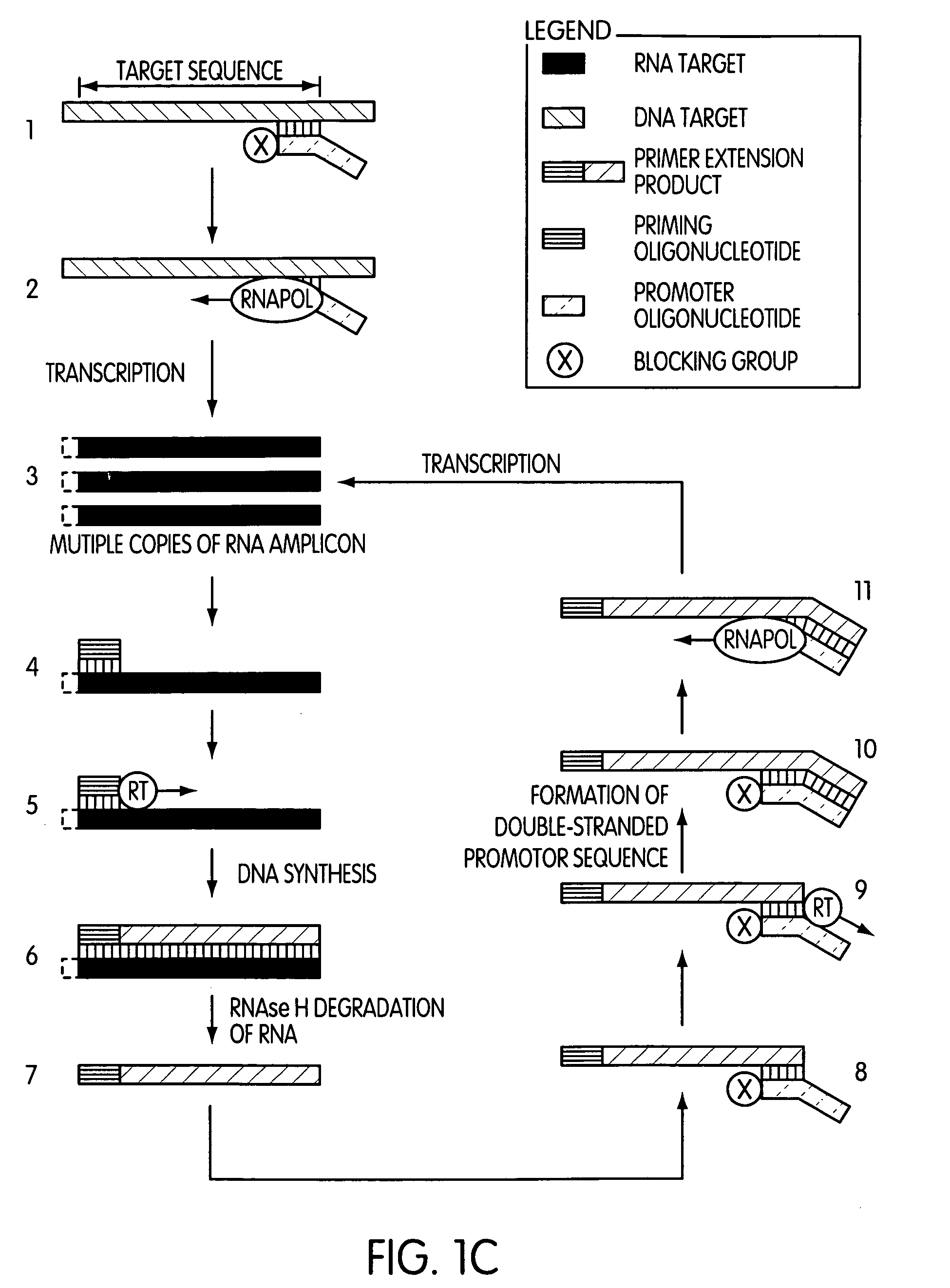
The present invention is directed to novel methods of synthesizing multiple copies of a target nucleic acid sequence which are autocatalytic (i.e., able to cycle automatically without the need to modify reaction conditions such as temperature, pH, or ionic strength and using the product of one cycle in the next one). In particular, the present invention discloses a method of nucleic acid amplification which is robust and efficient, while reducing the appearance of side-products. The method uses only one primer, the “priming oligonucleotide,” a promoter oligonucleotide modified to prevent polymerase extension from its 3′-terminus and, optionally, a means for terminating a primer extension reaction, to amplify RNA or DNA molecules in vitro, while reducing or substantially eliminating the formation of side-products. The method of the present invention minimizes or substantially eliminates the emergence of side-products, thus providing a high level of specificity. Furthermore, the appearance of side-products can complicate the analysis of the amplification reaction by various molecular detection techniques. The present invention minimizes or substantially eliminates this problem, thus providing an enhanced level of sensitivity.
Owner:GEN PROBE INC
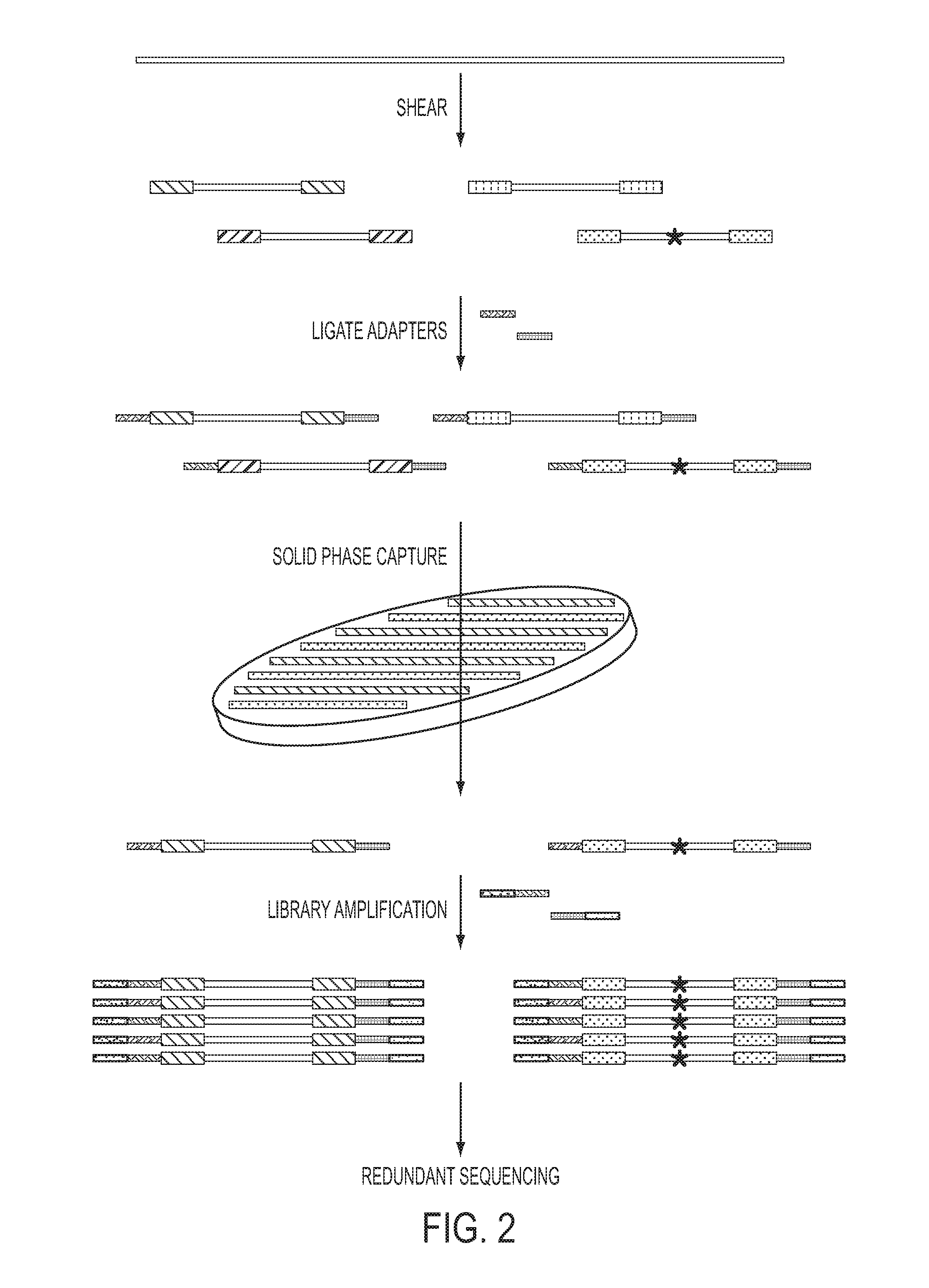
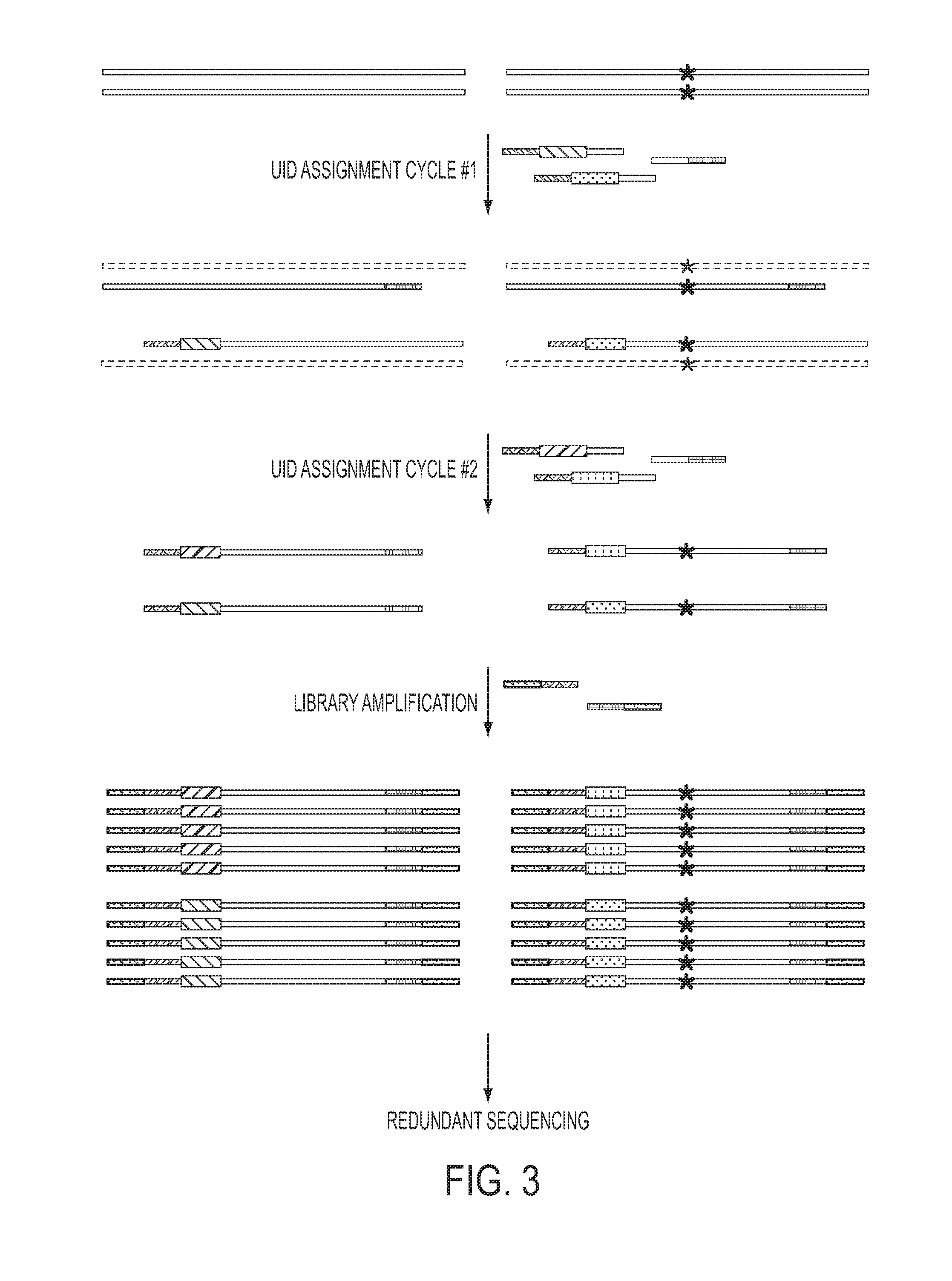
Safe sequencing system
ActiveUS20140227705A1Sensitively accurately determiningMicrobiological testing/measurementOligonucleotideInstrumentation
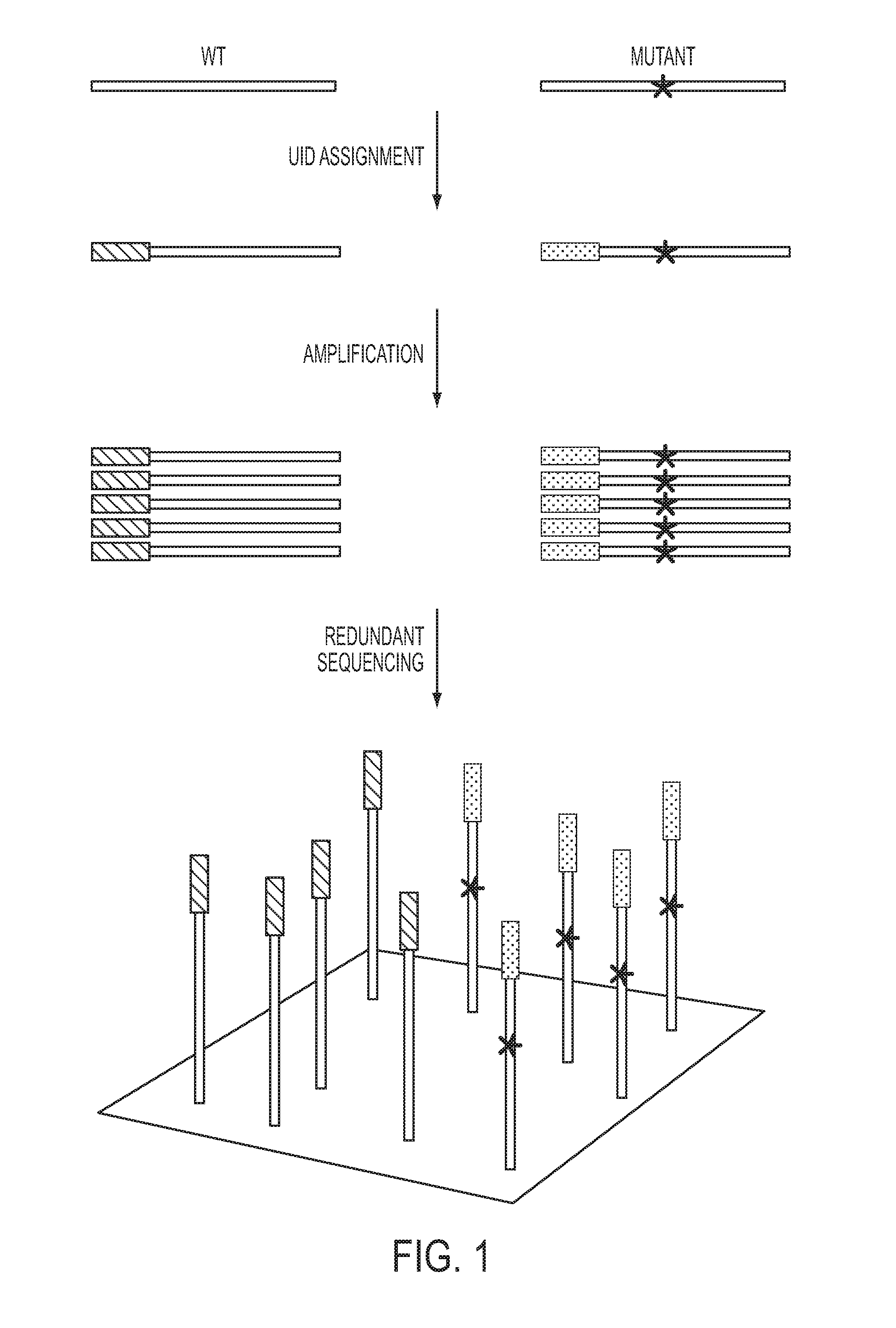
The identification of mutations that are present in a small fraction of DNA templates is essential for progress in several areas of biomedical research. Though massively parallel sequencing instruments are in principle well-suited to this task, the error rates in such instruments are generally too high to allow confident identification of rare variants. We here describe an approach that can substantially increase the sensitivity of massively parallel sequencing instruments for this purpose. One example of this approach, called “Safe-SeqS” for (Safe-Sequencing System) includes (i) assignment of a unique identifier (UID) to each template molecule; (ii) amplification of each uniquely tagged template molecule to create UID-families; and (iii) redundant sequencing of the amplification products. PCR fragments with the same UID are truly mutant (“super-mutants”) if ≧95% of them contain the identical mutation. We illustrate the utility of this approach for determining the fidelity of a polymerase, the accuracy of oligonucleotides synthesized in vitro, and the prevalence of mutations in the nuclear and mitochondrial genomes of normal cells.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Method for producing complex DNA methylation fingerprints
InactiveUS6214556B1Accurately determineSugar derivativesMicrobiological testing/measurementFingerprintGenomic DNA
Method for characterizing, classifying and differentiating tissues and cell types, for predicting the behavior of tissues and groups of cells, and for identifying genes with changed expression. The method involves obtaining genomic DNA from a tissue sample, the genomic DNA subsequently being subjected to shearing, cleaved by means of a restriction endonuclease or not treated by either one of these methods. The base cytosine, but not 5-methylcytosine, from the thus-obtained genomic DNA is then converted into uracil by treatment with a bisulfite solution. Fractions of the thus-treated genomic DNA are then amplified using either very short or degenerated oligonucleotides or oligonuclcotides which are complementary to adaptor oligonucleotides that have been ligated to the ends of the cleaved DNA. The quantity of the remaining cytosines on the guanine-rich DNA strand and / or the quantity of guanines on the cytosine-rich DNA strand from the amplified fractions are then detected by hybridization or polymerase reaction, which quantities are such that the data generated thereby and automatically applied to a processing algorithm allow the drawing of conclusions concerning the phenotype of the sample material.
