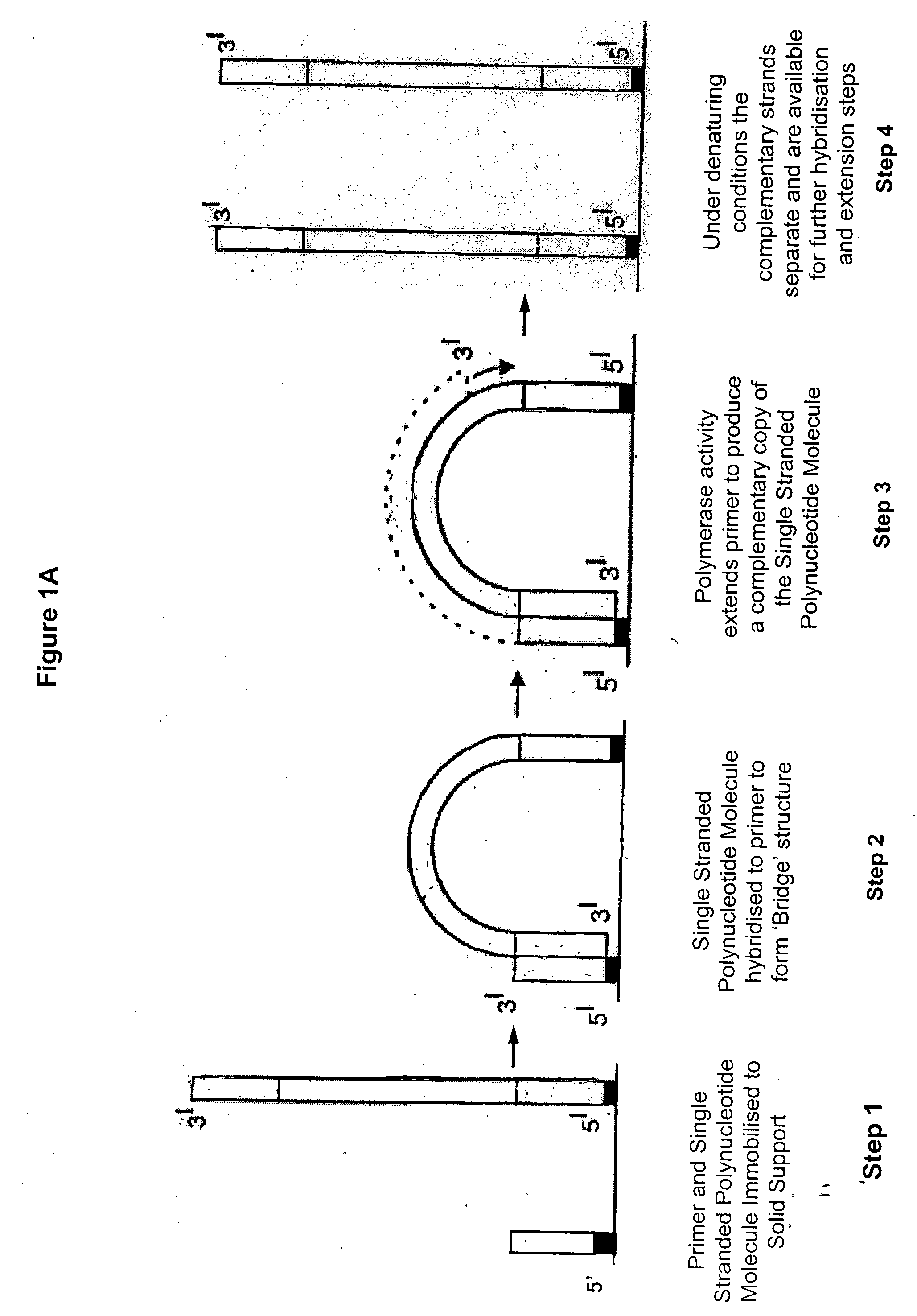
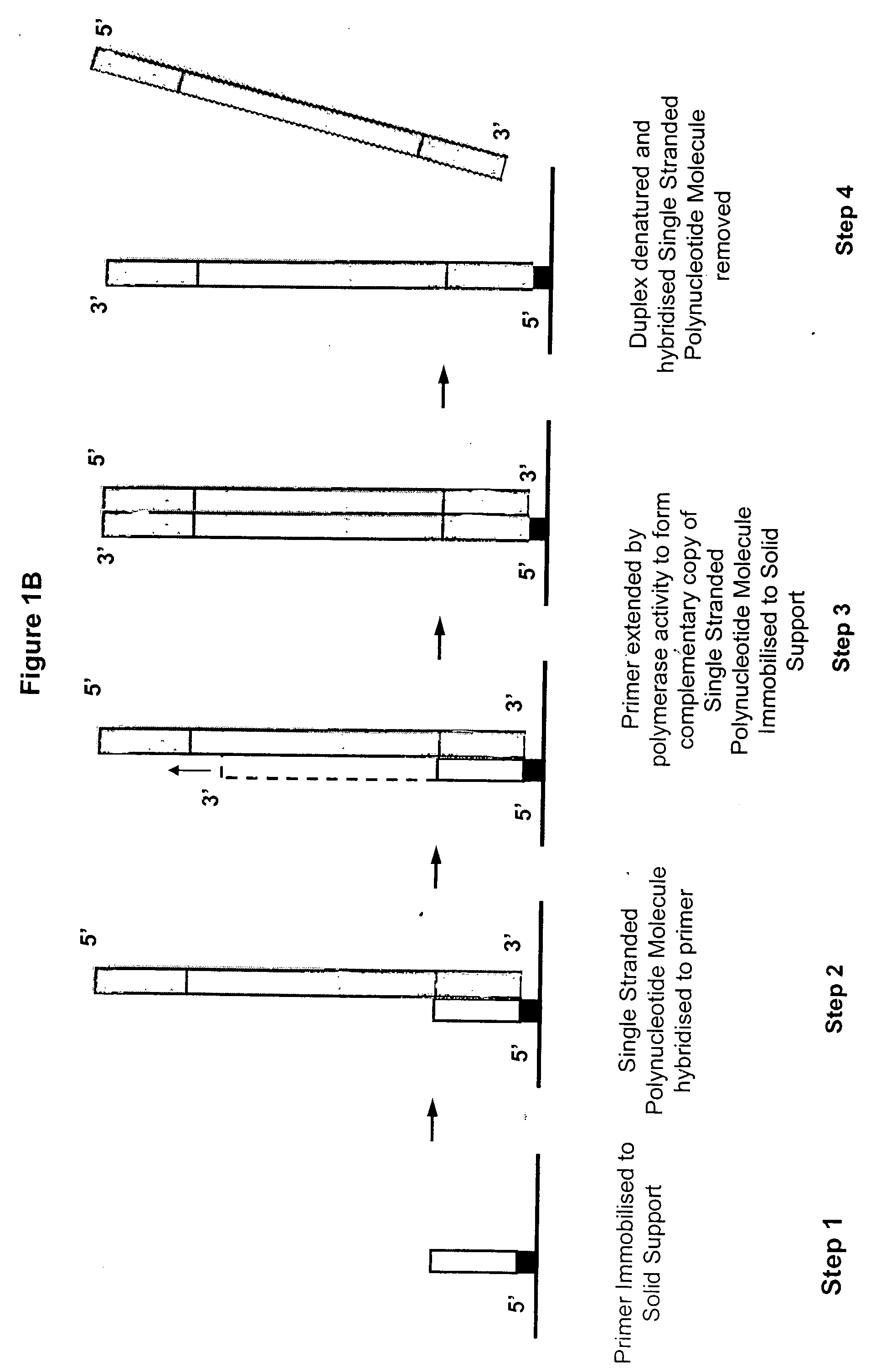
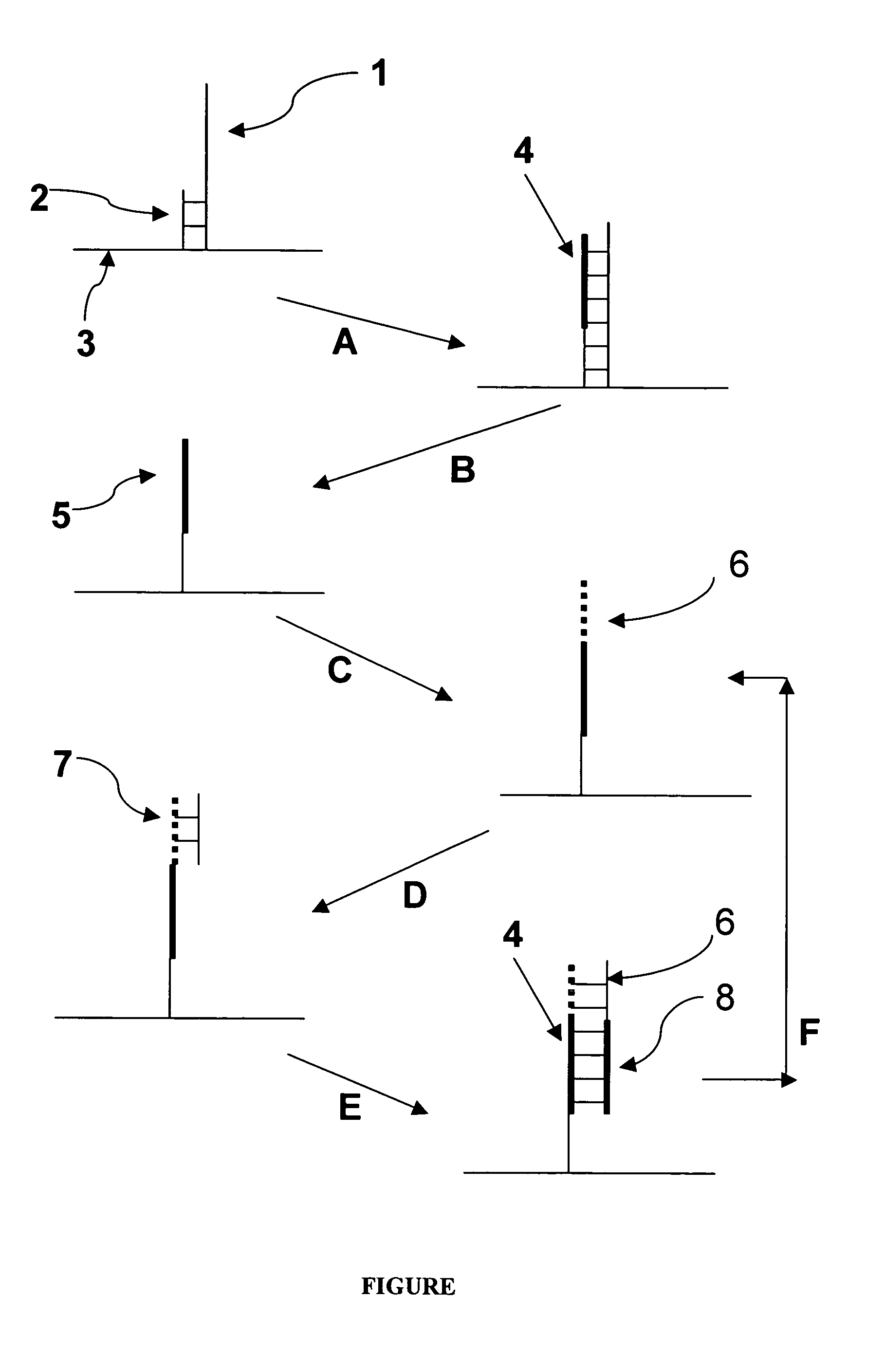
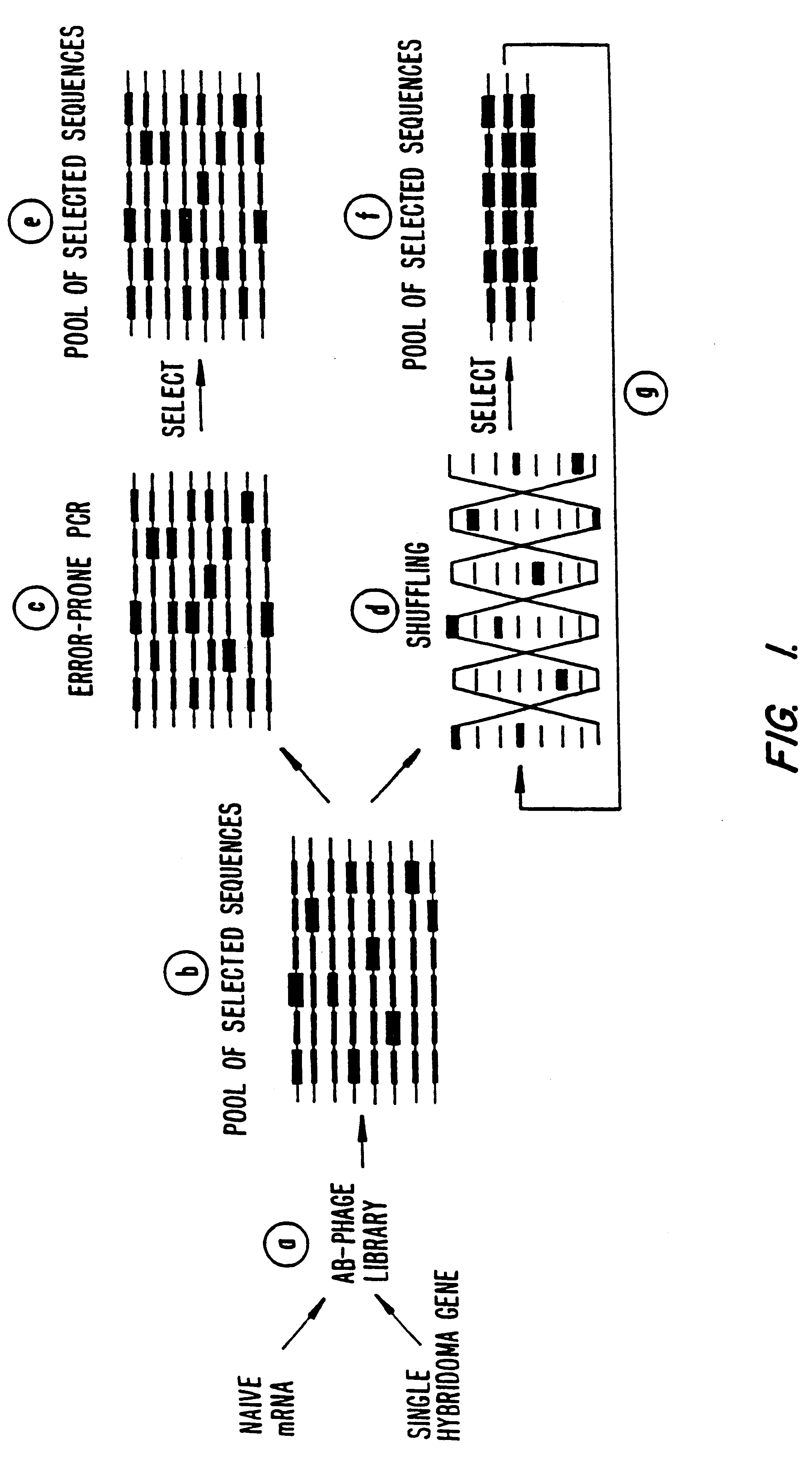
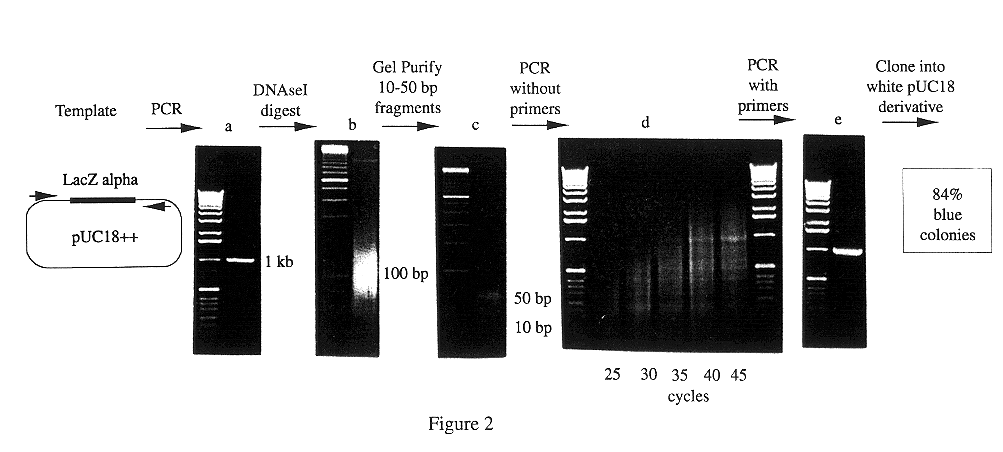
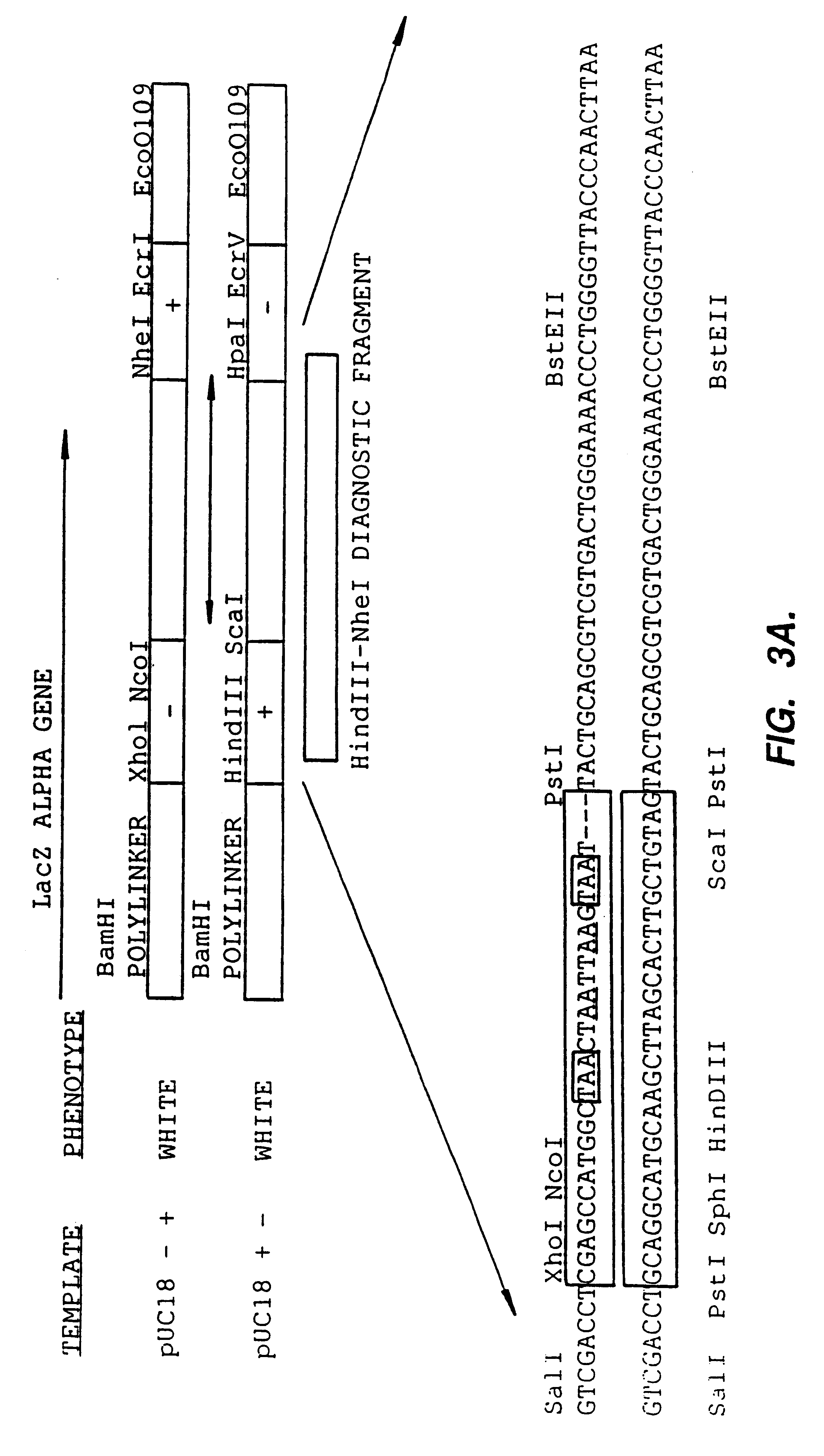
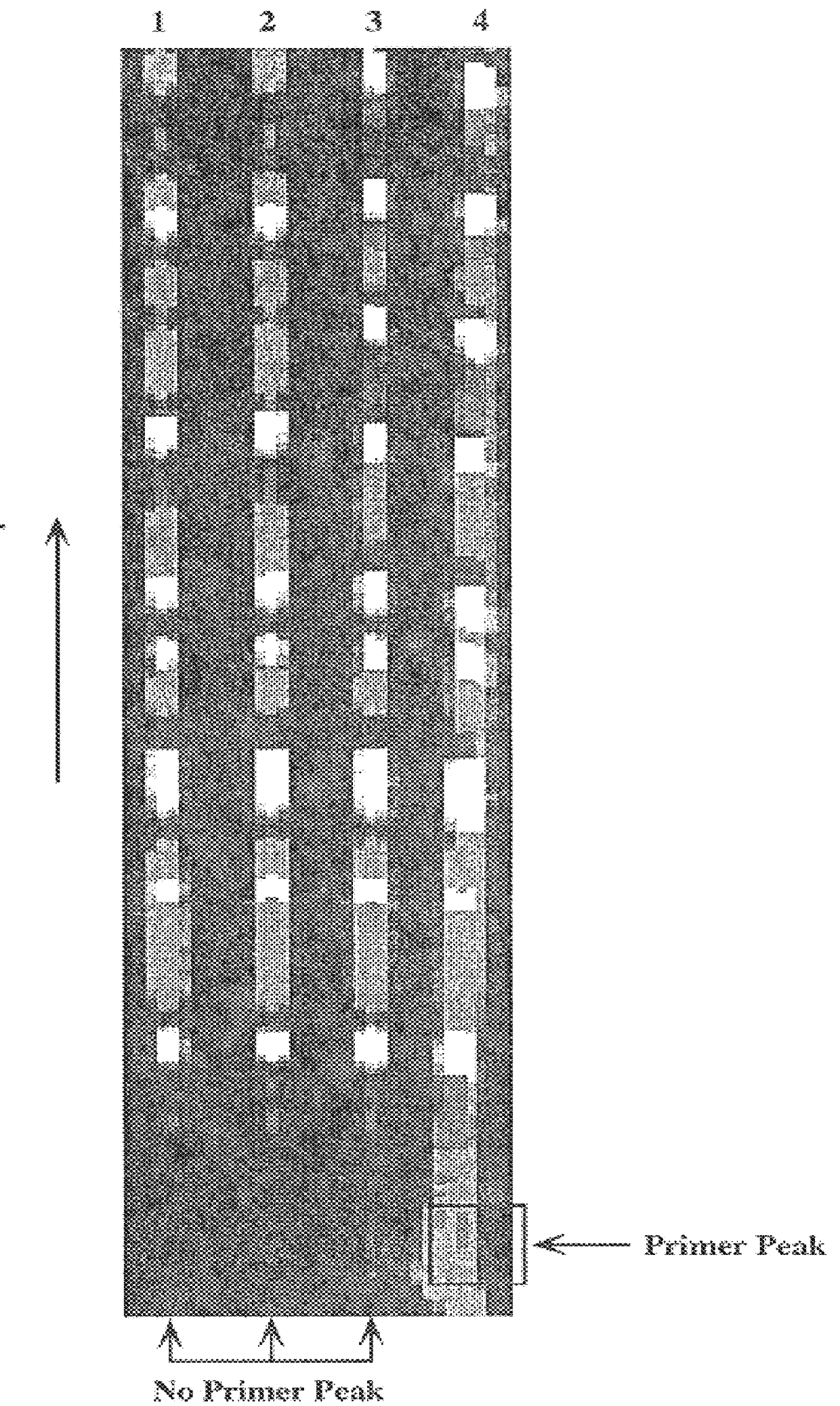
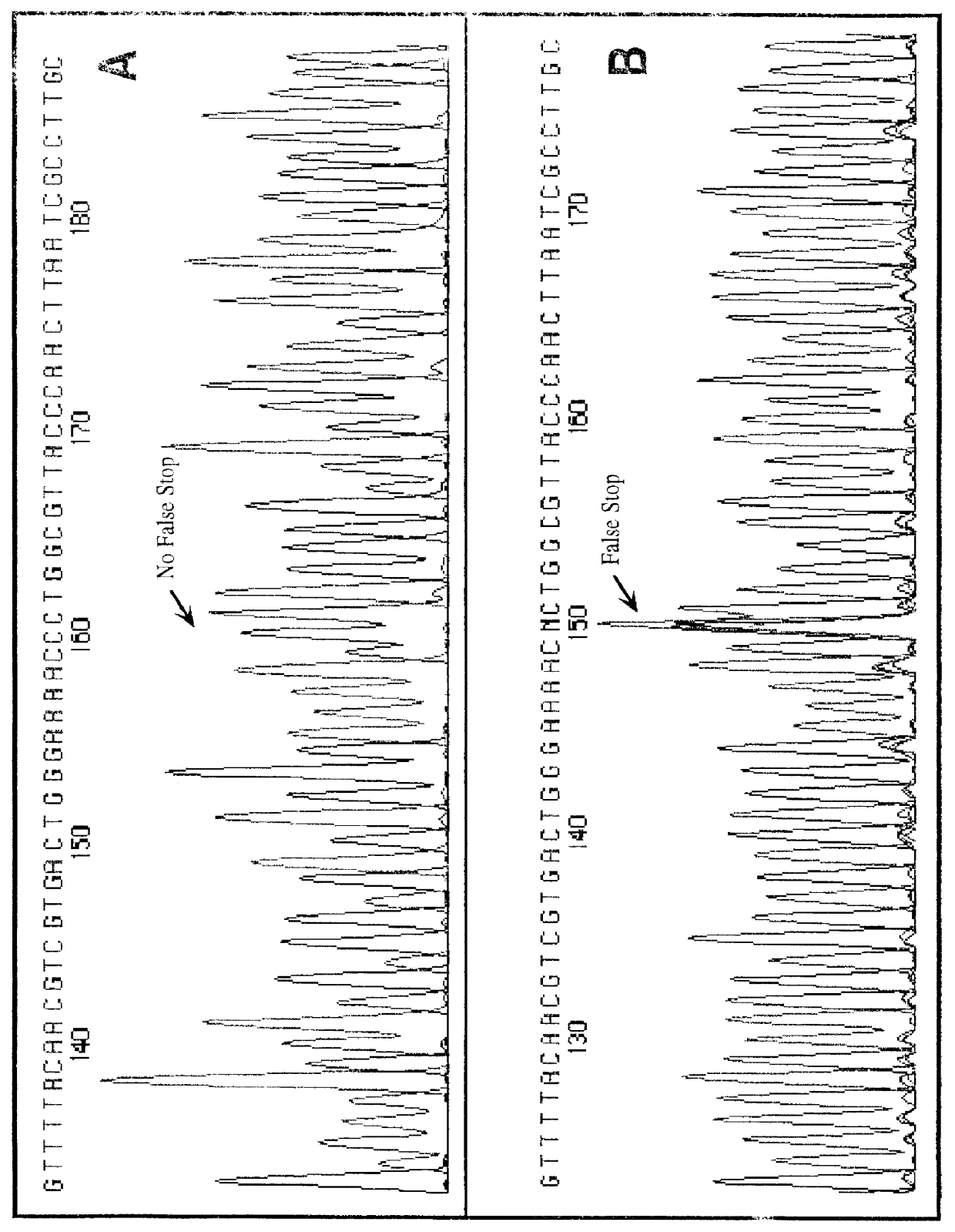
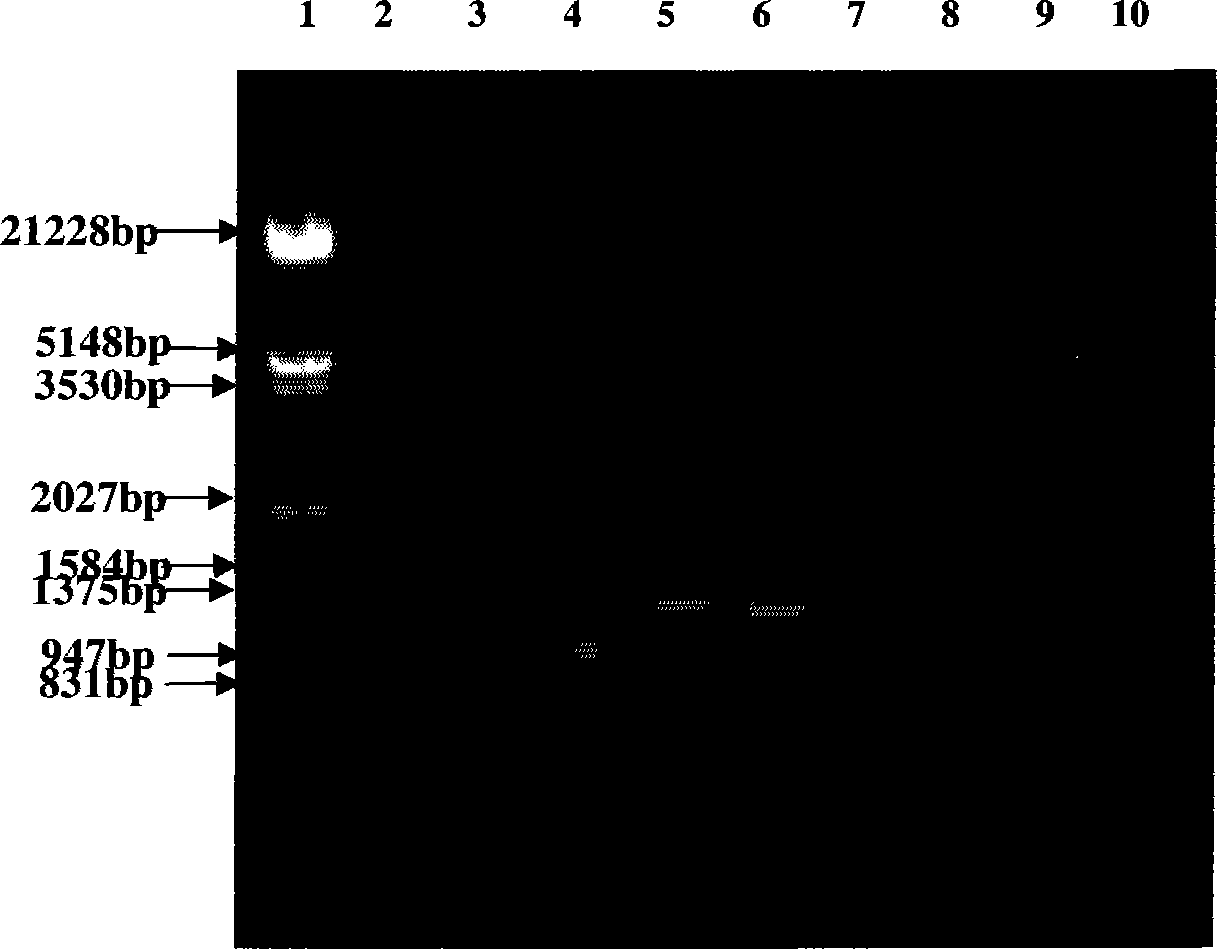
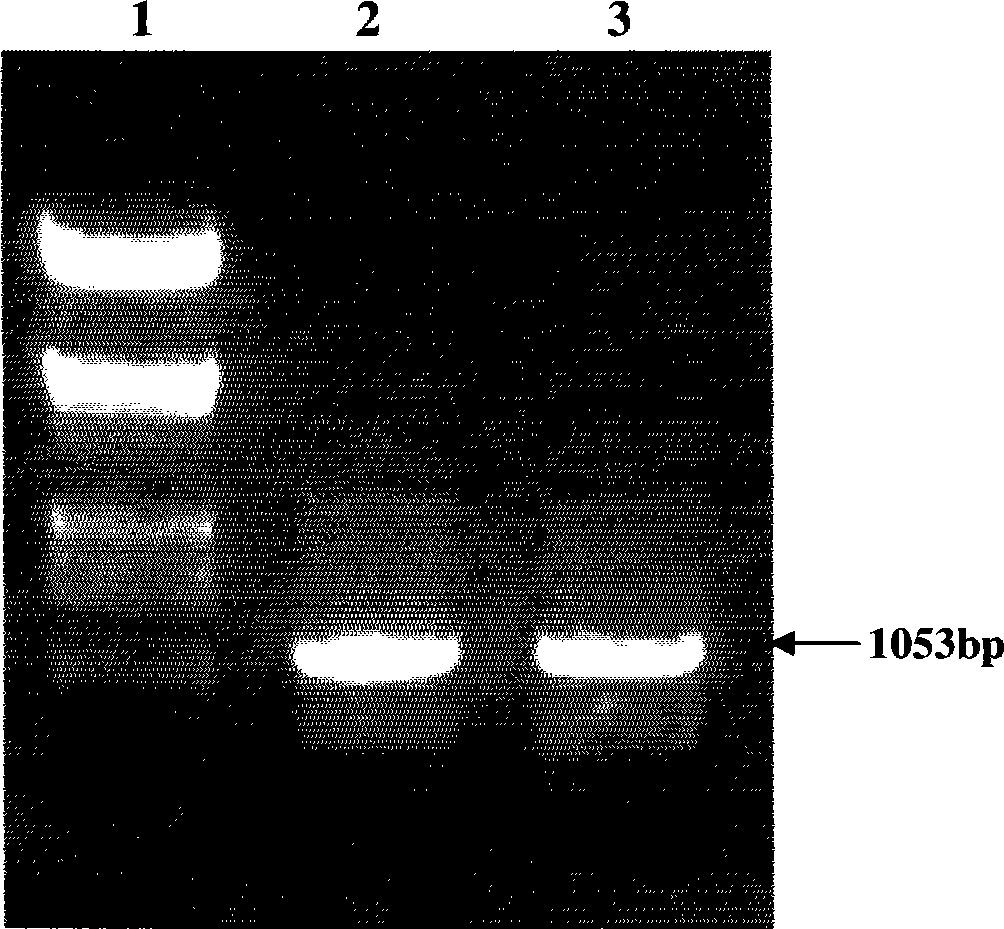
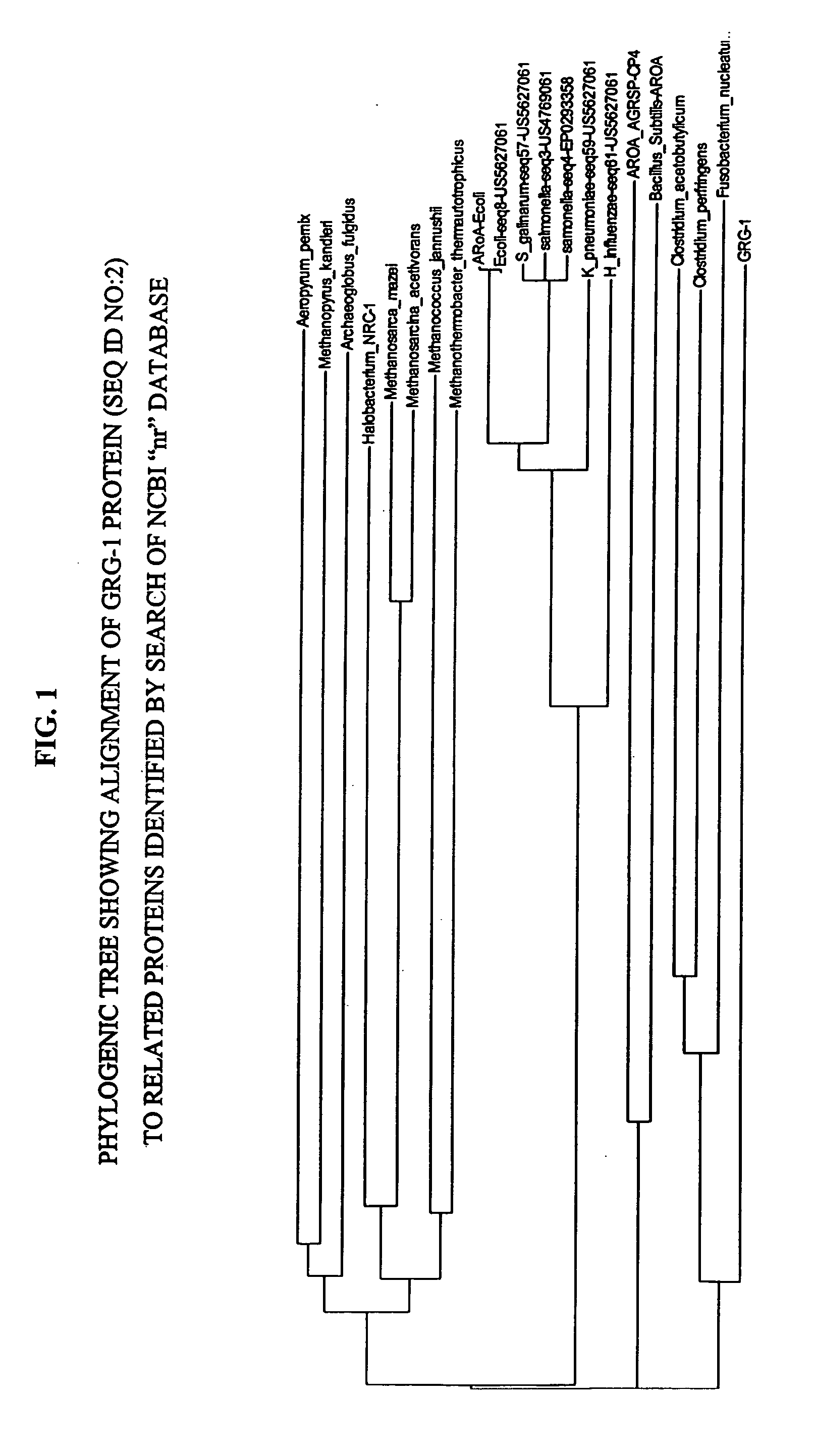
