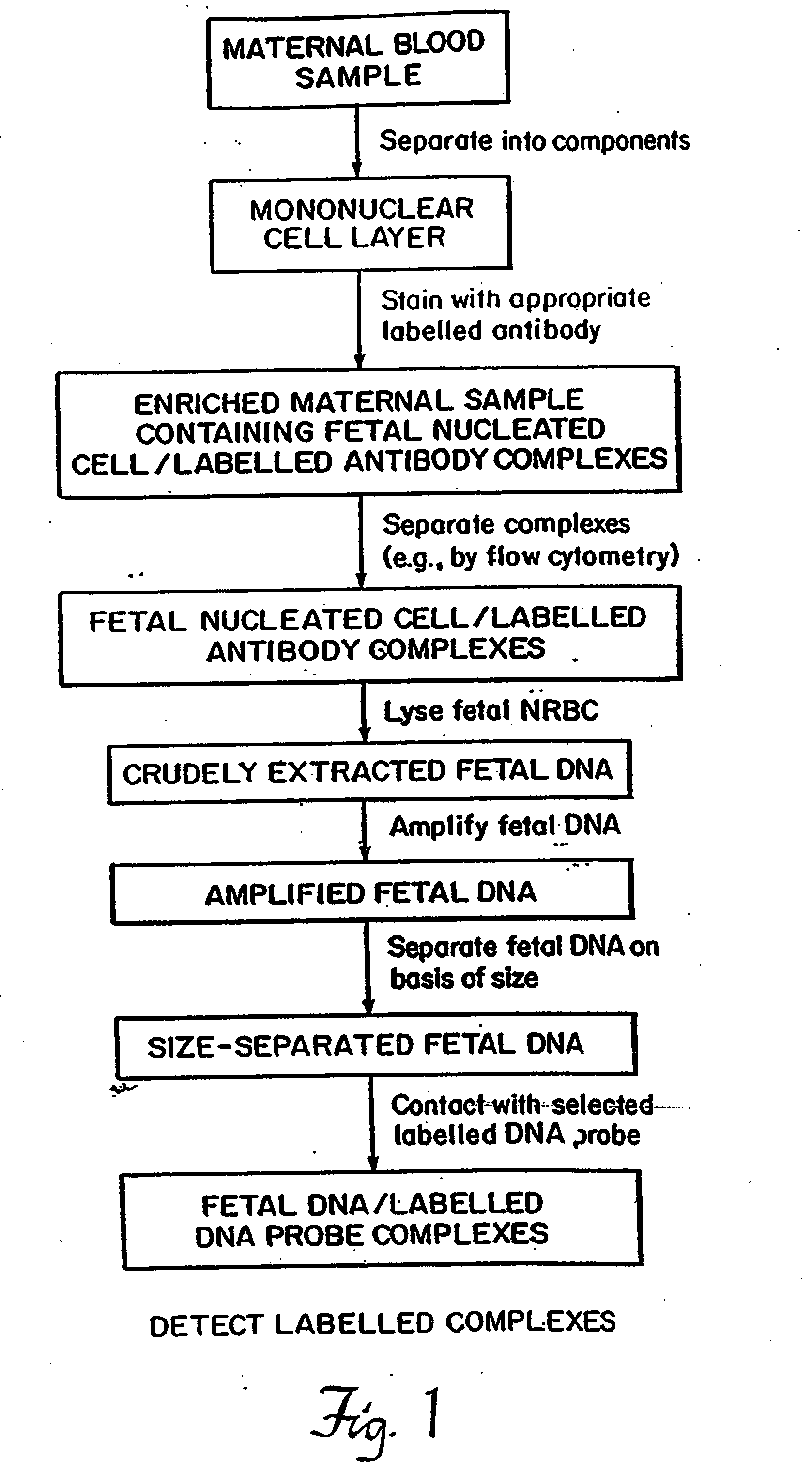
Non-invasive method for isolation and detection of fetal DNA
a fetal dna and non-invasive technology, applied in the field of non-invasive methods for isolation and detection of fetal dna, can solve the problems of not being able to distinguish fetal cells from maternal cells, not being able to confirm the presence of fetal dna in cells, and not being able to successfully isolate cells subsequently shown to contain fetal dna
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Antibody Selection for Isolation and Sorting of Fetal Nucleated Erythroctes (NRBCs)
Removal of Maternal Leucocytes from Maternal Blood Using Human Leucocyte Antigen (HLe-1)
[0074] The technique of fetal NRBC isolation began with an initial Ficoll-Hypaque density gradient centrifugation to remove the tremendously high number of non-nucleated erythrocytes in maternal blood. Peripheral blood was centrifuged and separated into a supernatant layer containing platelets, a mononuclear cell layer, and an agglutinated pellet consisting of non-nucleated erythrocytes and granulocytes. The mononuclear cell layer consisted of lymphocytes, monocytes, possible trophoblasts, and, due to their increased size and density, NRBCs and some reticulocytes. While the Ficoll-Hypaque centrifugation represented an initial enrichment in the proportion of fetal NRBCs present in the maternal sample, flow cytometry and cell sorting was used to improve the purity of the isolated cell population.