Owner:EPIGENOMICS AG
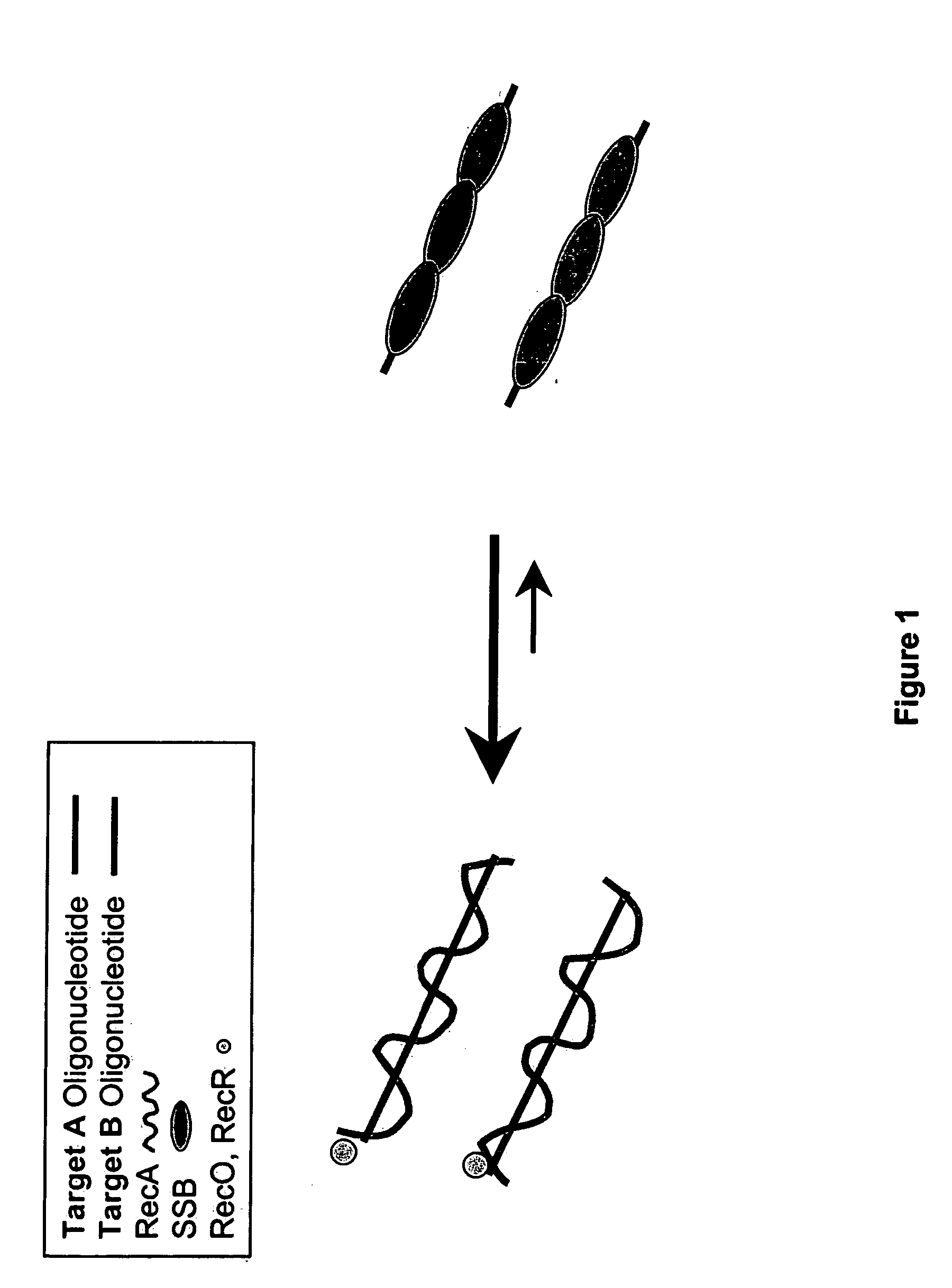
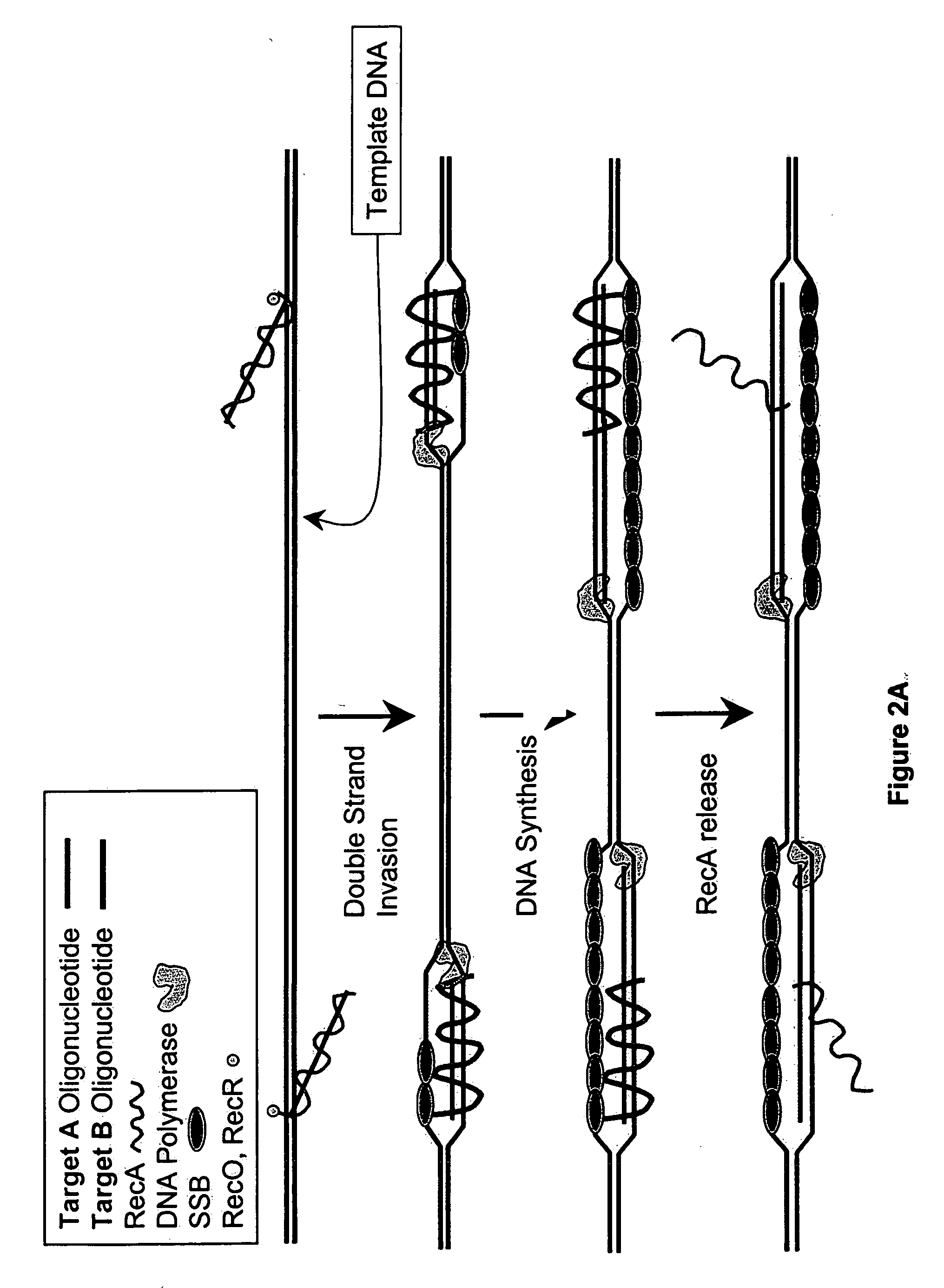
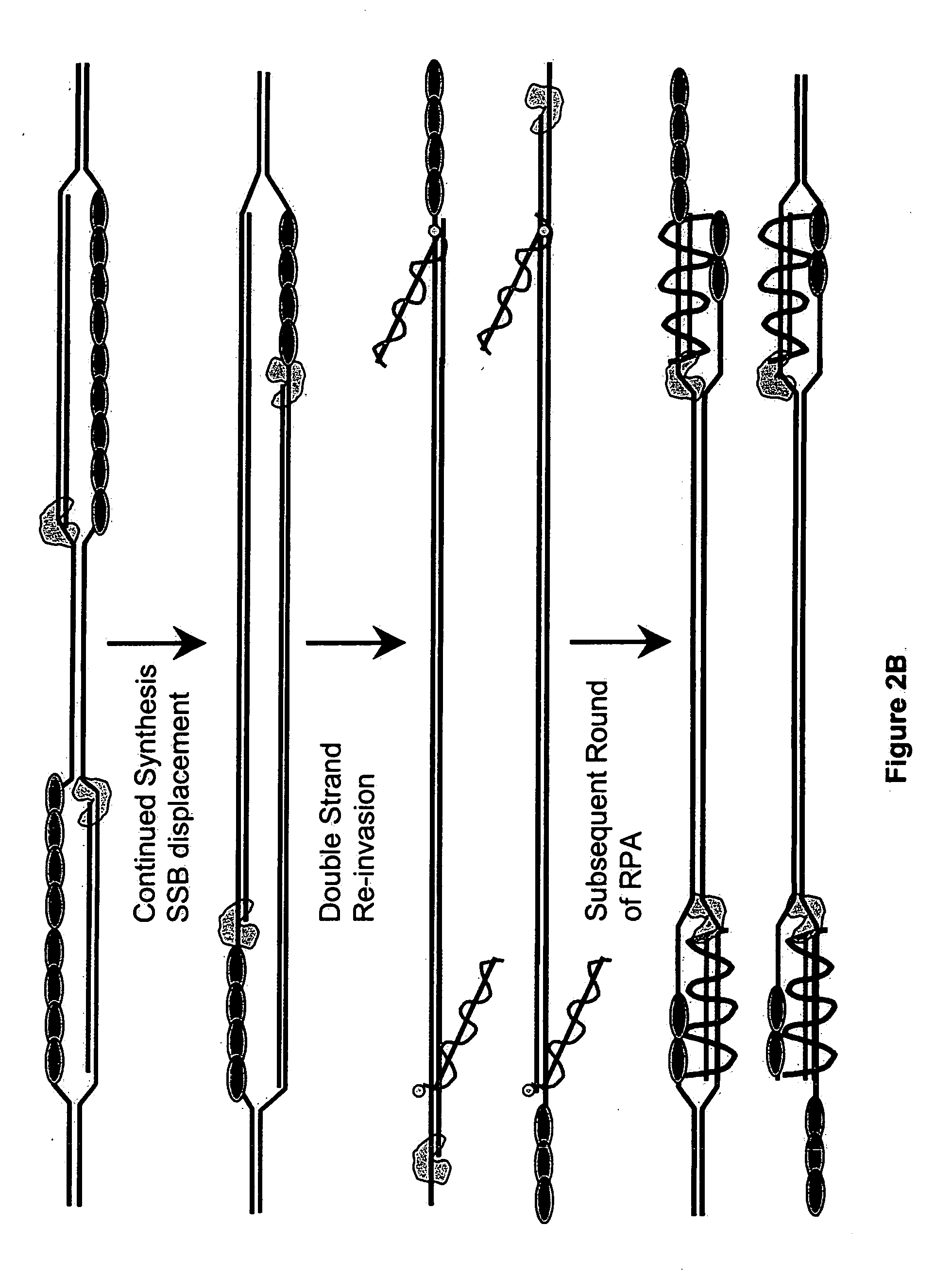
Recombinase polymerase amplification
ActiveUS20050112631A1HydrolasesMicrobiological testing/measurementRecombinase Polymerase AmplificationSingle strand
This disclosure describe three related novel methods for Recombinase-Polymerase Amplification (RPA) of a target DNA that exploit the properties of recombinase and related proteins, to invade double-stranded DNA with single stranded homologous DNA permitting sequence specific priming of DNA polymerase reactions. The disclosed methods have the advantage of not requiring thermocycling or thermophilic enzymes. Further, the improved processivity of the disclosed methods may allow amplification of DNA up to hundreds of megabases in length.
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
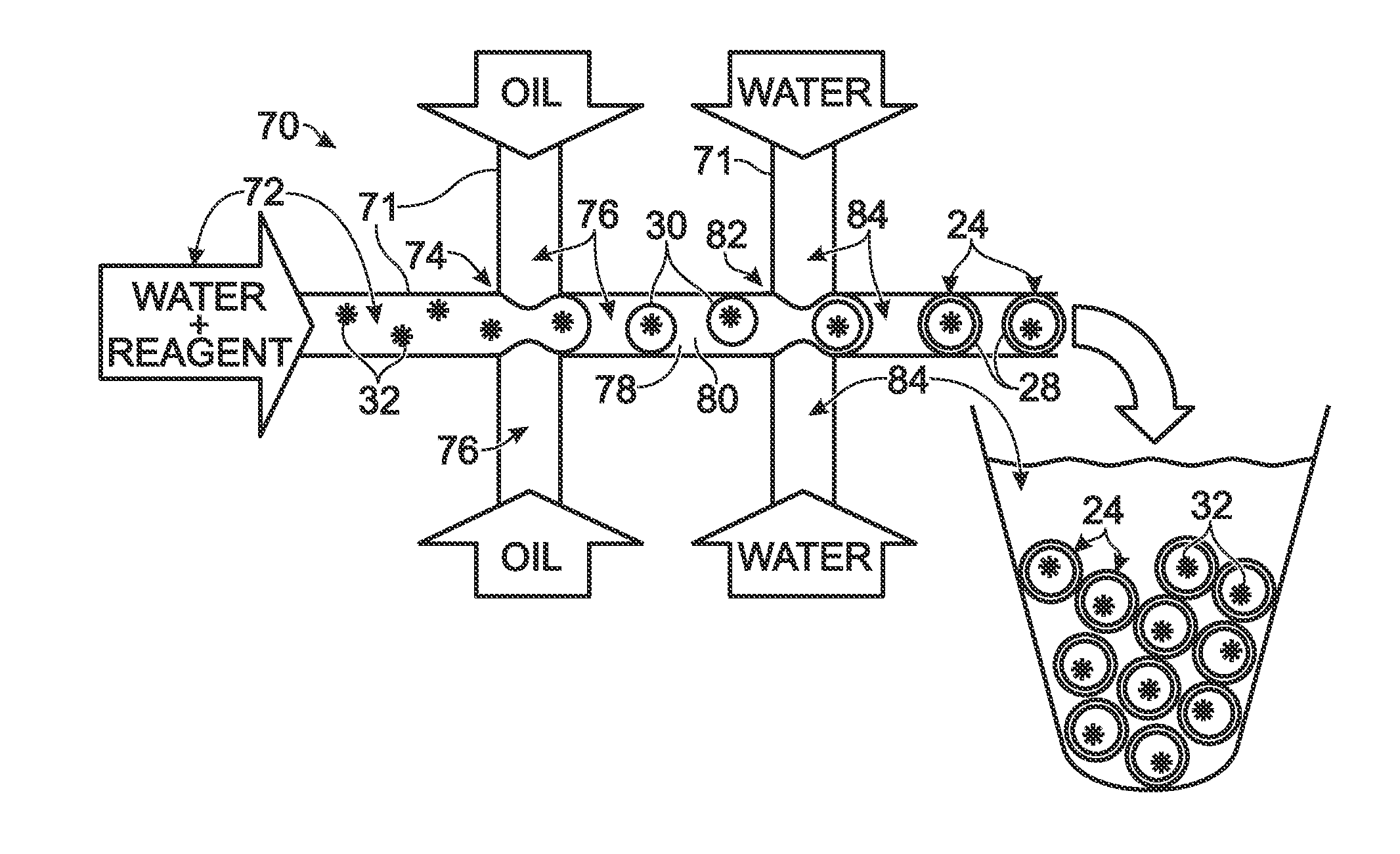
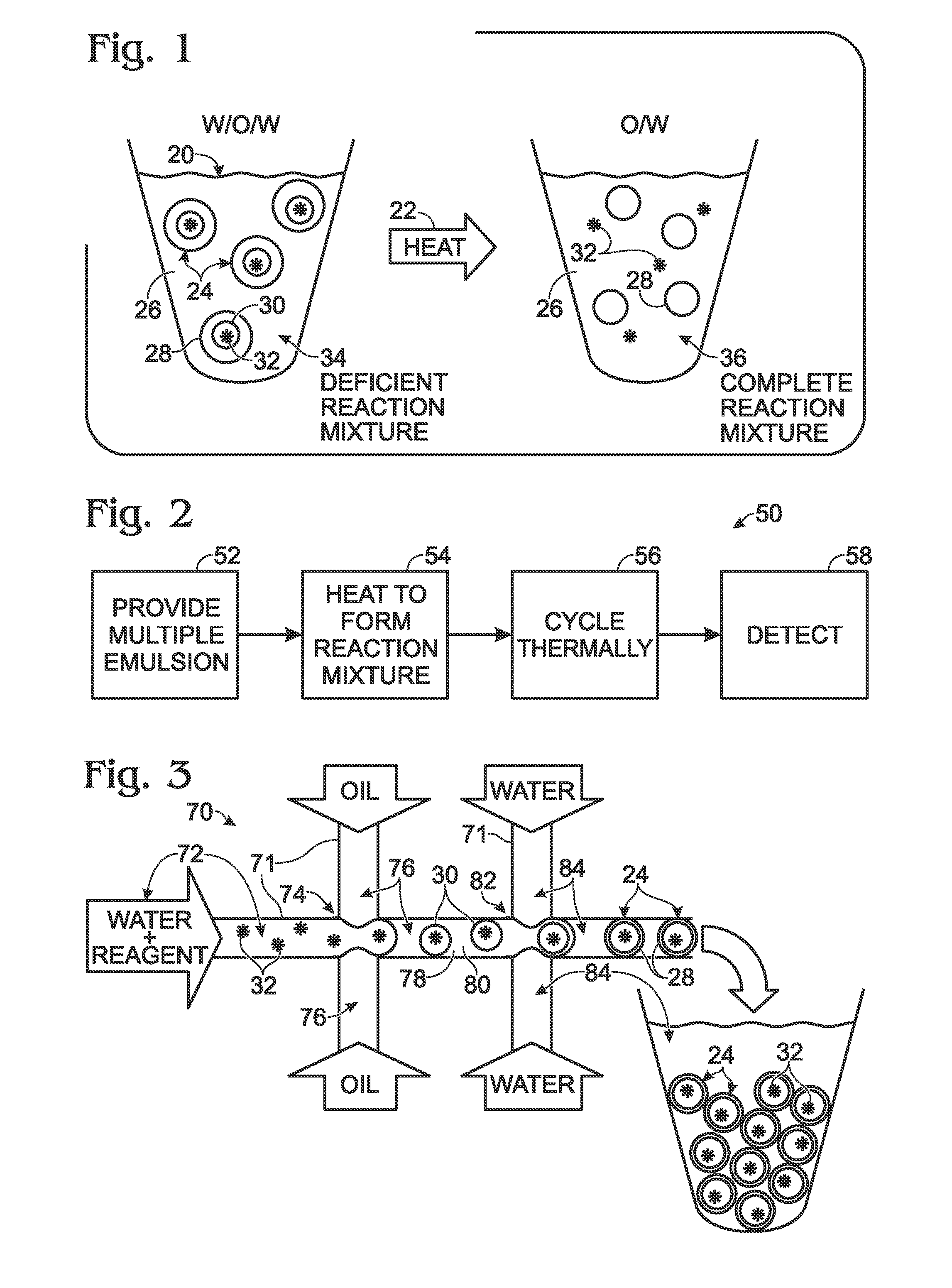
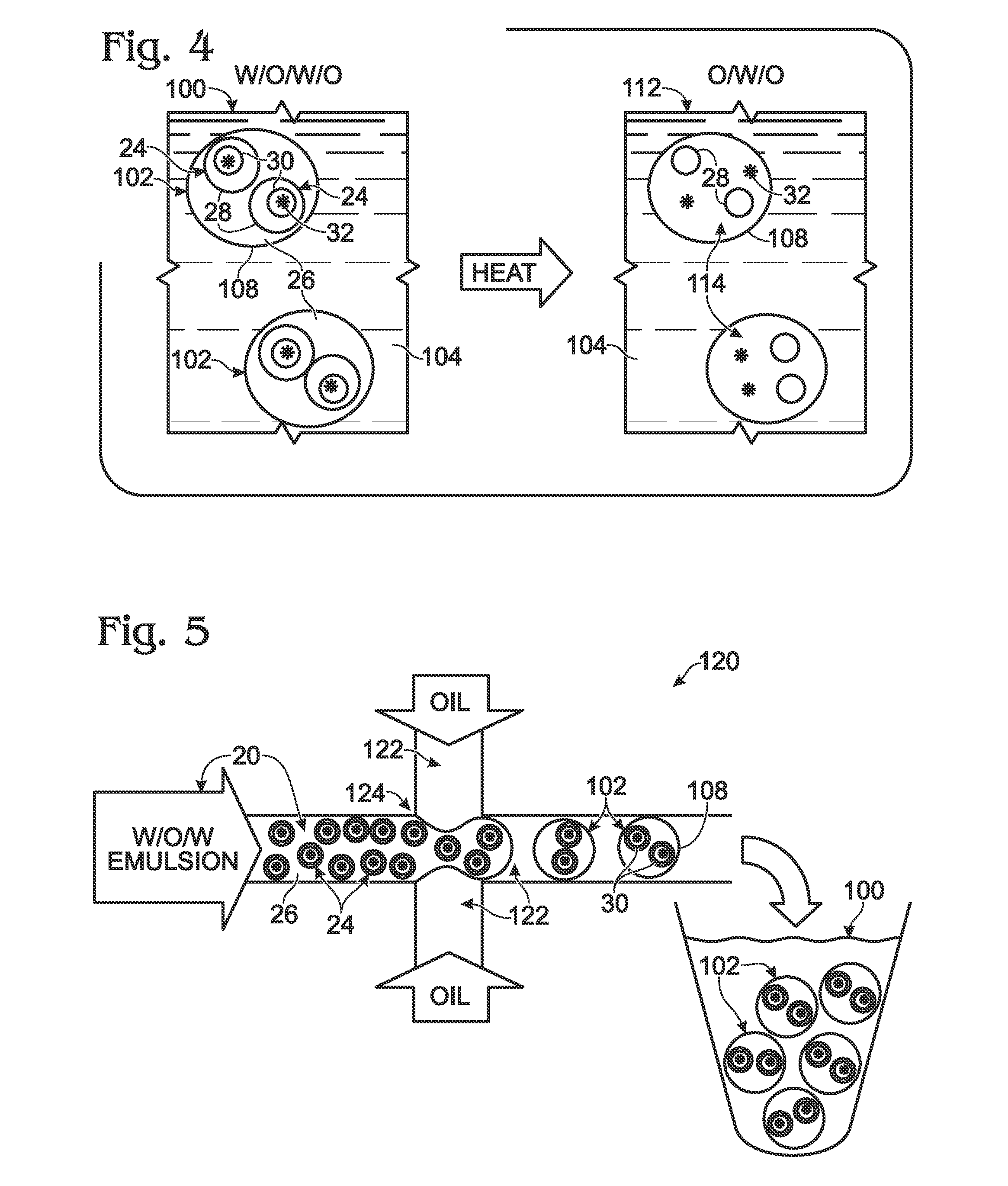
System for hot-start amplification via a multiple emulsion
System, including methods, apparatus, compositions, and kits, for making and using compound droplets of a multiple emulsion to supply an amplification reagent, such as a heat-stable DNA polymerase or DNA ligase, to an aqueous phase in which the compound droplets are disposed. The compound droplets may be induced to supply the amplification reagent by heating the multiple emulsion, to achieve hot-start amplification.
