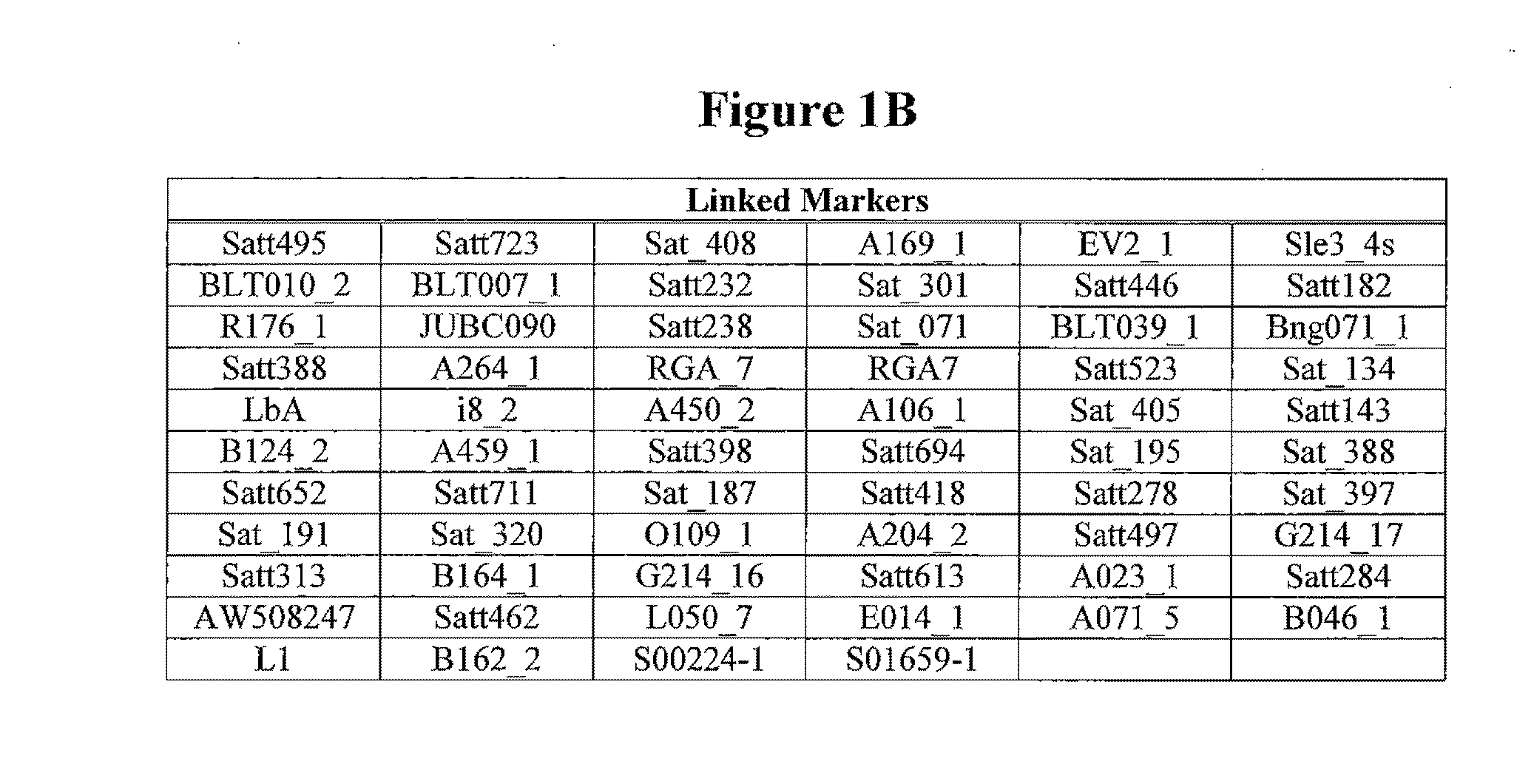
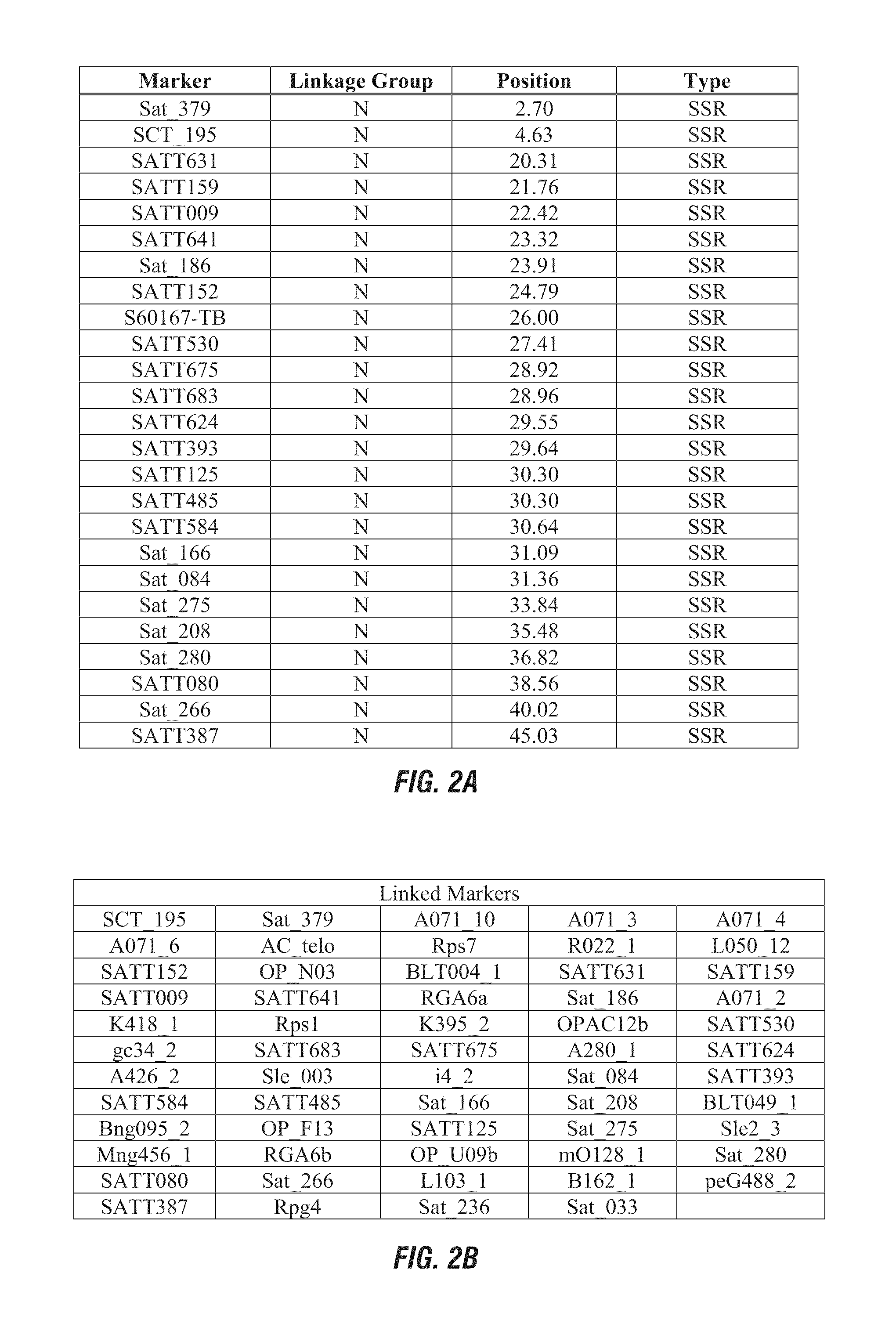
Patents
Literature
115 results about "Chromosomal region" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
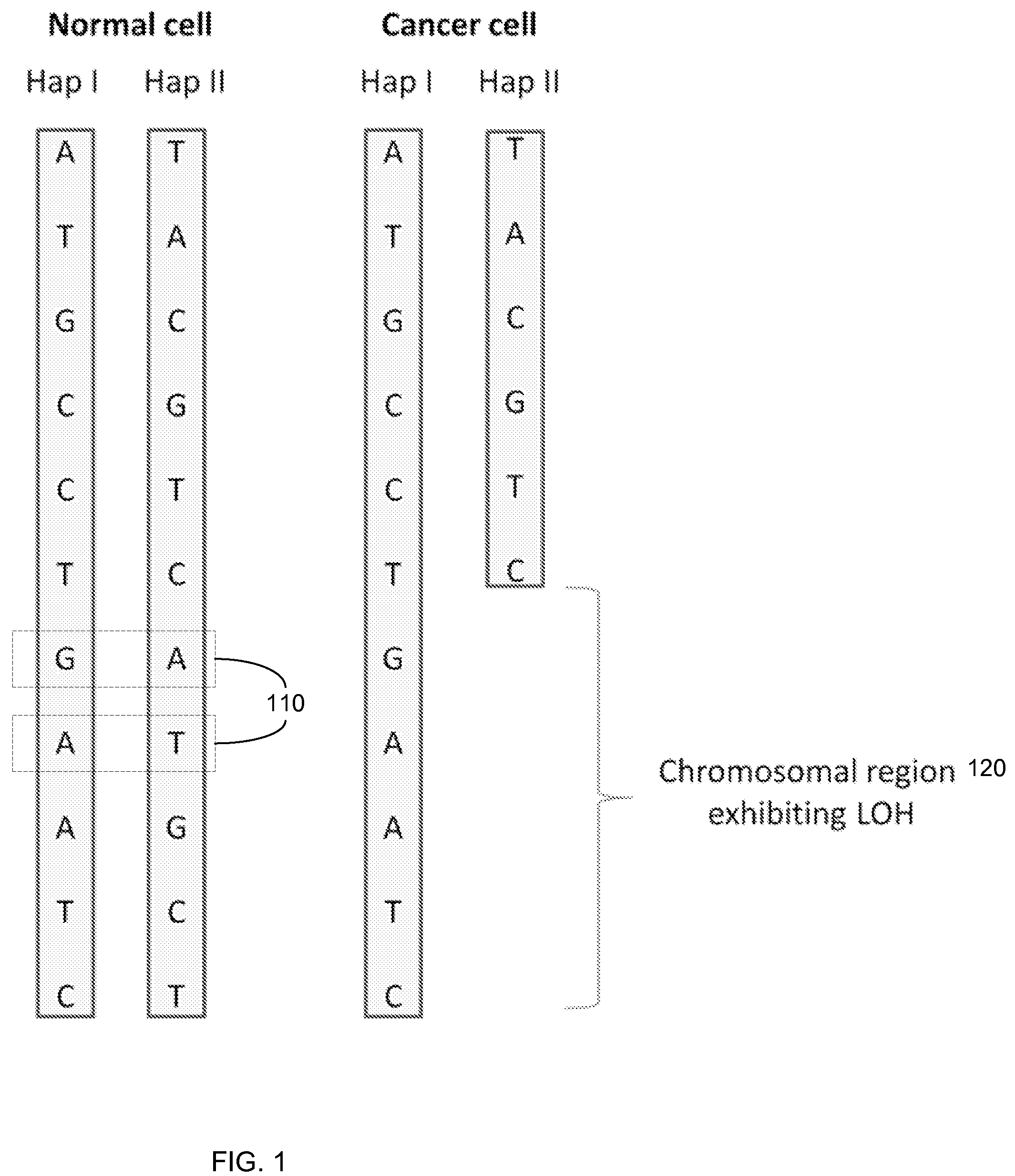
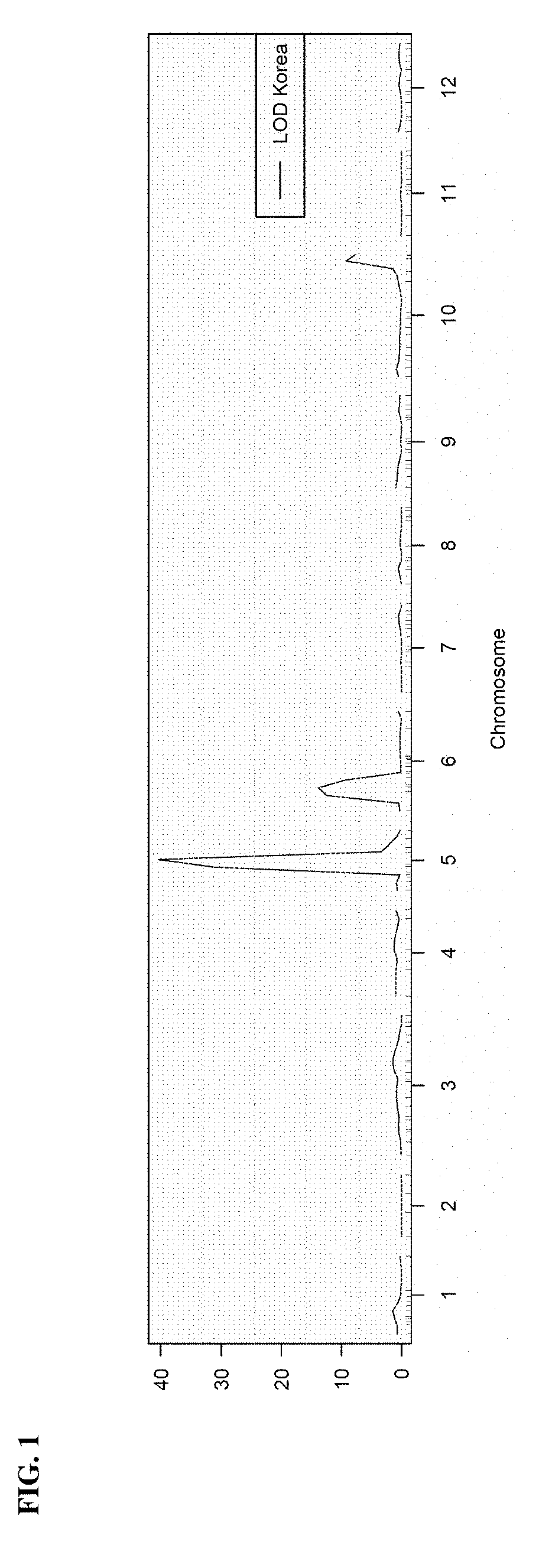
Any subdivision of a chromosome along its length. [GOC:dos]
Systems and methods to detect copy number variation
InactiveUS20120046877A1Correction biasMicrobiological testing/measurementProteomicsData fileWorkstation
In one aspect, a system for implementing a copy number variation analysis method, is disclosed. The system can include a nucleic acid sequencer and a computing device in communications with the nucleic acid sequencer. The nucleic acid sequencer can be configured to interrogate a sample to produce a nucleic acid sequence data file containing a plurality of nucleic acid sequence reads. In various embodiments, the computing device can be a workstation, mainframe computer, personal computer, mobile device, etc.The computing device can comprise a sequencing mapping engine, a coverage normalization engine, a segmentation engine and a copy number variation identification engine. The sequence mapping engine can be configured to align the plurality of nucleic acid sequence reads to a reference sequence, wherein the aligned nucleic acid sequence reads merge to form a plurality of chromosomal regions. The coverage normalization engine can be configured to divide each chromosomal region into one or more non-overlapping window regions, determine nucleic acid sequence read coverage for each window region and normalize the nucleic acid sequence read coverage determined for each window region to correct for bias. The segmentation engine can be configured to convert the normalized nucleic acid sequence read coverage for each window region to discrete copy number states. The copy number variation identification engine can be configured to identify copy number variation in the chromosomal regions by utilizing the copy number states of each window region.
