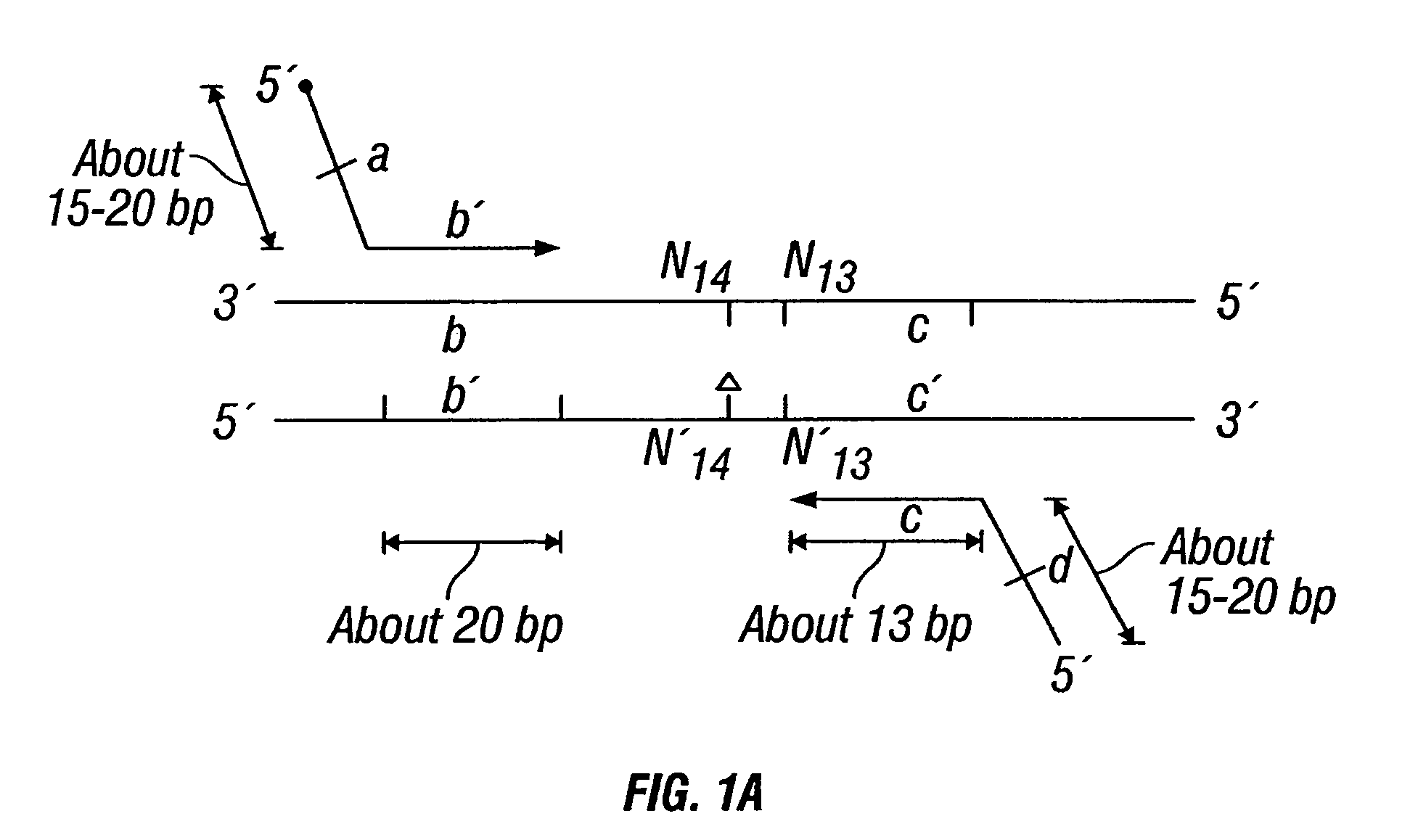
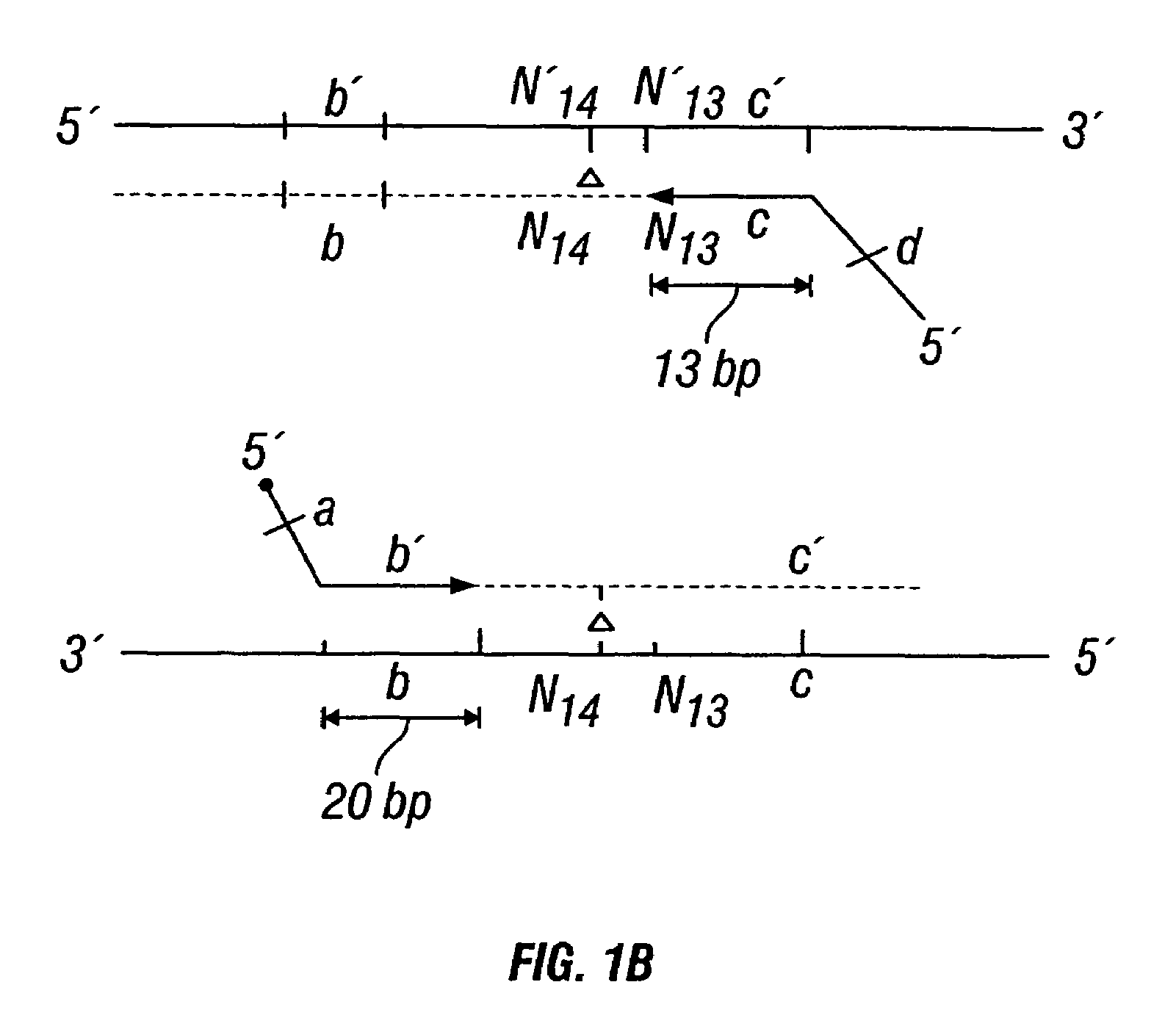
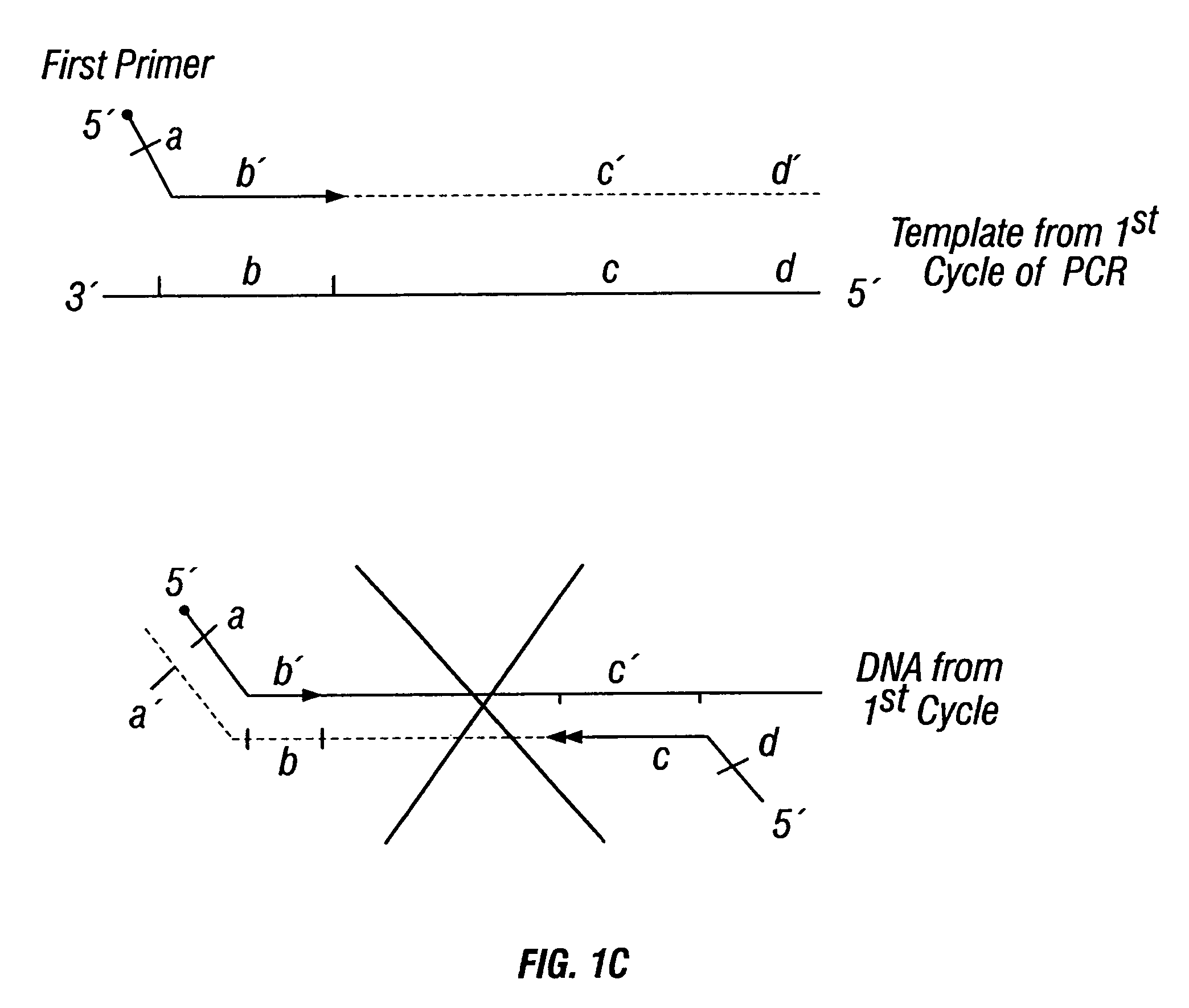
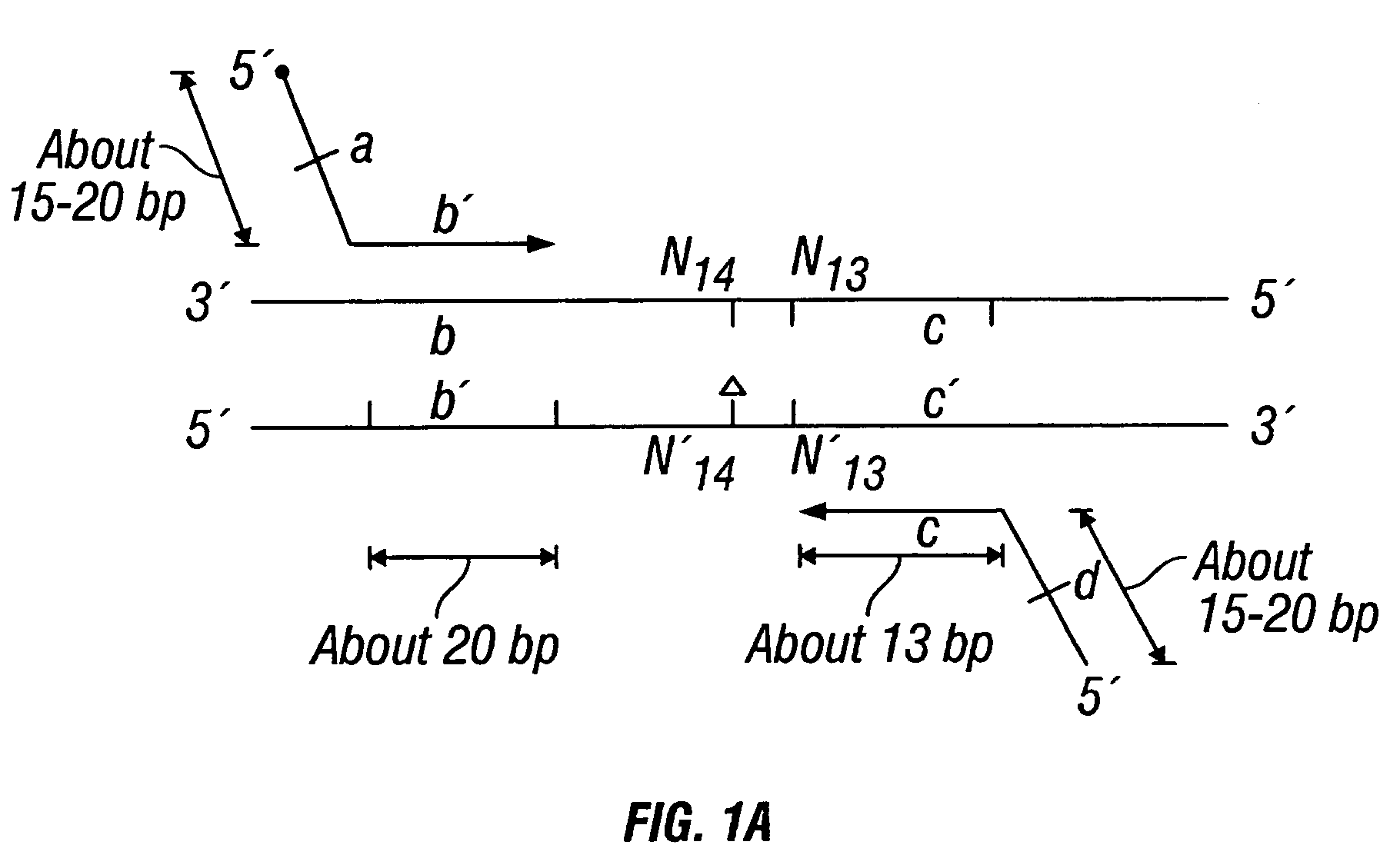
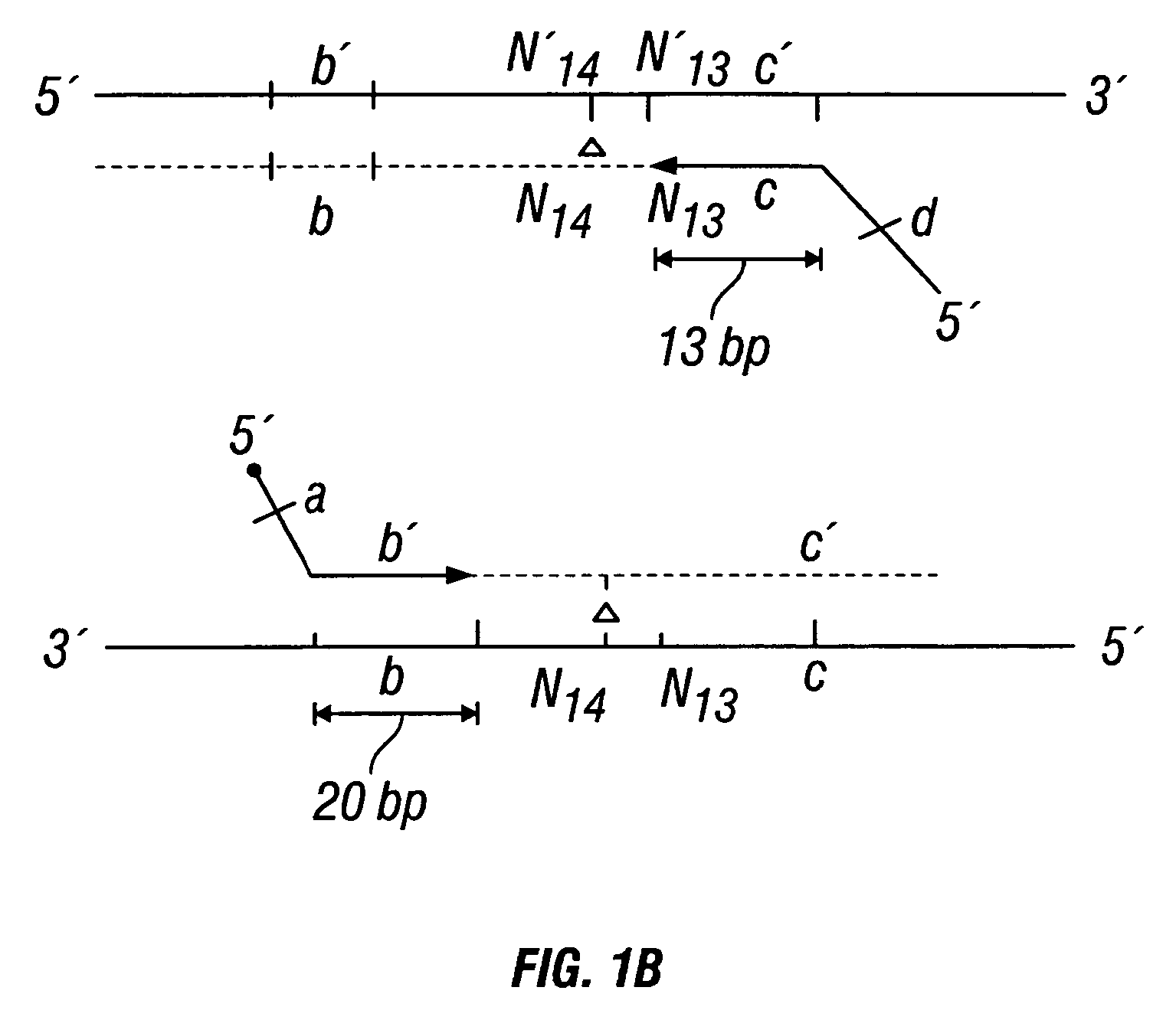
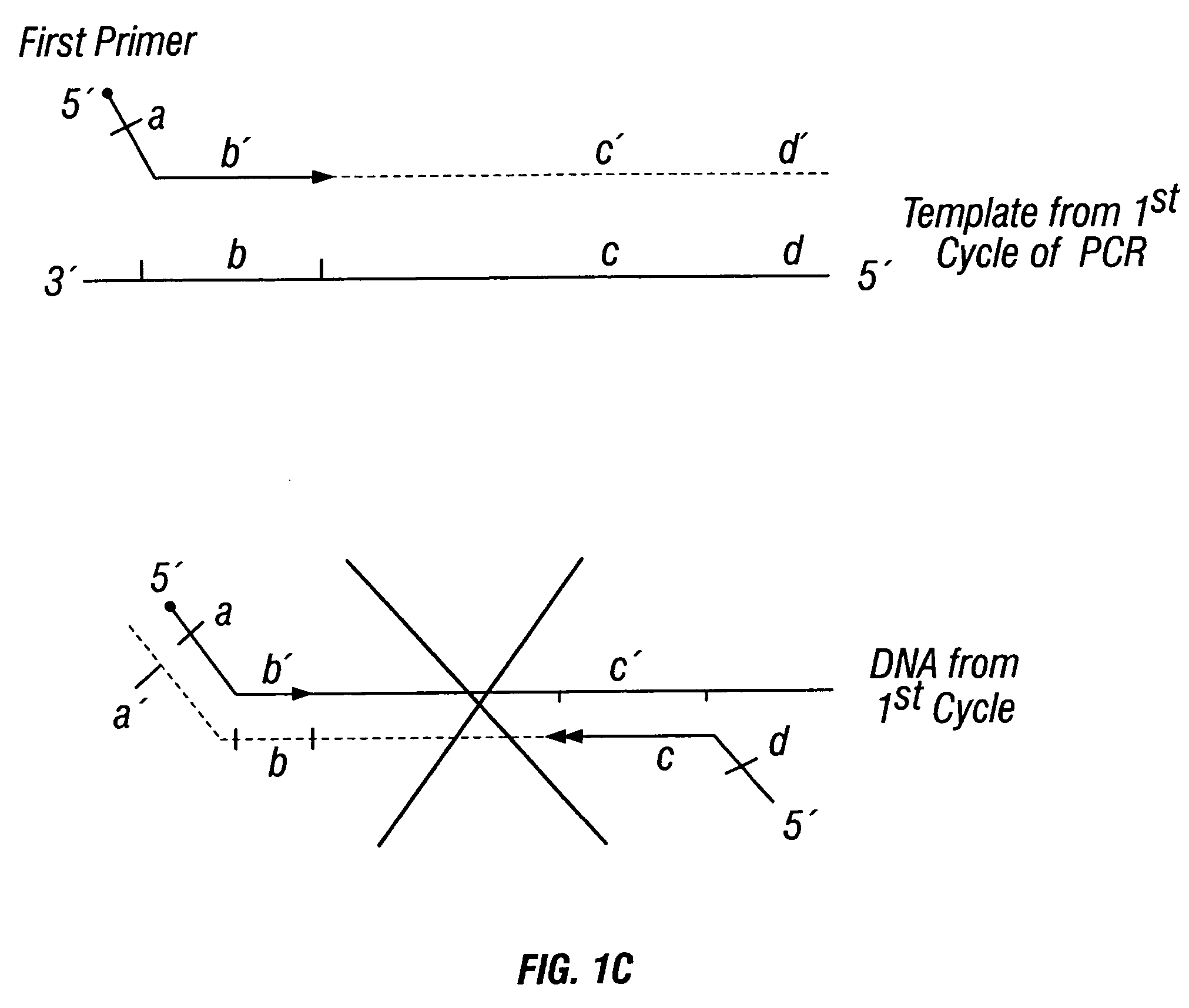
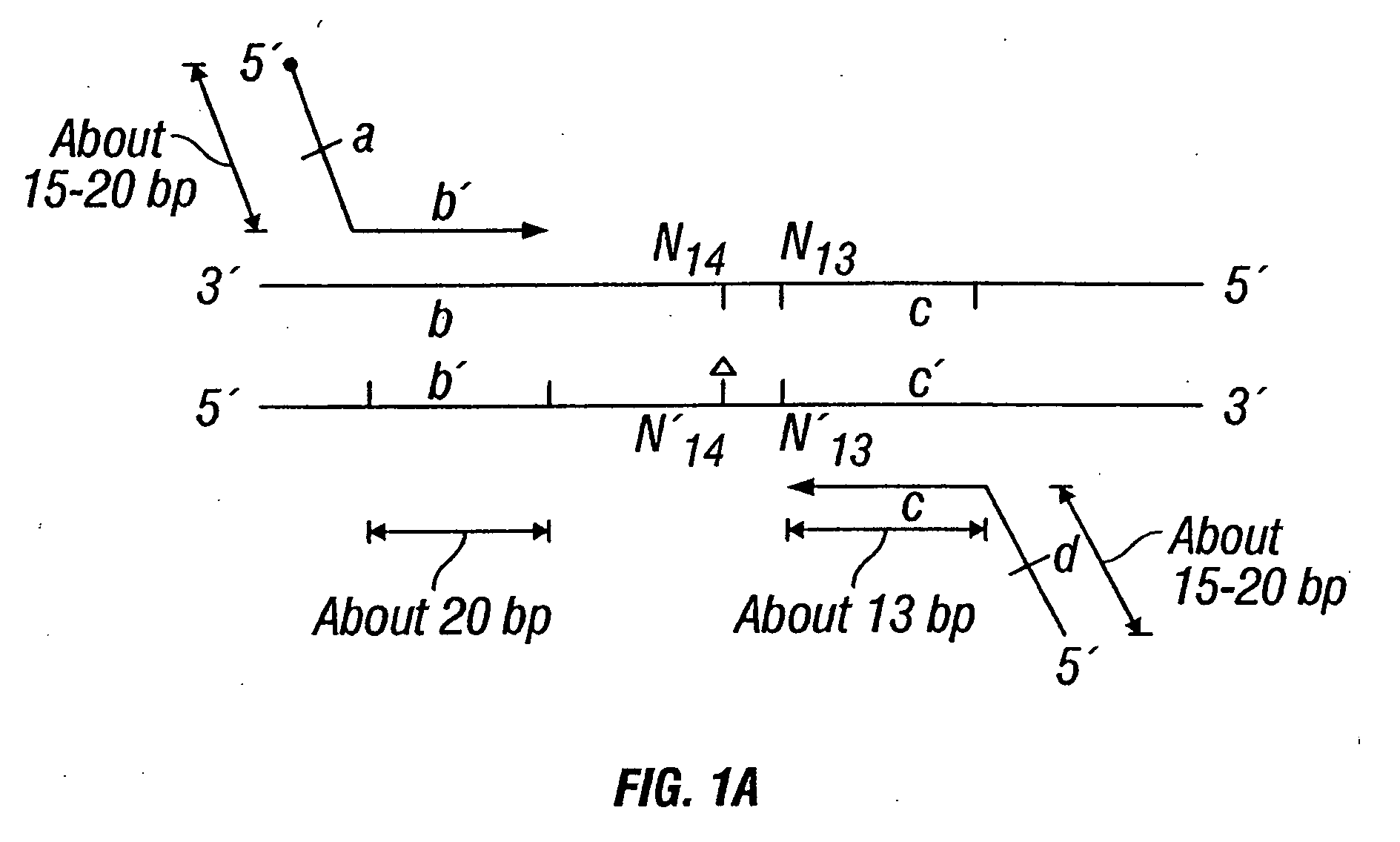
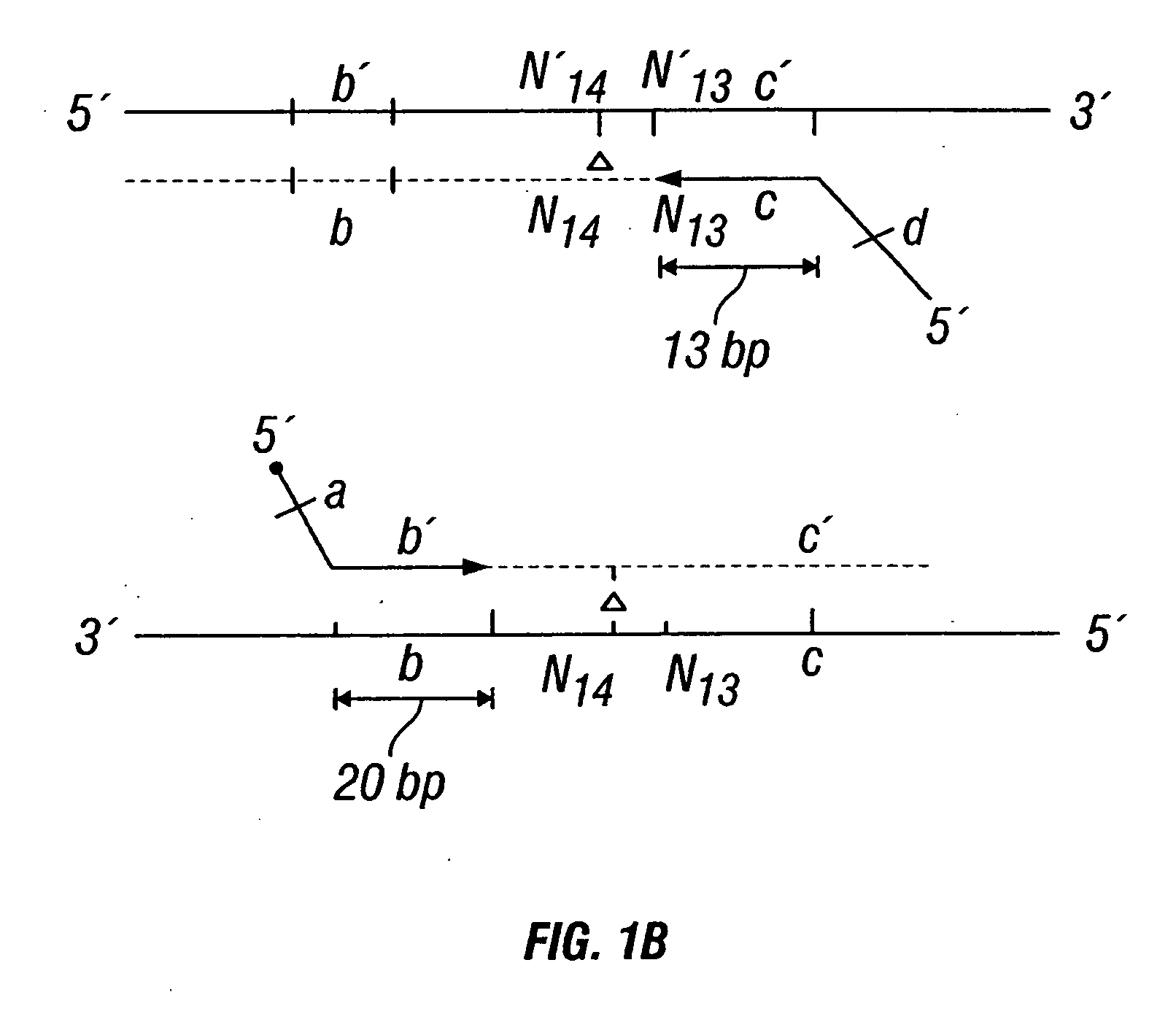