Owner:BIO RAD LAB INC
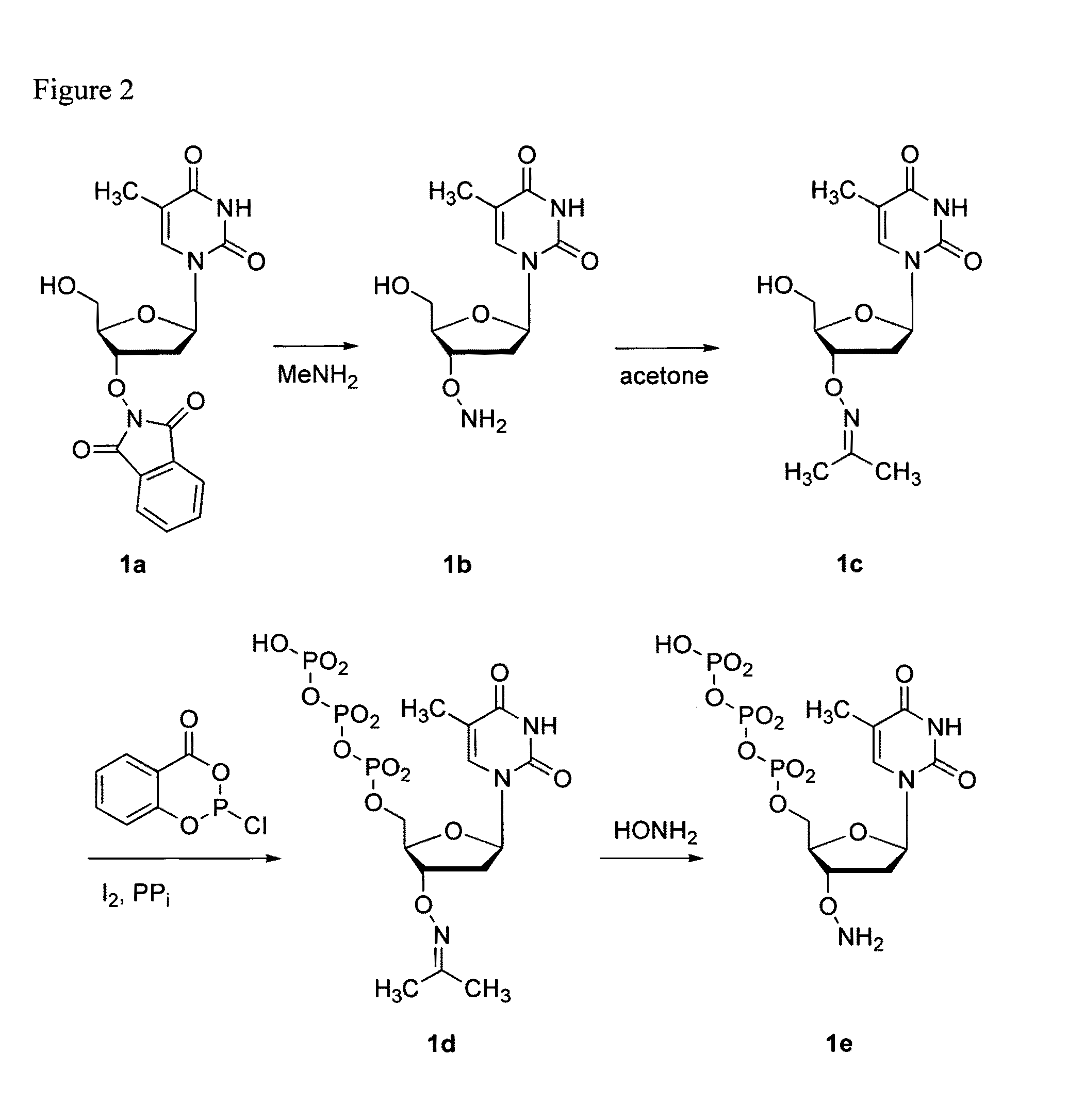
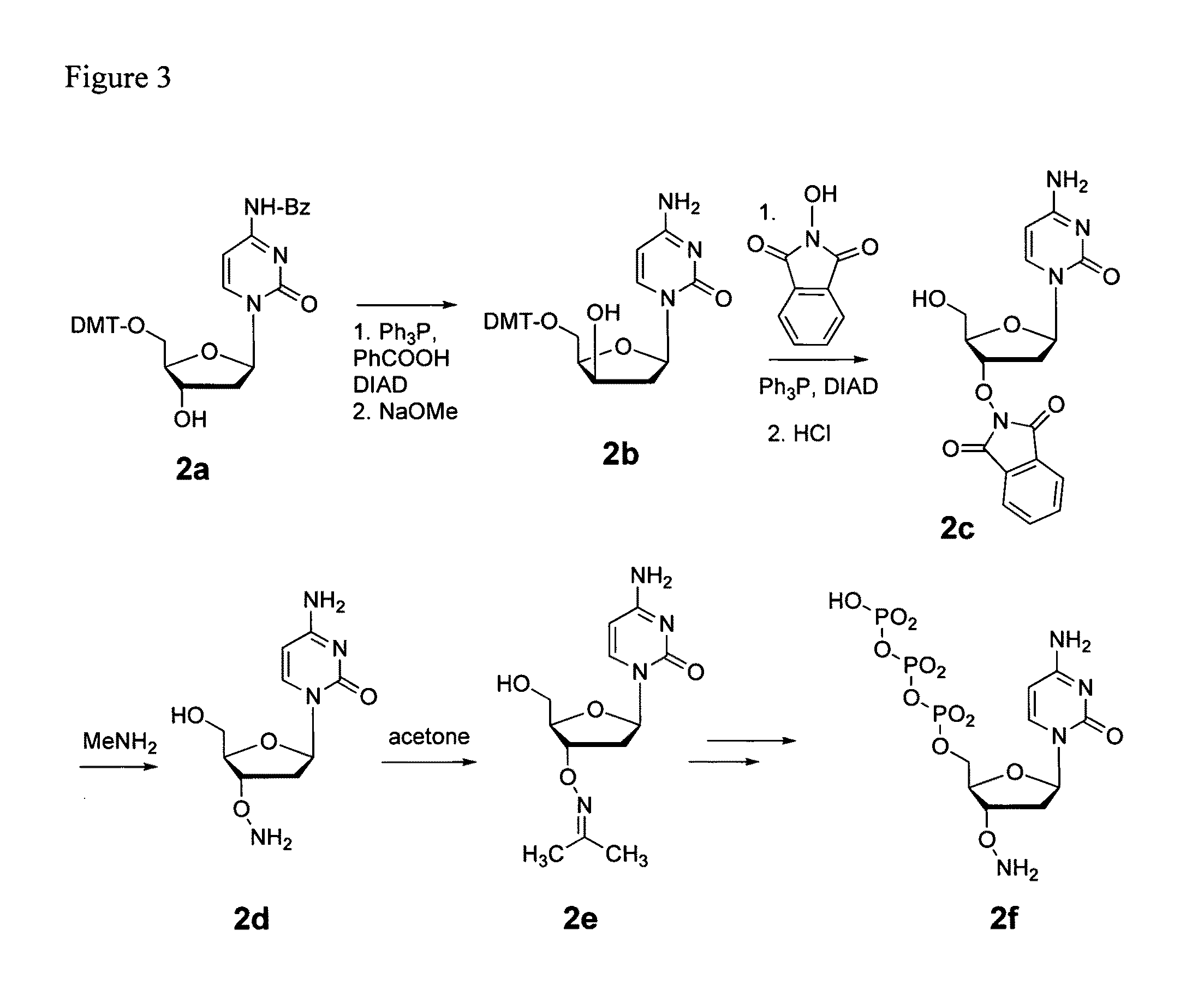
Nucleoside phosphoramidates
Disclosed herein are nucleoside phosphoramidates and their use as agents for treating viral diseases. These compounds are inhibitors of RNA-dependent RNA viral replication and are useful as inhibitors of HCV NS5B polymerase, as inhibitors of HCV replication and for treatment of hepatitis C infection in mammals.
Owner:GILEAD SCI INC
LED or laser enabled real-time PCR system and spectrophotometer
InactiveUS7122799B2Easy to useReduce entirely avoid costRadiation pyrometryHeating or cooling apparatusPhotodetectorUltraviolet
A system for conducting a polymerase chain reaction (PCR) assay upon a collection of samples is disclosed. The PCR assay is performed by absorption detection. The system includes a multi-well plate which is adapted to retain a collection of sample wells. This system includes a thermal cycler for the multi-well plate. The system additionally includes a collection of photodetectors, and a corresponding number of light sources. The light sources are positioned such that light emitted from each of the respective light sources passes through a corresponding well retained in the multi-well plate and to a corresponding photodetector. The system also includes a processor or other means for analyzing the output signals from the photodetectors. In certain versions of the system, ultra-violet light is used.