[0075] The mononu...
example 2
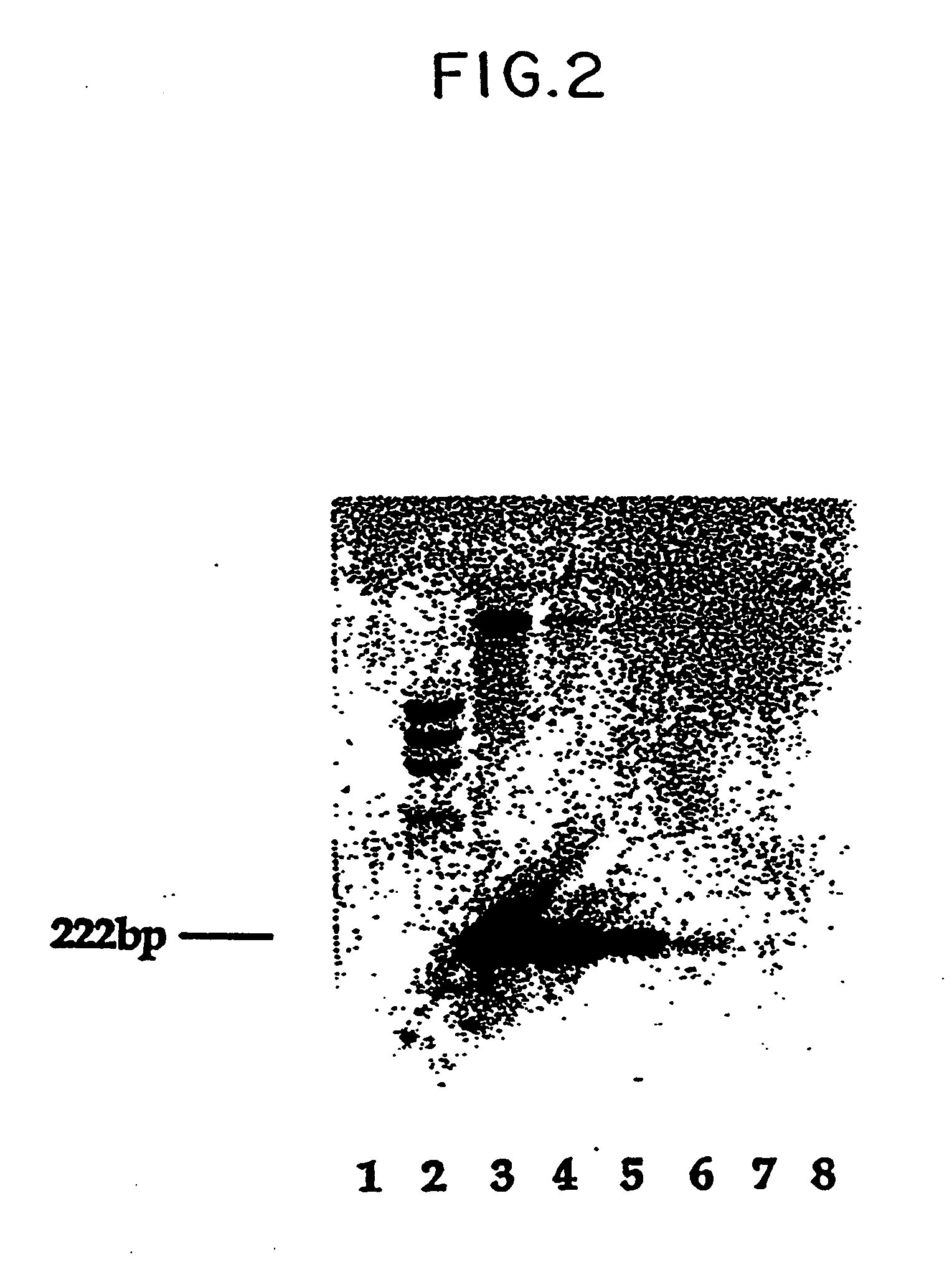
DNA Hybridization Studies in HLe-1 Negative Cells Sorted from Maternal Blood
[0082] To confirm fetal origin of the cells sorted as described in Example 1, Y chromosomal probes were used because it is the Y chromosome that is unquestionably fetal in origin. The assessments were designed to study whether the presence of Y chromosomal DNA in maternal blood as detected on autoradiographs performed antenatally correlated with the subsequent birth of a male infant.