Owner:LIFE TECH CORP
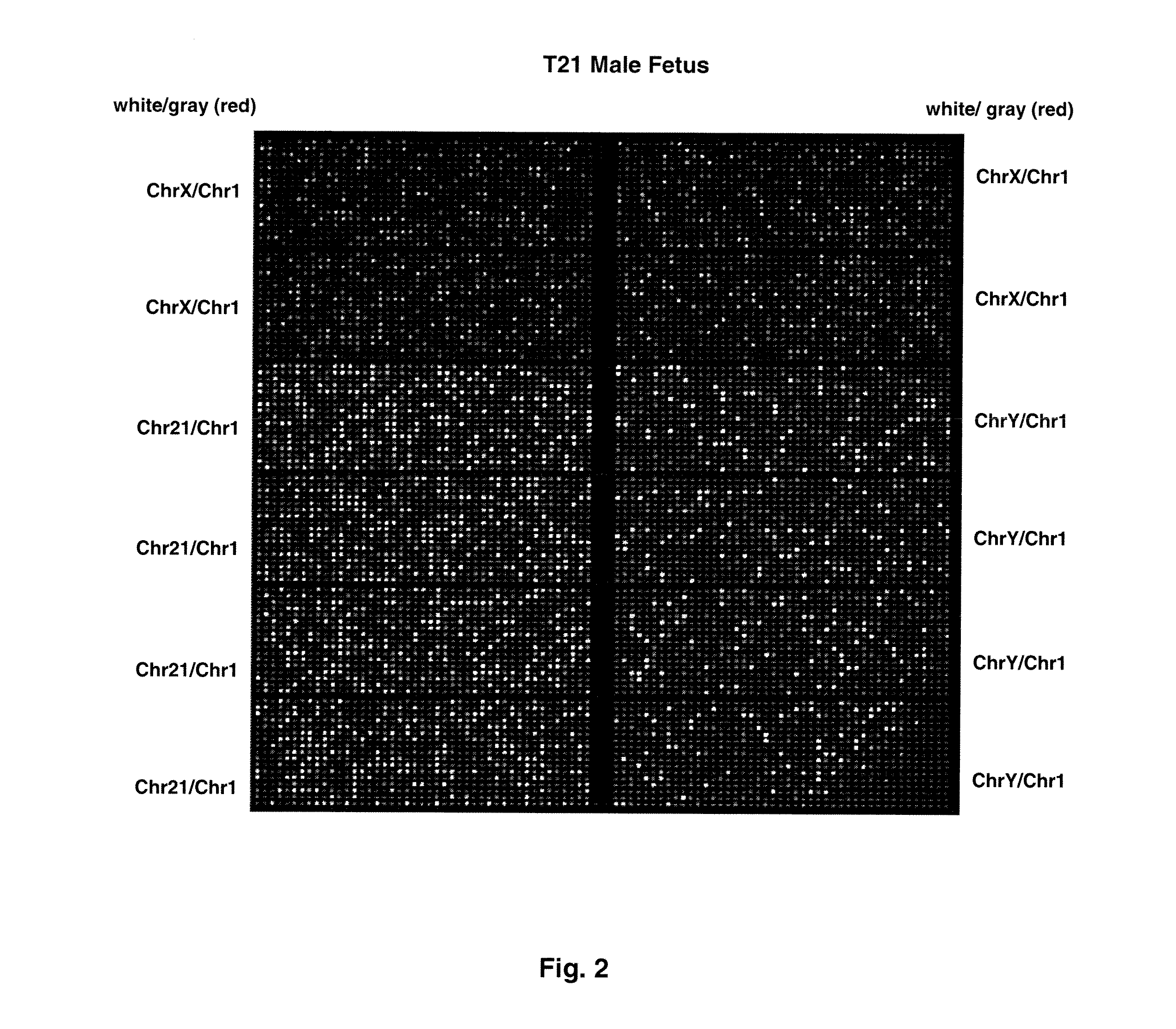
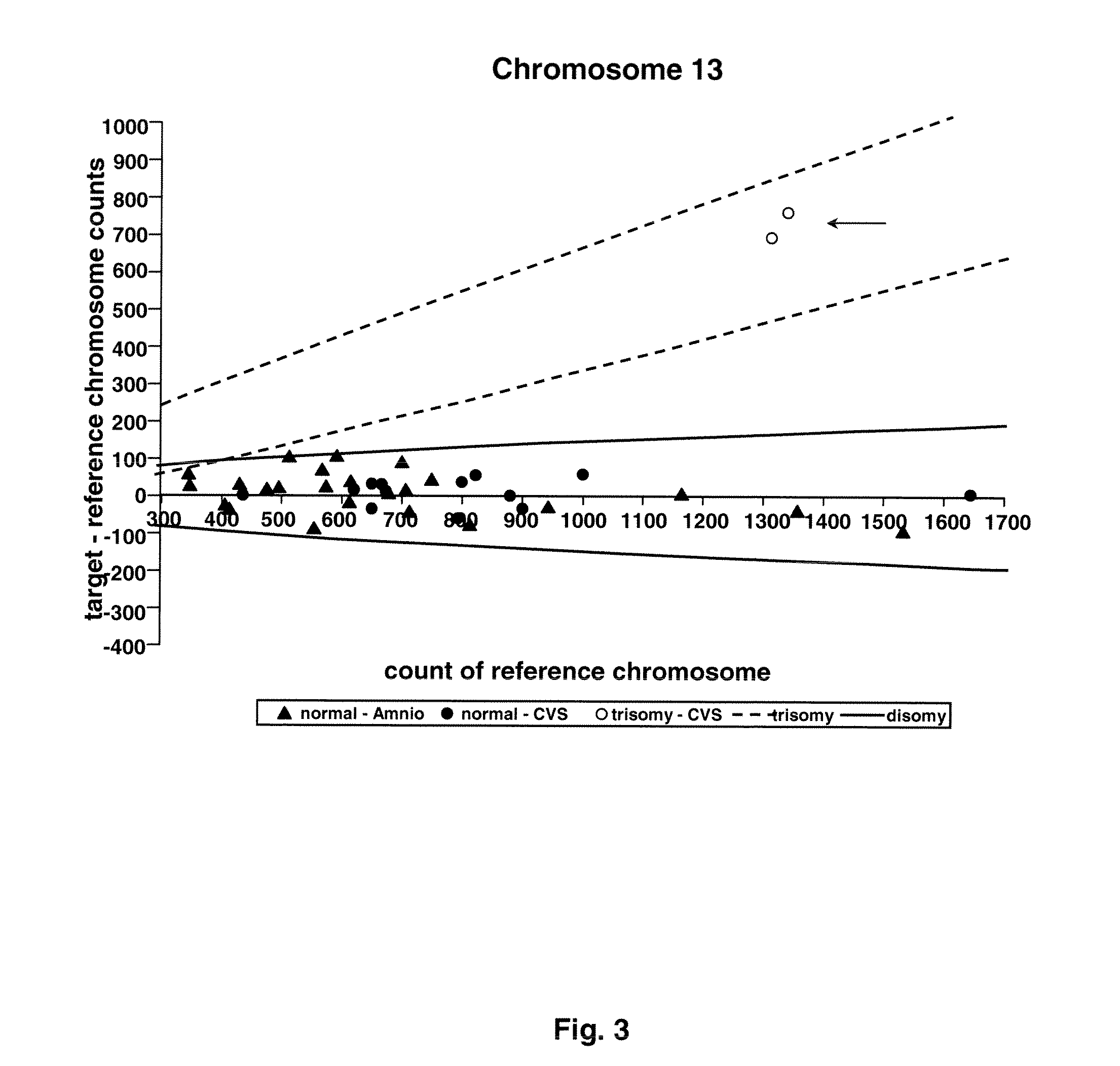
Direct Molecular Diagnosis of Fetal Aneuploidy
ActiveUS20110151442A1Addressing slow performanceSugar derivativesMicrobiological testing/measurementSpecific chromosomeFetal tissues
Methods and materials for detection of aneuploidy and other chromosomal abnormalities using fetal tissue are disclosed. Results can be obtained rapidly, without cell culture. The method uses digital PCR for amplification and detection of single target sequences, allowing an accurate count of a specific chromosome or chromosomal region. Specific polynucleic acid primers and probes are disclosed for chromosomes 1, 13, 18, 21, X and Y. These polynucleic acid sequences are chosen to be essentially invariant between individuals, so the test is not dependent on sequence differences between fetus and mother.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
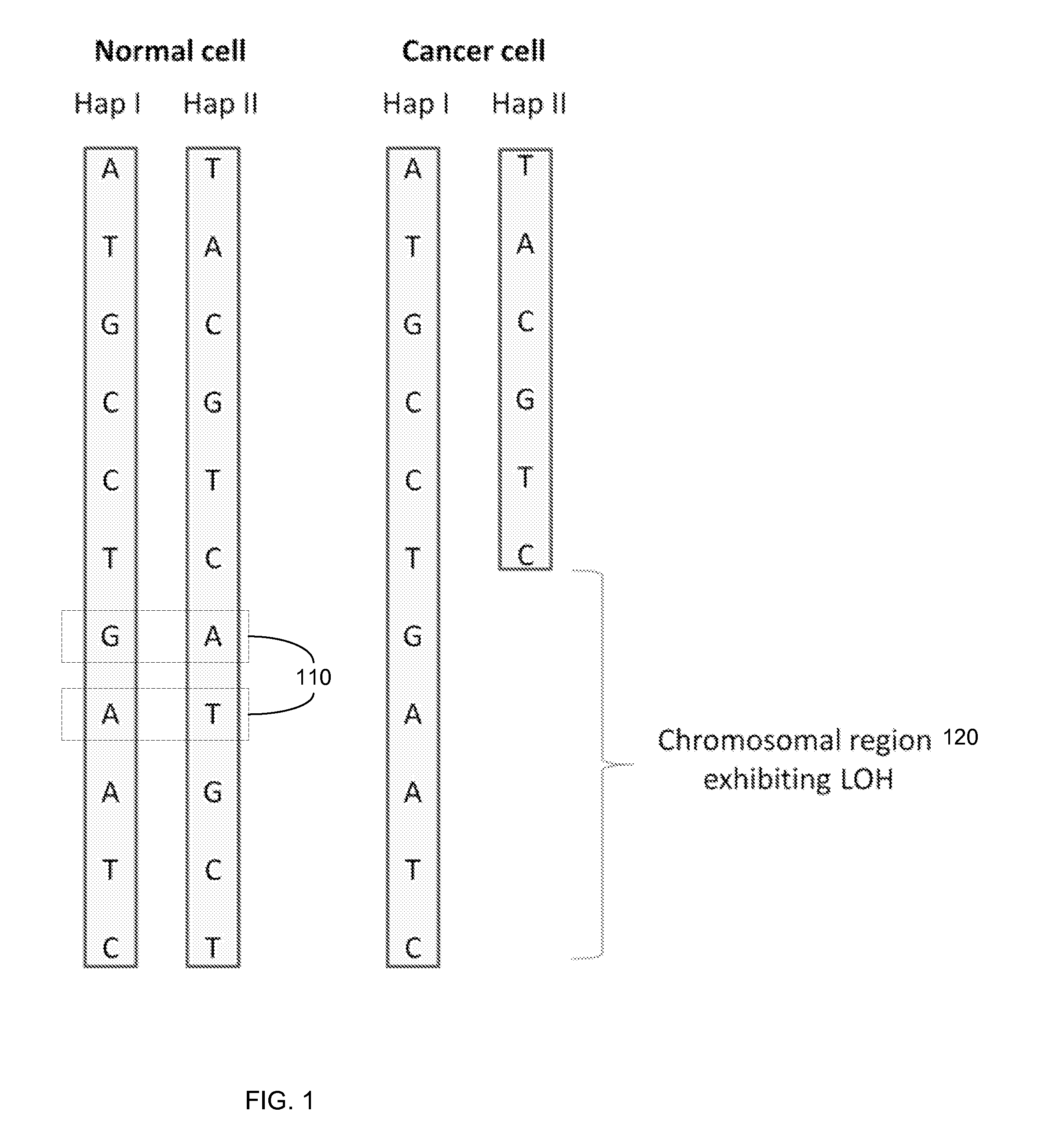
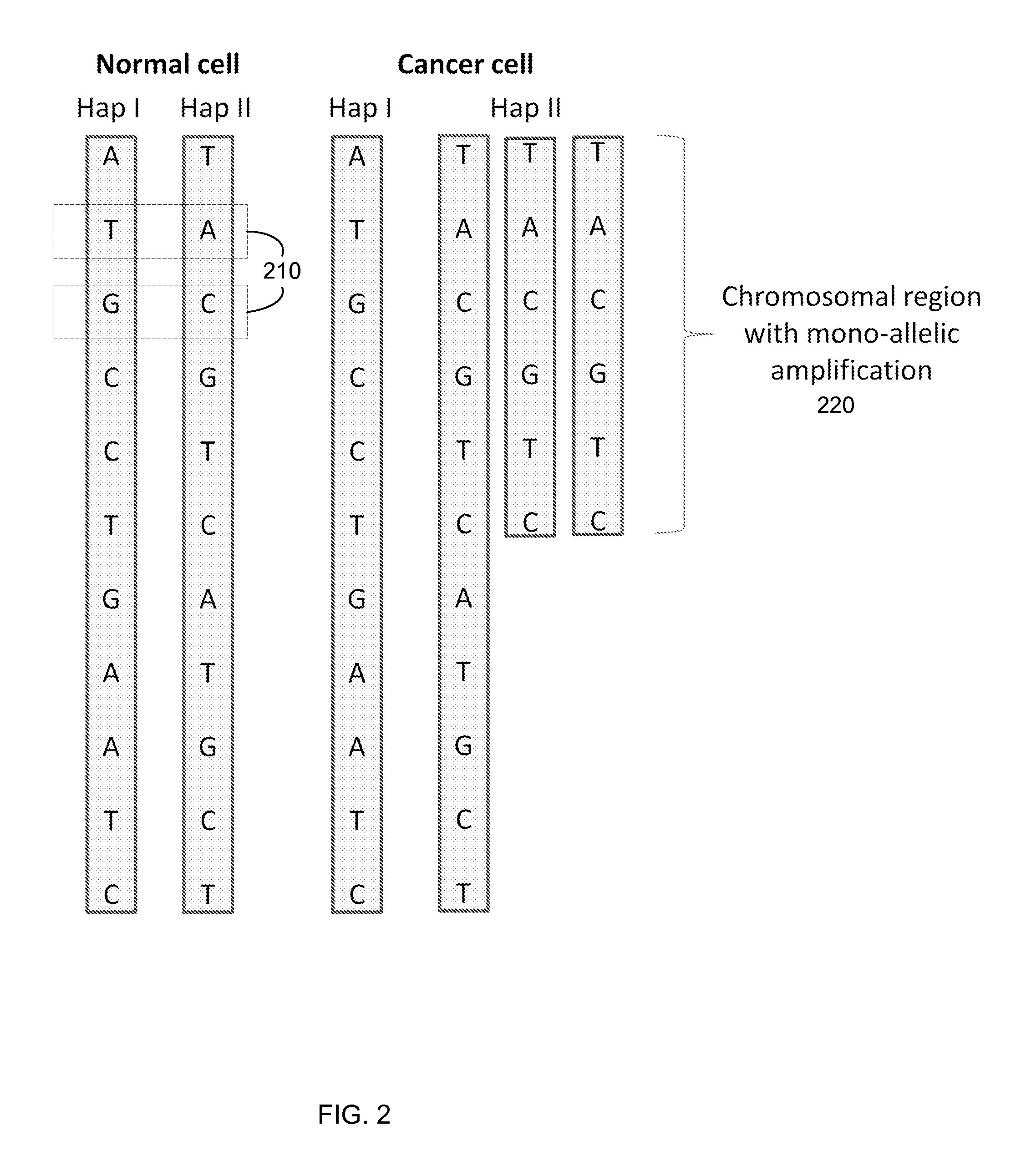
Detection of genetic or molecular aberrations associated with cancer
ActiveUS20130040824A1Improve accuracyImprove efficiencyMicrobiological testing/measurementLibrary screeningAbnormal tissue growthAfter treatment
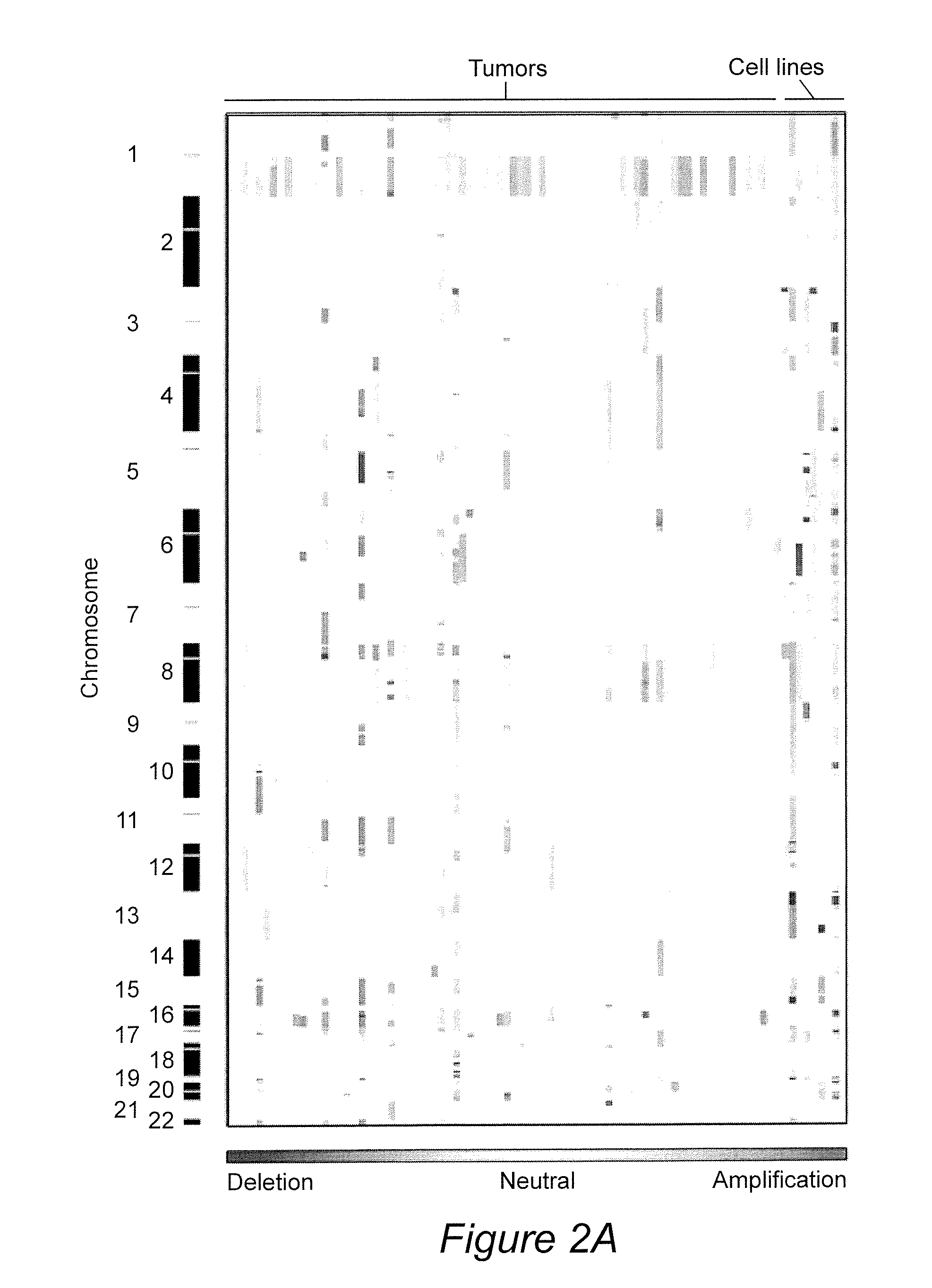
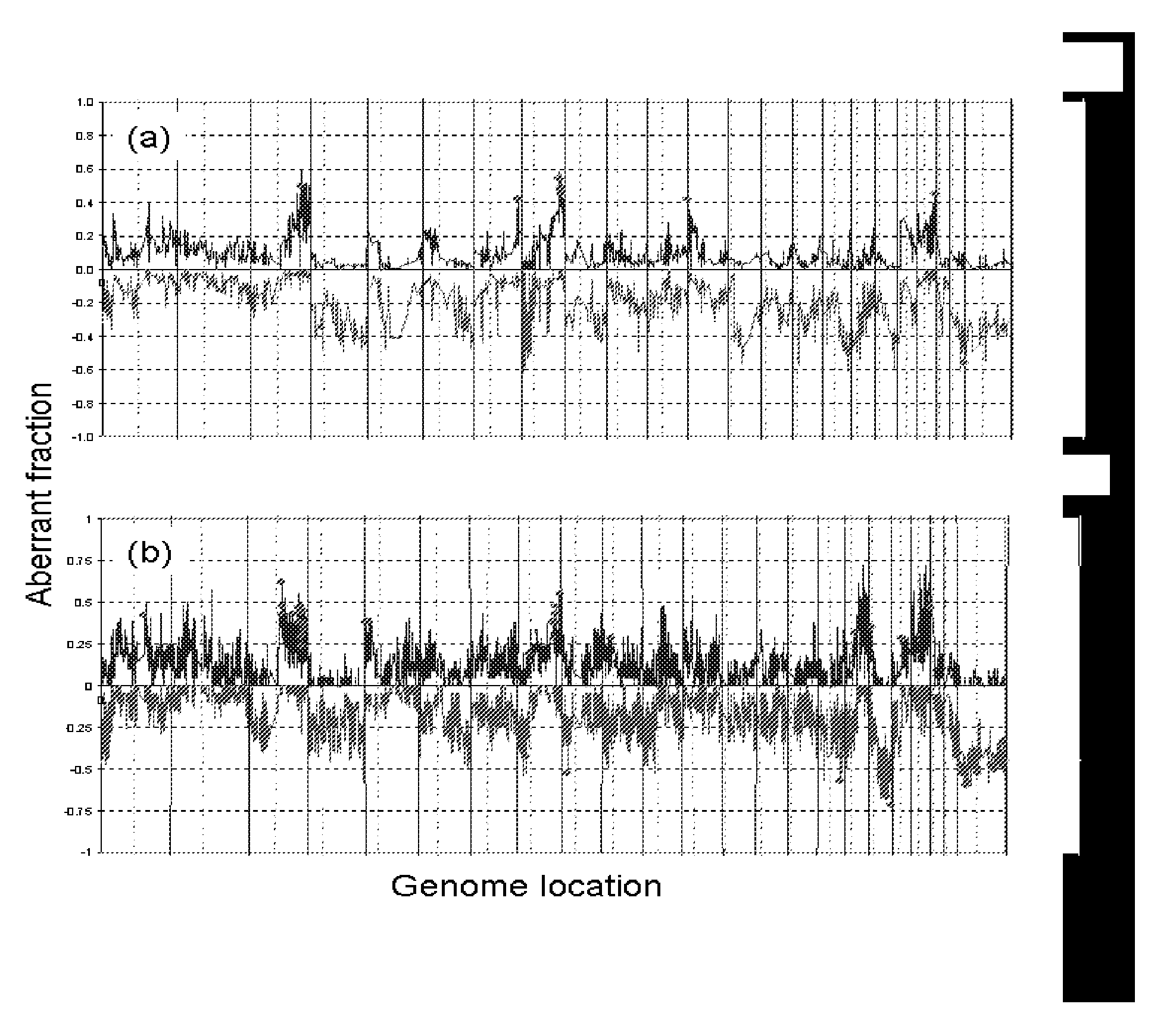
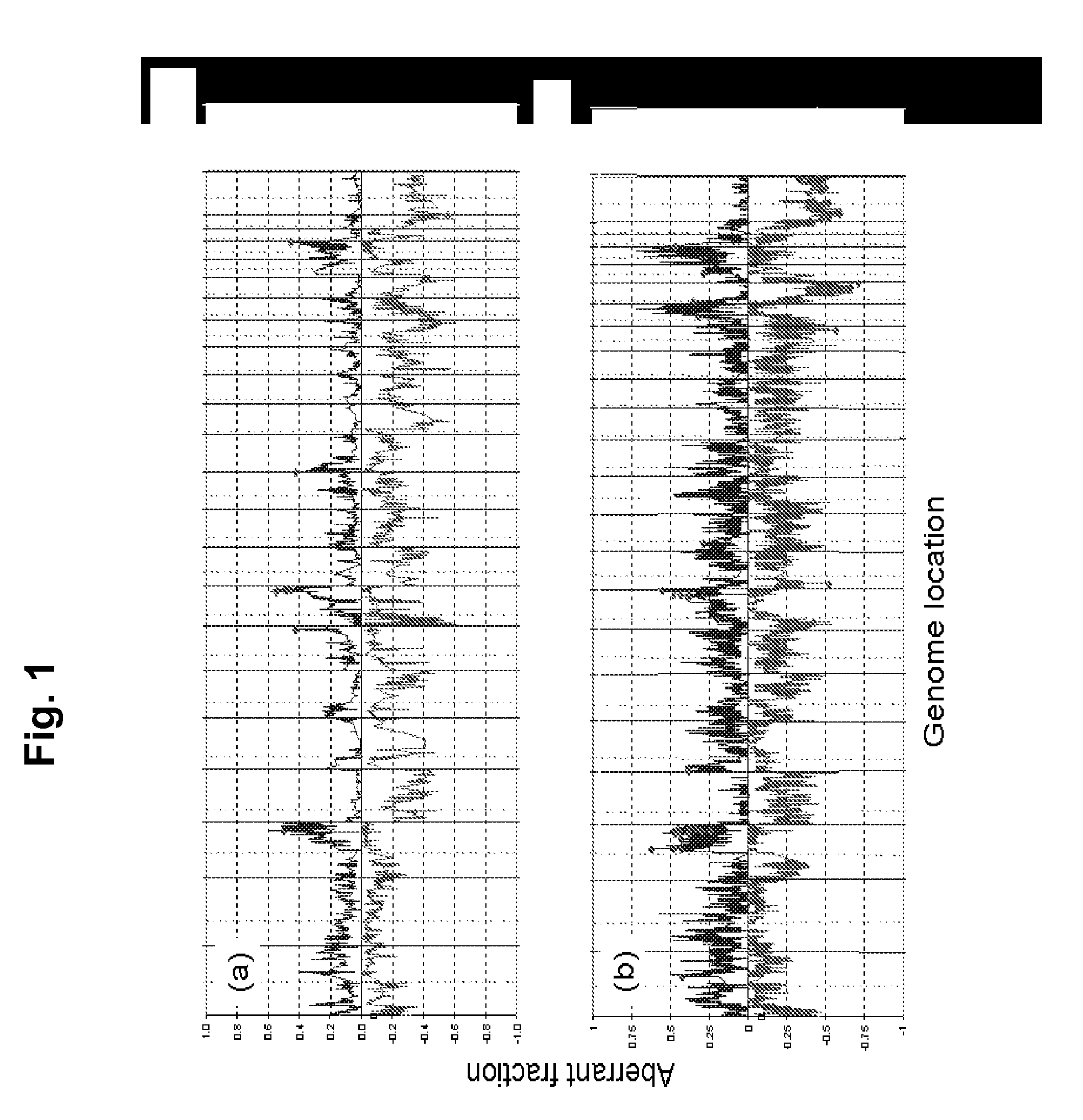
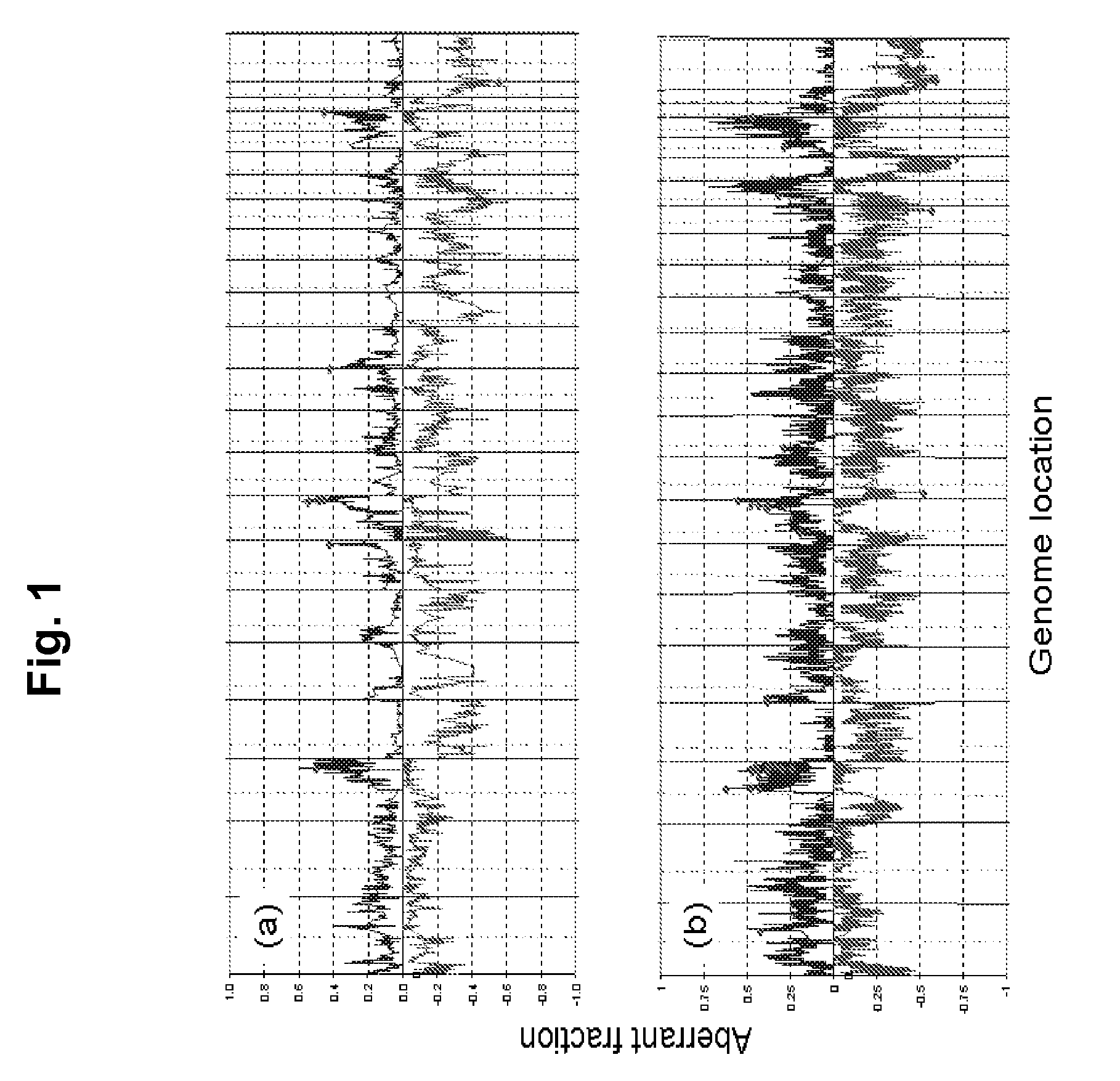
Biological samples including cell-free DNA fragments are analyzed to identify imbalances in chromosomal regions, e.g., due to deletions and / or amplifications in a tumor. Multiple loci are used for each chromosomal region. Such imbalances can then be used to diagnose (screen) a patient for cancer, as well as prognosticate a patient with cancer, or to detect the presence or to monitor the progress of a premalignant condition in a patient. The severity of an imbalance as well as the number of regions exhibiting an imbalance can be used. A systematic analysis of non-overlapping segments of a genome can provide a general screening tool for a sample. Additionally, a patient can be tested over time to track severity of each of one or more chromosomal regions and a number of chromosomal regions to enable screening and prognosticating, as well as monitoring of progress (e.g. after treatment).