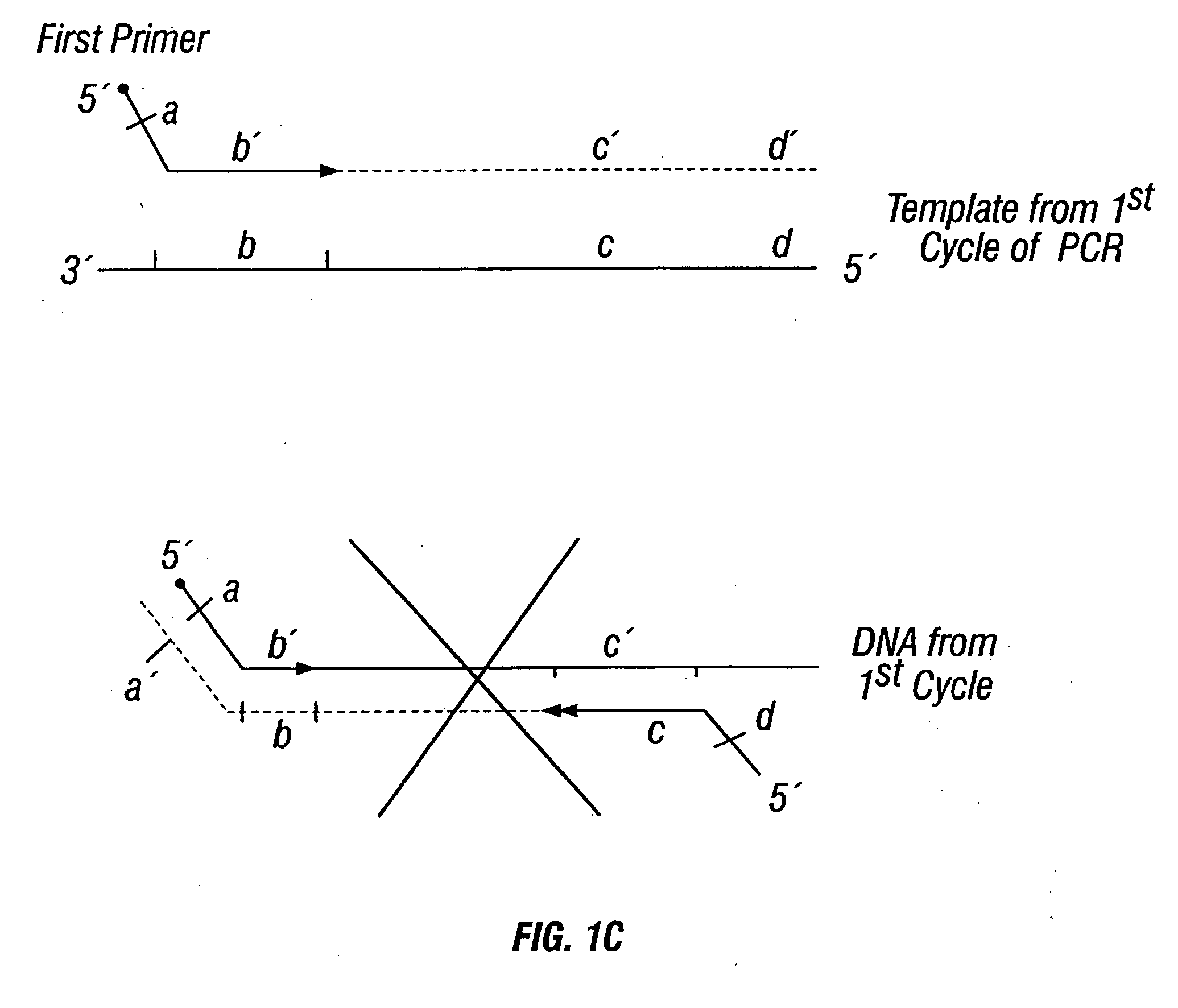
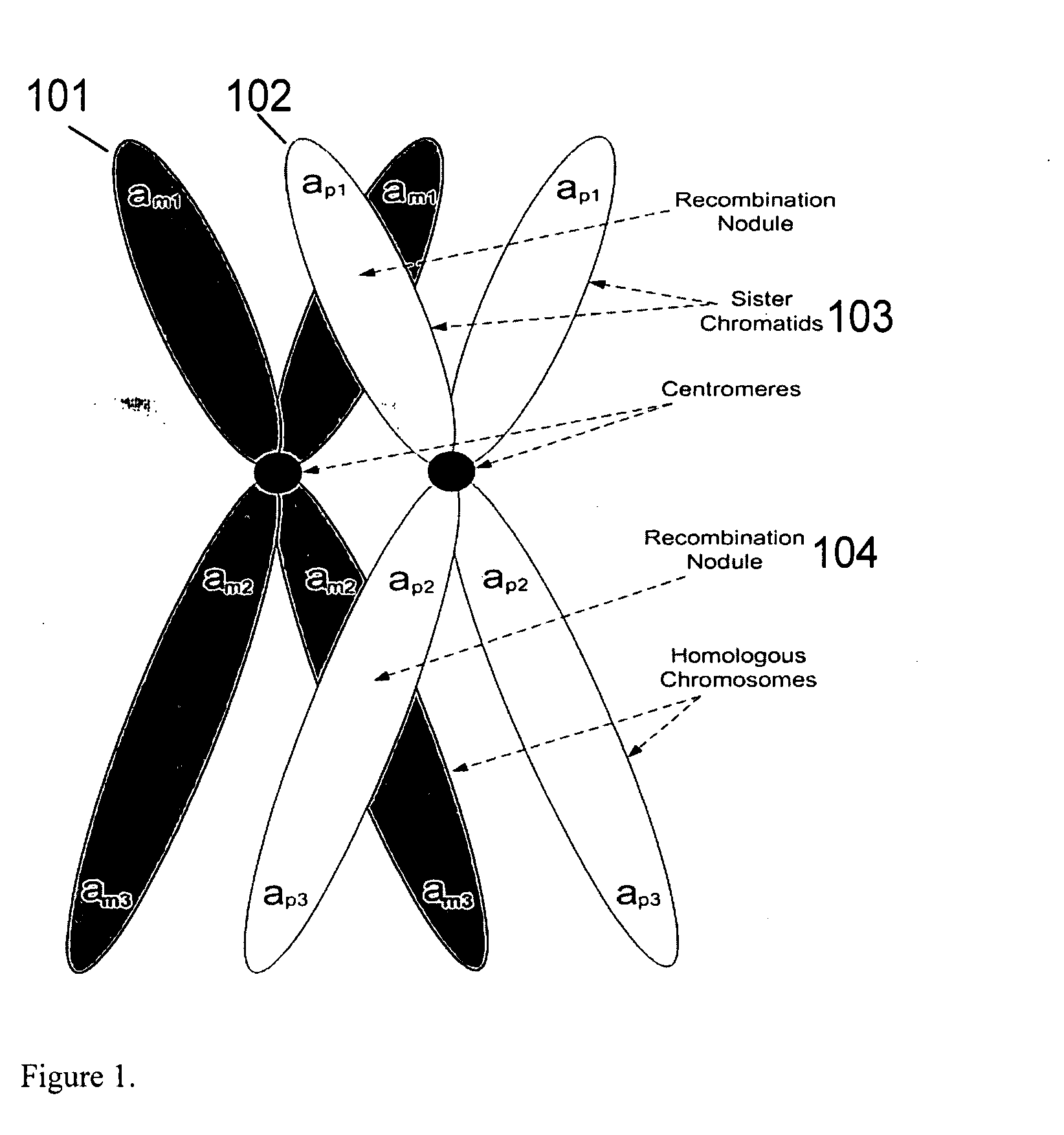
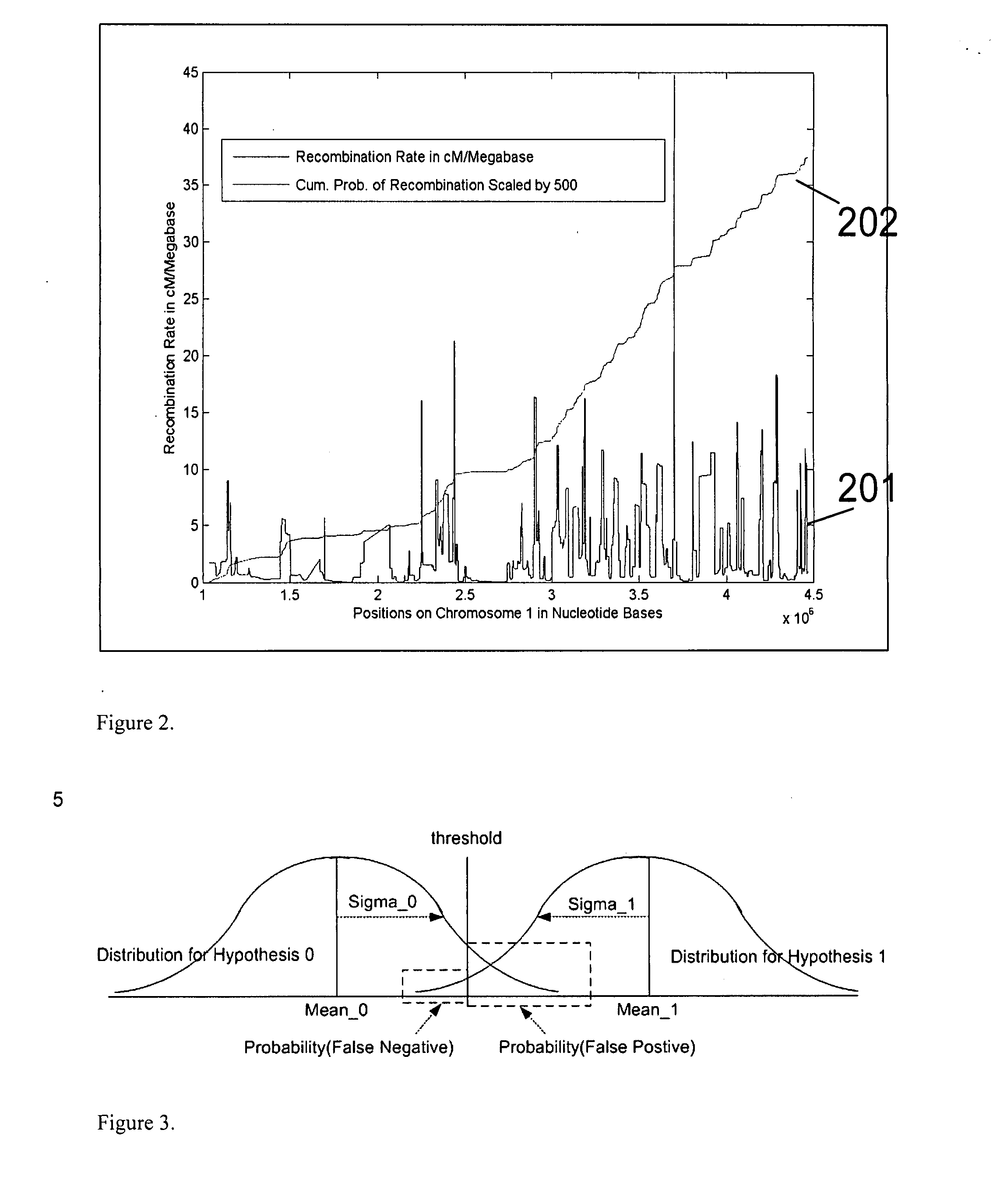
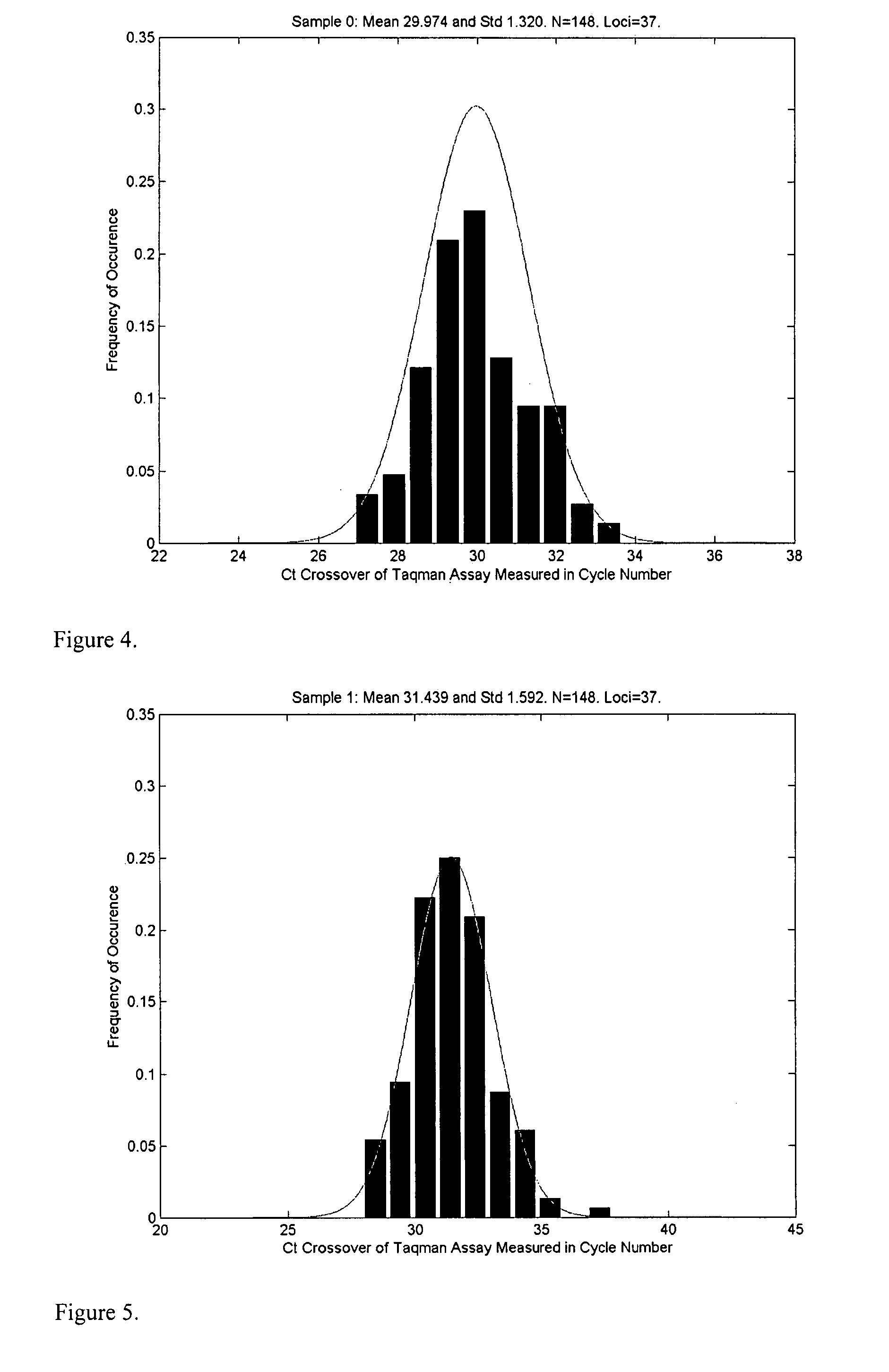
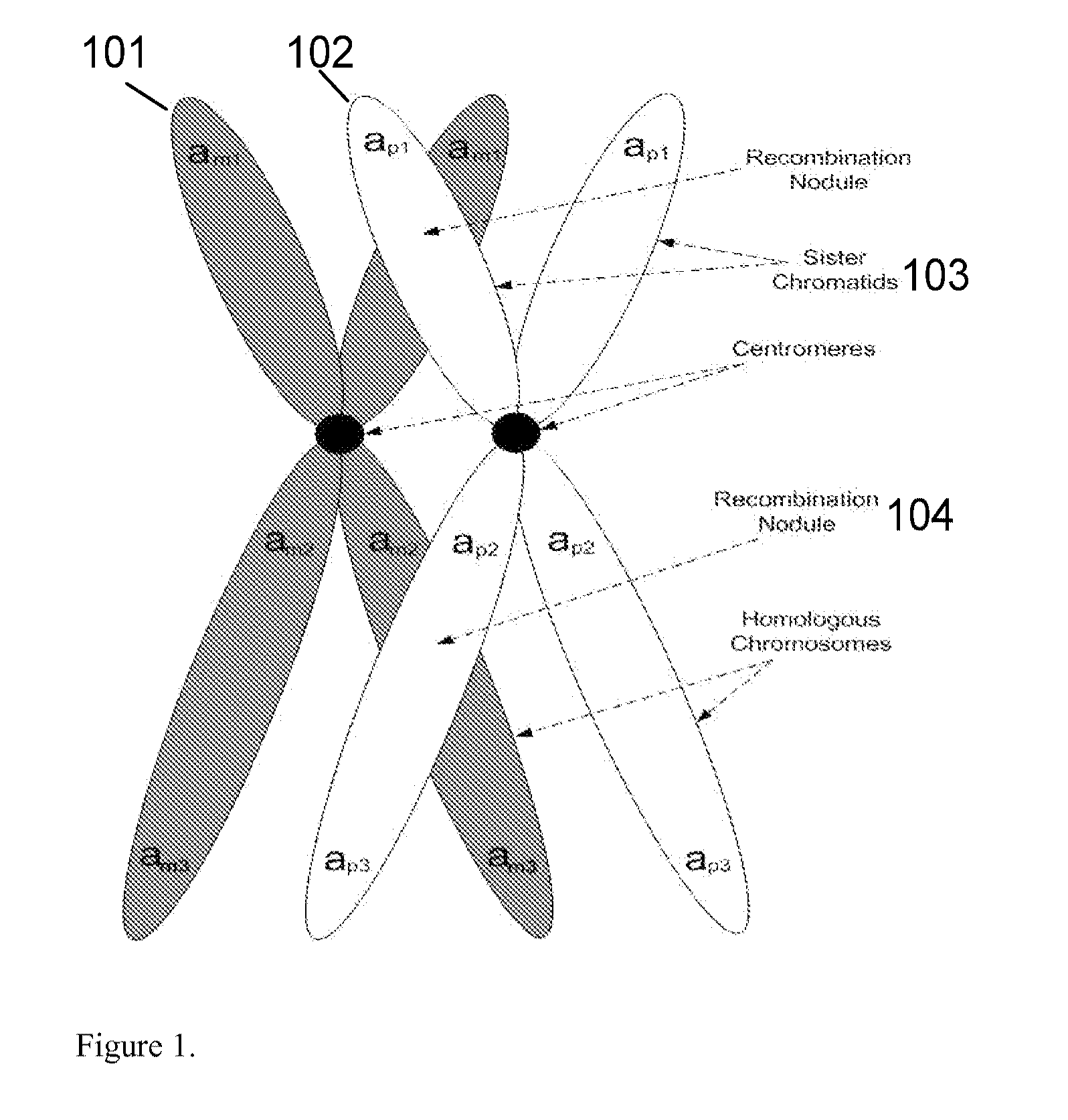
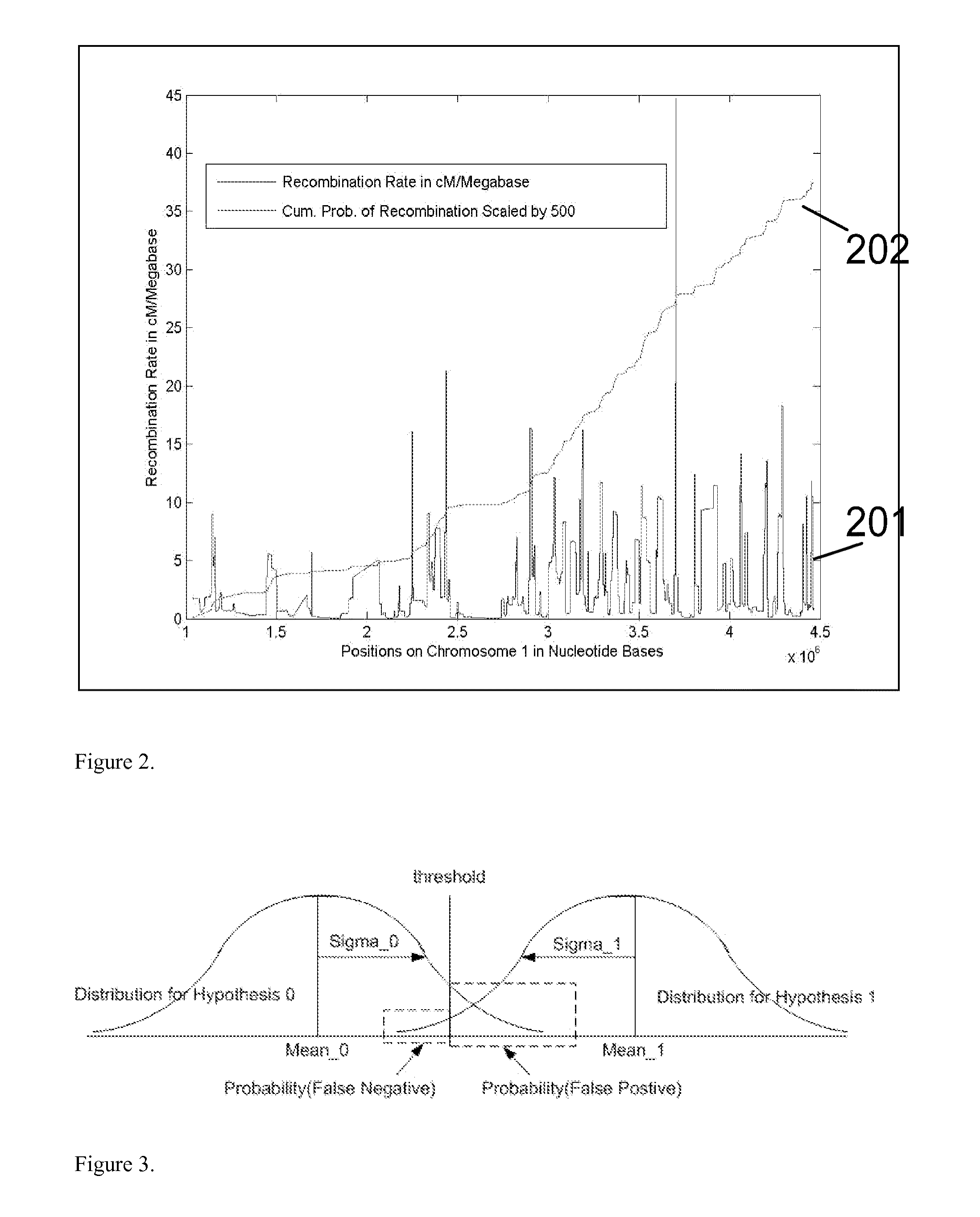
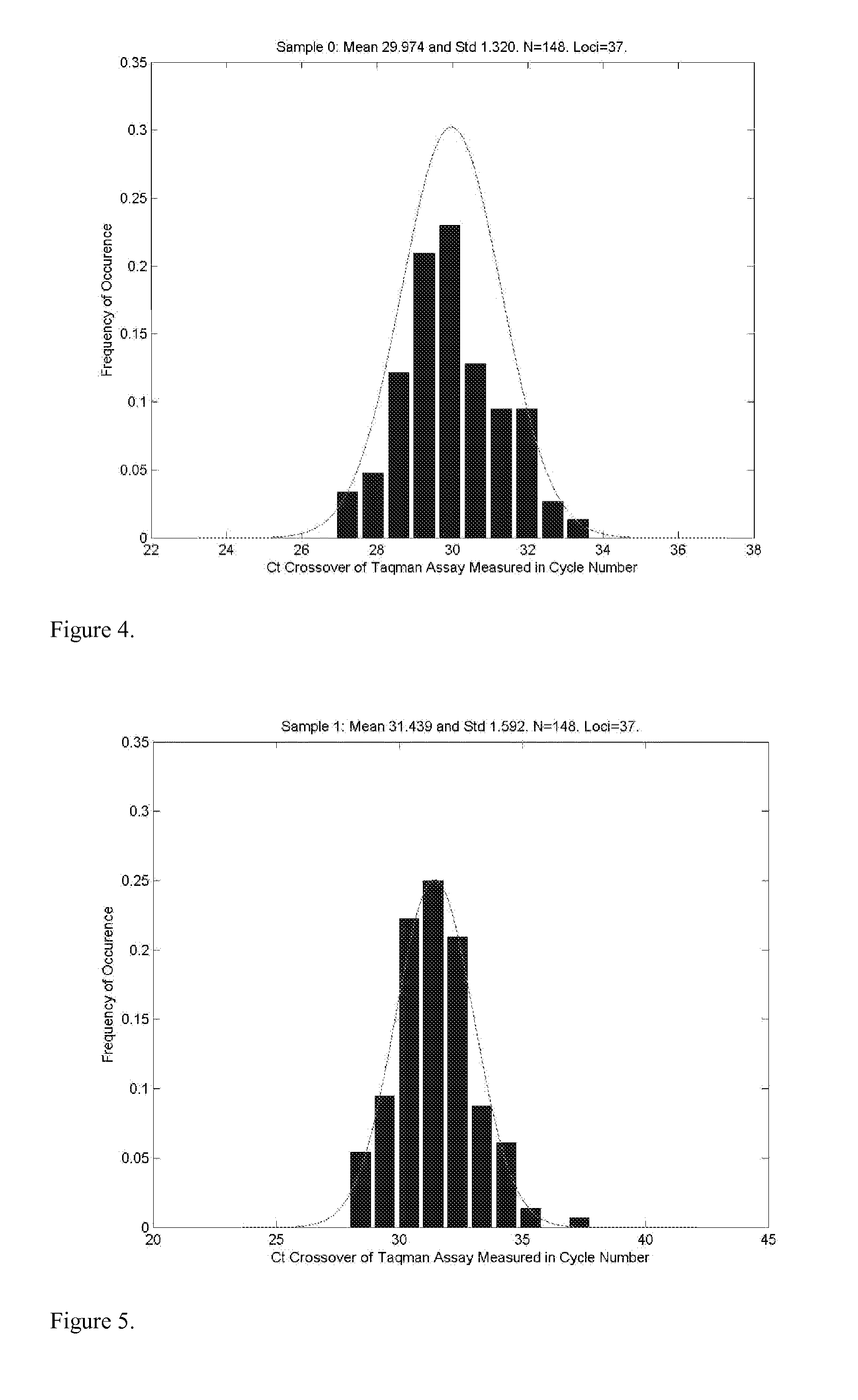
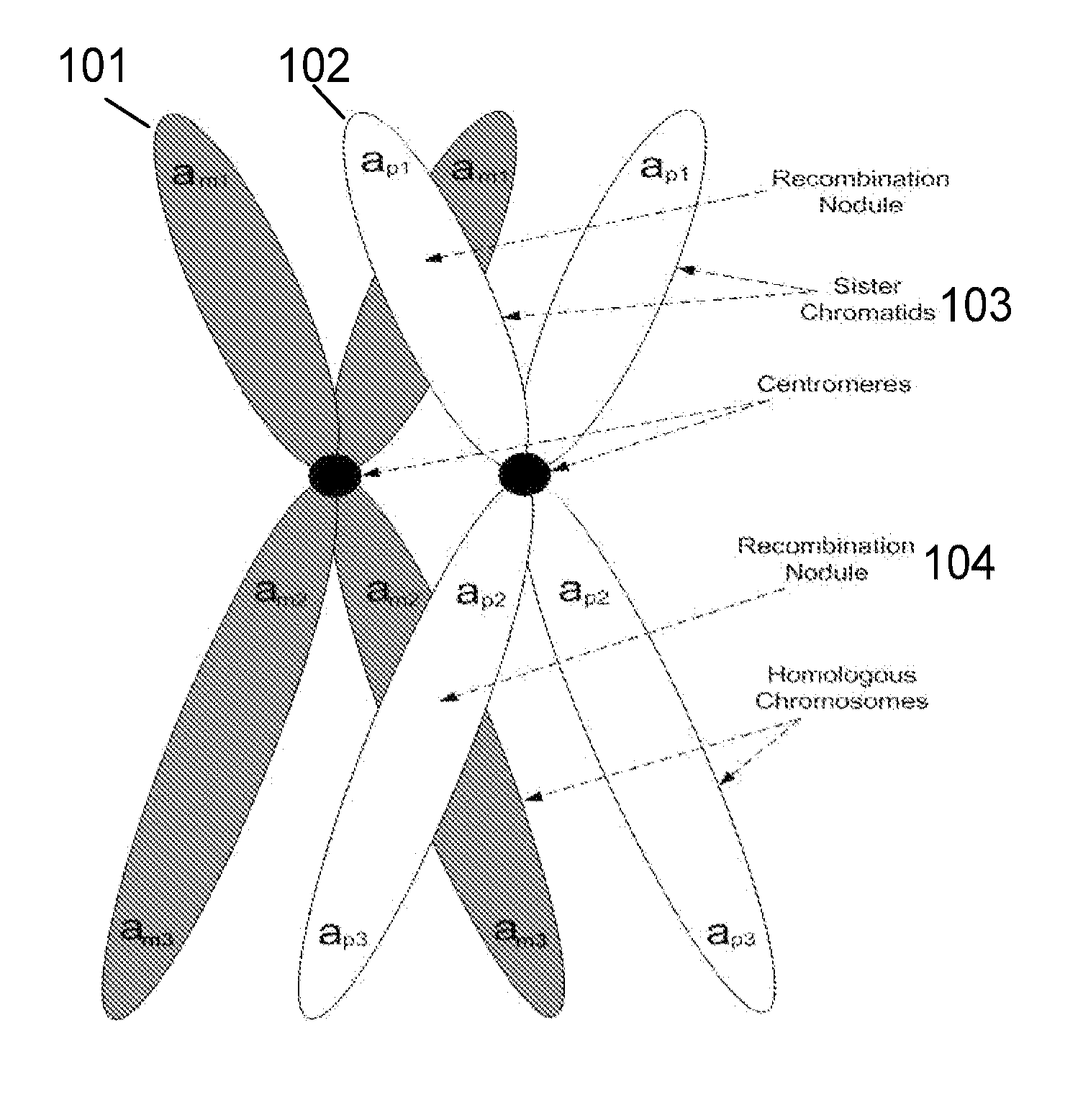
