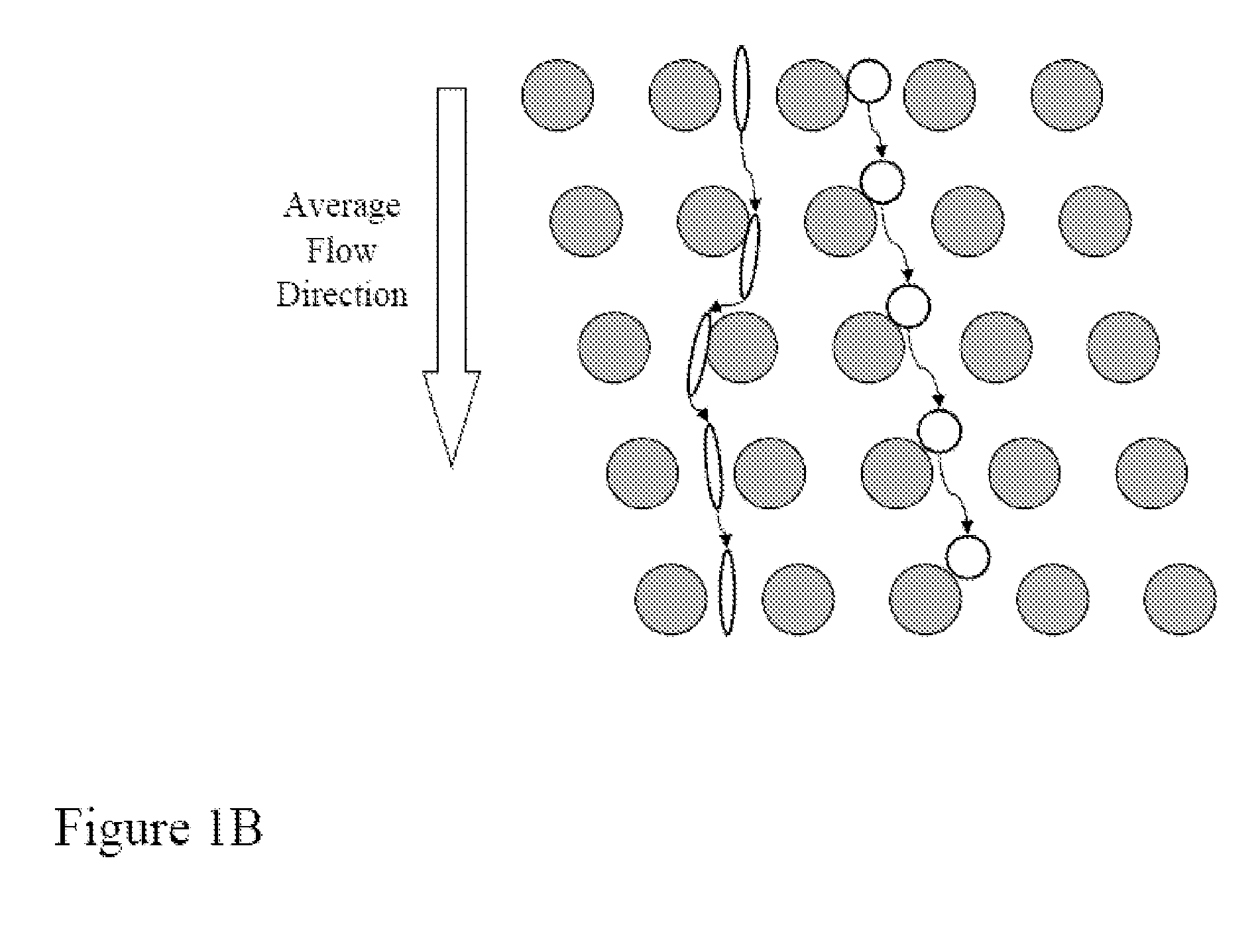Patents
Literature
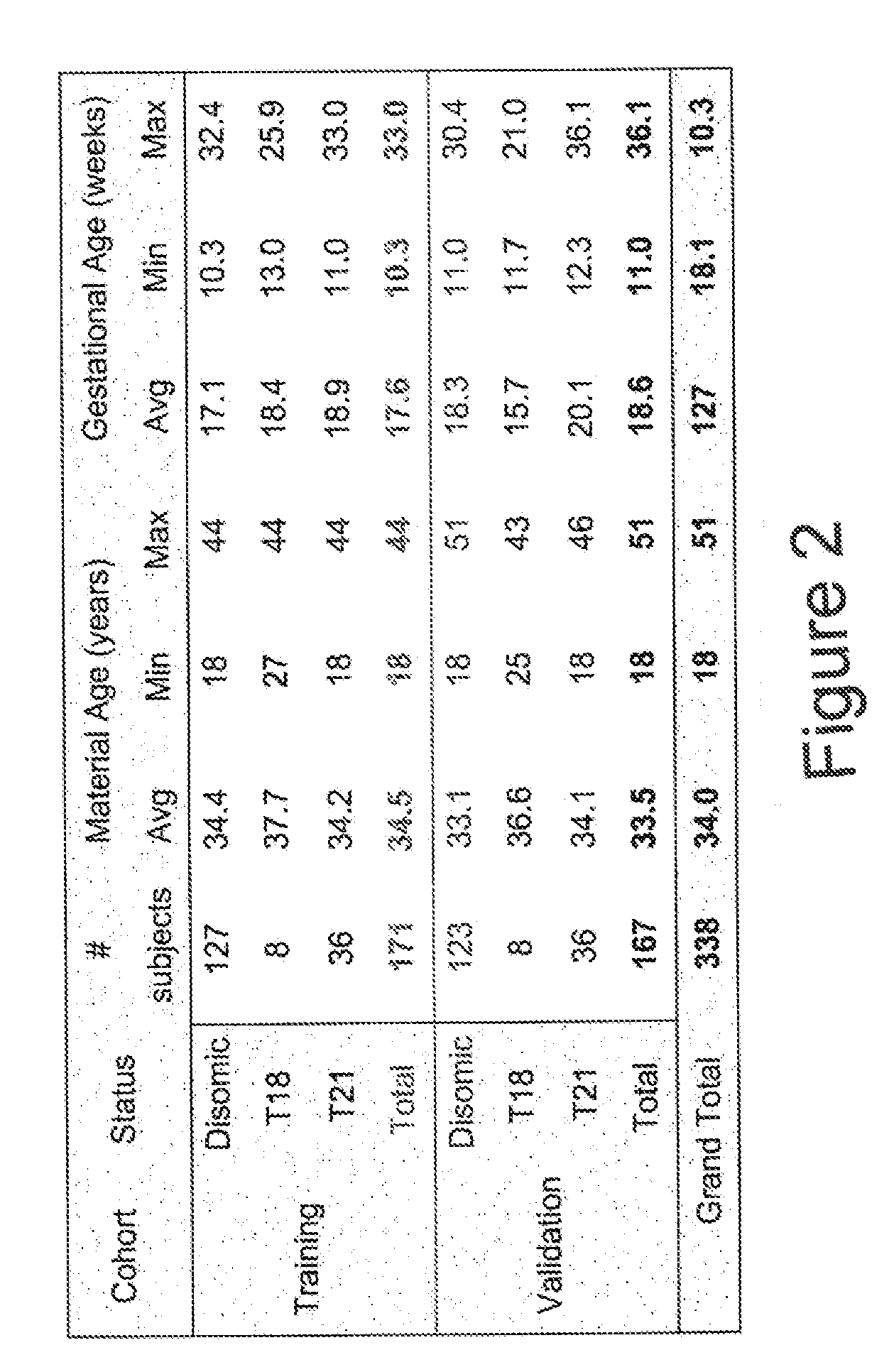
1130 results about "Fetus" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A fetus or foetus (/ˈfiːtəs/; plural fetuses, feti, foetuses, or foeti) is the unborn offspring of an animal that develops from an embryo. Following embryonic development the fetal stage of development takes place. In human prenatal development, fetal development begins from the ninth week after fertilisation (or eleventh week gestational age) and continues until birth. Prenatal development is a continuum, with no clear defining feature distinguishing an embryo from a fetus. However, a fetus is characterized by the presence of all the major body organs, though they will not yet be fully developed and functional and some not yet situated in their final anatomical location.
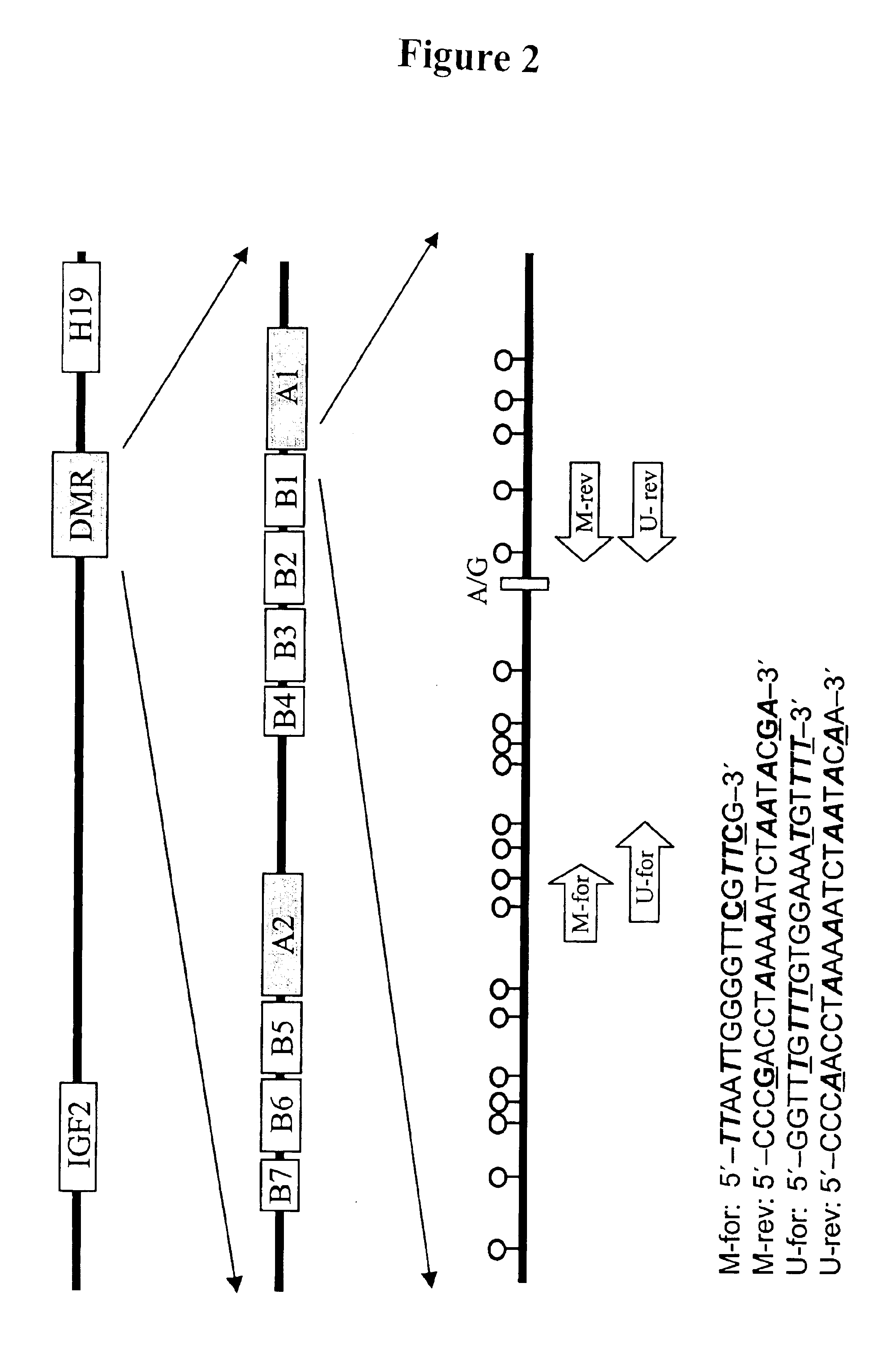
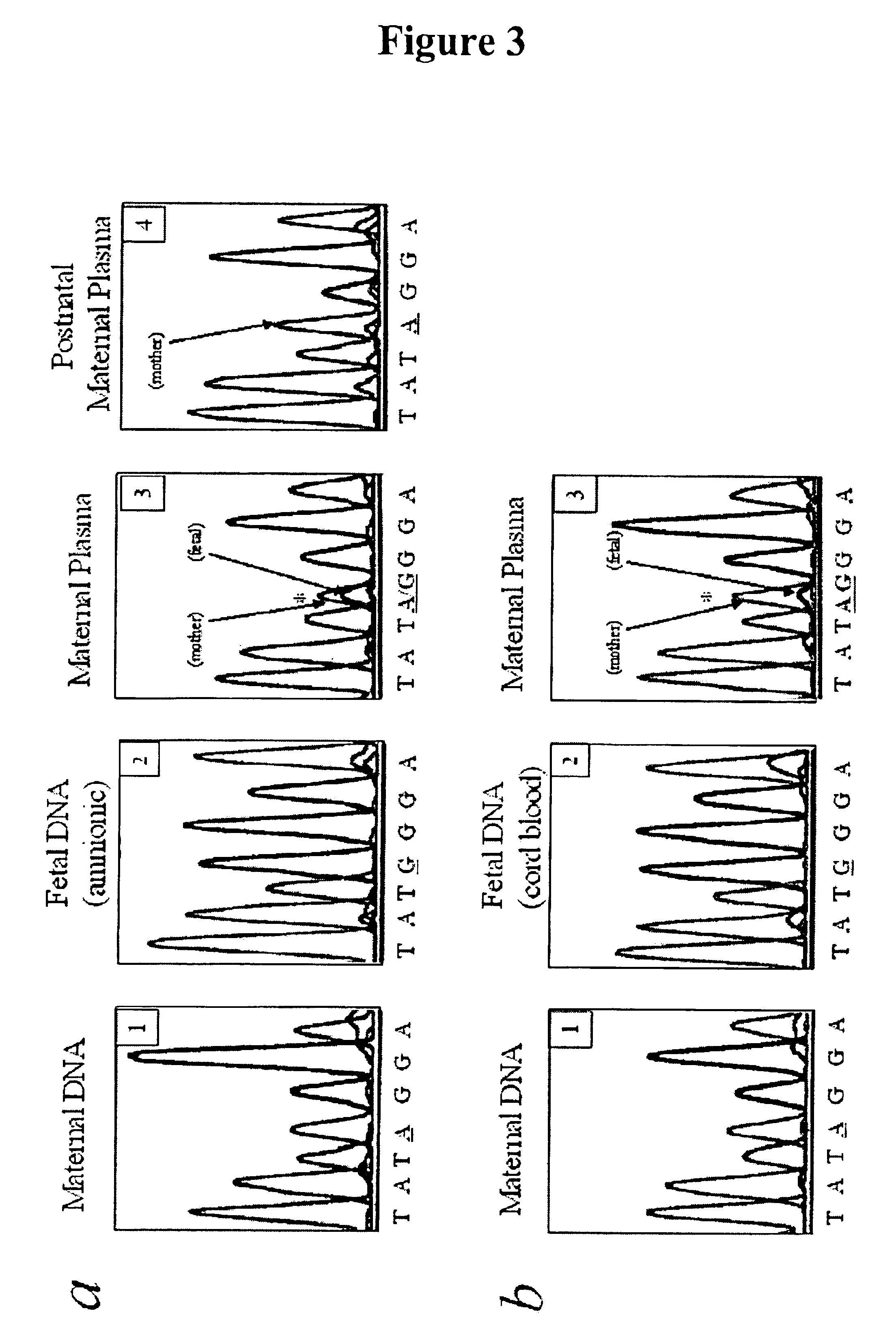
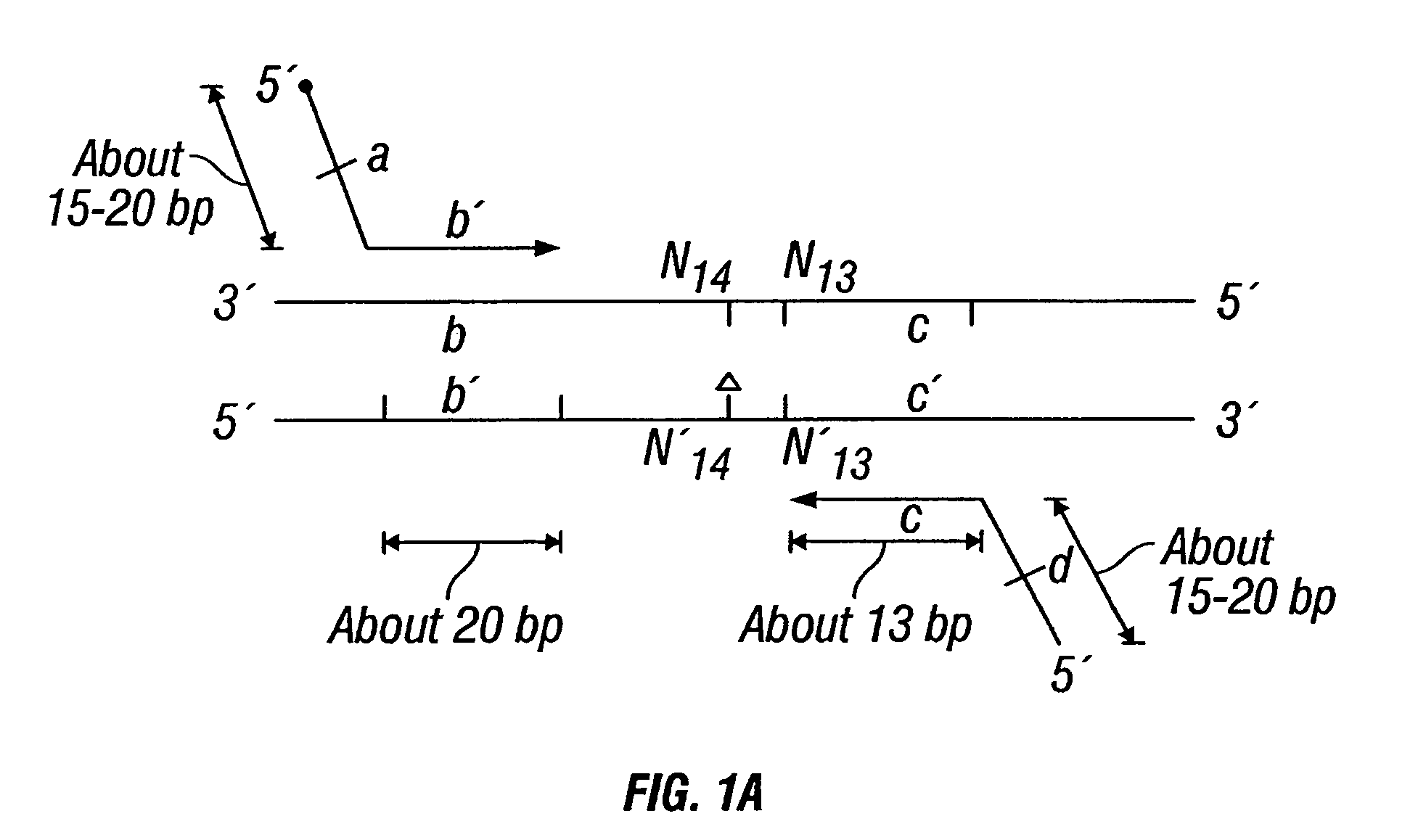
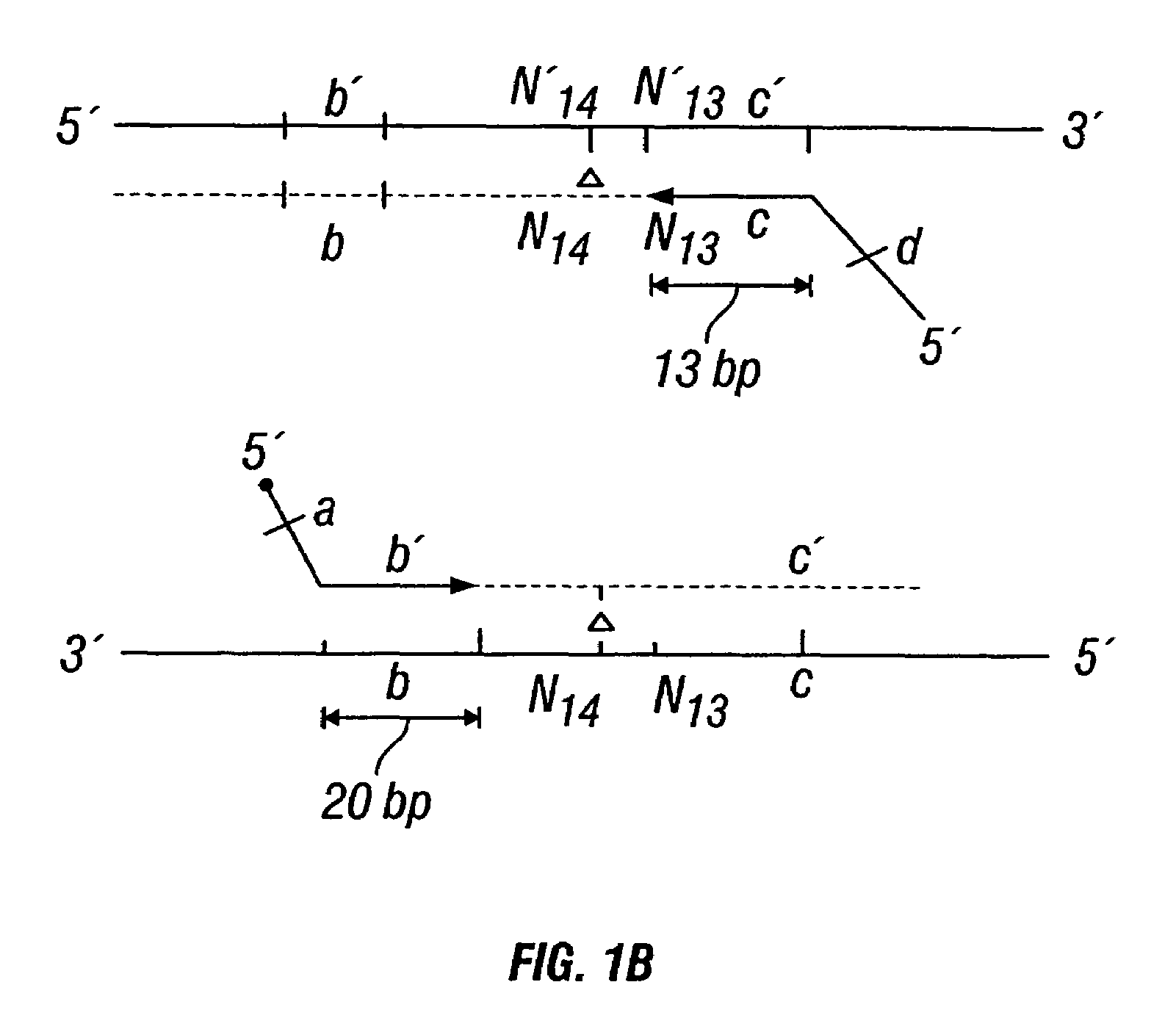
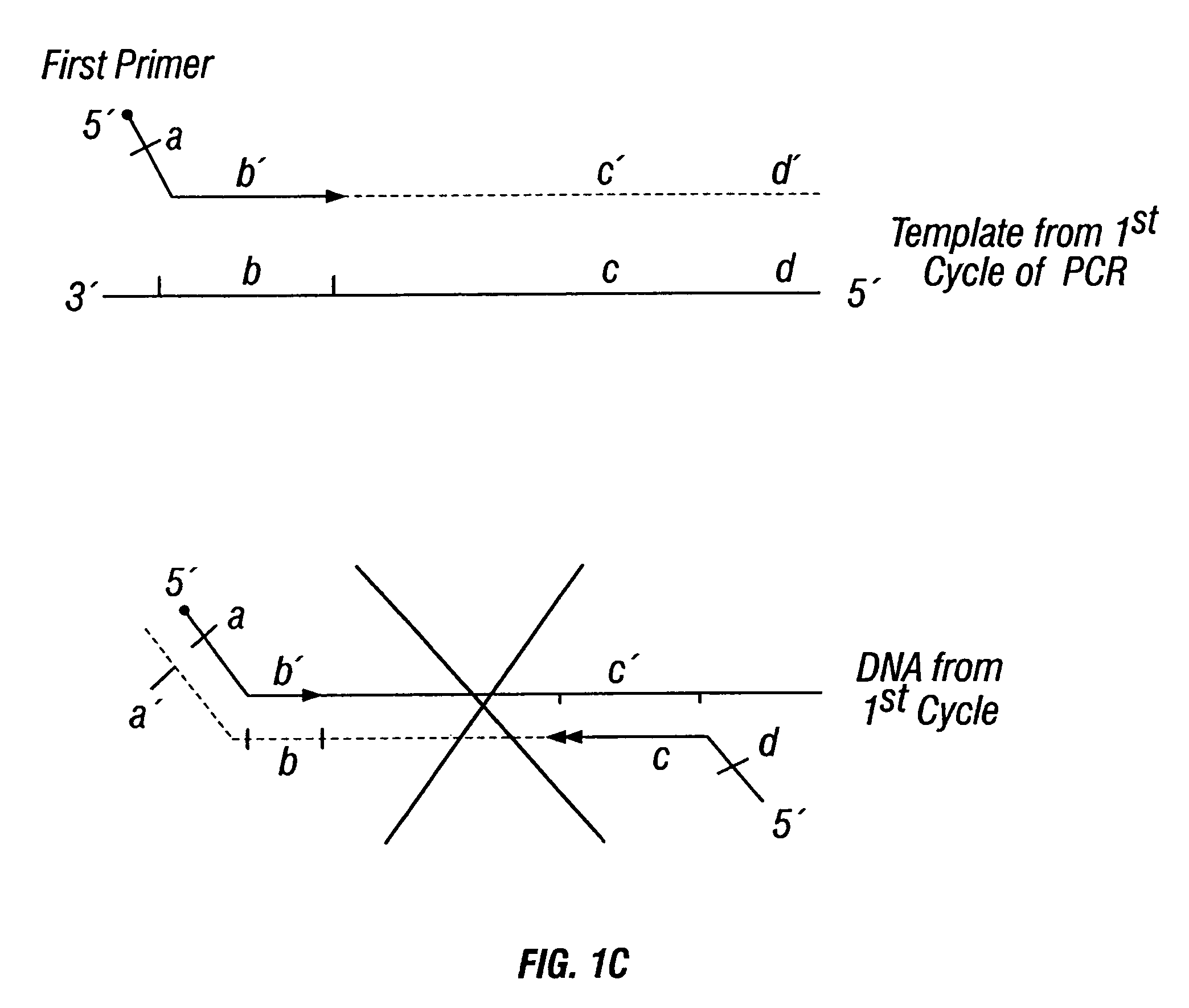
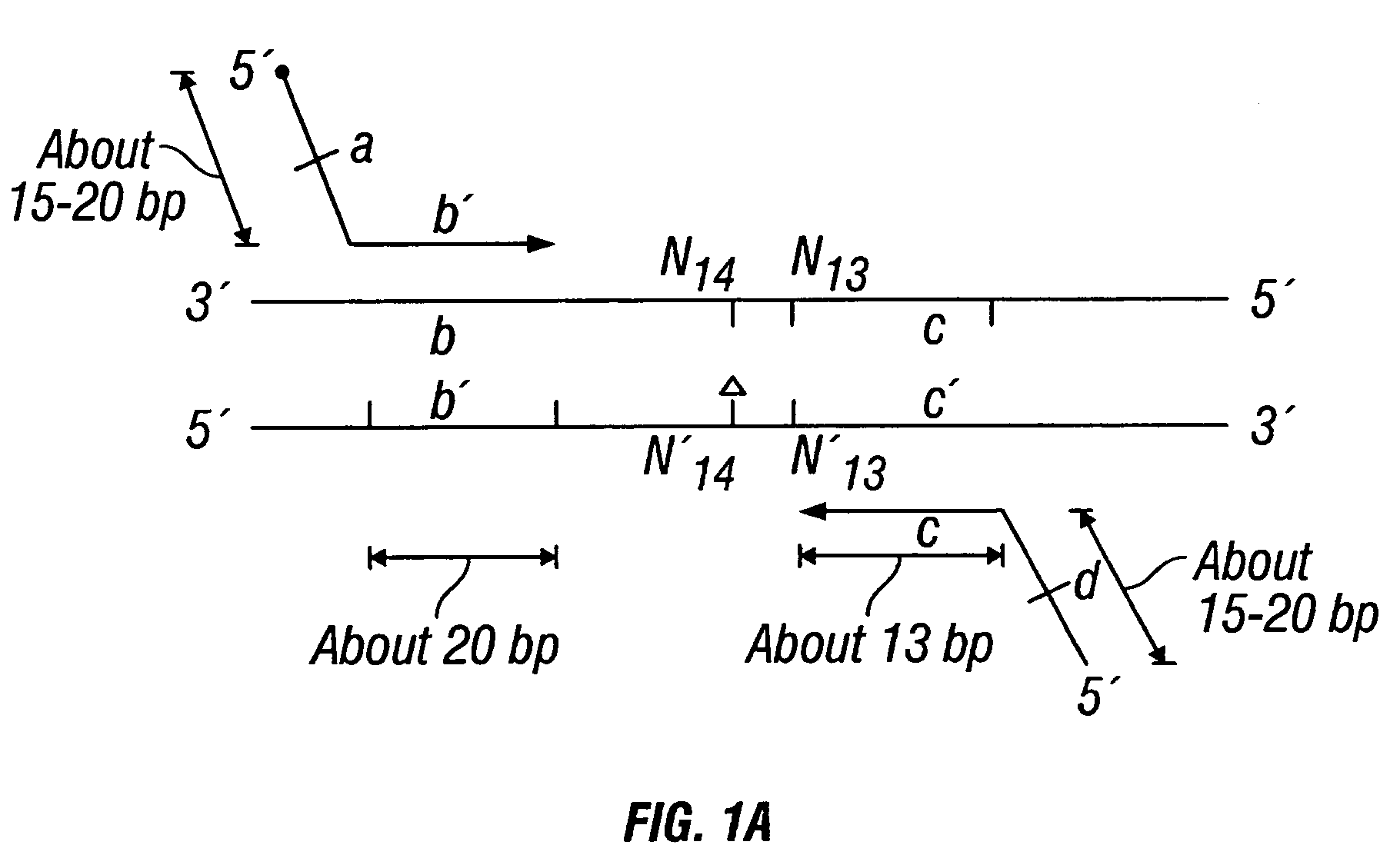
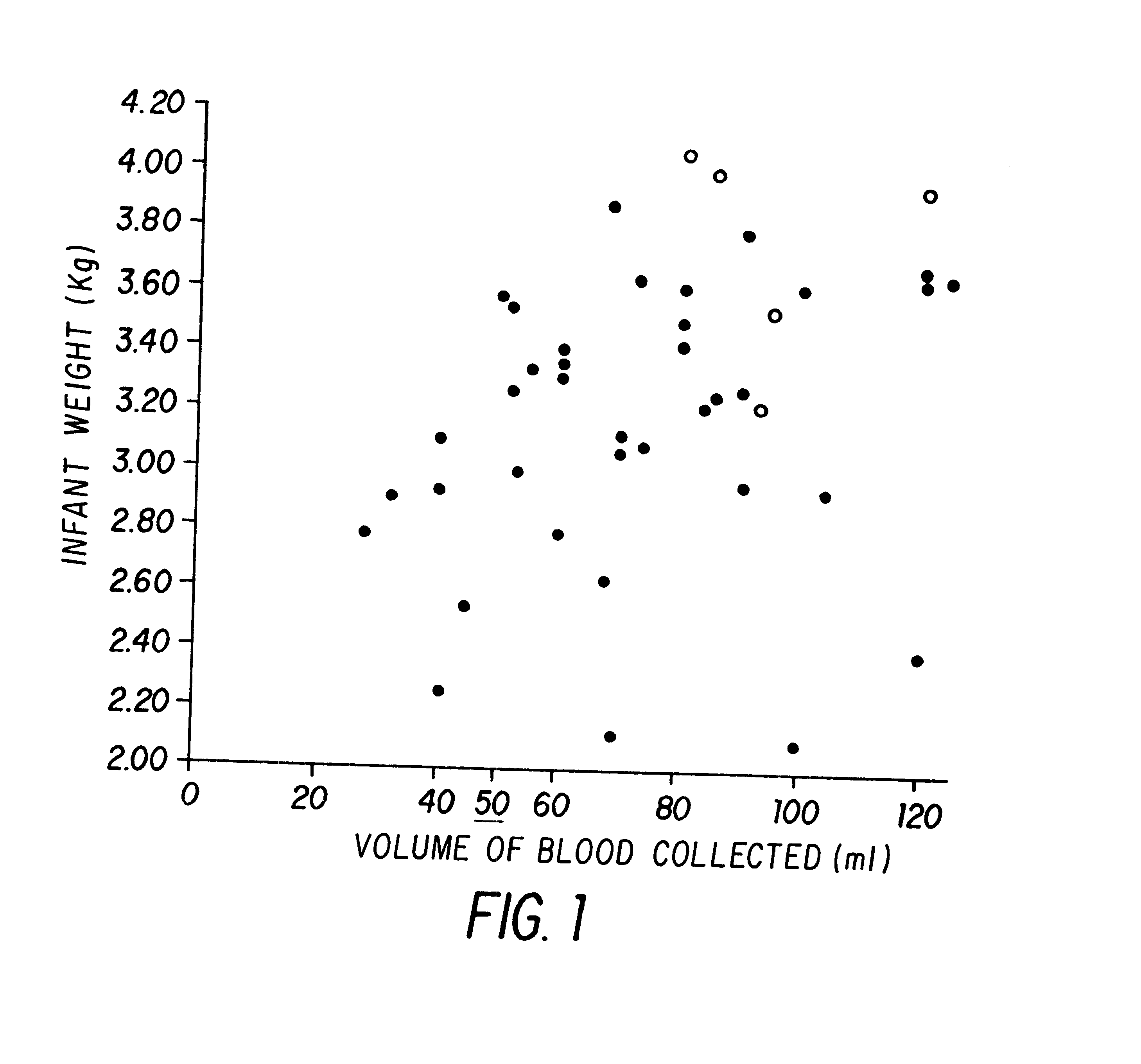
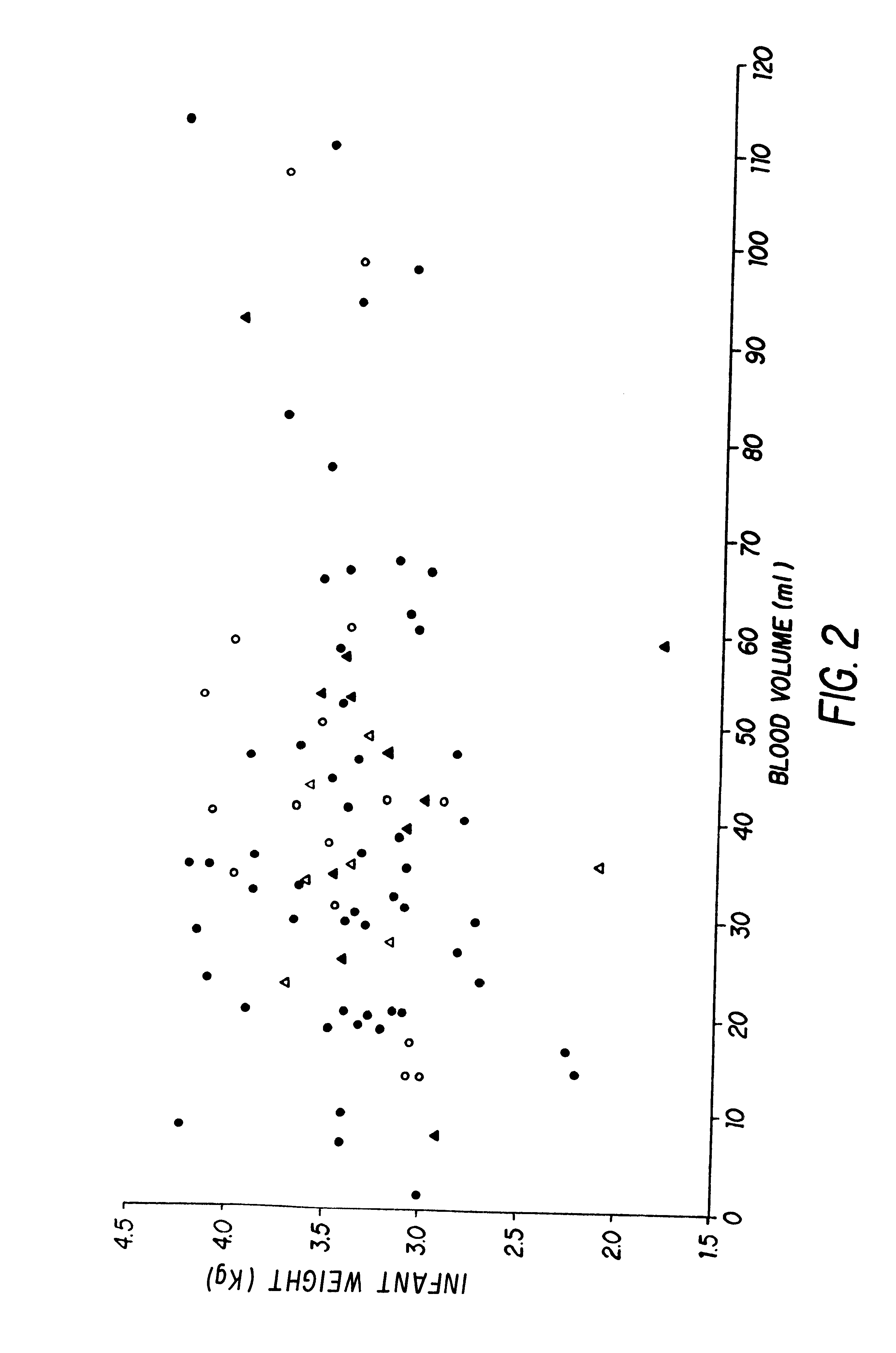
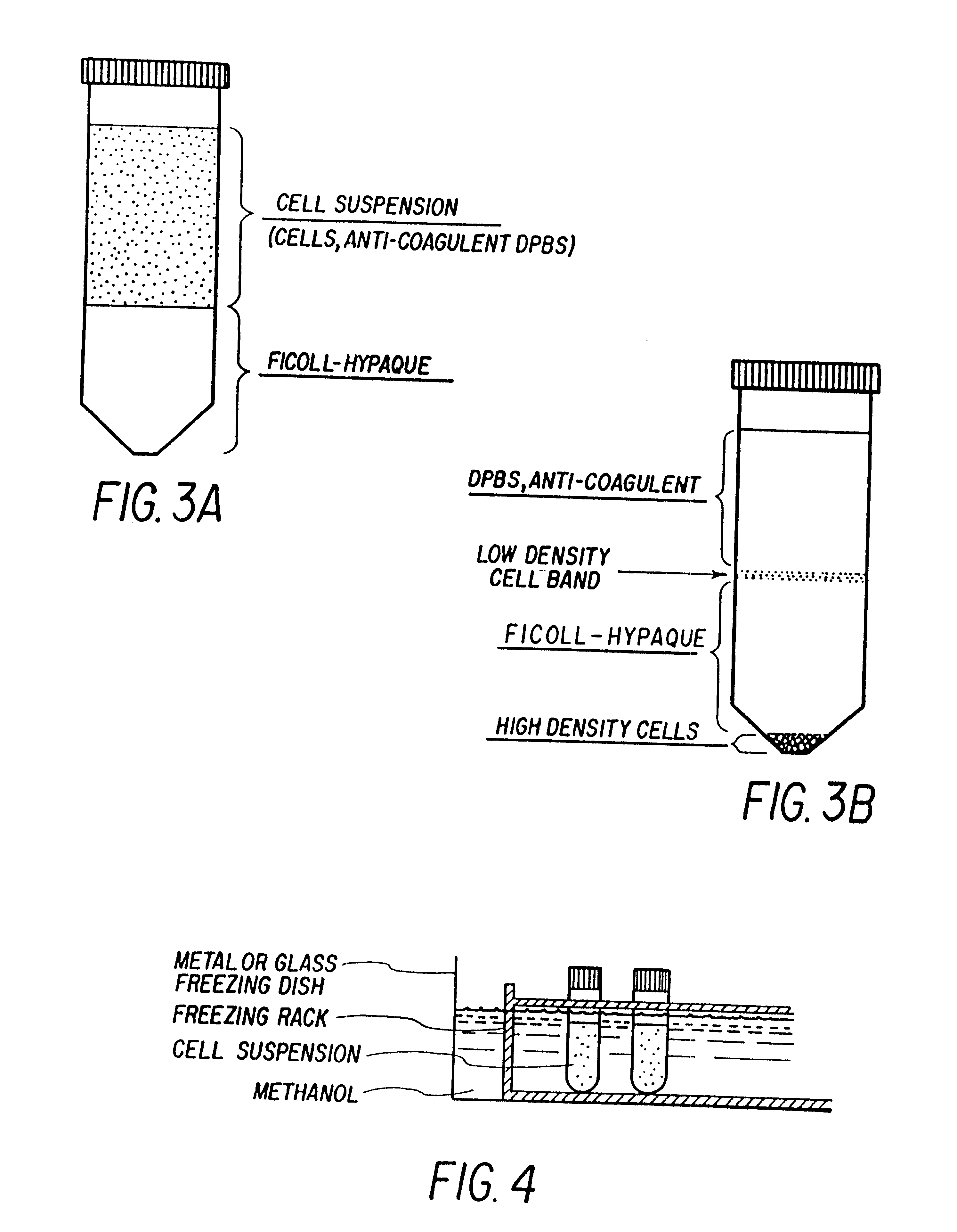
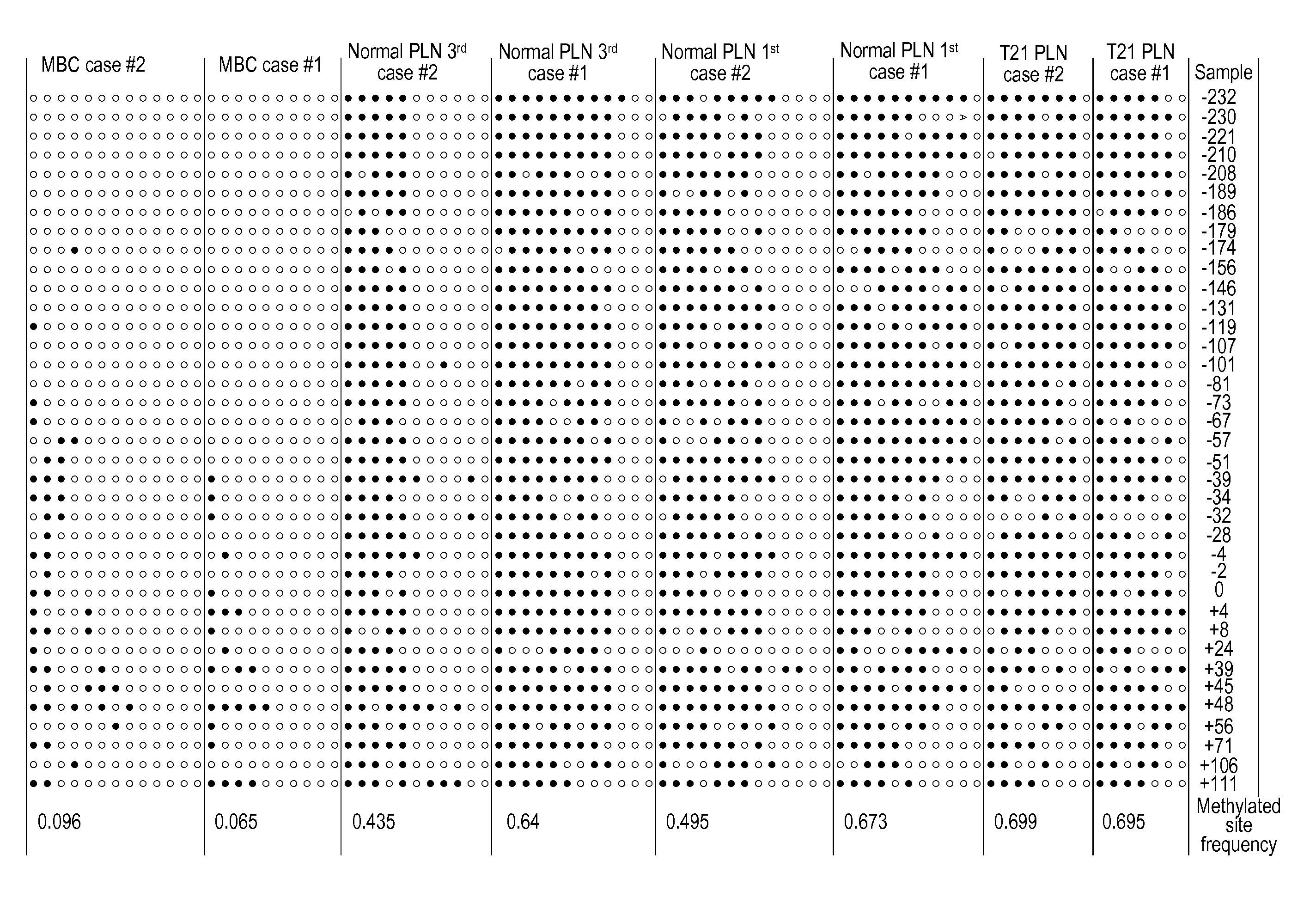
Non-invasive methods for detecting non-host DNA in a host using epigenetic differences between the host and non-host DNA
In a first aspect, the present invention features methods for differentiating DNA species originating from different individuals in a biological sample. These methods may be used to differentiate or detect fetal DNA in a maternal sample or to differentiate DNA of an organ donor from DNA of an organ recipient. In preferred embodiments, the DNA species are differentiated by observing epigenetic differences in the DNA species such as differences in DNA methylation. In a second aspect, the present invention features methods of detecting genetic abnormalities in a fetus by detecting fetal DNA in a biological sample obtained from a mother. In a third aspect, the present invention features methods for differentiating DNA species originating from an organ donor from those of an organ recipient. In a fourth aspect, the present invention features kits for differentiating DNA species originating from different individuals in a biological sample.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Methods for the Diagnosis of Fetal Abnormalities
ActiveUS20080138809A1Reliable and accurate clinical diagnosisAccurate and reliable diagnosisMicrobiological testing/measurementPreparing sample for investigationFetal abnormalityRare cell
The present invention relates to methods for detecting, enriching, and analyzing rare cells that are present in the blood, e.g. fetal cells. The invention further features methods of analyzing rare cell(s) to determine the presence of an abnormality, disease or condition in a subject, e.g. a fetus by analyzing a cellular sample from the subject.