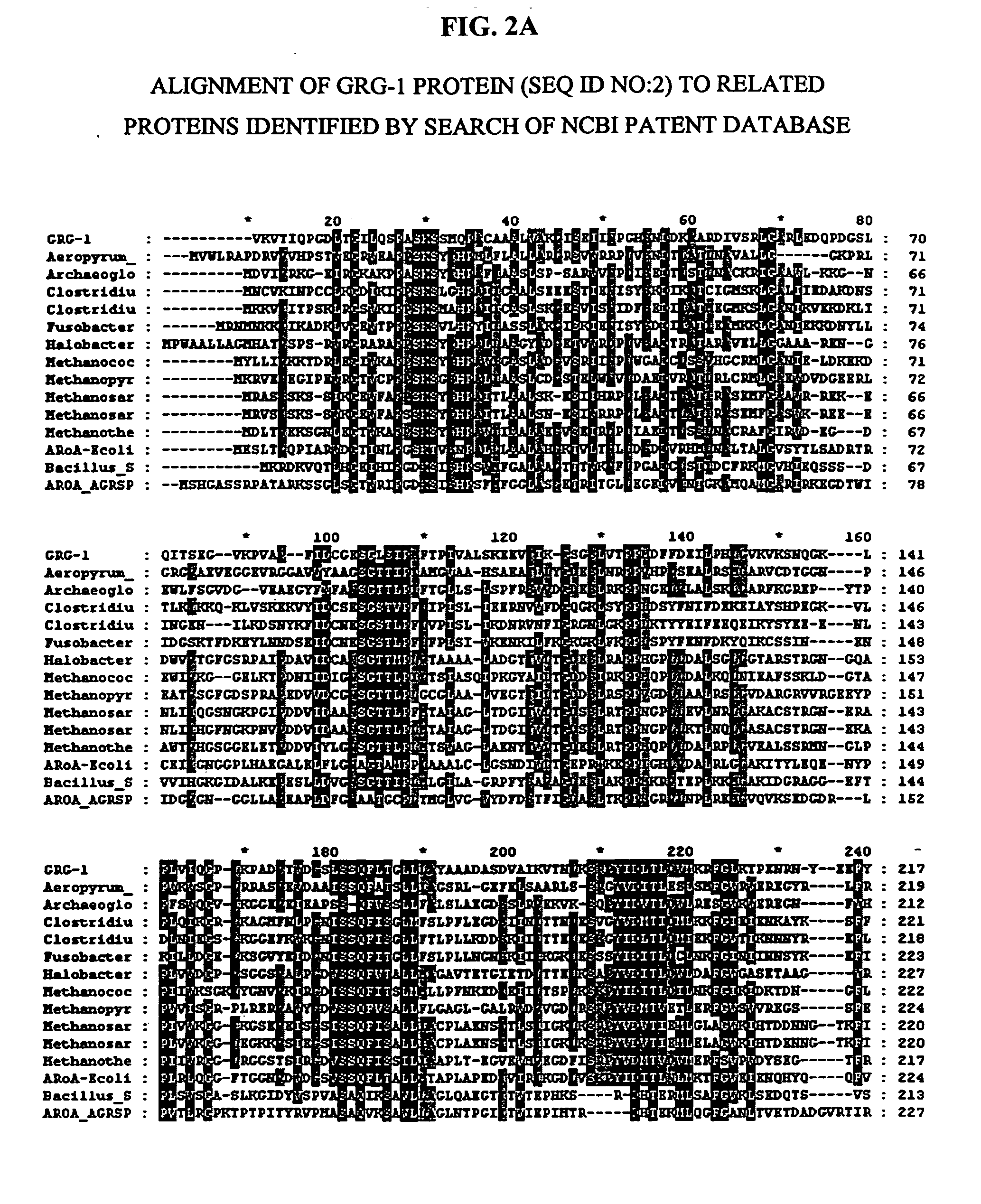
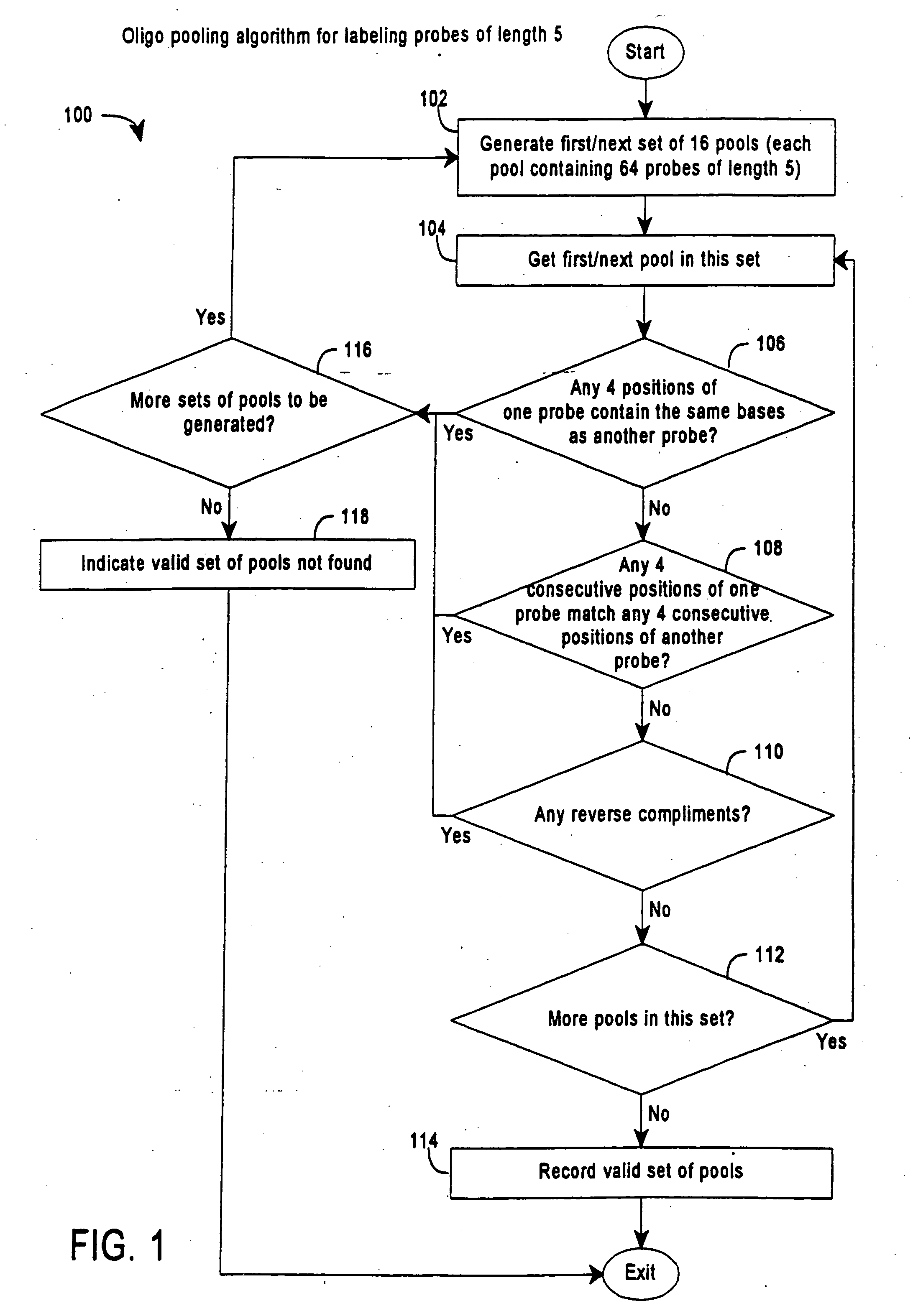
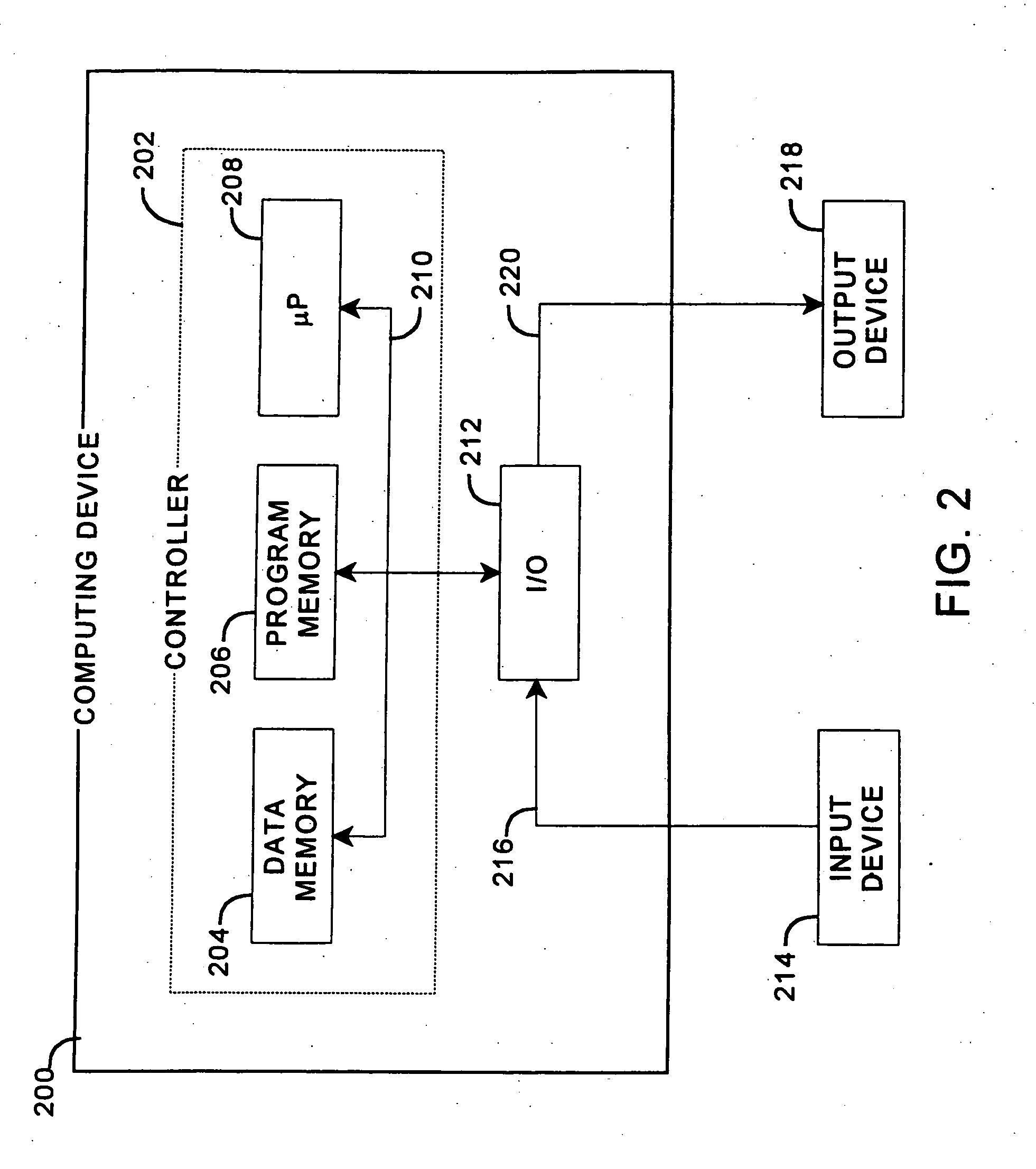
Patents
Literature
6995 results about "Nucleic acid sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
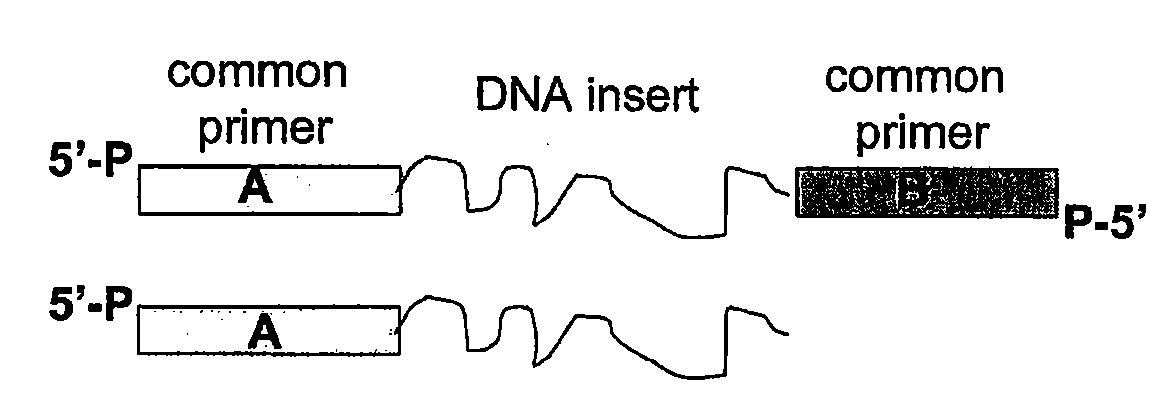
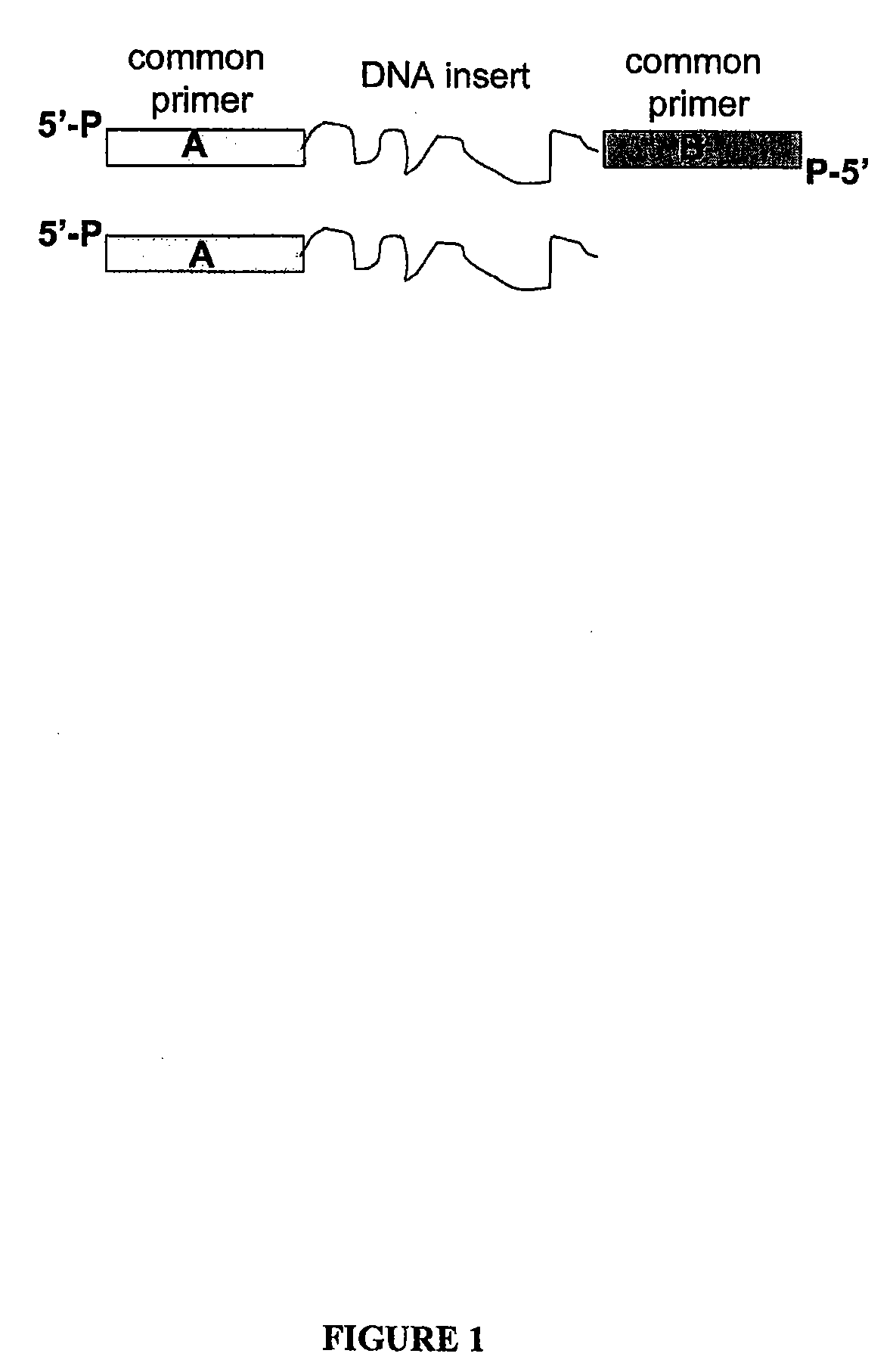
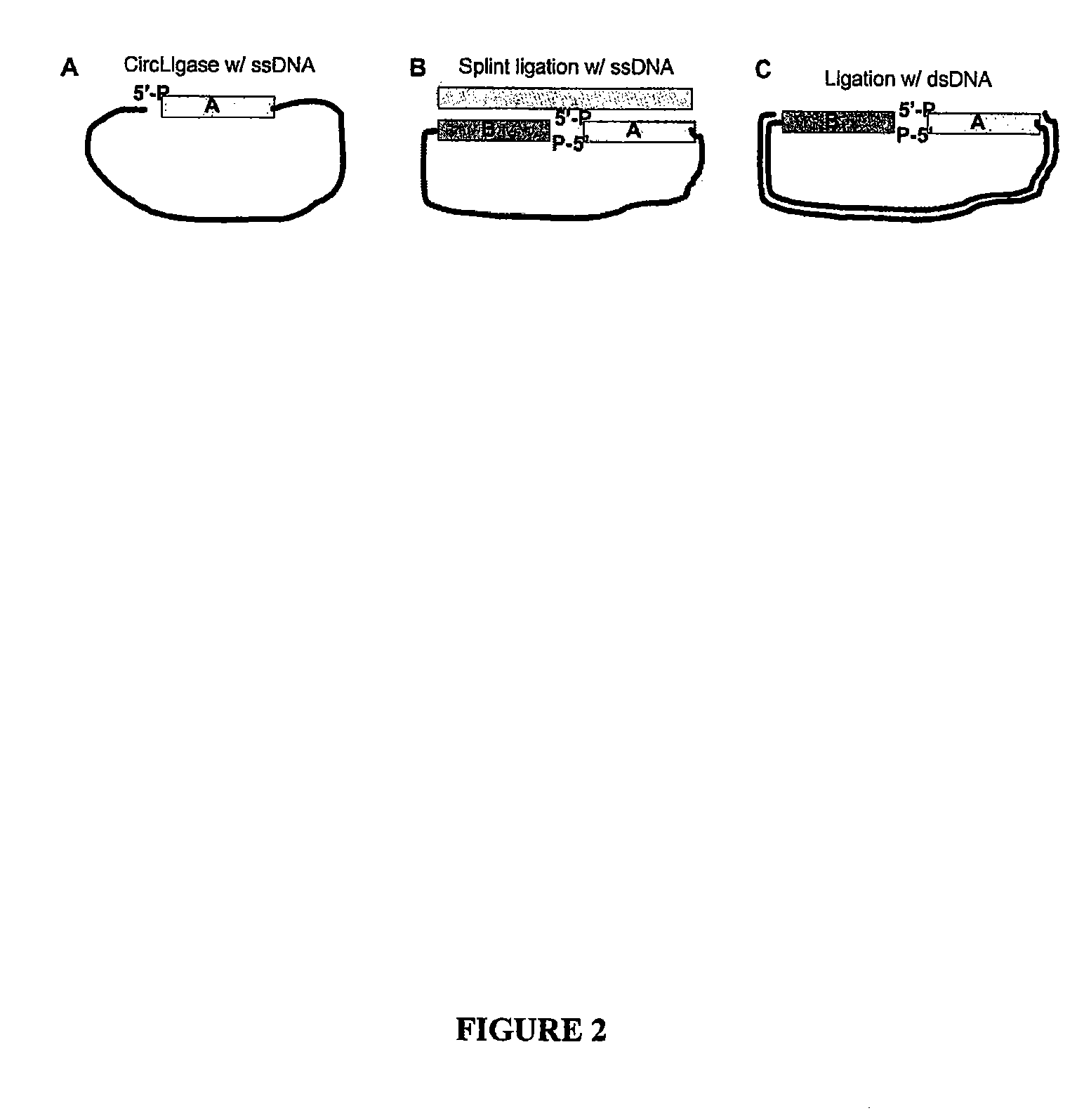
A nucleic acid sequence is a succession of letters that indicate the order of nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule.