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Pre-Implantation Genetic Diagnosis Test
InactiveUS20080182244A1Microbiological testing/measurementBiological testingChromosomal regionGenetic diagnosis
A method for determining viable normal blastomeres for implantation entailing labeling the blastomere with an antibody to hyperglycoslyated hCG and determining the binding of chromosomal probes directed to chromosomal regions of the chromosome.
Owner:IKONISYS INC
Methods and compositions for determining ploidy
The invention provides improved methods, compositions, and kits for detecting ploidy of chromosome regions, e.g. for detecting cancer or a chromosomal abnormality in a gestating fetus. The methods can utilize a set of more than 200 SNPs that are found within haploblocks and can include analyzing a series of target chromosomal regions related to cancer or a chromosomal abnormality in a gestating fetus. Finally the method may use knowledge about chromosome crossover locations or a best fit algorithm for the analysis. The compositions may comprise more than 200 primers located within haplotype blocks known to show CNV.
Owner:NATERA
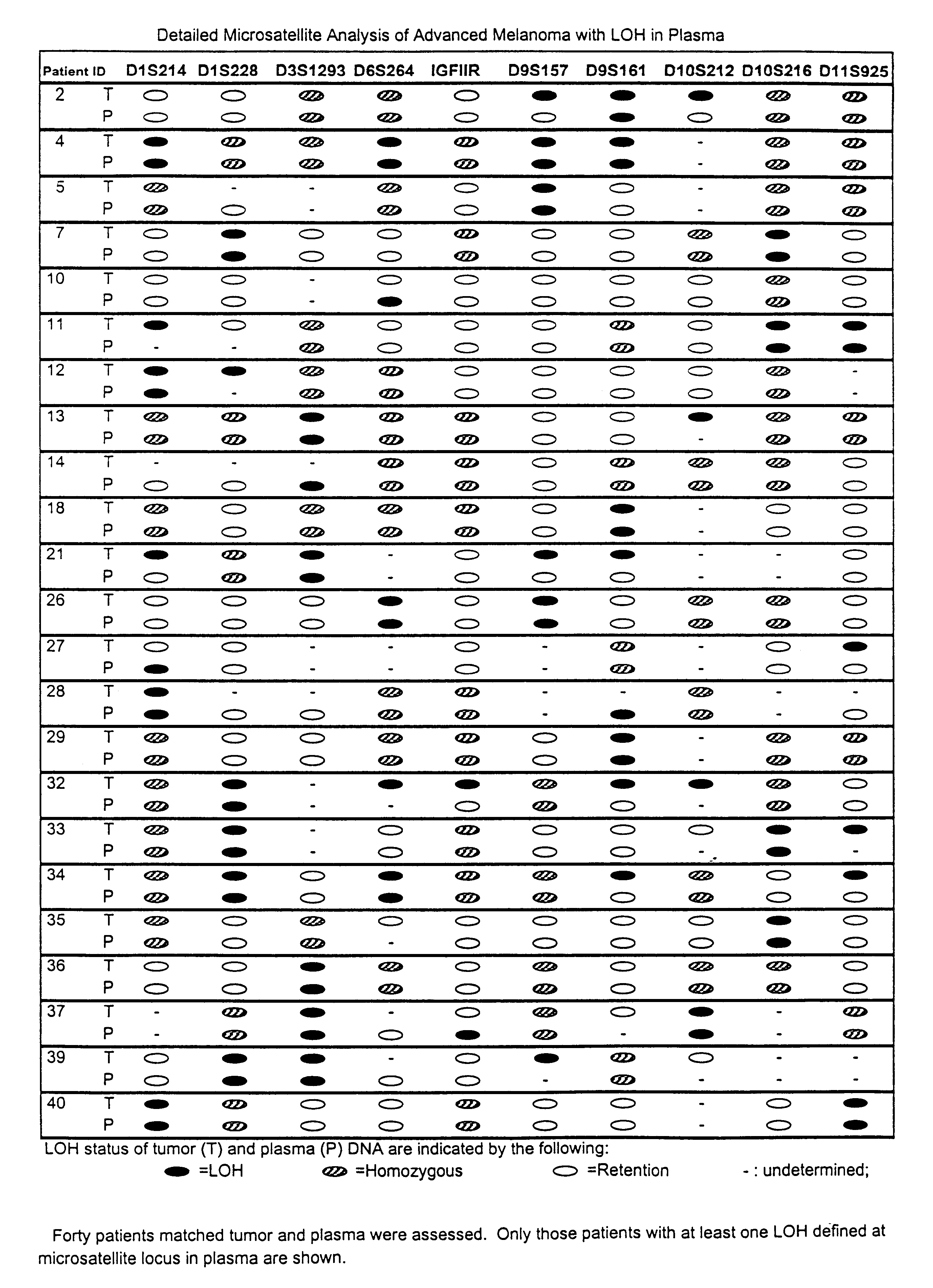
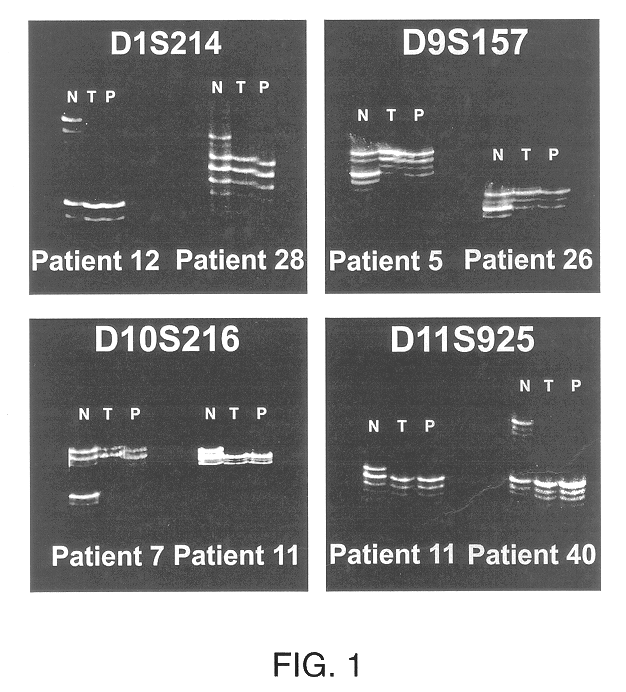
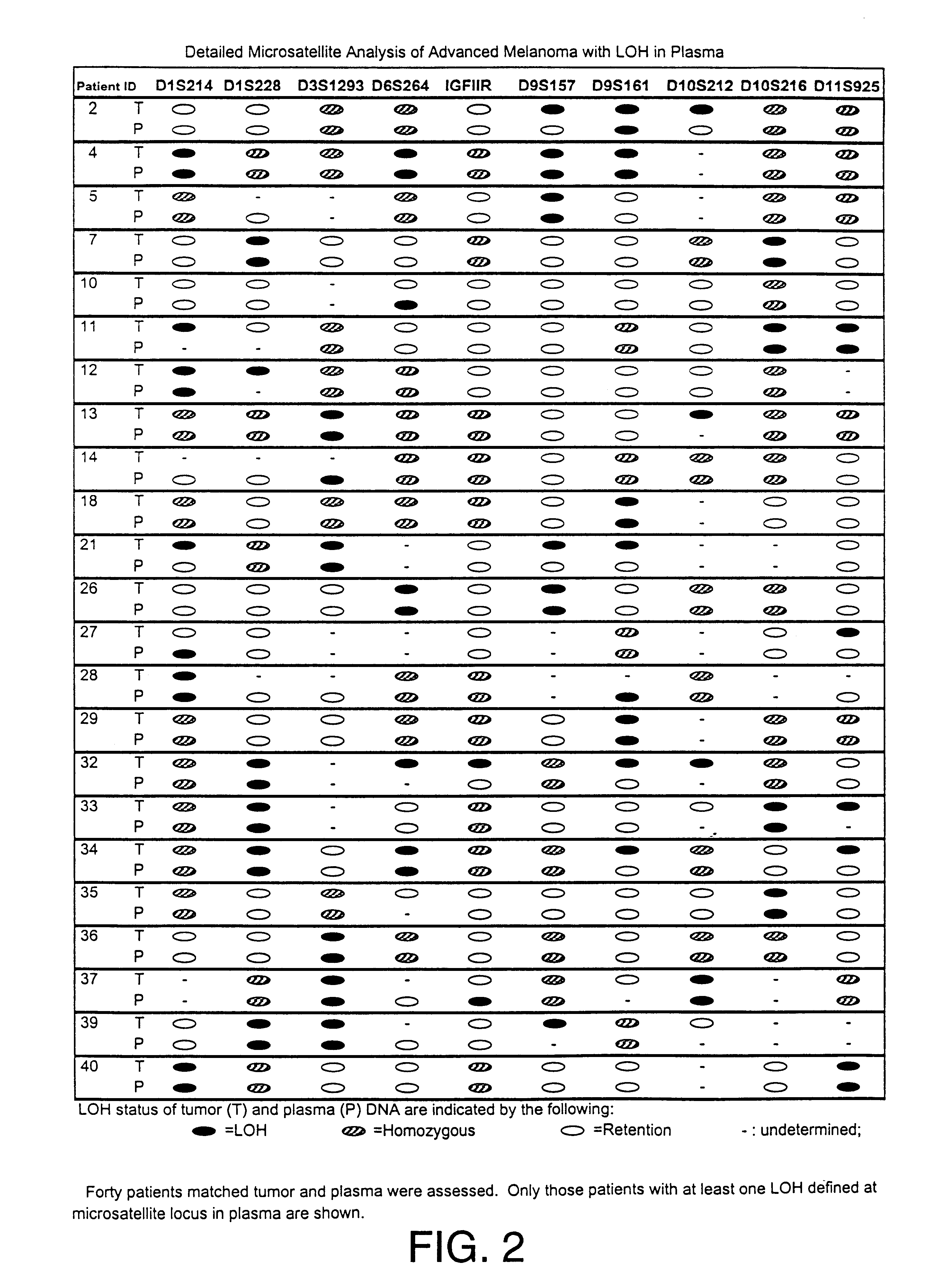
Detection of loss of heterozygosity in tumor and serum of melanoma patients
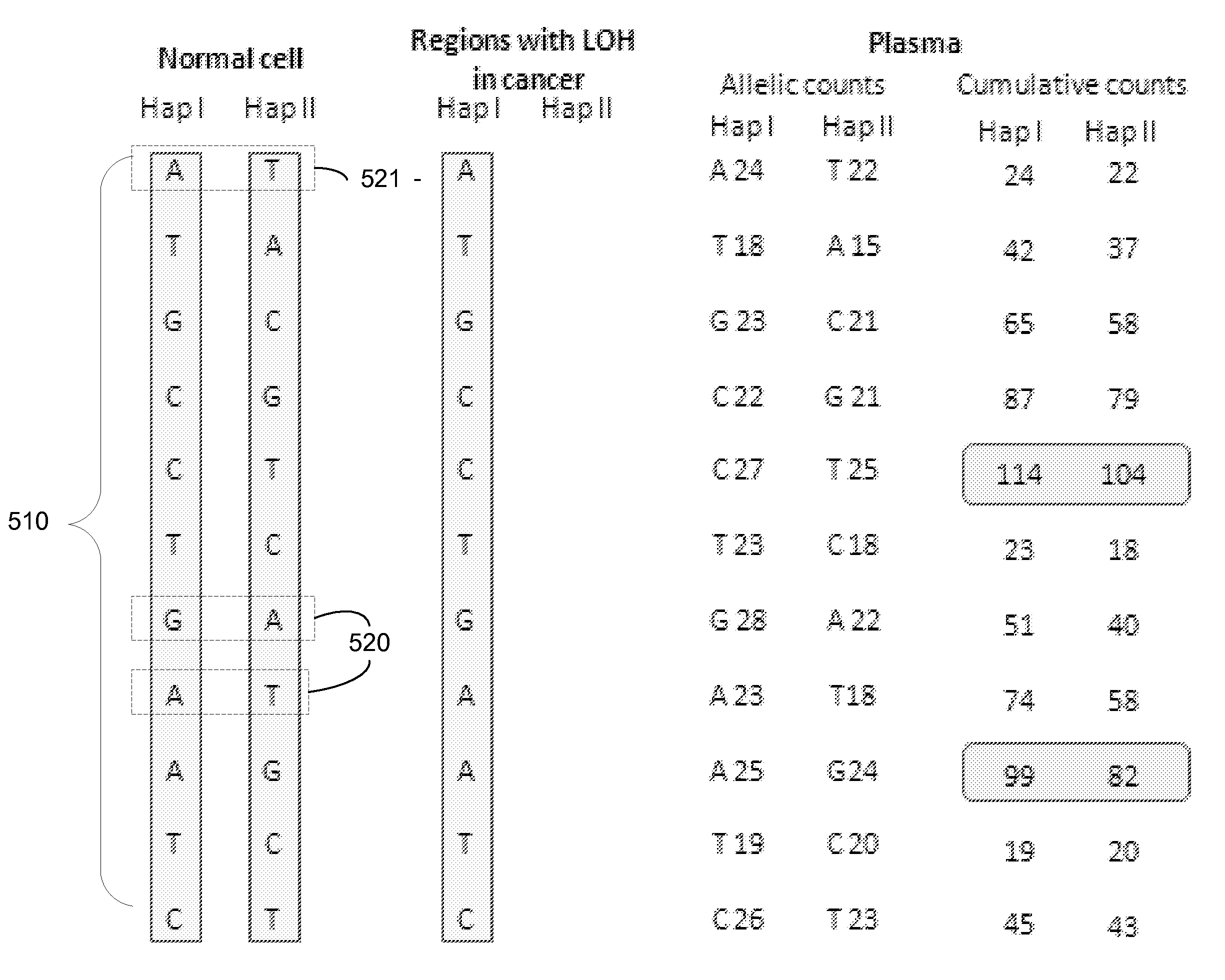
A method is provided for assessing allelic losses on specific chromosomal regions in melanoma patents. The method relies on the evidence that free DNA may be released in the plasma / serum of cancer patients allowing the detection of DNA with LOH in the plasma / serum of cancer patients by analysis for microsatellite markers. The amount of and specific allelic loss allows a prognosis to be made regarding tumor diagnosis and progression, tumor metastasis, tumor recurrence, and mortality.