Owner:VERINATA HEALTH INC +2
Fetal Genomic Analysis From A Maternal Biological Sample
ActiveUS20110105353A1Microbiological testing/measurementLibrary screeningDna concentrationPopulation
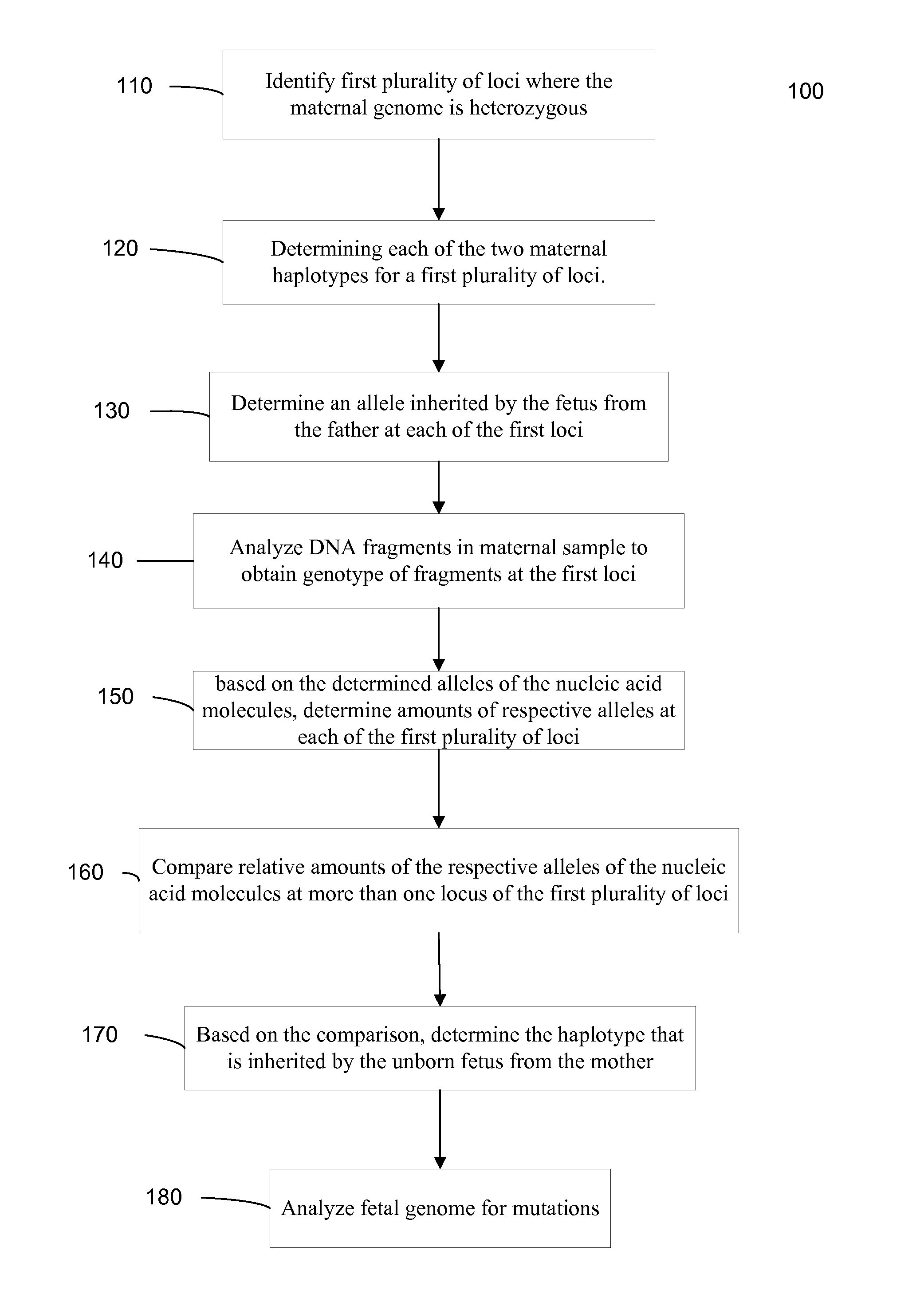
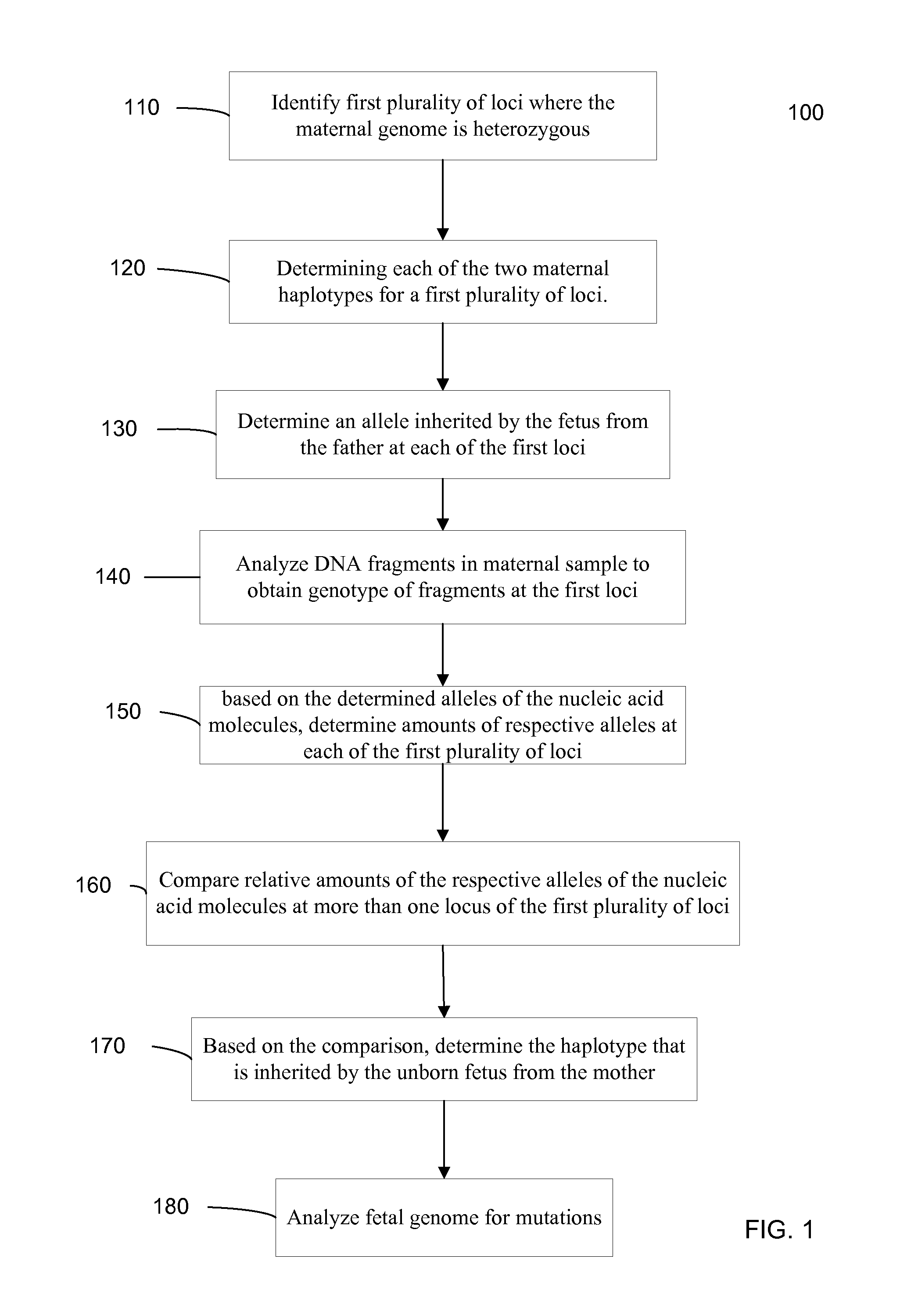
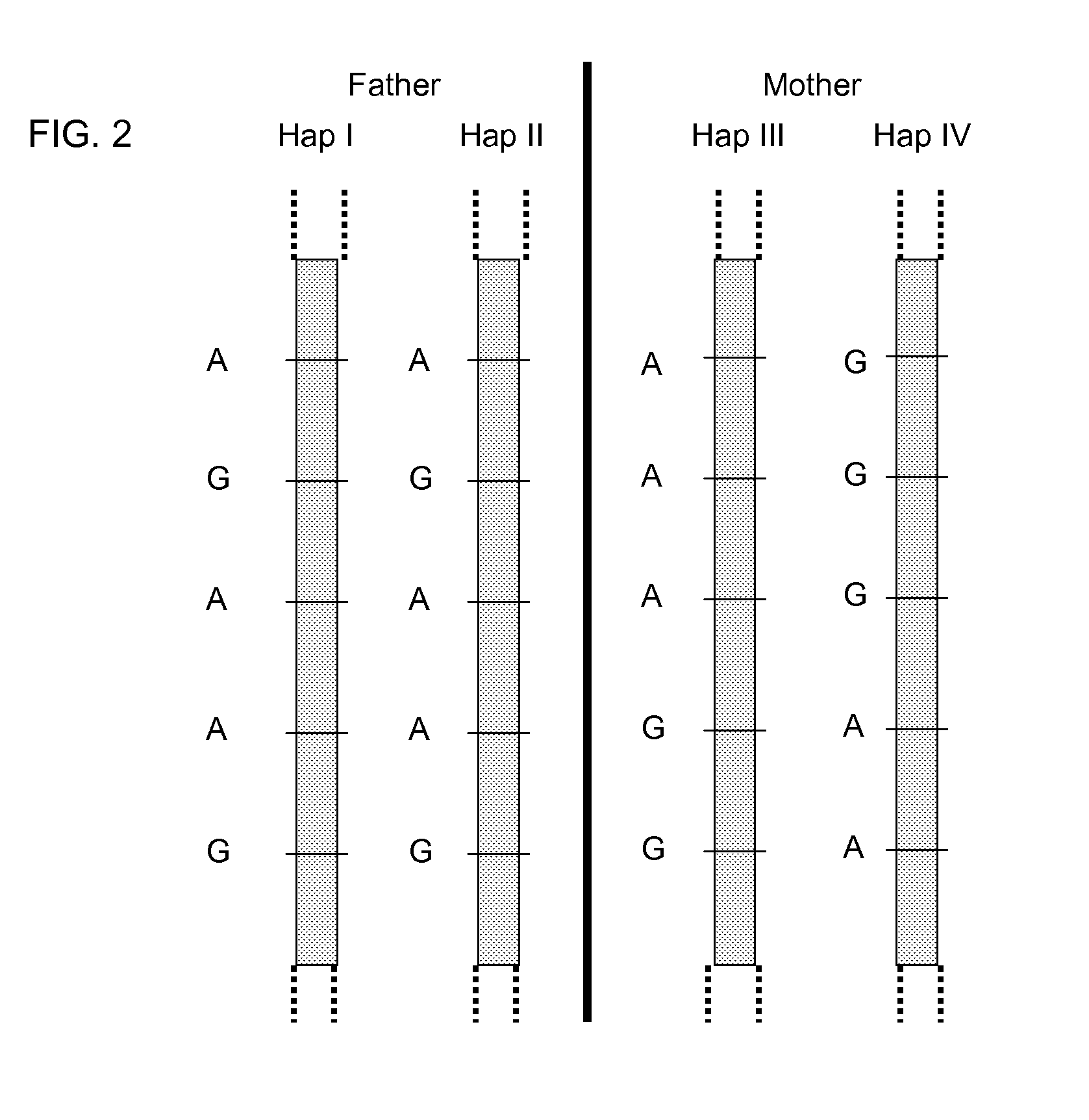
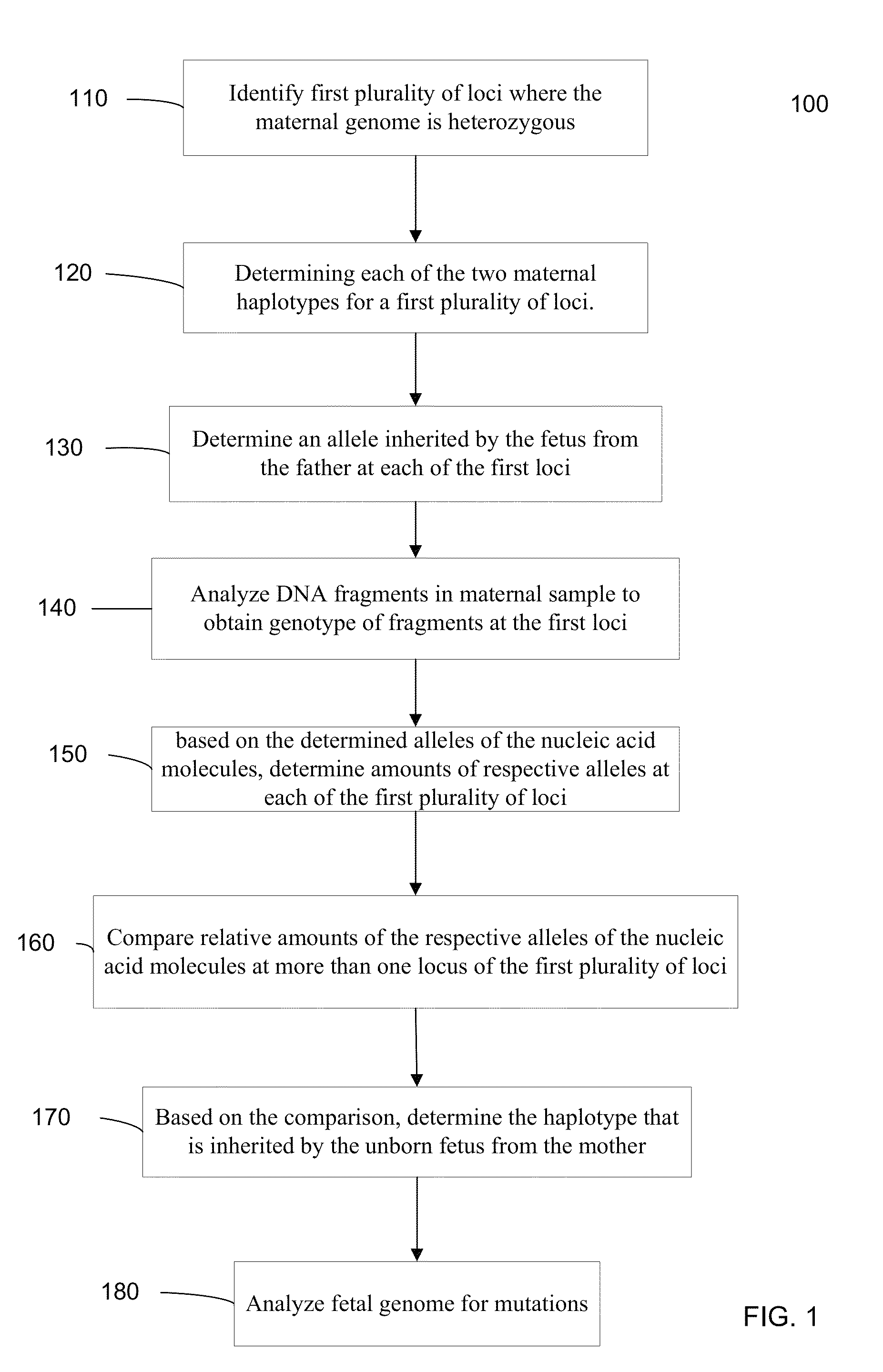
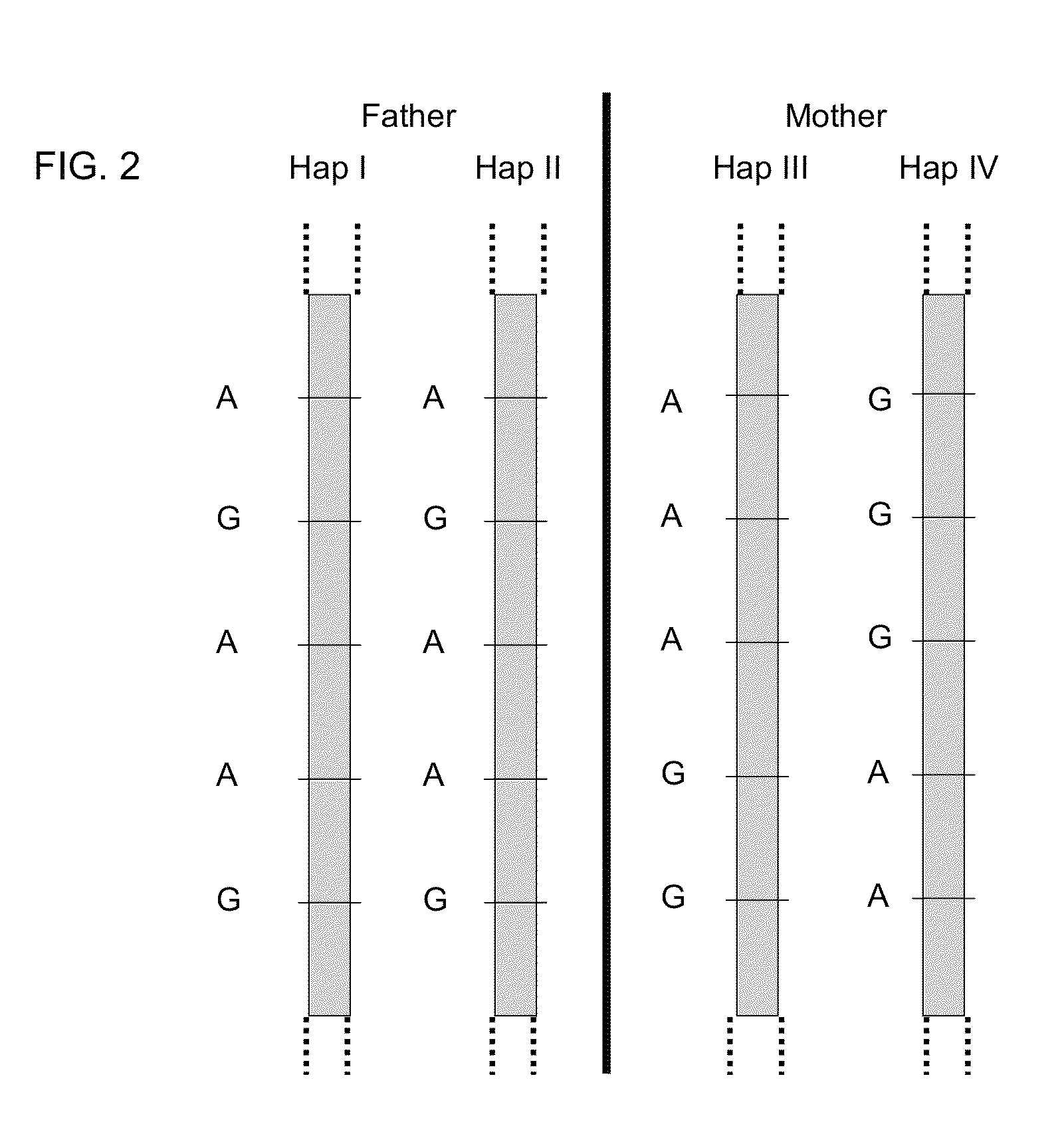
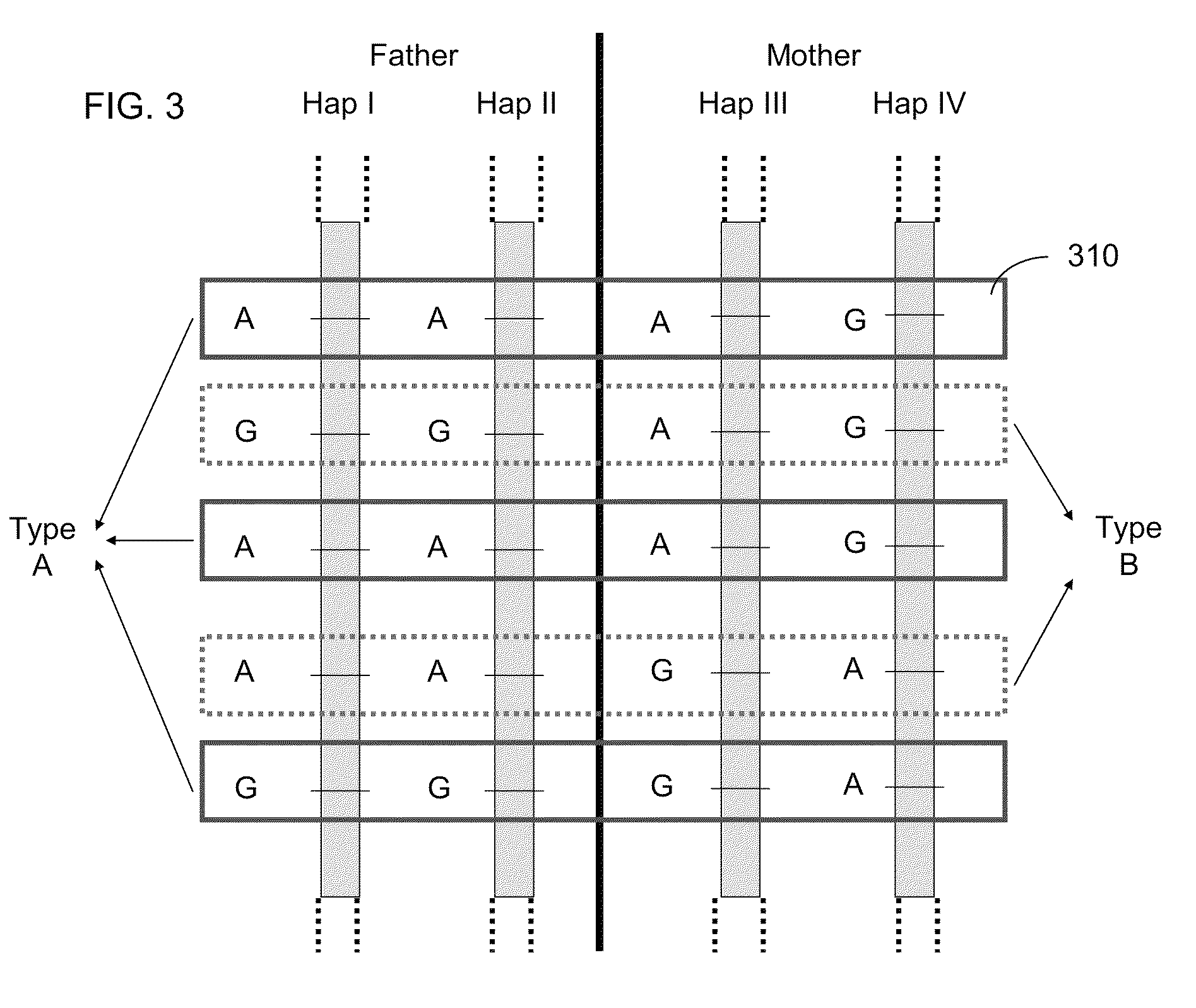
Systems, methods, and apparatus for determining at least a portion of fetal genome are provided. DNA fragments from a maternal sample (maternal and fetal DNA) can be analyzed to identify alleles at certain loci. The amounts of DNA fragments of the respective alleles at these loci can be analyzed together to determine relative amounts of the haplotypes for these loci and determine which haplotypes have been inherited from the parental genomes. Loci where the parents are a specific combination of homozygous and heterozygous can be analyzed to determine regions of the fetal genome. Reference haplotypes common in the population can be used along with the analysis of the DNA fragments of the maternal sample to determine the maternal and paternal genomes. Determination of mutations, a fractional fetal DNA concentration in a maternal sample, and a proportion of coverage of a sequencing of the maternal sample can also be provided.
Owner:SEQUENOM INC +1
Methods for detection of genetic disorders
InactiveUS7727720B2Maximize efficiencyIncrease delaySugar derivativesMicrobiological testing/measurementGeneticsAllele
The invention provides a method useful for detection of genetic disorders. The method comprises determining the sequence of alleles of a locus of interest, and quantitating a ratio for the alleles at the locus of interest, wherein the ratio indicates the presence or absence of a chromosomal abnormality. The present invention also provides a non-invasive method for the detection of chromosomal abnormalities in a fetus. The invention is especially useful as a non-invasive method for determining the sequence of fetal DNA. The invention further provides methods of isolation of free DNA from a sample.