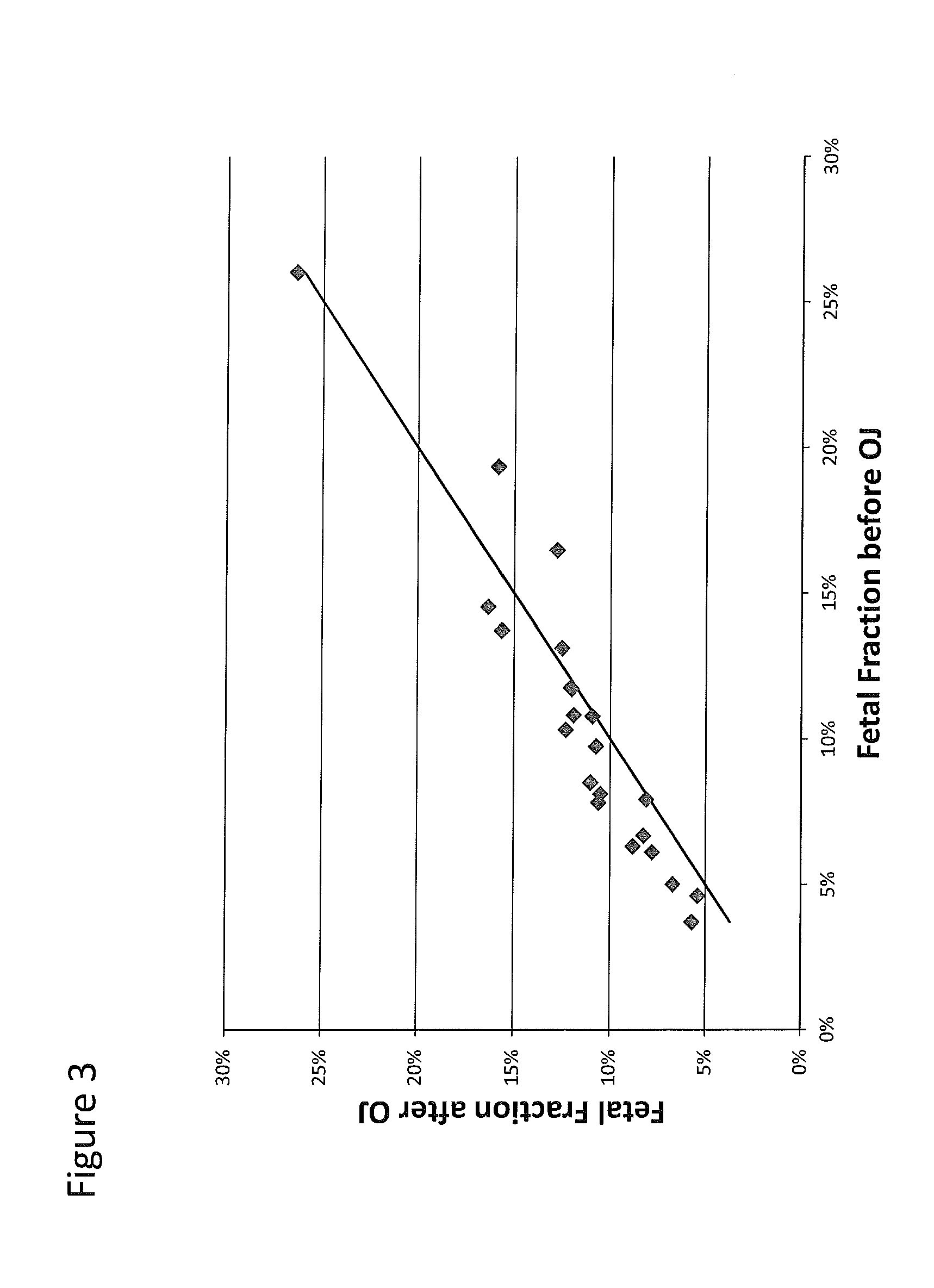
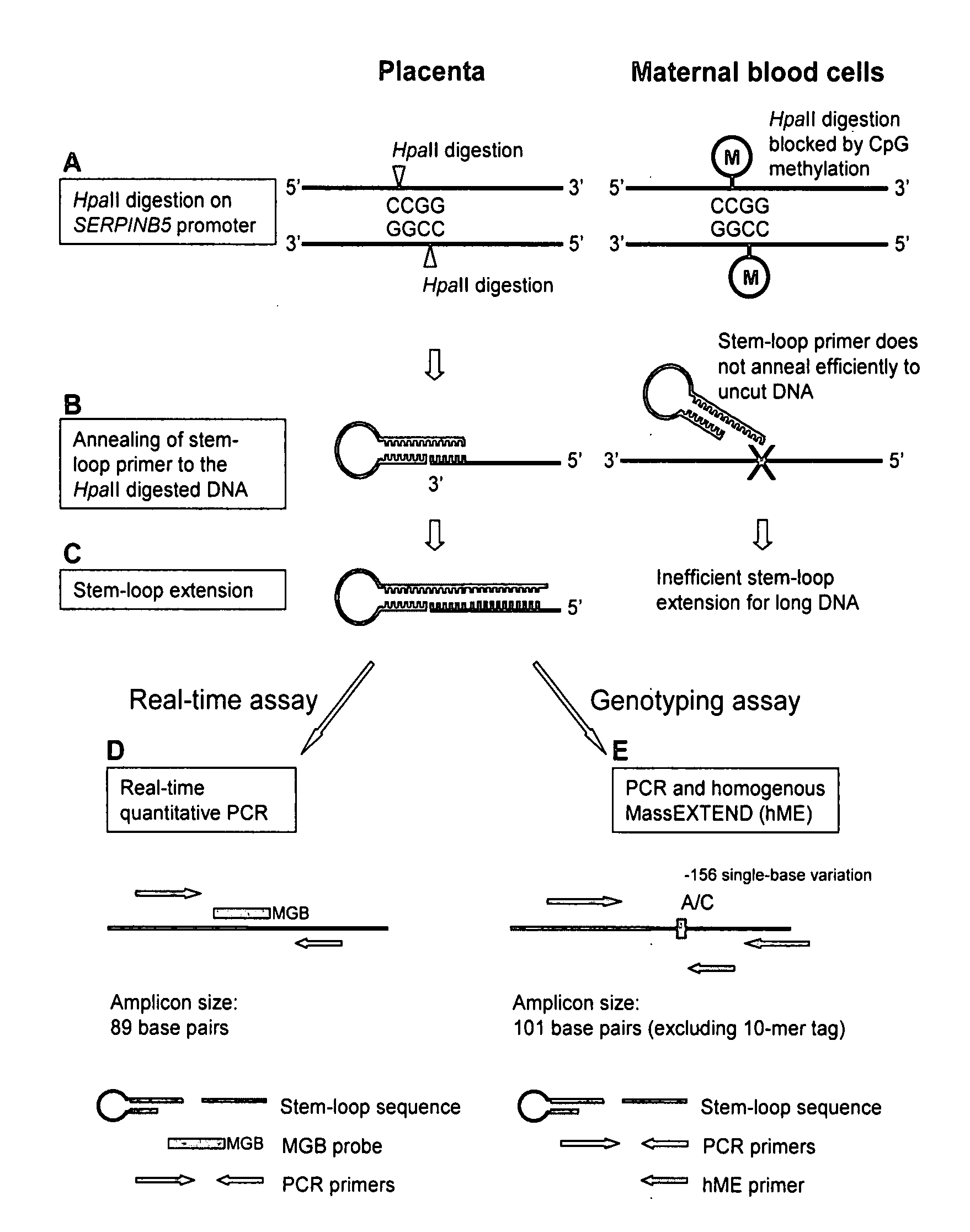
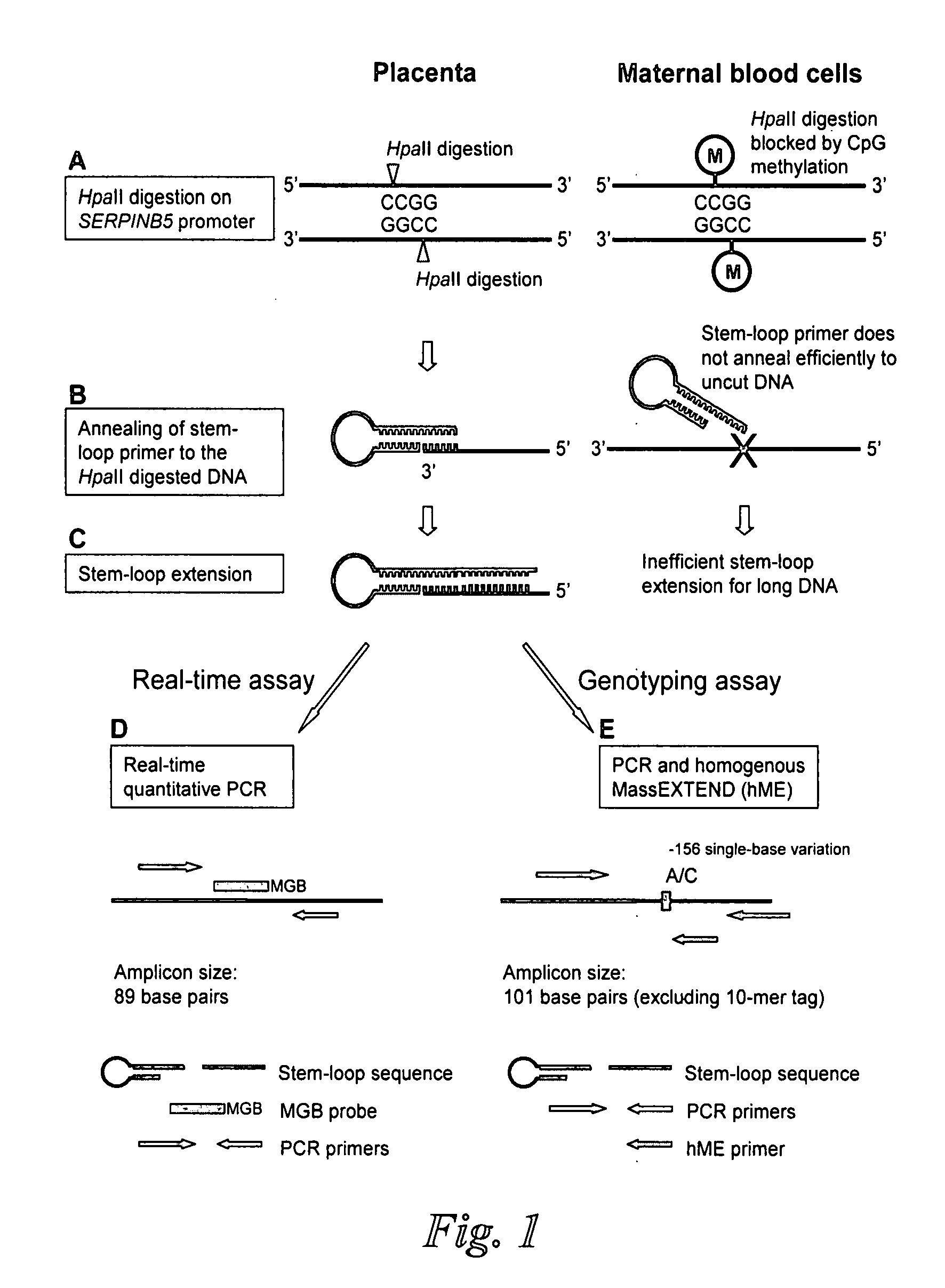
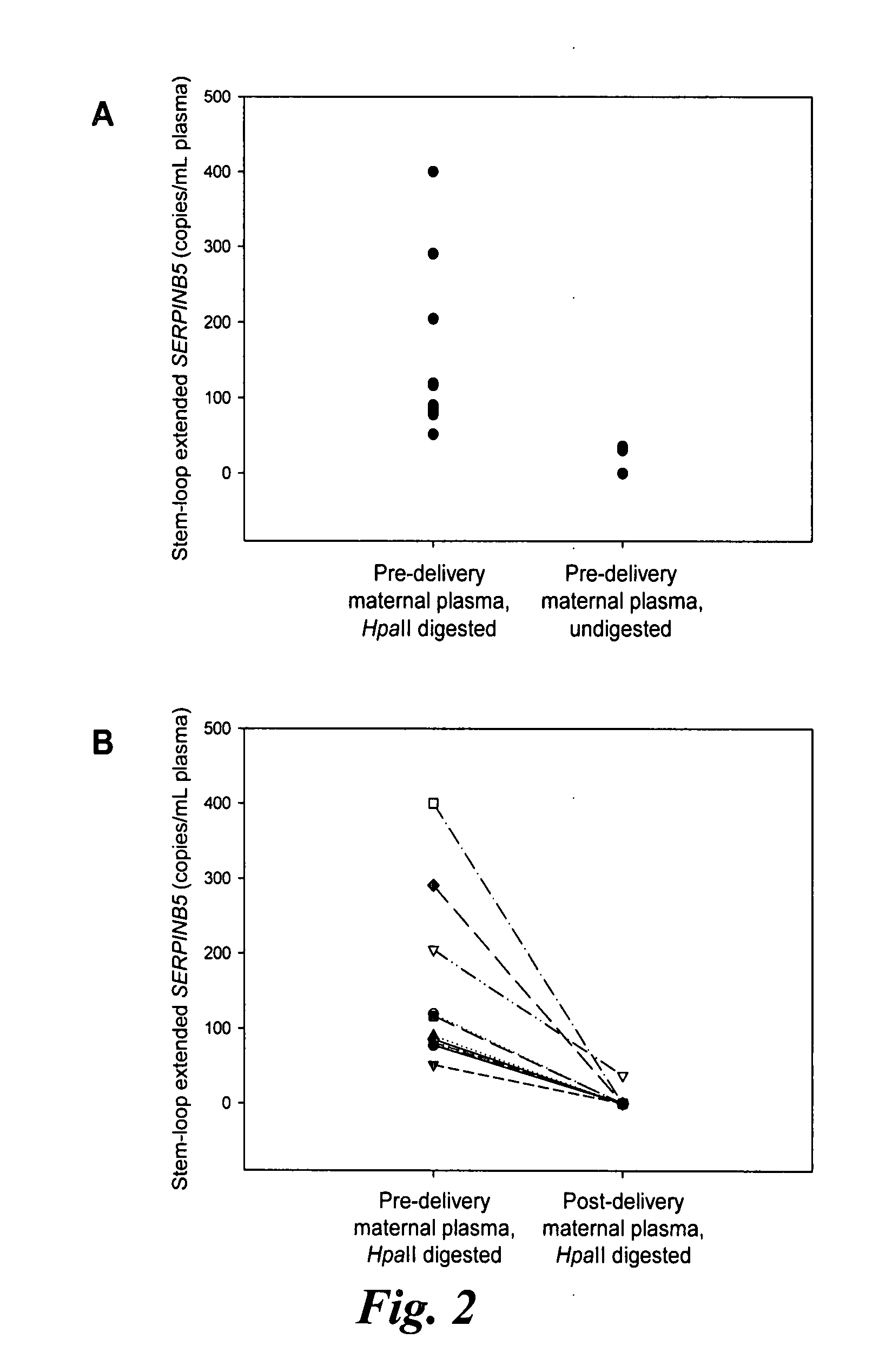
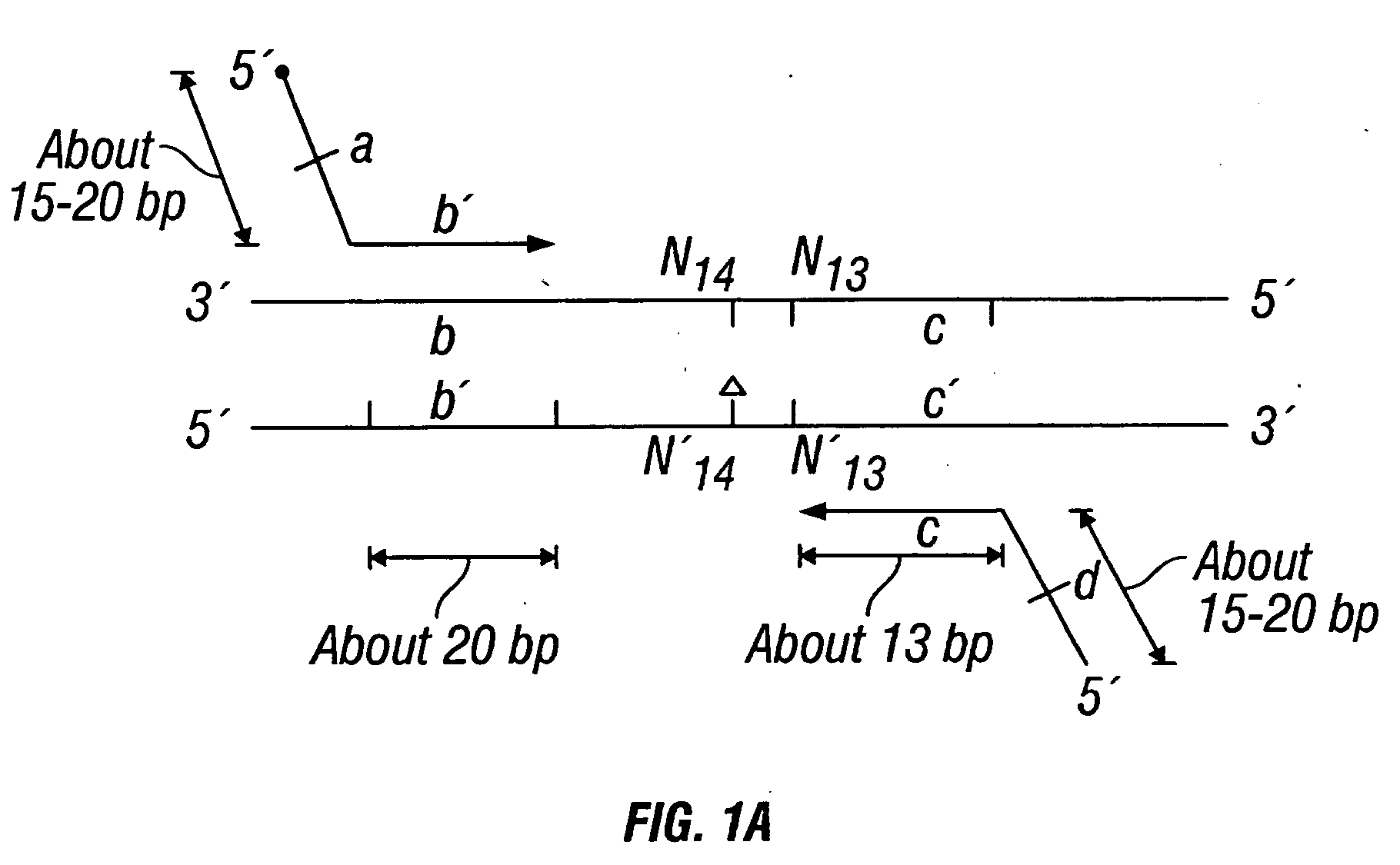
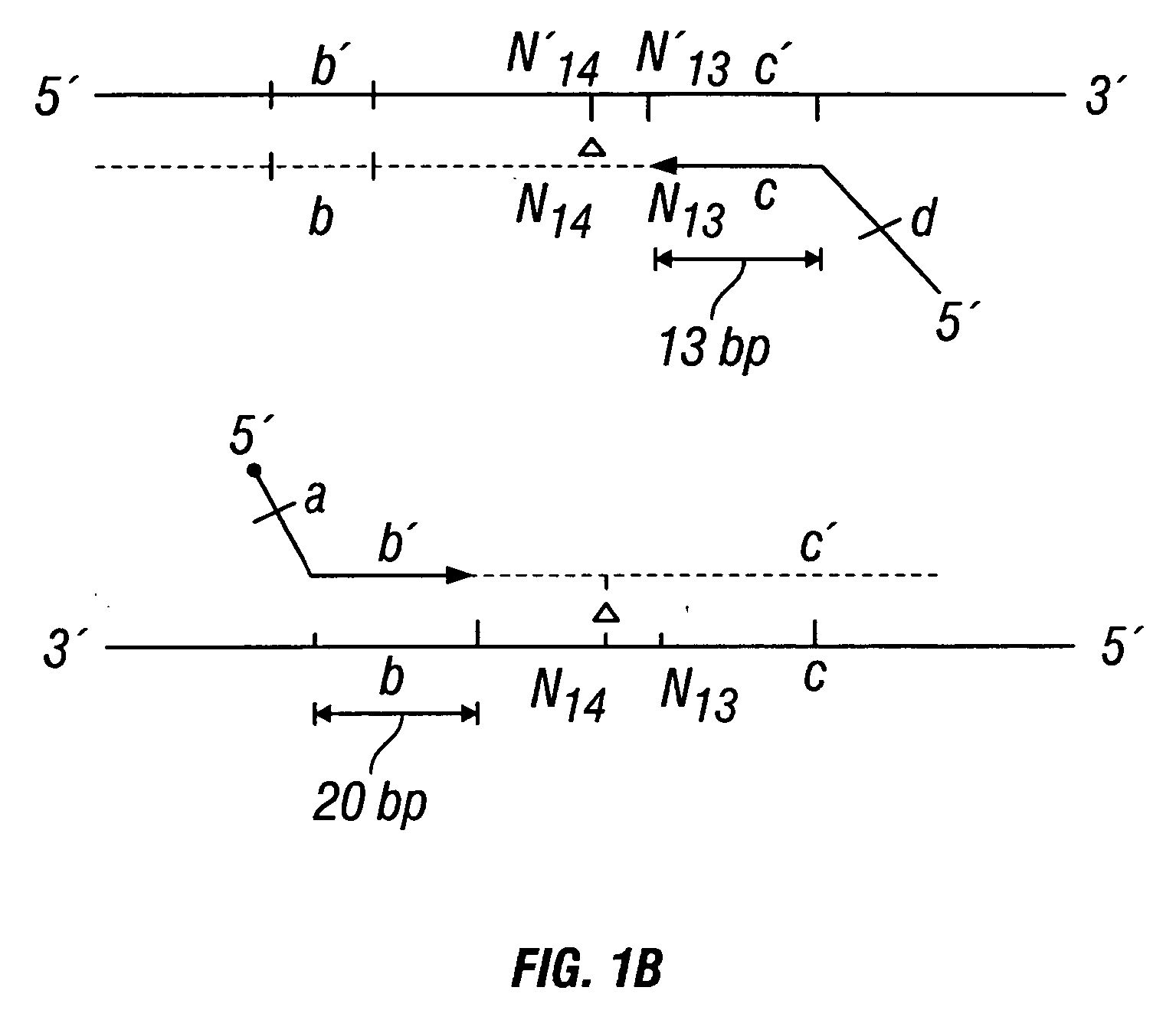
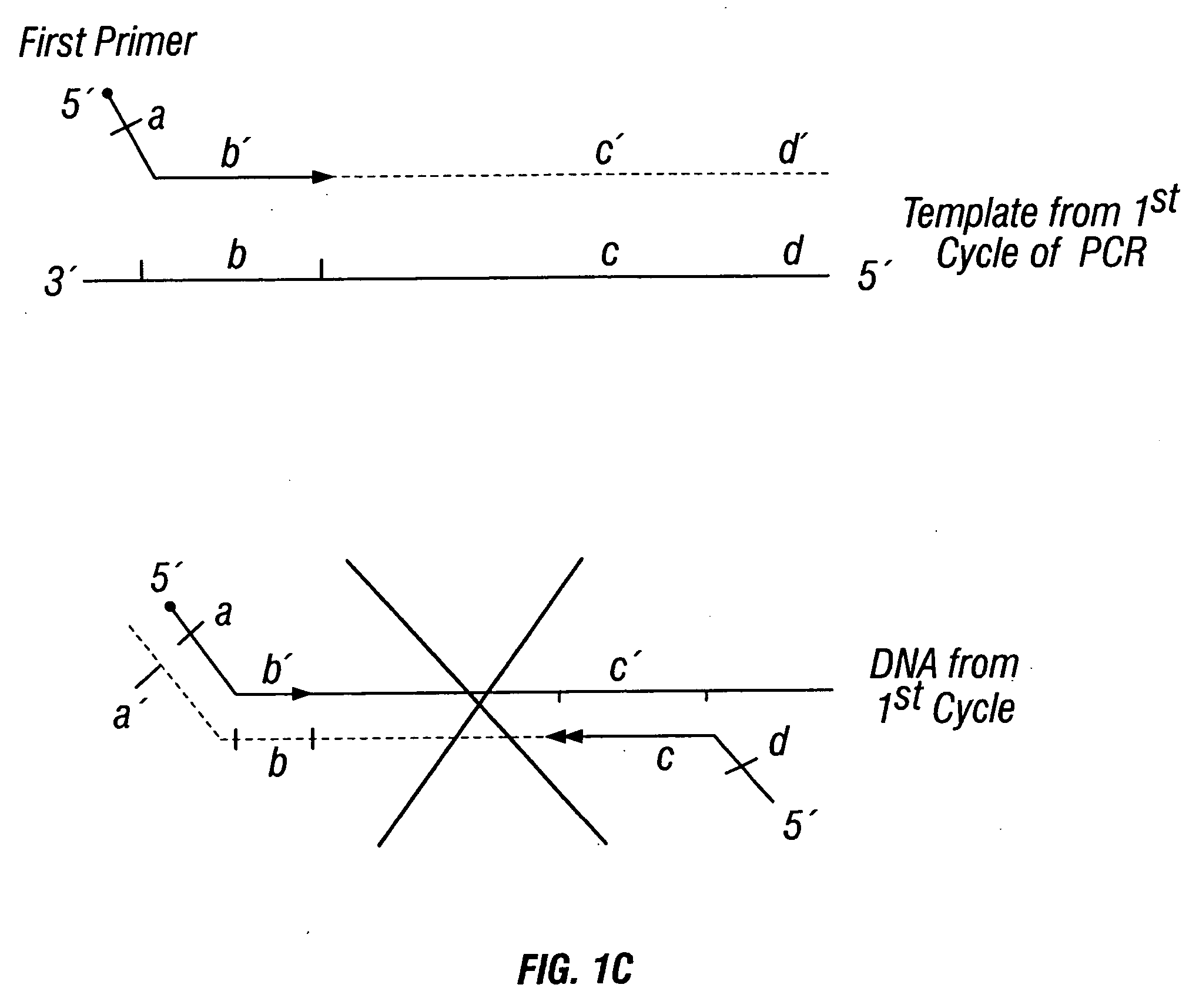
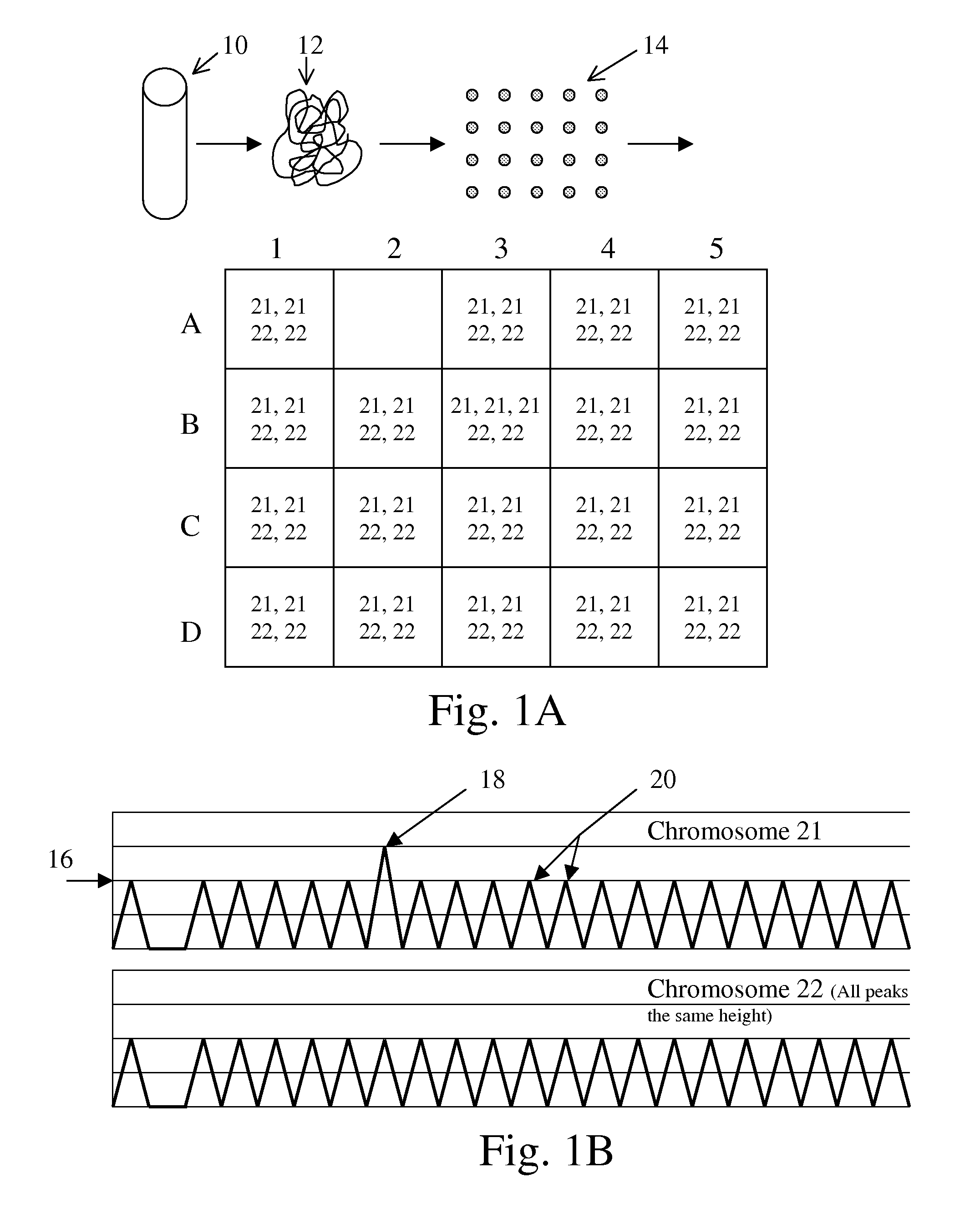
Patents
Literature
123 results about "Fetal dna" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Fetal DNA is a NIPS test that provides valuable insight into fetal chromosome health and can help avoid the risk of a miscarriage associated with invasive diagnostic prenatal procedures, such as an amniocentesis or chorionic villi sampling (CVS).