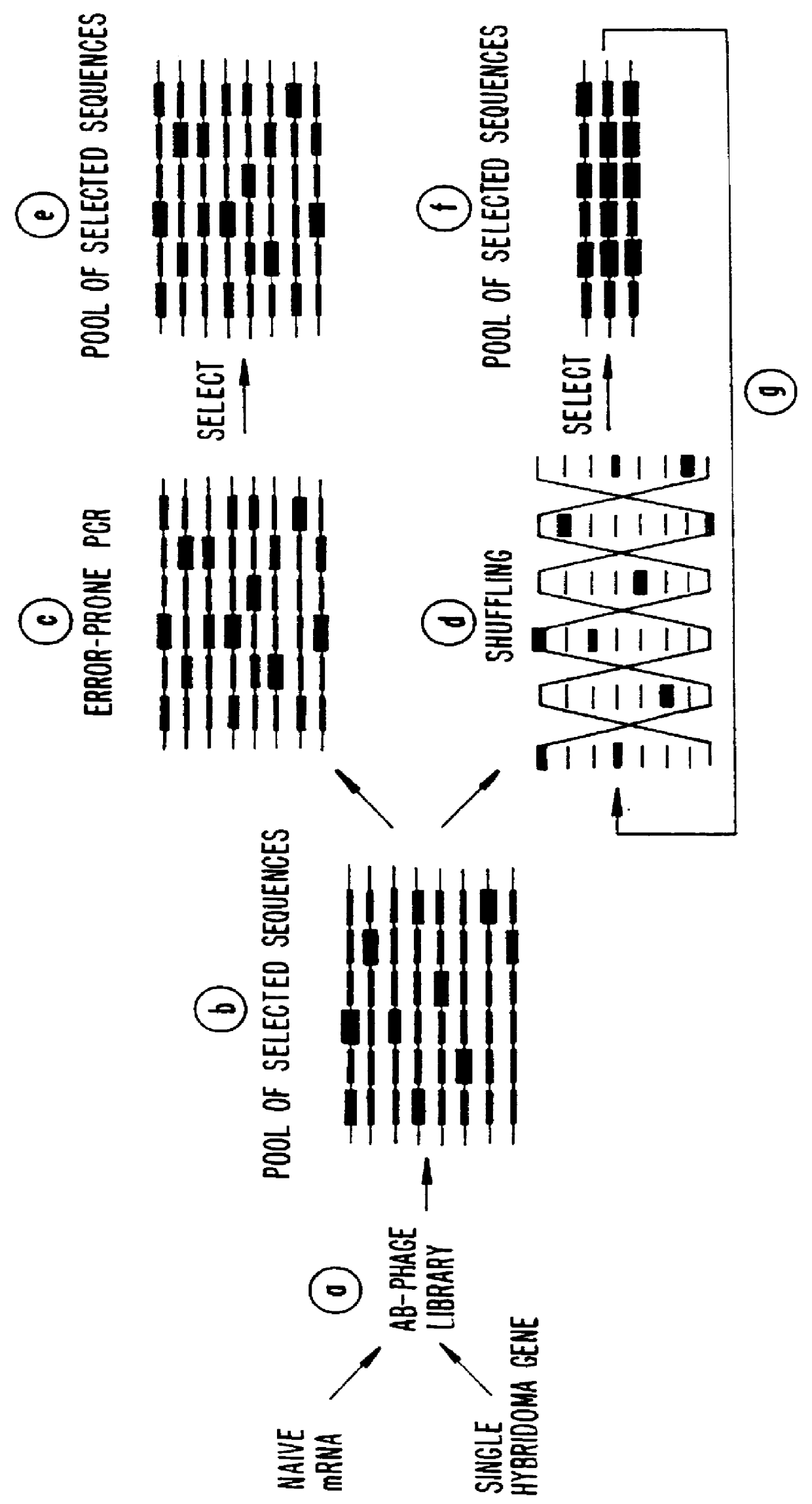
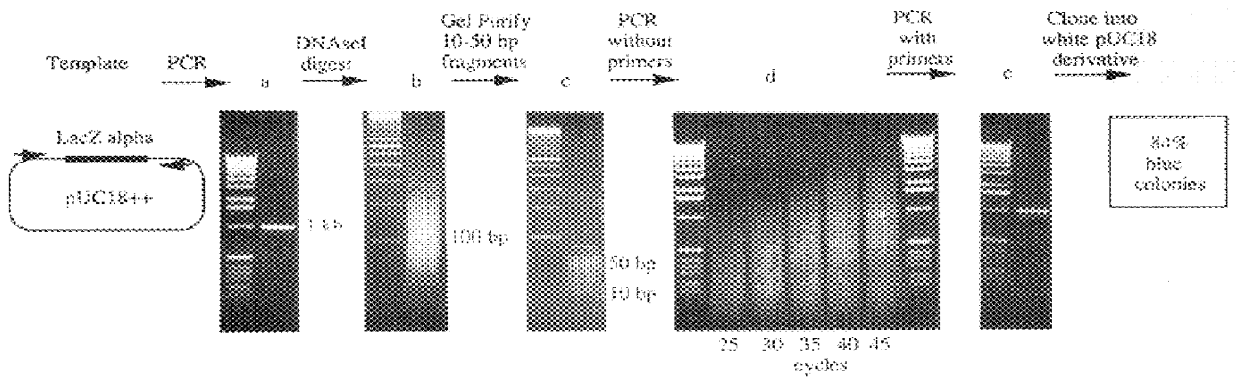
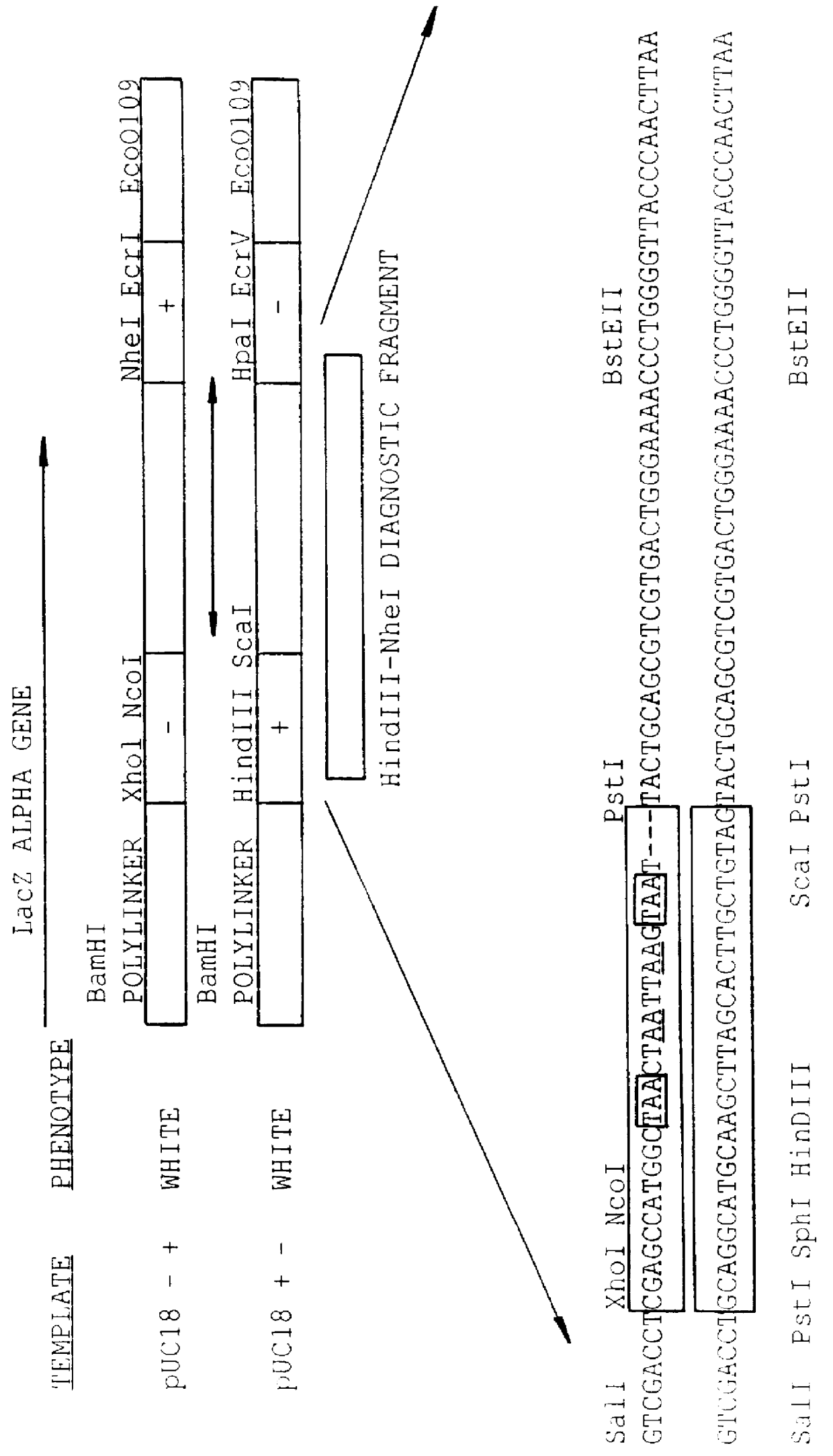
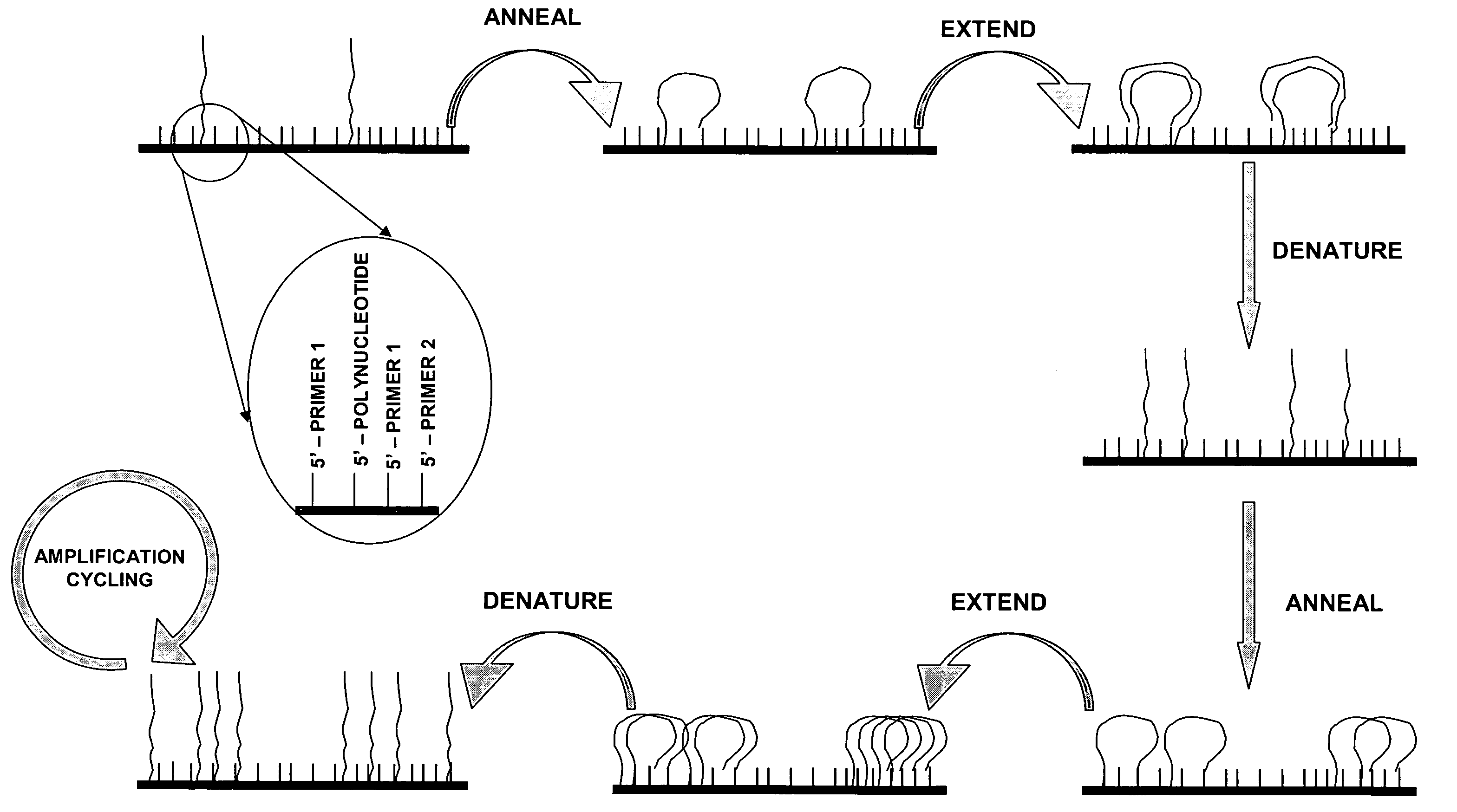
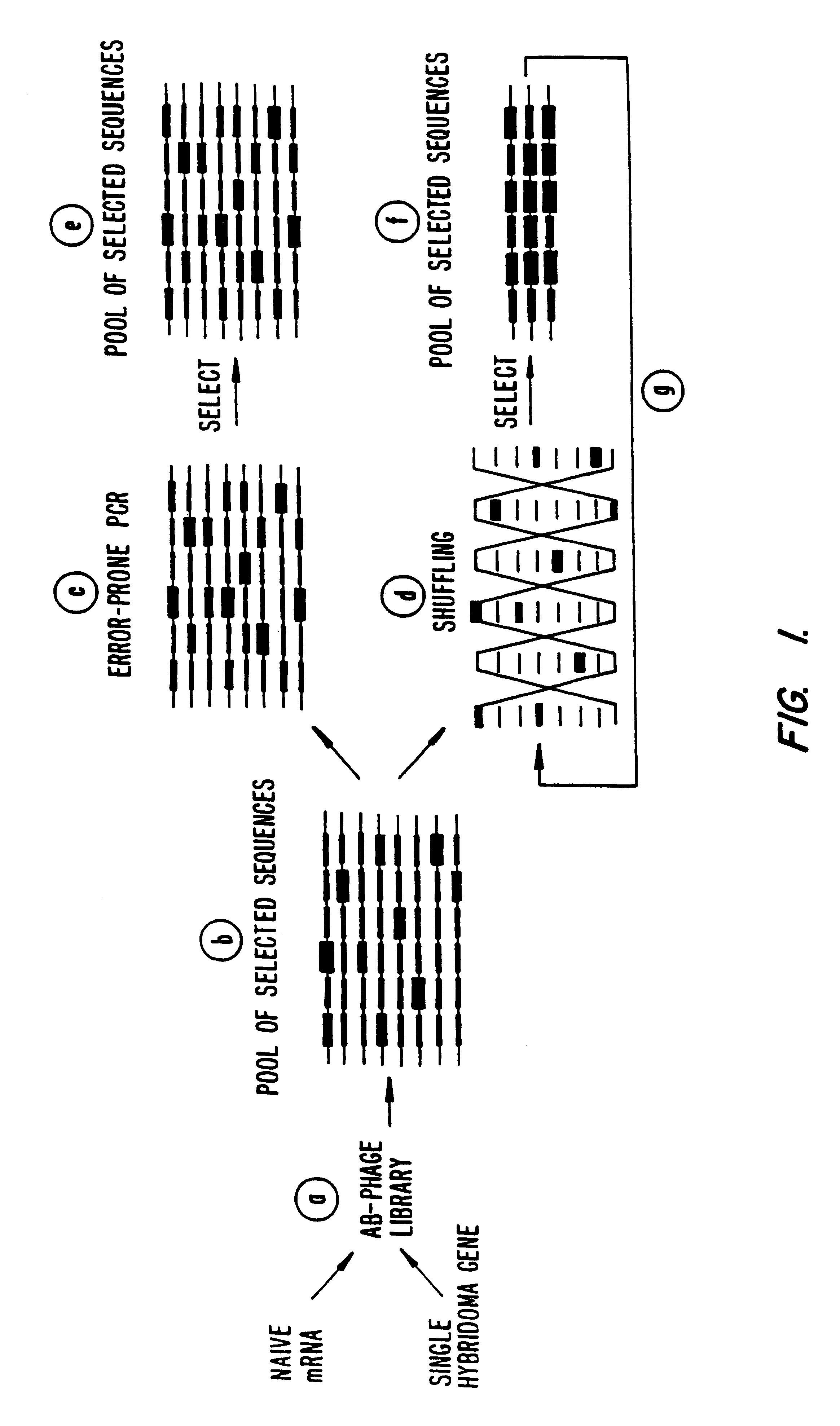
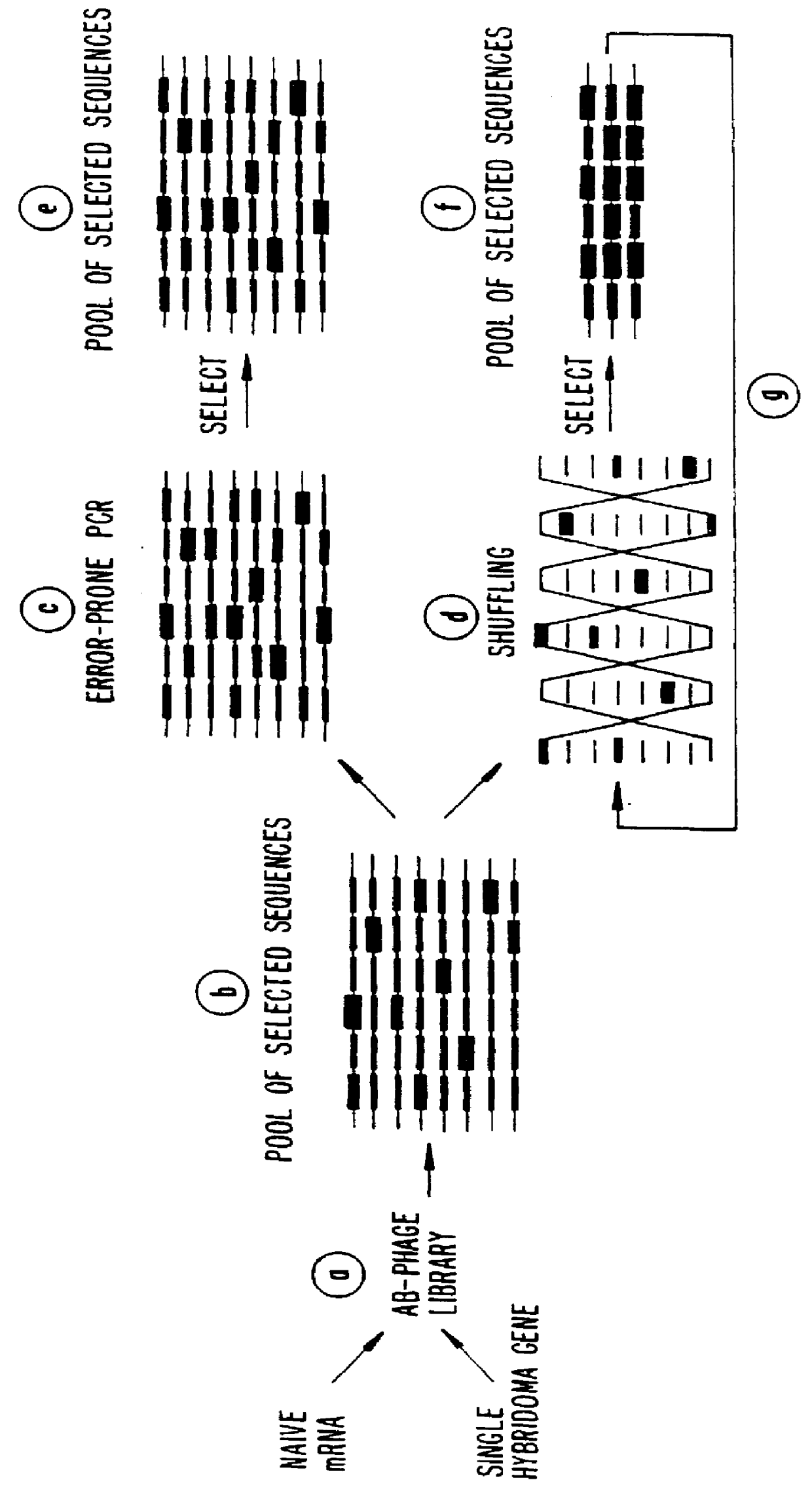
Methods for generating polynucleotides having desired characteristics by iterative selection and recombination
InactiveUS6117679ALess immunogenicLibrary screeningDirected macromolecular evolutionMutated proteinNucleic acid sequencing
A method for DNA reassembly after random fragmentation, and its application to mutagenesis of nucleic acid sequences by in vitro or in vivo recombination is described. In particular, a method for the production of nucleic acid fragments or polynucleotides encoding mutant proteins is described. The present invention also relates to a method of repeated cycles of mutagenesis, shuffling and selection which allow for the directed molecular evolution in vitro or in vivo of proteins.