Owner:JOHN WAYNE CANCER INST
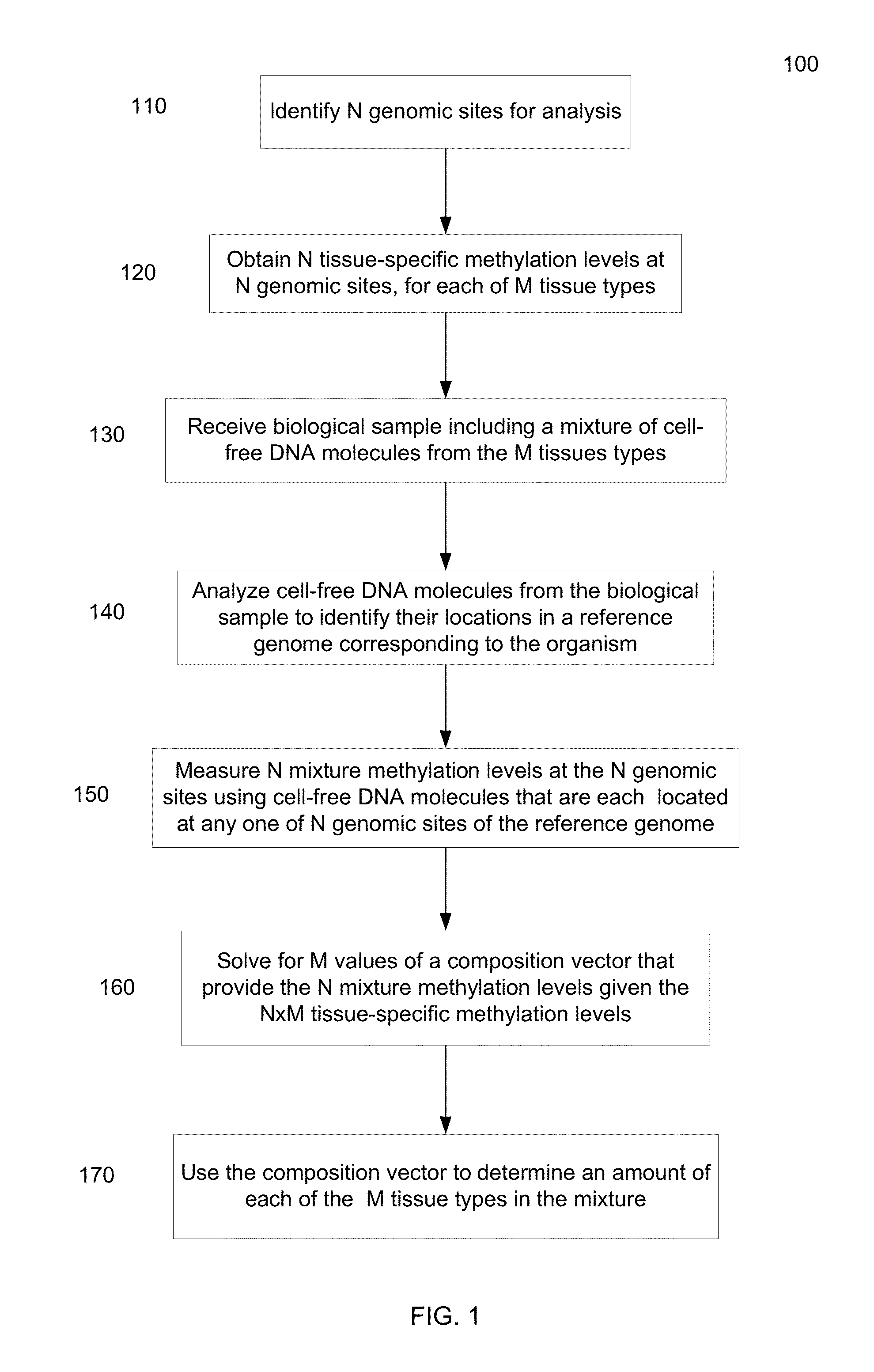
Methylation pattern analysis of tissues in a DNA mixture
ActiveUS20160017419A1Improve accuracyMicrobiological testing/measurementLibrary screeningAbnormal tissue growthSpecific chromosome
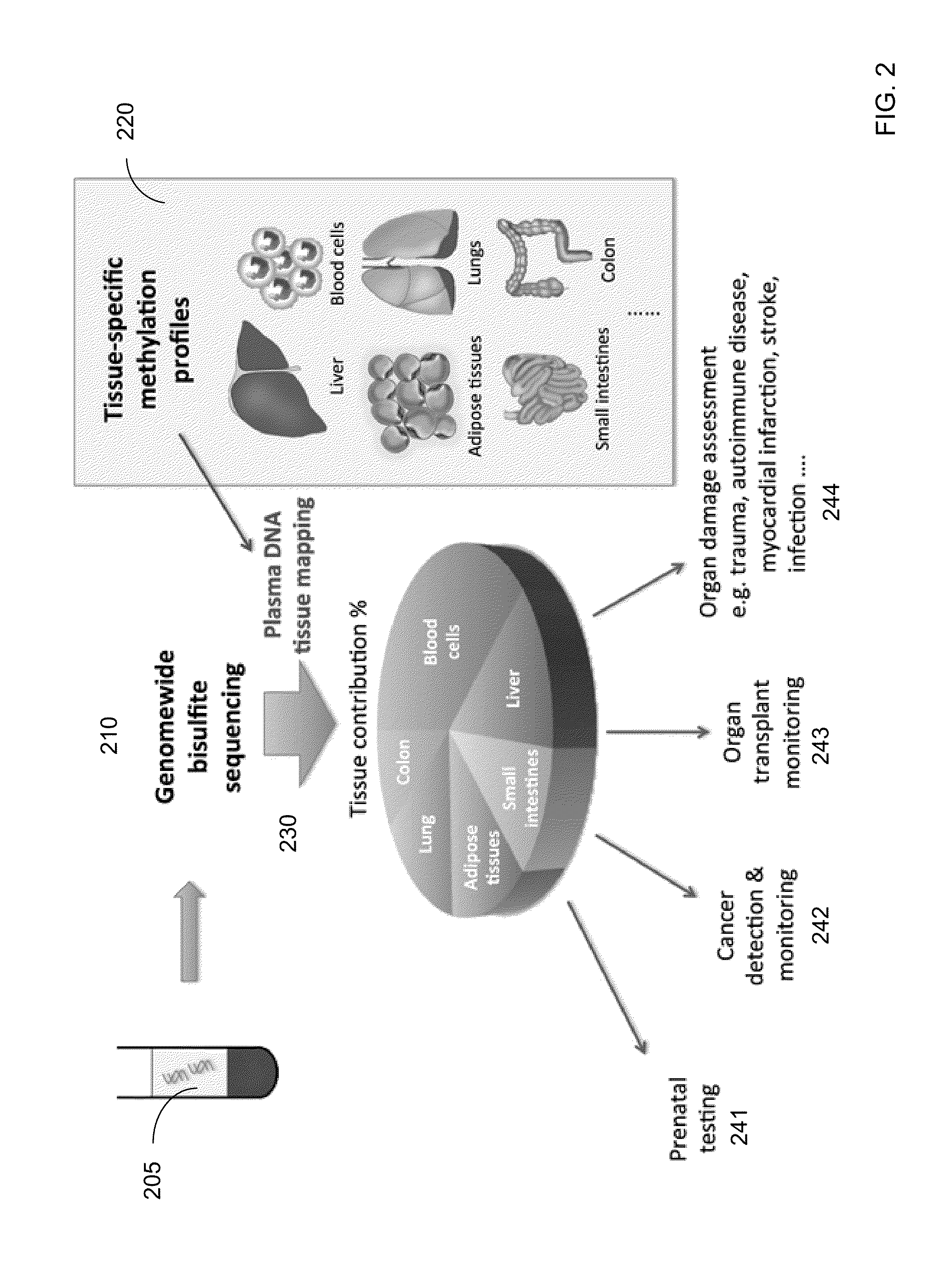
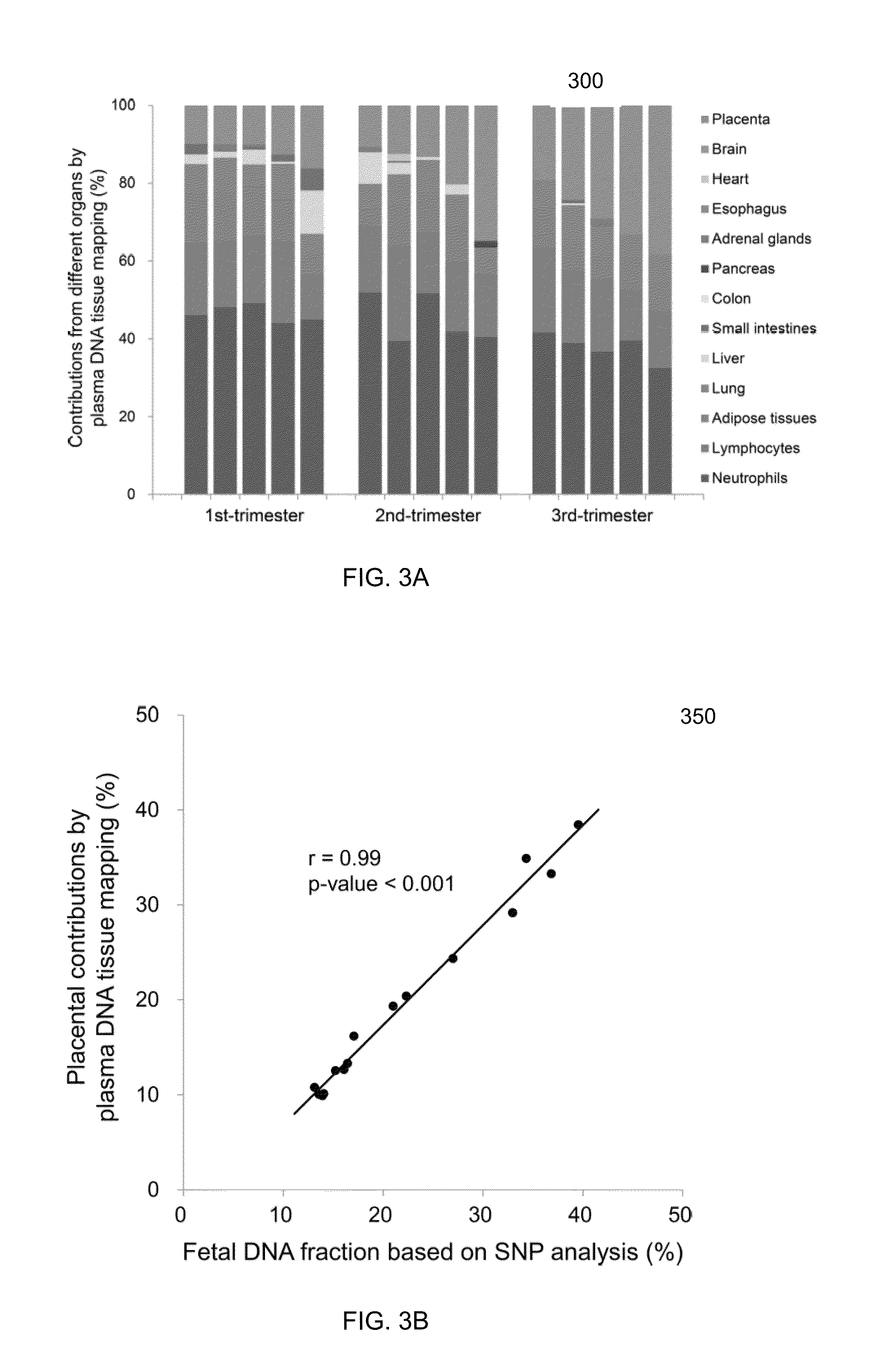
The contributions of different tissues to a DNA mixture are determined using methylation levels at particular genomic sites. Tissue-specific methylation levels of M tissue types can be used to deconvolve mixture methylation levels measured in the DNA mixture, to determine fraction contributions of each of the M tissue types. Various types of genomic sites can be chosen to have particular properties across tissue types and across individuals, so as to provide increased accuracy in determining contributions of the various tissue types. The fractional contributions can be used to detect abnormal contributions of a particular tissue, indicating a disease state for the tissue. A differential in fractional contributions for different sizes of DNA fragments can also be used to identify a diseased state of a particular tissue. A sequence imbalance for a particular chromosomal region can be detected in a particular tissue, e.g., identifying a location of a tumor.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Hppd-inhibitor herbicide tolerance
ActiveUS20110185445A1Negatively impactingBiocideMicrobiological testing/measurementNucleotidePlant cell
This invention relates generally to the detection of genetic differences among soybeans. More particularly, the invention relates to soybean quantitative trait loci (QTL) for tolerance or sensitivity to HPPD-inhibitor herbicides, such as mesotrione and isoxazole herbicides, to soybean plants possessing these QTLs, which map to a novel chromosomal region, and to genetic markers that are indicative of phenotypes associated with tolerance, improved tolerance, susceptibility, or increased susceptibility. Methods and compositions for use of these markers in genotyping of soybean and selection are also disclosed, as are methods and compositions for use of these markers in selection and use of herbicides for weed control. Also disclosed are isolated polynucleotides and polypeptides relating to such tolerance or sensitivity and methods of introgressing such tolerance into a plant by breeding or transgenically or by a combination thereof. Plant cells, plants, and seeds produced are also provided.