Non-invasive fetal genetic screening by digital analysis
ActiveUS20070202525A1Enriching fetal DNAMicrobiological testing/measurementFermentationChorionic villiMassively parallel
The present methods are exemplified by a process in which maternal blood containing fetal DNA is diluted to a nominal value of approximately 0.5 genome equivalent of DNA per reaction sample. Digital PCR is then be used to detect aneuploidy, such as the trisomy that causes Down Syndrome. Since aneuploidies do not present a mutational change in sequence, and are merely a change in the number of chromosomes, it has not been possible to detect them in a fetus without resorting to invasive techniques such as amniocentesis or chorionic villi sampling. Digital amplification allows the detection of aneuploidy using massively parallel amplification and detection methods, examining, e.g., 10,000 genome equivalents.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Non-invasive methods for detecting non-host DNA in a host using epigenetic differences between the host and non-host DNA
In a first aspect, the present invention features methods for differentiating DNA species originating from different individuals in a biological sample. These methods may be used to differentiate or detect fetal DNA in a maternal sample or to differentiate DNA of an organ donor from DNA of an organ recipient. In preferred embodiments, the DNA species are differentiated by observing epigenetic differences in the DNA species such as differences in DNA methylation. In a second aspect, the present invention features methods of detecting genetic abnormalities in a fetus by detecting fetal DNA in a biological sample obtained from a mother. In a third aspect, the present invention features methods for differentiating DNA species originating from an organ donor from those of an organ recipient. In a fourth aspect, the present invention features kits for differentiating DNA species originating from different individuals in a biological sample.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Non-invasive detection of fetal genetic traits
InactiveUS20050164241A1Facilitates non-invasive detectionComponent separationOther chemical processesPregnancyNon invasive
Blood plasma of pregnant women contains fetal and (generally>90%) maternal circulatory extracellular DNA. Most of said fetal DNA contains ≦500 base pairs, said maternal DNA having a greater size. Separation of circulatory extracellular DNA of <500 base pairs results in separation of fetal from maternal DNA. A fraction of a blood plasma or serum sample of a pregnant woman containing, due to size separation (e.g. by chromatography, density gradient centrifugation or nanotechnological methods), extracellular DNA substantially comprising ≦500 base pairs is useful for non-invasive detection of fetal genetic traits (including the fetal RhD gene in pregnancies at risk for HDN; fetal Y chromosome-specific sequences in pregnancies at risk for X chromosome-linked disorders; chromosomal aberrations; hereditary Mendelian genetic disorders and corresponding genetic markers; and traits decisive for paternity determination) by e.g. PCR, ligand chain reaction or probe hybridization techniques, or nucleic acid arrays.
Owner:SEQUENOM INC
Determining a nucleic acid sequence imbalance
ActiveUS20090087847A1Quantity minimizationMicrobiological testing/measurementDisease diagnosisNucleic acid sequencingNucleic acid sequence
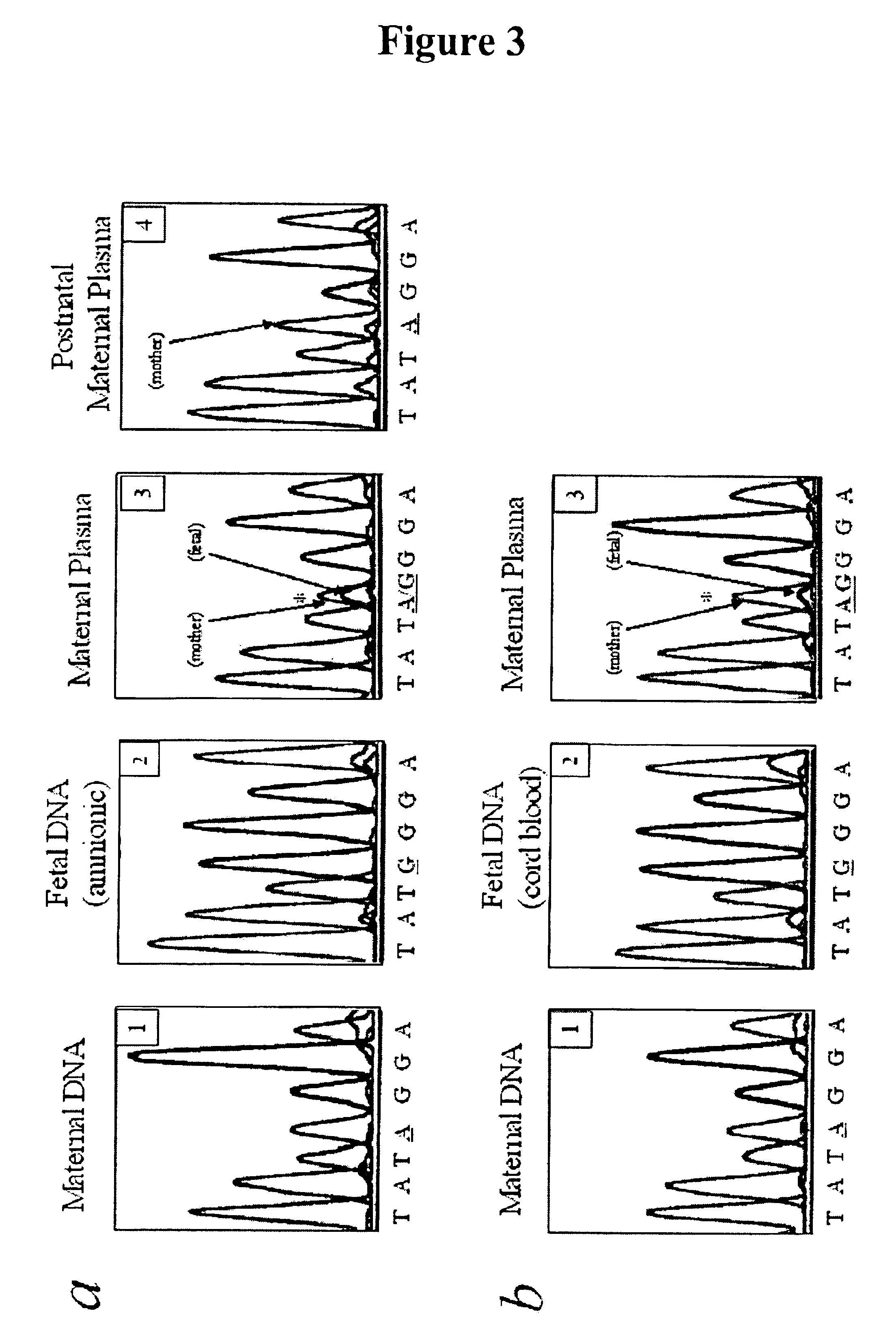
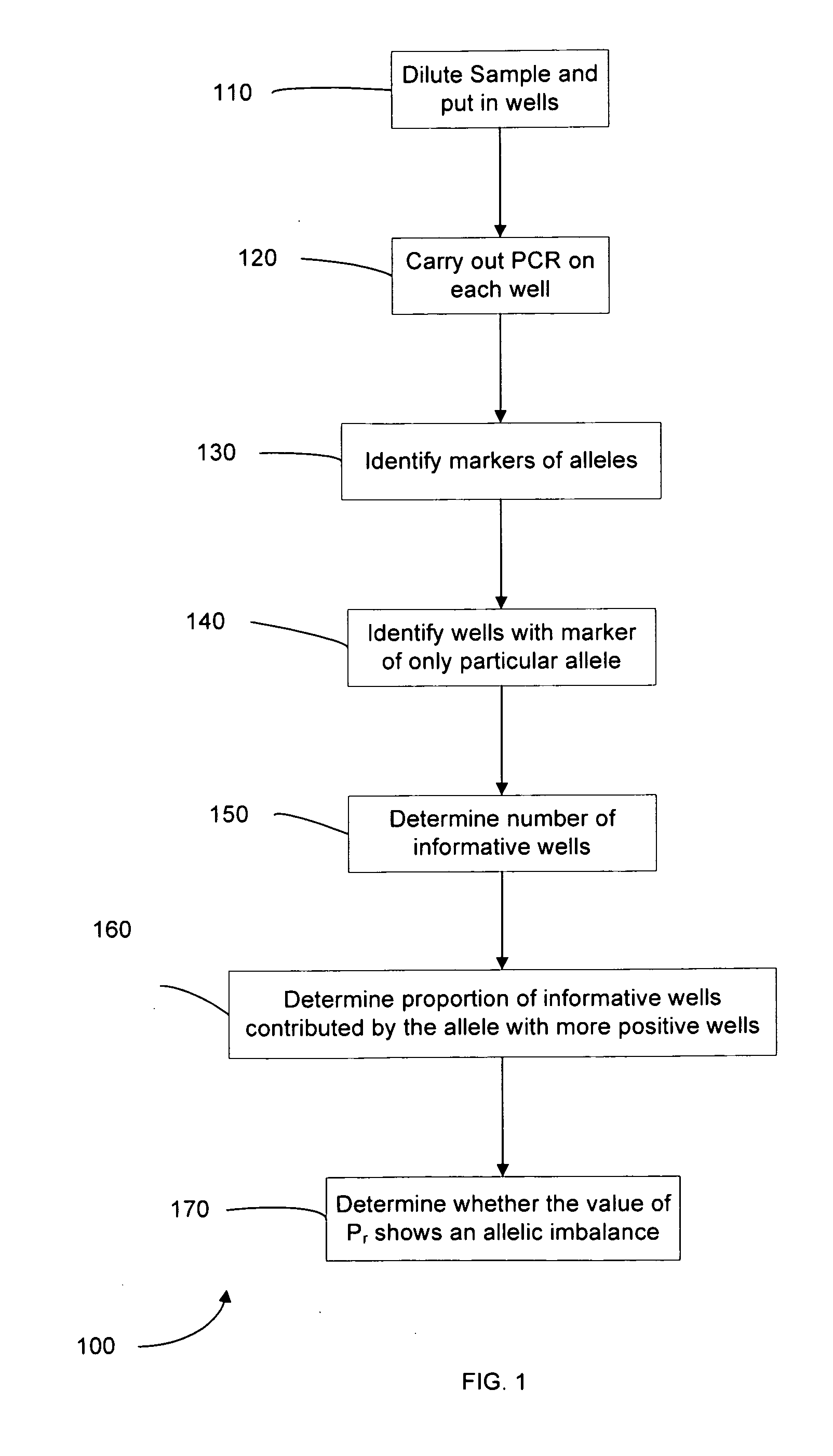
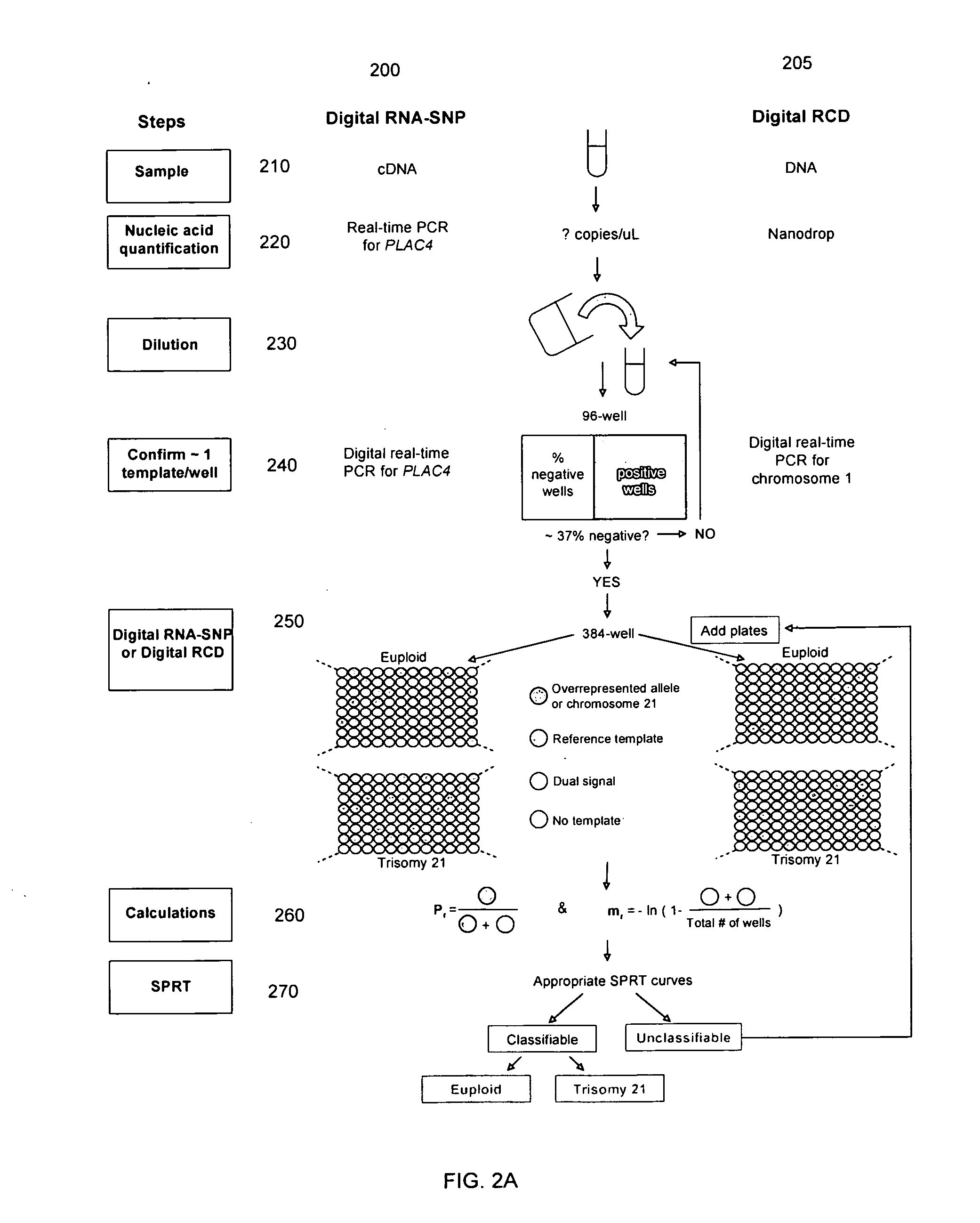
Methods, systems, and apparatus are provided for determining whether a nucleic acid sequence imbalance exists within a biological sample. One or more cutoff values for determining an imbalance of, for example, the ratio of the two sequences (or sets of sequences) are chosen. The cutoff value may be determined based at least in part on the percentage of fetal DNA in a sample, such as maternal plasma, containing a background of maternal nucleic acid sequences. The cutoff value may also be determined based on an average concentration of a sequence per reaction. In one aspect, the cutoff value is determined from a proportion of informative wells that are estimated to contain a particular nucleic acid sequence, where the proportion is determined based on the above-mentioned percentage and / or average concentration. The cutoff value may be determined using many different types of methods, such as sequential probability ratio testing (SPRT).
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Methods for detection of genetic disorders
InactiveUS7727720B2Maximize efficiencyIncrease delaySugar derivativesMicrobiological testing/measurementGeneticsAllele
The invention provides a method useful for detection of genetic disorders. The method comprises determining the sequence of alleles of a locus of interest, and quantitating a ratio for the alleles at the locus of interest, wherein the ratio indicates the presence or absence of a chromosomal abnormality. The present invention also provides a non-invasive method for the detection of chromosomal abnormalities in a fetus. The invention is especially useful as a non-invasive method for determining the sequence of fetal DNA. The invention further provides methods of isolation of free DNA from a sample.
Owner:RAVGEN INC
Methods for detection of genetic disorders
InactiveUS7442506B2Maximize efficiencyIncrease delaySugar derivativesMicrobiological testing/measurementGeneticsAllele
The invention provides a method useful for detection of genetic disorders. The method comprises determining the sequence of alleles of a locus of interest, and quantitating a ratio for the alleles at the locus of interest, wherein the ratio indicates the presence or absence of a chromosomal abnormality. The present invention also provides a non-invasive method for the detection of chromosomal abnormalities in a fetus. The invention is especially useful as a non-invasive method for determining the sequence of fetal DNA. The invention further provides methods of isolation of free DNA from a sample.
Owner:RAVGEN INC
Methods for detection of genetic disorders
InactiveUS20070178478A1Minimize disruptionMaximize efficiencyMicrobiological testing/measurementFermentationGeneticsAllele
The invention provides a method useful for detection of genetic disorders. The method comprises determining the sequence of alleles of a locus of interest, and quantitating a ratio for the alleles at the locus of interest, wherein the ratio indicates the presence or absence of a chromosomal abnormality. The present invention also provides a non-invasive method for the detection of chromosomal abnormalities in a fetus. The invention is especially useful as a non-invasive method for determining the sequence of fetal DNA. The invention further provides methods of isolation of free DNA from a sample.