Owner:CODEXIS MAYFLOWER HLDG LLC
Isothermal methods for creating clonal single molecule arrays
InactiveUS20080009420A1Efficient amplificationIncrease diversityNucleotide librariesMicrobiological testing/measurementMicrobiologyNucleic acid sequencing
The present invention is directed to a method for isothermal amplification of a plurality of different target nucleic acids, wherein the different target nucleic acids are amplified using universal primers and colonies produced thereby can be distinguished from each other. The method, therefore, generates distinct colonies of amplified nucleic acid sequences that can be analyzed by various means to yield information particular to each distinct colony.
Owner:SOLEXA
Methods for increasing accuracy of nucleic acid sequencing
InactiveUS7282337B1Improve accuracyImprove Sequencing AccuracyBioreactor/fermenter combinationsBiological substance pretreatmentsNucleic acid sequencingNucleic acid sequence
Owner:FLUIDIGM CORP
Methods for generating polynucleotides having desired characteristics by iterative selection and recombination
A method for DNA reassembly after random fragmentation, and its application to mutagenesis of nucleic acid sequences by in vitro or in vivo recombination is described. In particular, a method for the production of nucleic acid fragments or polynucleotides encoding mutant proteins is described. The present invention also relates to a method of repeated cycles of mutagenesis, shuffling and selection which allow for the directed molecular evolution in vitro or in vivo of proteins.
Owner:CODEXIS MAYFLOWER HLDG LLC
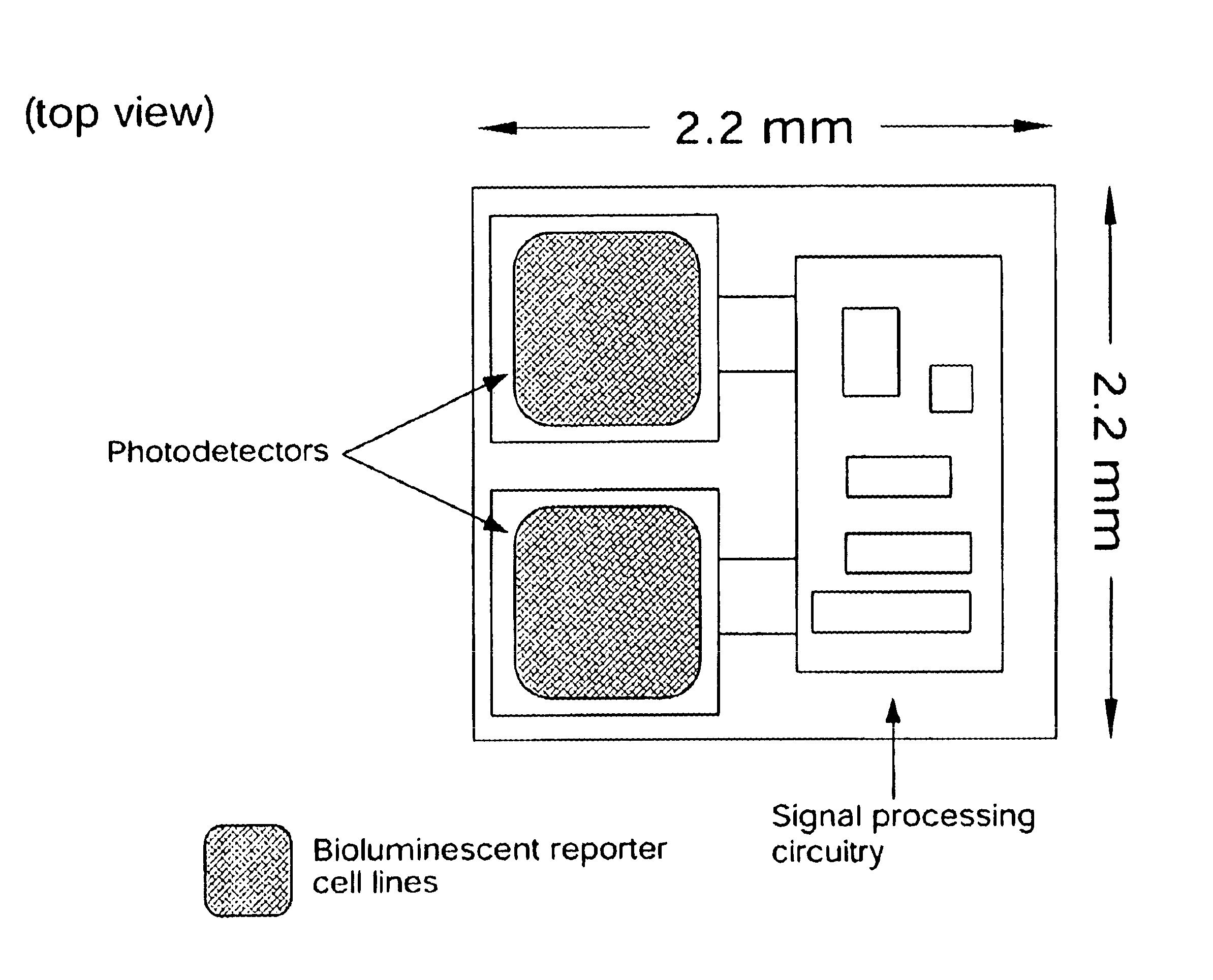
In vivo biosensor apparatus and method of use
InactiveUS6673596B1Less can be administeredCost-effective administration of drugBioreactor/fermenter combinationsBiological substance pretreatmentsIn vivoGenetically engineered
Disclosed are bioluminescent bioreporter integrated circuit devices that detect selected analytes in fluids when implanted in the body of an animal. The device comprises a bioreporter that has been genetically engineered to contain a nucleic acid segment that comprises a cis-activating response element that is responsive to the selected substance operably linked to a gene encoding a bioluminescent reporter polypeptide. In preferred embodiments, the target analyte is glucose, glucagons, or insulin. Exposure of the bioreporter to the target substance causes the response element to up-regulate the nucleic acid sequence encoding the reporter polypeptide to produce a luminescent response that is detected and quantitated. In illustrative embodiments, the bioreporter device is encapsulated on an integrated circuit that is capable of detecting the emitted light, processing the resultant signal, and then remotely reporting the results. Also disclosed are controlled drug delivery systems capable of being directly or indirectly controlled by the detection device that provide drugs such as insulin to the animal in reponse to the amount of target analyte present in the body fluids.
Owner:UNIV OF TENNESSEE RES FOUND +1
Ultra-fast nucleic acid sequencing device and a method for making and using the same
InactiveUS7001792B2Accurate identificationAccurately and effectively identifying bases of DNABioreactor/fermenter combinationsBiological substance pretreatmentsUltra fastNucleic acid sequencing
A system and method employing at least one semiconductor device, or an arrangement of insulating and metal layers, having at least one detecting region which can include, for example, a recess or opening therein, for detecting a charge representative of a component of a polymer, such as a nucleic acid strand, proximate to the detecting region, and a method for manufacturing such a semiconductor device. The system and method can thus be used for sequencing individual nucleotides or bases of ribonucleic acid (RNA) or deoxyribonucleic acid (DNA). The semiconductor device includes at least two doped regions, such as two n-typed regions implanted in a p-typed semiconductor layer or two p-typed regions implanted in an n-typed semiconductor layer. The detecting region permits a current to pass between the two doped regions in response to the presence of the component of the polymer, such as a base of a DNA or RNA strand. The current has characteristics representative of the component of the polymer, such as characteristics representative of the detected base of the DNA or RNA strand.
Owner:LIFE TECH CORP
Methods for generating polynucleotides having desired characteristics by iterative selection and recombination
InactiveUS6180406B1Less immunogenicLibrary screeningDirected macromolecular evolutionMutated proteinNucleic acid sequencing
A method for DNA reassembly after random fragmentation, and its application to mutagenesis of nucleic acid sequences by in vitro or in vivo recombination is described. In particular, a method for the production of nucleic acid fragments or polynucleotides encoding mutant proteins is described. The present invention also relates to a method of repeated cycles of mutagenesis, shuffling and selection which allow for the directed molecular evolution in vitro or in vivo of proteins.
Owner:CODEXIS MAYFLOWER HLDG LLC
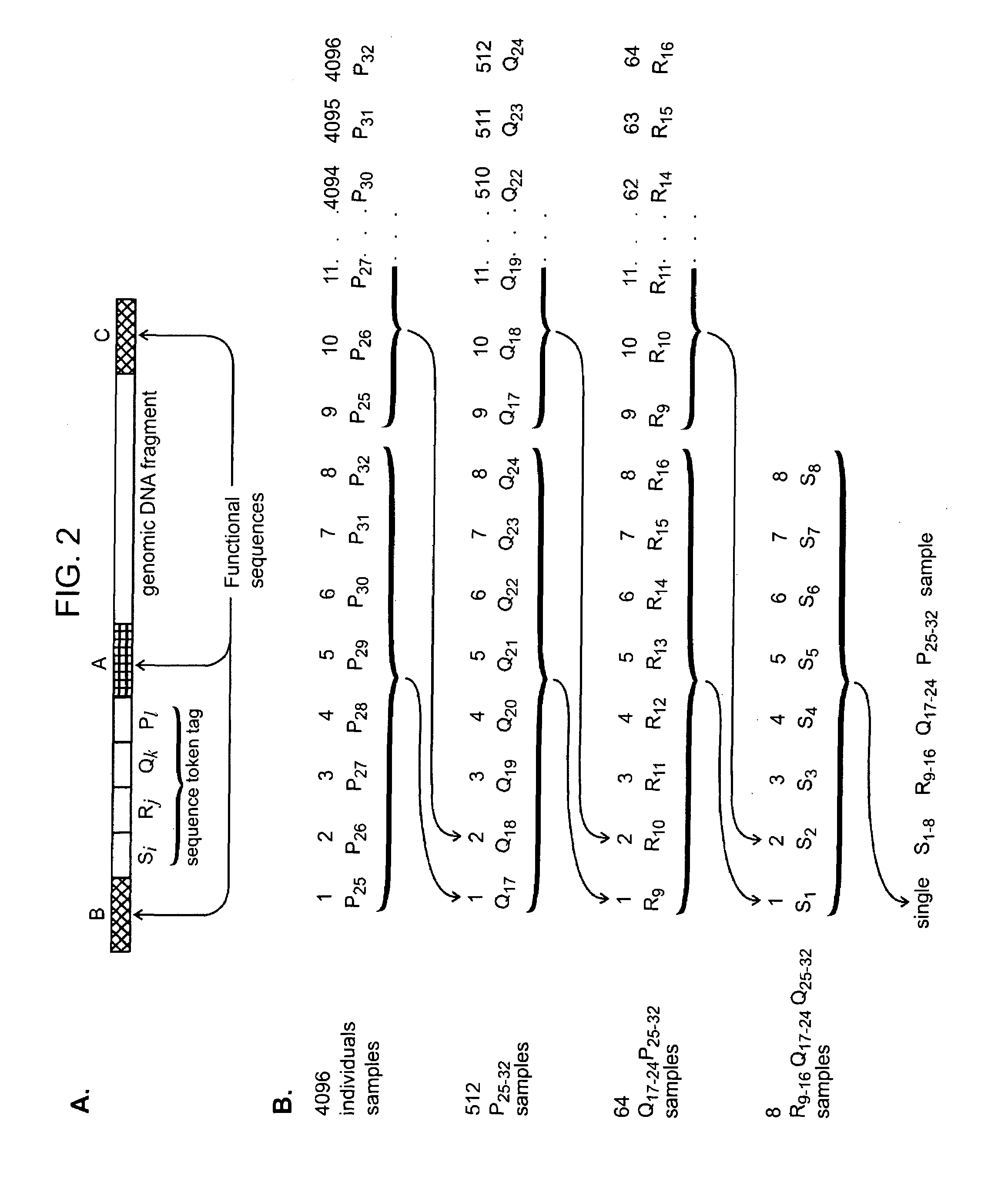
Nucleic acid analysis using sequence tokens
ActiveUS7544473B2Efficiently determine variations in nucleotide sequences in the associated nucleic acid sequence fragmentsBioreactor/fermenter combinationsBiological substance pretreatmentsDon't repeat yourselfNucleic acid sequencing
The present invention provides methods and compositions for tagging nucleic acid sequence fragments, e.g., a set of nucleic acid sequence fragments from a single genome, with one or more unique members of a collection of oligonucleotide tags, or sequence tokens, which, in turn, can be identified using a variety of readout platforms. As a general rule, a given sequence token is used once and only once in any tag sequence. In addition, the present invention also provides methods for using the sequence tokens to efficiently determine variations in nucleotide sequences in the associated nucleic acid sequence fragments.
Owner:PERSONAL GENOME DIAGNOSTICS INC
Methods for generating polynucleotides having desired characteristics by iterative selection and recombination
InactiveUS6165793ALess immunogenicDirected macromolecular evolutionImmunoglobulinsMutated proteinNucleotide
A method for DNA reassembly after random fragmentation, and its application to mutagenesis of nucleic acid sequences by in vitro or in vivo recombination is described. In particular, a method for the production of nucleic acid fragments or polynucleotides encoding mutant proteins is described. The present invention also relates to a method of repeated cycles of mutagenesis, shuffling and selection which allow for the directed molecular evolution in vitro or in vivo of proteins.
Owner:CODEXIS MAYFLOWER HLDG LLC
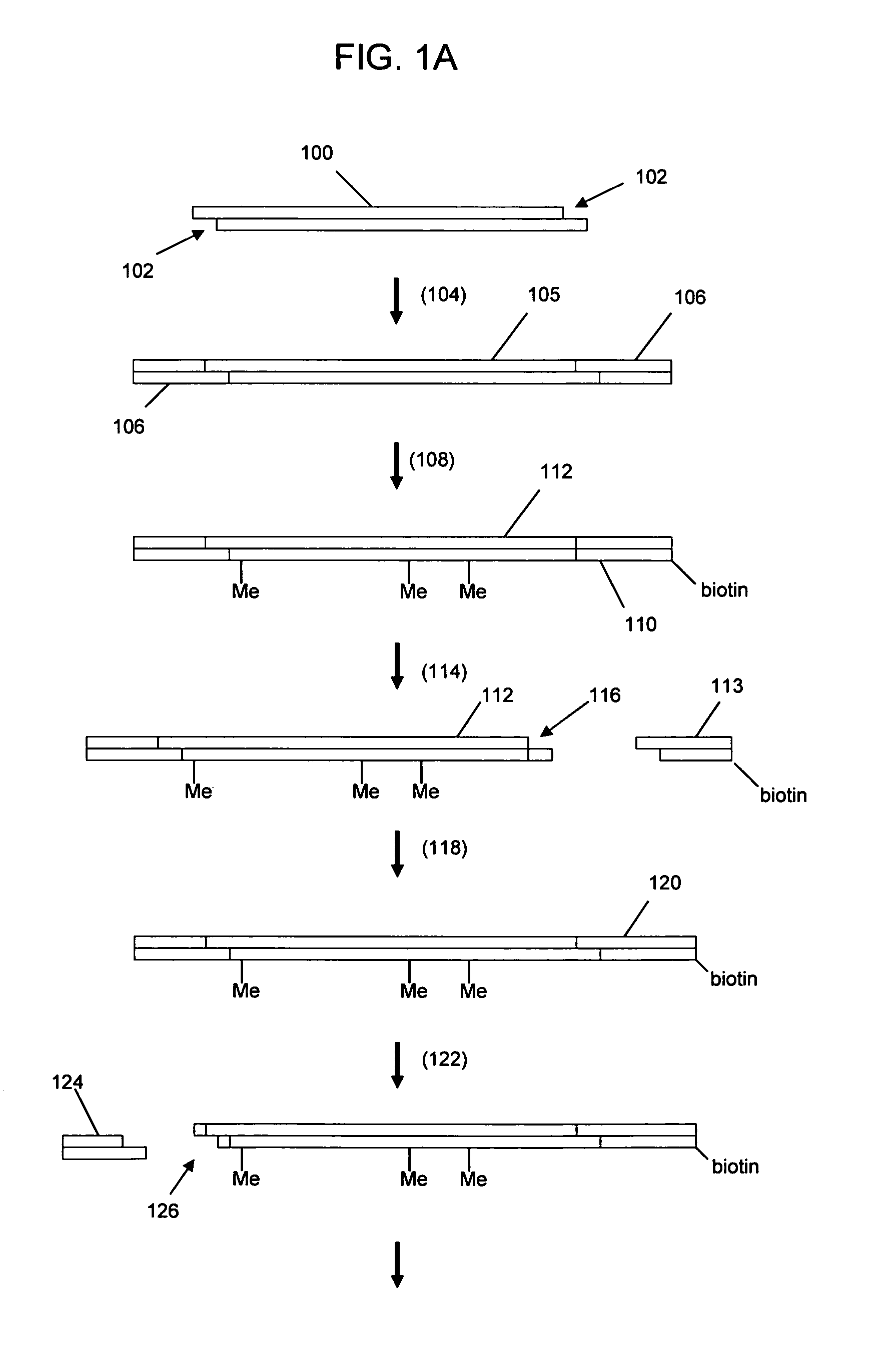
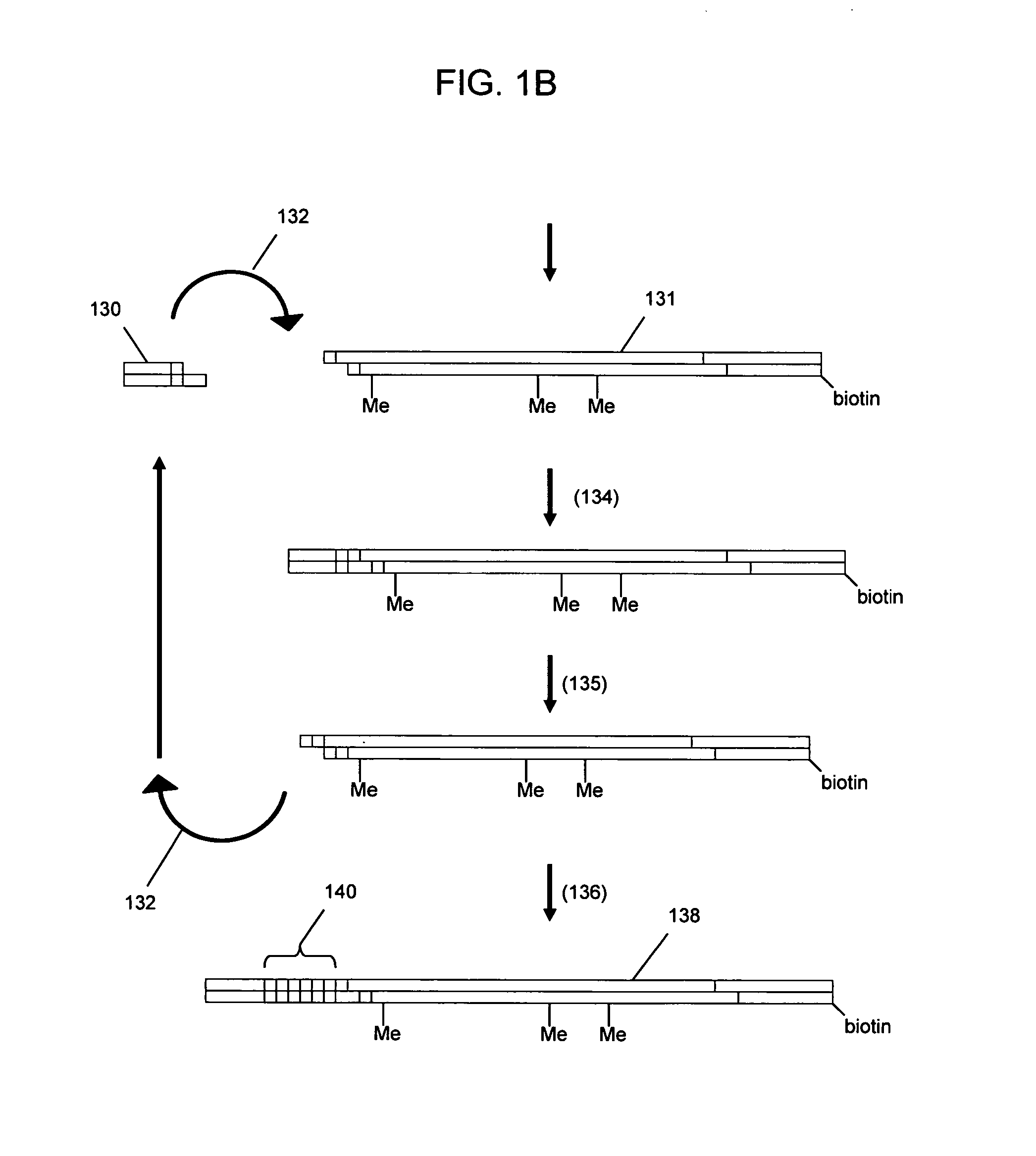
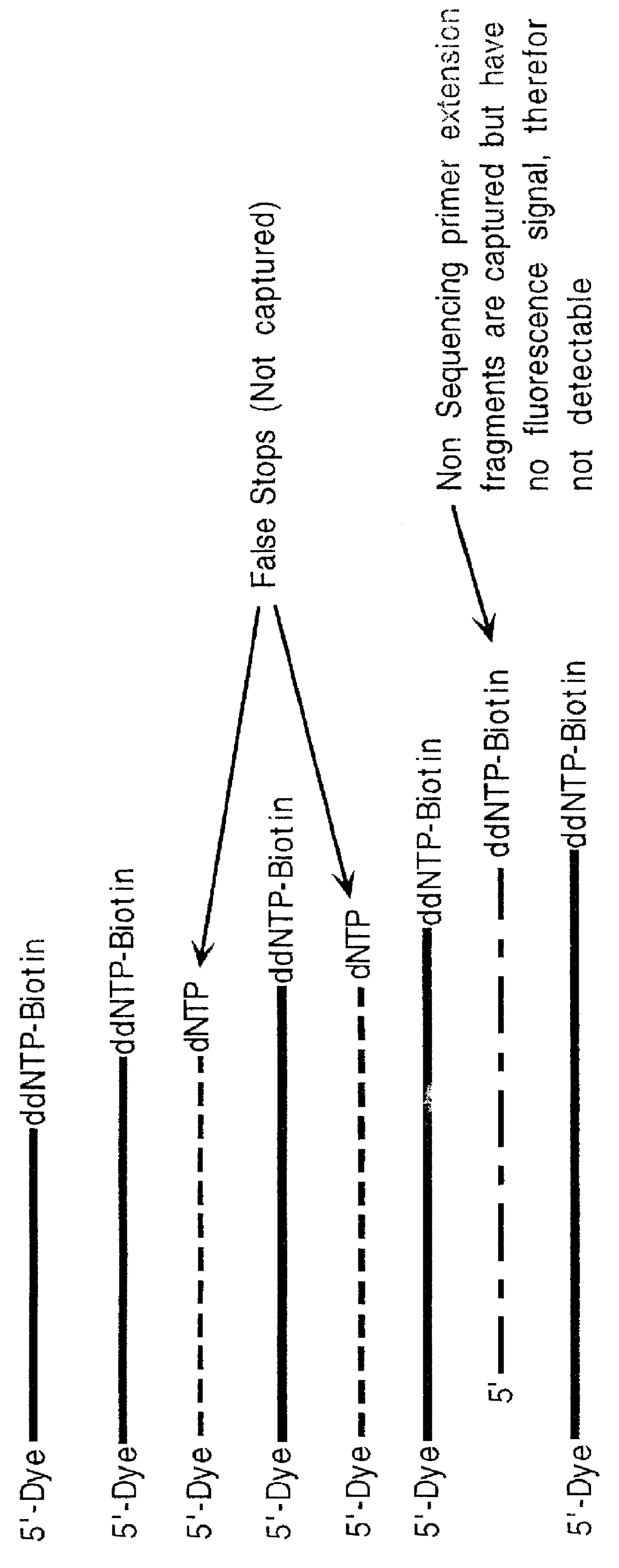
Nucleic acid sequencing with solid phase capturable terminators comprising a cleavable linking group
InactiveUS6046005ABioreactor/fermenter combinationsBiological substance pretreatmentsNucleic acid sequencingNucleic acid sequence
Methods of enzymatic nucleic acid sequencing are provided in which solid-phase capturable chain terminators are employed. In the subject methods, sequencing fragments are generated, where the fragments comprise capturable chain terminators. The fragments are then captured on a solid phase and separated from the remaining components of the sequencing reaction. The fragments are then released from the solid phase, size separated and detected to yield sequencing data from which the sequence of the nucleic acid is determined.
Owner:INCYTE PHARMA INC
Seed specific highly effective promoter and its application
The invention discloses a special promoter separated from millet, expressing carrier with nucleic acid sequence of SEQ ID No. 1 host with the expressing carrier and appliance of the promoter, which is characterized by the following: utilizing Tail-PCR (colored body step moving method); getting the special promoter from gene group DNA; possessing nucleic acid sequence of SEQ ID No. 1; ;linking downstream of the promoter to non-homologous or homologous gene; constructing plant expressing carrier; transferring host plant; driving the downstream gene to high effective and special express goal protein in the seed; realizing genetic modification of plant; or using as effective tool for studying plant and biological reactor.
