Safe sequencing system
a sequencing system and sequencing technology, applied in the field of nucleic acid sequencing, can solve the problem that mass parallel sequencing cannot be used to detect rare variants, and achieve the effect of sensitive and accurate determination of nucleic acid features or sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Endogenous UIDs
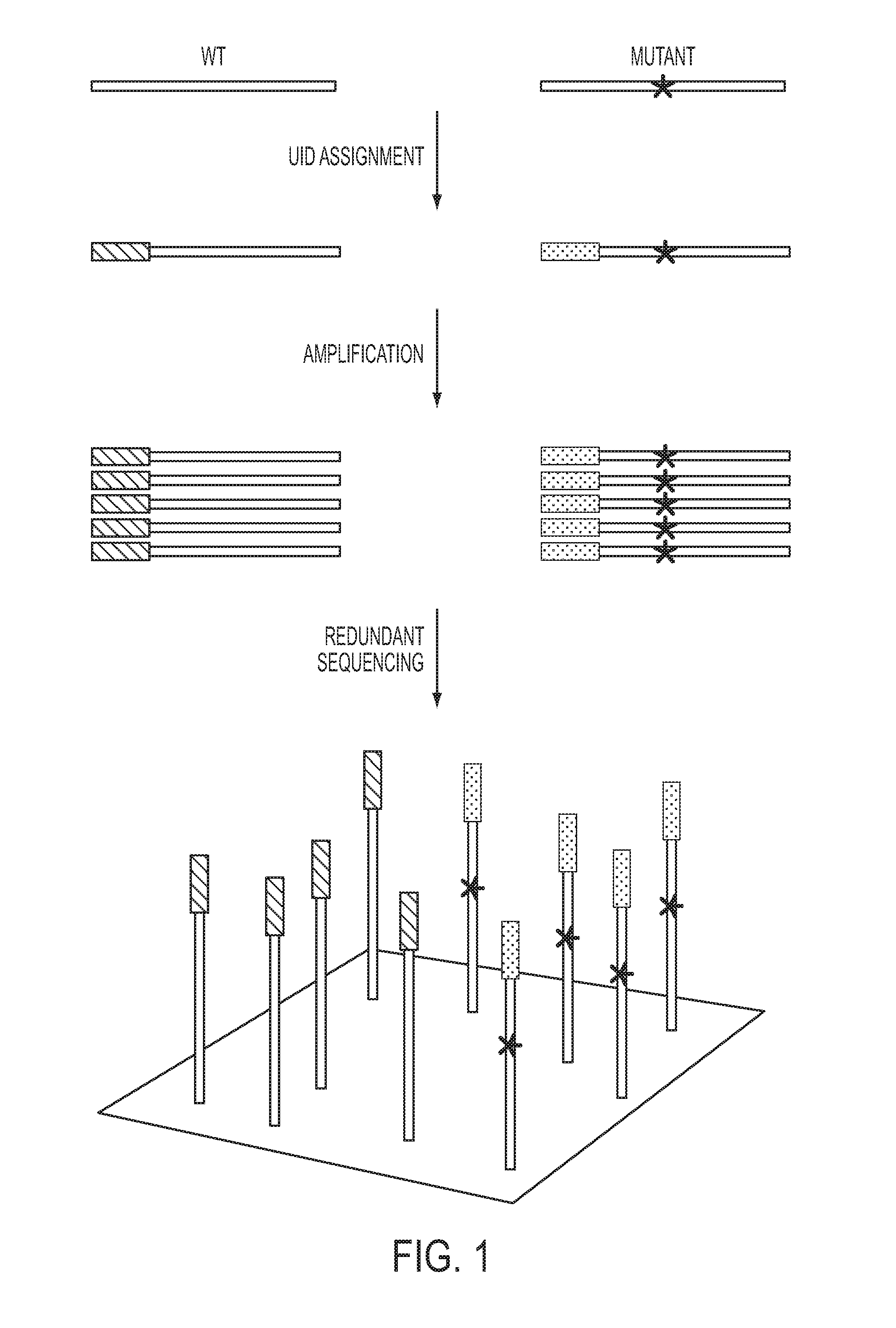
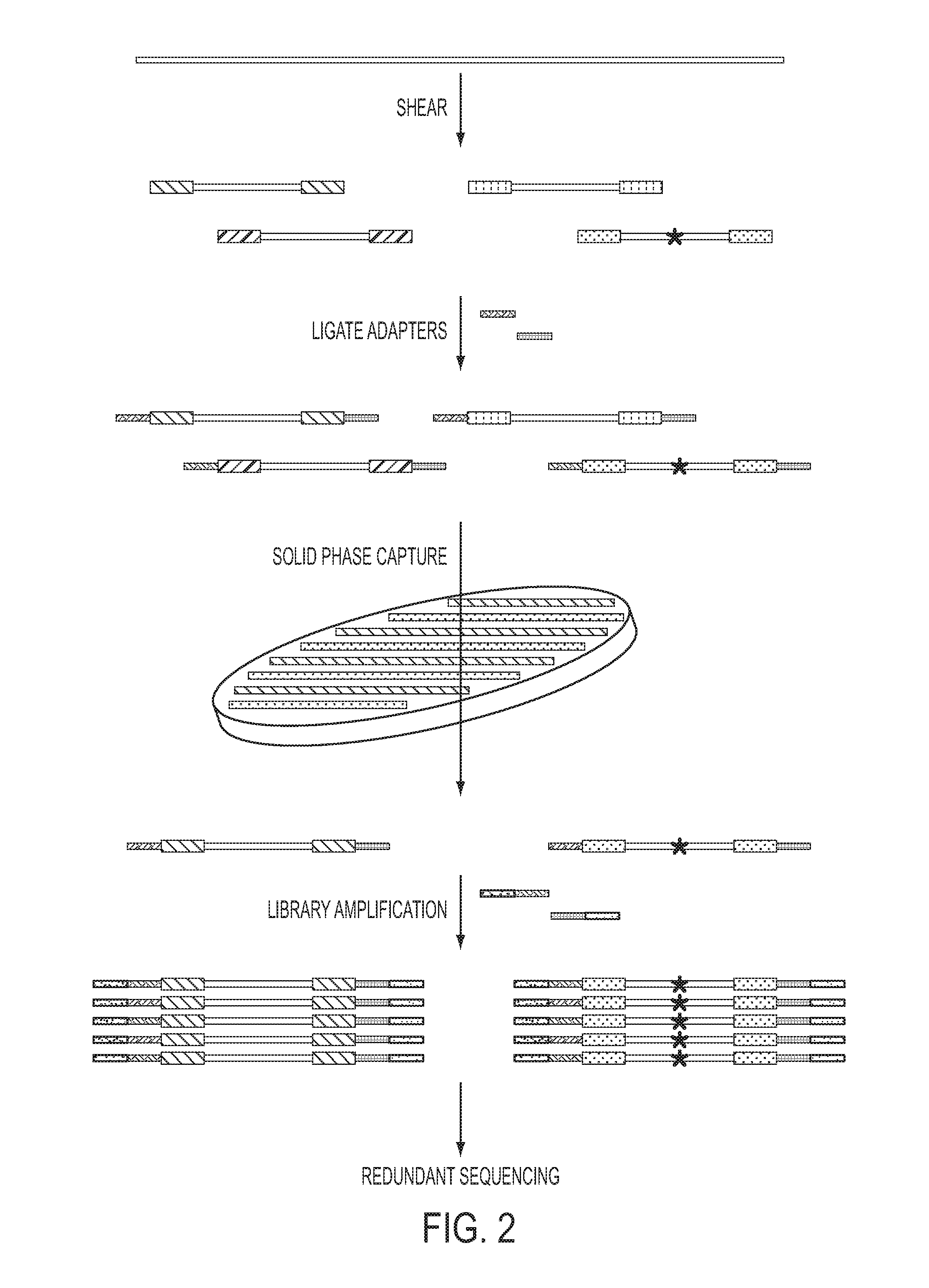
[0039]UIDs, sometimes called barcodes or indexes, can be assigned to nucleic acid fragments in many ways. These include the introduction of exogenous sequences through PCR (40, 41) or ligation (42, 43). Even more simply, randomly sheared genomic DNA inherently contains UIDs consisting of the sequences of the two ends of each sheared fragment (FIG. 2 and FIG. 5). Paired-end sequencing of these fragments yields UID-families that can be analyzed as described above. To employ such endogenous UIDs in Safe-SeqS, we used two separate approaches: one designed to evaluate many genes simultaneously and the other designed to evaluate a single gene fragment in depth (FIG. 2 and FIG. 5, respectively).
[0040]For the evaluation of multiple genes, we ligated standard Illumina sequencing adapters to the ends of sheared DNA fragments to produce a standard sequencing library, then captured genes of interest on a solid phase (44). In this experiment, a library made from the DNA of ˜15,000...
example 2
Exogenous UIDs
[0044]Though the results described above show that Safe-SeqS can increase the reliability of massively parallel sequencing, the number of different molecules that can be examined using endogenous UIDs is limited. For fragments sheared to an average size of 150 bp (range 125-175), 36 base paired-end sequencing can evaluate a maximum of ˜7,200 different molecules containing a specific mutation (2 reads×2 orientations×36 bases / read×50 base variation on either end of the fragment). In practice, the actual number of UIDs is smaller because the shearing process is not entirely random.