Owner:RAVGEN INC
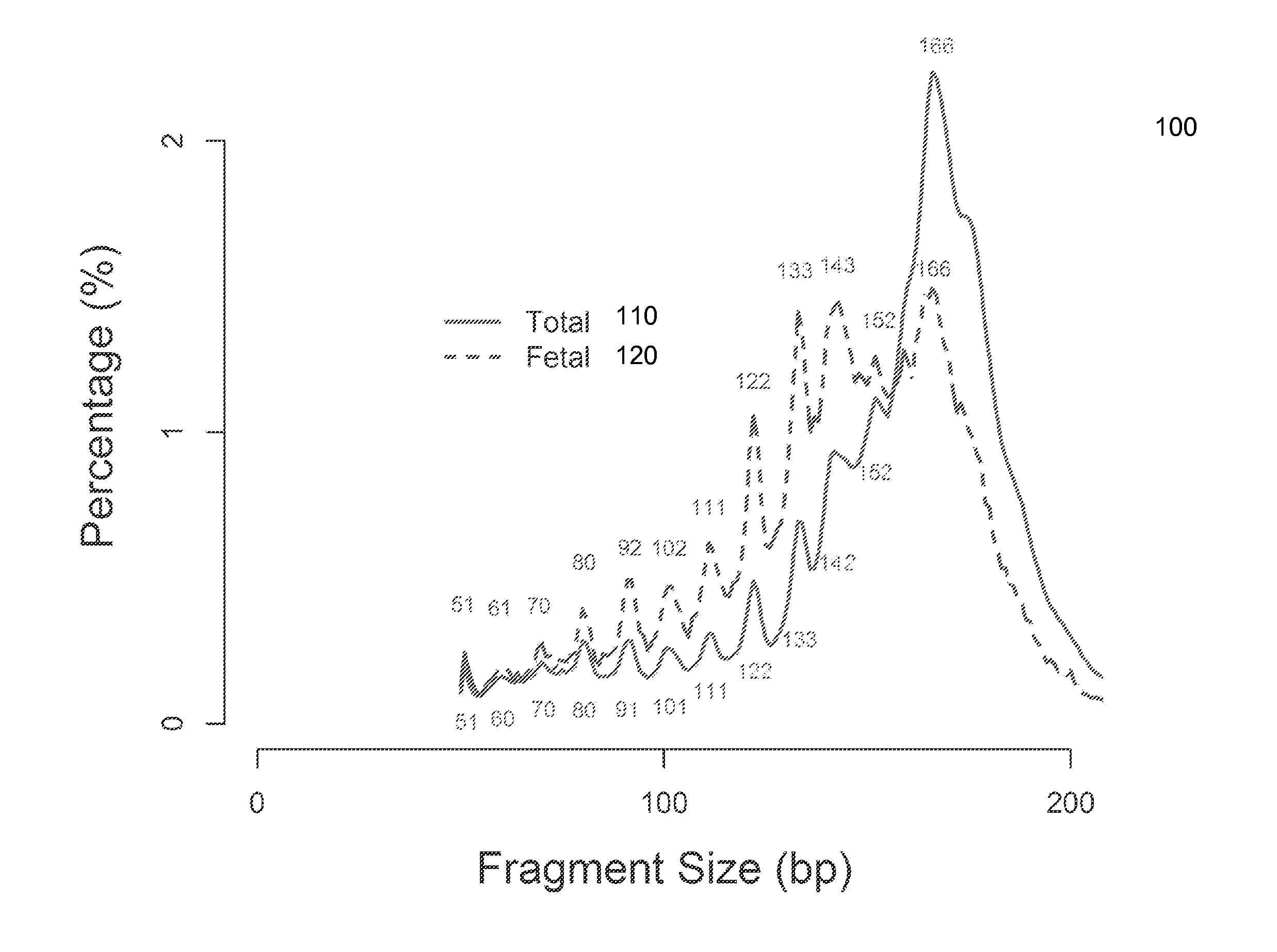
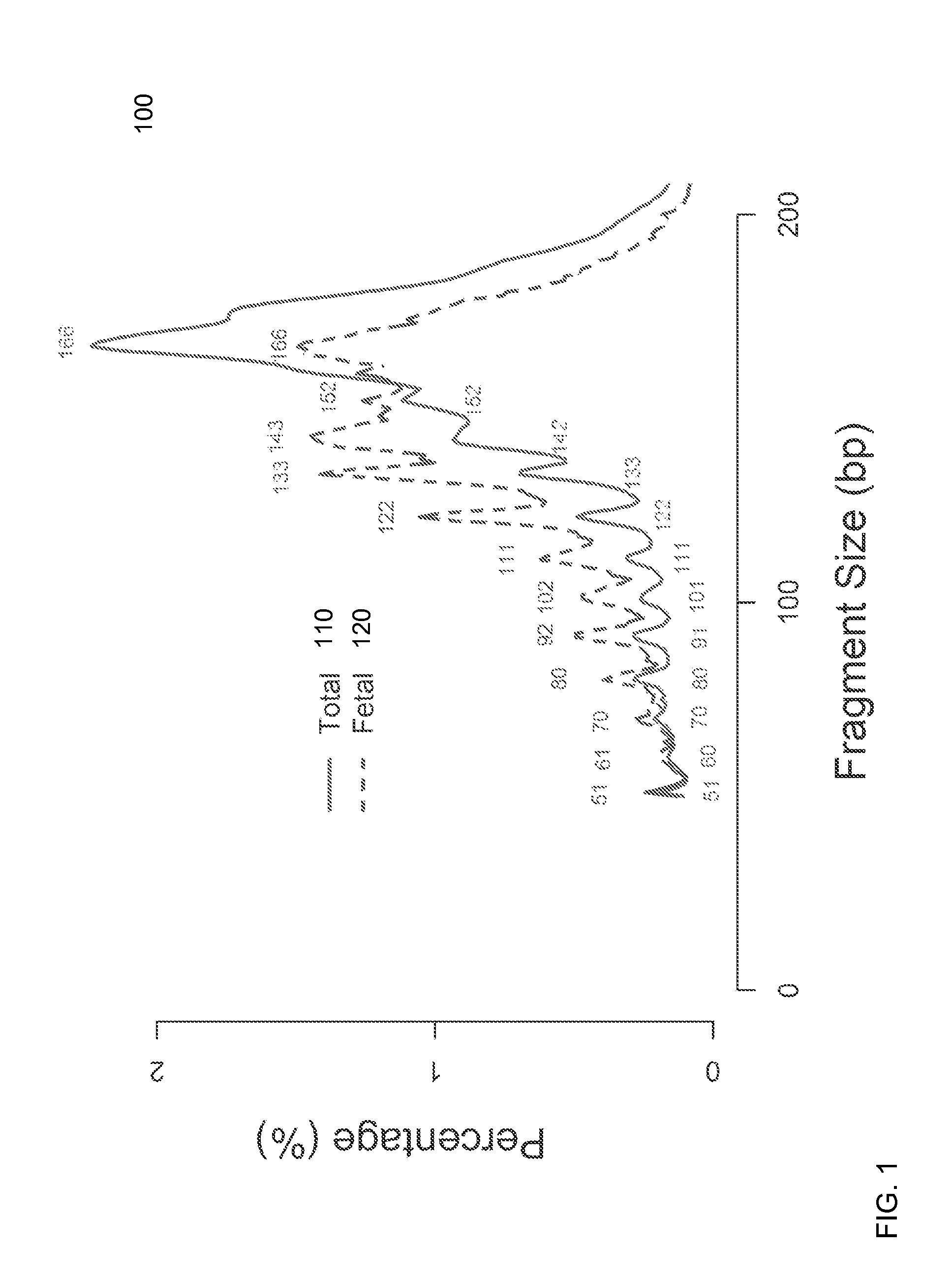
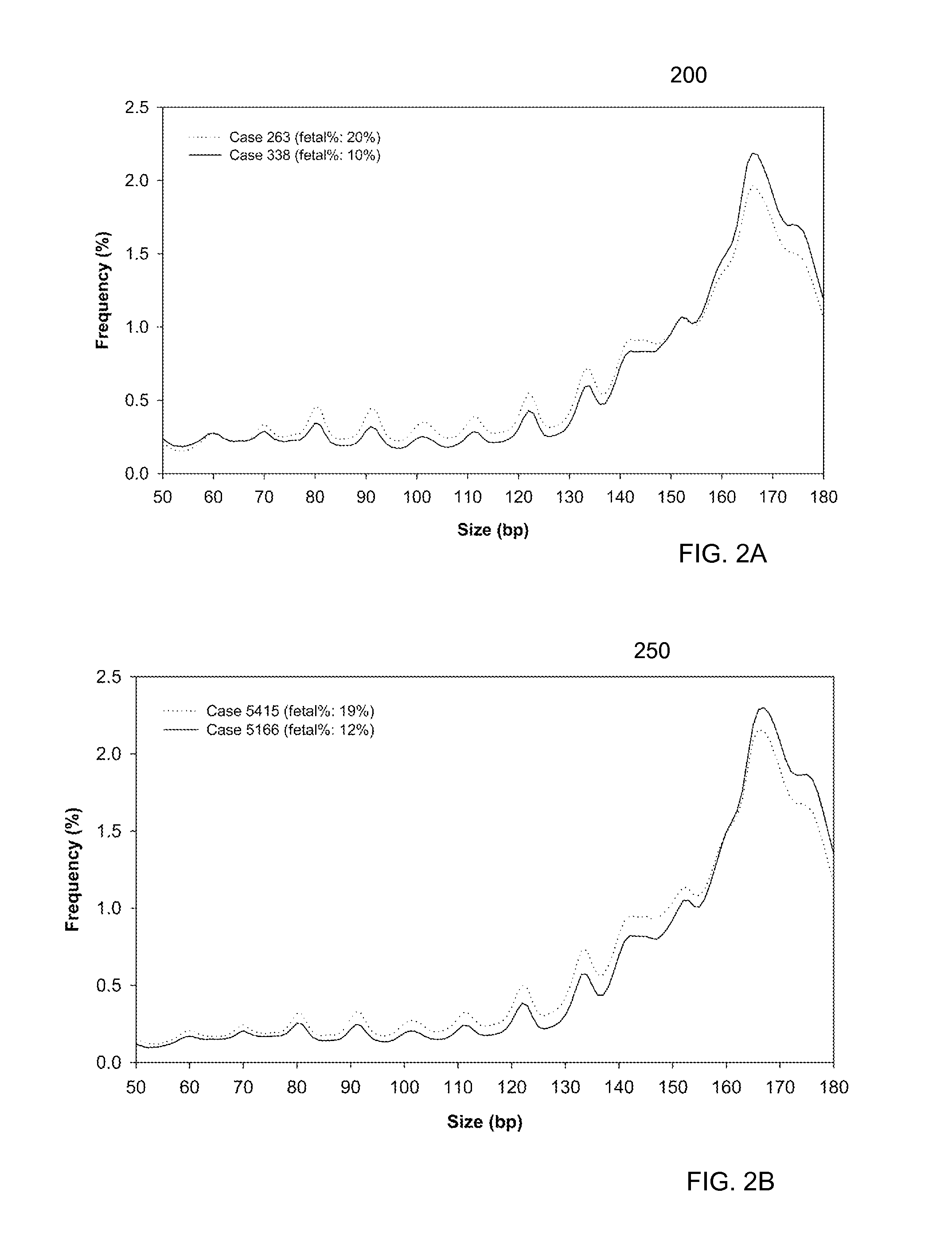
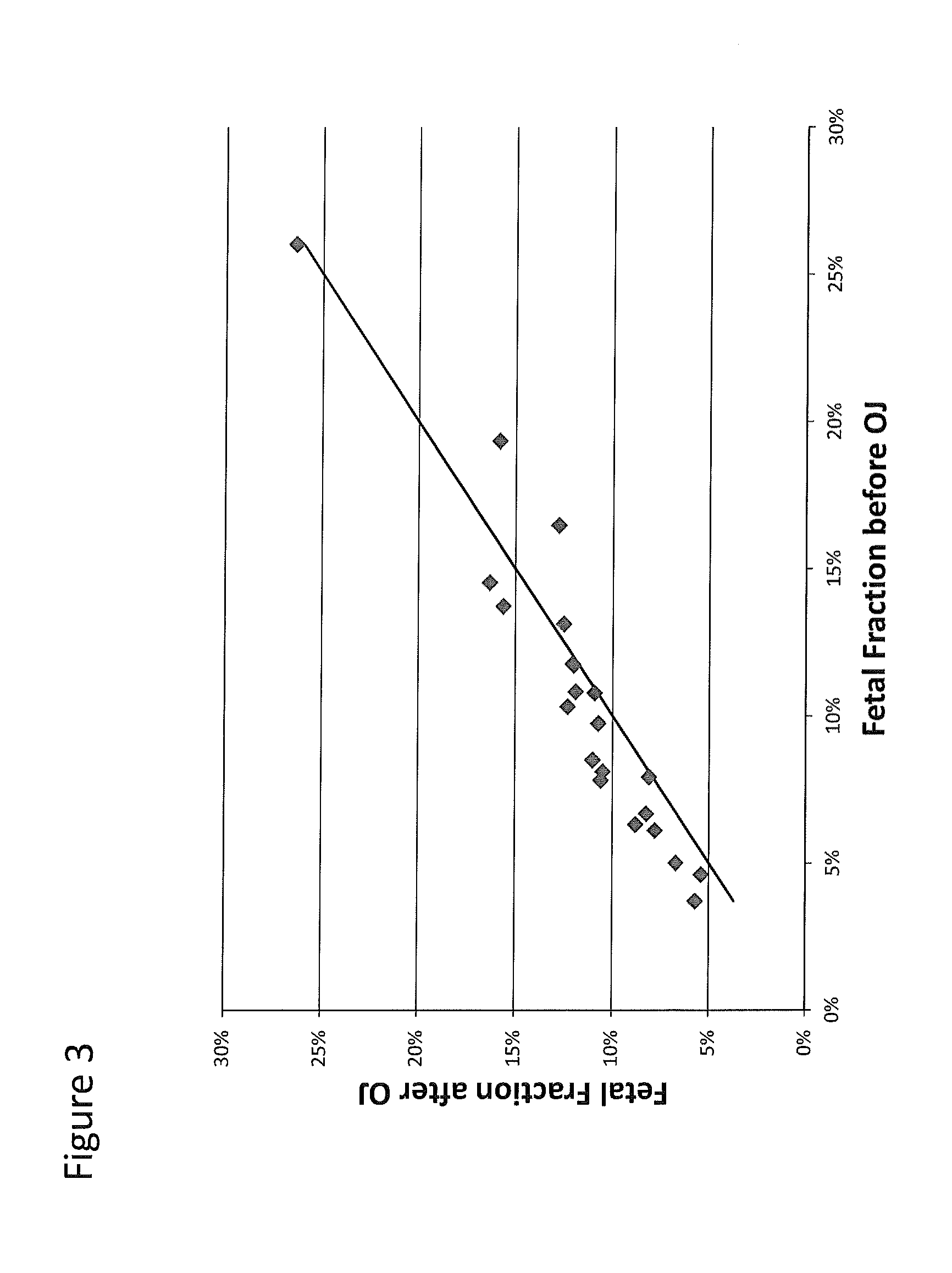
Size-based analysis of fetal DNA fraction in maternal plasma
ActiveUS20130237431A1Health-index calculationMicrobiological testing/measurementAbnormal tissue growthBlood plasma
A fractional concentration of clinically-relevant DNA in a mixture of DNA from a biological sample is determined based on amounts of DNA fragments at multiple sizes. For example, the fractional concentration of fetal DNA in maternal plasma or tumor DNA in a patient's plasma can be determined. The size of DNA fragments in a sample is shown to be correlated with a proportion of fetal DNA and a proportion of tumor DNA, respectively. Calibration data points (e.g., as a calibration function) indicate a correspondence between values of a size parameter and the fractional concentration of the clinically-relevant DNA. For a given sample, a first value of a size parameter can be determined from the sizes of DNA fragments in a sample. A comparison of the first value to the calibration data points can provide the estimate of the fractional concentration of the clinically-relevant DNA.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
ActiveUS20070184467A1Significant resultMicrobiological testing/measurementProteomicsUniparental disomyEmbryo
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell is reconstructed using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In accordance with another embodiment of the invention, incomplete genetic data from a fetus is acquired from fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In one embodiment, the genetic data can be reconstructed for the purposes of making phenotypic predictions. In another embodiment, the genetic data can be used to detect for aneuploides and uniparental disomy.
Owner:NATERA
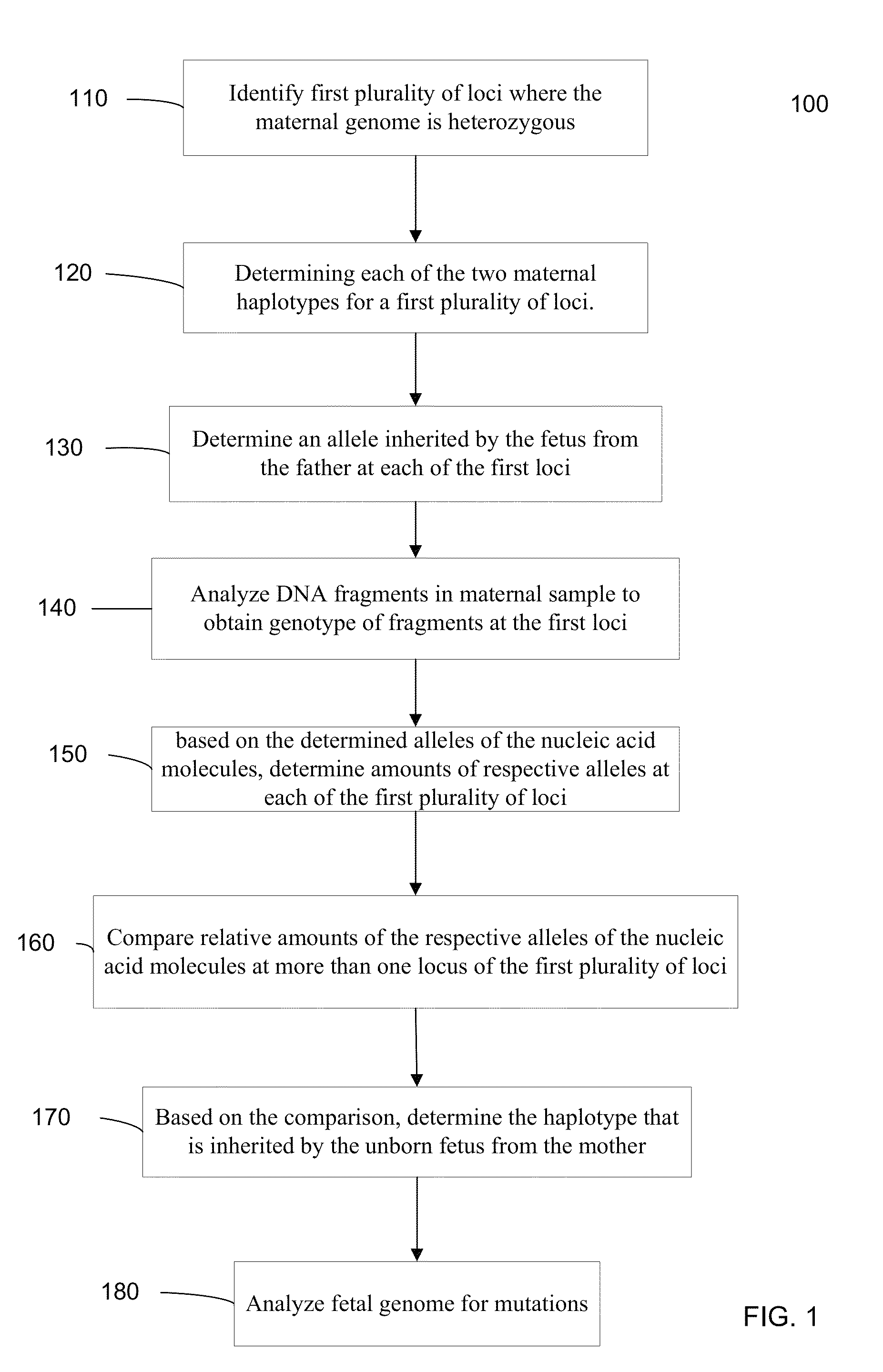
Fetal genomic analysis from a maternal biological sample
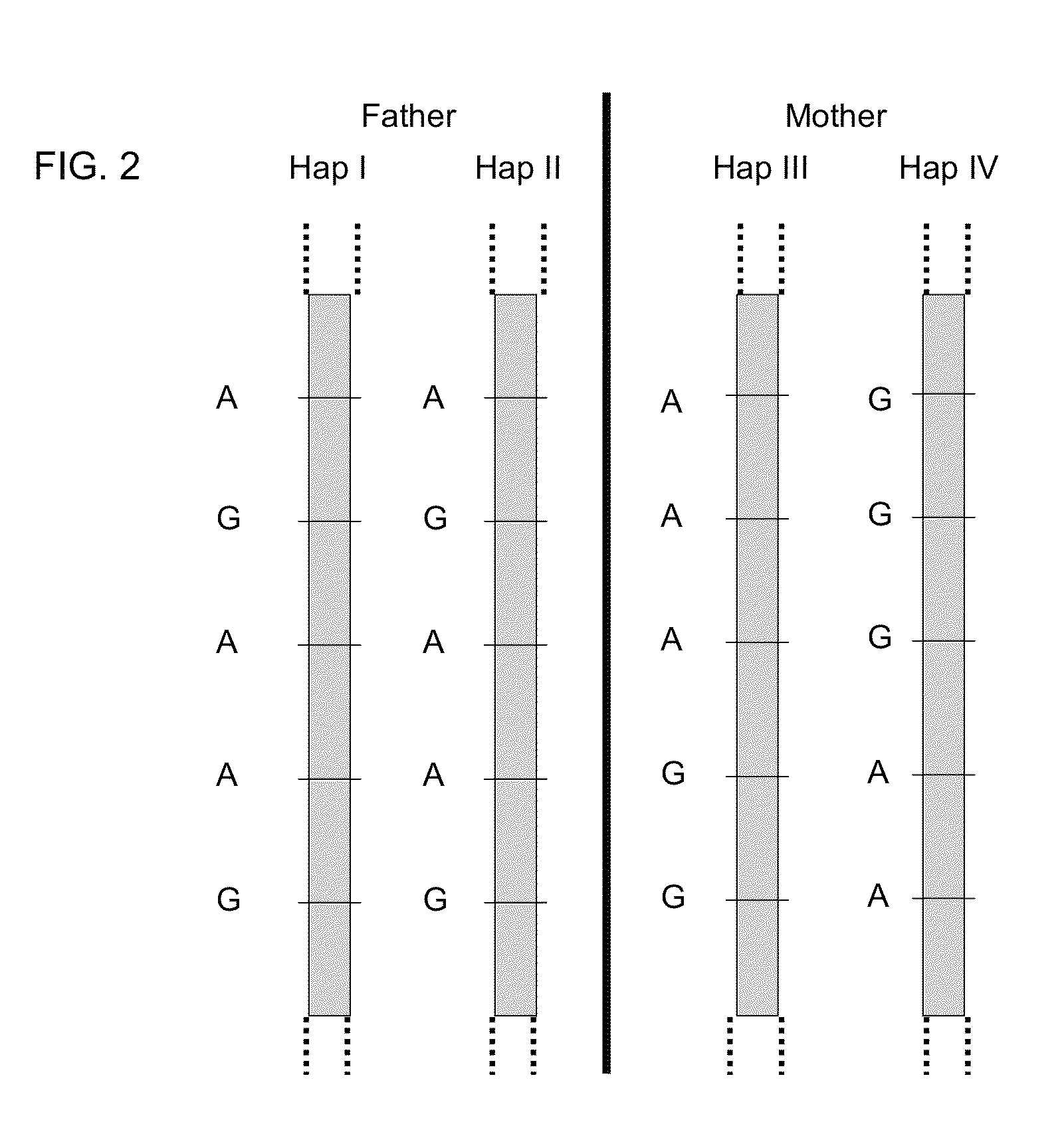
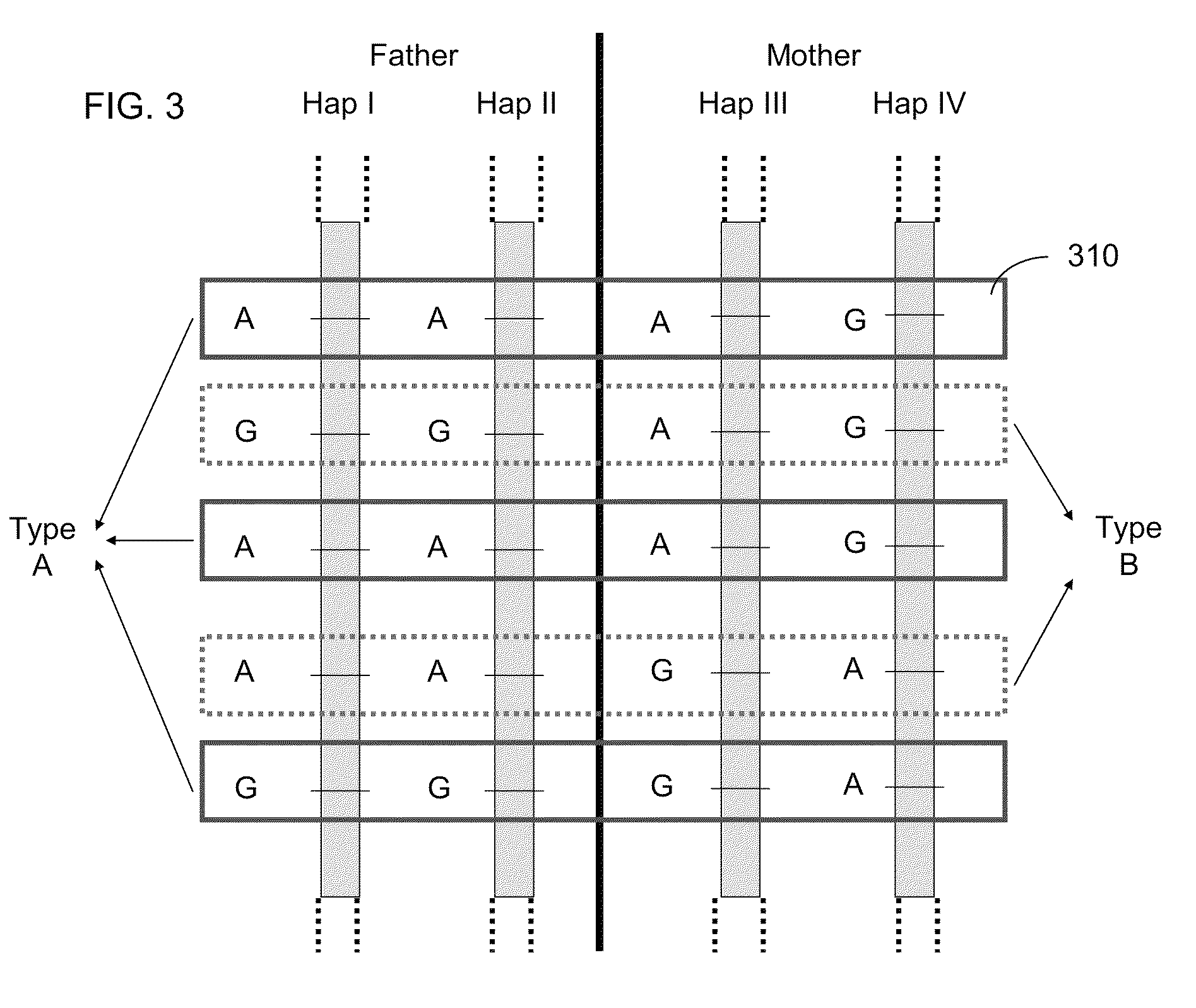
Systems, methods, and apparatus for determining at least a portion of fetal genome are provided. DNA fragments from a maternal sample (maternal and fetal DNA) can be analyzed to identify alleles at certain loci. The amounts of DNA fragments of the respective alleles at these loci can be analyzed together to determine relative amounts of the haplotypes for these loci and determine which haplotypes have been inherited from the parental genomes. Loci where the parents are a specific combination of homozygous and heterozygous can be analyzed to determine regions of the fetal genome. Reference haplotypes common in the population can be used along with the analysis of the DNA fragments of the maternal sample to determine the maternal and paternal genomes. Determination of mutations, a fractional fetal DNA concentration in a maternal sample, and a proportion of coverage of a sequencing of the maternal sample can also be provided.