Owner:RAVGEN INC
Methods for detection of genetic disorders
InactiveUS7442506B2Maximize efficiencyIncrease delaySugar derivativesMicrobiological testing/measurementGeneticsAllele
The invention provides a method useful for detection of genetic disorders. The method comprises determining the sequence of alleles of a locus of interest, and quantitating a ratio for the alleles at the locus of interest, wherein the ratio indicates the presence or absence of a chromosomal abnormality. The present invention also provides a non-invasive method for the detection of chromosomal abnormalities in a fetus. The invention is especially useful as a non-invasive method for determining the sequence of fetal DNA. The invention further provides methods of isolation of free DNA from a sample.
Owner:RAVGEN INC
Noninvasive Diagnosis of Fetal Aneuploidy by Sequencing
InactiveUS20100112575A1Maximum resultMicrobiological testing/measurementDisease diagnosisMassive parallel sequencingFetal aneuploidy
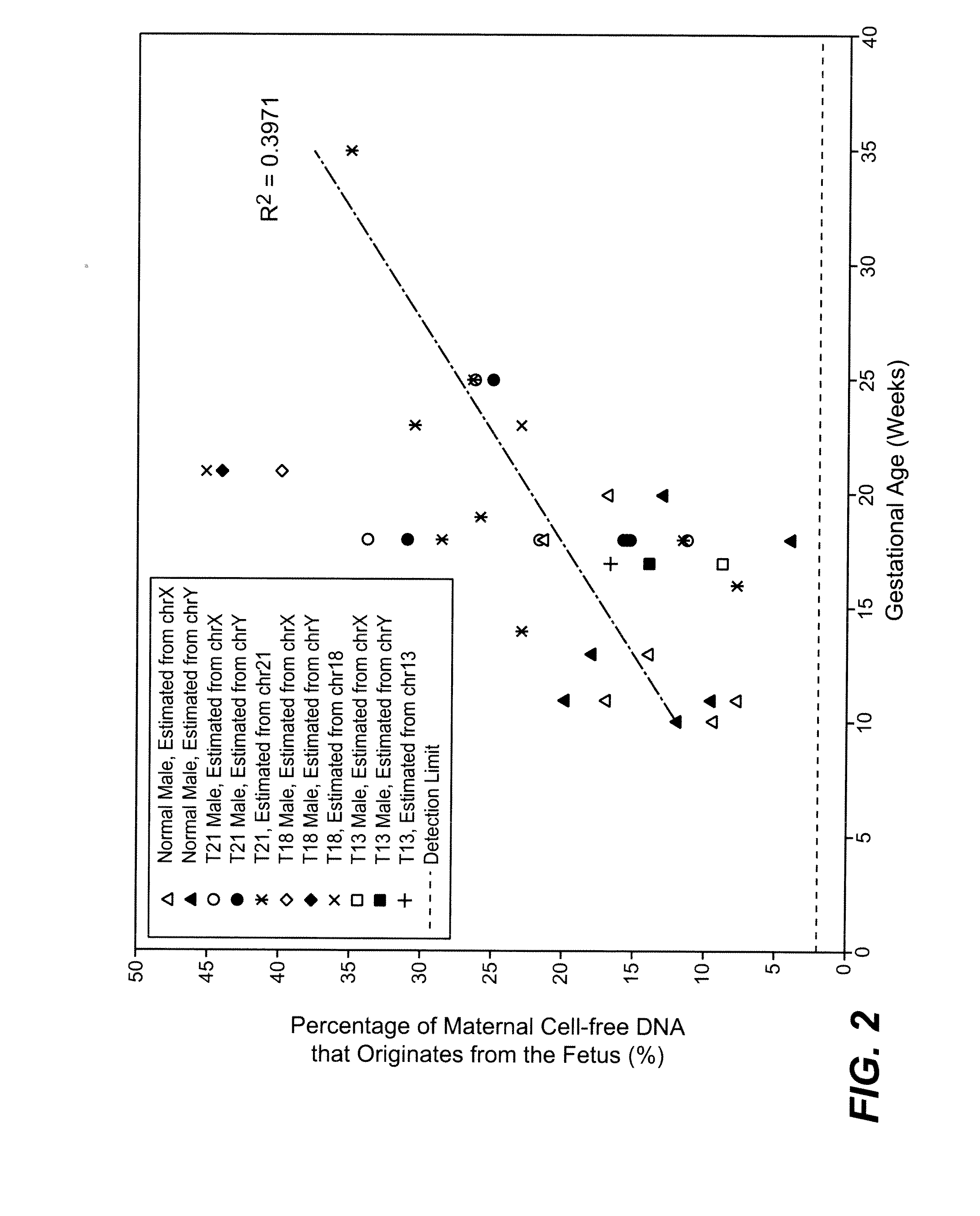
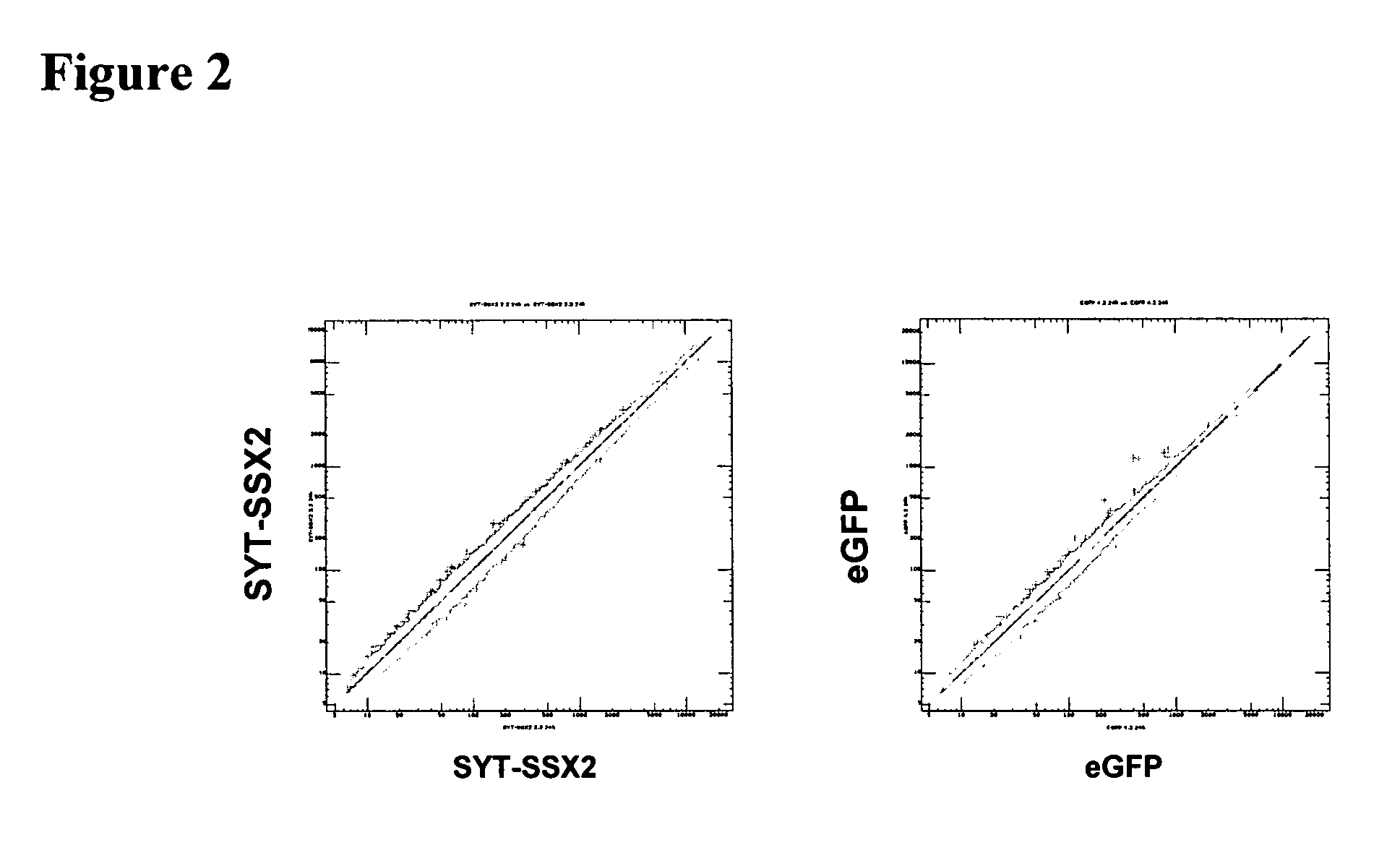
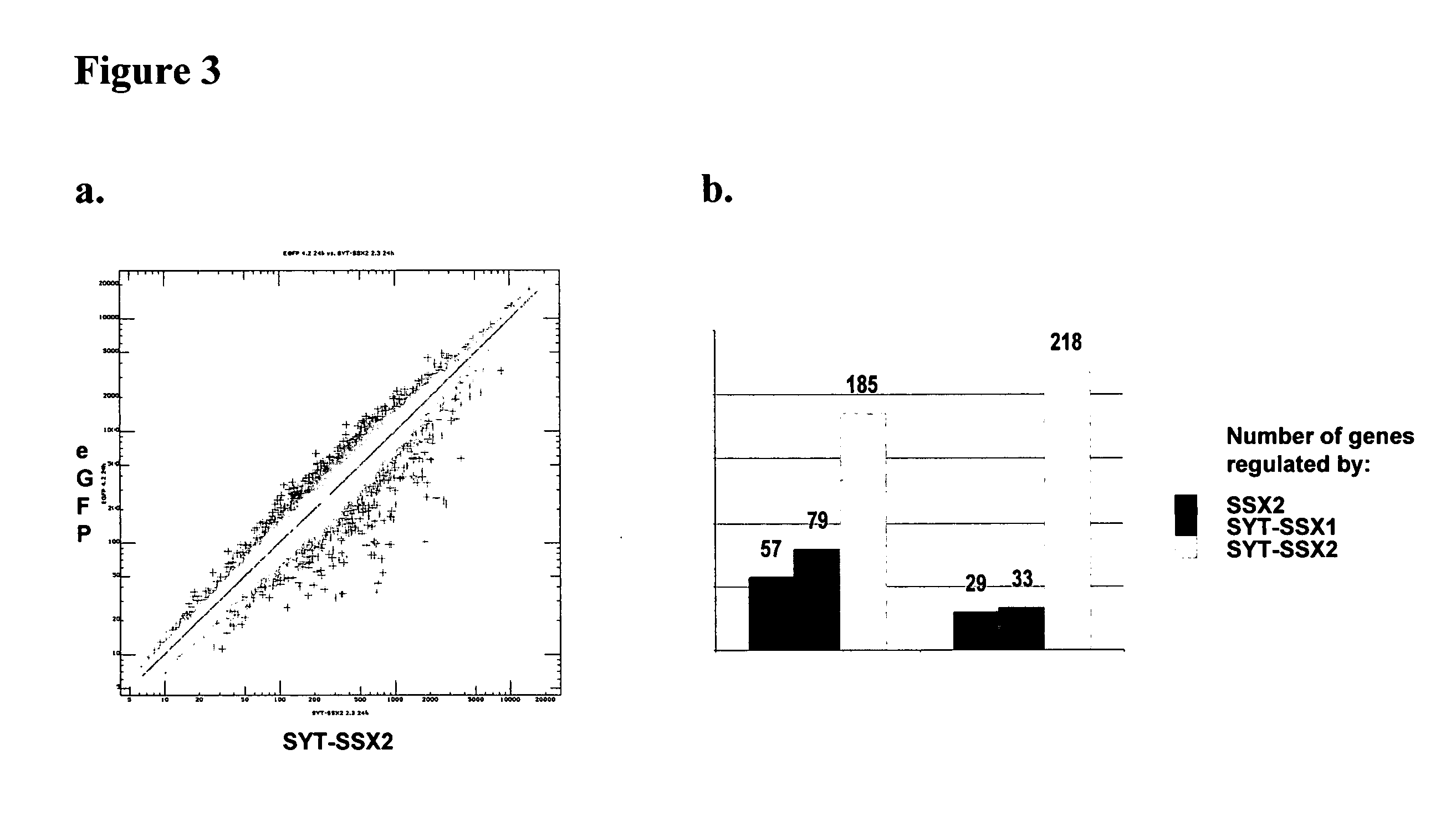
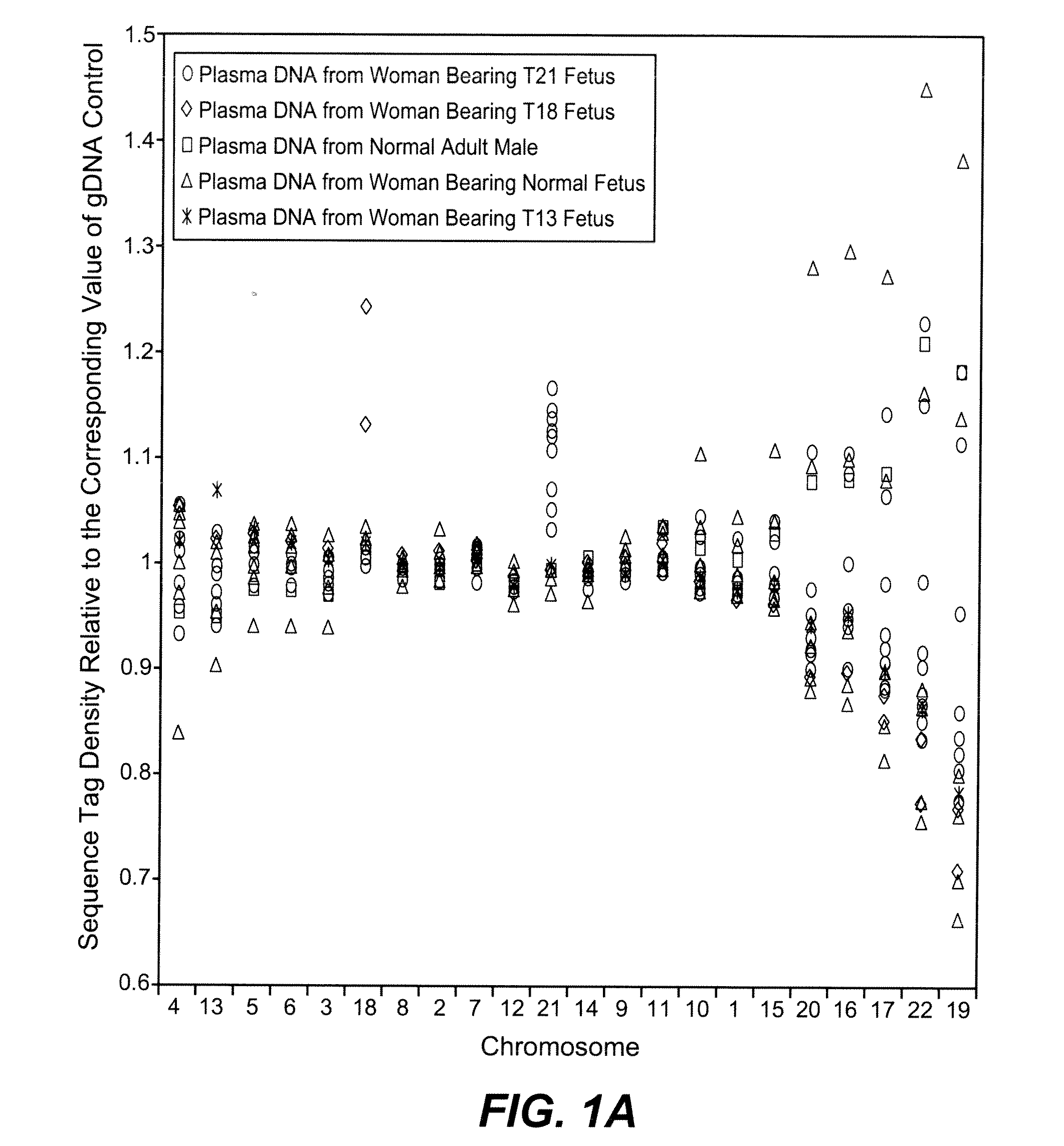
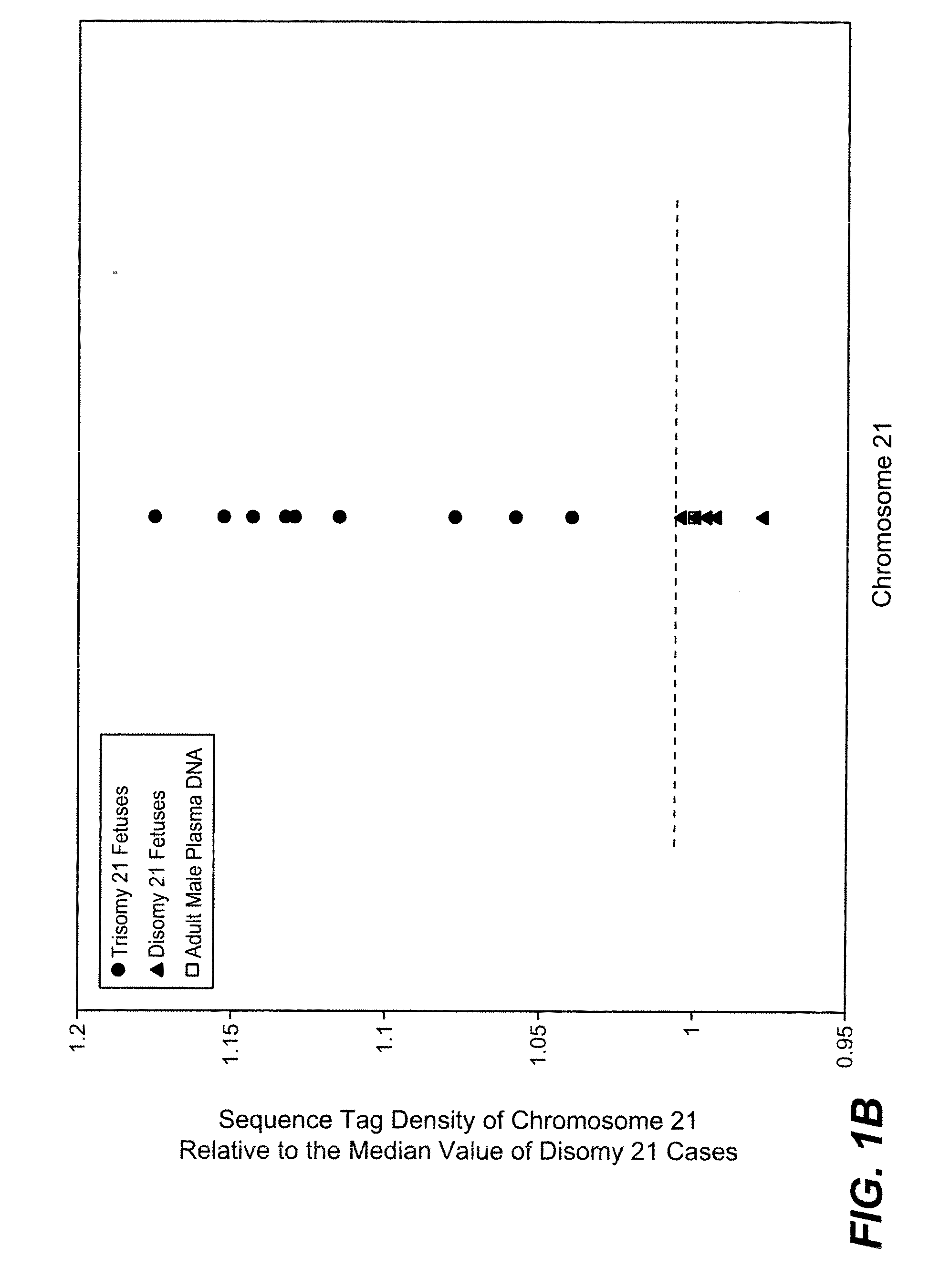
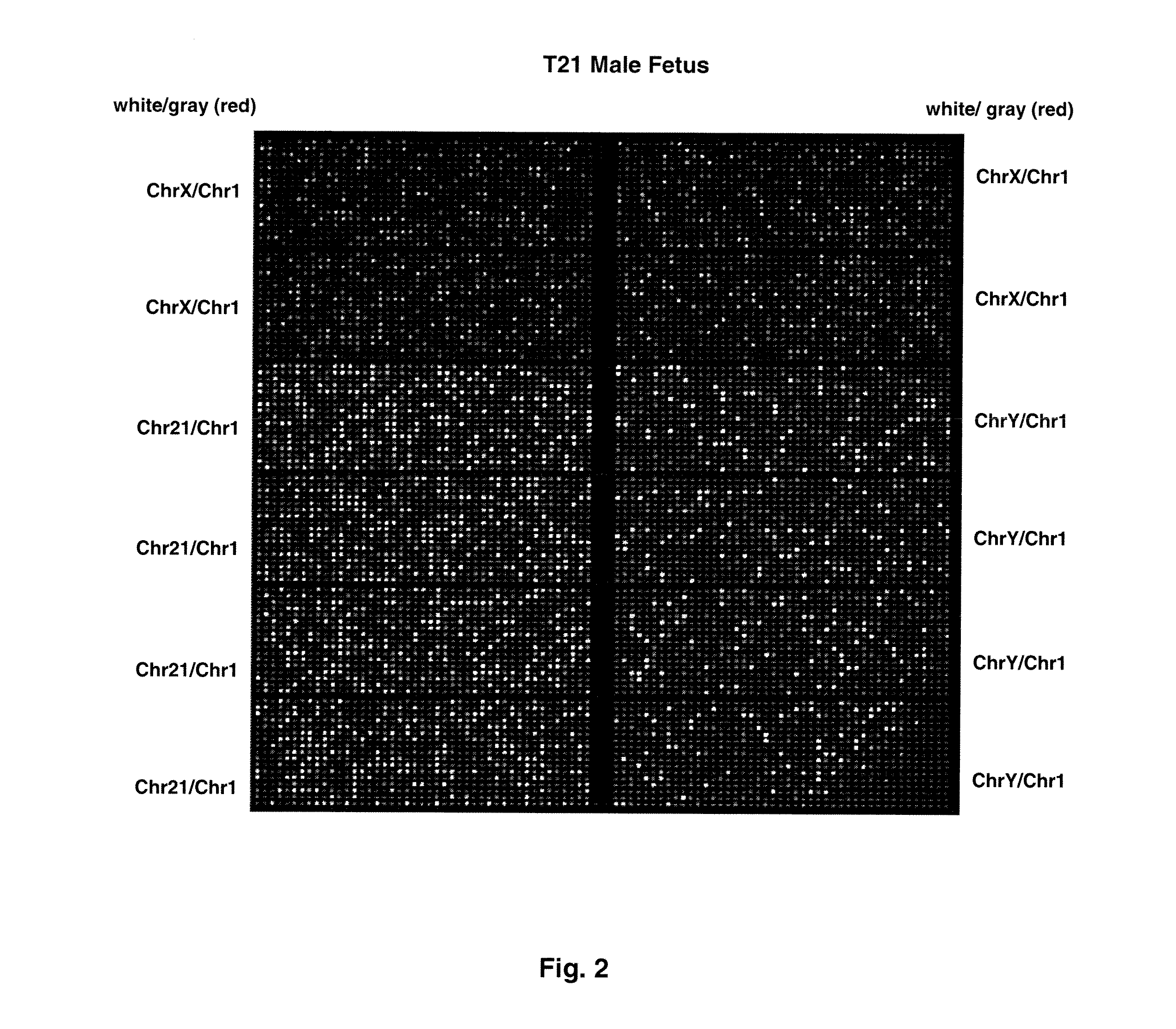
Disclosed is a method to achieve digital quantification of DNA (i.e., counting differences between identical sequences) using direct shotgun sequencing followed by mapping to the chromosome of origin and enumeration of fragments per chromosome. The preferred method uses massively parallel sequencing, which can produce tens of millions of short sequence tags in a single run and enabling a sampling that can be statistically evaluated. By counting the number of sequence tags mapped to a predefined window in each chromosome, the over- or under-representation of any chromosome in maternal plasma DNA contributed by an aneuploid fetus can be detected. This method does not require the differentiation of fetal versus maternal DNA. The median count of autosomal values is used as a normalization constant to account for differences in total number of sequence tags is used for comparison between samples and between chromosomes.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Methods for detection of genetic disorders