Owner:PALO ALTO RES CENT INC
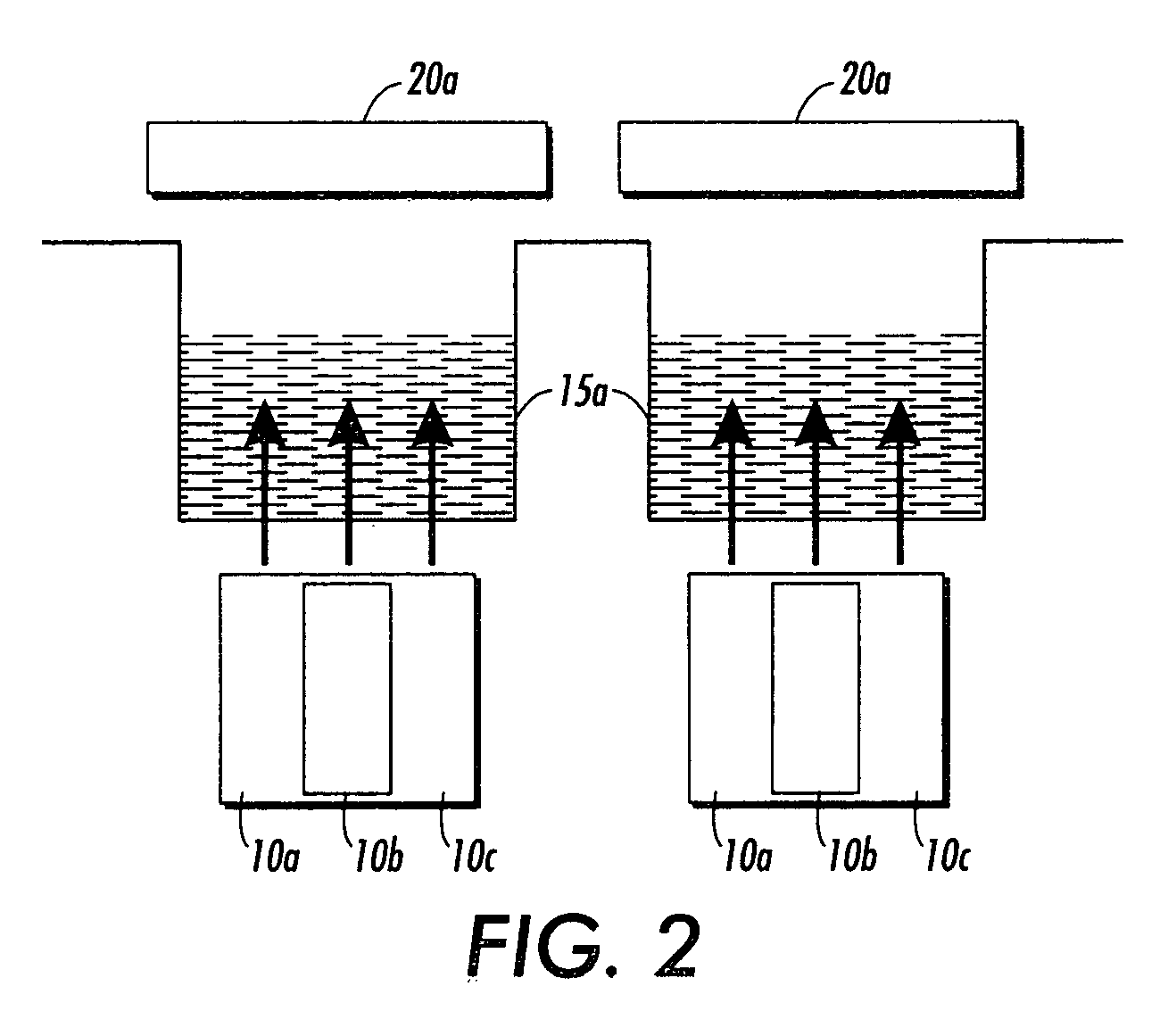
Analyte detection
A method of characterizing an analyte sample is provided that includes the steps of: (a) anchoring the analyte to a nucleic acid template of known sequence; (b) conducting a DNA polymerase reaction that includes the reaction of a template, a non-hydrolyzable primer, at least one terminal phosphate-labeled nucleotide, DNA polymerase, and an enzyme having 3'->5' exonuclease activity which reaction results in the production of labeled polyphosphate; (c) permitting the labeled polyphosphate to react with a phosphatase to produce a detectable species characteristic of the sample; (d) detecting the detectable species. The method may include the step of characterizing the nucleic acid sample based on the detection. Also provided are methods of analyzing multiple analytes in a sample, and kits for characterizing analyte samples.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
Method of determining the nucleotide sequence of oligonucleotides and DNA molecules
InactiveUS7875440B2Eliminate needHigh puritySugar derivativesMicrobiological testing/measurementOligonucleotide primersFluorescence
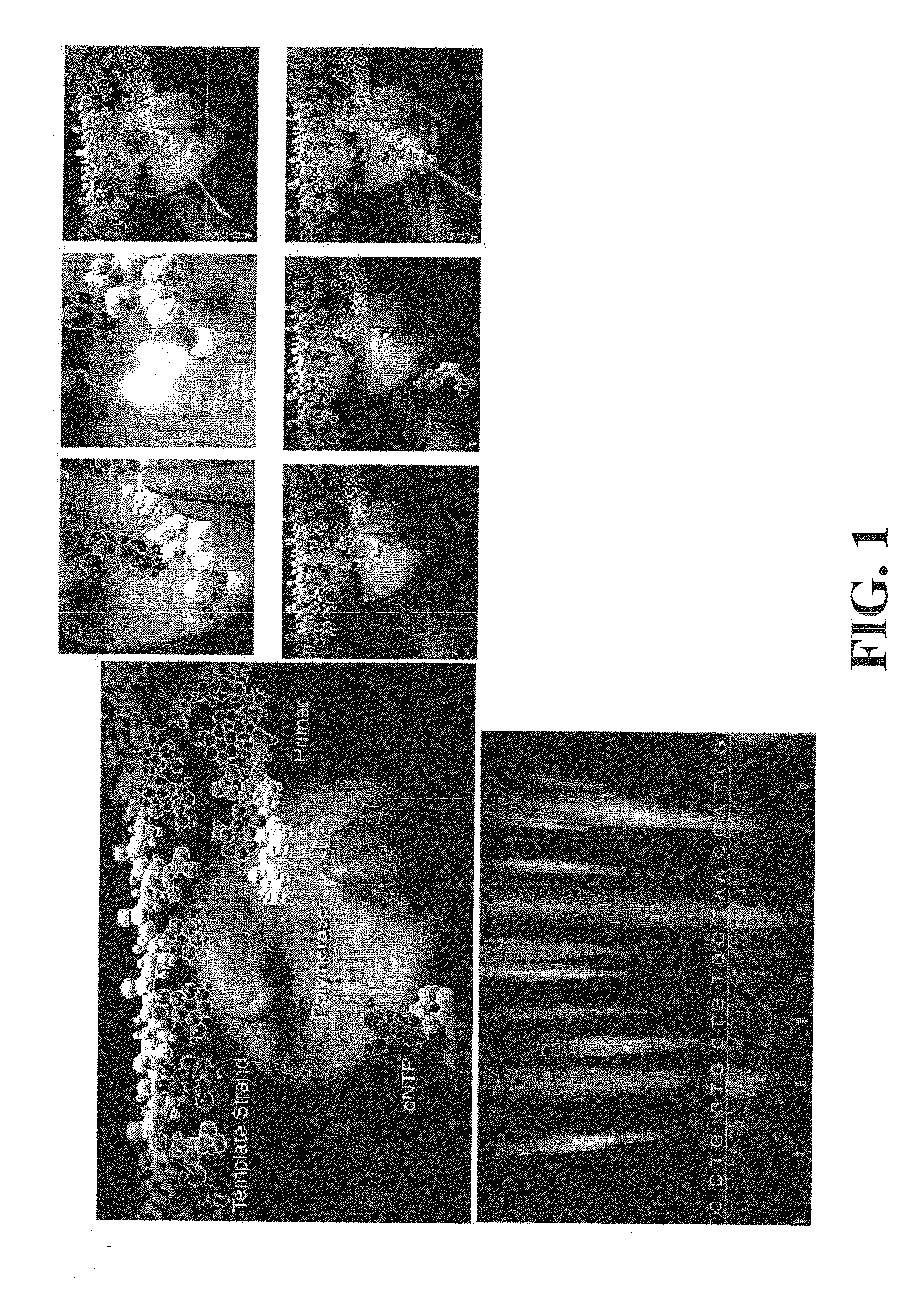
The present invention relates to a novel method for analyzing nucleic acid sequences based on real-time detection of DNA polymerase-catalyzed incorporation of each of the four nucleotide bases, supplied individually and serially in a microfluidic system, to a reaction cell containing a template system comprising a DNA fragment of unknown sequence and an oligonucleotide primer. Incorporation of a nucleotide base into the template system can be detected by any of a variety of methods including but not limited to fluorescence and chemiluminescence detection. Alternatively, microcalorimetic detection of the heat generated by the incorporation of a nucleotide into the extending template system using thermopile, thermistor and refractive index measurements can be used to detect extension reactions.
Owner:LIFE TECH CORP
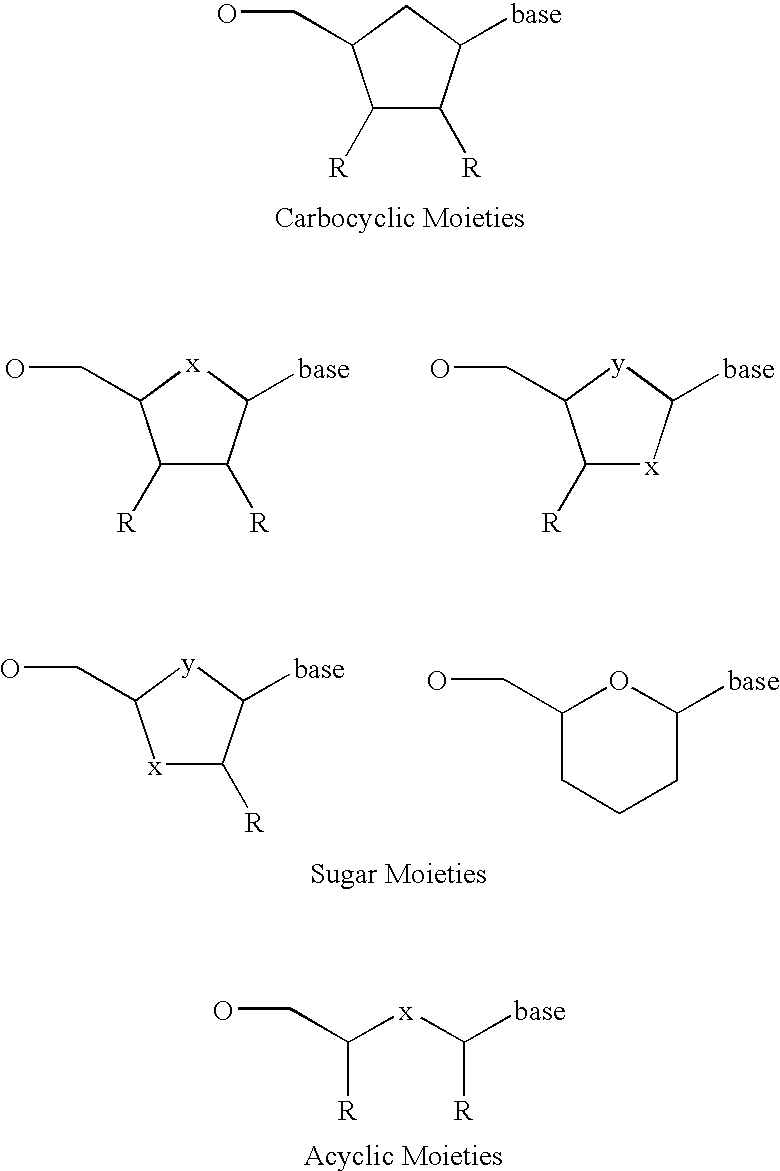
Substituted nucleosides, preparation thereof and use as inhibitors of RNA viral polymerases
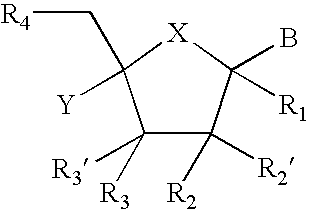
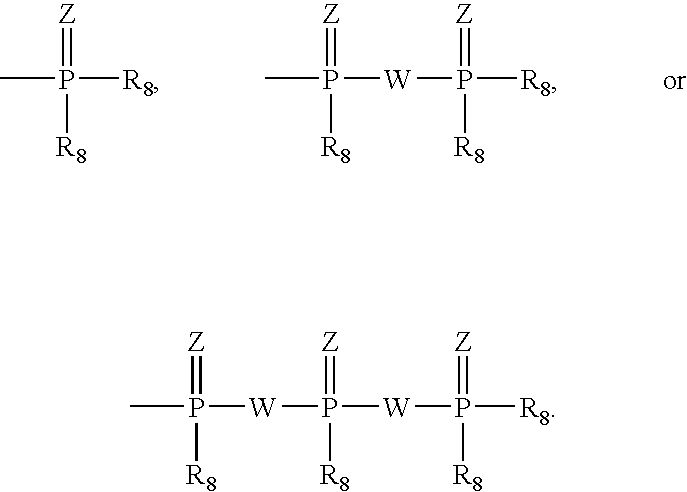
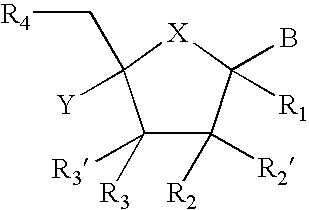
Provided are compounds represented by: X is O, S or NR6, R1 is H or (CH2)mR5, R2, R2', R3 and R3' are independently NO2, N3 or (CH2)mR5, OH R4 is H, OR6, SR6, NR6R6a, CN, C(O)OR6, C(O)NR6R6a, R6, OR7 or (CH2)nR7, R5 is H, halo, OR6, SR6, NR6R6a, CN, C(O)OR6, C(O)NR6R6a, R6, OR7 or (CH2)mR7, R6 and R6a are individually H, alkyl, substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted alkynyl, aryl or substituted aryl, R7 is: R8 is H, F, SR9 or OR9, R9 is H, alkyl, alkenyl, alkynyl, aryl or hydroxyprotecting group, Y is H, CH3 or (CH2)mR5, Z is O or S W is CH2, CF2, CHF or O, m is 0-4, B is adenine, guanine, cytosine, uracil, thymine, modified purines and pyrimidines substituted pyridines, five membered heterocycles substituted by at least one of amines, substituted amines, amides, substituted amides, esters, halogens, alkyls, ethers; and pharmaceutically acceptable salts thereof and prodrugs thereof. These ring systems may be substituted.
Owner:BIOCRYST PHARM INC
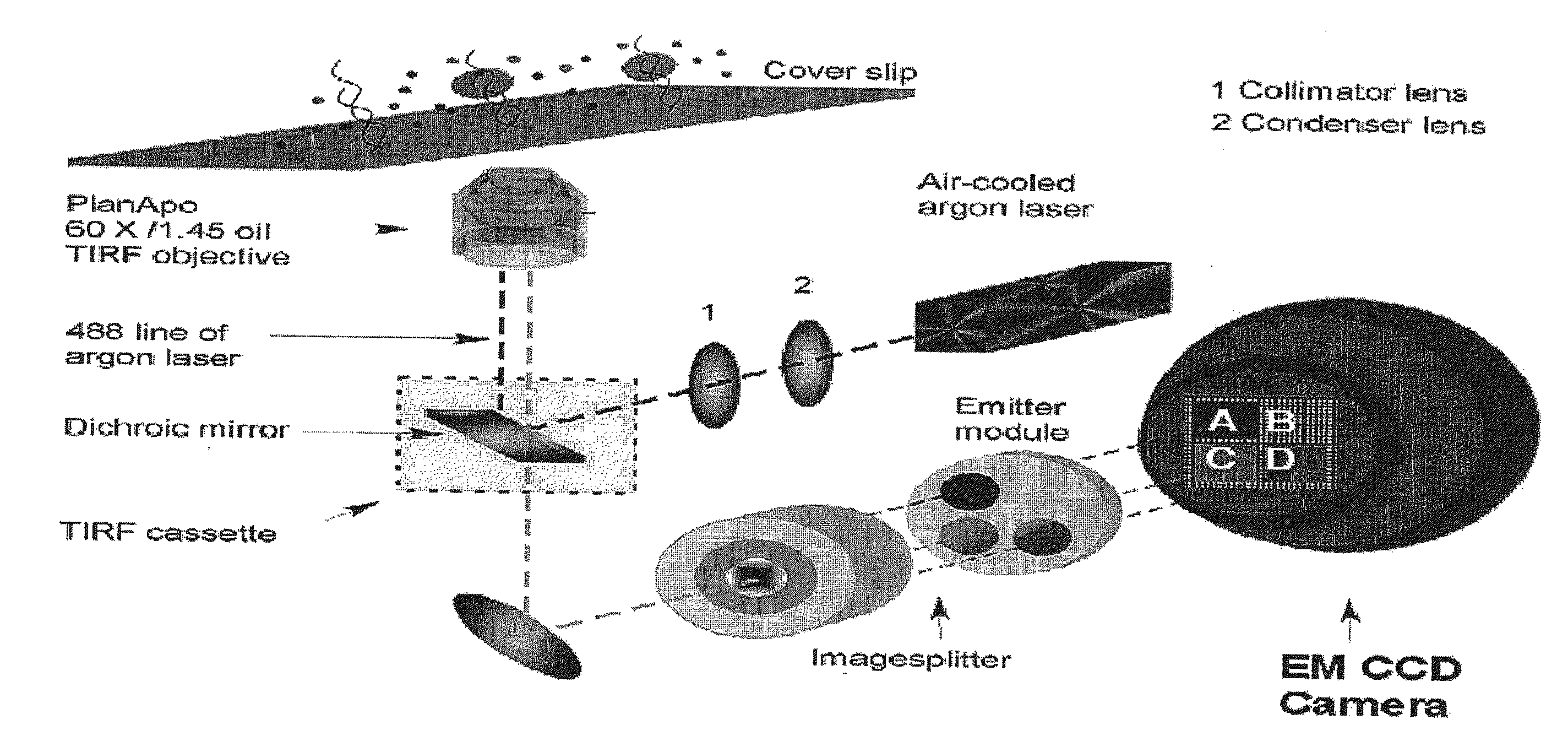
Compositions, methods and systems for single molecule sequencing
InactiveUS20110165652A1Enhanced signalReduce background noiseSugar derivativesMicrobiological testing/measurementNucleotideSynthesis methods
Compositions, systems and methods of sequencing are disclosed, where the compositions and systems include polymerase enzymes that have been genetically modified to more efficiently incorporate nucleotides including labels having a detectable properties that are released during incorporation, to augment a rate of labeled nucleotide incorporation, to augment a rate of pyrophosphate release, or to augment two or more of these properties and rates. Also disclosed are terminally labeled and dual labeled nucleotides, and click-chemistry based methods of synthesizing the same.
Owner:LIFE TECH CORP
Varietal counting of nucleic acids for obtaining genomic copy number information
A method for obtaining from genomic material genomic copy number information unaffected by amplification distortion, comprising obtaining segments of the genomic material, tagging the segments with substantially unique tags to generate tagged nucleic acid molecules, such that each tagged nucleic acid molecule comprises one segment of the genomic material and a tag, subjecting the tagged nucleic acid molecules to polymerase chain reaction (PCR) amplification, generating tag associated sequence reads by sequencing the product of the PCR reaction, assigning each tagged nucleic acid molecule to a location on a genome associated with the genomic material by mapping the subsequence of each tag associated sequence read corresponding to a segment of the genomic material to a location on the genome, and counting the number of tagged nucleic acid molecules assigned to the same location on the genome having a different tag, thereby obtaining genomic copy number information unaffected by amplification distortion.
Owner:COLD SPRING HARBOR LAB INC
Reagents for reversibly terminating primer extension
Processes are disclosed that use 3′-reversibly terminated nucleoside triphosphates to analyze DNA for purposes other than sequencing using cyclic reversible termination. These processes are based on the unexpected ability of terminal transferase to accept these triphosphates as substrates, the unexpected ability of polymerases to add reversibly and irreversibly terminated triphosphates in competition with each other, the development of cleavage conditions to remove the terminating group rapidly, in high yield, and without substantial damage to the terminated oligonucleotide product, and the ability of reversibly terminated primer extension products to capture groups. The presently preferred embodiments of the disclosed processes use a triphosphate having its 3′-OH group blocked as a 3′-ONH2 group, which can be removed in buffered NaNO2 and use variants of Taq DNA polymerase, including one that has a replacement (L616A).