Owner:SEQUENOM INC +1
Marker for prenatal diagnosis and monitoring
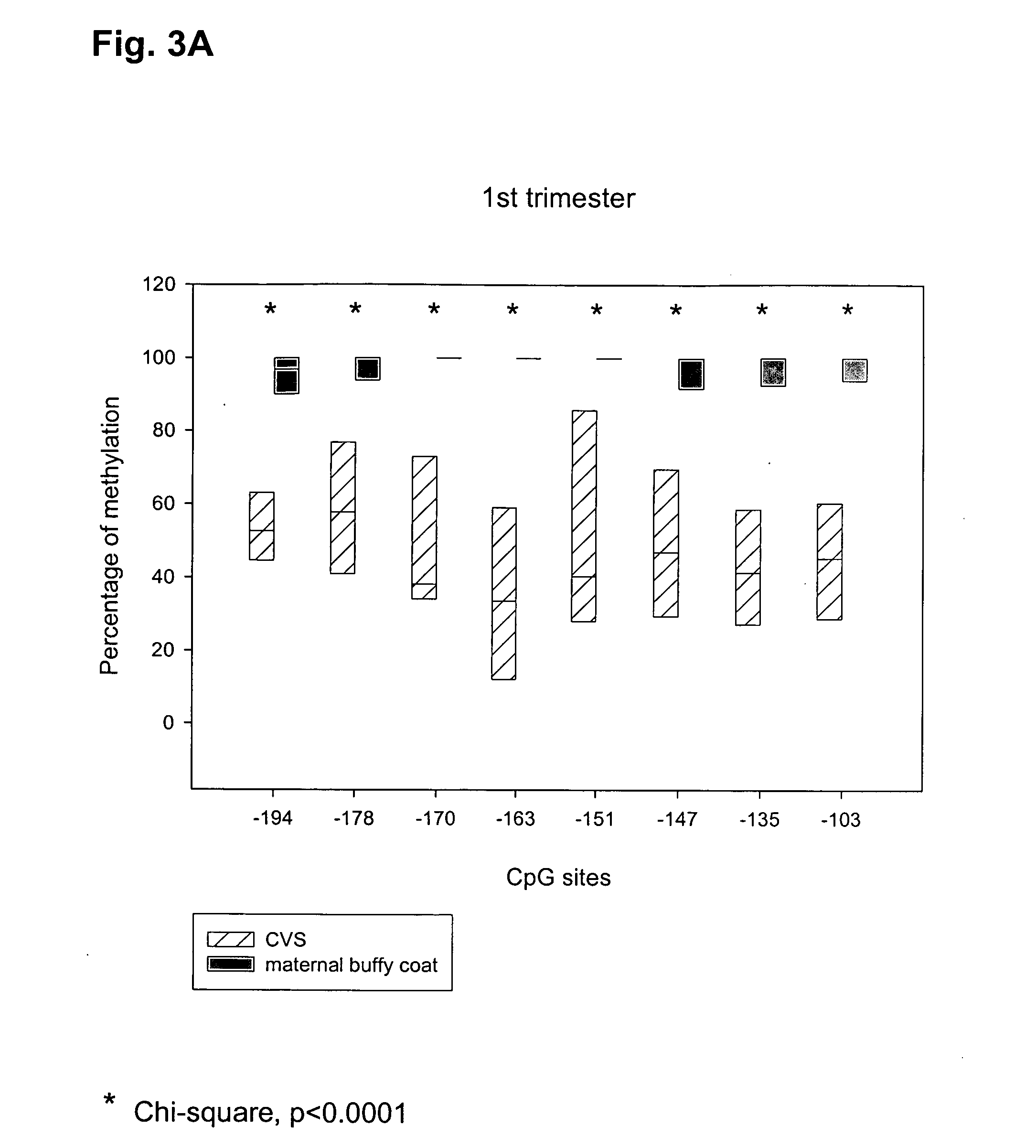
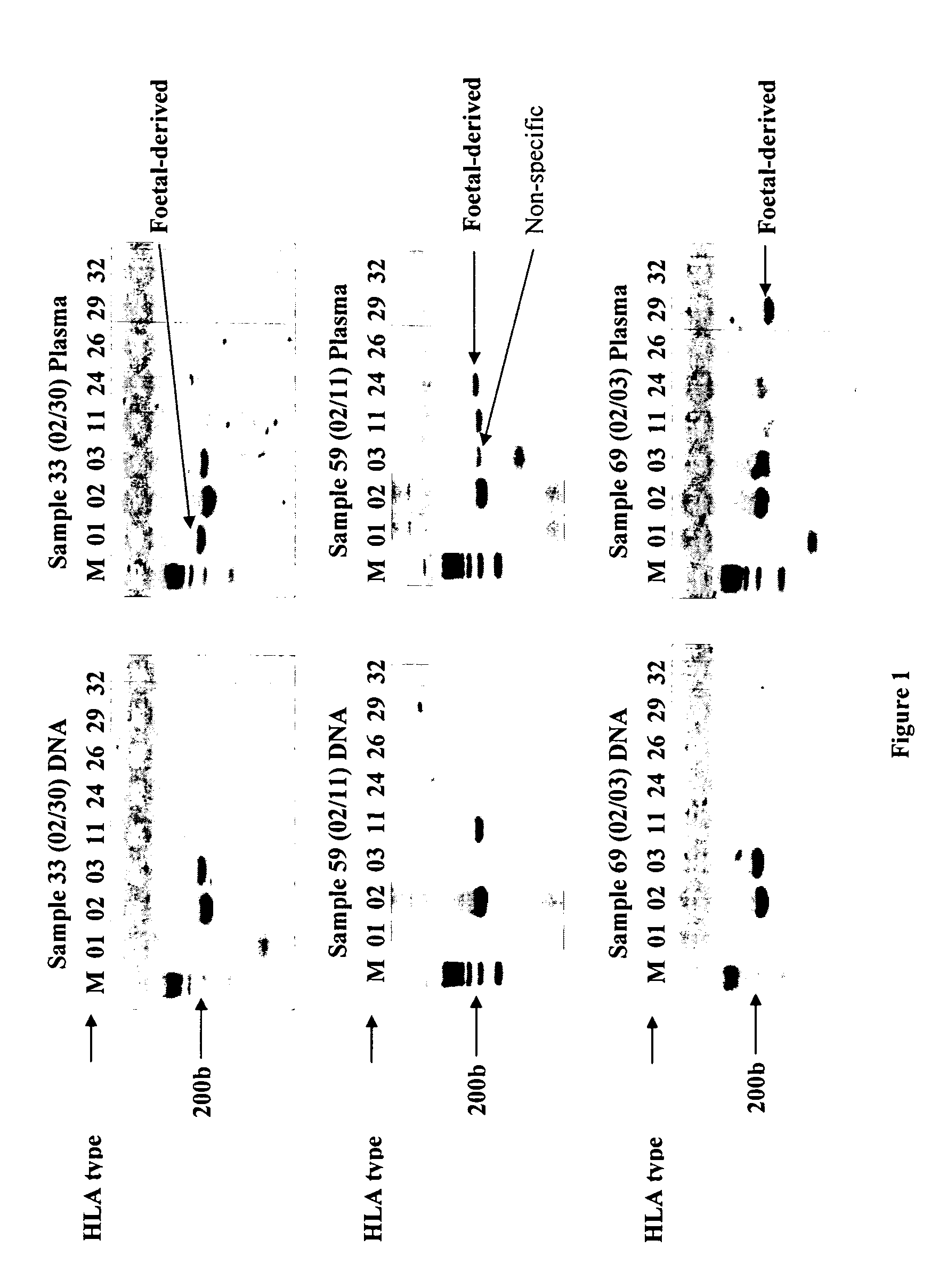
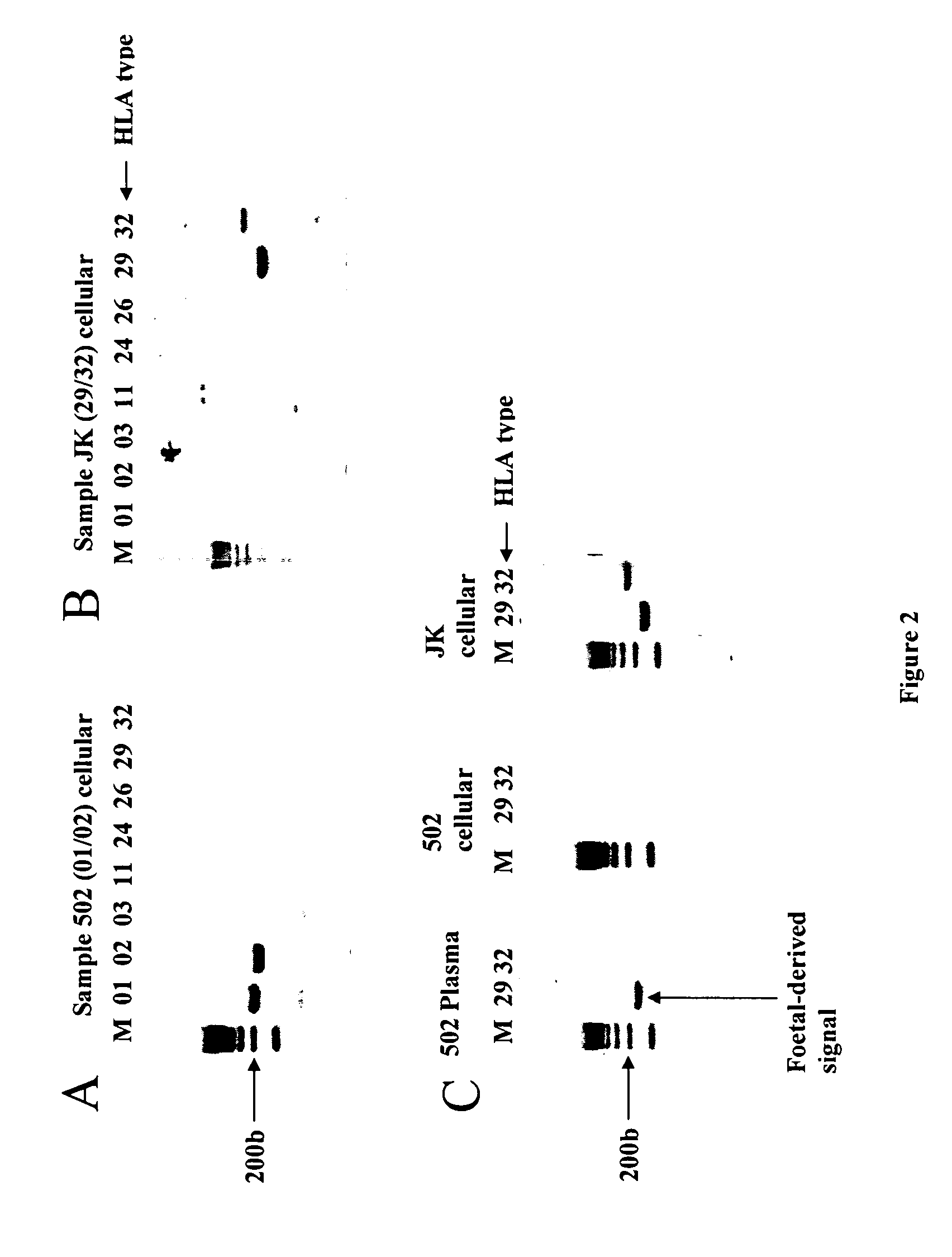
The present invention relates to new methods for diagnosing a pregnancy-associated disorder by analyzing fetal DNA present in the mother's blood. More specifically, this invention relies on the discovery that the maspin gene is differentially methylated in fetal DNA and in maternal DNA and provides these new diagnostic methods, which distinguish fetal DNA from maternal DNA and detect prenatal disorders based on abnormalities in fetal DNA level and methylation status.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Non-invasive detection of fetal genetic traits
ActiveUS20080071076A1Facilitates non-invasive detectionSugar derivativesOther chemical processesPregnancyNon invasive
Blood plasma of pregnant women contains fetal and (generally>90%) maternal circulatory extracellular DNA. Most of said fetal DNA contains .Itoreq.500 base pairs, said maternal DNA having a greater size. Separation of circulatory extracellular DNA of .Itoreq.500 base pairs results in separation of fetal from maternal DNA. A fraction of a blood plasma or serum sample of a pregnant woman containing, due to size separation (e.g. by chromatography, density gradient centrifugation or nanotechnological methods), extracellular DNA substantially comprising .Itoreq.500 base pairs is useful for non-invasive detection of fetal genetic traits (including the fetal RhD gene in pregnancies at risk for HDN; fetal Y chromosome-specific sequences in pregnancies at risk for X chromosome-linked disorders; chromosomal aberrations; hereditary Mendelian genetic disorders and corresponding genetic markers; and traits decisive for paternity determination) by e.g. PCR, ligand chain reaction or probe hybridization techniques, or nucleic acid arrays.
Owner:SEQUENOM INC
Methods for prenatal diagnosis of chromosomal abnormalities
ActiveUS20070059707A1Rapid productionAccurate detectionMicrobiological testing/measurementFermentationDiseaseNon invasive
Chromosomal abnormalities are responsible for a significant number of birth defects, including mental retardation. The present invention is related to methods for non-invasive and rapid, prenatal diagnosis of chromosomal abnormalities based on analysis of a maternal blood sample. The invention exploits the differences in DNA between the mother and fetus, for instance differences in their methylation states, as a means to enrich for fetal DNA in maternal plasma sample. The methods described herein can be used to detect chromosomal DNA deletions and duplications. In a preferred embodiment, the methods are used to diagnose chromosomal aneuploidy and related disorders, such as Down's and Turner's Syndrome.
Owner:TRUSTEES OF BOSTON UNIV
Enrichment of circulating fetal DNA
InactiveUS20070243549A1High accuracy of resultsMicrobiological testing/measurementImmunoglobulins against animals/humansNon invasiveMolecular diagnostics
A non-invasive screening or diagnostic method for determining the likelihood of a fetus with a genetic abnormality or a potential pregnancy complication, which utilizes a liquid blood sample from a pregnant woman. Antibodies specific to a section of histone 3.1 which is exposed to a far greater extent in chromatin of fetal origin than in chromatin of maternal origin are used to sequester and isolate such fetal nucleosomes including the associated fetal DNA. Following isolation / enrichment of such fetal DNA, genetic analysis is carried out using known molecular diagnostics.
Owner:BAYLOR COLLEGE OF MEDICINE
Identification of fetal dna and fetal cell markers in maternal plasma or serum
InactiveUS20070134658A1Reduce in quantityMicrobiological testing/measurementTissue cultureAntigenAllele
The present invention relates to the identification of fetal specific nucleic acids and fetal cell markers in maternal plasma or serum. In particular, the present invention relates to methods which rely on the analysis of polymorphic alleles of a population to determine an allele which is possessed by the fetus but absent from the mother. Fetal specific alleles identified using the methods of the invention can be used to quantify fetal DNA from maternal plasma or serum. In addition, antigens encoded by alleles identified using the methods of the invention can be targeted in methods of isolating or detecting fetal cells.
Owner:GENETIC TECHNOLOGIES LIMTIED
Marker for Prenatal Diagnosis and Monitoring
The present invention relates to new methods for diagnosing a pregnancy-associated disorder by analyzing fetal DNA present in the mother's blood. More specifically, this invention relies on the discovery that the maspin gene is differentially methylated in fetal DNA and in maternal DNA and provides these new diagnostic methods, which distinguish fetal DNA from maternal DNA and detect prenatal disorders based on abnormalities in fetal DNA level and methylation status.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Methods for prenatal diagnosis of chromosomal abnormalities
ActiveUS7655399B2Rapid productionAccurate detectionMicrobiological testing/measurementPlasma samplesNon invasive
Chromosomal abnormalities are responsible for a significant number of birth defects, including mental retardation. The present invention is related to methods for non-invasive and rapid, prenatal diagnosis of chromosomal abnormalities based on analysis of a maternal blood sample. The invention exploits the differences in DNA between the mother and fetus, for instance differences in their methylation states, as a means to enrich for fetal DNA in maternal plasma sample. The methods described herein can be used to detect chromosomal DNA deletions and duplications. In a preferred embodiment, the methods are used to diagnose chromosomal aneuploidy and related disorders, such as Down's and Turner's Syndrome.
Owner:TRUSTEES OF BOSTON UNIV
Prenatal diagnosis using cell-free fetal DNA in amniotic fluid
InactiveUS20070111233A1Improved and rapid methodHigh recovery rateSugar derivativesMicrobiological testing/measurementCell-free fetal DNAAmniotic fluid
The present invention relates to improved methods of prenatal diagnosis, screening, monitoring and / or testing. The inventive methods include the analysis by array-based hybridization of cell-free fetal DNA isolated from amniotic fluid. In addition to allowing the prenatal diagnosis of a variety of diseases and conditions, and the assessment of fetal characteristics such as fetal sex and chromosomal abnormalities, the new inventive methods provide substantially more information about the fetal genome in less time than it takes to perform a conventional metaphase karyotype analysis. In particular, the enhanced molecular karyotype methods provided by the present invention allow the detection of chromosomal aberrations that are not often detected prenatally such as microdeletions, microduplications and subtelomeric rearrangements. Also provided are improved methods of extraction of fetal DNA from amniotic fluid.
Methods for increasing fetal fraction in maternal blood
InactiveUS20140065621A1High copy numberMicrobiological testing/measurementEdible oils/fatsBlood plasmaBiology
The invention provides methods of increasing the fetal fraction in maternal blood and plasma. This increase in fetal fraction improves the accuracy and decreases the “no call” rate for prenatal testing that measures fetal DNA in maternal blood.