InactiveUS20070178478A1Minimize disruptionMaximize efficiencyMicrobiological testing/measurementFermentationGeneticsAllele
The invention provides a method useful for detection of genetic disorders. The method comprises determining the sequence of alleles of a locus of interest, and quantitating a ratio for the alleles at the locus of interest, wherein the ratio indicates the presence or absence of a chromosomal abnormality. The present invention also provides a non-invasive method for the detection of chromosomal abnormalities in a fetus. The invention is especially useful as a non-invasive method for determining the sequence of fetal DNA. The invention further provides methods of isolation of free DNA from a sample.
Owner:RAVGEN INC
Methods for Non-Invasive Prenatal Ploidy Calling
ActiveUS20120270212A1Fill in the blanksMathematical modelsMicrobiological testing/measurementDistribution patternGenotype
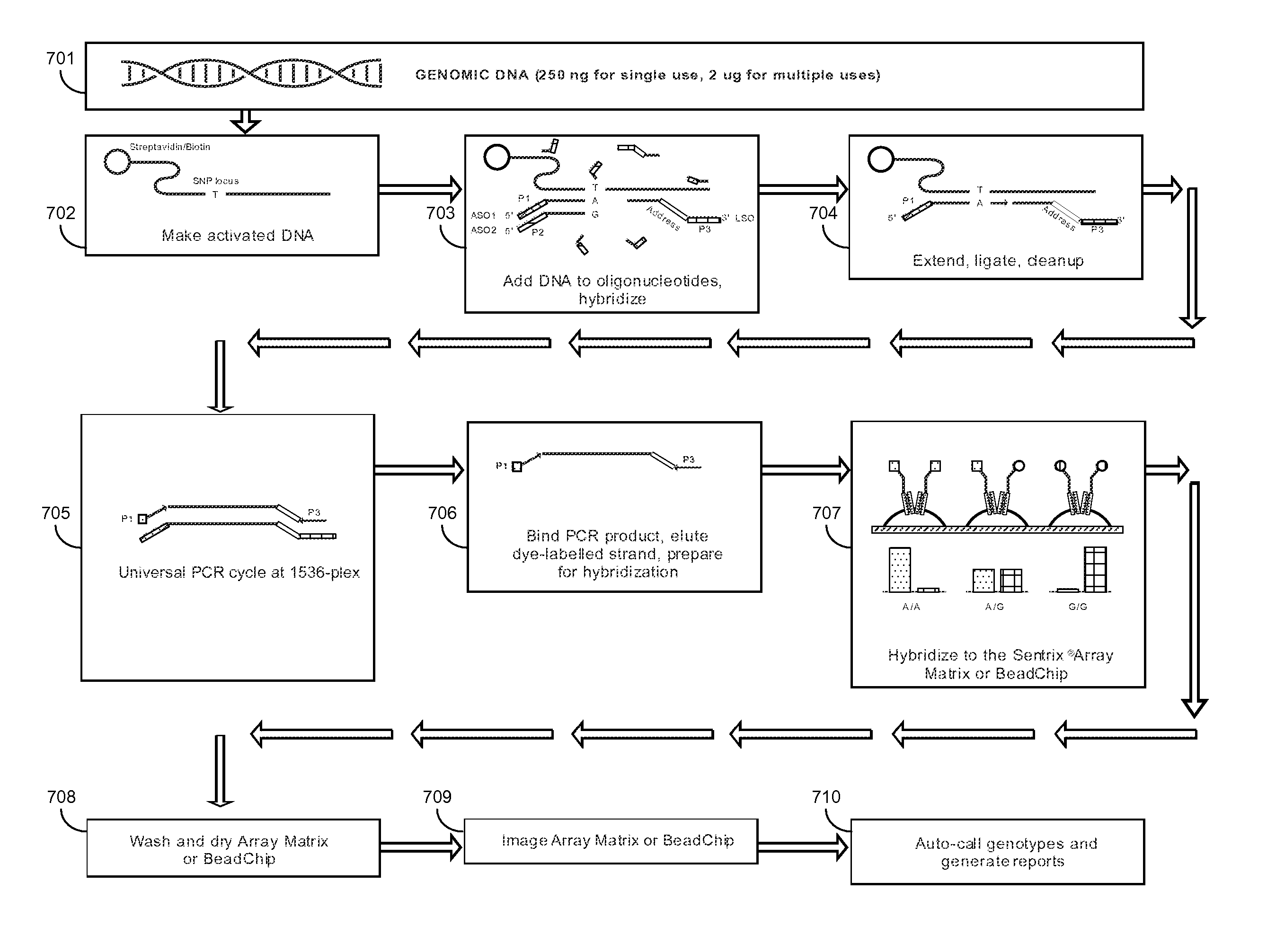
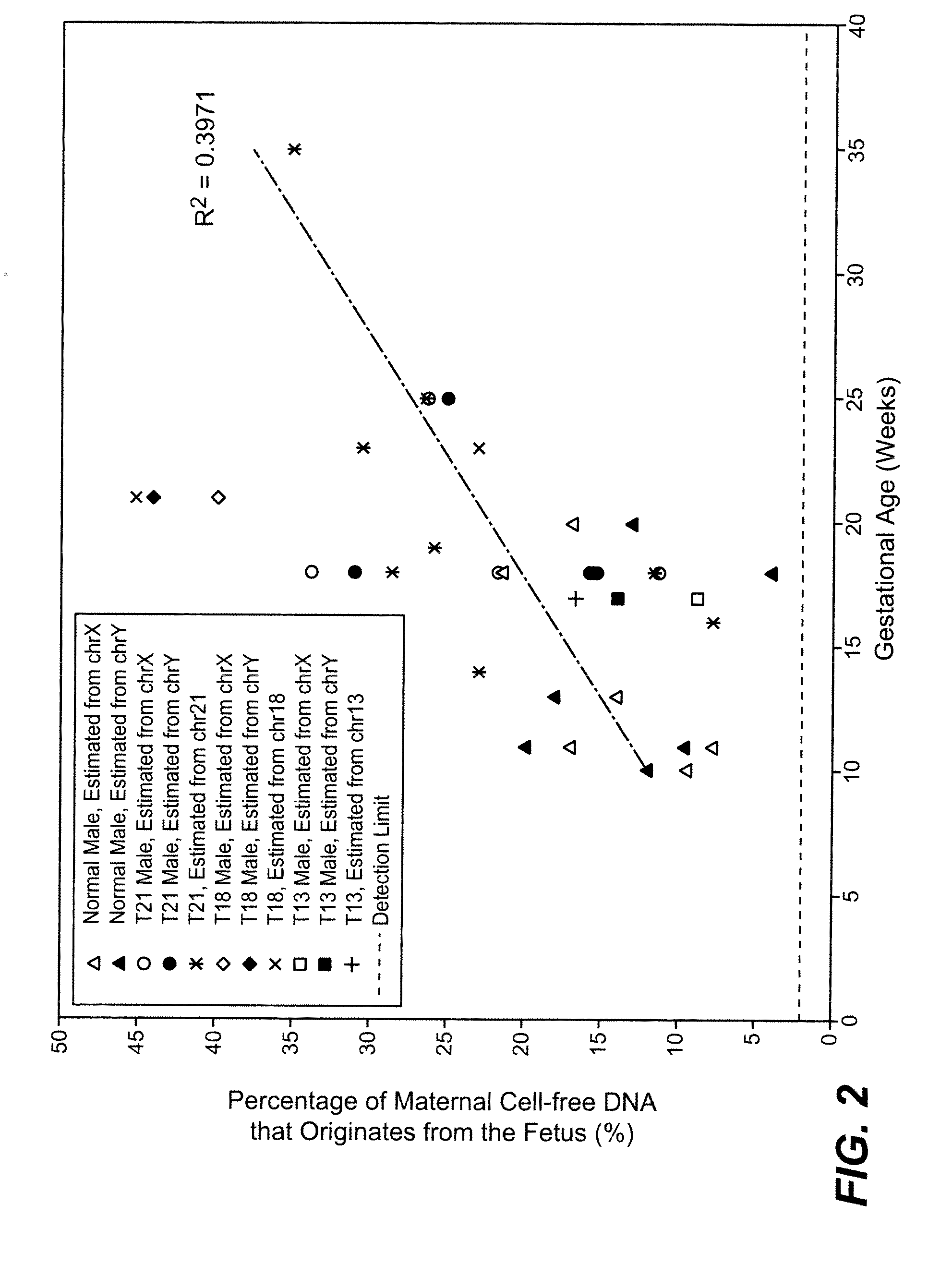
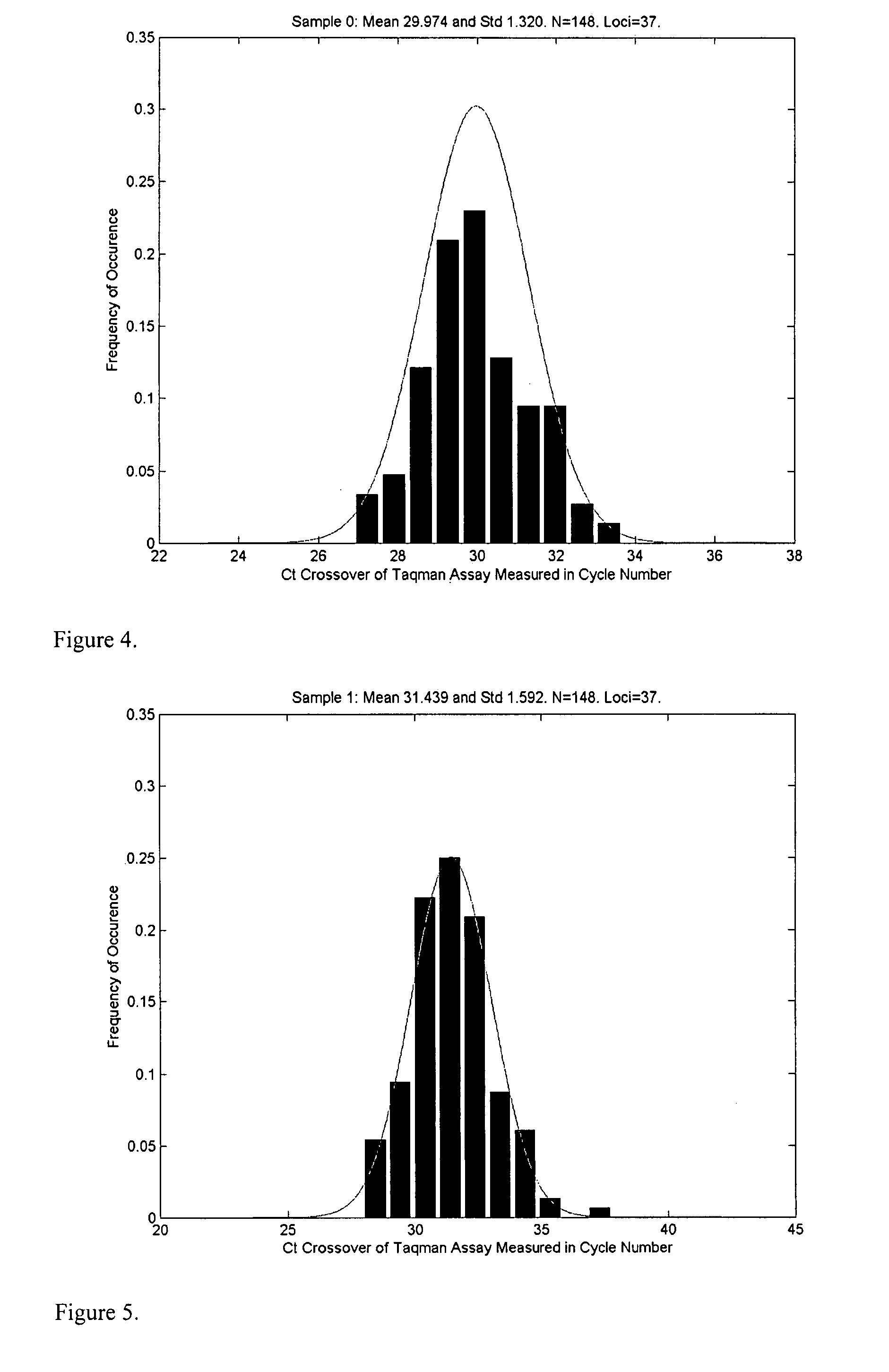
The present disclosure provides methods for determining the ploidy status of a chromosome in a gestating fetus from genotypic data measured from a mixed sample of DNA comprising DNA from both the mother of the fetus and from the fetus, and optionally from genotypic data from the mother and father. The ploidy state is determined by using a joint distribution model to create a plurality of expected allele distributions for different possible fetal ploidy states given the parental genotypic data, and comparing the expected allelic distributions to the pattern of measured allelic distributions measured in the mixed sample, and choosing the ploidy state whose expected allelic distribution pattern most closely matches the observed allelic distribution pattern. The mixed sample of DNA may be preferentially enriched at a plurality of polymorphic loci in a way that minimizes the allelic bias, for example using massively multiplexed targeted PCR.