Owner:PIONEER HI BRED INT INC
System, Method and computer program product for integrated analysis and visualization of genomic data
Described is a system for analysis and visualization of genomic data. The system allows a user to select at least one individual sample. The sample has chromosomal data representing a genome with a chromosome and also includes chromosomal measurements of at least one event at a particular location on the chromosome. A frequency of event is generated based on the selected sample. The frequency of event is a frequency of occurrence of the event in the selected sample. At least one annotation can be selected that includes chromosomal region specific information as related to the chromosome. Finally, the chromosomal data, the annotation, and the frequency of event on a display can all be simultaneously displayed, thereby allowing a user to view chromosomal region specific information with respect to a particular chromosomal event.
Owner:BIODISCOVERY
Detection of genetic or molecular aberrations associated with cancer
ActiveUS8741811B2Improve accuracyImprove efficiencyMicrobiological testing/measurementLibrary screeningCell freeAfter treatment
Biological samples including cell-free DNA fragments are analyzed to identify imbalances in chromosomal regions, e.g., due to deletions and / or amplifications in a tumor. Multiple loci are used for each chromosomal region. Such imbalances can then be used to diagnose (screen) a patient for cancer, as well as prognosticate a patient with cancer, or to detect the presence or to monitor the progress of a premalignant condition in a patient. The severity of an imbalance as well as the number of regions exhibiting an imbalance can be used. A systematic analysis of non-overlapping segments of a genome can provide a general screening tool for a sample. Additionally, a patient can be tested over time to track severity of each of one or more chromosomal regions and a number of chromosomal regions to enable screening and prognosticating, as well as monitoring of progress (e.g. after treatment).
Owner:THE CHINESE UNIVERSITY OF HONG KONG
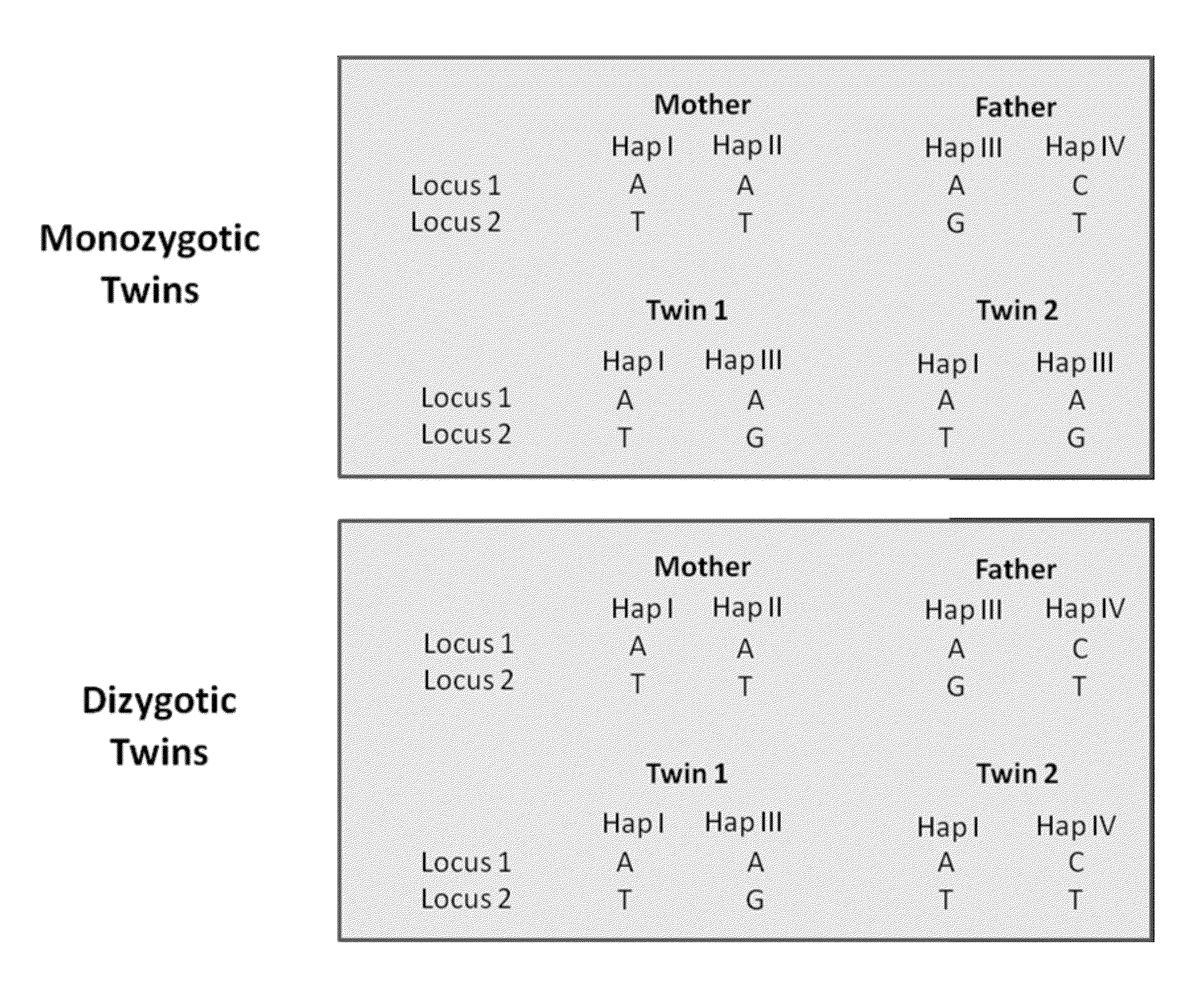