Owner:BENNER STEVEN ALBERT +3
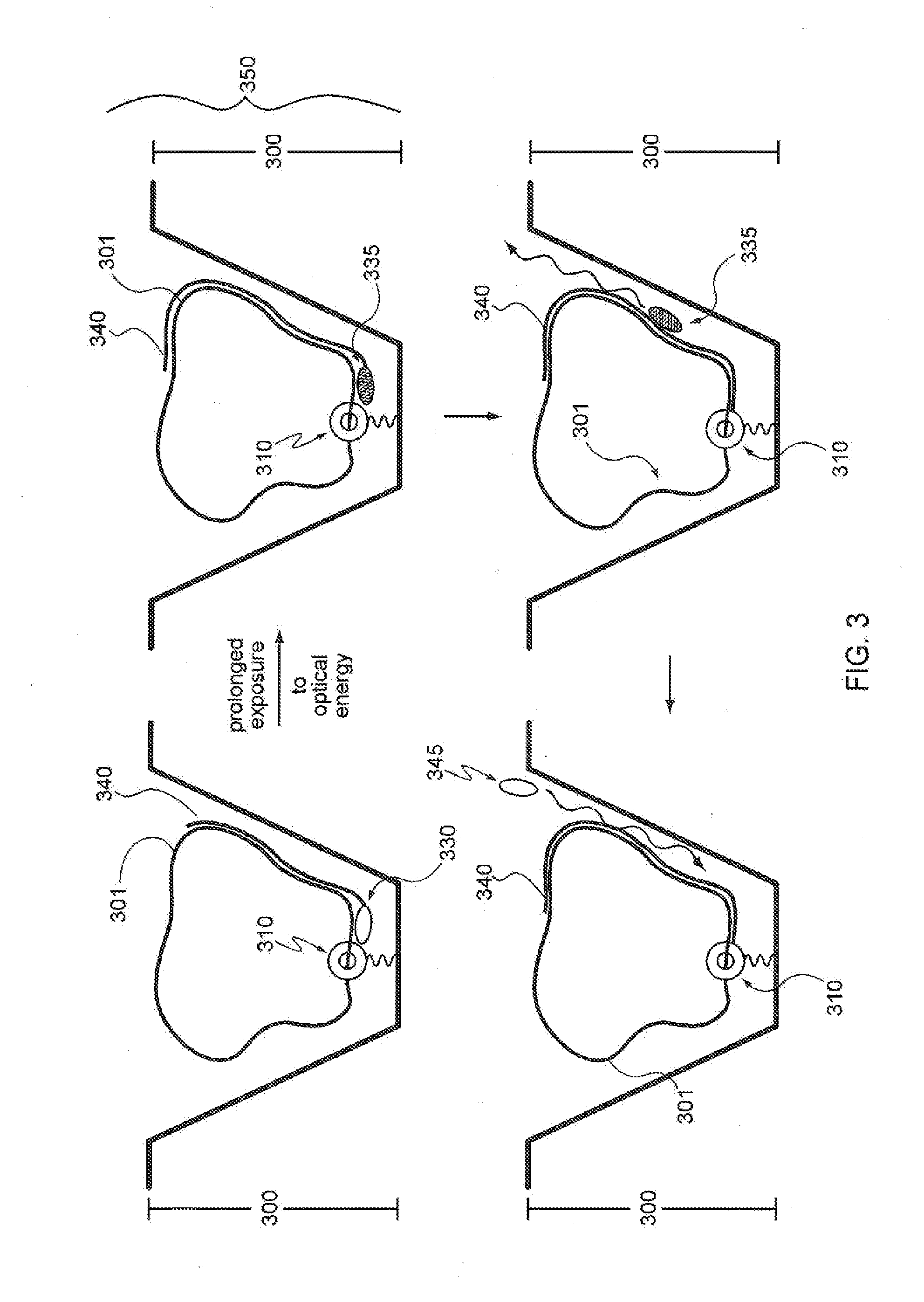
Enzymes resistant to photodamage
ActiveUS20100093555A1Reduce oxidationImprove the immunitySugar derivativesMicrobiological testing/measurementModified dnaPolymerase L
Provided are compositions comprising modified DNA polymerases that exhibit improved photostability compared to the parental polymerases from which they were derived. Provided are methods for generating enzymes, such as DNA polymerases, with the aforementioned phenotype. Provided are methods of using polymerases with increased resistance to photodamage to make a DNA or to sequence a DNA template.
Owner:PACIFIC BIOSCIENCES
Single nucleotide detection using degradation of a fluorescent sequence
Methods and compositions are provided for detecting single nucleotide polymorphisms using a pair of oligonucleotides, a primer and a snp detection sequence, where the snp detection sequence hybridizes to the target DNA downstream from the primer and in the direction of primer extension. The snp detection sequence is characterized by having a nucleotide complementary to the snp and adjacent nucleotide complementary to adjacent nucleotides in the target and an electophoretic tag bonded to the 5'-nucleotide. The pair of oligonucleotides is combined with the target DNA under primer extension conditions, where the polymerase has 5'-3' exonuclease activity. When the snp is present, the electophoretic tag is released from the snp detection sequence, and can be detected by electrophoresis as indicative of the presence of the snp in the target DNA.
Owner:MONOGRAM BIOSCIENCES
Pyrazolopyrimidines as protein kinase inhibitors
In its many embodiments, the present invention provides a novel class of pyrazolo[1,5-a]pyrimidine compounds as inhibitors of protein and / or checkpoint kinases, methods of preparing such compounds, pharmaceutical compositions including one or more such compounds, methods of preparing pharmaceutical formulations including one or more such compounds, and methods of treatment, prevention, inhibition, or amelioration of one or more diseases associated with the protein or checkpoint kinases using such compounds or pharmaceutical compositions. The invention also relates to the inhibition of hepatitis C virus (HCV) replication. In particular, embodiments of the invention provide compounds and methods for inhibiting HCV RNA-dependent RNA polymerase enzymatic activity. The invention also provides compositions and methods for the prophylaxis and treatment of HCV infection.
Owner:MERCK SHARP & DOHME LLC
Compositions and methods for the detection of a nucleic acid using a cleavage reaction