Owner:NATERA
Cultivation of primate embryonic stem cells
InactiveUS20050148070A1Improve cloning efficiencyAvoid variabilityOrganic active ingredientsCulture processMammalFeeder Layer
The invention relates to methods for culturing human embryonic stem cells by culturing the stem cells in an environment essentially free of mammalian fetal serum and in a stem cell culture medium including amino acids, vitamins, salts, minerals, transferring, insulin, albumin, and a fibroblast growth factor that is supplied from a source other than just a feeder layer the medium. Also disclosed are compositions capable of supporting the culture and proliferation of human embryonic stem cells without the need for feeder cells or for exposure of the medium to feeder cells.
Owner:WISCONSIN ALUMNI RES FOUND
Cultivation of primate embryonic stem cells
InactiveUS20050244962A1Maintain normalMaintaining the karyotype of the stem cellsCulture processArtificial cell constructsFeeder LayerStem cell culture
The invention relates to methods for culturing human embryonic stem cells by culturing the stem cells in an environment essentially free of mammalian fetal serum and in a stem cell culture medium including amino acids, vitamins, salts, minerals, transferring, insulin, albumin, and a fibroblast growth factor that is supplied from a source other than just a feeder layer the medium. Also disclosed are compositions capable of supporting the culture and proliferation of human embryonic stem cells without the need for feeder cells or for exposure of the medium to feeder cells.
Owner:WICELL RES INST
CICM cells and non-human mammalian embryos prepared by nuclear transfer of a proliferating differentiated cell or its nucleus
InactiveUS6235970B1Simple procedureSimplifying and facilitating procedureNervous disorderMuscular disorderPresent methodNuclear transfer
An improved method of nuclear transfer involving the transplantation of donor differentiated cell nuclei into enucleated oocytes of the same species as the donor cell is provided. The resultant nuclear transfer units are useful for multiplication of genotypes and transgenic genotypes by the production of fetuses and offspring, and for production of isogenic CICM cells, including human isogenic embryonic or stem cells. Production of genetically engineered or transgenic mammalian embryos, fetuses and offspring is facilitated by the present method since the differentiated cell source of the donor nuclei can be genetically modified and clonally propagated.
Owner:UNIVERSITY OF MASSACHUSETTS AMHERST
Use of RNA for reprogramming somatic cells
InactiveUS20110065103A1Promotes reprogrammingBlood/immune system cellsDrug compositionsReprogrammingSomatic cell
The present invention provides methods for de-differentiating somatic cells into stem-like cells without generating embryos or fetuses. More specifically, the present invention provides methods for effecting the de-differentiation of somatic cells to cells having stem cell characteristics, in particular pluripotency, by introducing RNA encoding factors inducing the de-differentiation of somatic cells into the somatic cells and culturing the somatic cells allowing the cells to de-differentiate.
Owner:BIONTECH AG +2
Noninvasive Diagnosis of Fetal Aneuploidy by Sequencing
ActiveUS20100138165A1Maximum resultMicrobiological testing/measurementDisease diagnosisMassive parallel sequencingFetal aneuploidy
Disclosed is a method to achieve digital quantification of DNA (i.e., counting differences between identical sequences) using direct shotgun sequencing followed by mapping to the chromosome of origin and enumeration of fragments per chromosome. The preferred method uses massively parallel sequencing, which can produce tens of millions of short sequence tags in a single run and enabling a sampling that can be statistically evaluated. By counting the number of sequence tags mapped to a predefined window in each chromosome, the over- or under-representation of any chromosome in maternal plasma DNA contributed by an aneuploid fetus can be detected. This method does not require the differentiation of fetal versus maternal DNA. The median count of autosomal values is used as a normalization constant to account for differences in total number of sequence tags is used for comparison between samples and between chromosomes.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
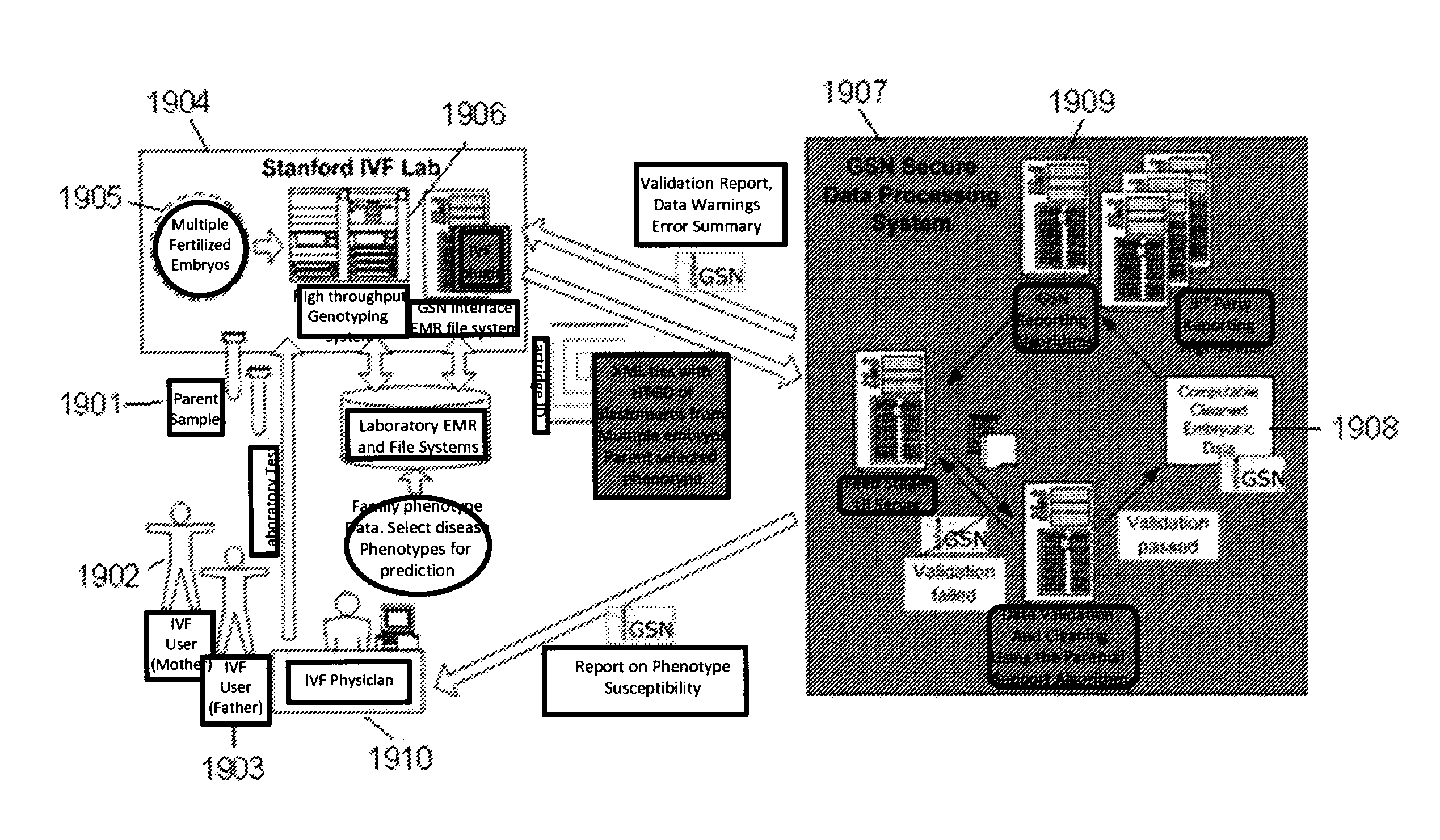
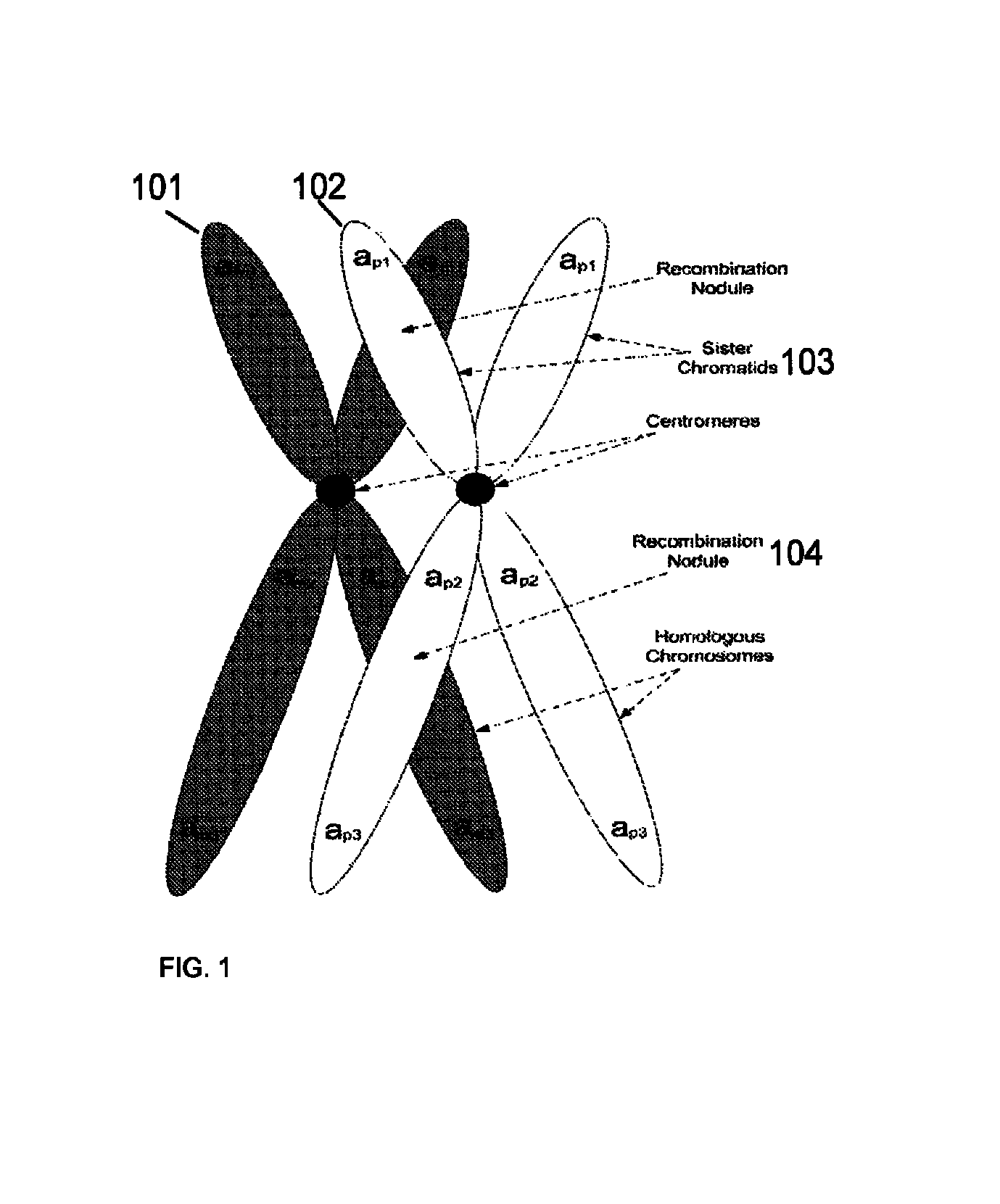
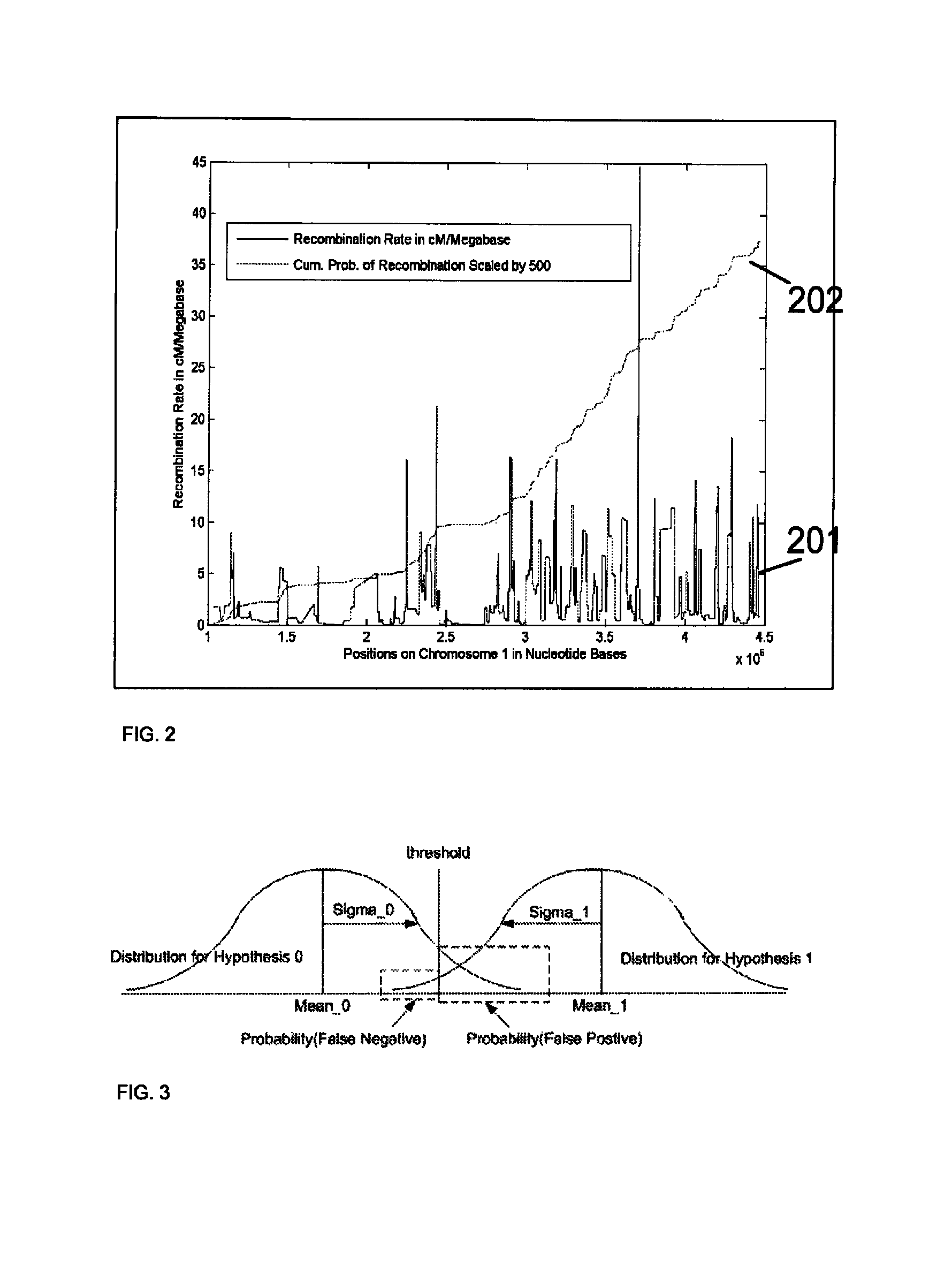
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
ActiveUS20070184467A1Significant resultMicrobiological testing/measurementProteomicsUniparental disomyEmbryo
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell is reconstructed using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In accordance with another embodiment of the invention, incomplete genetic data from a fetus is acquired from fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In one embodiment, the genetic data can be reconstructed for the purposes of making phenotypic predictions. In another embodiment, the genetic data can be used to detect for aneuploides and uniparental disomy.
Owner:NATERA
Fetal genomic analysis from a maternal biological sample
Systems, methods, and apparatus for determining at least a portion of fetal genome are provided. DNA fragments from a maternal sample (maternal and fetal DNA) can be analyzed to identify alleles at certain loci. The amounts of DNA fragments of the respective alleles at these loci can be analyzed together to determine relative amounts of the haplotypes for these loci and determine which haplotypes have been inherited from the parental genomes. Loci where the parents are a specific combination of homozygous and heterozygous can be analyzed to determine regions of the fetal genome. Reference haplotypes common in the population can be used along with the analysis of the DNA fragments of the maternal sample to determine the maternal and paternal genomes. Determination of mutations, a fractional fetal DNA concentration in a maternal sample, and a proportion of coverage of a sequencing of the maternal sample can also be provided.
Owner:SEQUENOM INC +1
Enrichment of circulating fetal DNA
InactiveUS20070243549A1High accuracy of resultsMicrobiological testing/measurementImmunoglobulins against animals/humansNon invasiveMolecular diagnostics
A non-invasive screening or diagnostic method for determining the likelihood of a fetus with a genetic abnormality or a potential pregnancy complication, which utilizes a liquid blood sample from a pregnant woman. Antibodies specific to a section of histone 3.1 which is exposed to a far greater extent in chromatin of fetal origin than in chromatin of maternal origin are used to sequester and isolate such fetal nucleosomes including the associated fetal DNA. Following isolation / enrichment of such fetal DNA, genetic analysis is carried out using known molecular diagnostics.