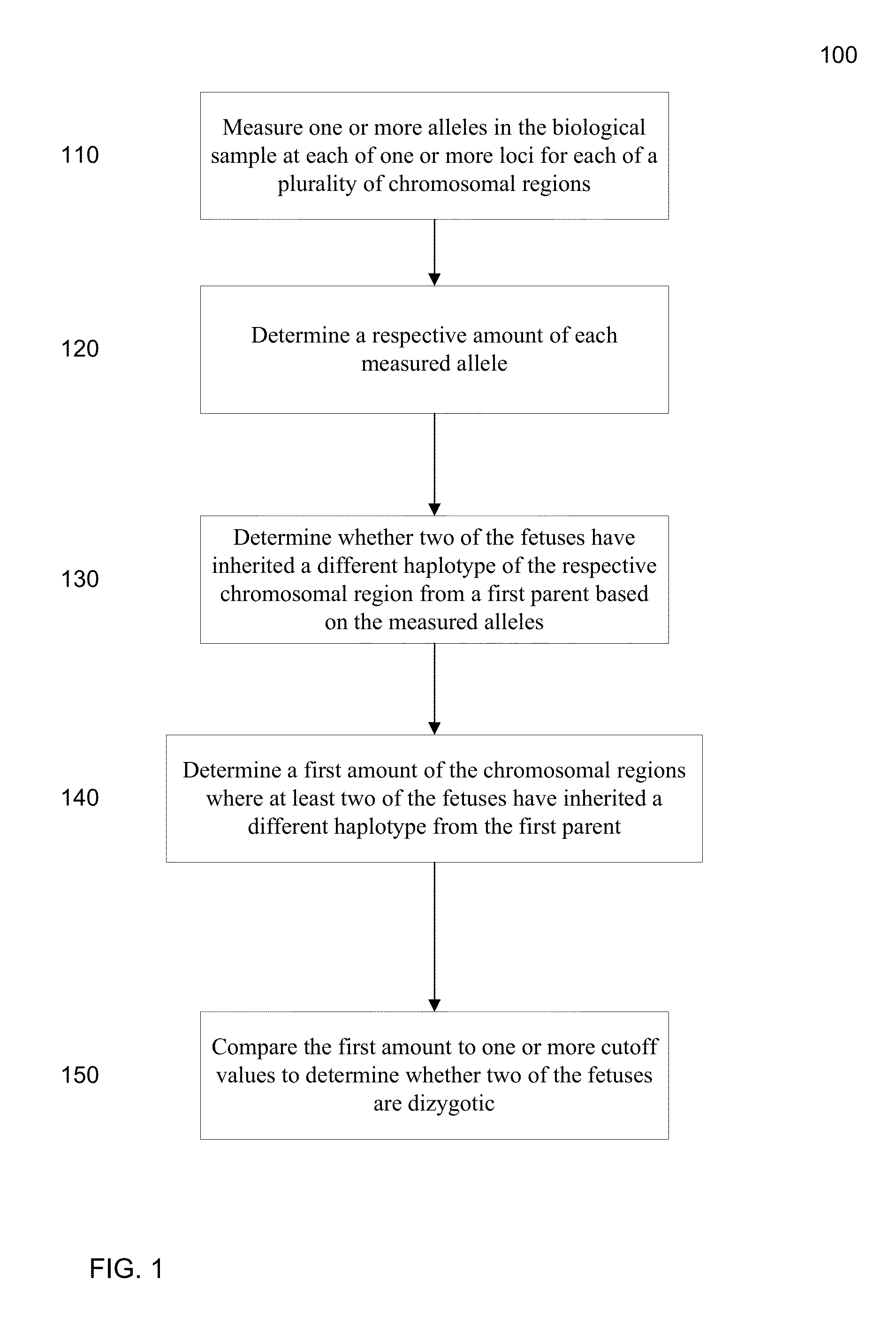
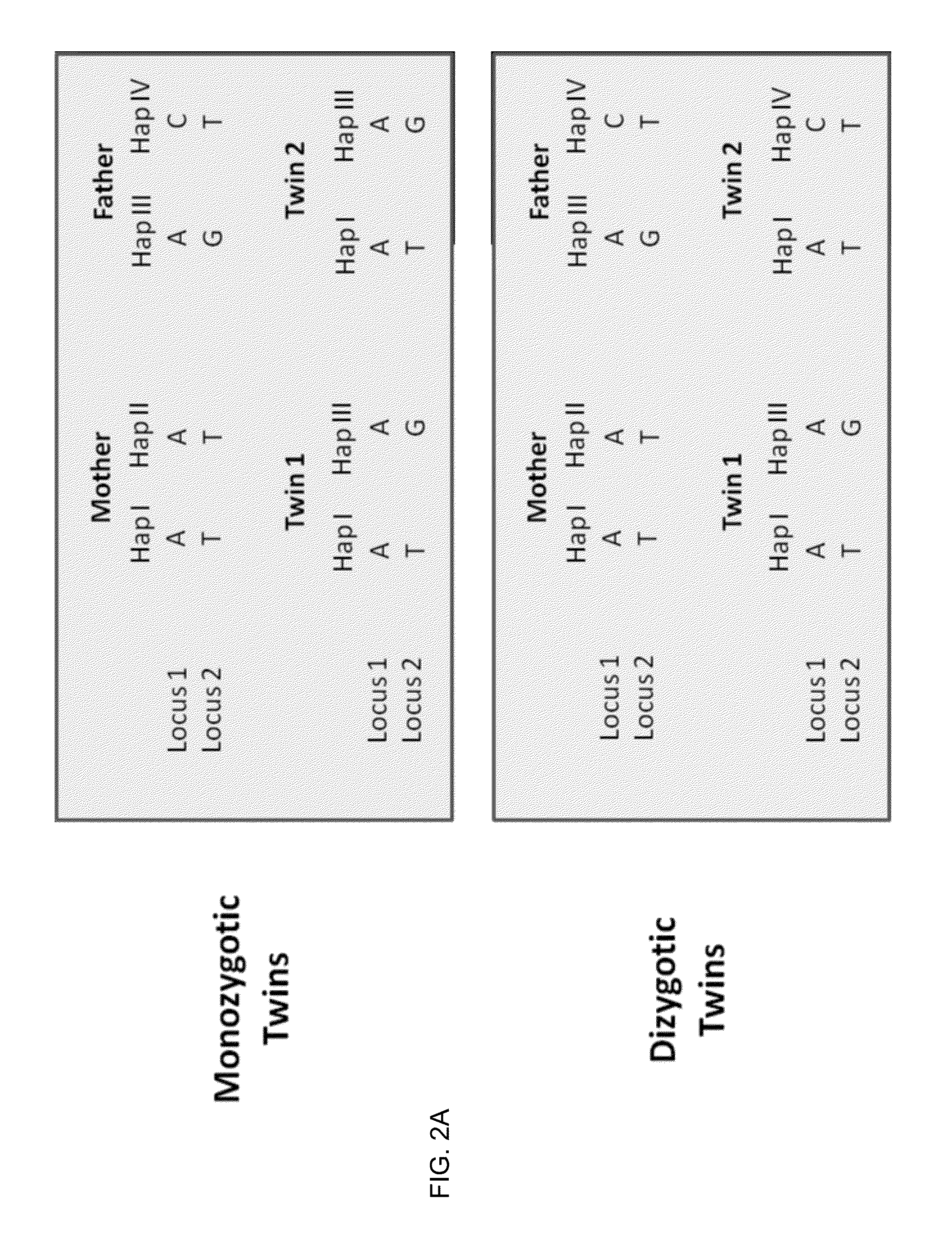
Molecular testing of multiple pregnancies
ActiveUS20130059733A1Non invasiveMicrobiological testing/measurementLibrary screeningSpecific chromosomeGenetic divergence
Methods, systems, and apparatus are provided for determining zygosity of a multiple-fetus pregnancy using a biological sample taken from the mother. The fetal and maternal DNA in the sample (e.g. plasma) can be analyzed for a particular chromosomal region to identify genetic differences in the fetuses. For example, a normalized parameter for the measure of a primary or secondary allele can show variances for different chromosomal regions when fetuses are dizygotic. Such a variance can be determined relative to an expected value if the fetuses were genetically identical. Statistical methods are provided for analyzing the variation of the normalized parameters to determine fetal DNA concentration and the maternal-fetal mixed genotype at various loci. Parental genotype and haplotype information can also be used to identify inheritance of different parental haplotypes to indicate genetic differences among the fetuses.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Pepper plants with improved disease resistance
ActiveUS20180171353A1Vector-based foreign material introductionPlant genotype modificationDiseaseGermplasm
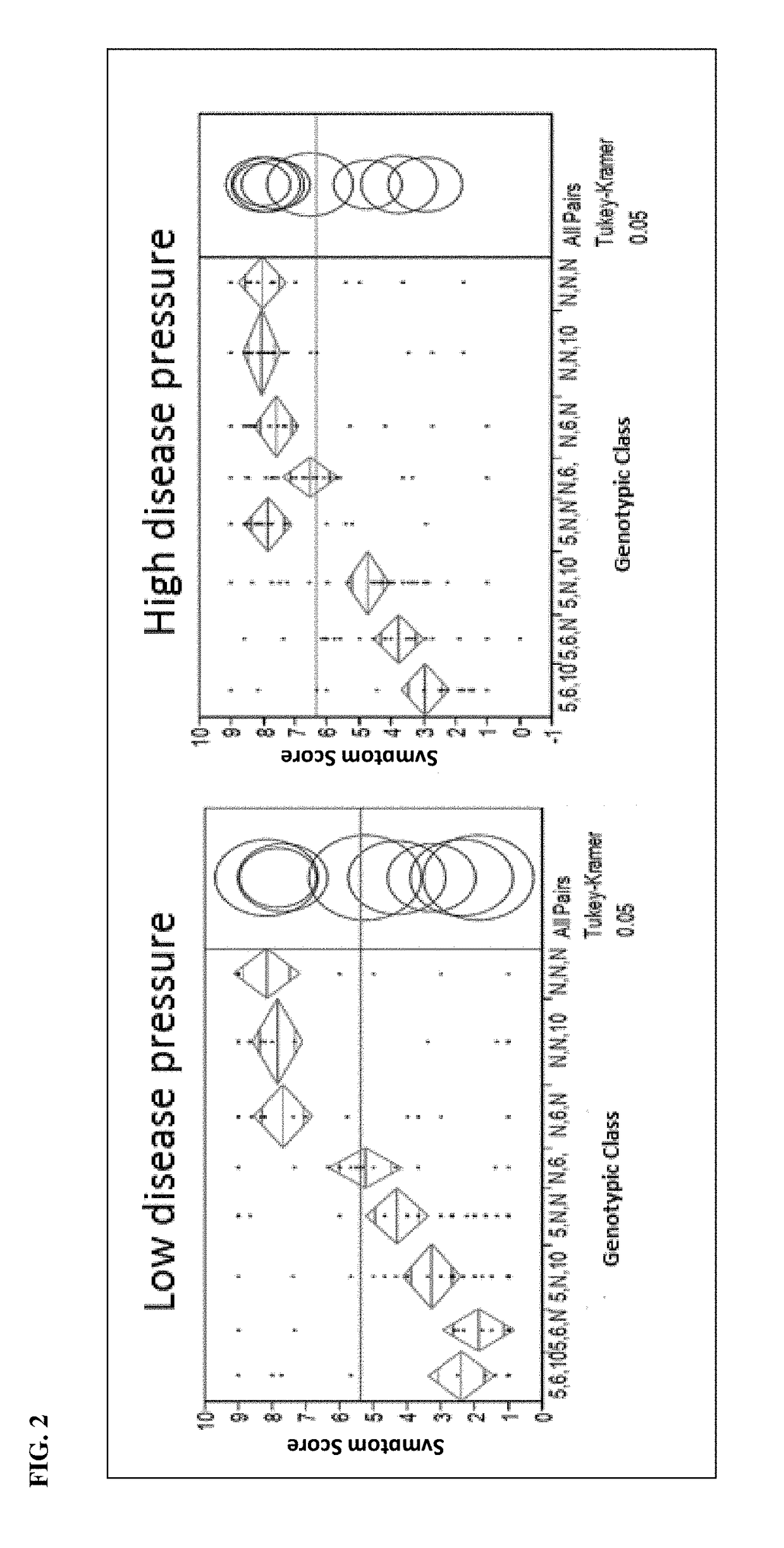
The present disclosure provides pepper plants exhibiting resistance to Phytophthora capsici. Such plants may comprise novel introgressed chromosomal regions associated with disease resistance. In certain aspects, compositions, including novel polymorphic markers and methods for producing, breeding, identifying, and selecting plants or germplasm with a disease resistance phenotype are provided.
Owner:SEMINIS VEGETABLE SEEDS
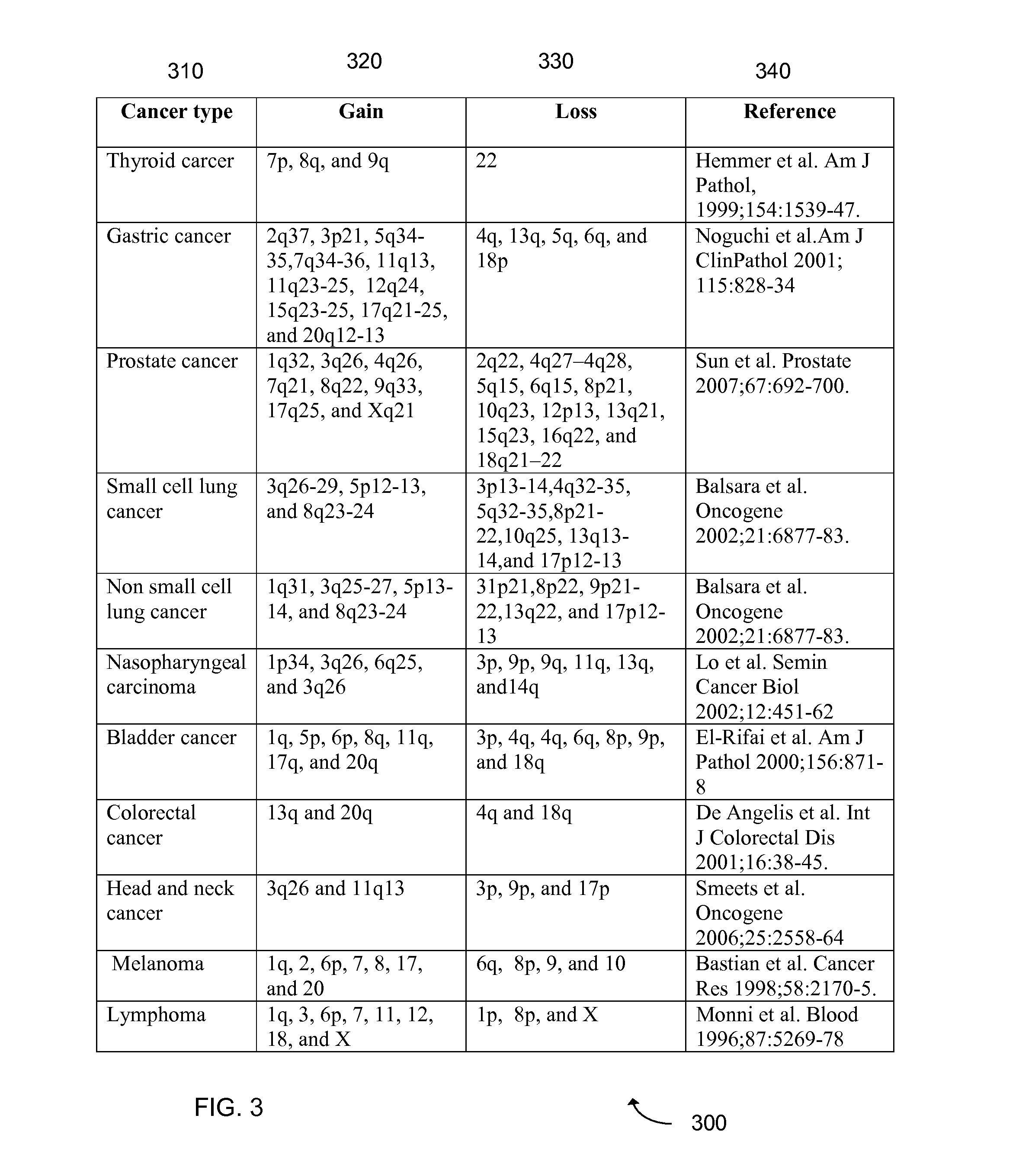
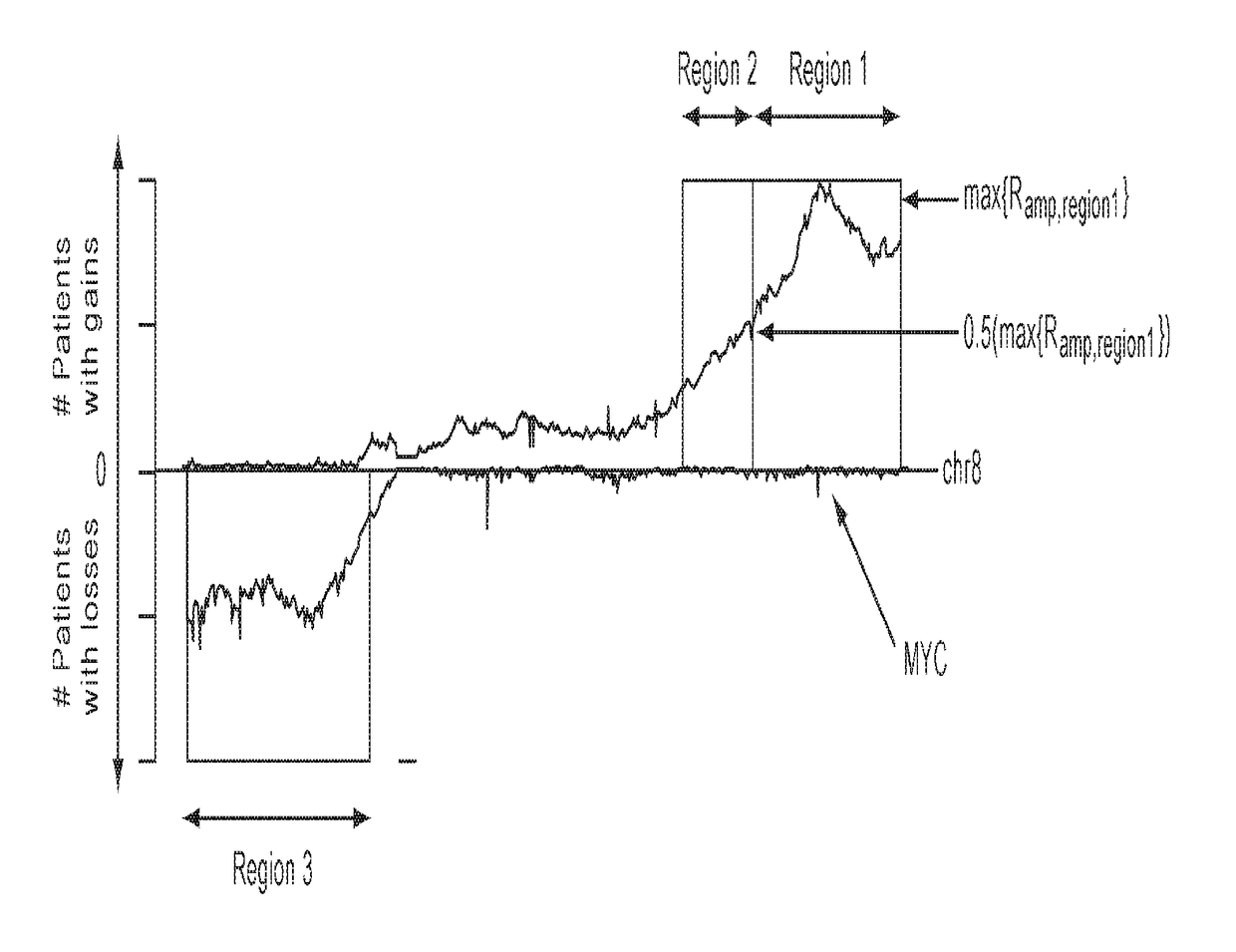
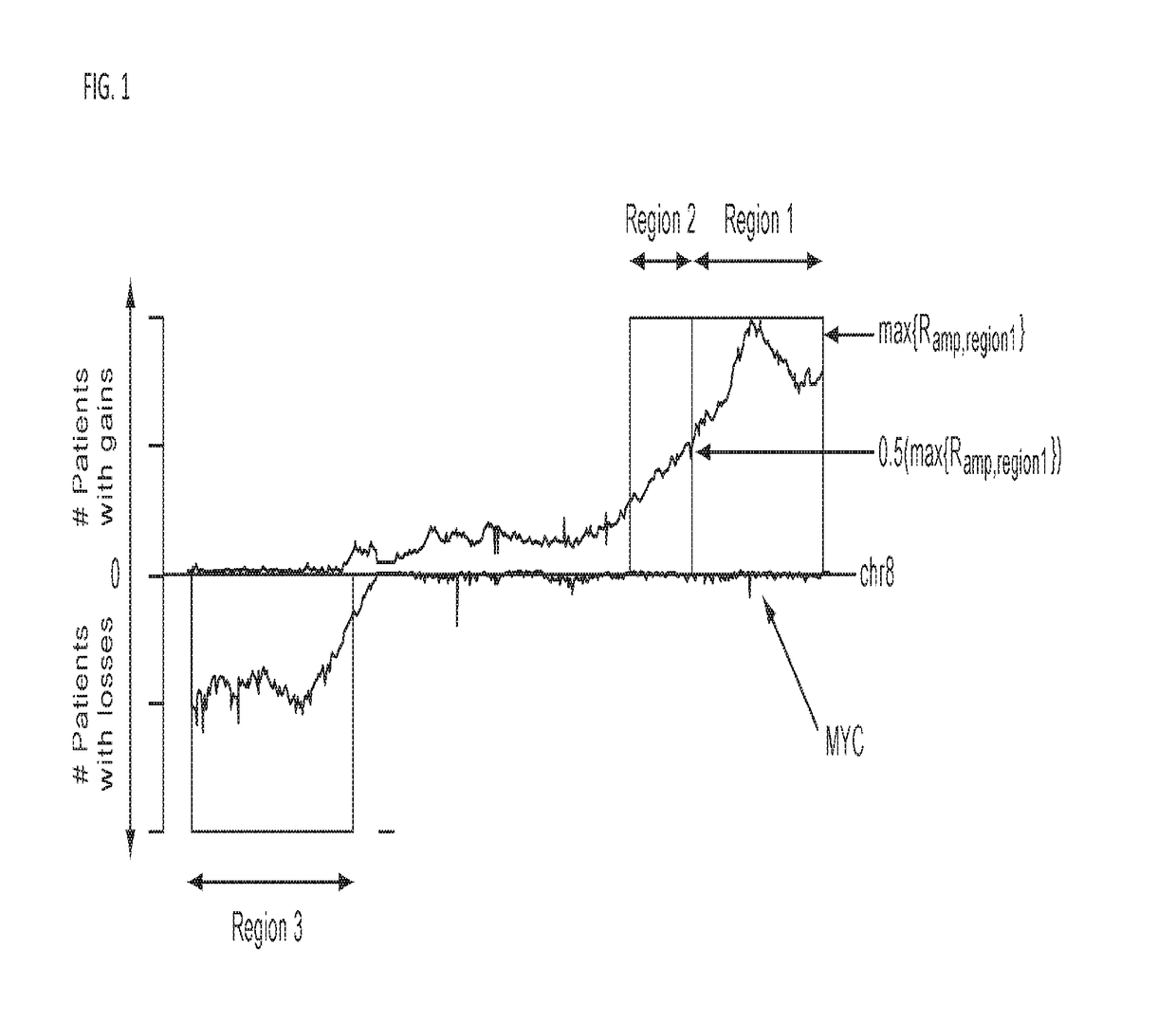
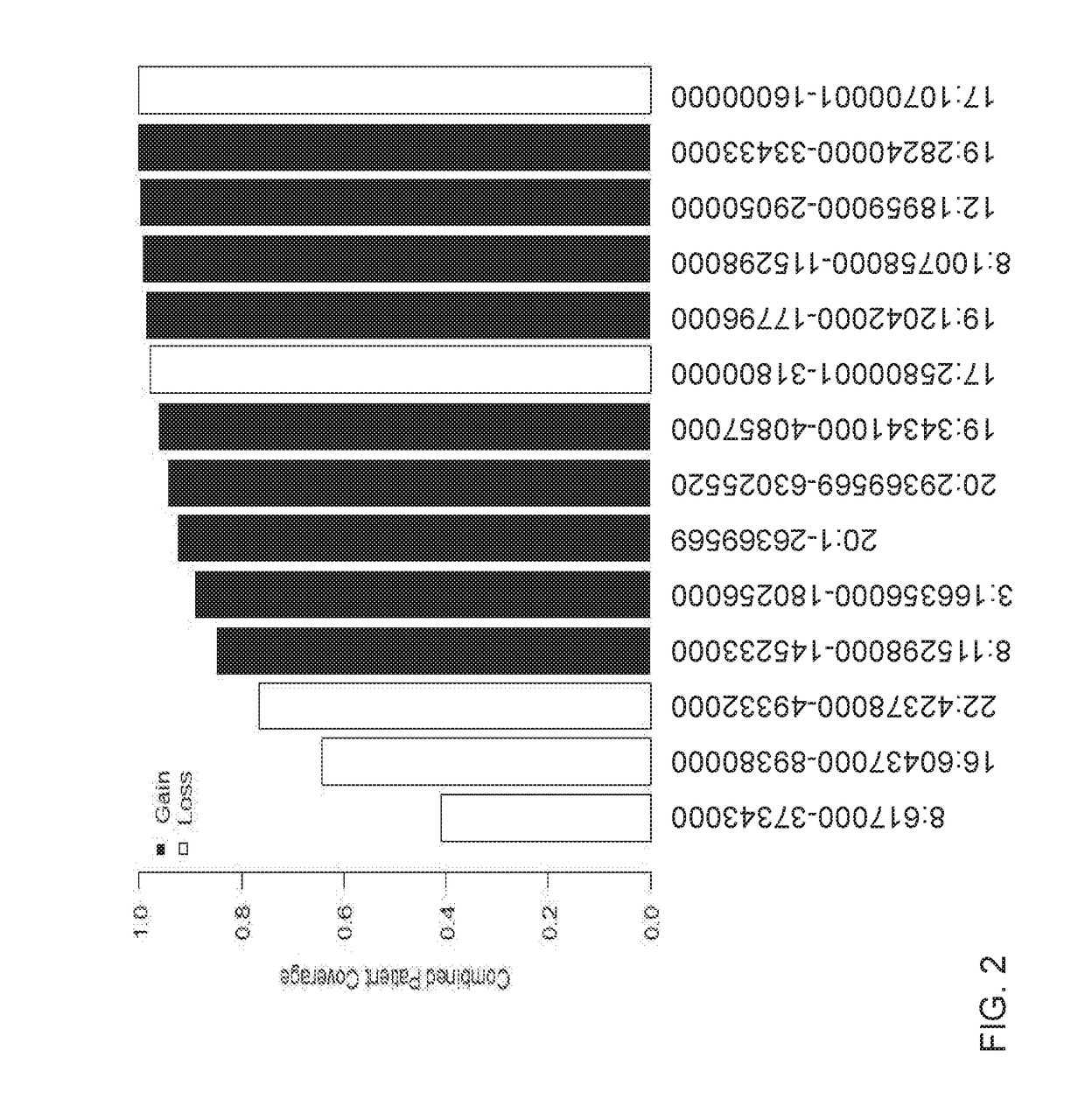
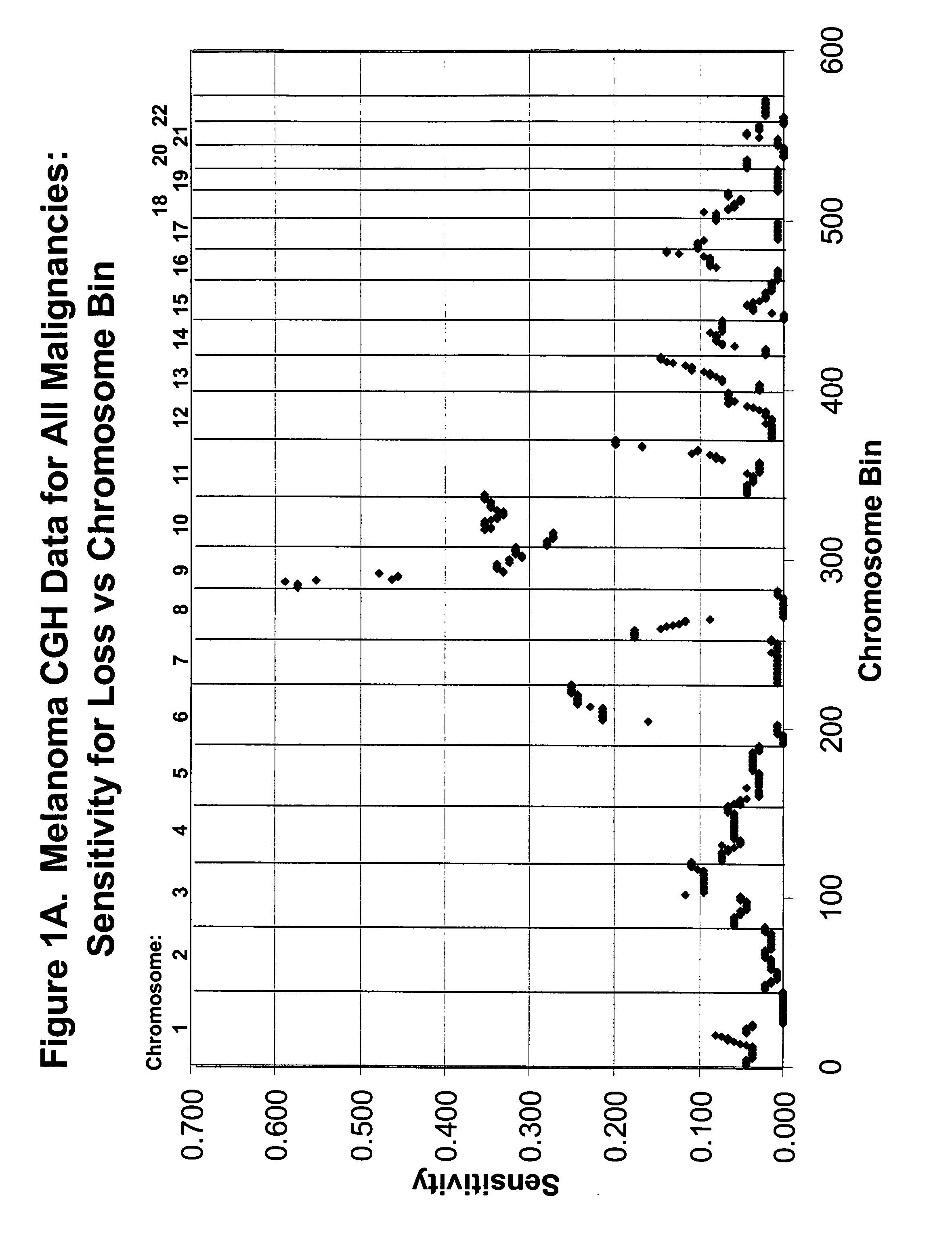
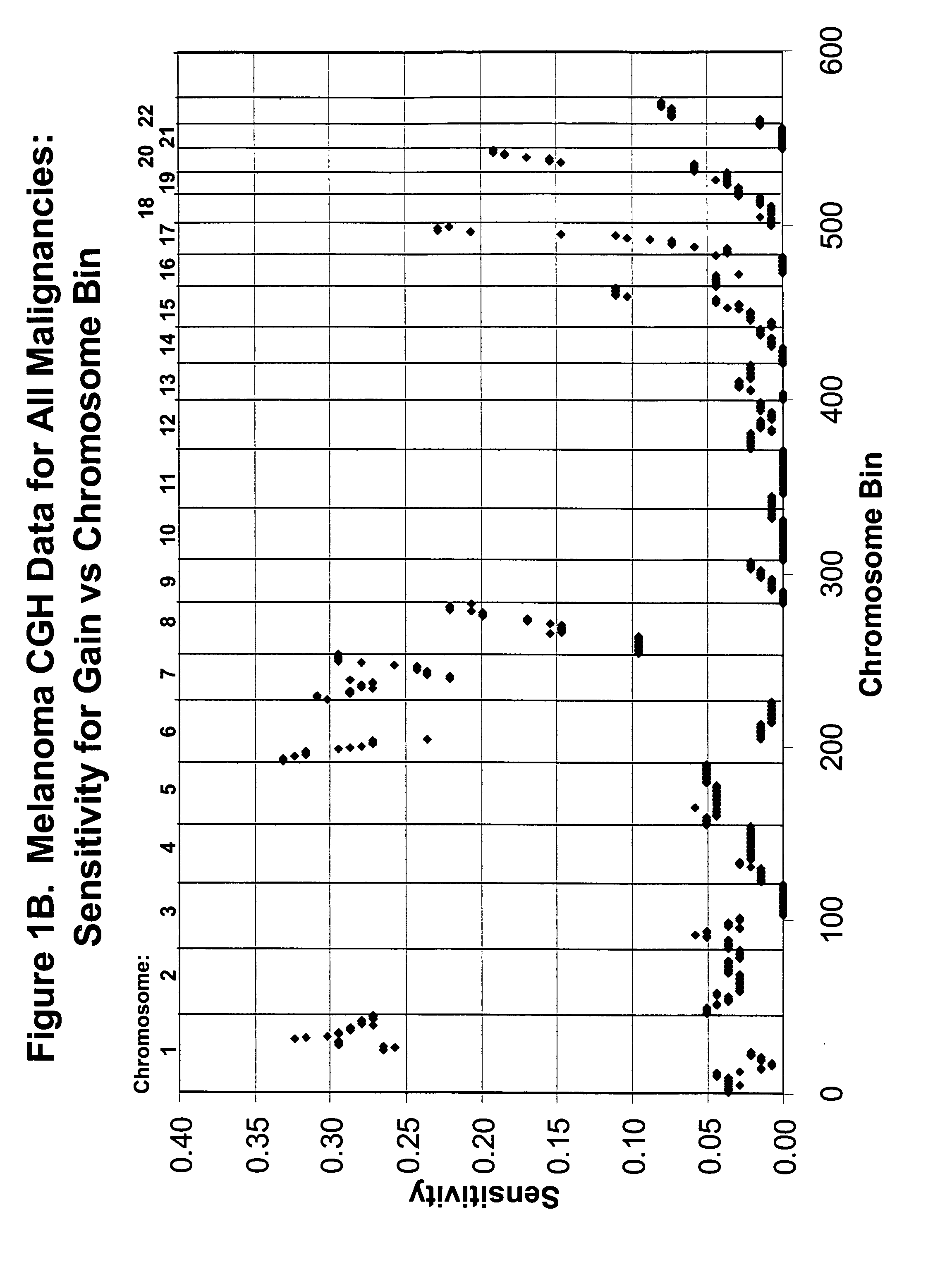
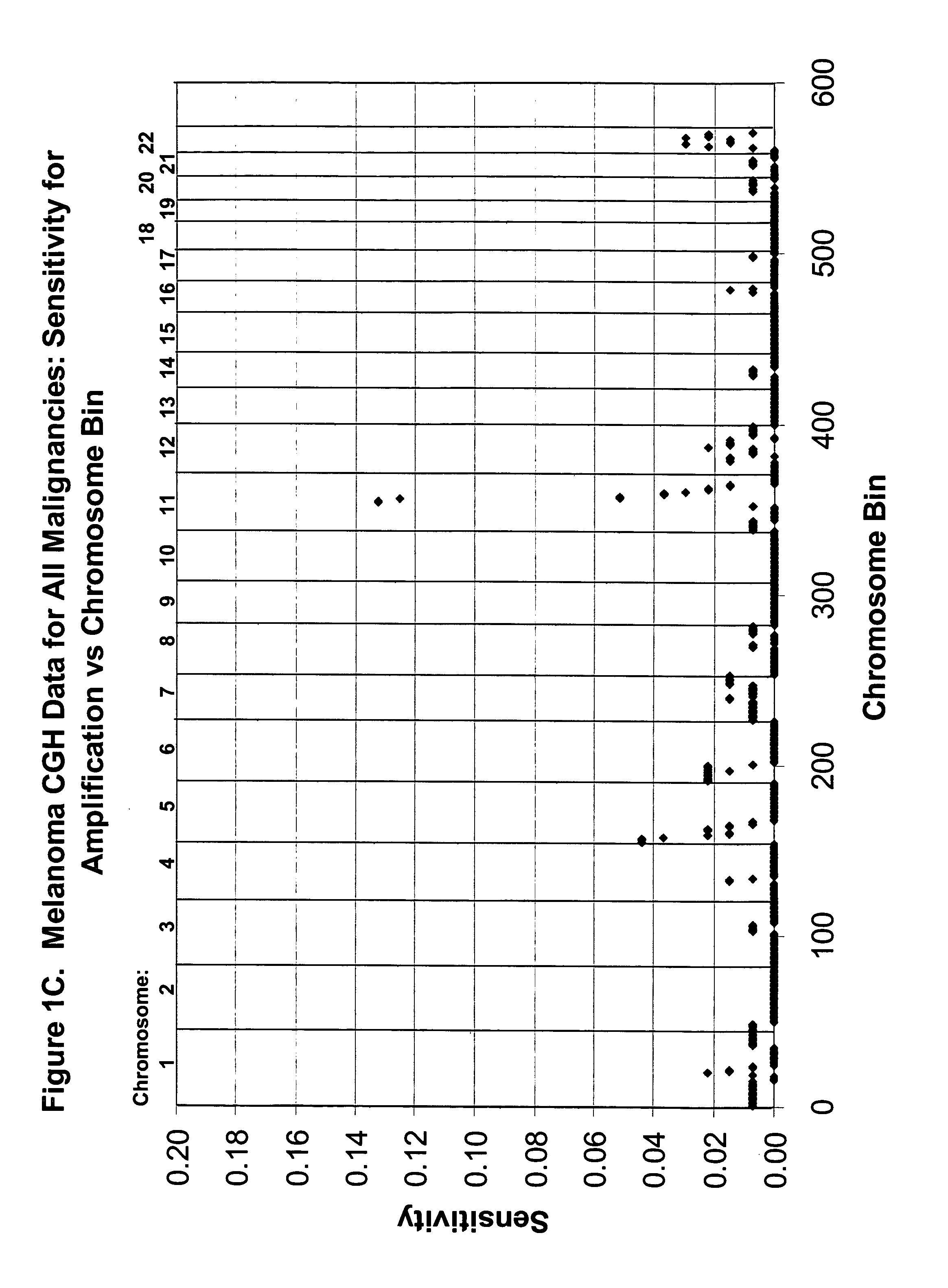
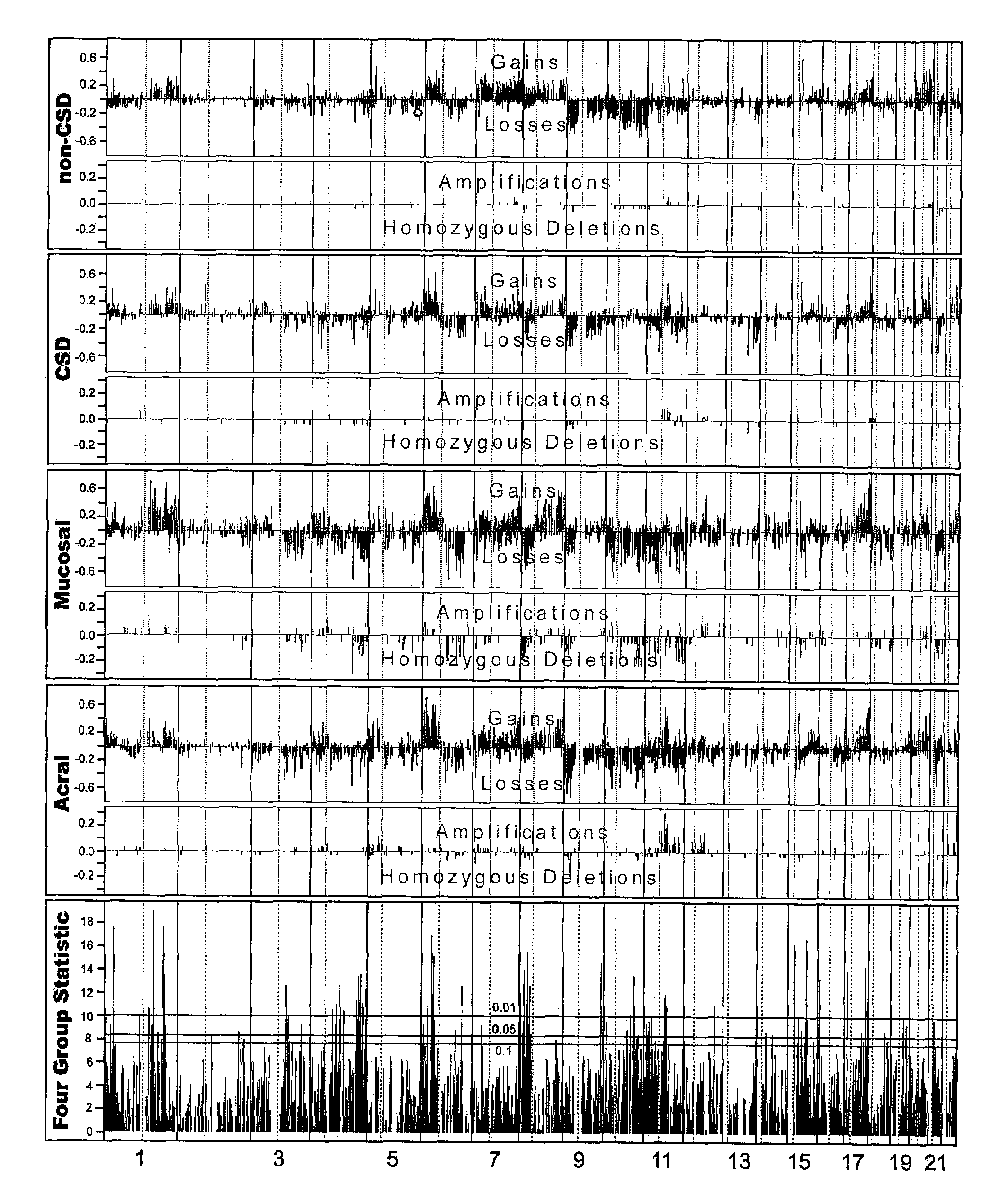
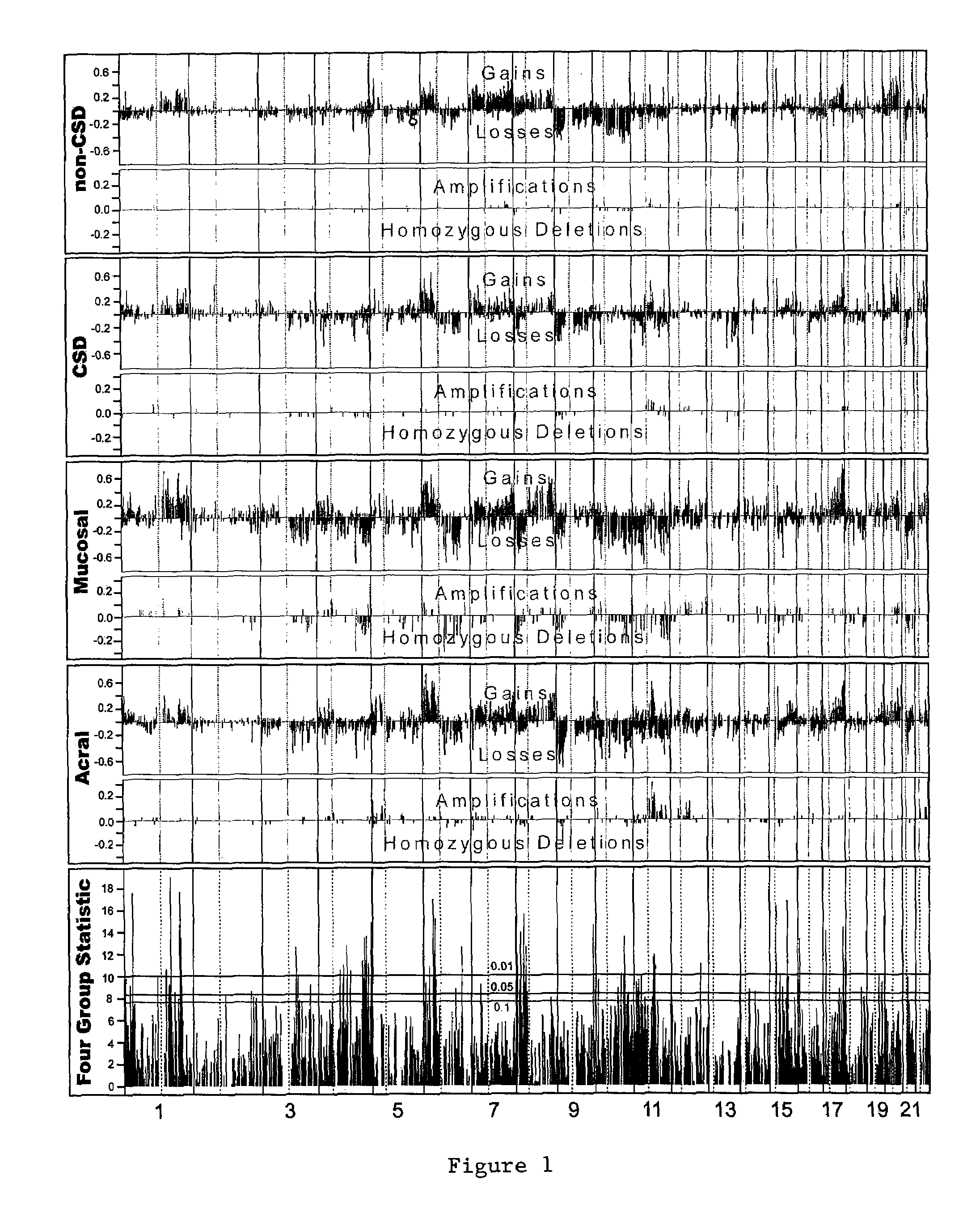
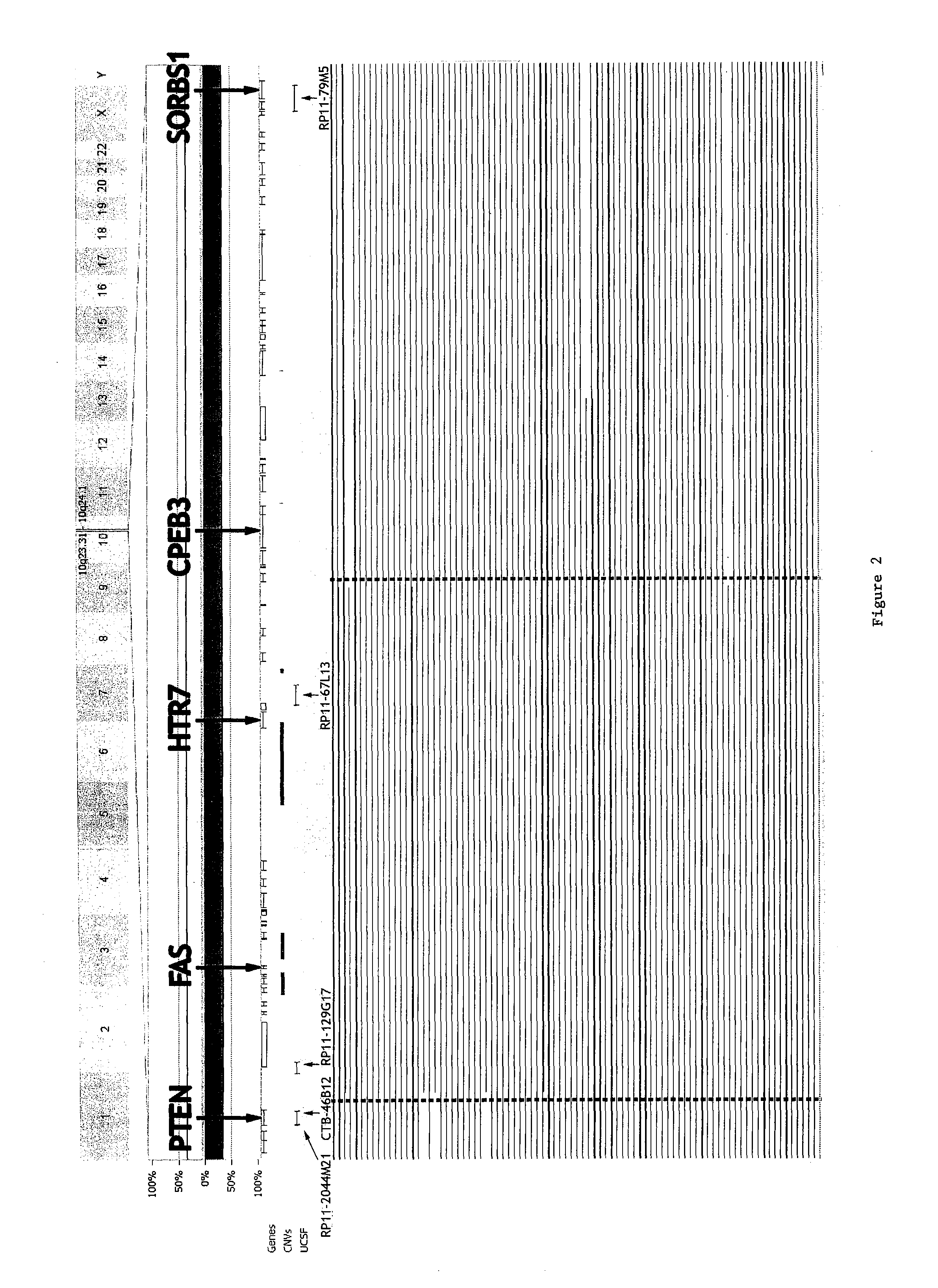
Detection of chromosomal region copy number changes to diagnose melanoma