DNA Isolation
[0083] HLe-1 negative cells from cord blood and pregnant women were sorted into test tubes. Conventional methods of DNA isolation as well as modification of cruder methods (Lau, Y-F, et al. Lancet, 1:14-16 (1984); McCabe, E. R. B., et al., Hum Genet., 75:213-216 (1987) were attempted without success in detecting Y chromosome derived bands on Southern Blots. All were limited by the small numbers of cells present.
example 3
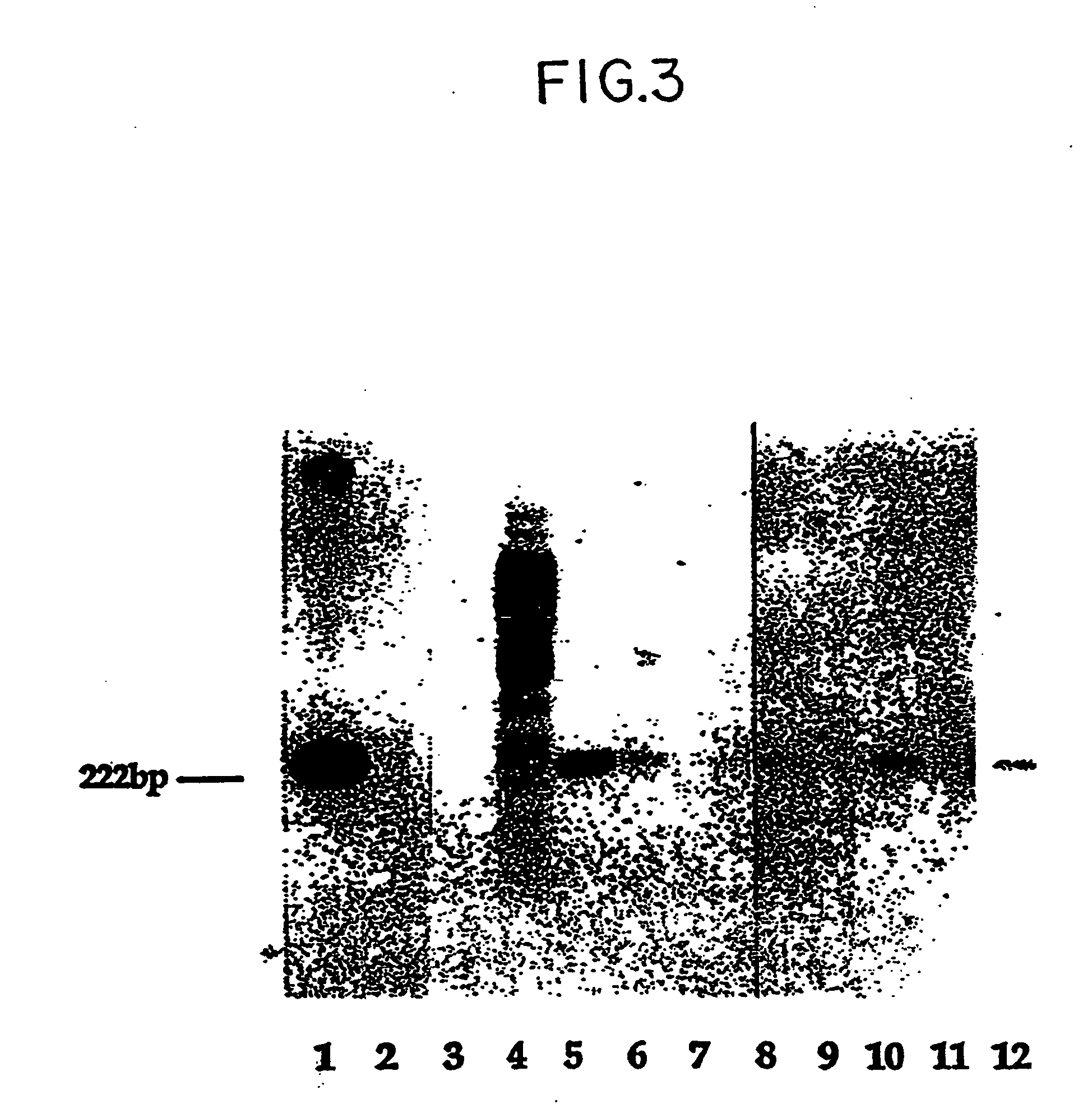
Direct Hybridization to Cells Deposited on Filters
[0084] In order to circumvent technical problems associated wtih DNA isolation, a method of direct DNA hybridization to cells flow sorted onto nitrocellulose filters was developed (Bianchi, D. W., et al., Cytometry, 8:197-202 (1987)). In control experiments, the sex of a newborn was determined from as few as 50 sorted cord blood leucocytes or 5,000 HLe-1 negative cells (a mixture of nucleated and non-nucleated cells).
[0085] The methodology was then applied to detection of Y chromosomal sequences in HLe-1 negative cells sorted from peripheral blood samples in 40 women between 8½ and 38 weeks gestation. Results were the following:
Dot BlotDeliveredDeliveredLost ToHybridization with YMaleFemaleFollow-Chromosomal ProbeInfantInfantup+ 3 20−21122
[0086] It was concluded that hybridization with this probe was not predictive of male pregnancy. The possibility exists that there was fetal DNA present on the filters where DNA hybridization oc...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Time | aaaaa | aaaaa |
| Density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com