InactiveUS7381532B2Sugar derivativesMicrobiological testing/measurementNucleic acid sequencingNuclease
Owner:HOLOGIC INC
Nucleic acid sequence analysis
ActiveUS20120115736A1Improve accuracyImprove processivityMicrobiological testing/measurementPeptidesNucleic acid detectionPolymerase L
Provided are methods for sequencing a nucleic acid with a sequencing enzyme, e.g., a polymerase or exonuclease. The sequencing enzyme can optionally be exchanged with a second sequencing enzyme, which continues the sequencing of the nucleic acid. In certain embodiments, a template is fixed to a surface through a template localizing moiety. The template localizing moiety can optionally anneal with the nucleic acid and / or associate with the sequencing enzyme. Also provided are compositions comprising a nucleic acid and a first sequencing enzyme, which can sequence the nucleic acid and optionally exchange with a second sequencing enzyme present in the composition. Compositions in which a template localizing moiety is immobilized on a surface are provided. Also provided are methods for using data from analytical reactions wherein two different enzymes are employed, e.g., at a same or different reaction regions.
Owner:PACIFIC BIOSCIENCES
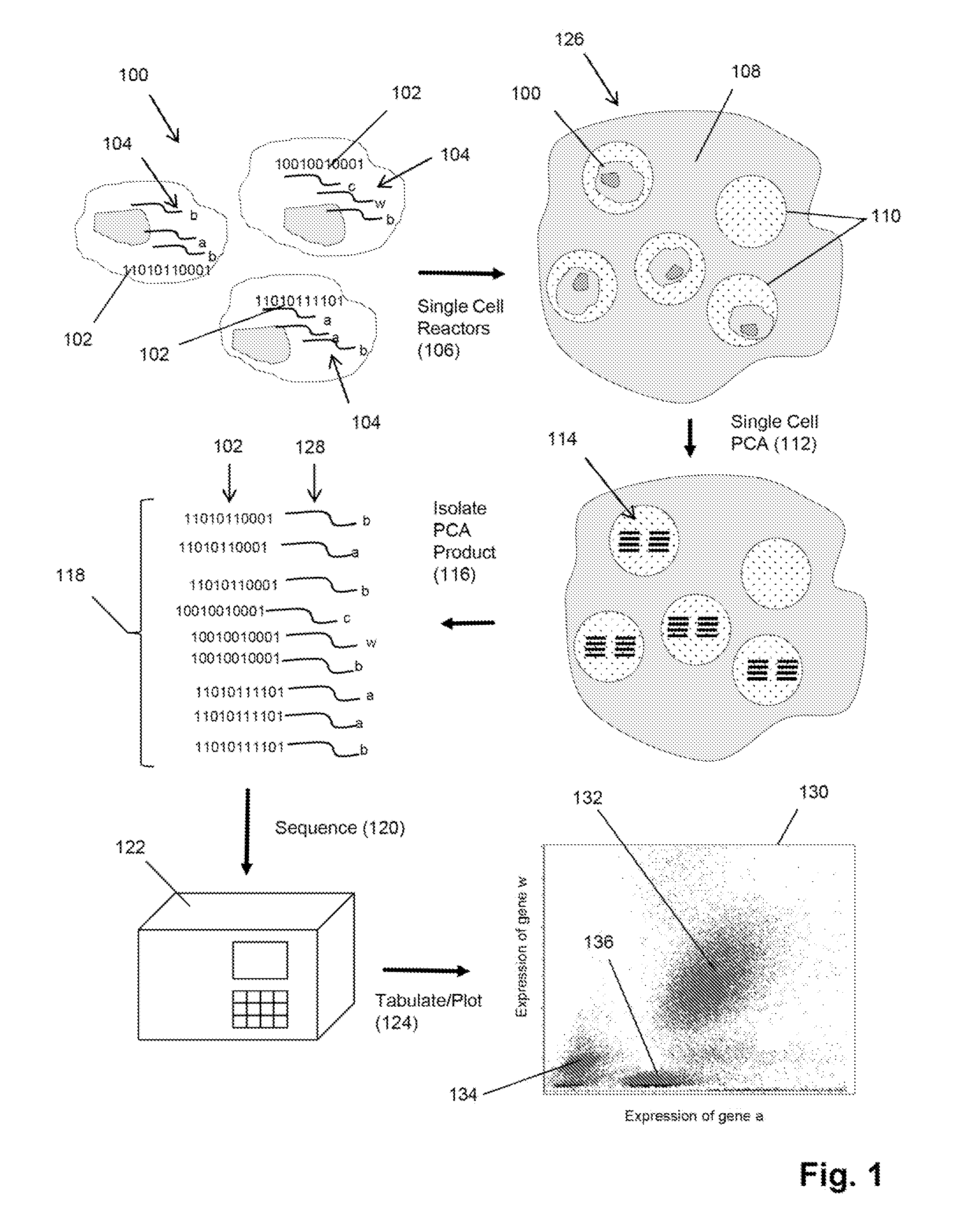
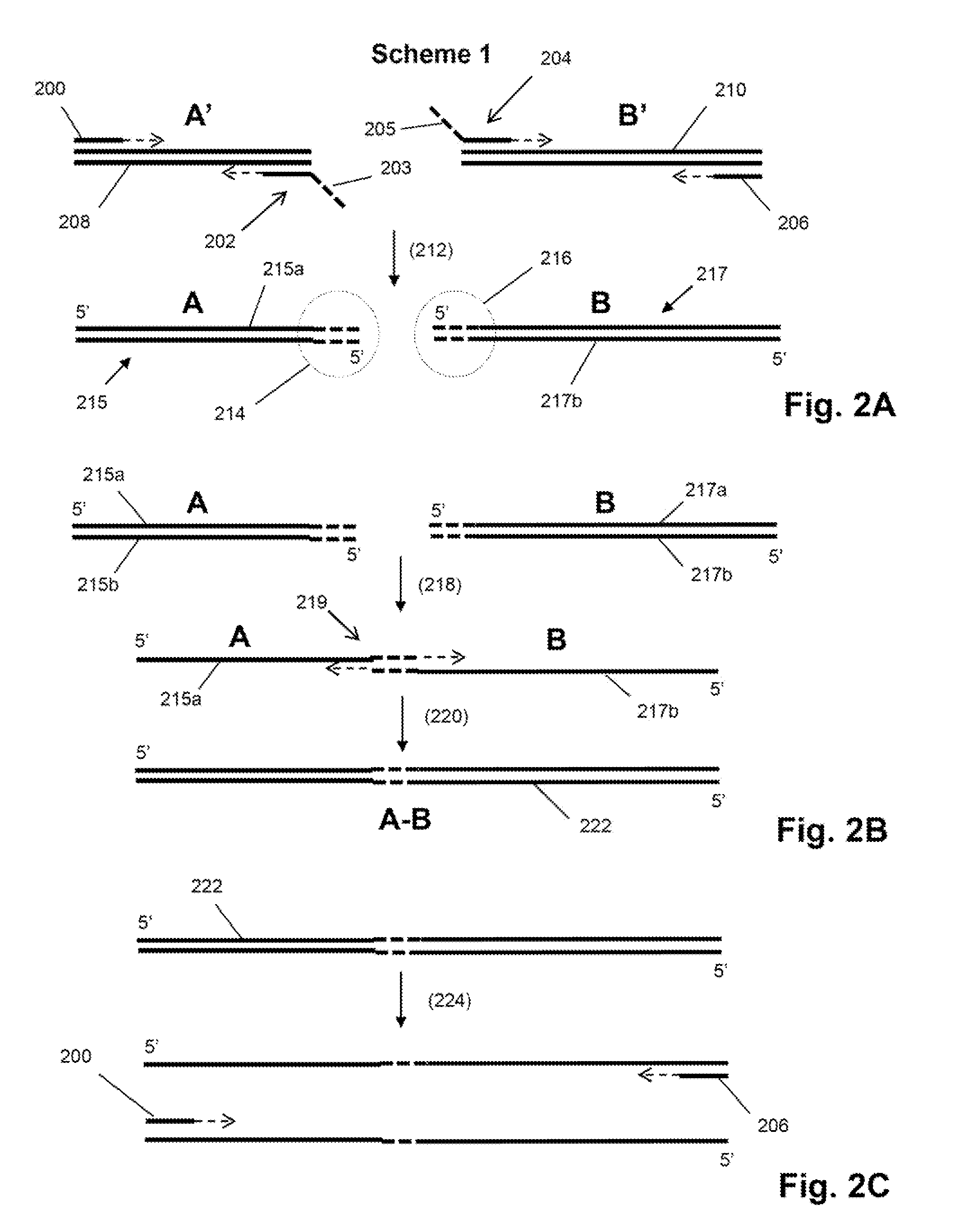
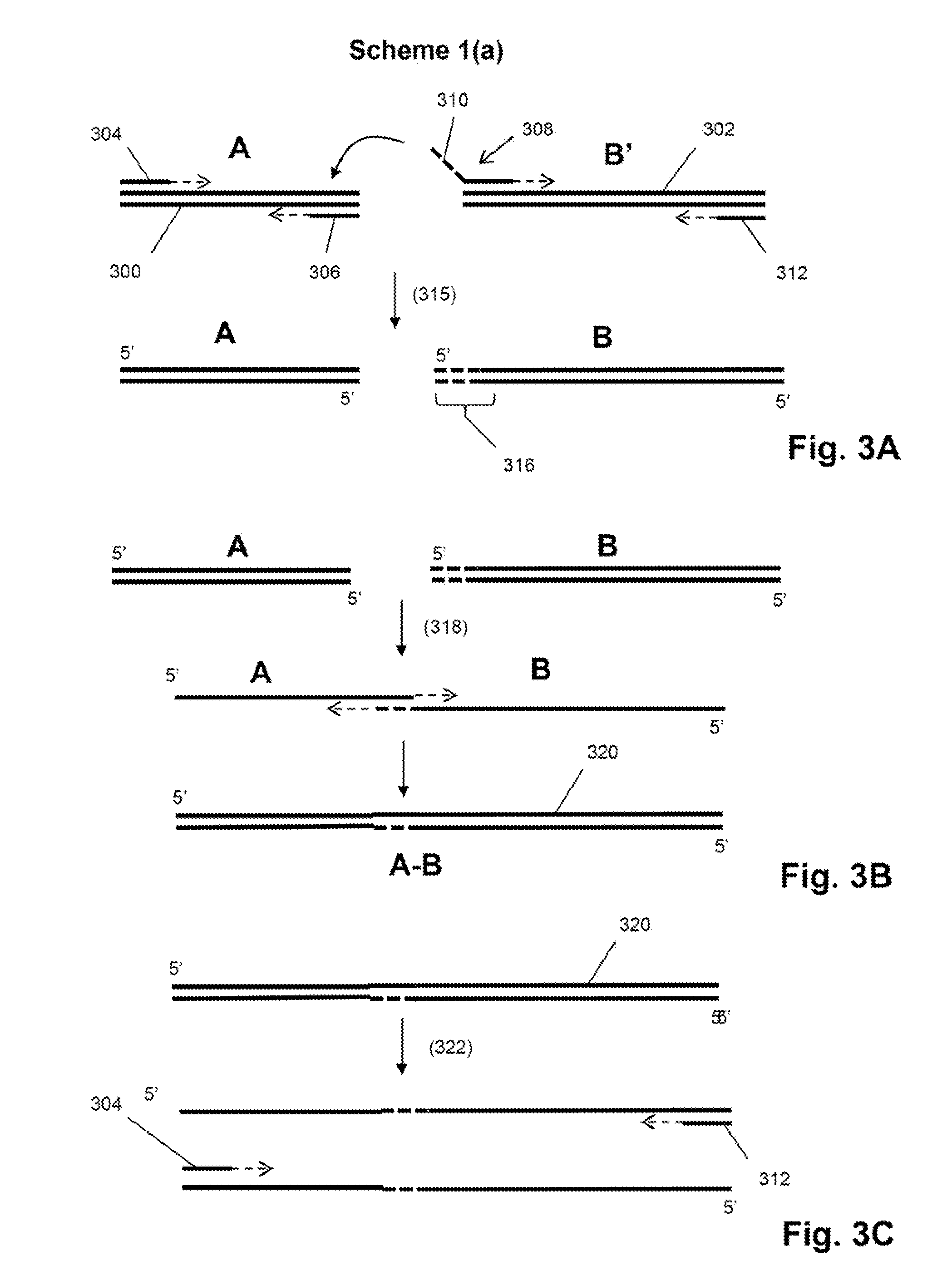
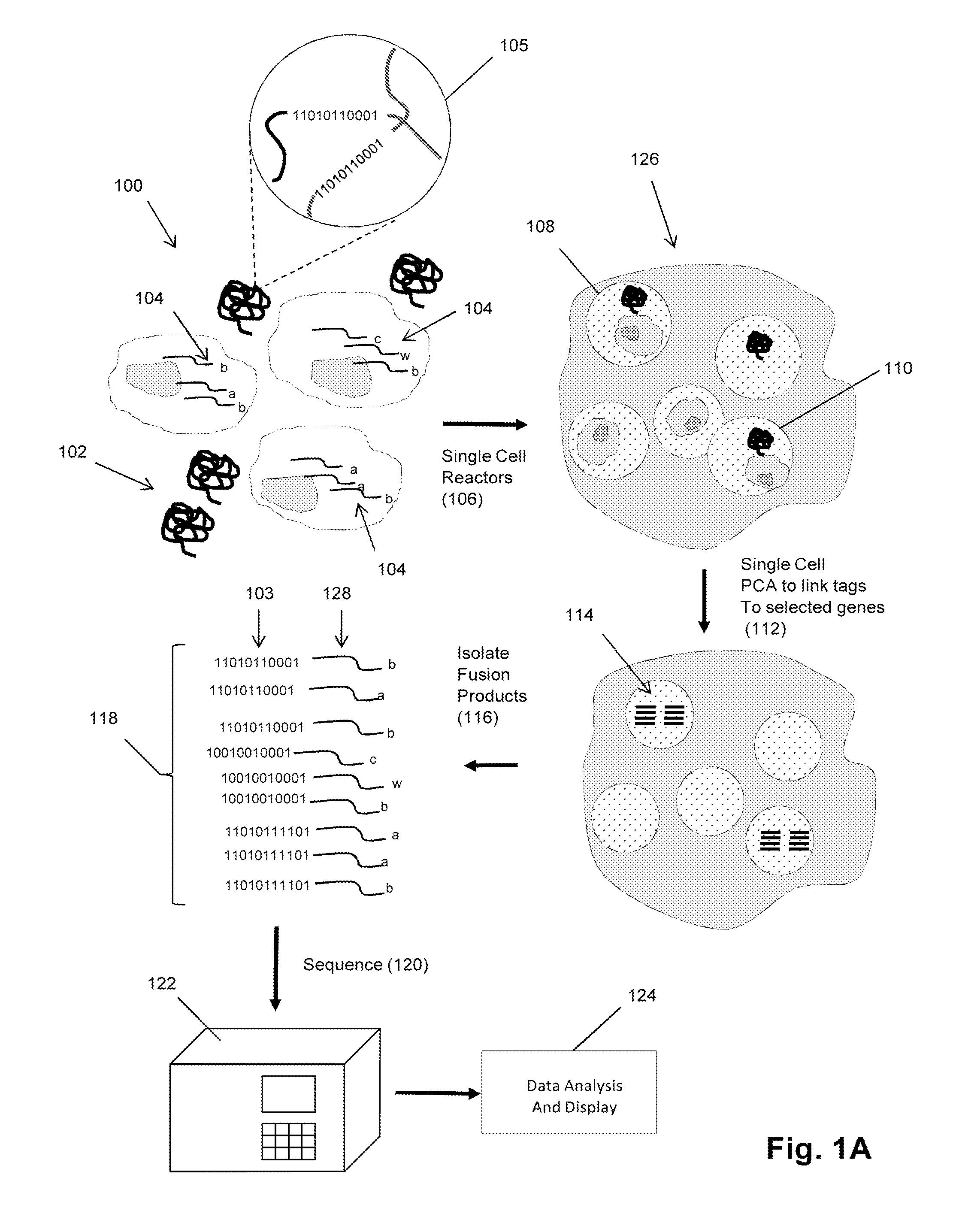
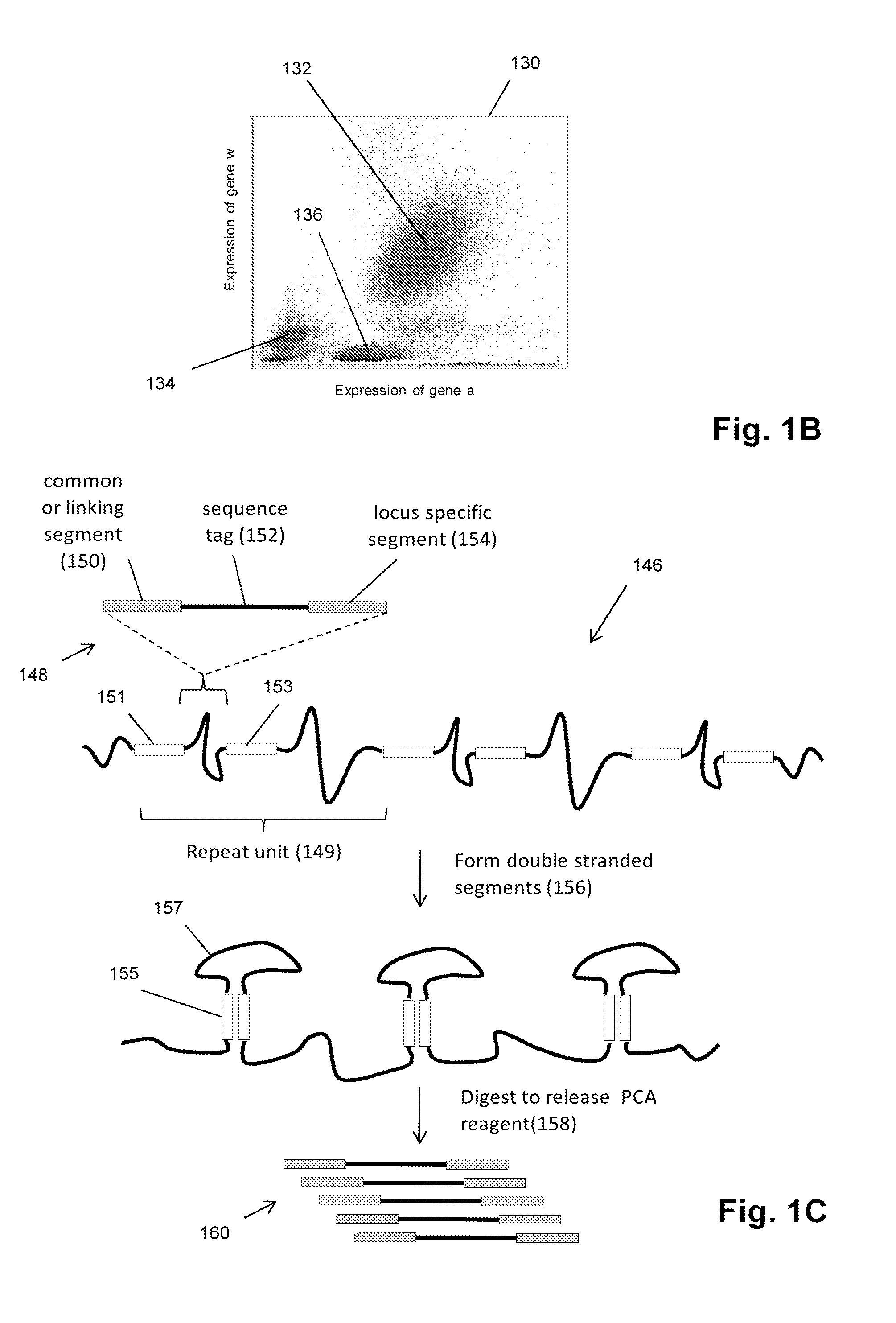
Single cell analysis by polymerase cycling assembly
ActiveUS20110207617A1Microbiological testing/measurementLibrary screeningNucleic acid sequencingPolymerase cycling assembly
The invention provides a method of making measurements on individual cells of a population, particularly cells that have identifying nucleic acid sequences, such as lymphoid cells. In one aspect, the invention provides a method of making multiparameter measurements on individual cells of such a population by carrying out a polymerase cycling assembly (PCA) reaction to link their identifying nucleic acid sequences to other cellular nucleic acids of interest. The fusion products of such PCA reaction are then sequenced and tabulated to generate multiparameter data for cells of the population.
Owner:ADAPTIVE BIOTECH
DNA polymerases and related methods
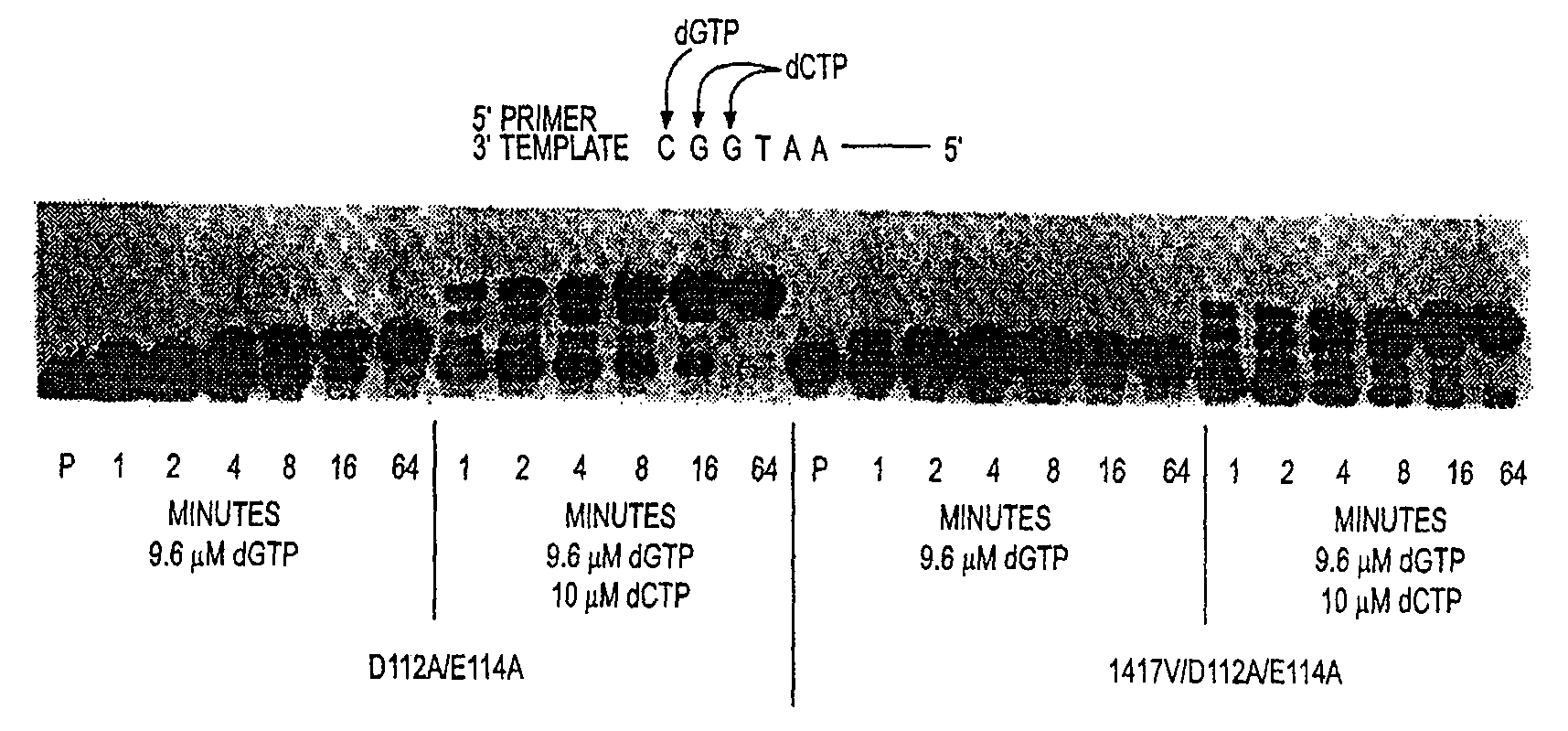
ActiveUS20090148891A1Improved nucleic acid extension rateIncrease chanceBacteriaSugar derivativesPolymerase LDNA polymerase
Disclosed are mutant DNA polymerases having improved extension rates relative to a corresponding, unmodified polymerase. The mutant polymerases are useful in a variety of disclosed primer extension methods. Also disclosed are related compositions, including recombinant nucleic acids, vectors, and host cells, which are useful, e.g., for production of the mutant DNA polymerases.