Owner:RGT UNIV OF CALIFORNIA
Prognostic Marker for Endometrial Carcinoma
InactiveUS20110217701A1Microbiological testing/measurementDisease diagnosisNon small lung cancerRegimen
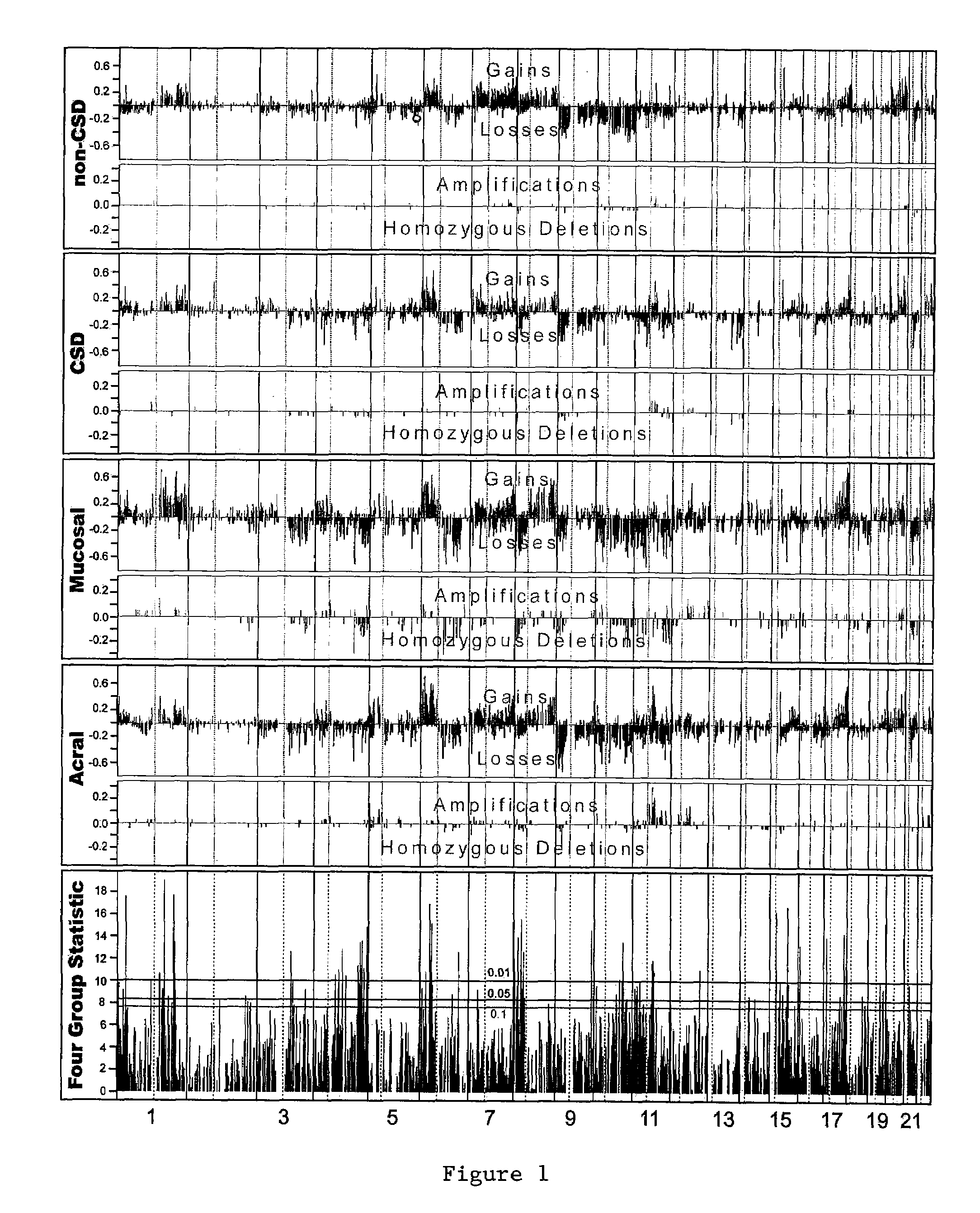
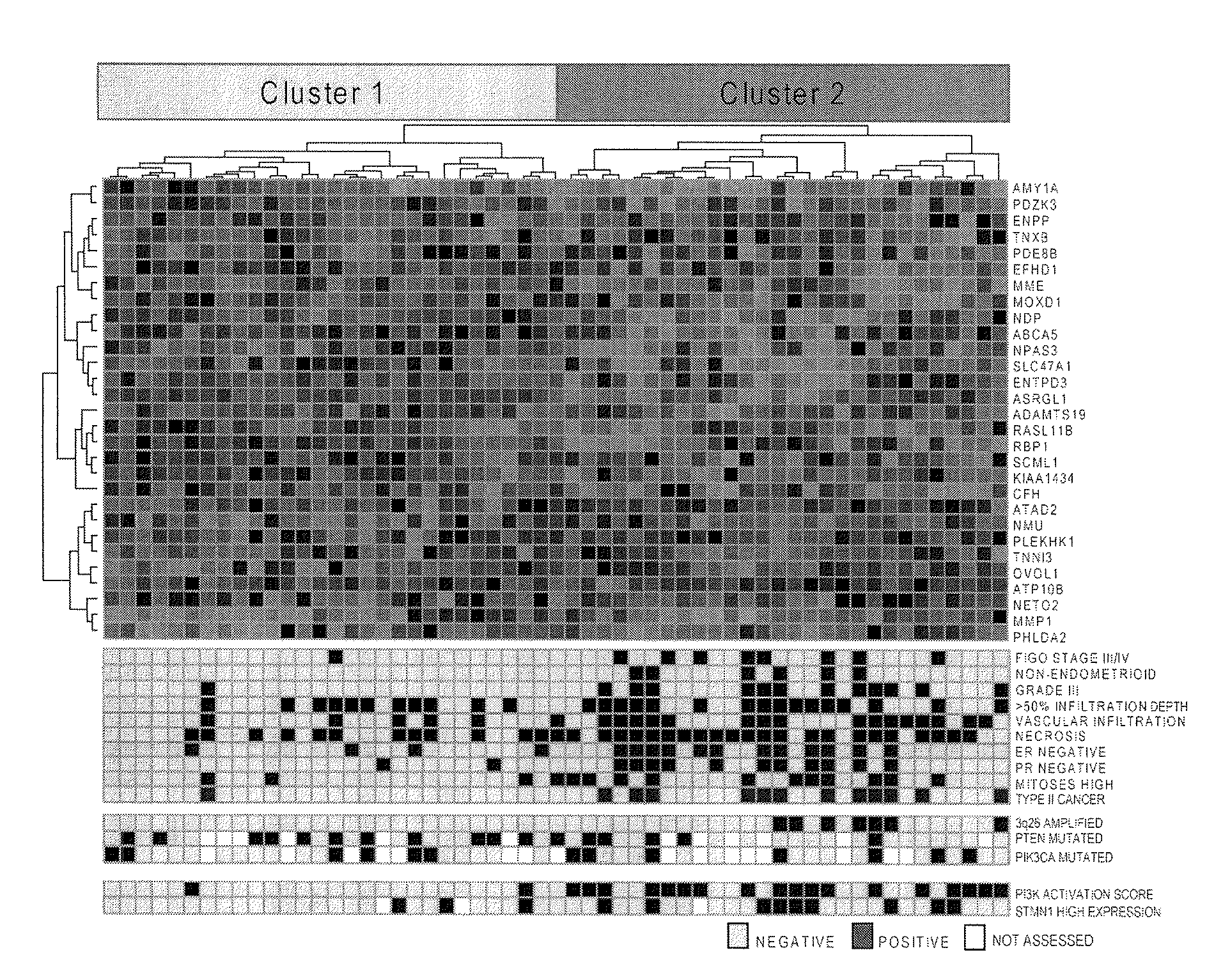
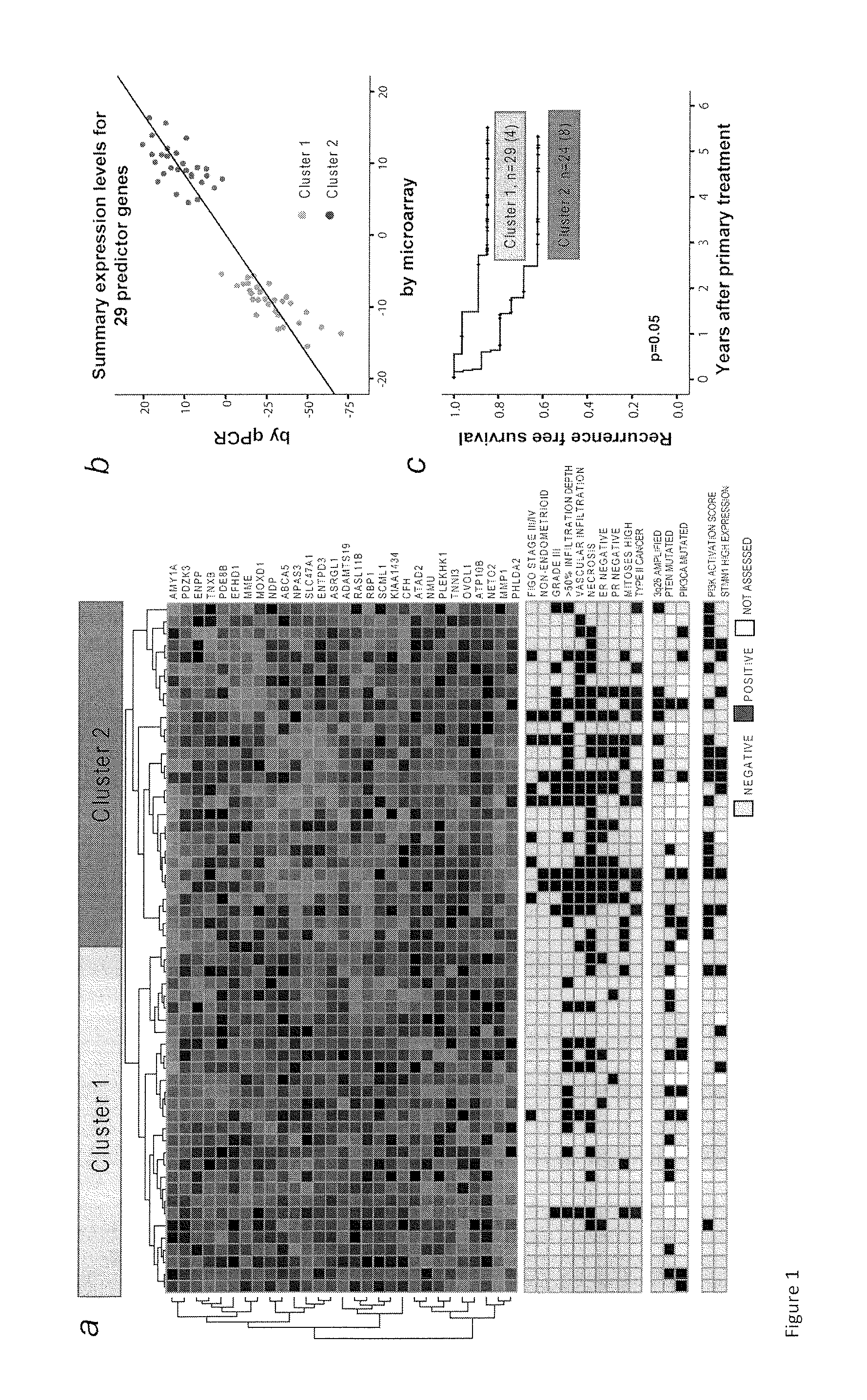
The present invention relates to a method for diagnosis of different stages of endometrial cancer in an individual. Further, the present invention relates to a method for evaluating the probability of survival for an individual suffering from endometrial carcinoma. In another aspect, the present invention relates to the stratification of therapy regimen of endometrial tumor, ovarian cancer, breast cancer, non-small lung cancer or hormone refractory prostate cancer therapy in an individual or monitoring therapeutic efficacy in an individual suffering from the same based on the expression status of STMN1 gene or protein. Moreover, the present invention relates to a kit for use in any of the above referenced methods comprising a means for determining amplifications and deletions of chromosomal regions 3q26.32 and 12p12.1, determining alterations of the gene expression profile of the genes (gene signature): upregulation of the genes PLEKHK1, ATP10B, NMU, MMP1, ATAD2, NETO2, TNNI3, PHLDA2, OVOL1 and down-regulation of the genes: NDP, KIAA1434, MME, CFH, MOXD1, SLC47A1, RBP1, PDE8B, ASRGL1, ADAMTS19, EFHD1, ABCA5, NPAS3, SCML1, TNXB, ENTPD3, AMY1A, ENPP, RASL11B, PDZK3, or the expression status of the STMN1 gene or protein, respectively. Finally, the present invention provides a method for predicting the response to taxanes in an individual suffering from a disease treated with the taxanes based on the expression status of the STMN1 gene or protein.
Owner:BERGEN TEKNOLOGIOVERFORING
Predictive and Therapeutic Markers in Ovarian Cancer
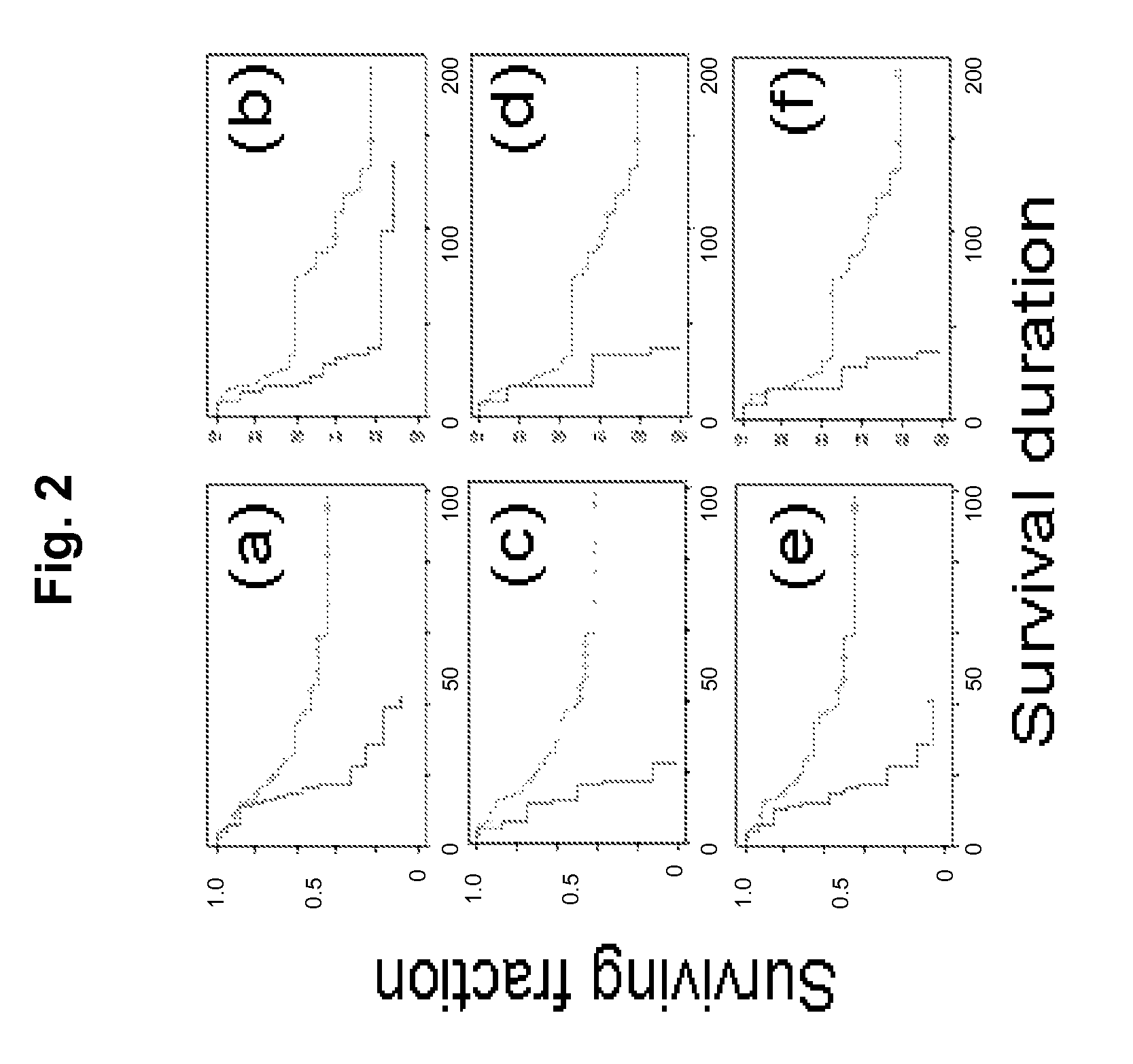
ActiveUS20080312096A1Reduced survival durationImprove the level ofSugar derivativesMicrobiological testing/measurementApoptosisIn vivo
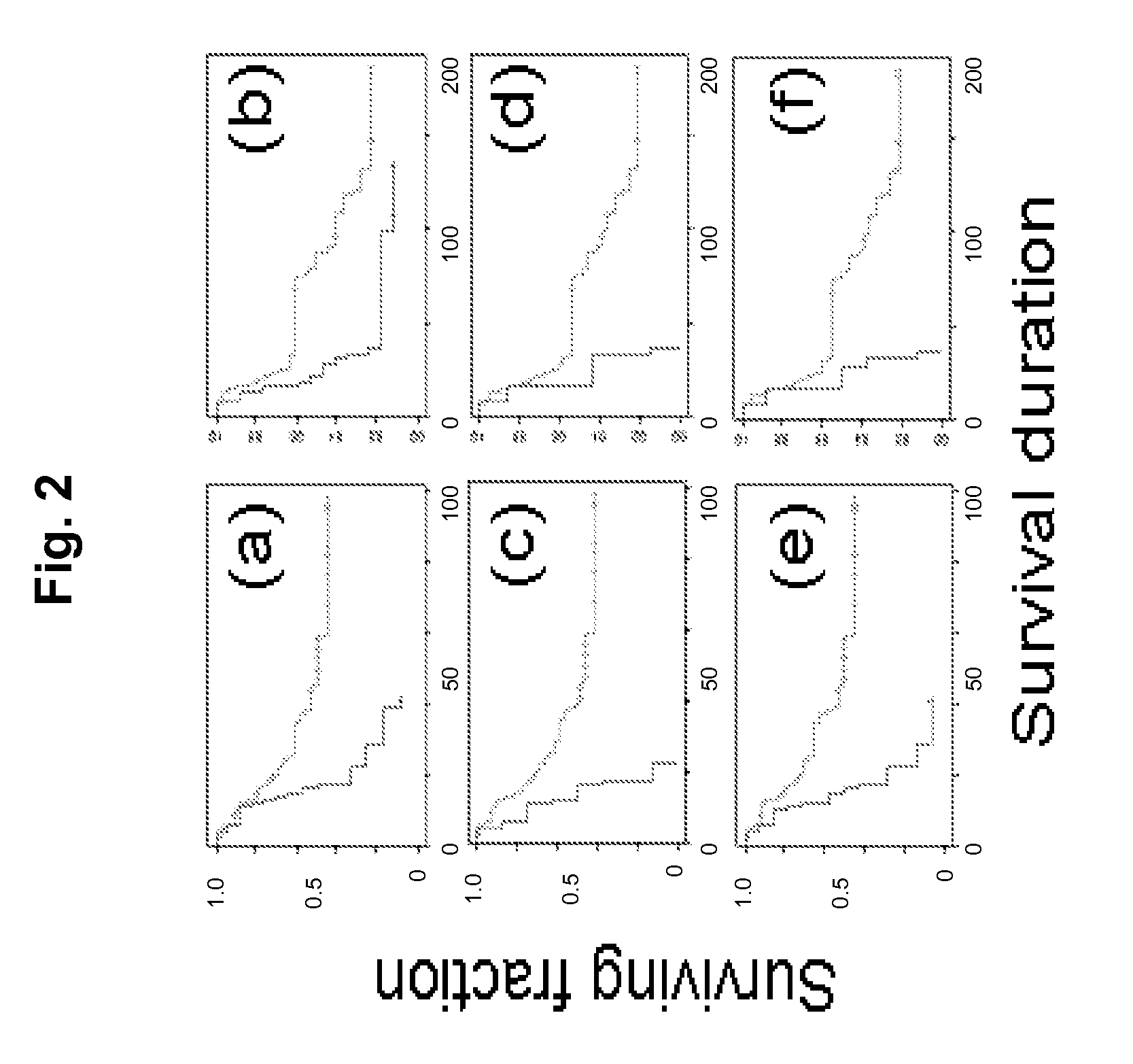