Owner:CHINA AGRI UNIV
Seed specificity highly effective promoter and its application
InactiveCN101063139AReduce adverse effectsFermentationVector-based foreign material introductionHeterologousNucleotide
The invention discloses a special promoter separated from millet, expressing carrier with nucleic acid sequence of SEQ ID No. 1 host with the expressing carrier and appliance of the promoter, which is characterized by the following: utilizing Tail-PCR (colored body step moving method); getting the special promoter from gene group DNA; possessing nucleic acid sequence of SEQ ID No. 1; ;linking downstream of the promoter to non-homologous or homologous gene; constructing plant expressing carrier; transferring host plant; driving the downstream gene to high effective and special express goal protein in the seed; realizing genetic modification of plant; or using as effective tool for studying plant and biological reactor.
Owner:CHINA AGRI UNIV
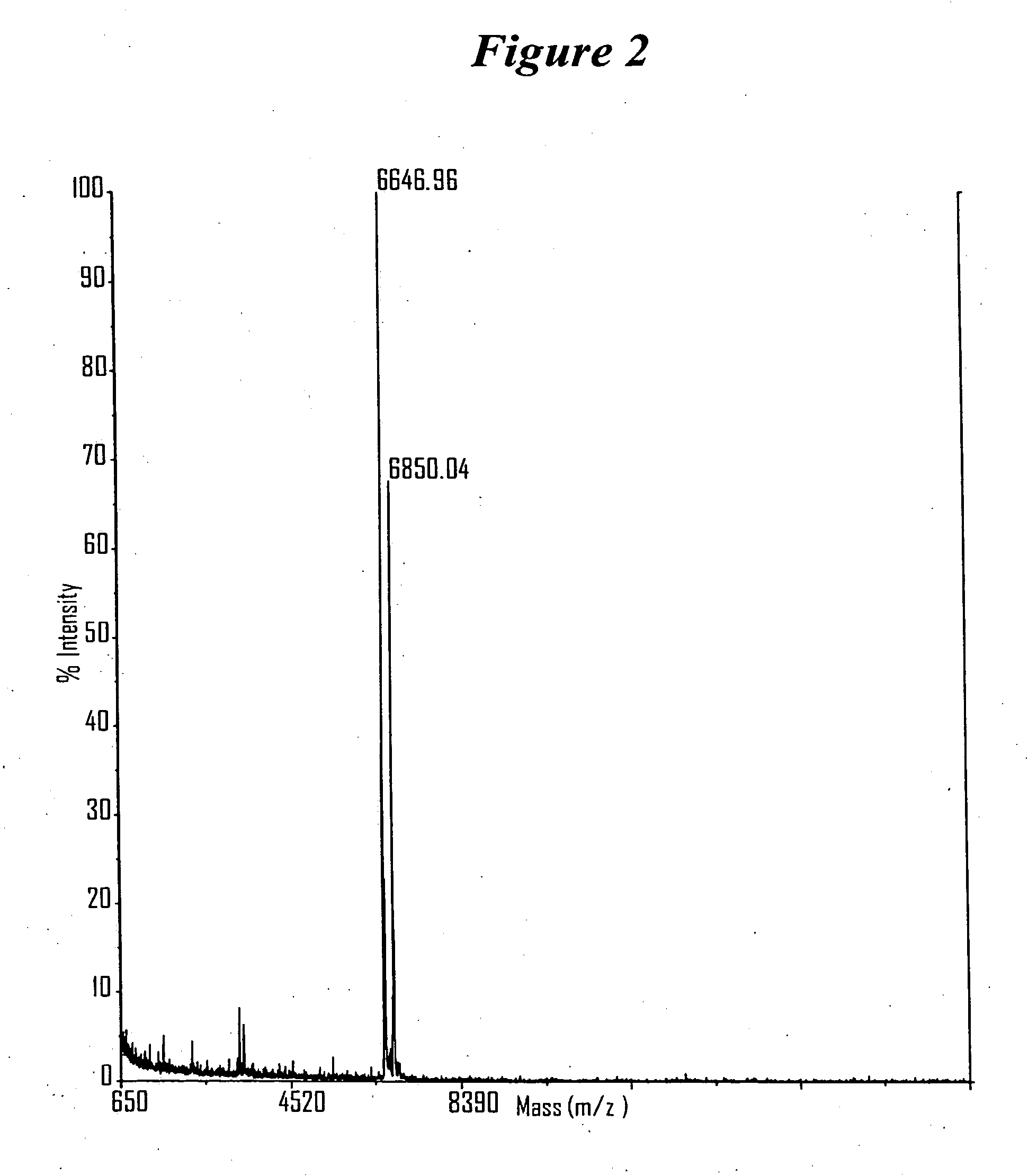
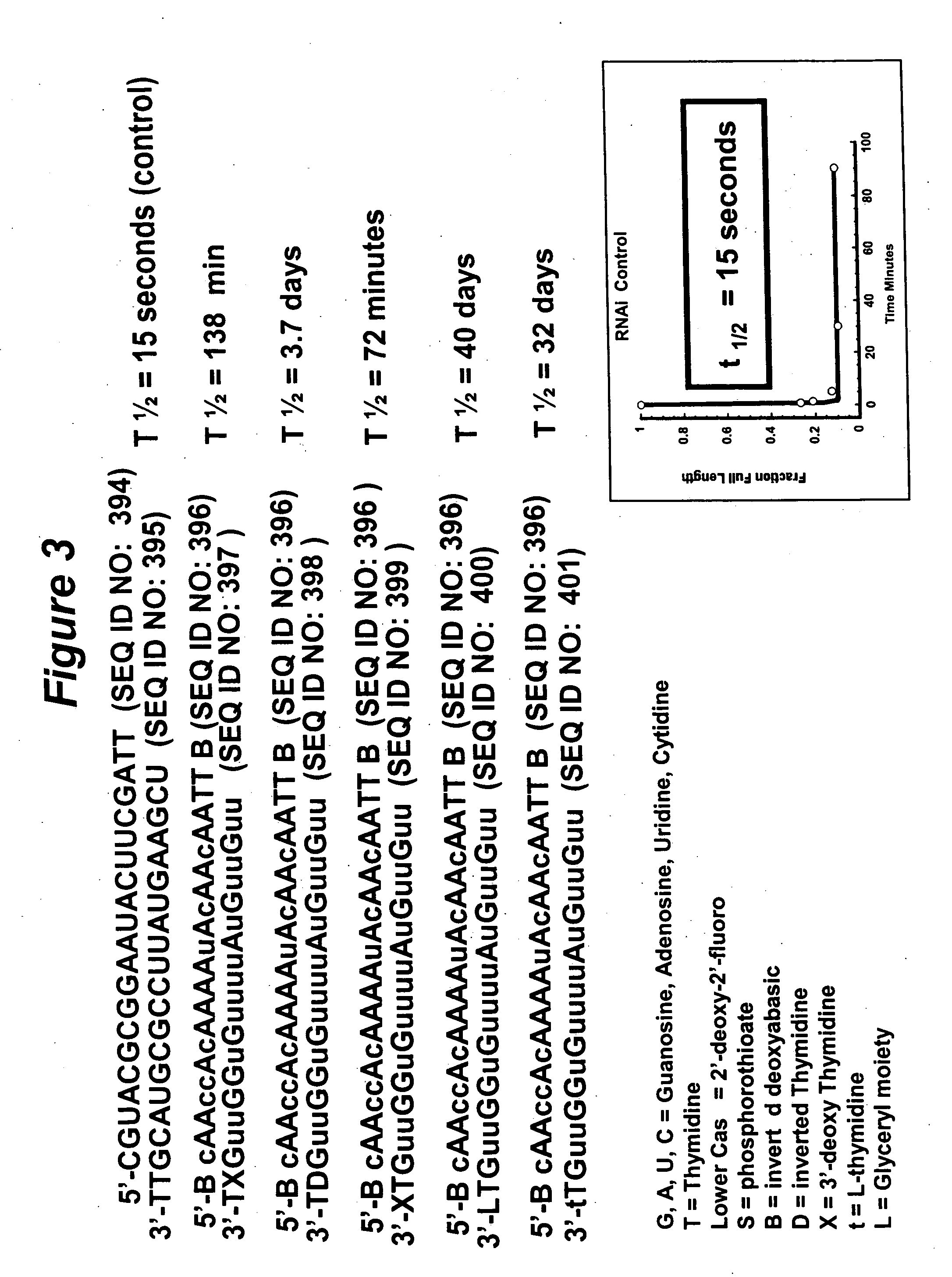
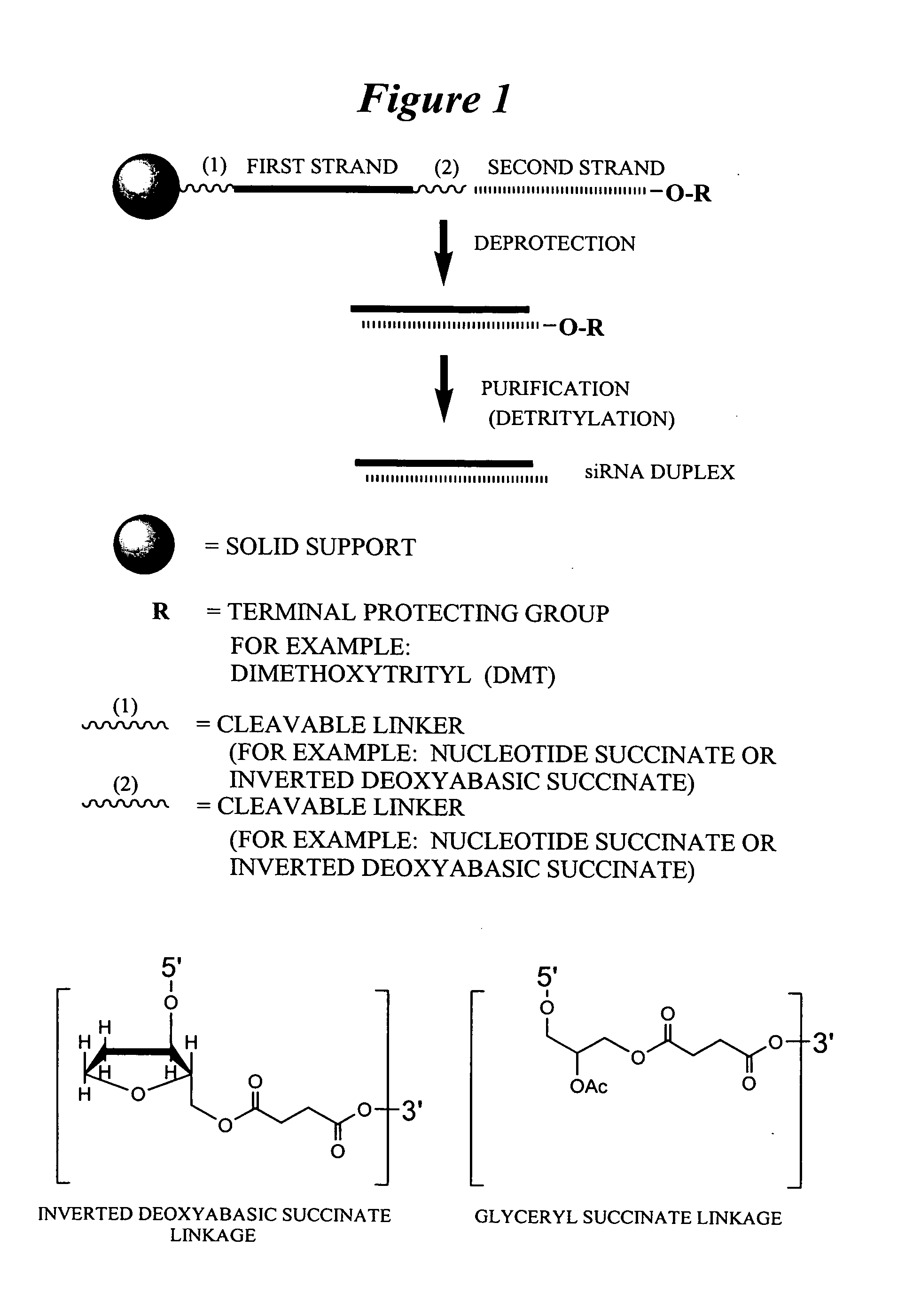
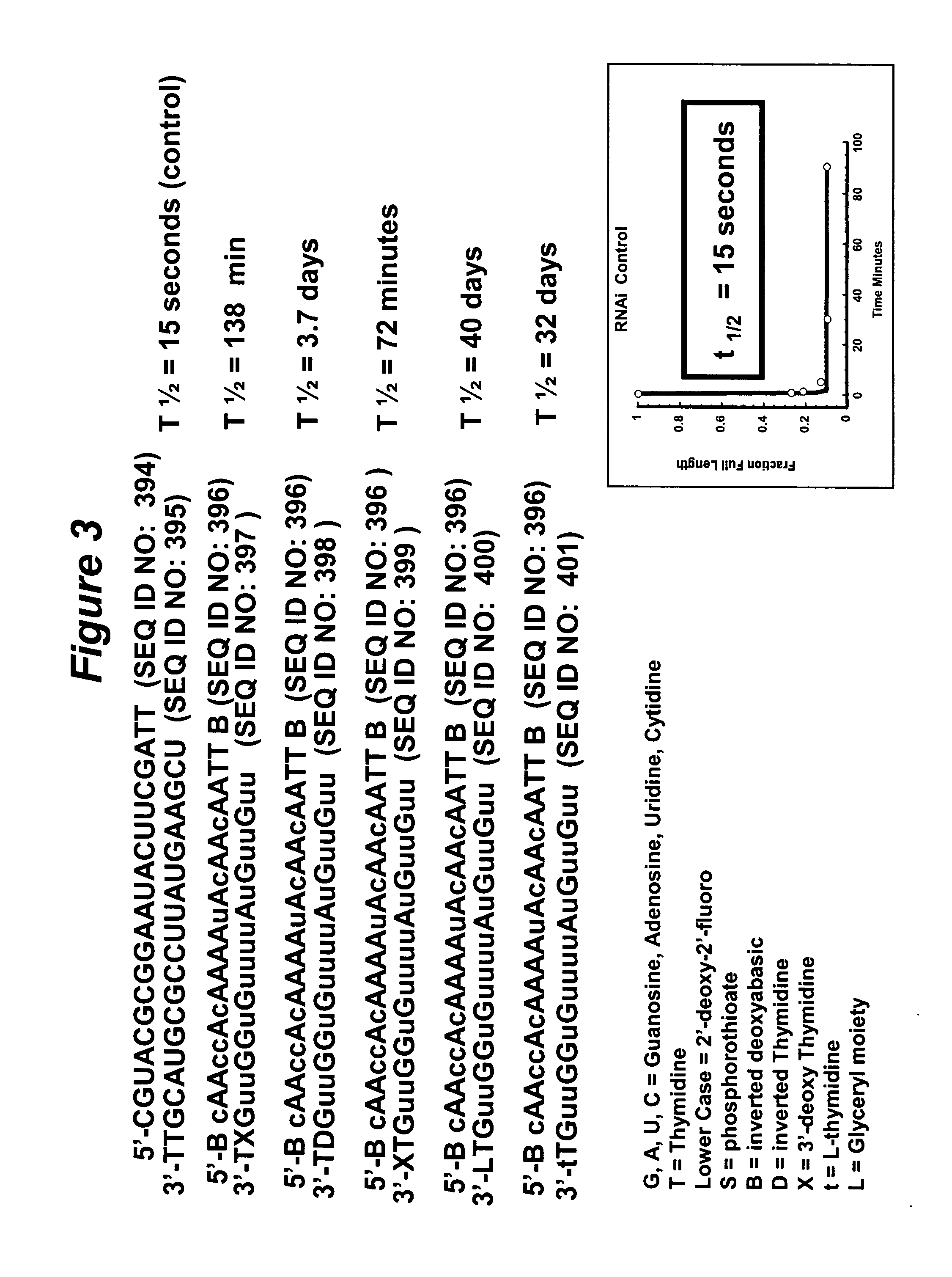
RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA)
InactiveUS20050020525A1Improve various propertyModulate its functionCompounds screening/testingSugar derivativesDouble strandOrganism
The present invention concerns methods and reagents useful in modulating gene expression in a variety of applications, including use in therapeutic, diagnostic, target validation, and genomic discovery applications. Specifically, the invention relates to synthetic chemically modified small nucleic acid molecules, such as short interfering nucleic acid (siNA), short interfering RNA (siRNA), double-stranded RNA (dsRNA), micro-RNA (miRNA), and short hairpin RNA (shRNA) molecules capable of mediating RNA interference (RNAi) against target nucleic acid sequences. The small nucleic acid molecules are useful in the treatment of any disease or condition that responds to modulation of gene expression or activity in a cell, tissue, or organism.
Owner:SIMA THERAPEUTICS ICN
Polypeptide compositions toxic to coleopteran insects
InactiveUS6063597AHigh expressionStably occupyBiocideSugar derivativesBiotechnologyNucleic acid sequencing
Disclosed are Coleopteran-toxic B. thuringiensis delta -endotoxins, nucleic acid sequences, and transgenic plants expressing these genes. Methods of making and using these genes and proteins are disclosed as well as methods for the recombinant expression, and transformation of suitable host cells.
Owner:MONSANTO TECH LLC
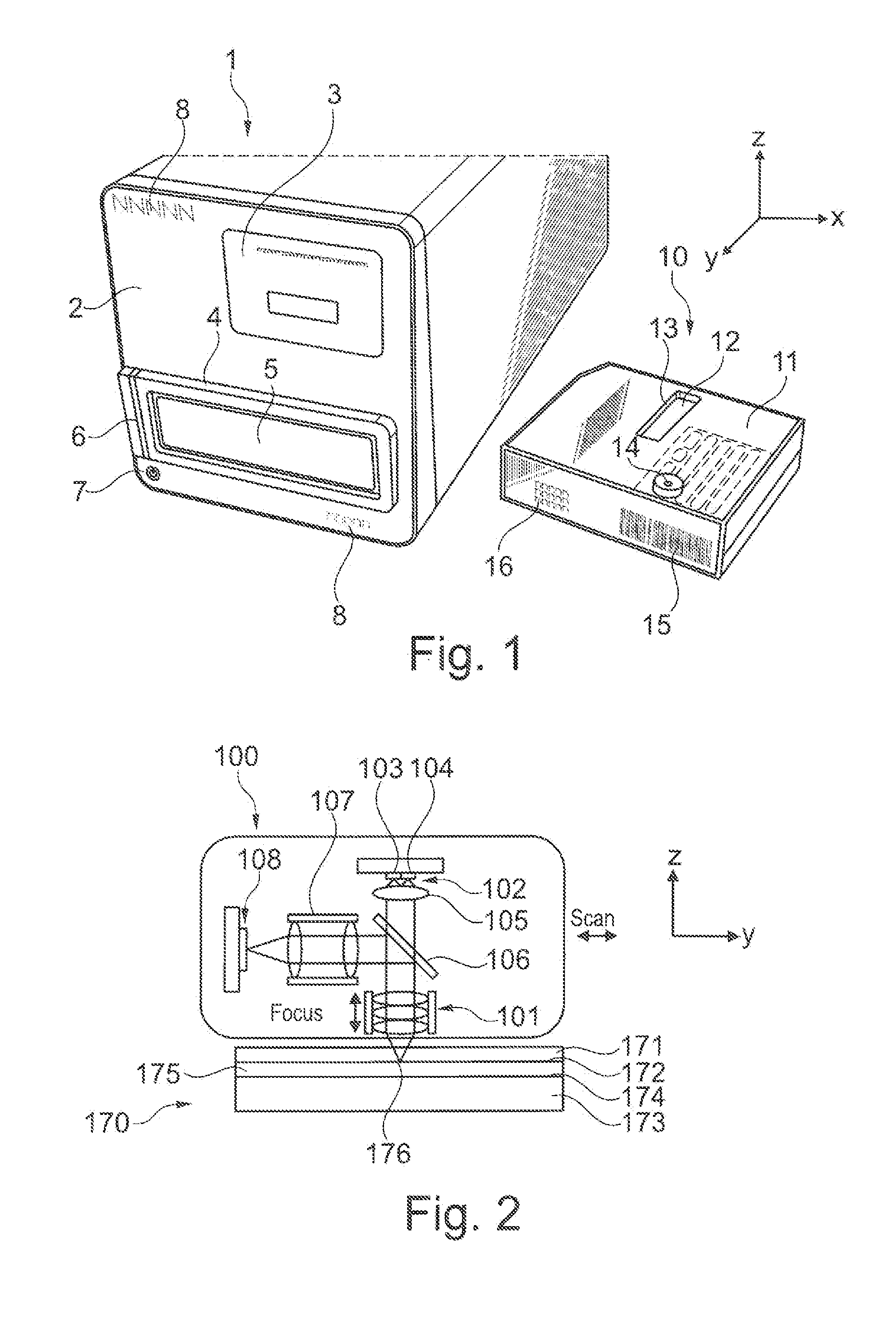
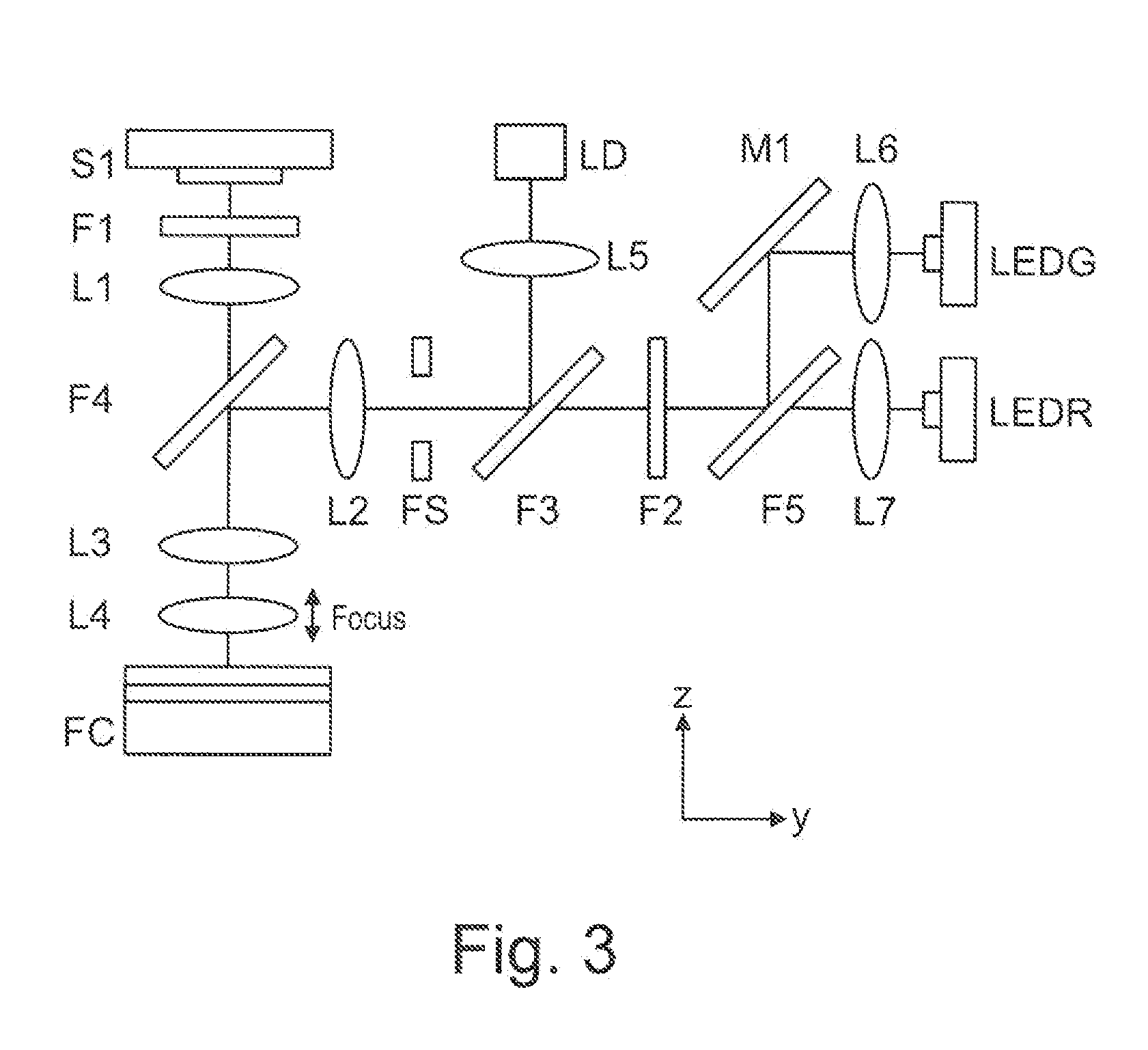
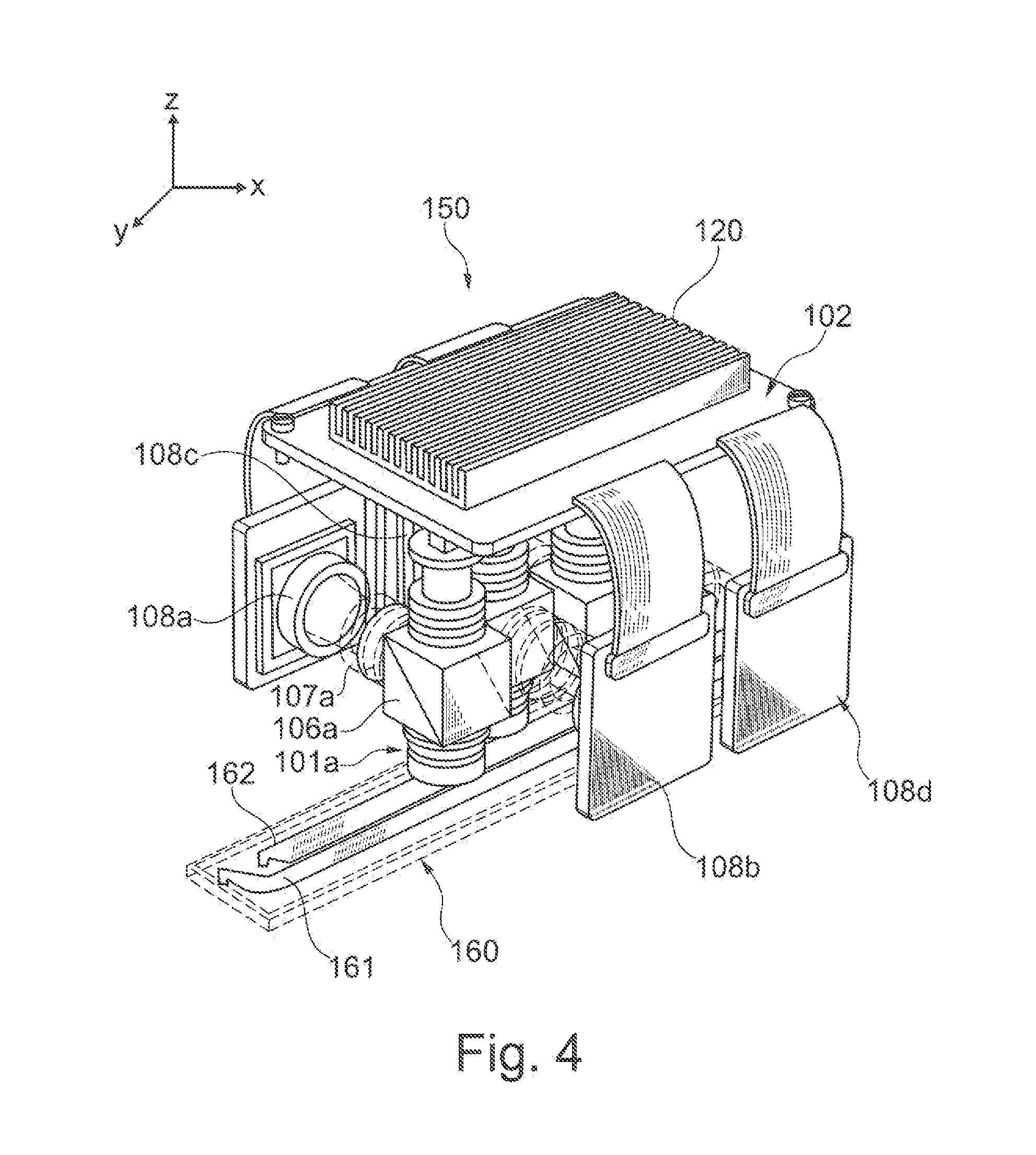
Integrated optoelectronic read head and fluidic cartridge useful for nucleic acid sequencing
ActiveUS20130260372A1Bioreactor/fermenter combinationsBiological substance pretreatmentsWide fieldEngineering
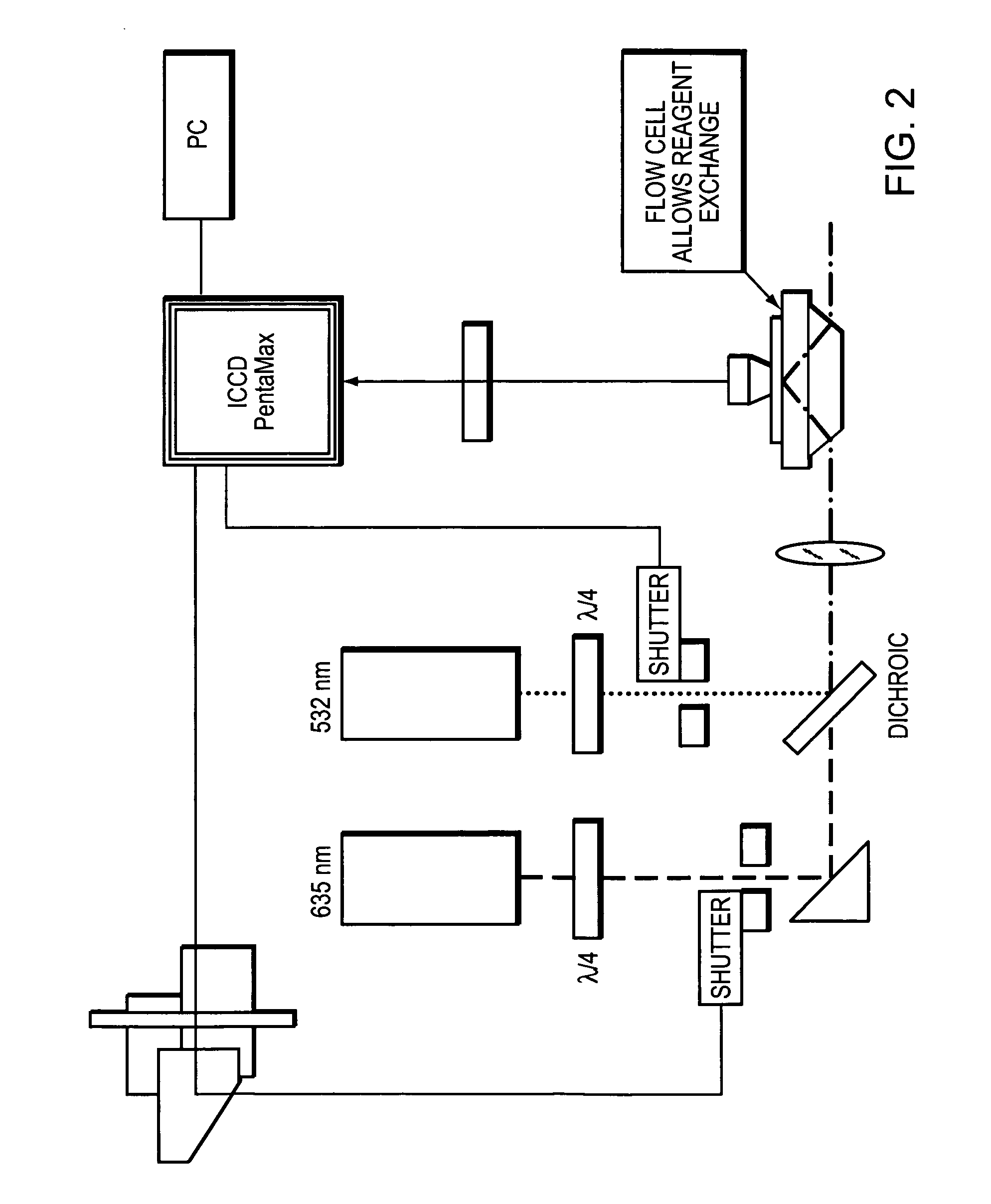
A detection apparatus having a read head including a plurality of microfluorometers positioned to simultaneously acquire a plurality of the wide-field images in a common plane; and (b) a translation stage configured to move the read head along a substrate that is in the common plane. The substrate can be a flow cell that is included in a cartridge, the cartridge also including a housing for (i) a sample reservoir; (ii) a fluidic line between the sample reservoir and the flow cell; (iii) several reagent reservoirs in fluid communication with the flow cell, (iv) at least one valve configured to mediate fluid communication between the reservoirs and the flow cell; and (v) at least one pressure source configured to move liquids from the reservoirs to the flow cell. The detection apparatus and cartridge can be used together or independent of each other.