Owner:BAYLOR COLLEGE OF MEDICINE
Direct Molecular Diagnosis of Fetal Aneuploidy
ActiveUS20110151442A1Addressing slow performanceSugar derivativesMicrobiological testing/measurementSpecific chromosomeFetal tissues
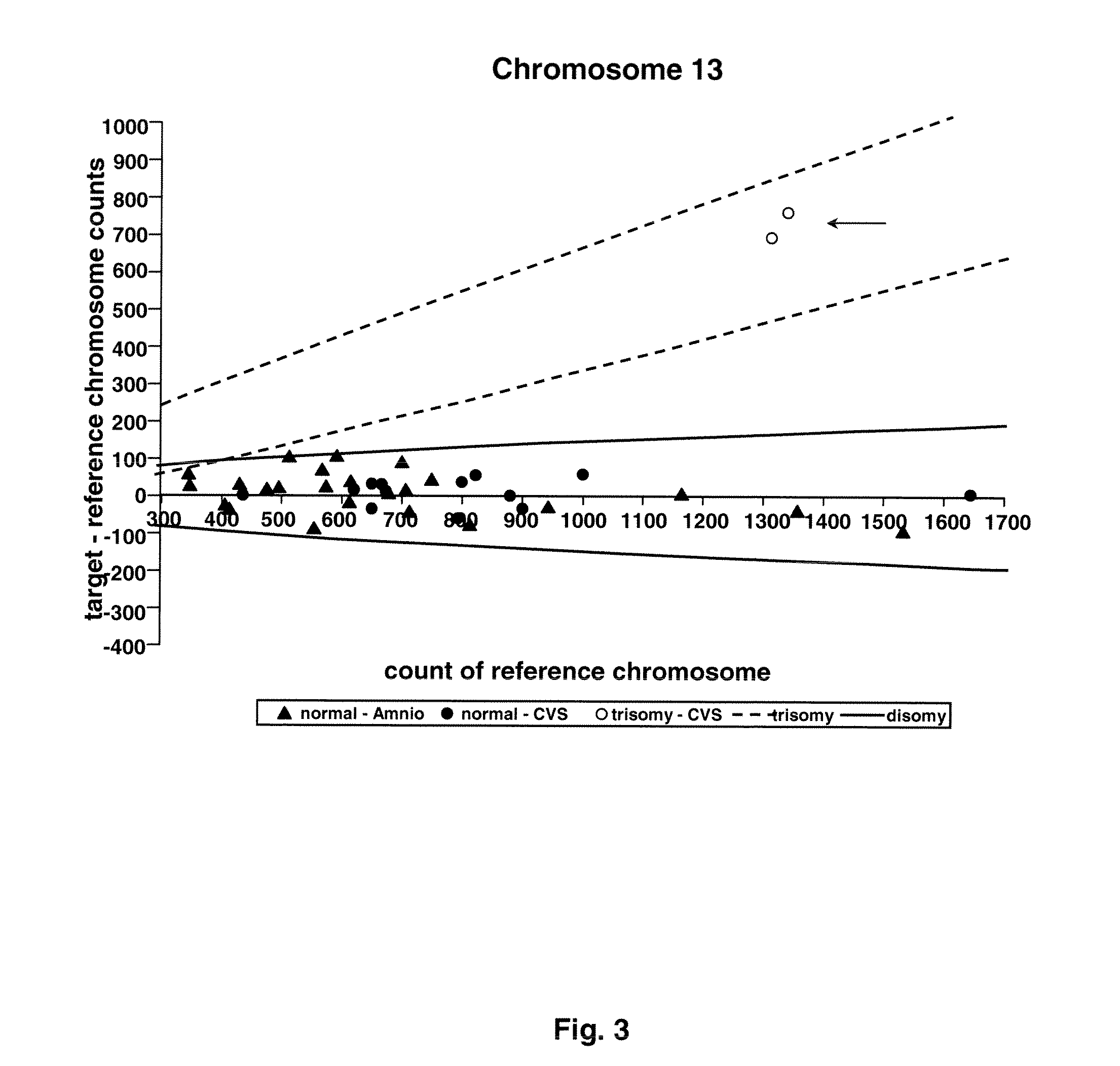
Methods and materials for detection of aneuploidy and other chromosomal abnormalities using fetal tissue are disclosed. Results can be obtained rapidly, without cell culture. The method uses digital PCR for amplification and detection of single target sequences, allowing an accurate count of a specific chromosome or chromosomal region. Specific polynucleic acid primers and probes are disclosed for chromosomes 1, 13, 18, 21, X and Y. These polynucleic acid sequences are chosen to be essentially invariant between individuals, so the test is not dependent on sequence differences between fetus and mother.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Identification of fetal dna and fetal cell markers in maternal plasma or serum
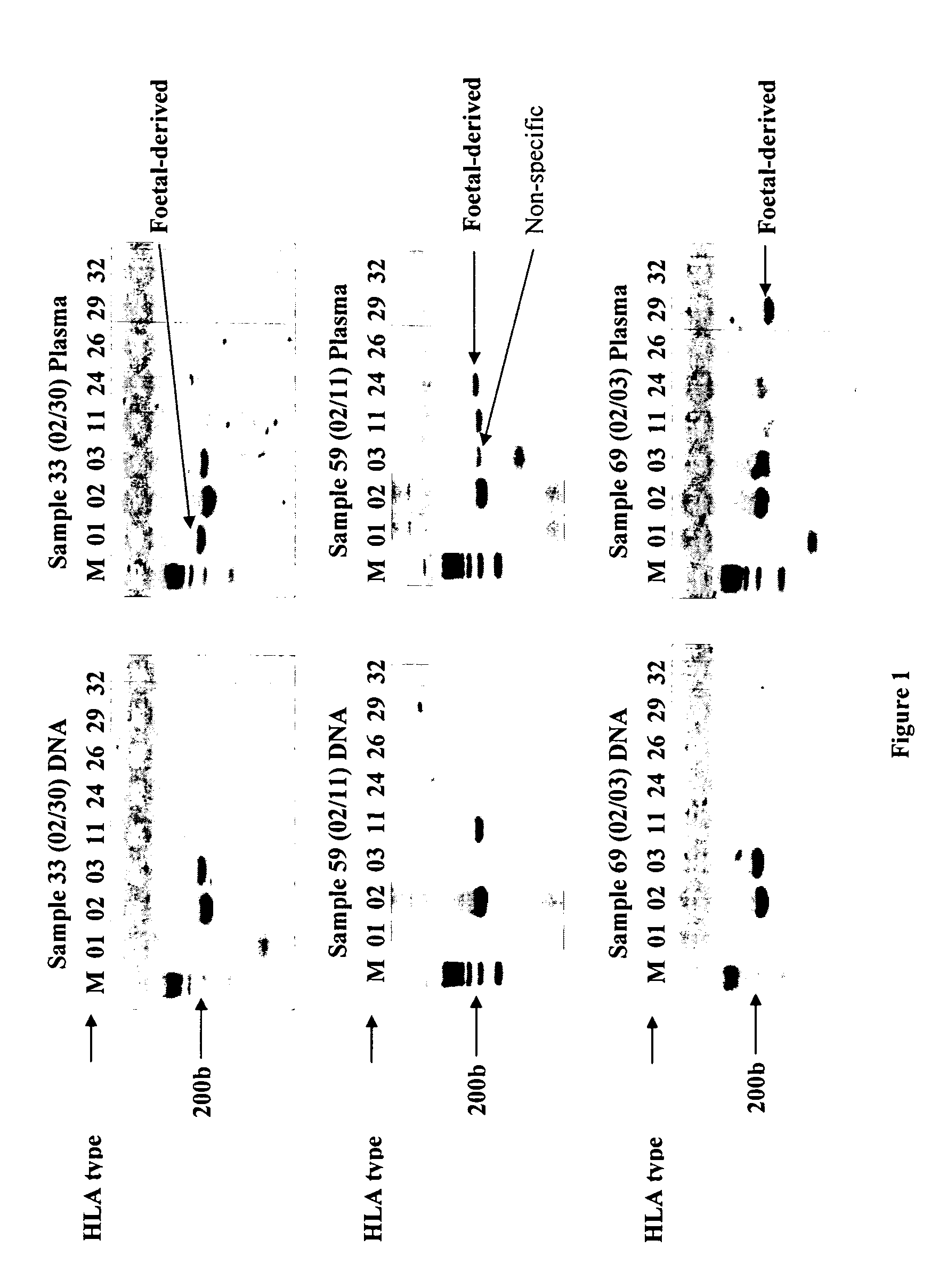
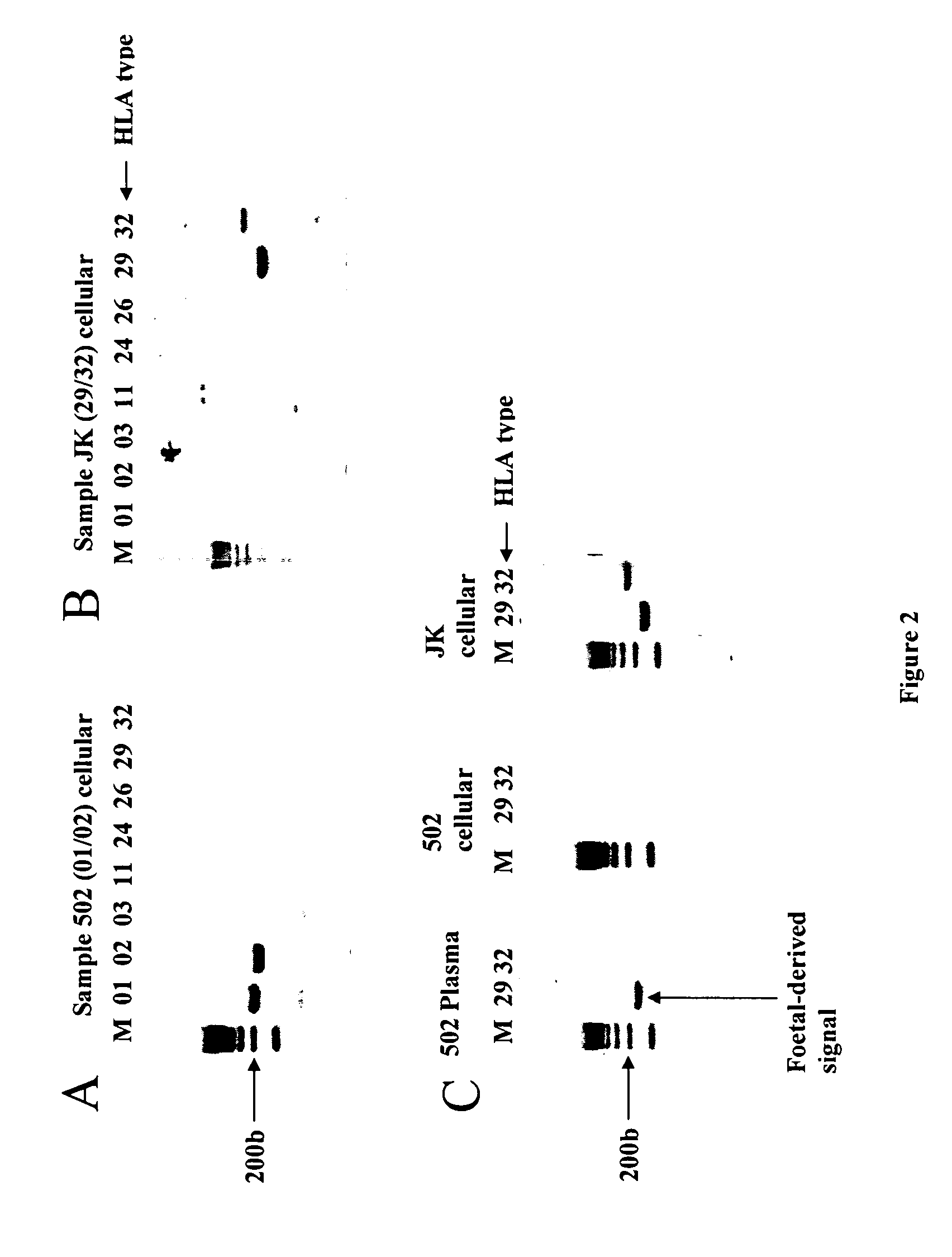
InactiveUS20070134658A1Reduce in quantityMicrobiological testing/measurementTissue cultureAntigenAllele
The present invention relates to the identification of fetal specific nucleic acids and fetal cell markers in maternal plasma or serum. In particular, the present invention relates to methods which rely on the analysis of polymorphic alleles of a population to determine an allele which is possessed by the fetus but absent from the mother. Fetal specific alleles identified using the methods of the invention can be used to quantify fetal DNA from maternal plasma or serum. In addition, antigens encoded by alleles identified using the methods of the invention can be targeted in methods of isolating or detecting fetal cells.
Owner:GENETIC TECHNOLOGIES LIMTIED
Isolation and preservation of fetal and neonatal hematopoietic stem and progenitor cells of the blood
The present invention relates to hematopoietic stem and progenitor cells of neonatal or fetal blood that are cryopreserved, and the therapeutic uses of such stem and progenitor cells upon thawing. In particular, the present invention relates to the therapeutic use of fetal or neonatal stem cells for hematopoietic (or immune) reconstitution. Hematopoietic reconstitution with the cells of the invention can be valuable in the treatment or prevention of various diseases and disorders such as anemias, malignancies, autoimmune disorders, and various immune dysfunctions and deficiencies. In another embodiment, fetal or neonatal hematopoietic stem and progenitor cells which contain a heterologous gene sequence can be used for hematopoietic reconstitution in gene therapy. In a preferred embodiment of the invention, neonatal or fetal blood cells that have been cryopreserved and thawed can be used for autologous (self) reconstitution.
Owner:PHARMASTEM THERAPEUTICS
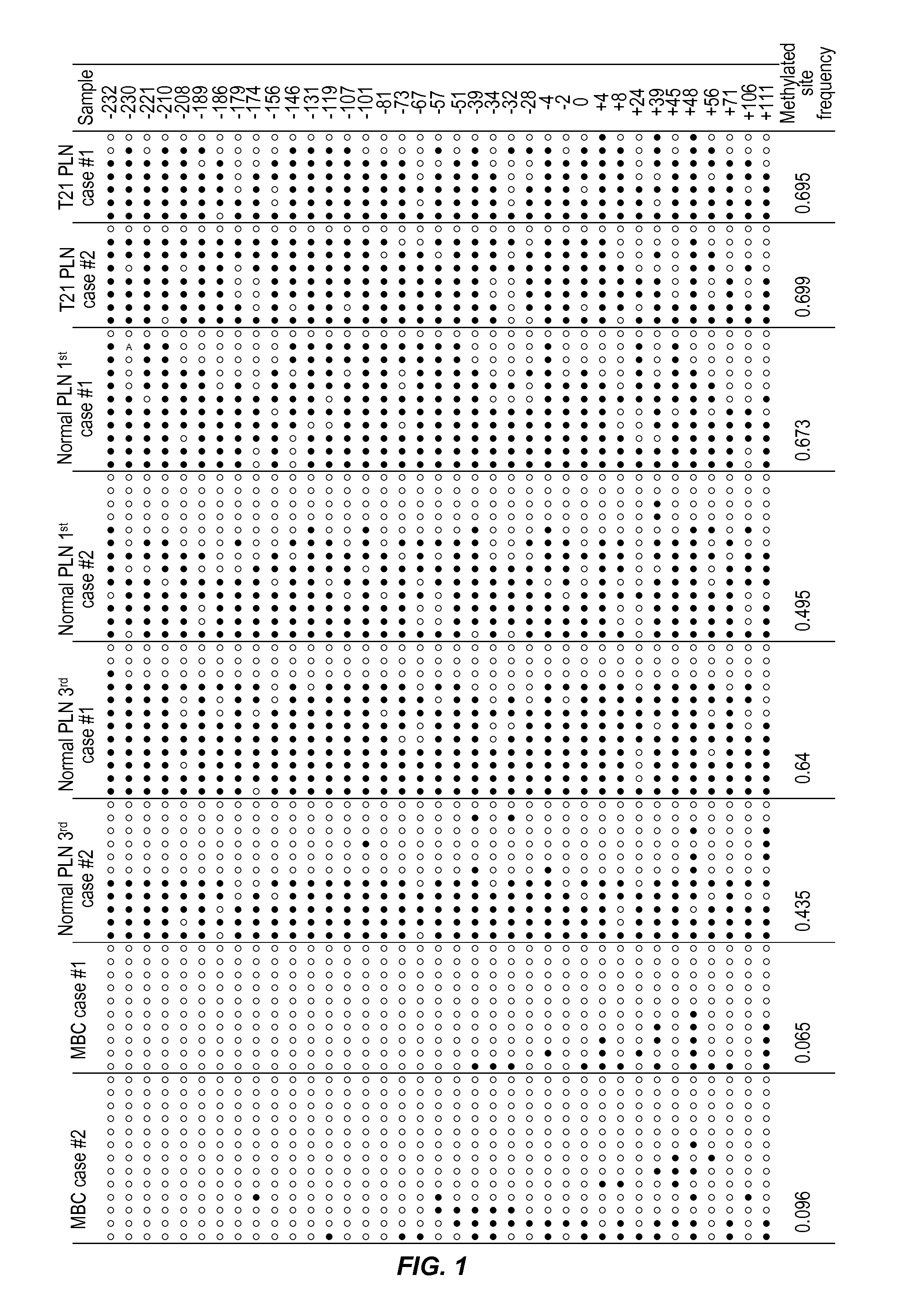
Method for detecting chromosomal aneuploidy
The present invention relates to a new, non-invasive method for detecting chromosomal aneuploidy by analyzing a sample from a pregnant woman. The detection is based on the ratio between the amount of a fetal methylation marker located on a chromosome relevant to the aneuploidy and the amount of a fetal genetic marker located on a reference chromosome, offering improved accuracy.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Risk calculation for evaluation of fetal aneuploidy
The present invention provides processes for determining accurate risk probabilities for fetal aneuploidies. Specifically, the invention provides non-invasive evaluation of genomic variations through chromosome-selective sequencing and non-host fraction data analysis of maternal samples.
Owner:ROCHE MOLECULAR SYST INC
Method for determining the number of copies of a chromosome in the genome of a target individual using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell is reconstructed using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In accordance with another embodiment of the invention, incomplete genetic data from a fetus is acquired from fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In one embodiment, the genetic data can be reconstructed for the purposes of making phenotypic predictions. In another embodiment, the genetic data can be used to detect for aneuploides and uniparental disomy.