Owner:ROCHE MOLECULAR SYST INC
Coupled polymerase chain reaction-restriction-endonuclease digestion-ligase detection reaction process
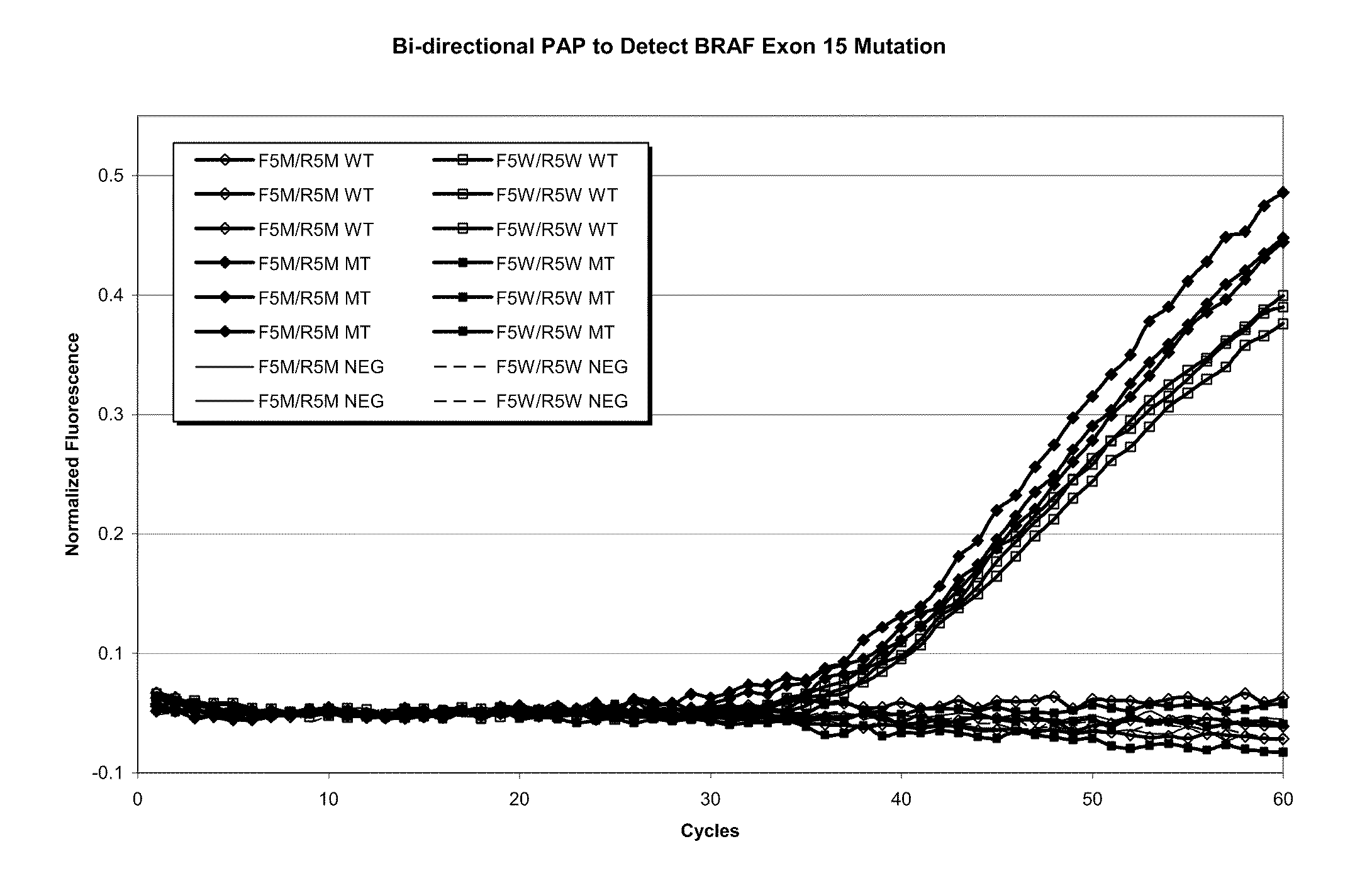
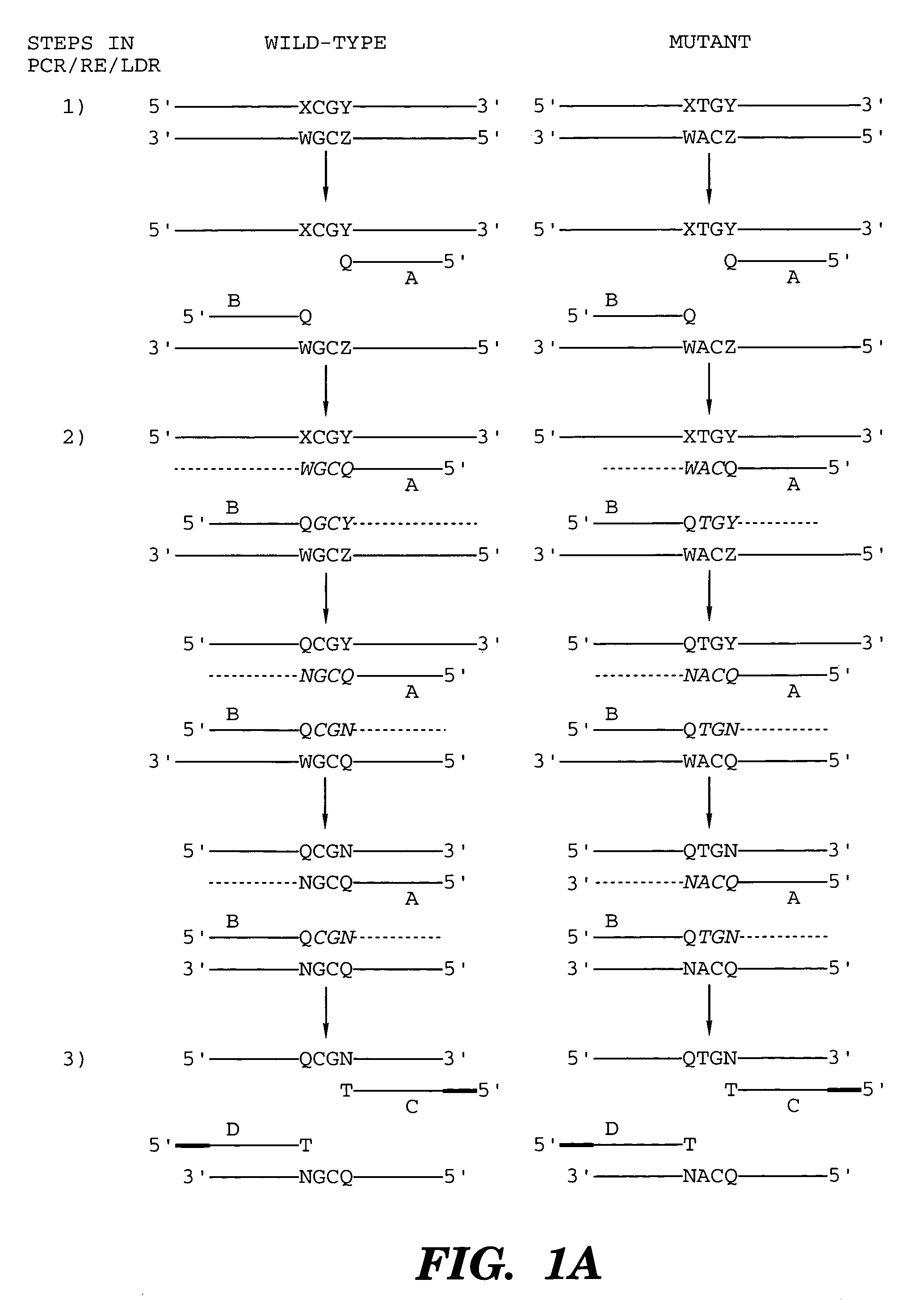
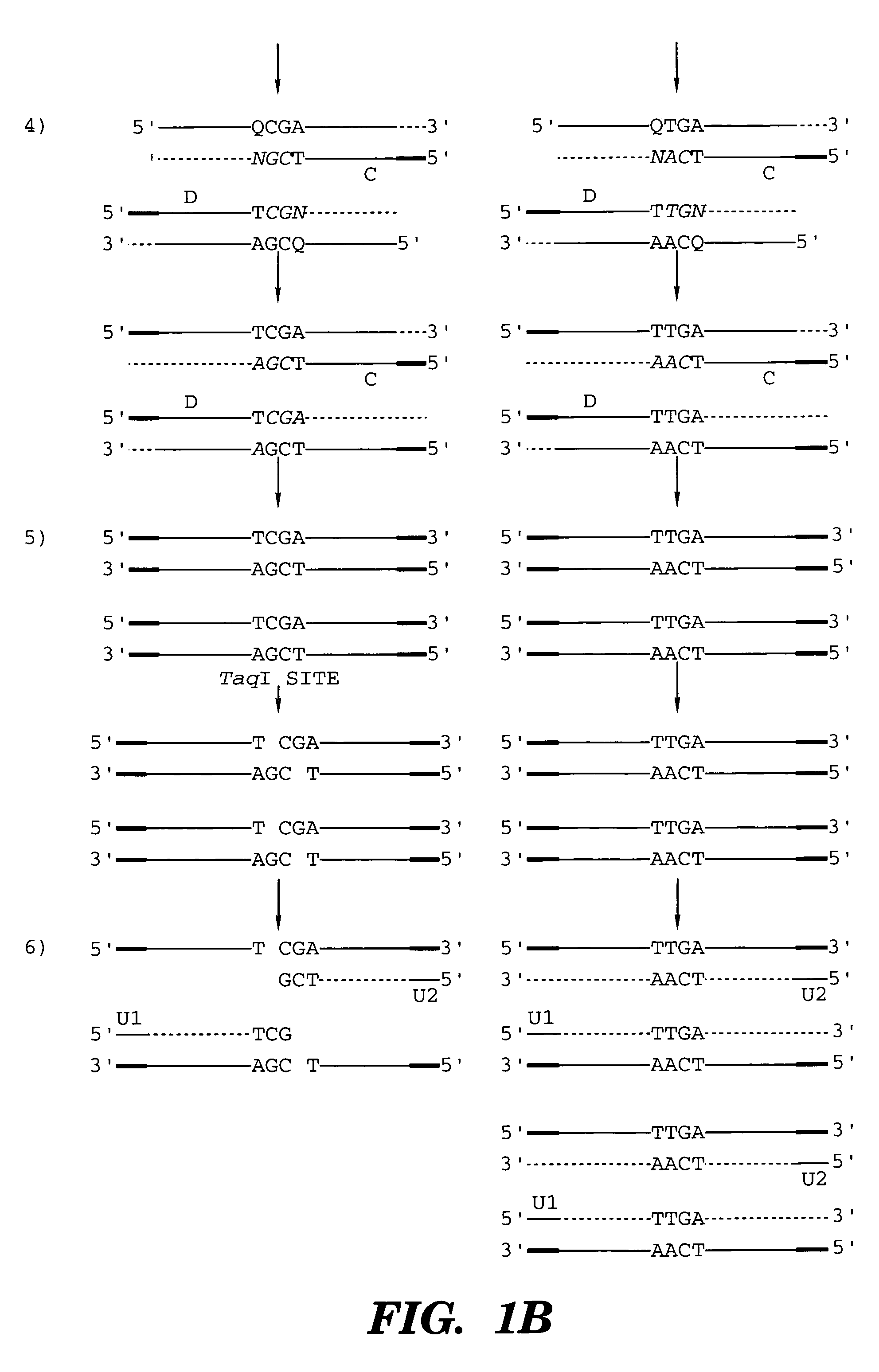
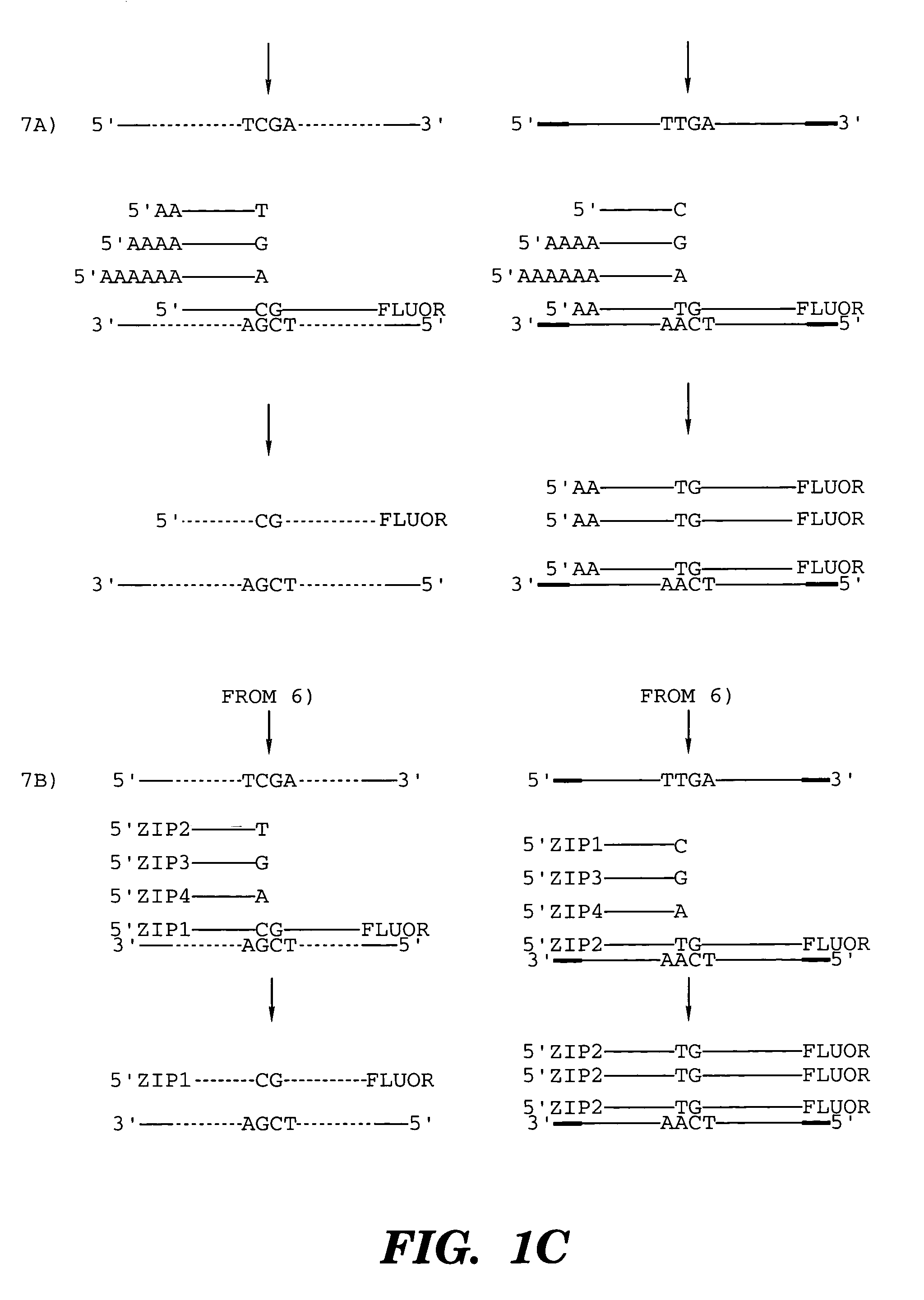
InactiveUS7014994B1Sensitive highOptimizationSugar derivativesMicrobiological testing/measurementNucleotideWild type
The present invention provides a method for identifying one or more low abundance sequences differing by one or more single-base changes, insertions, or deletions, from a high abundance sequence in a plurality of target nucleotide sequences. The high abundance wild-type sequence is selectively removed using high fidelity polymerase chain reaction analog conversion, facilitated by optimal buffer conditions, to create a restriction endonuclease site in the high abundance wild-type gene, but not in the low abundance mutant gene. This allows for digestion of the high abundance DNA. Subsequently the low abundant mutant DNA is amplified and detected by the ligase detection reaction assay. The present invention also relates to a kit for carrying out this procedure.
Owner:LOUISIANA STATE UNIV +1
Devices and methods for the performance of miniaturized in vitro amplification assays
InactiveUS20040259237A1Temperature controlEfficient use ofBioreactor/fermenter combinationsBiological substance pretreatmentsAssayBiomedical engineering
This invention relates to methods and apparatus for performing microanalytic and microsynthetic analyses and procedures. The invention provides a microsystem platform and a micromanipulation device for manipulating the platform that utilizes the centripetal force resulting from rotation of the platform to motivate fluid movement through microchannels. The invention specifically provides devices and methods for performing miniaturized in vitro amplification assays such as the polymerase chain reaction. Methods specific for the apparatus of the invention for performing PCR are provided.
Owner:TECAN TRADING AG
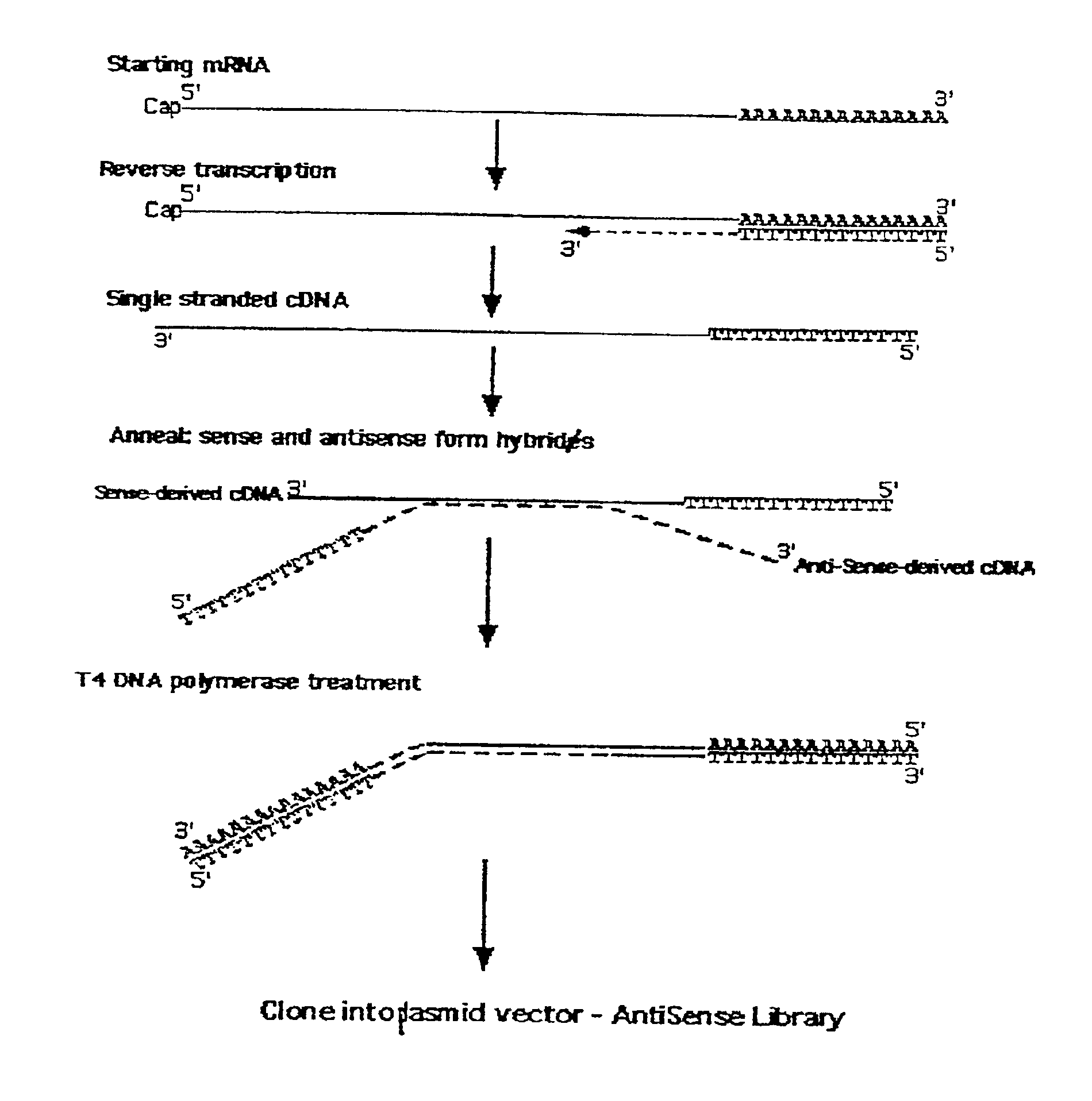
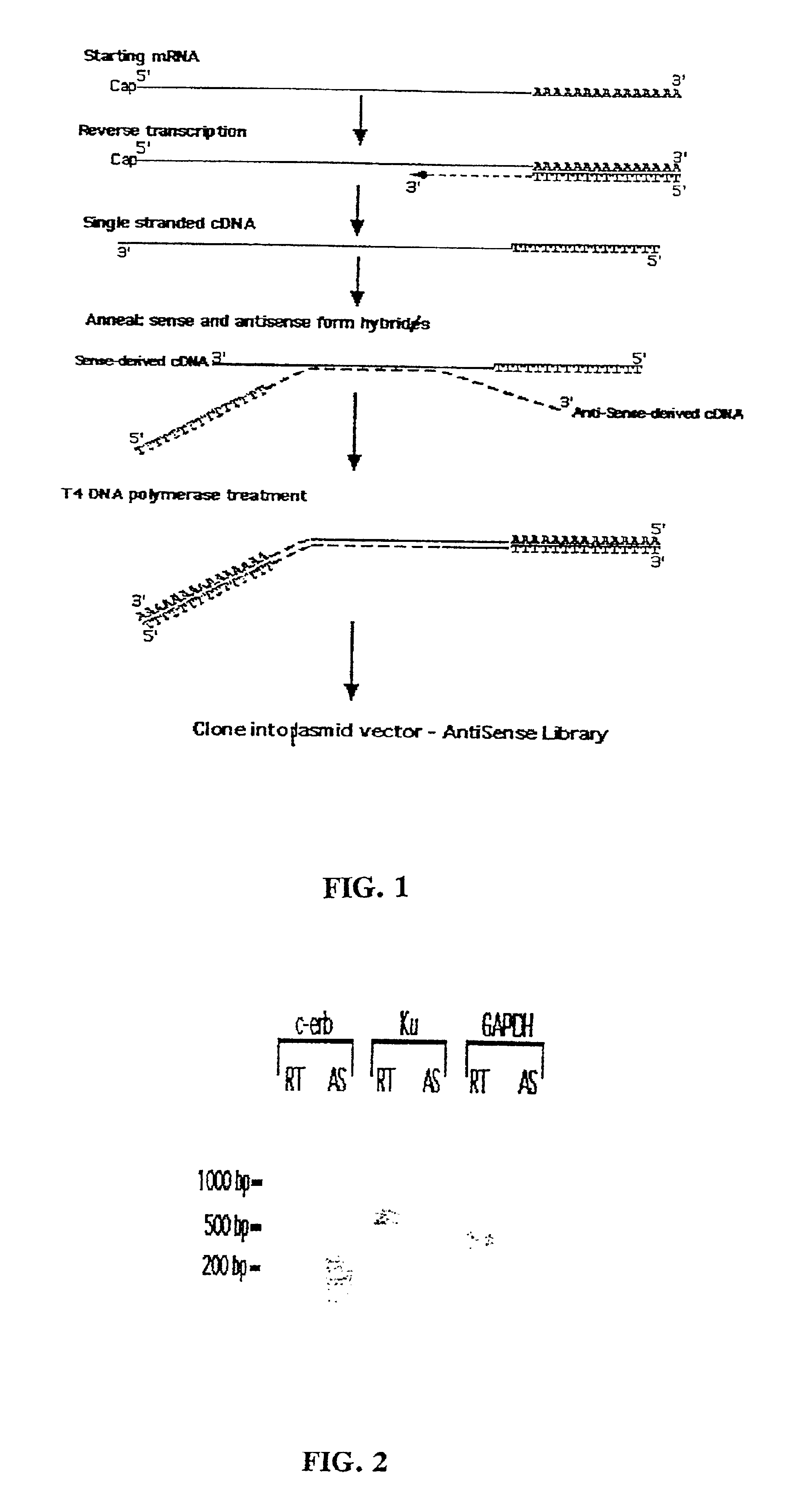
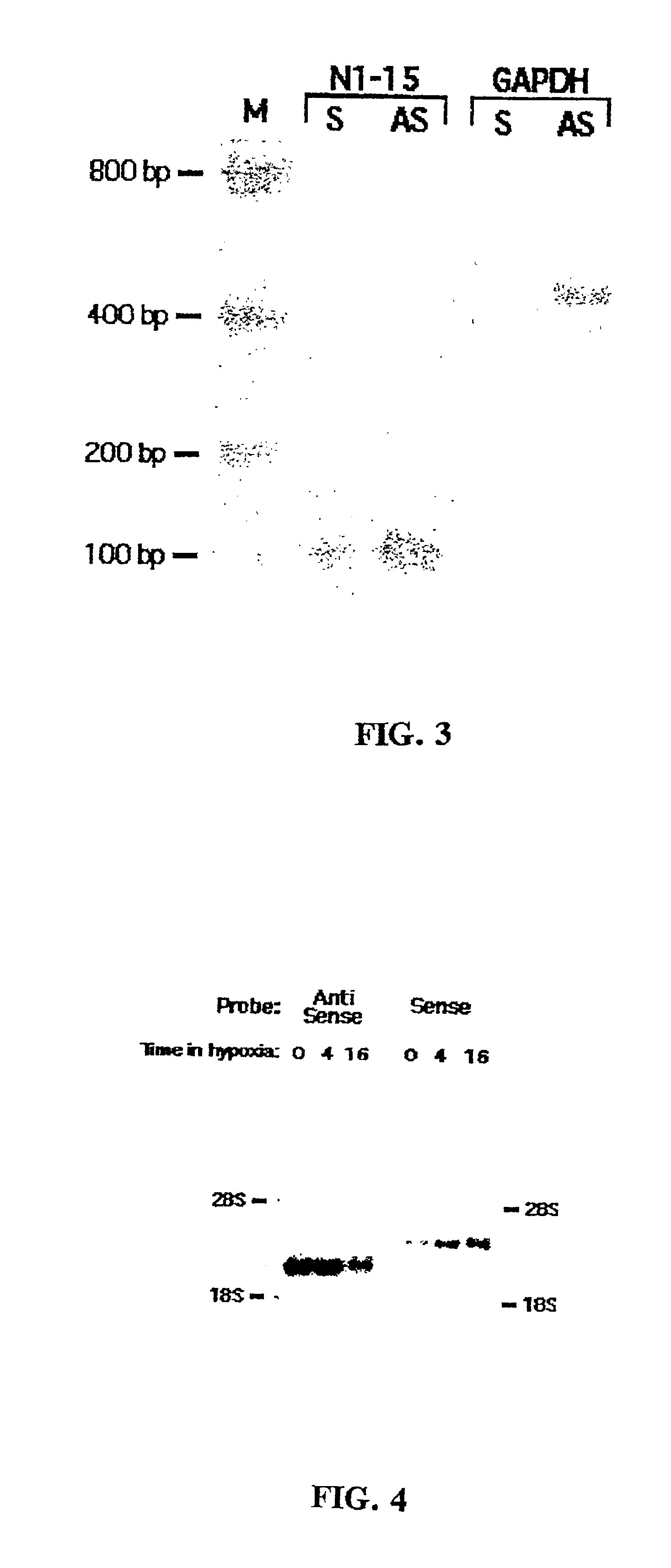
Method for enrichment of natural antisense messenger RNA
A method for enrichment of natural antisense mRNA which involves hybridization of cDNA obtained from sense RNA with cDNA obtained from antisense RNA, followed by DNA polymerase treatment of the sense-antisense hybrid DNA molecule. A natural antisense library can be generated by cloning of sense-antisense hybrid DNA molecules in a vector.