Owner:ILLUMINA INC
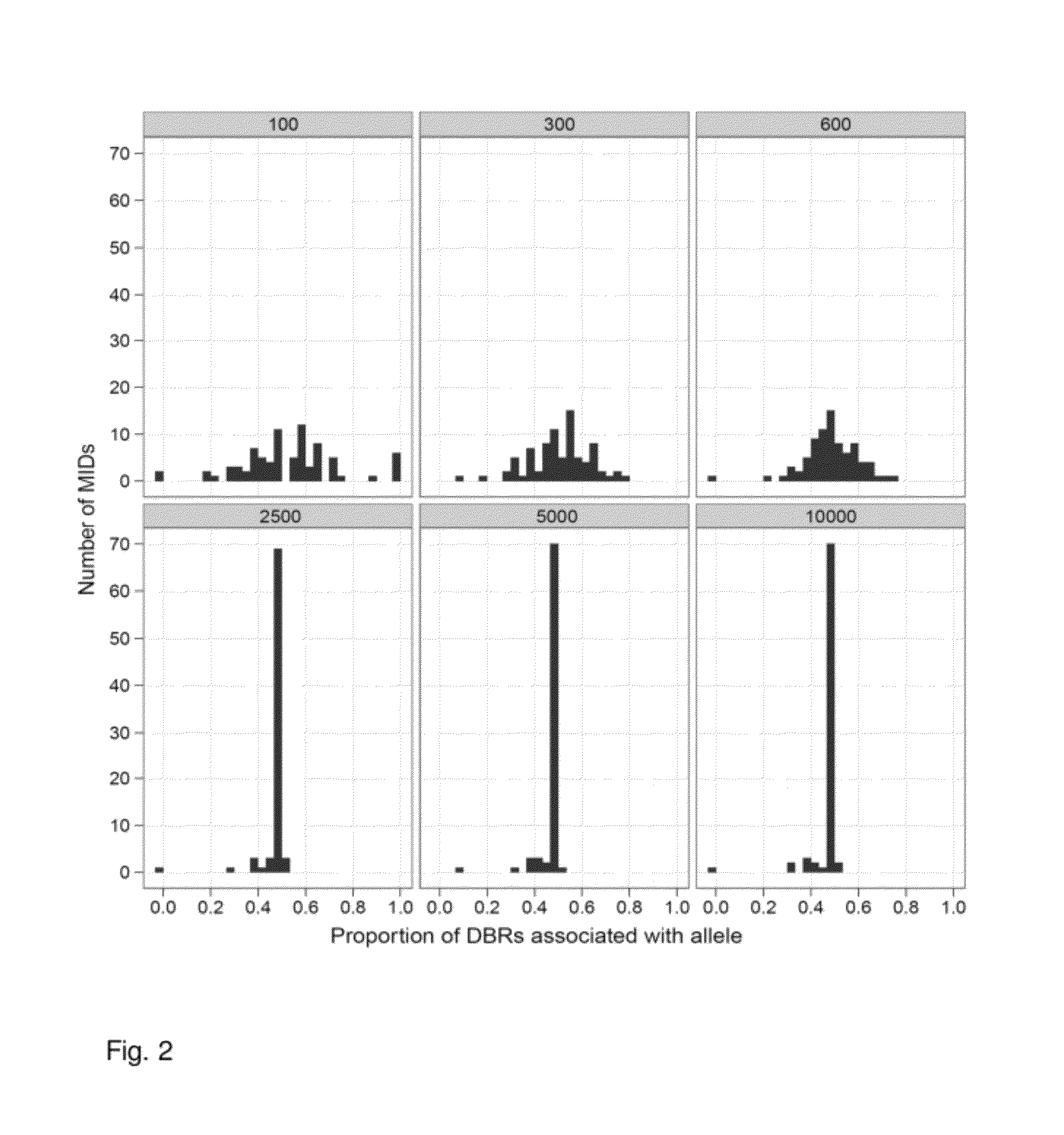
Increasing confidence of allele calls with molecular counting
ActiveUS8481292B2Easy to analyzeSugar derivativesMicrobiological testing/measurementSequence analysisNucleotide
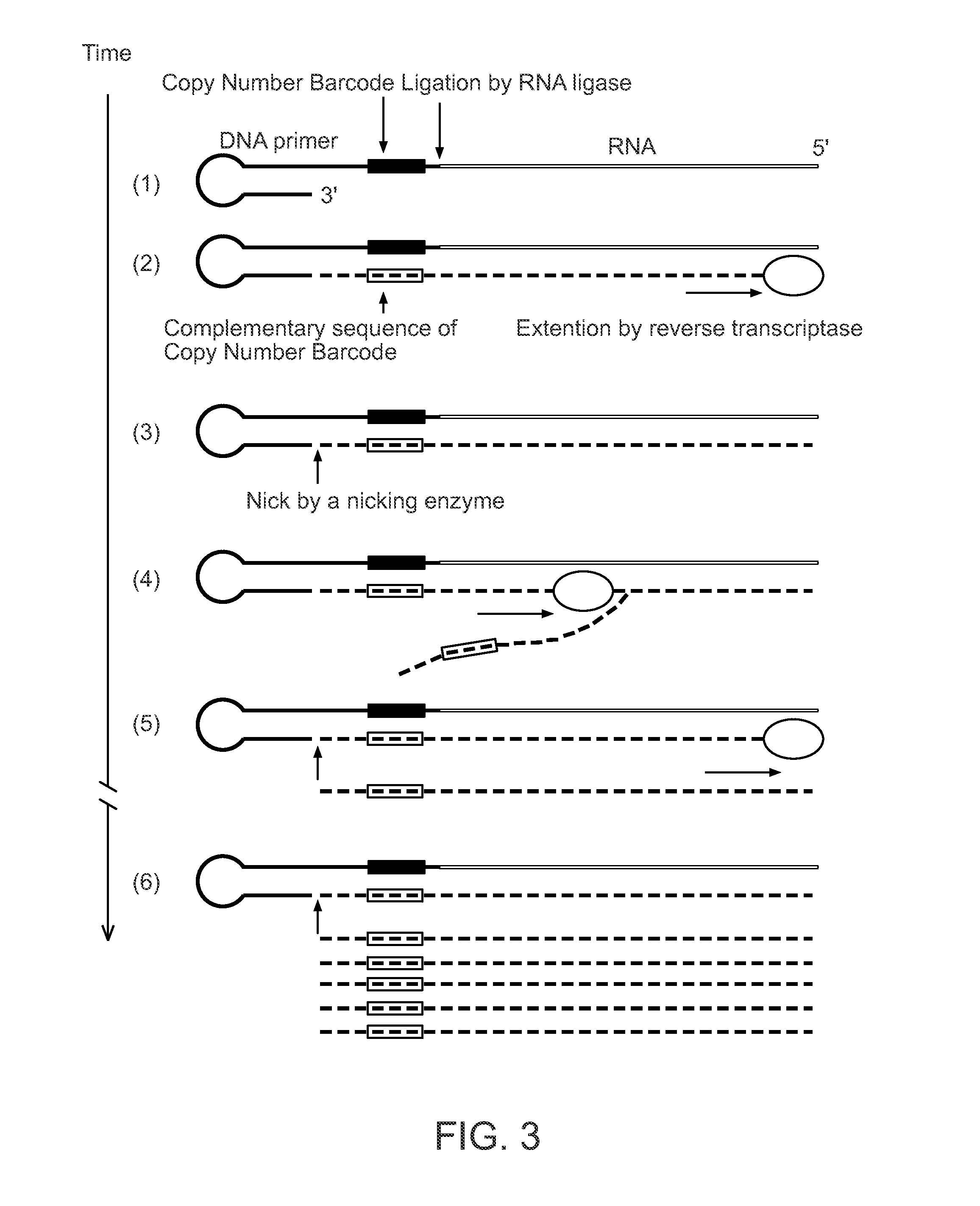
Aspects of the present invention include methods and compositions for determining the number of individual polynucleotide molecules originating from the same genomic region of the same original sample that have been sequenced in a particular sequence analysis configuration or process. In these aspects of the invention, a degenerate base region (DBR) is attached to the starting polynucleotide molecules that are subsequently sequenced (e.g., after certain process steps are performed, e.g., amplification and / or enrichment). The number of different DBR sequences present in a sequencing run can be used to determine / estimate the number of different starting polynucleotides that have been sequenced. DBRs can be used to enhance numerous different nucleic acid sequence analysis applications, including allowing higher confidence allele call determinations in genotyping applications.
Owner:AGILENT TECH INC
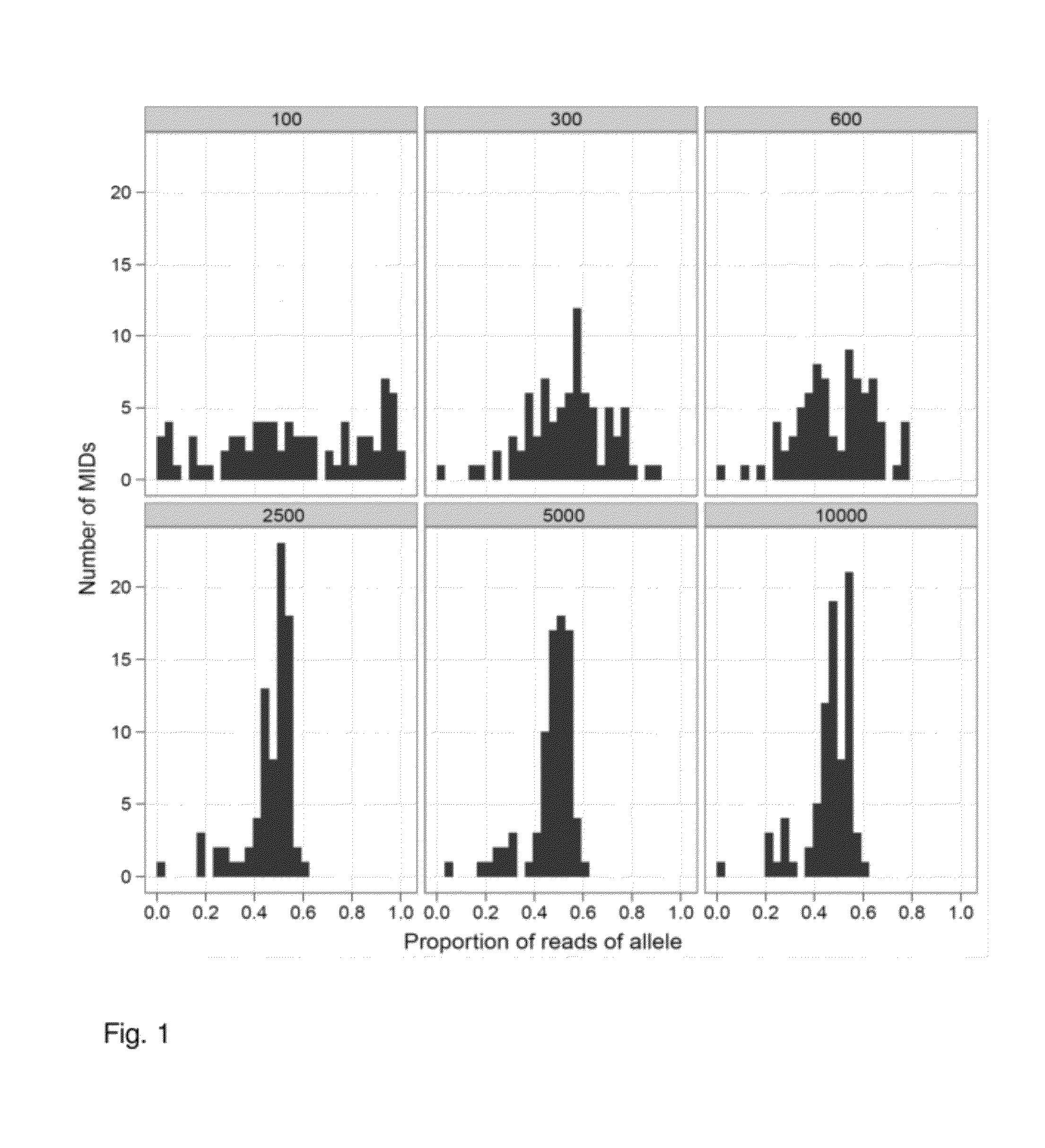
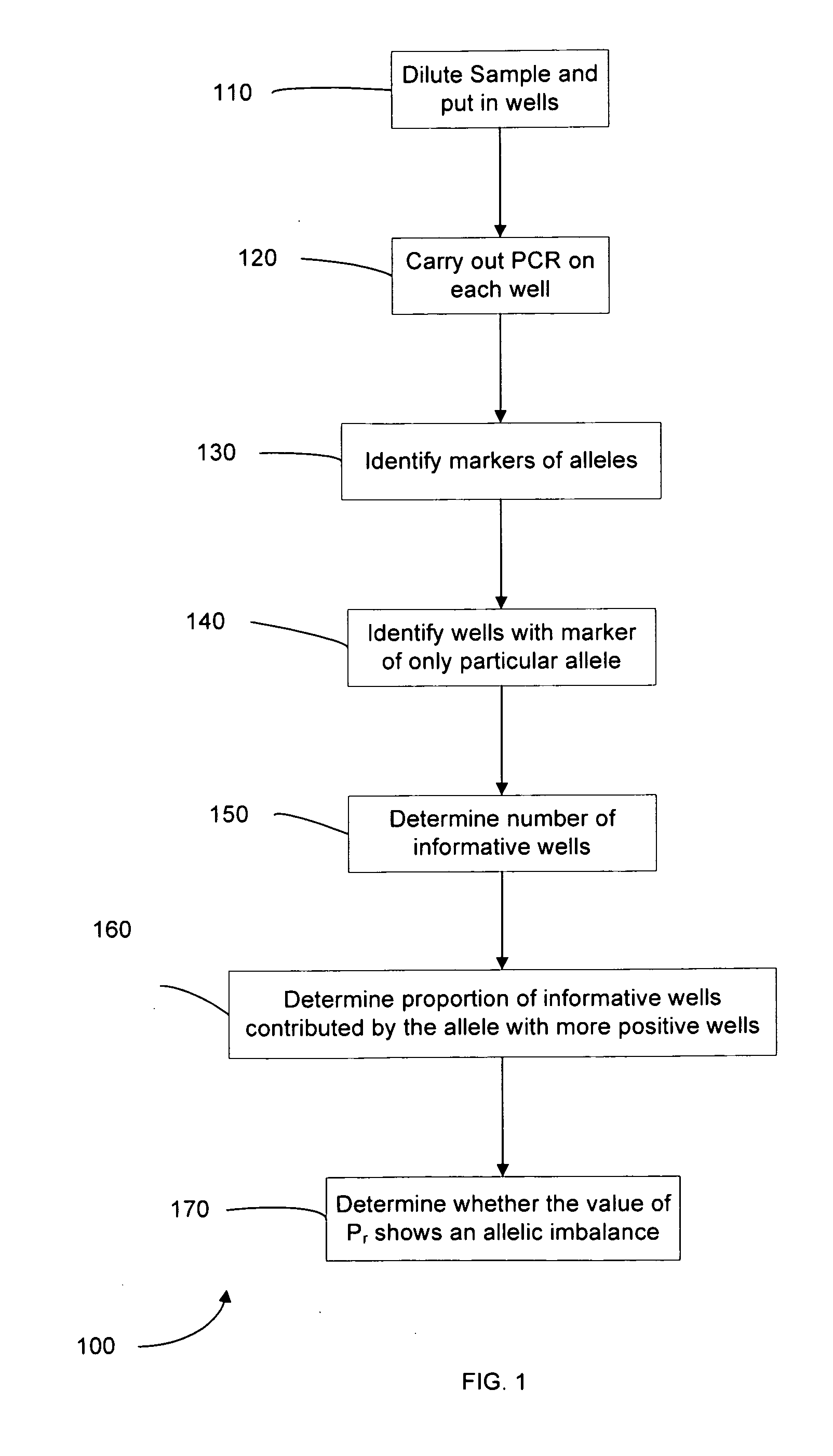
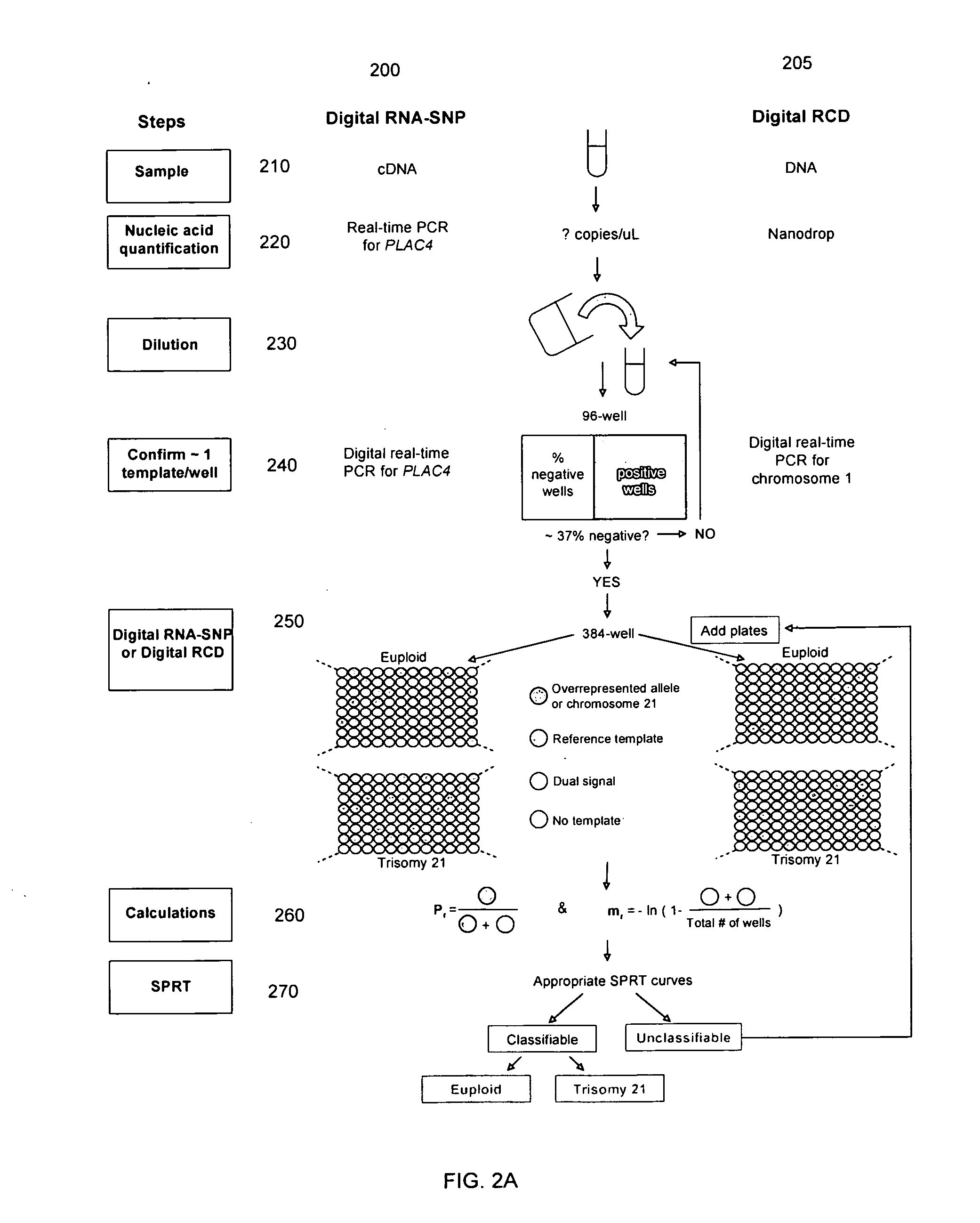
Determining a nucleic acid sequence imbalance
ActiveUS20090087847A1Quantity minimizationMicrobiological testing/measurementDisease diagnosisNucleic acid sequencingNucleic acid sequence
Methods, systems, and apparatus are provided for determining whether a nucleic acid sequence imbalance exists within a biological sample. One or more cutoff values for determining an imbalance of, for example, the ratio of the two sequences (or sets of sequences) are chosen. The cutoff value may be determined based at least in part on the percentage of fetal DNA in a sample, such as maternal plasma, containing a background of maternal nucleic acid sequences. The cutoff value may also be determined based on an average concentration of a sequence per reaction. In one aspect, the cutoff value is determined from a proportion of informative wells that are estimated to contain a particular nucleic acid sequence, where the proportion is determined based on the above-mentioned percentage and / or average concentration. The cutoff value may be determined using many different types of methods, such as sequential probability ratio testing (SPRT).