Owner:NATERA
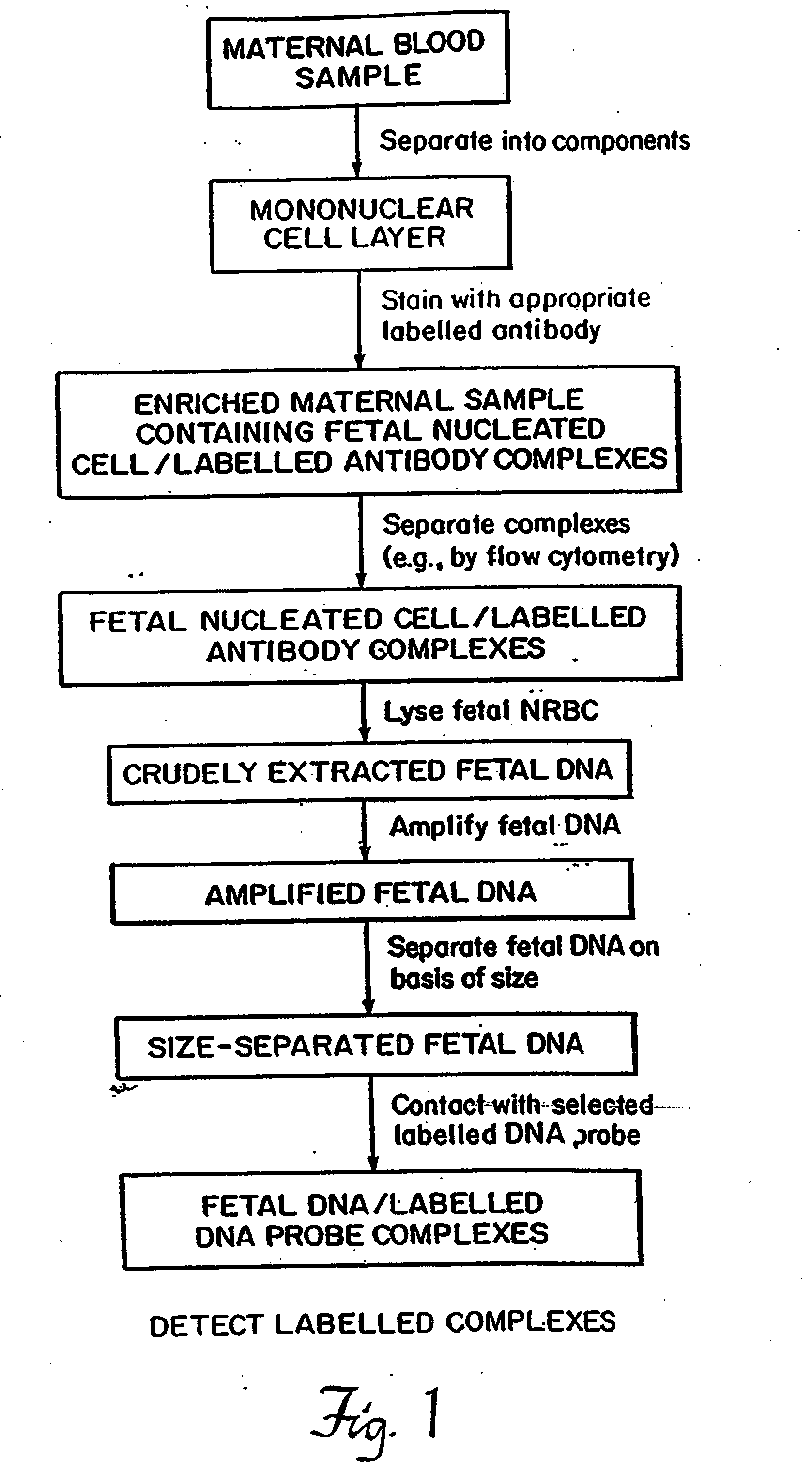
Non-invasive method for isolation and detection of fetal DNA
A method of detecting the presence or absence of the fetal DNA sequence of interest in fetal DNA derived from a sample of peripheral blood obtained from a pregnant woman is described. The method involves obtaining a sample peripheral blood from a pregnant woman, treating the sample of peripheral blood such that the fetal DNA present in the fetal nucleated cells is made available for detection and detecting the presence or absence of the fetal DNA sequence of interest in the available fetal DNA. The proportion of fetal nucleated cells present in the sample of peripheral blood can be increased forming a sample enriched in fetal nucleated cells prior to the detection step. The fetal DNA sequence of interest can be detected by treating the peripheral blood sample such that fetal DNA present in the sample is made available for hybridization with a DNA probe and subsequently contacting the available fetal DNA with a DNA probe hybridizable to fetal DNA of interest under hybridization conditions. The presence or absence of hybridization between the DNA probe and the fetal DNA of interest is detected as an indication of the presence or absence of the fetal DNA of interest.
Owner:CHILDRENS HOSPITAL
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available, are disclosed. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data is acquired from embryonic cells, fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals.
Owner:NATERA
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available, are disclosed. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention incomplete genetic data is acquired from embryonic cells, fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals.
Owner:NATERA
Methods for increasing fetal fraction in maternal blood
InactiveUS20150232938A1High copy numberMicrobiological testing/measurementEdible oils/fatsBlood plasmaBiology
The invention provides methods of increasing the fetal fraction in maternal blood and plasma. This increase in fetal fraction improves the accuracy and decreases the “no call” rate for prenatal testing that measures fetal DNA in maternal blood.
Owner:NATERA
Methods and kits for selectively amplifying, detecting or quantifying target DNA with specific end sequences
ActiveUS20090061425A1Microbiological testing/measurementFermentationPlasma samplesRestriction enzyme digestion
Disclosed herein are methods and kits for selectively amplifying, detecting or quantifying a DNA fragment with a specific end sequence, especially generated following restriction enzyme digestion. This method can be used, for example, to detect a hypomethylated DNA fragment. This methods and kits are especially useful in detecting or quantifying a hypomethylated fetal DNA fragment in a maternal plasma sample containing a corresponding hypermethylated maternal DNA fragment.
Owner:THE CHINESE UNIVERSITY OF HONG KONG +1
Methods for detection of genetic disorders
InactiveUS20060160105A1Minimize disruptionMaximize efficiencySugar derivativesMicrobiological testing/measurementGeneticsAllele
Owner:RAVGEN INC
Non-Invasive Fetal Genetic Screening by Digital Analysis
The present methods are exemplified by a process in which maternal blood containing fetal DNA is diluted to a nominal value of approximately 0.5 genome equivalent of DNA per reaction sample. Digital PCR is then be used to detect aneuploidy, such as the trisomy that causes Down Syndrome. Since aneuploidies do not present a mutational change in sequence, and are merely a change in the number of chromosomes, it has not been possible to detect them in a fetus without resorting to invasive techniques such as amniocentesis or chorionic villi sampling. Digital amplification allows the detection of aneuploidy using massively parallel amplification and detection methods, examining, e.g., 10,000 genome equivalents.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Fetal methylation markers
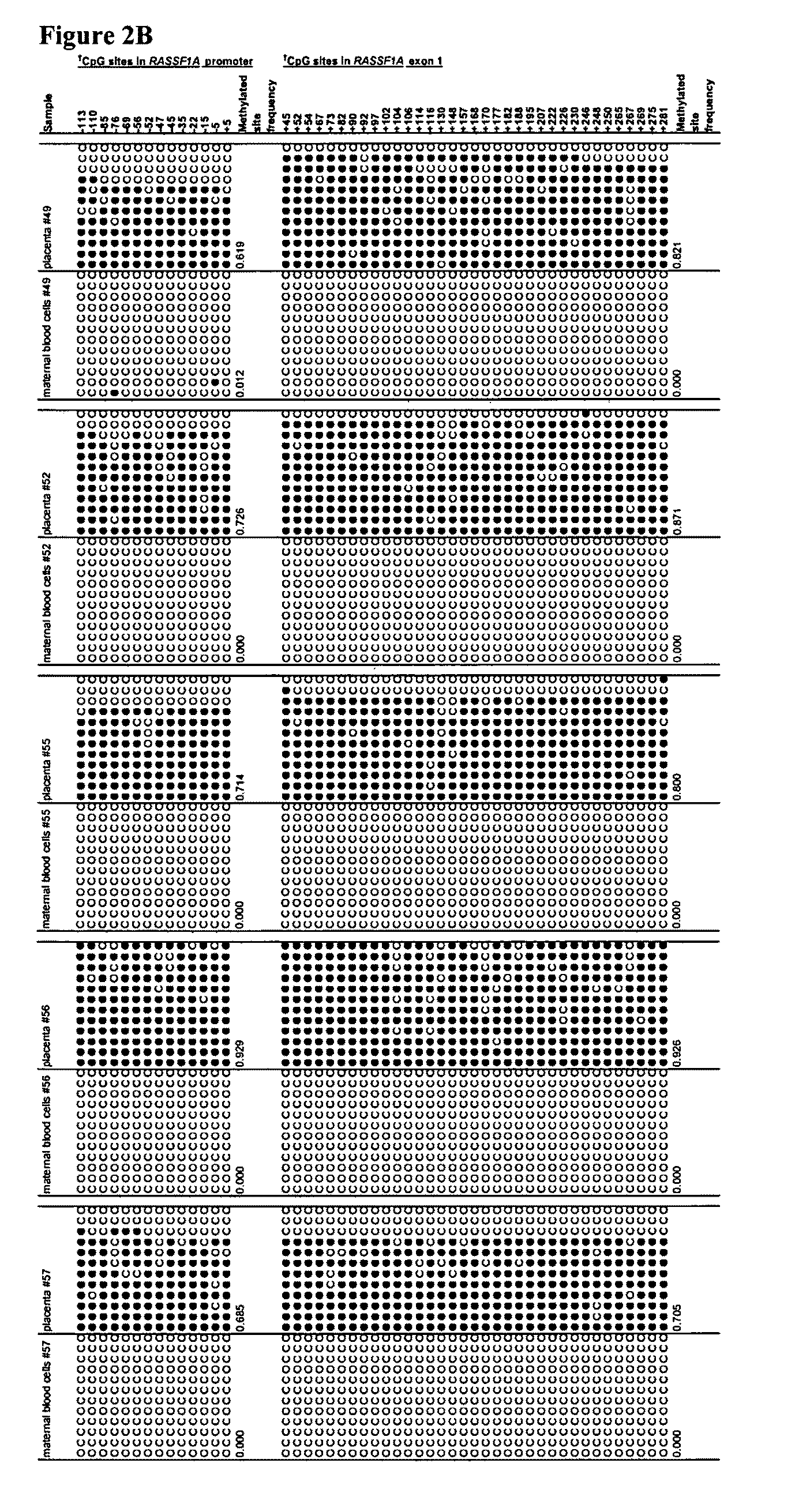
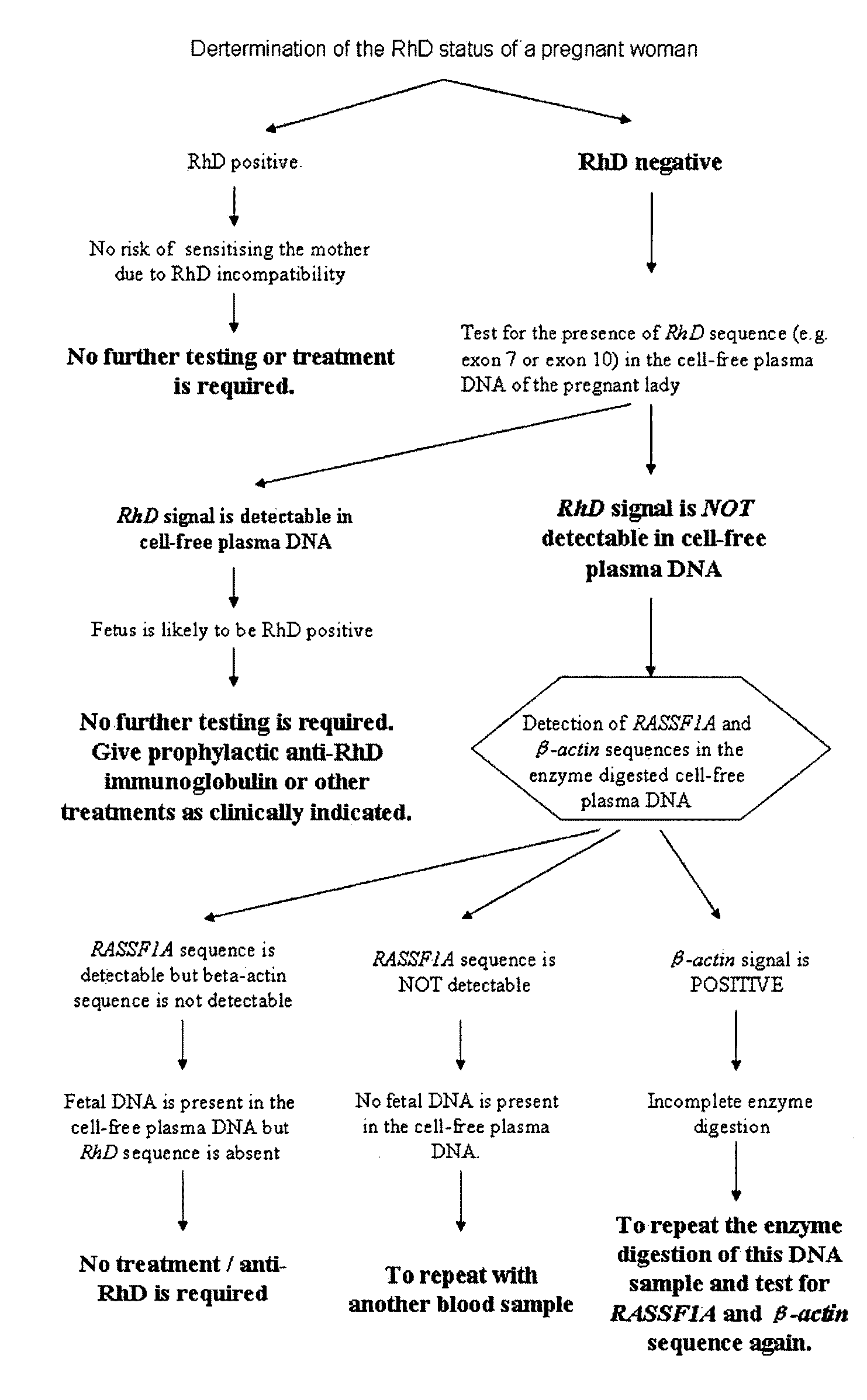
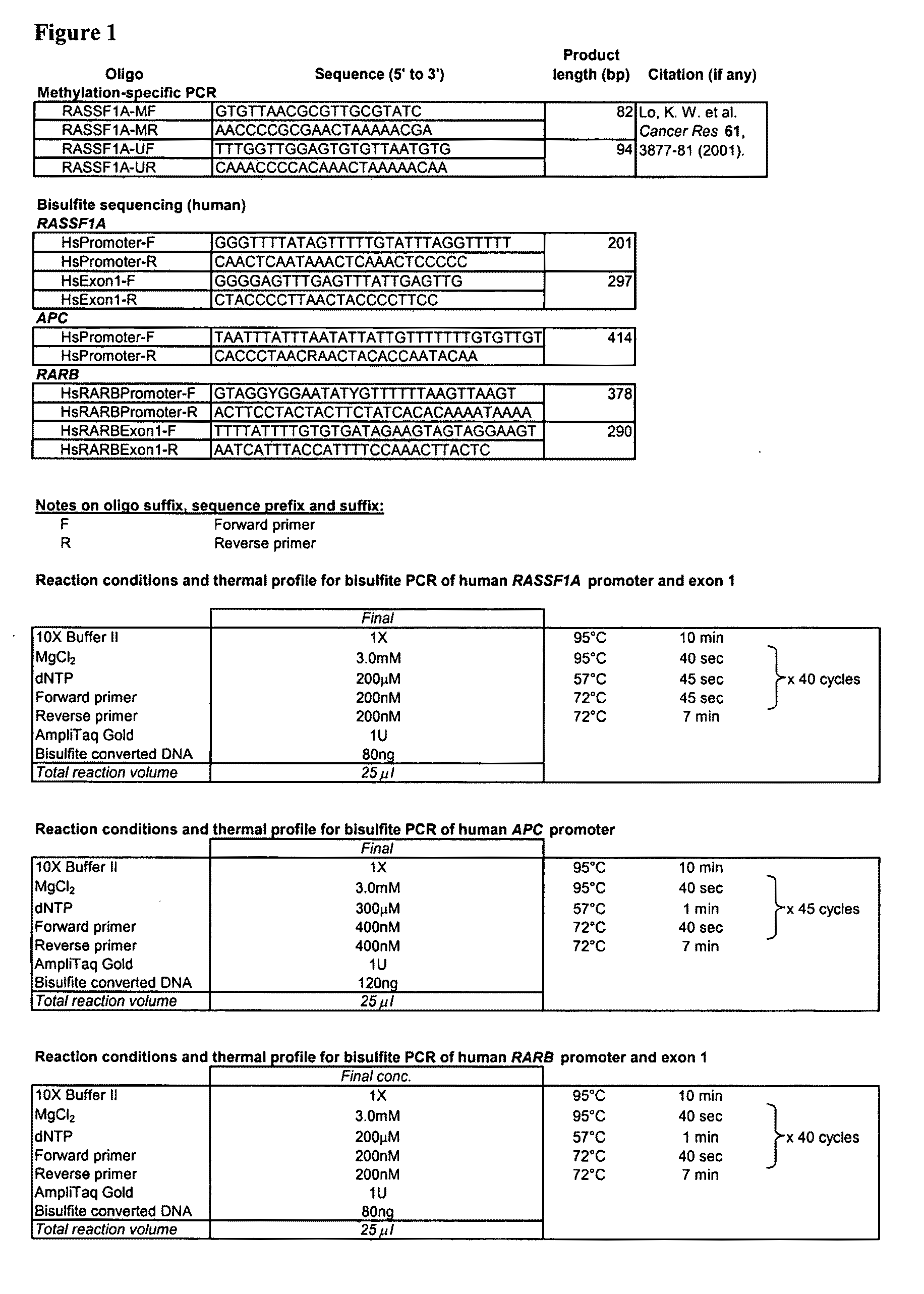
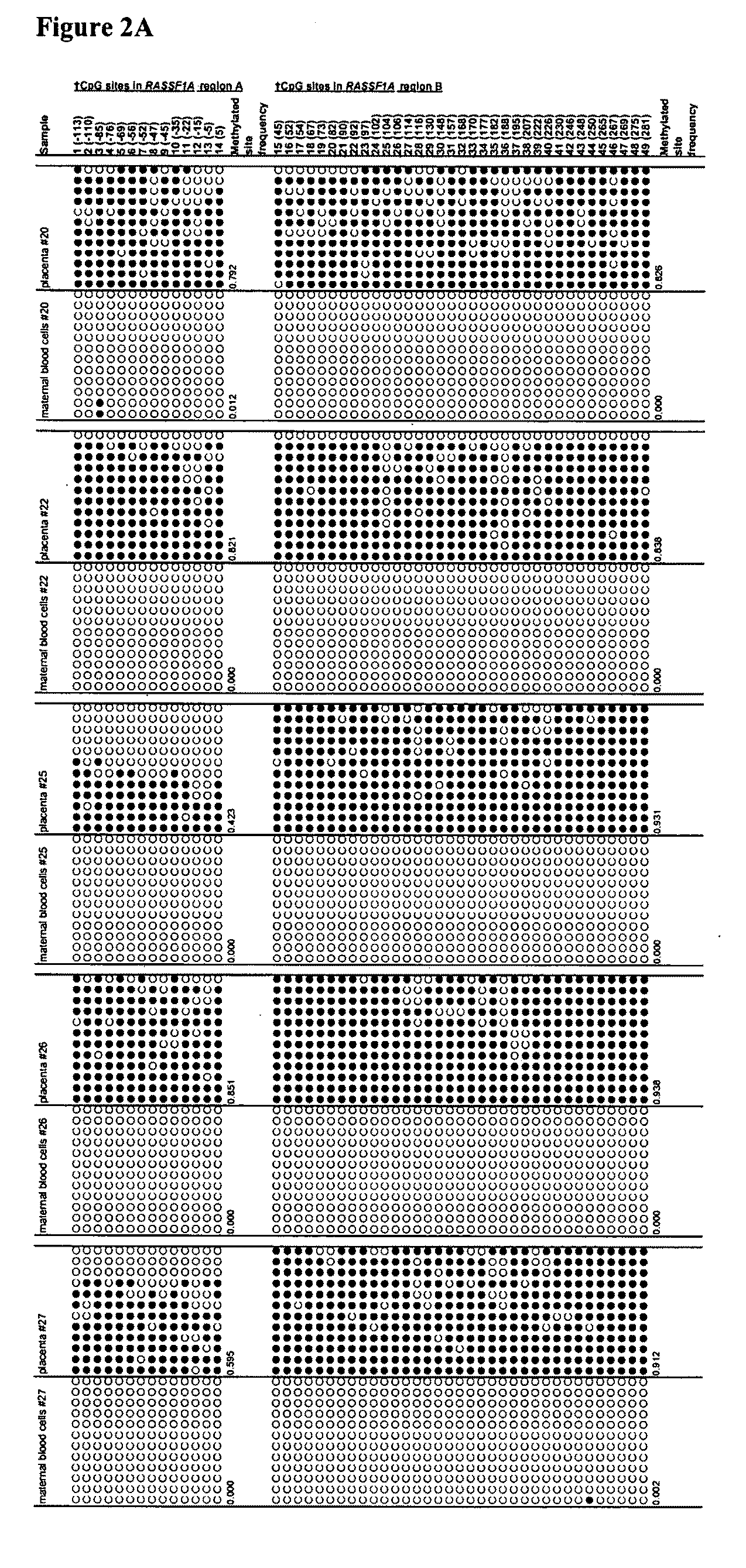
This application describes the discovery that, in a pregnant woman, certain genes (such as RASSF1A, APC, CASP8, RARB, SCGB3A1, DAB2IP, PTPN6, THY1, TMEFF2, and PYCARD) originated from a fetus are highly methylated, whereas the same genes of maternal origin are unmethylated. This discovery allows the easy detection of one or more of these methylated fetal genes in a biological sample from a pregnant woman, serving as a universal indicator of the presence of fetal DNA in the sample. These fetal methylation markers are particularly useful as positive controls for a non-invasive analytical process during which the quality and quantity of fetal DNA are monitored. These newly identified fetal markers can also be measured directly for diagnosis of certain pregnancy-related conditions.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Methods for detecting DNA originating from different individuals
InactiveUS20050282185A1Microbiological testing/measurementRecombinant DNA-technologyDNA methylationOrganism
In a first aspect, the present invention features methods for differentiating DNA species originating from different individuals in a biological sample. These methods may be used to differentiate or detect fetal DNA in a maternal sample or to differentiate DNA of an organ donor from DNA of an organ recipient. In preferred embodiments, the DNA species are differentiated by observing epigenetic differences in the DNA species such as differences in DNA methylation. In a second aspect, the present invention features methods of detecting genetic abnormalities in a fetus by detecting fetal DNA in a biological sample obtained from a mother. In a third aspect, the present invention features methods for differentiating DNA species originating from an organ donor from those of an organ recipient. In a fourth aspect, the present invention features kits for differentiating DNA species originating from different individuals in a biological sample.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Fetal methylation markers
ActiveUS20090155776A1Increased riskSugar derivativesMicrobiological testing/measurementASC proteinPositive control
This application describes the discovery that, in a pregnant woman, certain genes (such as RASSF1A, APC, CASP8, RARB, SCGB3A1, DAB2IP, PTPN6, THY1, TMEFF2, and PYCARD) originated from a fetus are highly methylated, whereas the same genes of maternal origin are unmethylated. This discovery allows the easy detection of one or more of these methylated fetal genes in a biological sample from a pregnant woman, serving as a universal indicator of the presence of fetal DNA in the sample. These fetal methylation markers are particularly useful as positive controls for a non-invasive analytical process during which the quality and quantity of fetal DNA are monitored. These newly identified fetal markers can also be measured directly for diagnosis of certain pregnancy-related conditions.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Assays for the detection of genotype, mutations, and/or aneuploidy
InactiveUS20140186827A1Increase percentageMicrobiological testing/measurementDisease diagnosisAssayGenotype
The present invention provides amplification-based methods for detection of genotype, mutations, and / or aneuploidy. These methods have broad applicability, but are particularly well-suited to detecting and quantifying target nucleic acids in free fetal DNA present in a maternal bodily fluid sample.
Owner:FLUIDIGM INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com