Owner:QUARK FARMACUITIKALS INC
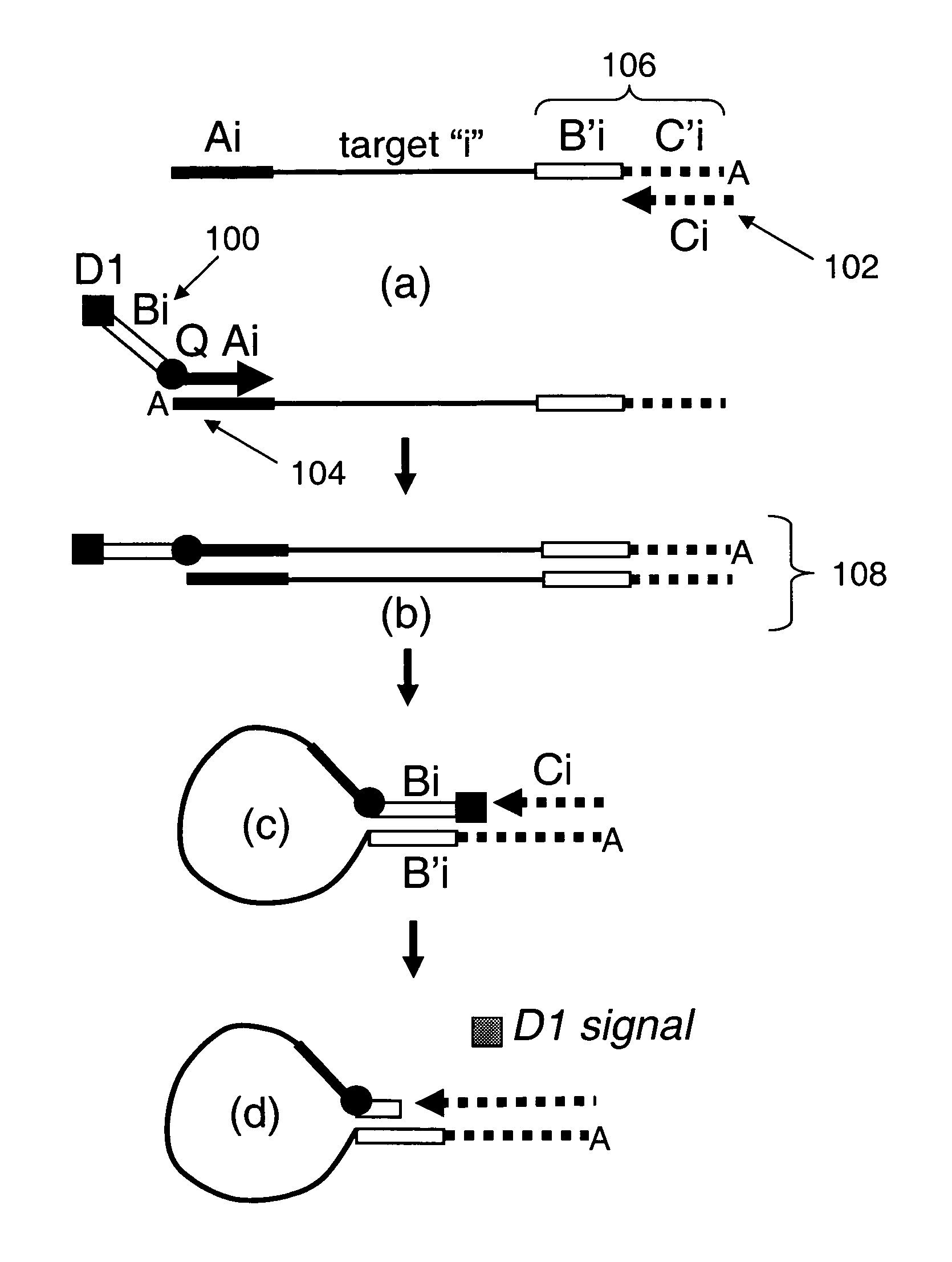
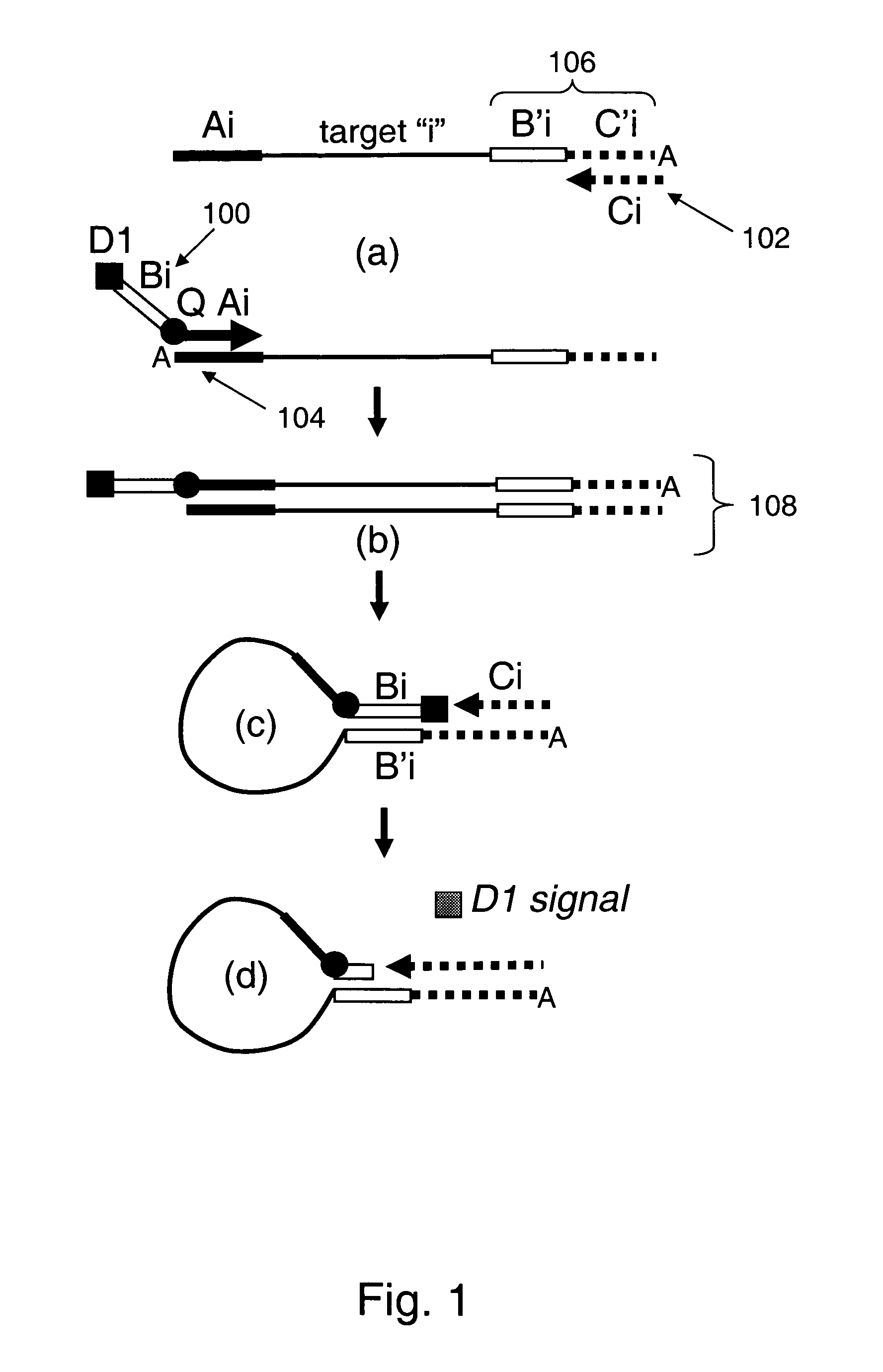
Methods and compositions for universal detection of nucleic acids
ActiveUS20110212846A1Increase distanceMicrobiological testing/measurementLibrary screeningPolymerase chain reactionClinical settings
Provided are methods and compositions for detecting the presence or amount of one or more target nucleic acids in a sample. Methods of the present invention include linking universal nucleic acid segments into a single molecule in a linking reaction dependent on a target nucleic acid of interest. A variety of universal segment linking strategies are provided, including preamplification by polymerase chain reaction, ligation-based strategies, reverse transcription and linear polymerase extension. Linking the universal segments into a single molecule generates a tagged target nucleic acid which is detected in a manner dependent on an intramolecular interaction between one universal segment and a second portion of the tagged target nucleic acid. In certain embodiments, the intramolecular interaction includes the formation of a hairpin having a stem between a universal segment at one end of the tagged target nucleic acid and a second universal segment at the opposite end of the tagged target nucleic acid. A variety of detection formats are provided, including solution-phase and surface-based formats. The methods and compositions are well-suited for highly multiplexed nucleic acid detection, and are applicable for the detection of any target nucleic acid of interest in both research and clinical settings.
Owner:UNITAQ BIO
Nano-PCR: methods and devices for nucleic acid amplification and detection
ActiveUS20060019274A1Significant improvementMaterial nanotechnologyHeating or cooling apparatusNucleic acid sequencingPcr method
Methods, devices, and compositions are described that provide for amplification of nucleic acid sequences without reliance upon temperature cycling, thus freeing the methods from conventional benchtop thermal cycling devices. Denaturation of double stranded nucleic acids, primer annealing, and precision control over primer extension by polymerase can be accomplished by applying stress to a nucleic acid. These methods can provide one ore more benefits over conventional PCR methods including: precision control over the PCR process; generally improved fidelity; improved accuracy over problematic sequences such as GC-rich or tandem repeat regions; greater sequence length; increased reaction yield; reduced experimental time; greater efficiency; lower cost; greater portability; and, robustness to various environmental parameters, such as temperature, pH, and ionic strengths.
Owner:NANOBIOSYM INC
Method of determining the nucleotide sequence of oligonucleotides and DNA molecules
InactiveUS20020137062A1Eliminate needHigh purityMicrobiological testing/measurementRecombinant DNA-technologyFluorescenceOligonucleotide primers
The present invention relates to a novel method for analyzing nucleic acid sequences based on real-time detection of DNA polymerase-catalyzed incorporation of each of the four nucleotide bases, supplied individually and serially in a microfluidic system, to a reaction cell containing a template system comprising a DNA fragment of unknown sequence and an oligonucleotide primer. Incorporation of a nucleotide base into the template system can be detected by any of a variety of methods including but not limited to fluorescence and chemiluminescence detection. Alternatively, microcalorimetic detection of the heat generated by the incorporation of a nucleotide into the extending template system using thermopile, thermistor and refractive index measurements can be used to detect extension reactions.
Owner:LIFE TECH CORP
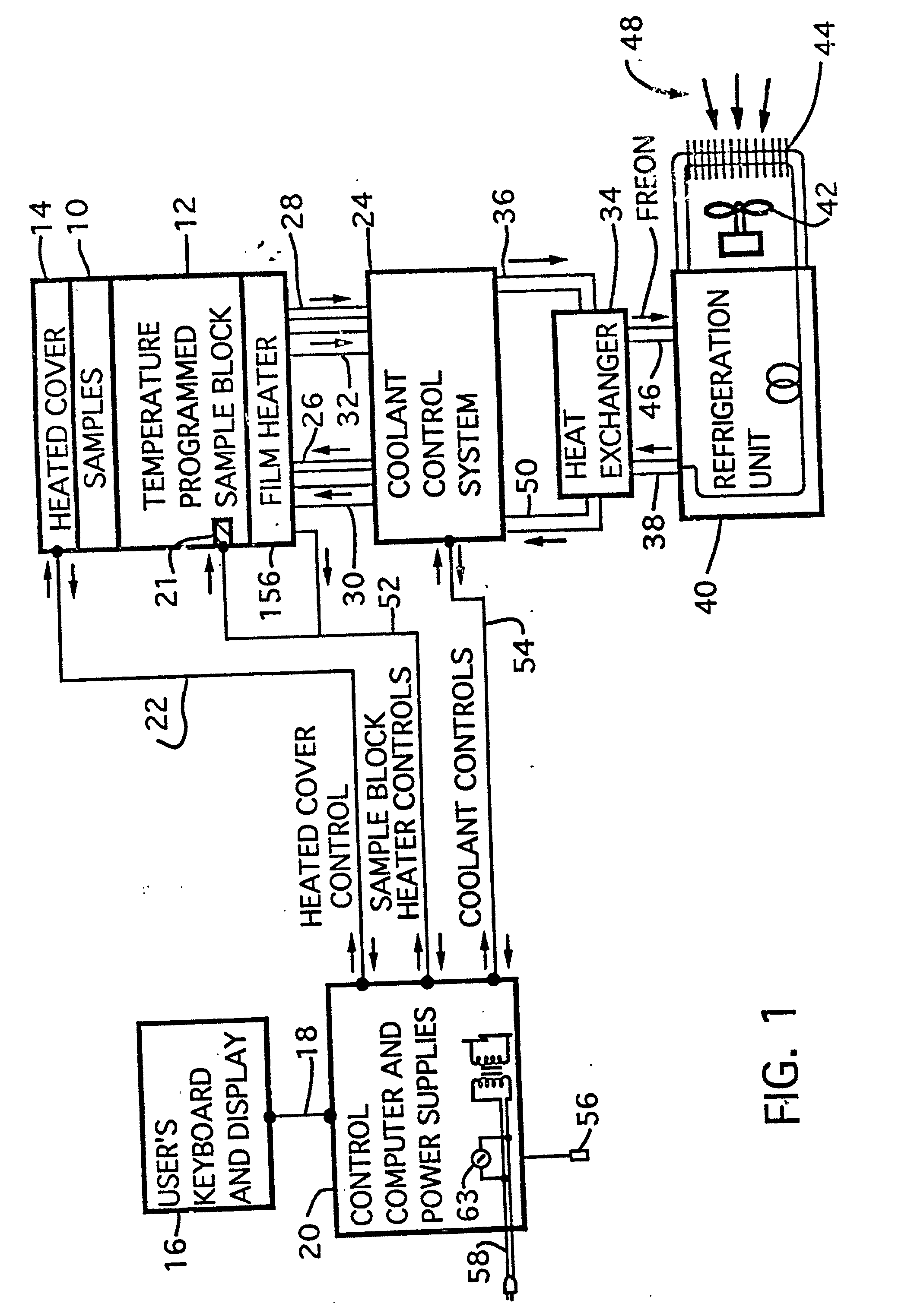
Thermal cycler for automatic performance of the polymerase chain reaction with close temperature control
InactiveUS20020072112A1Shorten cycle timeLow cost of reagentsBioreactor/fermenter combinationsBiological substance pretreatmentsTemperature controlSolenoid valve
An instrument for performing highly accurate PCR employing a sample block in microtiter tray format. The sample block has local balance and local symmetry. A three zone film heater controlled by a computer and ramp cooling solenoid valves also controlled by the computer for gating coolant flow through the block controls the block temperature. Constant bias cooling is used for small changes. Sample temperature is calculated instead of measured. A platen deforms plastic caps to apply a minimum acceptable threshold force for seating the tubes and thermally isolates them. A cover isolates the block. The control software includes diagnostics. An install program tests and characterizes the instrument. A new user interface is used. Disposable, multipiece plastic microtiter trays to give individual freedom to sample tubes are taught.
Owner:APPL BIOSYSTEMS INC
Single cell analysis using sequence tags
InactiveUS20150247182A1Microbiological testing/measurementLibrary member identificationNucleic acid sequencingPolymerase cycling assembly
The invention provides a method of making measurements on individual cells of a population by forming reactors containing single cells and a predetermined number, usually one, homogeneous sequence tag. In one aspect, the invention provides a method of making multiparameter measurements on individual cells of such a population by carrying out a polymerase cycling assembly (PCA) reaction to link their identifying nucleic acid sequences, such as sequence tag copies derived from a homogeneous sequence tag, to other cellular nucleic acids of interest, thereby forming fusion products. The fusion products of such PCA reactions are then sequenced and tabulated to generate multiparameter data for cells of the population.
Owner:ADAPTIVE BIOTECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com