Owner:THE CHINESE UNIVERSITY OF HONG KONG
RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (SiNA)
InactiveUS20050032733A1Improve various propertyModulate its functionCompounds screening/testingSpecial deliveryBiological bodyNucleic acid sequencing
The present invention concerns methods and reagents useful in modulating gene expression in a variety of applications, including use in therapeutic, diagnostic, target validation, and genomic discovery applications. Specifically, the invention relates to synthetic chemically modified small nucleic acid molecules, such as short interfering nucleic acid (siNA), short interfering RNA (siRNA), double-stranded RNA (dsRNA), micro-RNA (miRNA), and short hairpin RNA (shRNA) molecules capable of mediating RNA interference (RNAi) against target nucleic acid sequences. The small nucleic acid molecules are useful in the treatment of any disease or condition that responds to modulation of gene expression or activity in a cell, tissue, or organism.
Owner:SIRNA THERAPEUTICS INC
Method of determining the nucleotide sequence of oligonucleotides and DNA molecules
InactiveUS7037687B2Eliminate needBioreactor/fermenter combinationsBiological substance pretreatmentsOligonucleotide primersThermopile
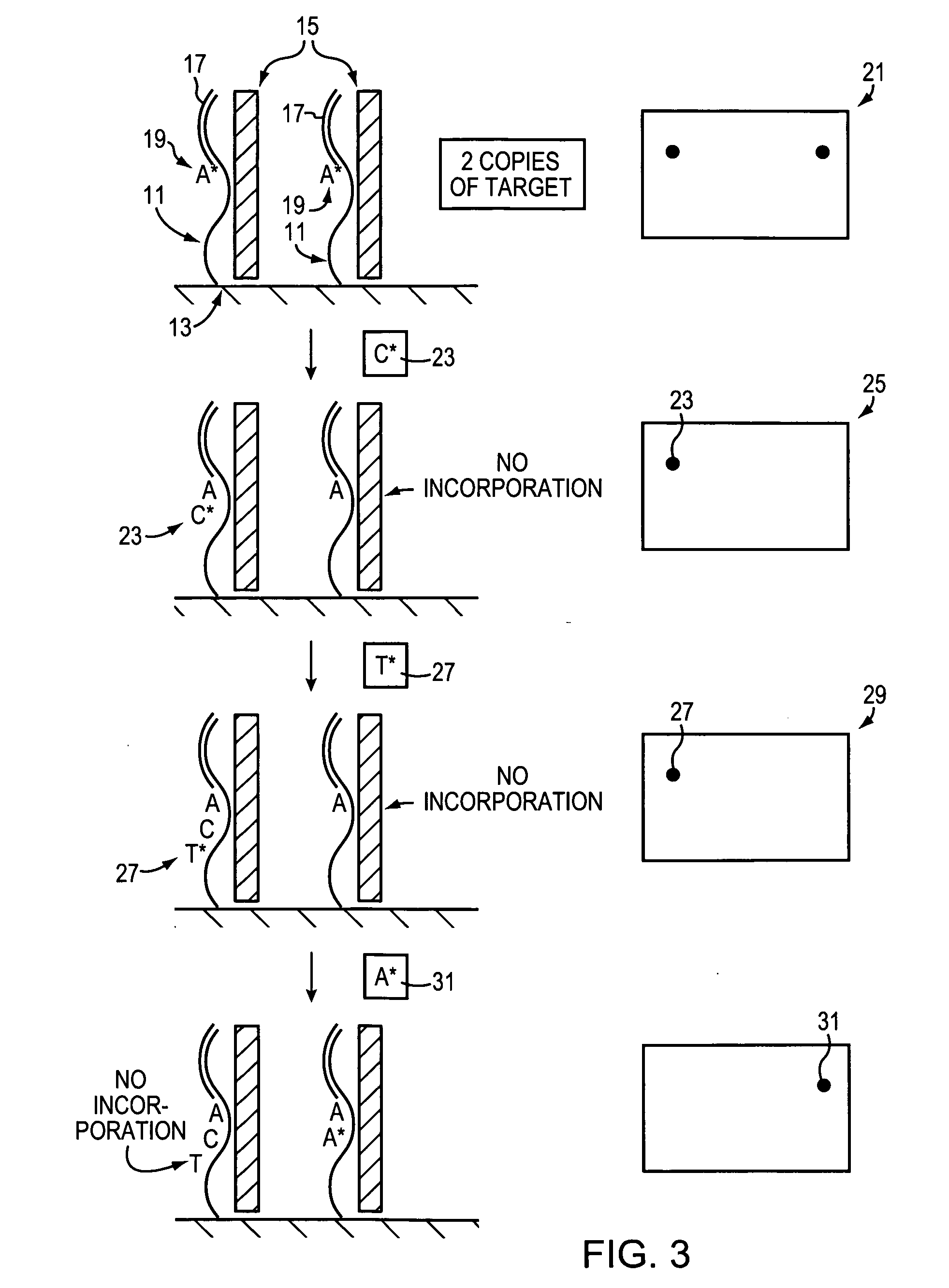
The present invention relates to a novel method for analyzing nucleic acid sequences based on real-time detection of DNA polymerase-catalyzed incorporation of each of the four nucleotide bases, supplied individually and serially in a microfluidic system, to a reaction cell containing a template system comprising a DNA fragment of unknown sequence and an oligonucleotide primer. Incorporation of a nucleotide base into the template system can be detected by any of a variety of methods including but not limited to fluorescence and chemiluminescence detection. Alternatively, microcalorimetic detection of the heat generated by the incorporation of a nucleotide into the extending template system using thermopile, thermistor and refractive index measurements can be used to detect extension reactions.
Owner:ALBERTA UNIV OF +1
Genes conferring herbicide resistance
Compositions and methods for conferring herbicide resistance to plants, plant cells, tissues and seeds are provided. Compositions comprising a coding sequence for a polypeptide that confers resistance or tolerance to glyphosate herbicides are provided. The coding sequences can be used in DNA constructs or expression cassettes for transformation and expression in plants. Compositions also comprise transformed plants, plant cells, tissues, and seeds. In particular, isolated nucleic acid molecules corresponding to glyphosate resistant nucleic acid sequences are provided. Additionally, amino acid sequences corresponding to the polynucleotides are encompassed. In particular, the present invention provides for isolated nucleic acid molecules comprising nucleotide sequences encoding the amino acid sequence shown in SEQ ID NO:2 or the nucleotide sequence set forth in SEQ ID NO:1.
Owner:BASF AGRICULTURAL SOLUTIONS SEED LLC
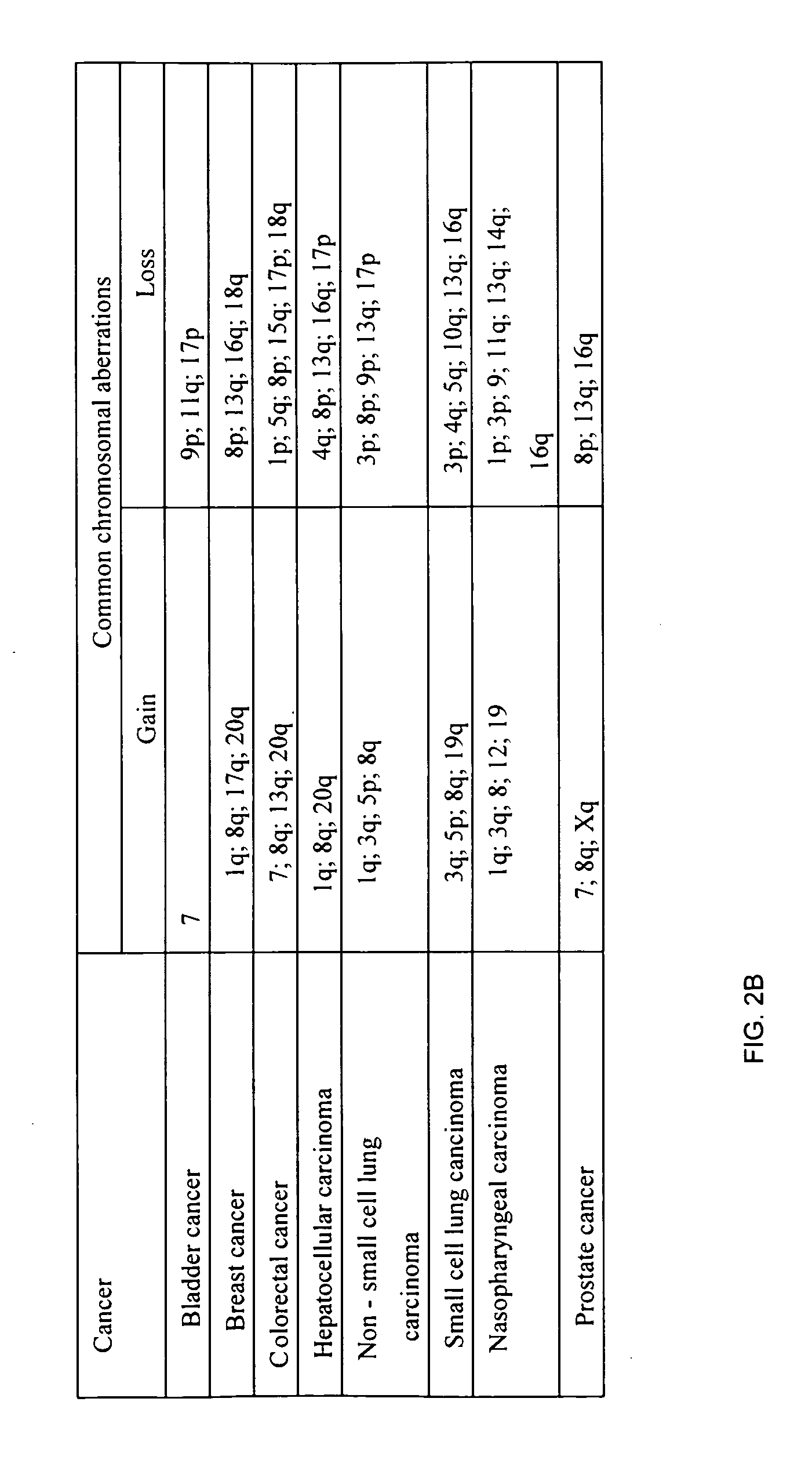
Bacillus thuringiensis CryET33 and CryET34 compositions and uses therefor
Disclosed are Bacillus thuringiensis strains comprising novel crystal proteins which exhibit insecticidal activity against coleopteran insects including red flour beetle larvae (Tribolium castaneum) and Japanese beetle larvae (Popillia japonica). Also disclosed are novel B. thuringiensis crystal toxin genes, designated cryET33 and cryET34, which encode respectively the coleopteran-toxic proteins, CryET33 (29-kDa) crystal protein, and CryET34 (14-kDa) crystal protein. Also disclosed are methods of making and using transgenic cells comprising the novel nucleic acid sequences of the invention.
Owner:MONSANTO TECH LLC
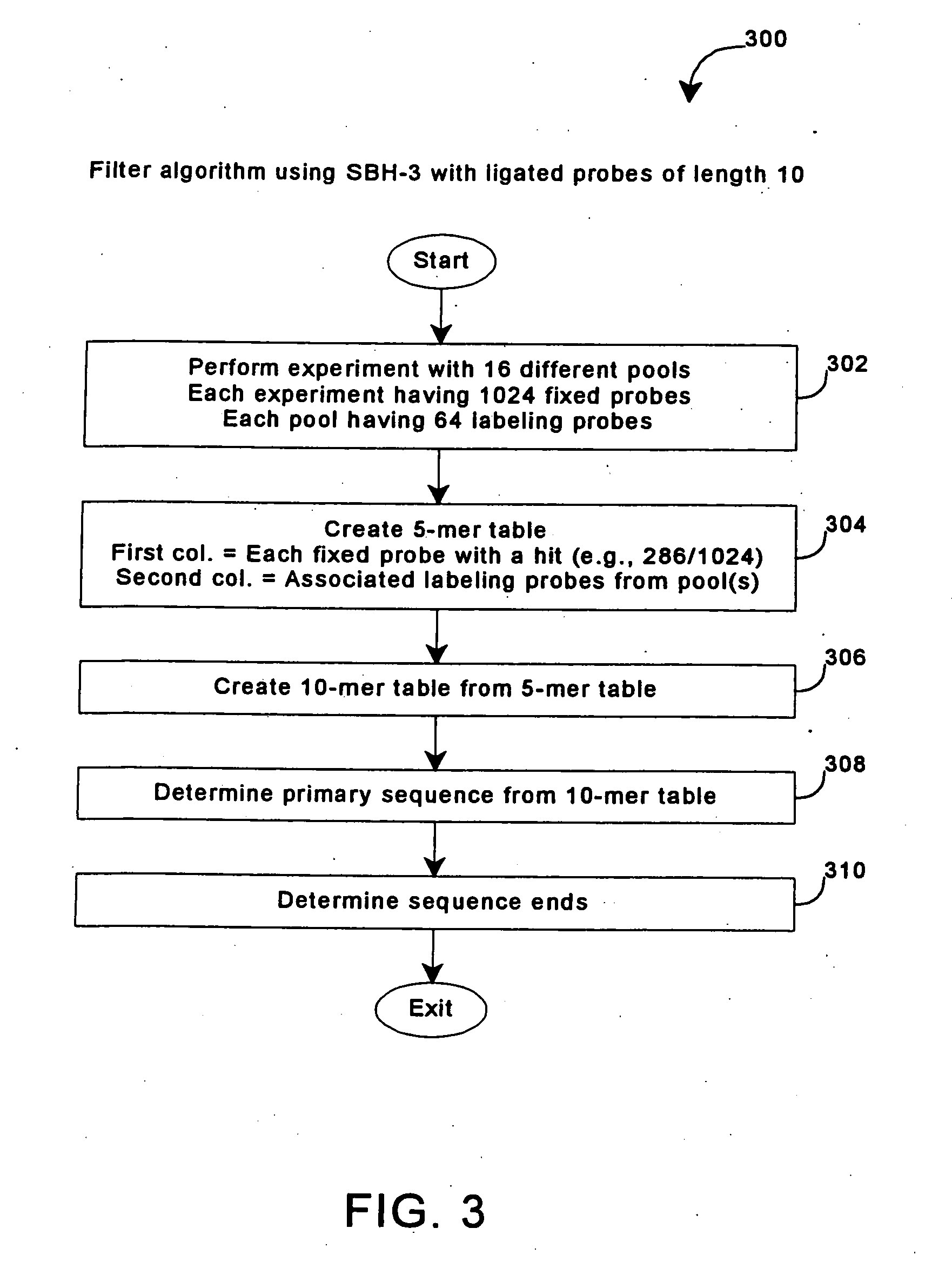
Enhanced sequencing by hybridization using pools of probes
InactiveUS20050191656A1Enhances SBH methodReduce redundancySugar derivativesMicrobiological testing/measurementNucleic acid sequencingNucleic acid sequence
The invention provides methods for sequencing by hybridization (SBH) using pools of probes that allow greater efficiency in conducting SBH by reducing the number of separate measurements of hybridization signals required to identify each particular nucleotide in a target nucleic acid sequence. The invention also provides pools and sets of pools of probes, as well as methods of generating pools of probes.
Owner:COMPLETE GENOMICS INC
Method of determining the nucleotide sequence of oligonucleotides and DNA molecules
InactiveUS6780591B2Eliminate needHigh purityMicrobiological testing/measurementRecombinant DNA-technologyFluorescenceOligonucleotide primers
The present invention relates to a novel method for analyzing nucleic acid sequences based on real-time detection of DNA polymerase-catalyzed incorporation of each of the four nucleotide bases, supplied individually and serially in a microfluidic system, to a reaction cell containing a template system comprising a DNA fragment of unknown sequence and an oligonucleotide primer. Incorporation of a nucleotide base into the template system can be detected by any of a variety of methods including but not limited to fluorescence and chemiluminescence detection. Alternatively, microcalorimetic detection of the heat generated by the incorporation of a nucleotide into the extending template system using thermopile, thermistor and refractive index measurements can be used to detect extension reactions.
Owner:LIFE TECH CORP
Methods for generating amplified nucleic acid arrays
InactiveUS20080242560A1Sequential/parallel process reactionsMicrobiological testing/measurementNucleic acid sequencingNucleic acid sequence
The present invention relates to methods for generating an array of amplified nucleic acid sequences. The methods can utilize amplicons that form nucleic acid balls that can be arrayed on a solid support. The invention additionally provides methods for obtaining targeted nucleic acid sequences.
Owner:ILLUMINA INC
Targeted modification of rat genome
Compositions and methods are provided for modifying a rat genomic locus of interest using a large targeting vector (LTVEC) comprising various endogenous or exogenous nucleic acid sequences as described herein. Compositions and methods for generating a genetically modified rat comprising one or more targeted genetic modifications in their germline are also provided. Compositions and methods are provided which comprise a genetically modified rat or rat cell comprising a targeted genetic modification in the rat interleukin-2 receptor gamma locus, the rat ApoE locus, the rat Rag2 locus, the rat Rag1 locus and / or the rat Rag2 / Rag1 locus. The various methods and compositions provided herein allows for these modified loci to be transmitted through the germline.
Owner:REGENERON PHARM INC
Single Cell Nucleic Acid Detection and Analysis
ActiveUS20140155274A1High amplification efficiencyEasy to detectMicrobiological testing/measurementScreening processNucleic acid detectionNucleic acid sequencing
Methods and compositions for digital profiling of nucleic acid sequences present in a sample are provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Use of single-stranded nucleic acid binding proteins in sequencing
InactiveUS20060024678A1Stabilizing sequencingIncrease speedMicrobiological testing/measurementFermentationNucleic acid sequencingSingle strand
The invention provides methods for stabilizing a nucleic acid sequencing reaction. Generally, methods of the invention include exposing a target nucleic acid to a single-stranded nucleic acid binding protein and performing a sequencing reaction.
Owner:FLUIDIGM CORP
Method for nucleic acid amplification that results in low amplification bias
InactiveUS7297485B2Consistent outputQuick buildMicrobiological testing/measurementFermentationNucleic acid sequencingAmplification bias
Disclosed are compositions and methods for amplification of nucleic acid sequences of interest. It has been discovered that amplification reactions can produce amplification products of high quality, such as low amplification bias, if performed on an amount of nucleic acid at or over a threshold amount and / or on nucleic acids at or below a threshold concentration. The threshold amount and concentration can vary depending on the nature and source of the nucleic acids to be amplified and the type of amplification reaction employed. Disclosed is a method of determining the threshold amount and / or threshold concentration of nucleic acids that can be used with nucleic acid samples of interest in amplification reactions of interest. Because amplification reactions can produce high quality amplification products, such as low bias amplification products, below the threshold amount and / or concentration of nucleic acid, such below-threshold amounts and / or concentrations can be used in amplification reactions.