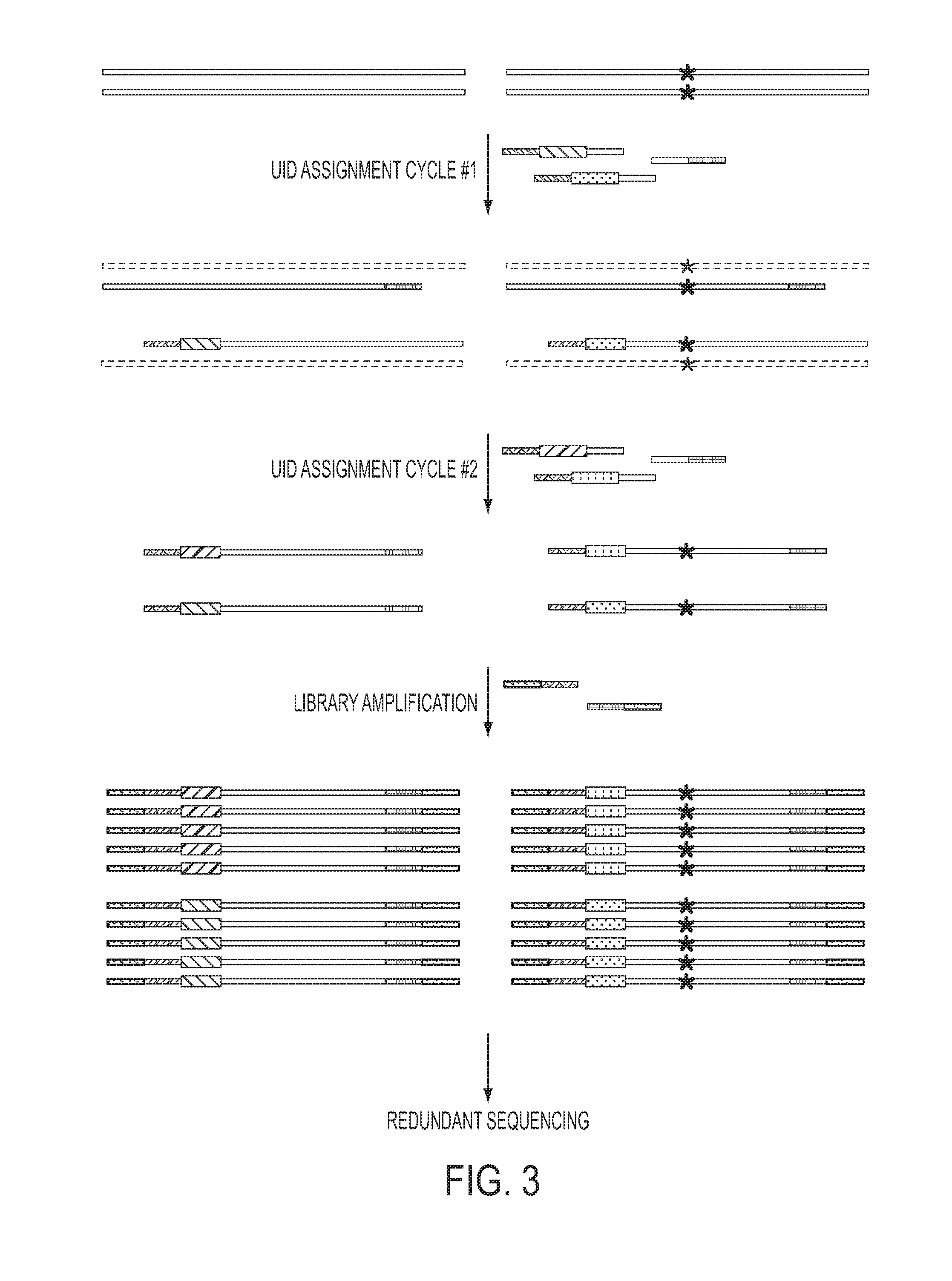
[0045]To make more efficient use of the original templates, we developed a Safe-SeqS strategy that employed a minimum number of enzymatic steps. This strategy also permitted the use of degraded or damaged DNA, such as found in clinical specimens or after bisulfite-treatment for the examination of cytosine methylation (45). As depicted in FIG. 3, this strategy employs two sets of PCR primers. The...
example 3
Analysis of DNA Polymerase Fidelity
[0046]Measurement of the error rates of DNA polymerases is essential for their characterization and dictates the situations in which these enzymes can be used. We chose to measure the error rate of Phusion polymerase, as this polymerase has one of the lowest reported error frequencies of any commercially available enzyme and therefore poses a particular challenge for an in vitro-based approach. We first amplified a single human DNA template molecule, comprising a segment of an arbitrarily chosen human gene, through 19 rounds of PCR. The PCR products from these amplifications, in their entirety, were used as templates for Safe-SeqS as described in FIG. 3. In seven independent experiments of this type, the number of UID-families identified by sequencing was 624,678±421,274, which is consistent with an amplification efficiency of 92±9.6% per round of PCR.
[0047]The error rate of Phusion polymerase, estimated through cloning of PCR products encoding β-g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com