Owner:NATERA
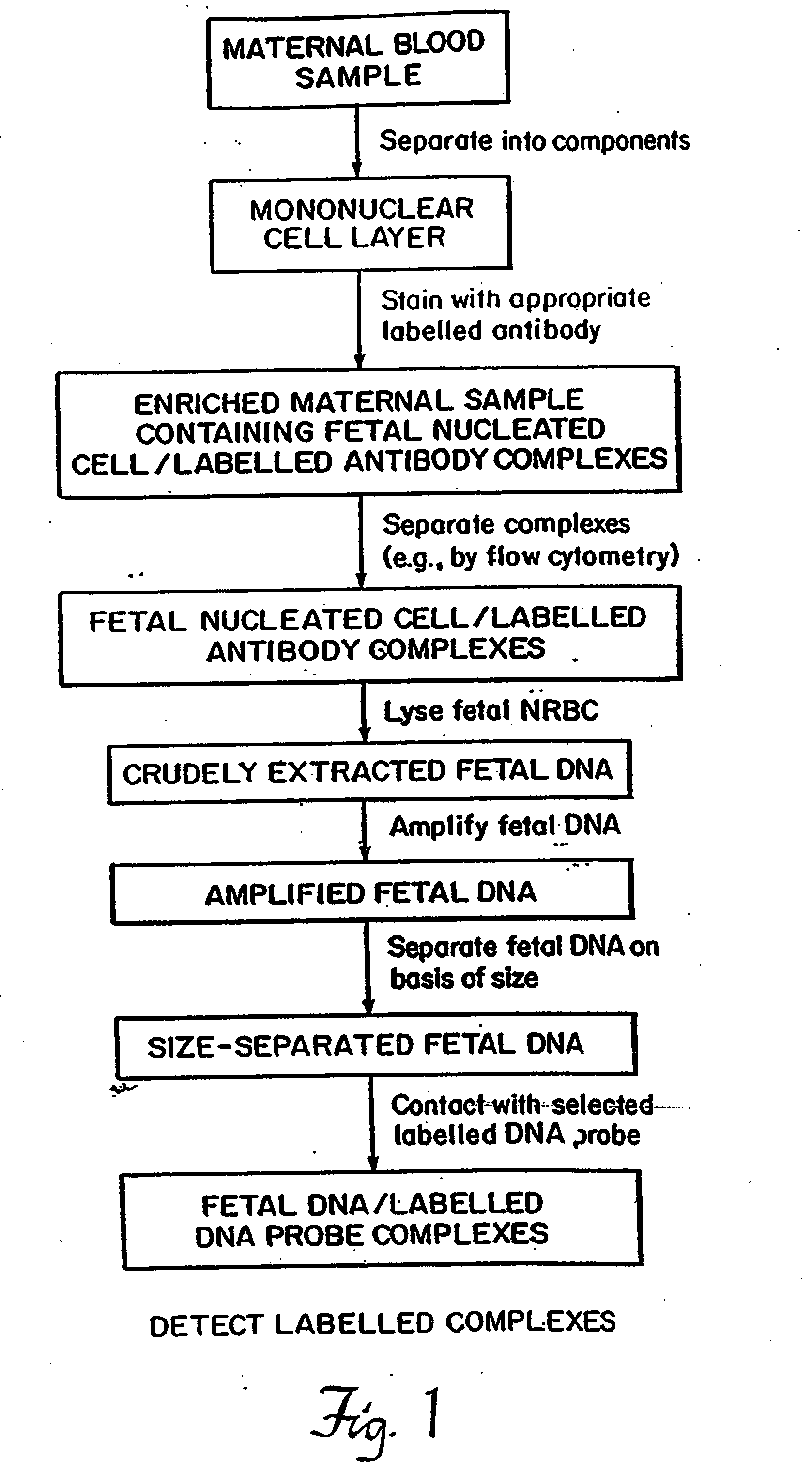
Non-invasive method for isolation and detection of fetal DNA
A method of detecting the presence or absence of the fetal DNA sequence of interest in fetal DNA derived from a sample of peripheral blood obtained from a pregnant woman is described. The method involves obtaining a sample peripheral blood from a pregnant woman, treating the sample of peripheral blood such that the fetal DNA present in the fetal nucleated cells is made available for detection and detecting the presence or absence of the fetal DNA sequence of interest in the available fetal DNA. The proportion of fetal nucleated cells present in the sample of peripheral blood can be increased forming a sample enriched in fetal nucleated cells prior to the detection step. The fetal DNA sequence of interest can be detected by treating the peripheral blood sample such that fetal DNA present in the sample is made available for hybridization with a DNA probe and subsequently contacting the available fetal DNA with a DNA probe hybridizable to fetal DNA of interest under hybridization conditions. The presence or absence of hybridization between the DNA probe and the fetal DNA of interest is detected as an indication of the presence or absence of the fetal DNA of interest.
Owner:CHILDRENS HOSPITAL
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available, are disclosed. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data is acquired from embryonic cells, fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals.
Owner:NATERA
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available, are disclosed. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention incomplete genetic data is acquired from embryonic cells, fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals.
Owner:NATERA
Cloning pigs using donor nuclei from non-quiescent differentiated cells
InactiveUS6235969B1Superior genotypes of pigsSpeed up genetic progressNervous disorderPeptide/protein ingredientsBiotechnologyPresent method
An improved method of nuclear transfer involving the transplantation of donor differentiated pig cell nuclei into enucleated pig oocytes is provided. The resultant nuclear transfer units are useful for multiplication of genotypes and transgenic genotypes by the production of fetuses and offspring. Production of genetically engineered or transgenic pig embryos, fetuses and offspring is facilitated by the present method since the differentiated cell source of the donor nuclei can be genetically modified and clonally propagated.
Owner:MASSACHUSETTS UNIV OF
Apparatus and method for distributed ultrasound diagnostics
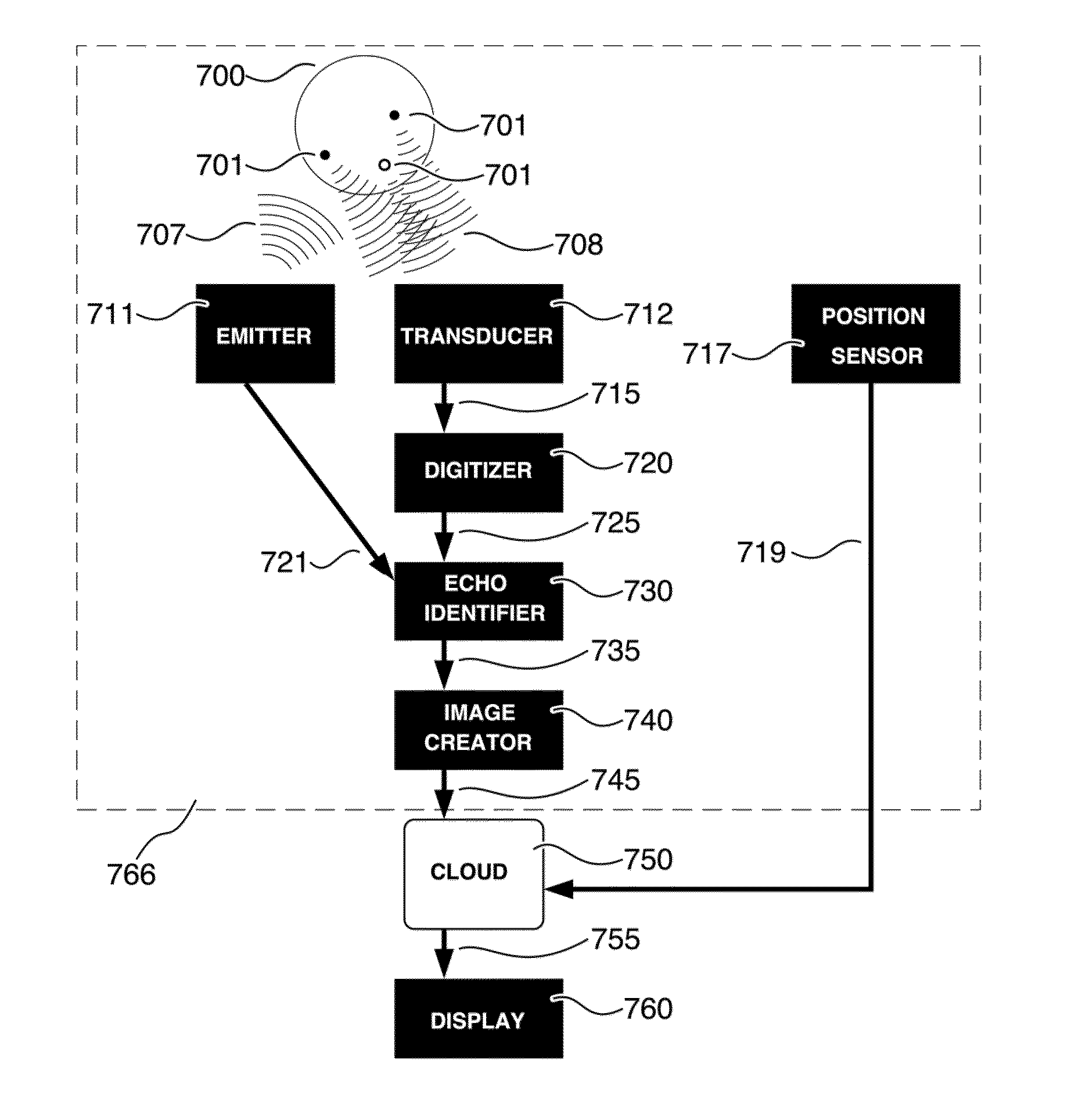
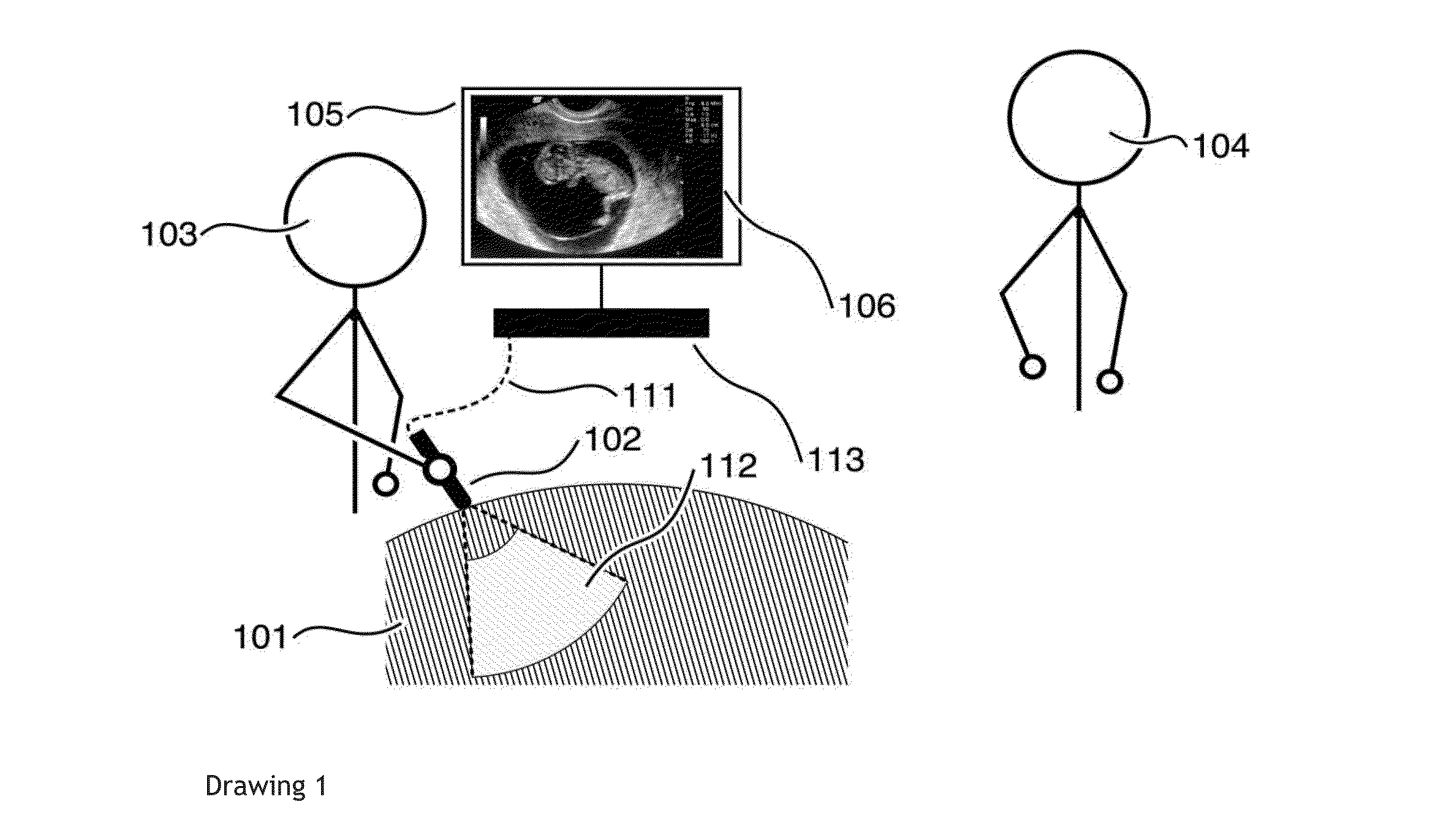
ActiveUS20150173715A1Correction of artifactSolve the lack of densityImage analysisOrgan movement/changes detectionSonificationImaging quality
A local user obtains data on the response of internal tissues of a subject to a non-invasive imaging system, choosing sensor positions according to a geometric display. The data obtained are evaluated with respect to predefined quantitative values. The process of obtaining and processing the internal tissue response data is repeated until an image meeting predetermined image quality characteristics is obtained. Specific or general content of the image may be restricted by the local processor, with a distal processor receiving the obtained image data, so as to limit specific or general types of image information from being viewed by the local user, such as image data that can be used to identify the sex of a fetus within the subject.
Owner:RAGHAVAN RAGHU +1
Risk calculation for evaluation of fetal aneuploidy
InactiveUS20120190557A1Microbiological testing/measurementLibrary member identificationNon invasiveFetal aneuploidy
The present invention provides processes for determining accurate risk probabilities for chromosome dosage abnormalities. Specifically, the invention provides non-invasive evaluation of genomic variations through chromosome-selective sequencing and non-host fraction data analysis of maternal samples.
Owner:ROCHE MOLECULAR SYST INC
Methods and compositions for determining ploidy
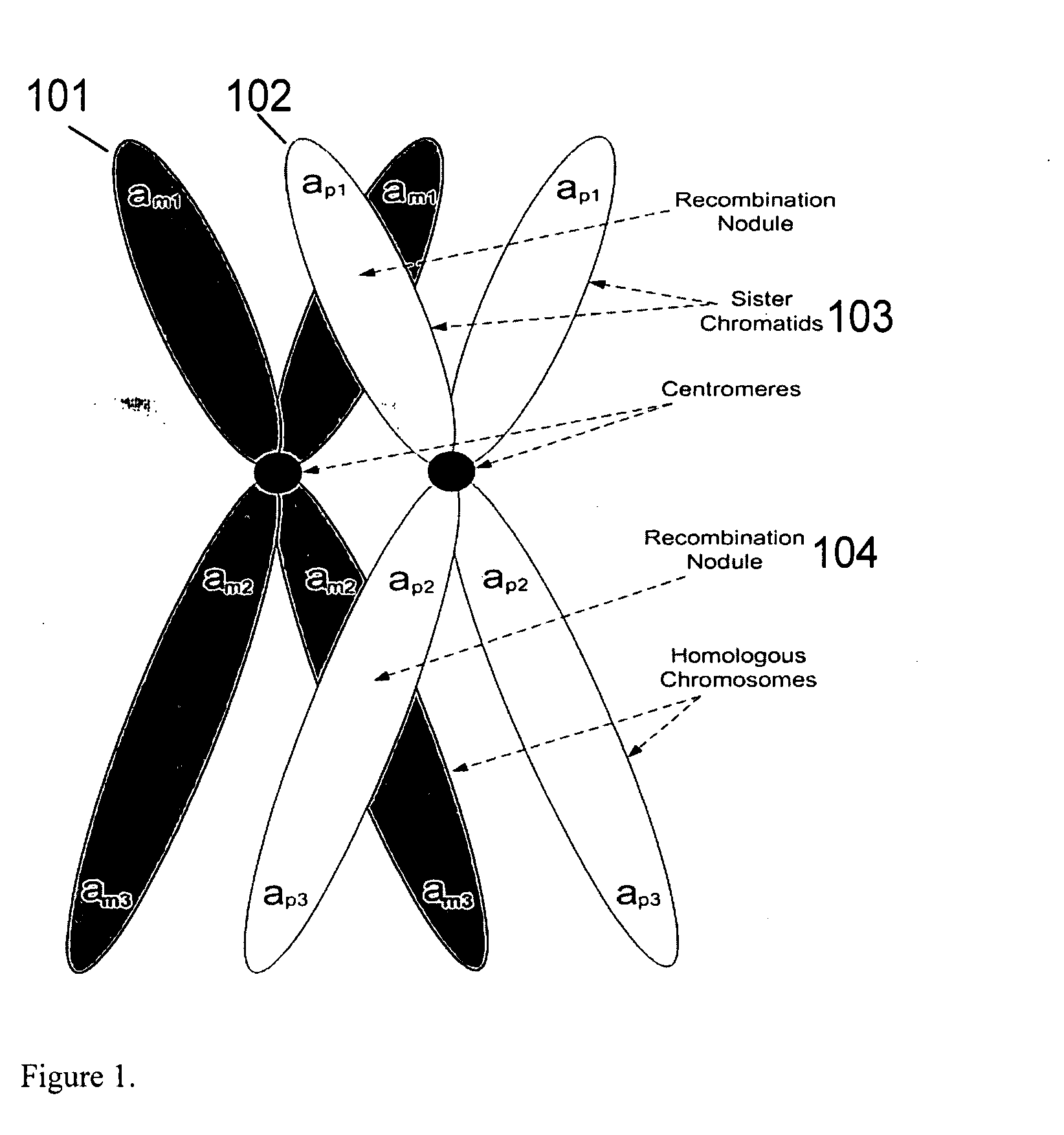
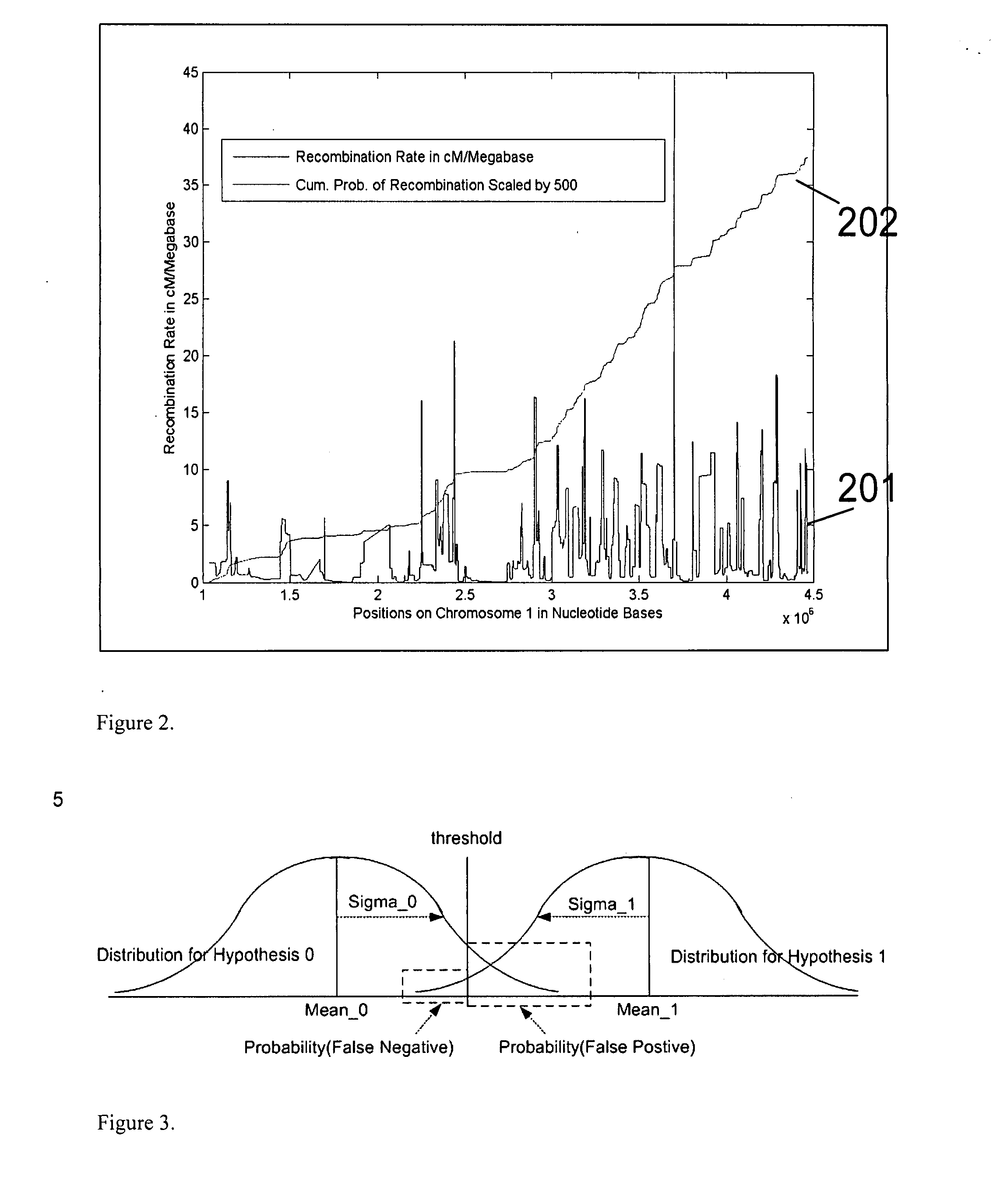
The invention provides improved methods, compositions, and kits for detecting ploidy of chromosome regions, e.g. for detecting cancer or a chromosomal abnormality in a gestating fetus. The methods can utilize a set of more than 200 SNPs that are found within haploblocks and can include analyzing a series of target chromosomal regions related to cancer or a chromosomal abnormality in a gestating fetus. Finally the method may use knowledge about chromosome crossover locations or a best fit algorithm for the analysis. The compositions may comprise more than 200 primers located within haplotype blocks known to show CNV.
Owner:NATERA
Methods for delivering a drug to a patient while preventing the exposure of a foetus or other contraindicated individual to the drug
InactiveUS6908432B2Avoiding terrible birth defectAdvantageous and effectiveDrug and medicationsSurgeryPharmacyMale patient
Novel methods for delivering a drug to a patient while preventing the exposure of a foetus or other contraindicated individual to the drug. Embodiments are provided in which the involved prescribers, pharmacies and patients are registered in one or more computer databases. Embodiments are also provided in which registered patients receive counseling information concerning the risks attendant to foetal exposure to the drug. Male patients and female patients who are not pregnant may, in certain circumstances, receive the drug.
Owner:CELGENE CORP
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com