Owner:QIAGEN GMBH
Single-primer nucleic acid amplification methods
ActiveUS7374885B2Reduce appearanceHigh levelMicrobiological testing/measurementFermentationNucleotideNucleic acid sequencing
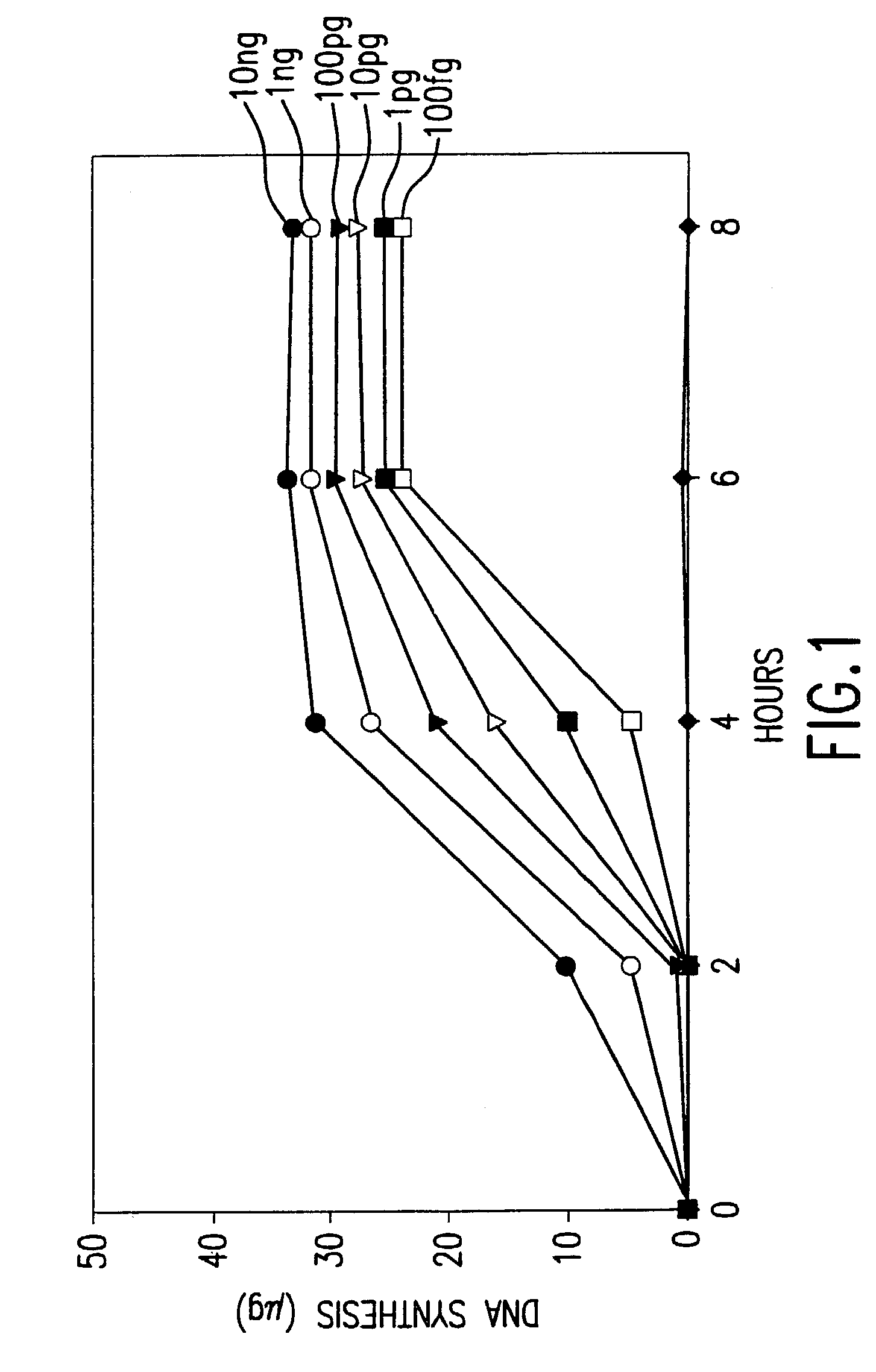
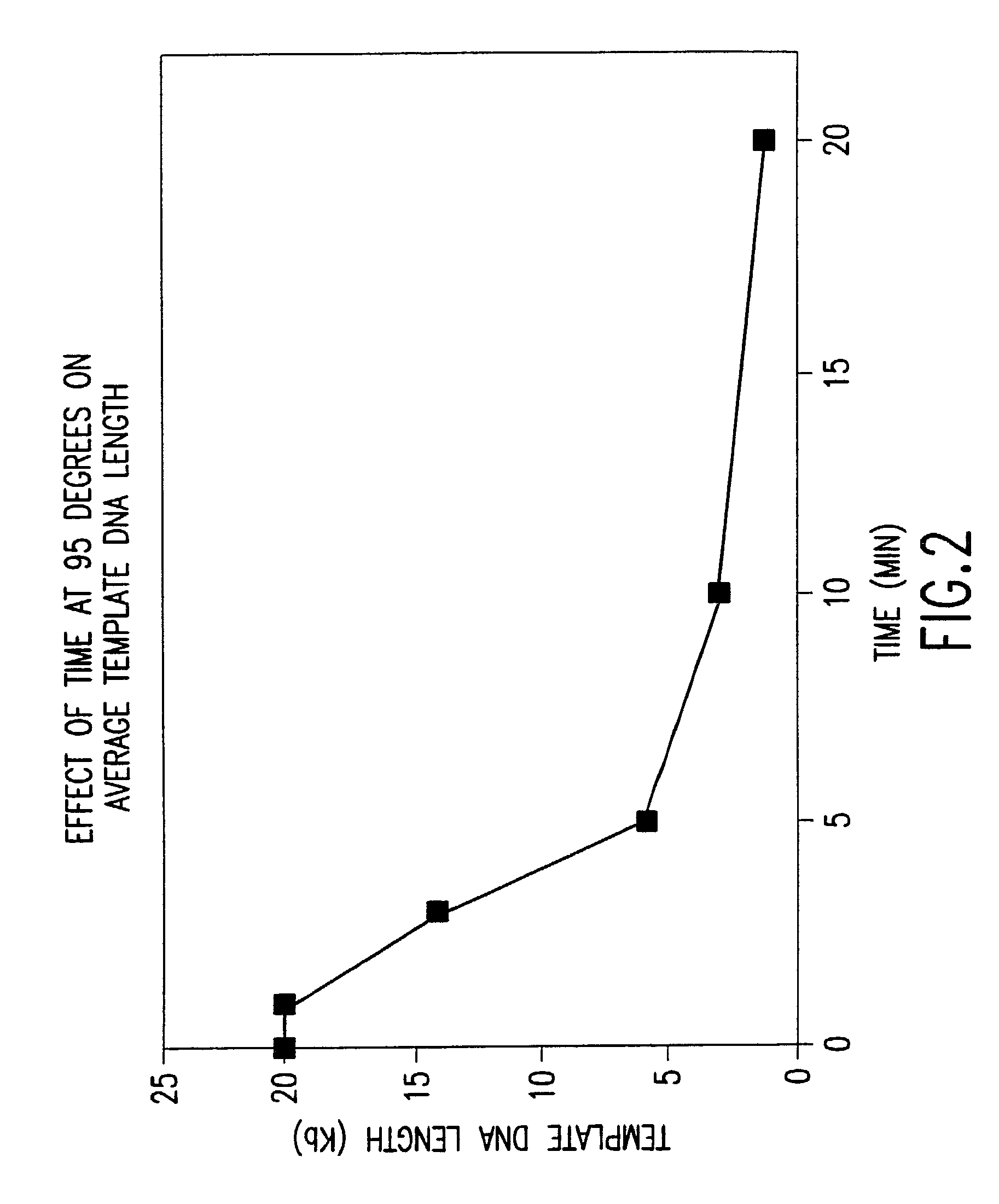
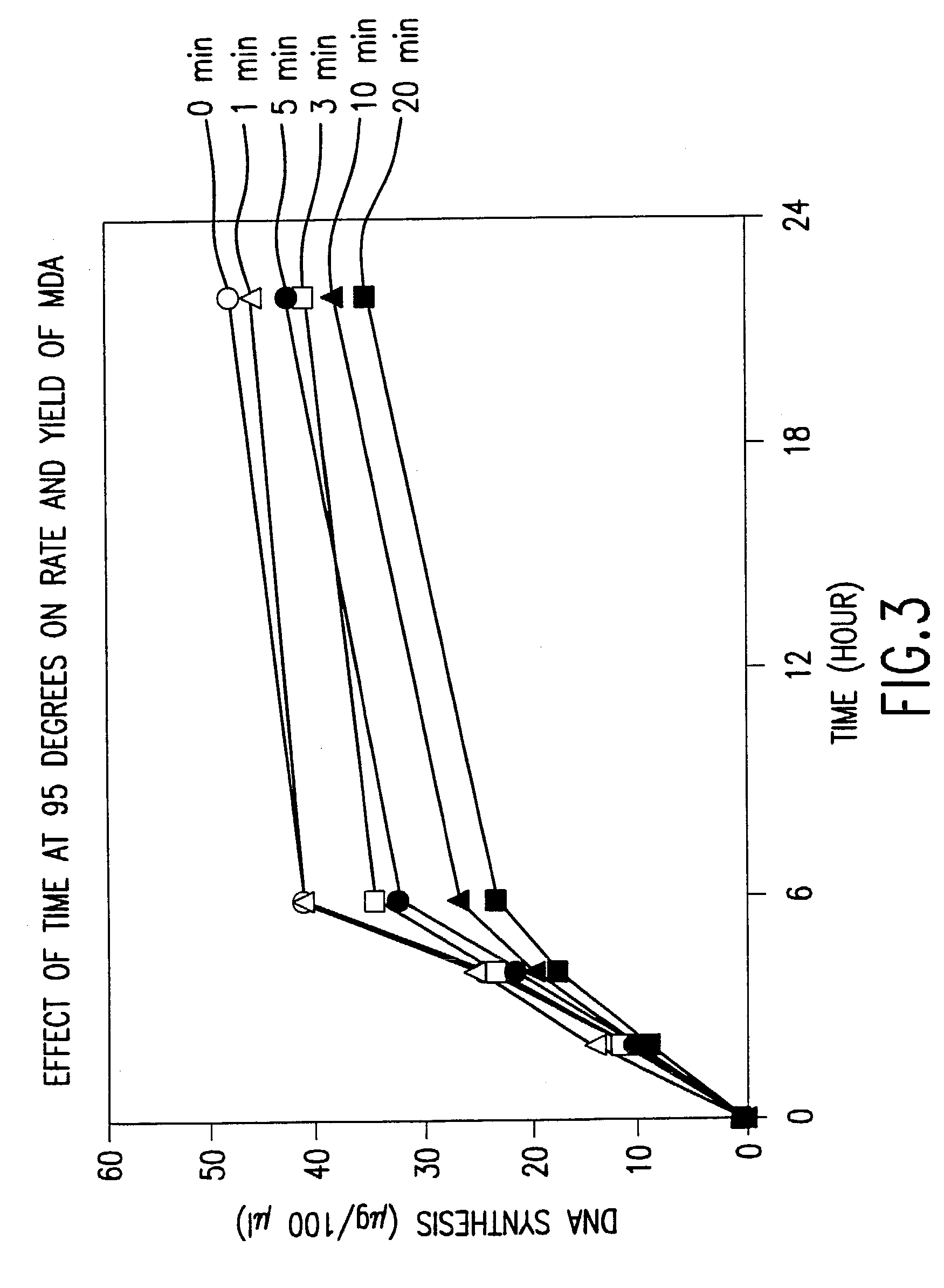
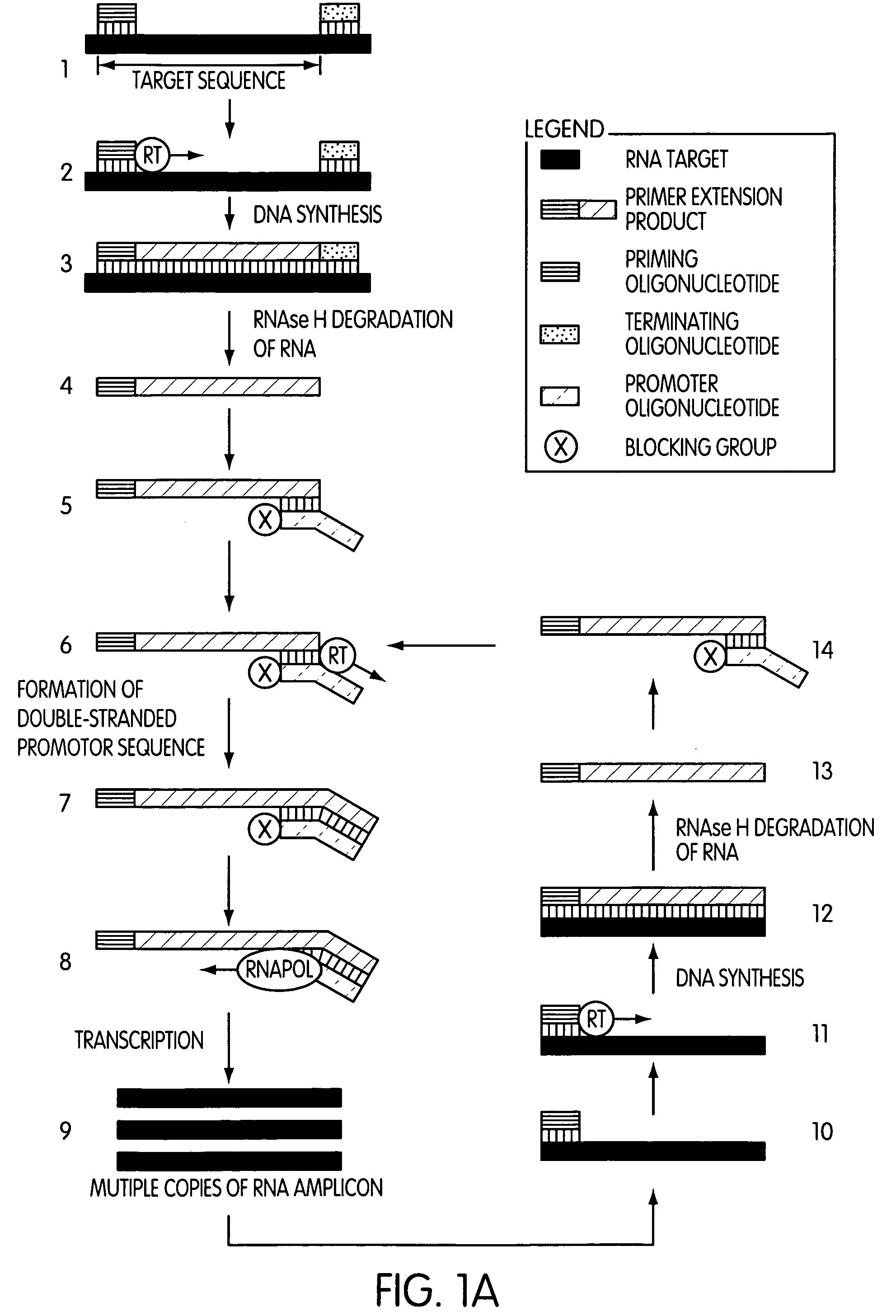
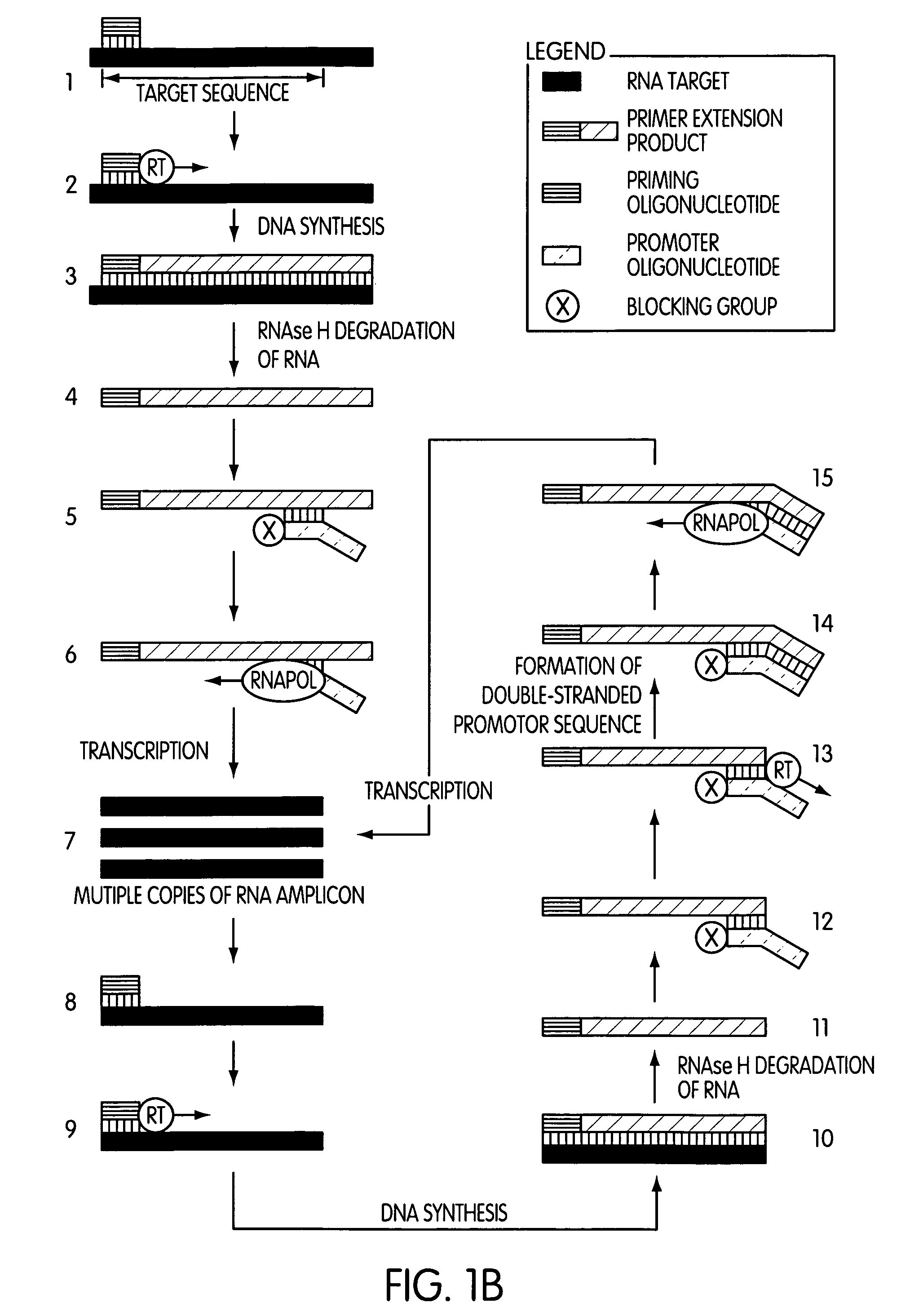
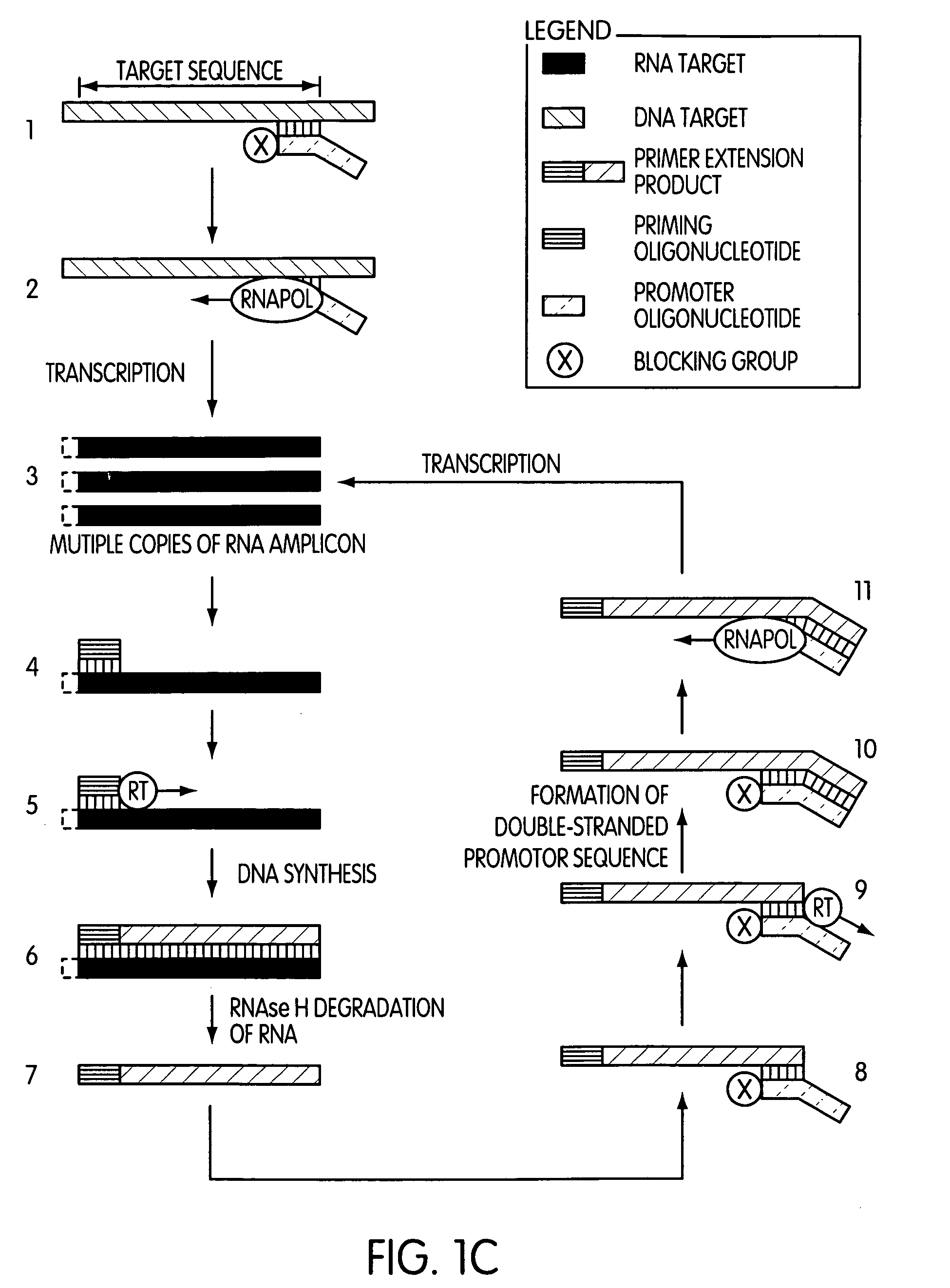
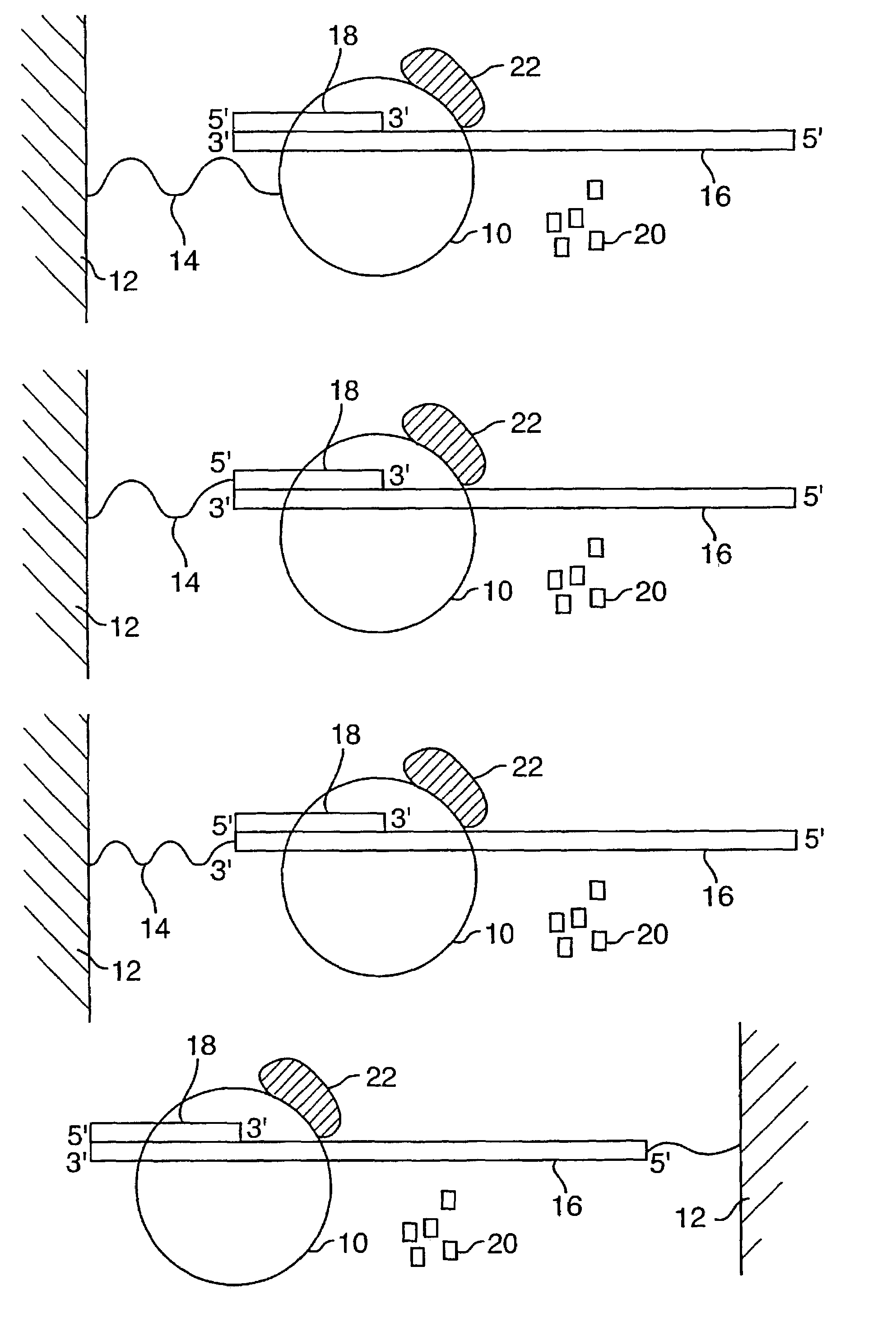
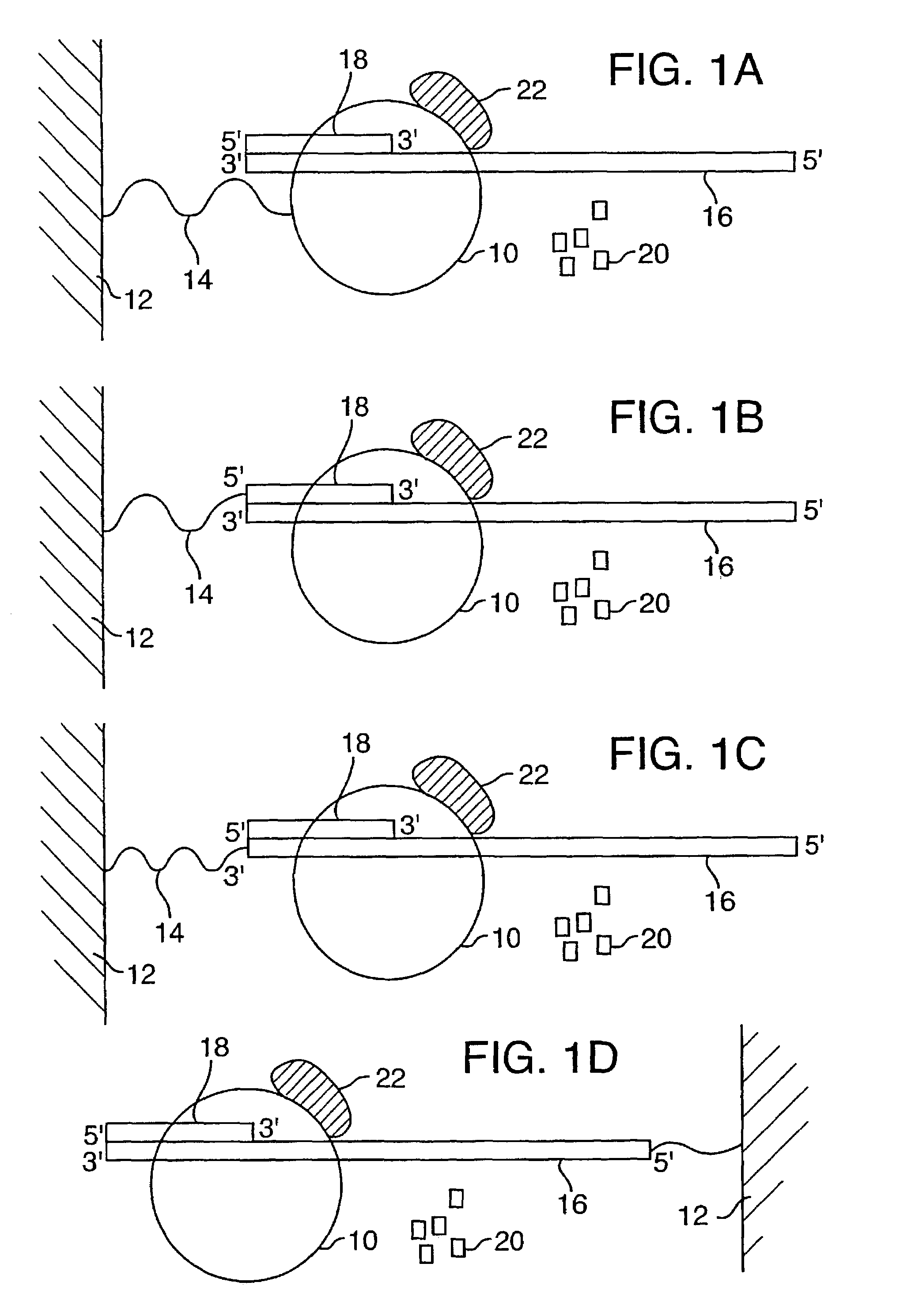
The present invention is directed to novel methods of synthesizing multiple copies of a target nucleic acid sequence which are autocatalytic (i.e., able to cycle automatically without the need to modify reaction conditions such as temperature, pH, or ionic strength and using the product of one cycle in the next one). In particular, the present invention discloses a method of nucleic acid amplification which is robust and efficient, while reducing the appearance of side-products. The method uses only one primer, the “priming oligonucleotide,” a promoter oligonucleotide modified to prevent polymerase extension from its 3′-terminus and, optionally, a means for terminating a primer extension reaction, to amplify RNA or DNA molecules in vitro, while reducing or substantially eliminating the formation of side-products. The method of the present invention minimizes or substantially eliminates the emergence of side-products, thus providing a high level of specificity. Furthermore, the appearance of side-products can complicate the analysis of the amplification reaction by various molecular detection techniques. The present invention minimizes or substantially eliminates this problem, thus providing an enhanced level of sensitivity.
Owner:GEN PROBE INC
High speed parallel molecular nucleic acid sequencing
InactiveUS6982146B1Minimize shearImprove automationSugar derivativesMicrobiological testing/measurementNucleotidePolymerase L
A method and device is disclosed for high speed, automated sequencing of nucleic acid molecules. A nucleic acid molecule to be sequenced is exposed to a polymerase in the presence of nucleotides which are to be incorporated into a complementary nucleic acid strand. The polymerase carries a donor fluorophore, and each type of nucleotide (e.g. A, T / U, C and G) carries a distinguishable acceptor fluorophore characteristic of the particular type of nucleotide. As the polymerase incorporates individual nucleic acid molecules into a complementary strand, a laser continuously irradiates the donor fluorophore, at a wavelength that causes it to emit an emission signal (but the laser wavelength does not stimulate the acceptor fluorophore). In particular embodiments, no laser is needed if the donor fluorophore is a luminescent molecule or is stimulated by one. The emission signal from the polymerase is capable of stimulating any of the donor fluorophores (but not acceptor fluorophores), so that as a nucleotide is added by the polymerase, the acceptor fluorophore emits a signal associated with the type of nucleotide added to the complementary strand. The series of emission signals from the acceptor fluorophores is detected, and correlated with a sequence of nucleotides that correspond to the sequence of emission signals.
Owner:GOVERNMENT OF US SEC THE DEPT OF HEALTH & HUMAN SERVICES THE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com