Cancer markers may be developed to detect diseases characterized by increased expression of apoptosis-suppressing genes, such as aggressive cancers. Genes in the human chromosomal regions, 8q24, 11q13, 20q11-q13, were found to be amplified indicating in vivo drug resistance in diseases such as ovarian cancer. Diagnosis and assessment of amplification levels certain genes shown to be amplified, including PVT1, can be useful in prediction of poor outcome of patient's response and drug resistance in ovarian cancer patients with low survival rates. Certain genes were found to be high priority therapeutic targets by the identification of recurrent aberrations involving genome sequence, copy number and / or gene expression are associated with reduced survival duration in certain diseases and cancers, specifically ovarian cancer. Therapeutics to inhibit amplification and inhibitors of one of these genes, PVT1, target drug resistance in ovarian cancer patients with low survival rates is described.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST +1
Method for gene mapping from chromosome and phenotype data
InactiveUS6909971B2Data processing applicationsMicrobiological testing/measurementGene mappingNucleotide
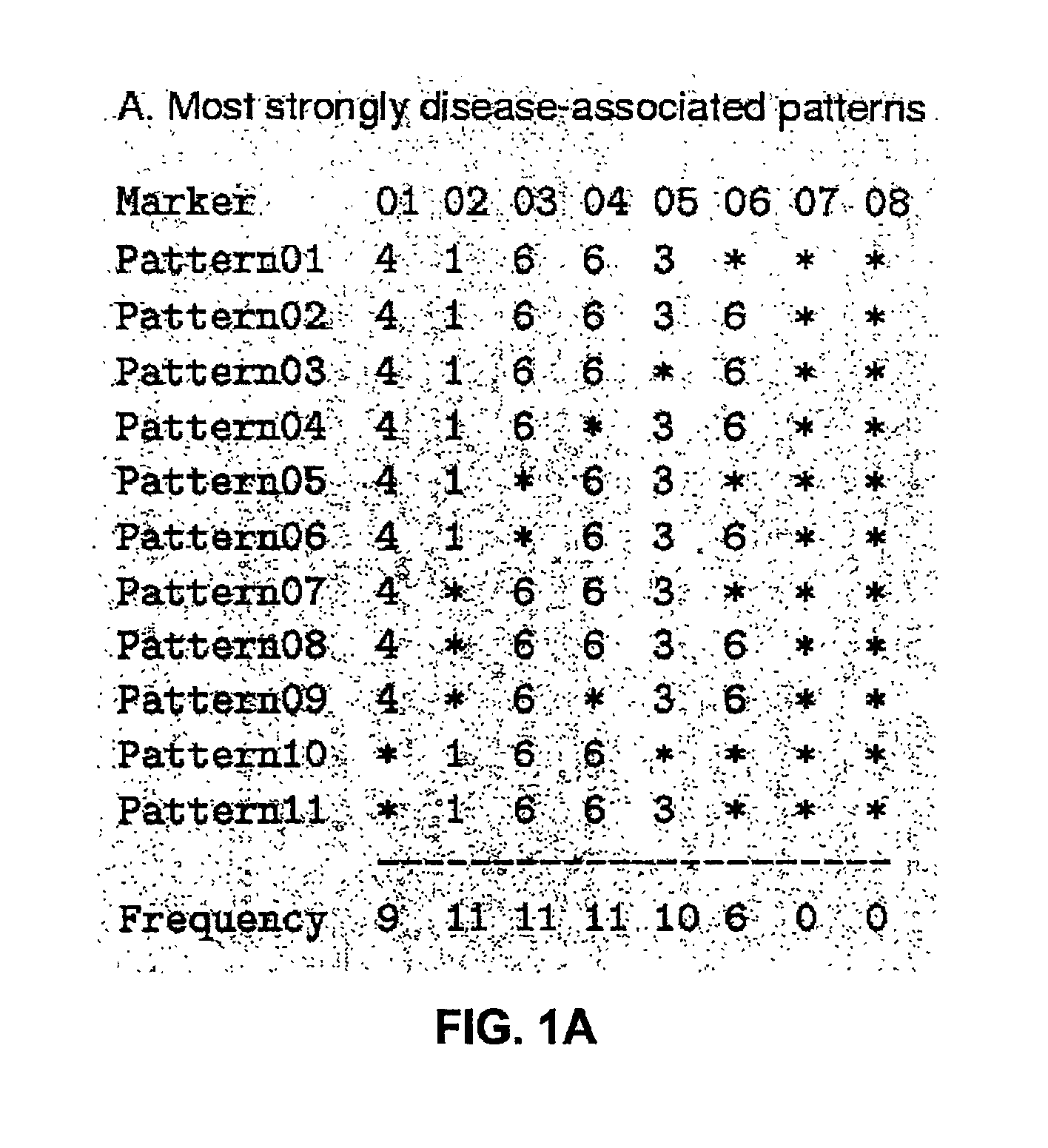
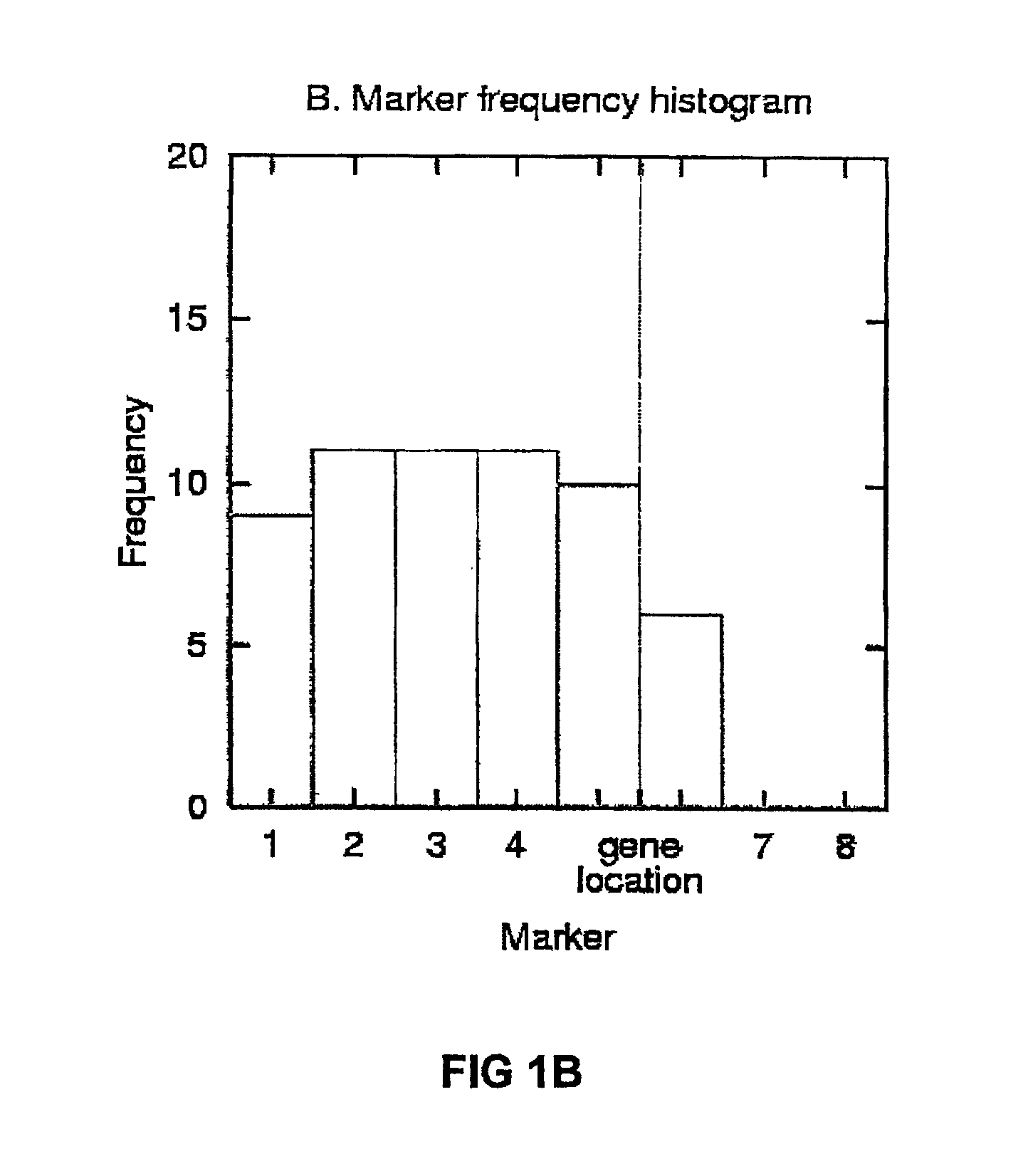
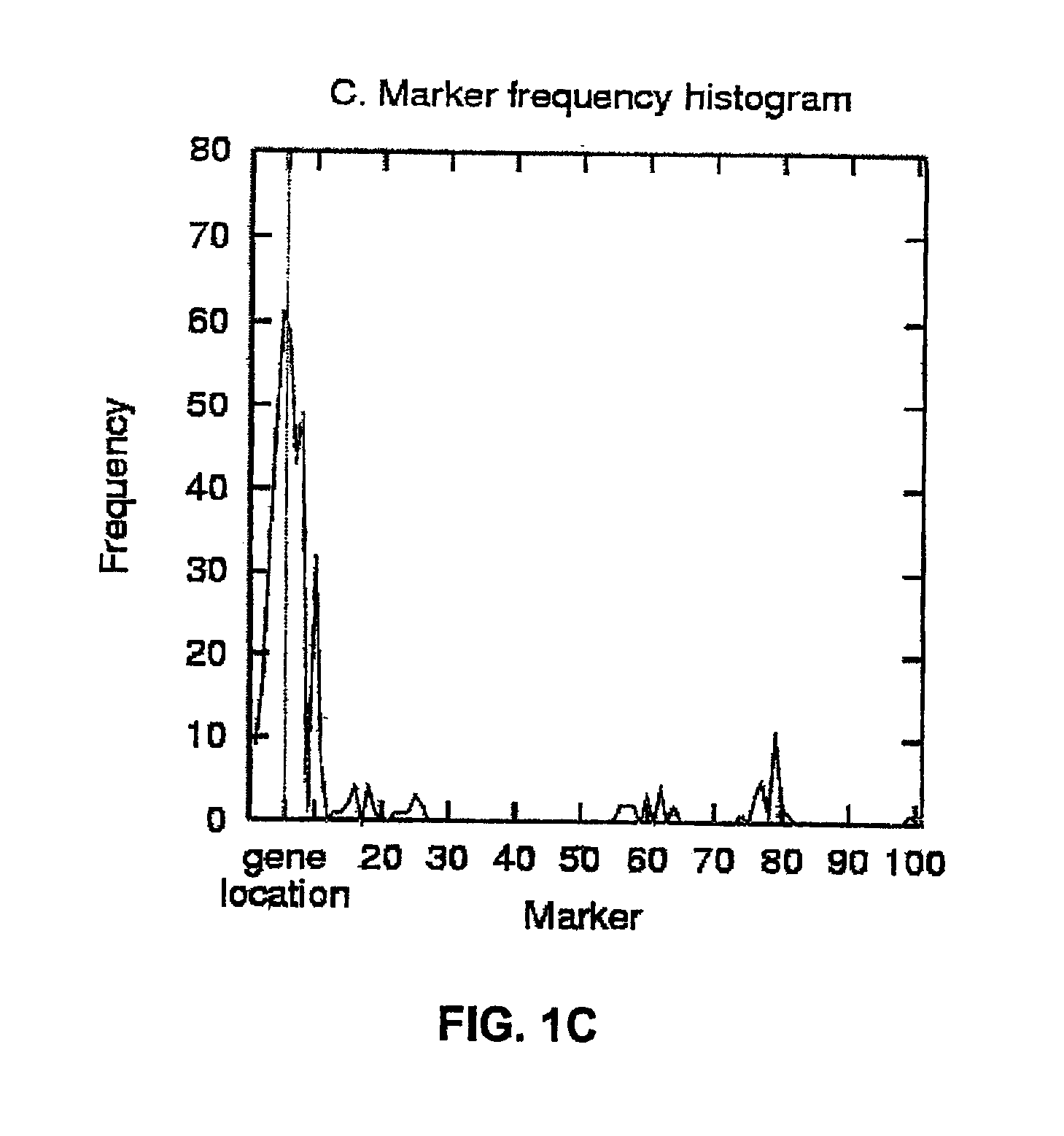
The present invention relates to a method for gene mapping from chromosome and phenotype data, which utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. All marker patterns P that satisfy a certain pattern evaluation function e(P) are searched from the data, each marker mi of the data is scored by a marker score and the location of the gene is predicted as a function of the scores s(mi) of all the markers mi in the data.
Owner:LICENTIA OY
Method for improving plant variety
InactiveUS20200008387A1Reliable methodMicrobiological testing/measurementPlant genotype modificationChromosomal regionGenetic stock
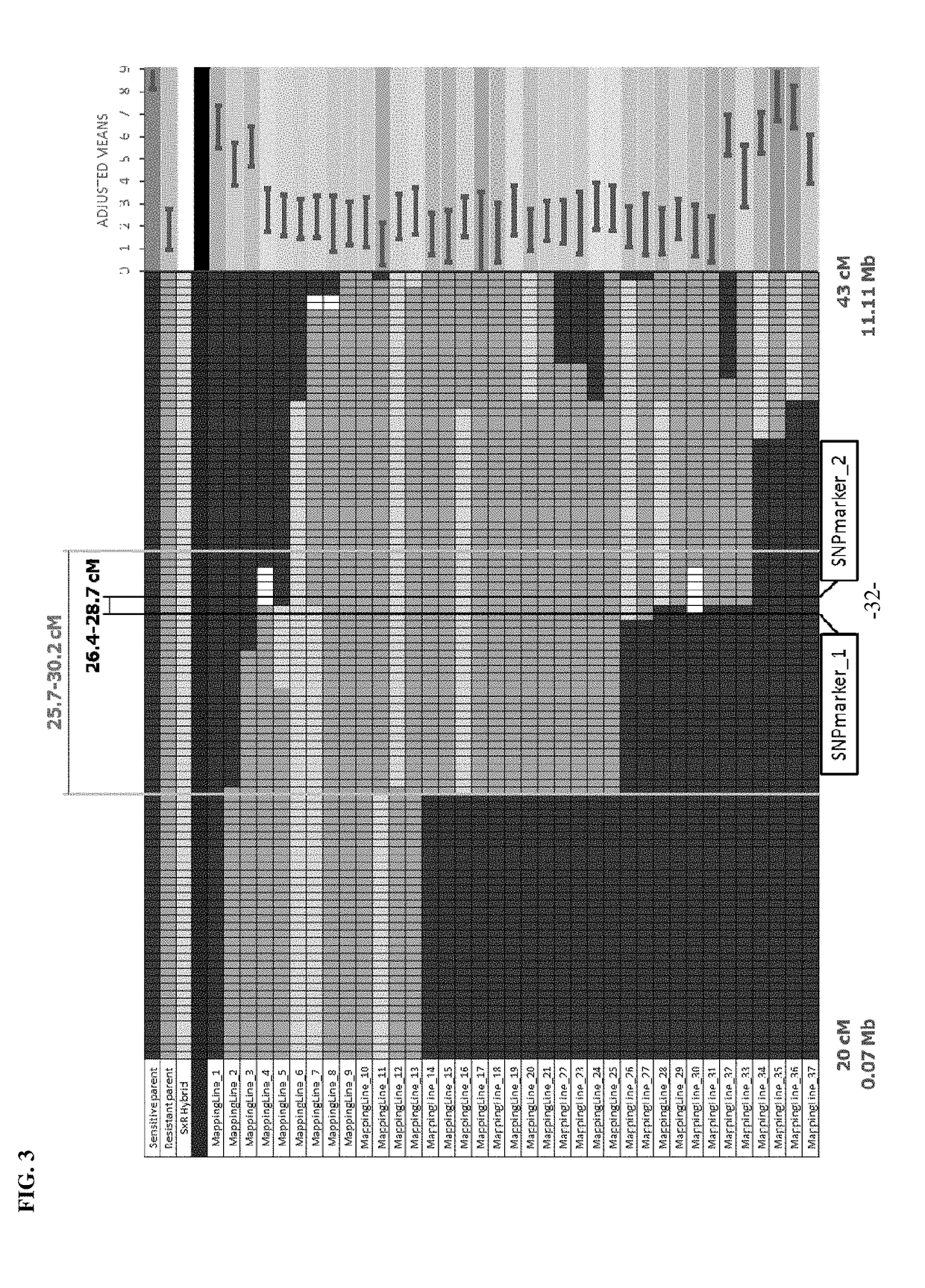
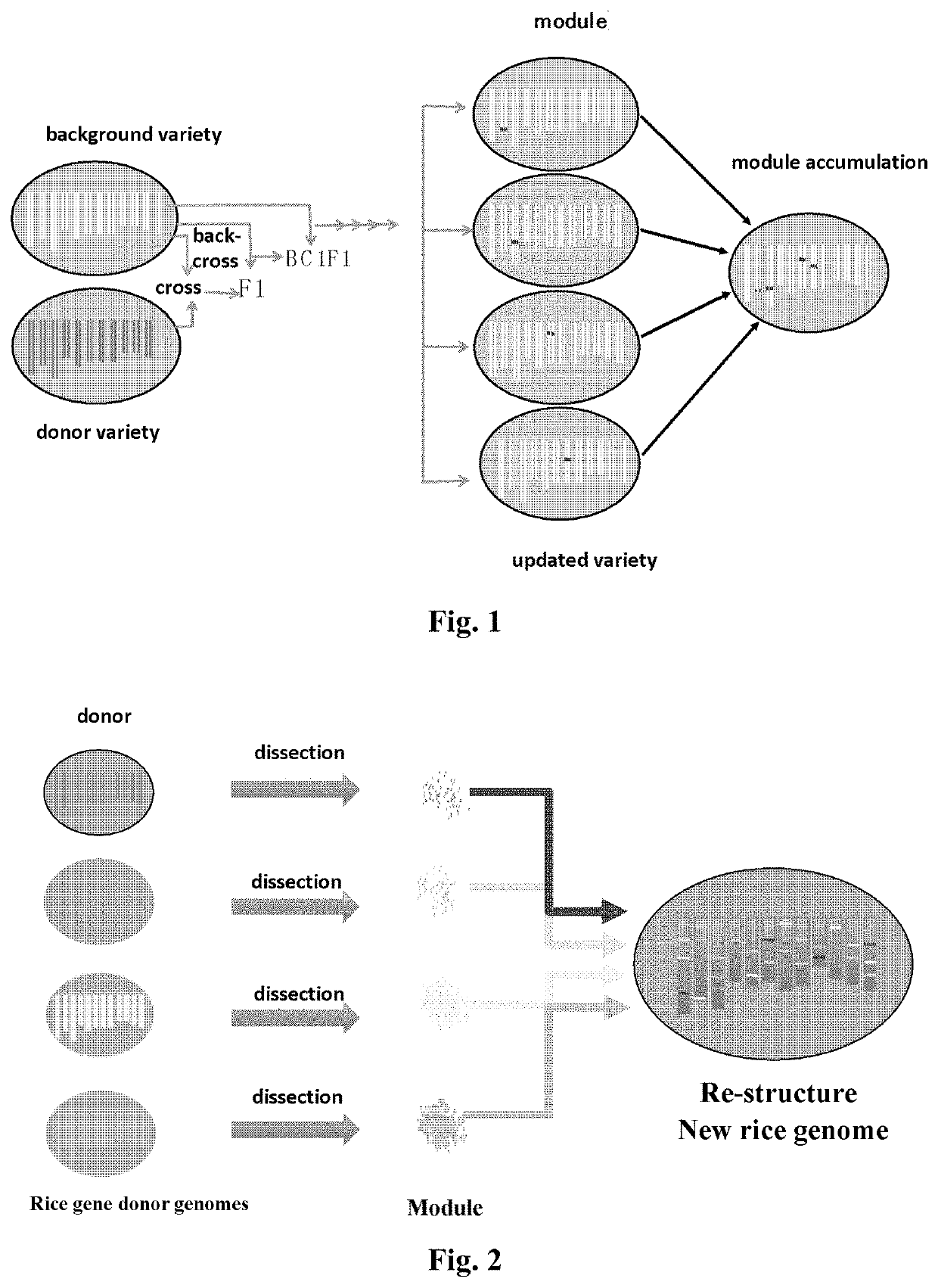
The present disclosure relates to a method for improving a plant variety. The present disclosure relates to a method of plant breeding, the method comprising the following steps:1) Selecting a background variety and a donor variety,2) Comparing the background variety and the donor variety to identify a module or locus to be improved,3) Crossing the background variety and the donor variety to obtain a hybrid progeny, backcrossing the hybrid progeny to the background variety to obtain a backcross progeny, and constructing a genetic population using the backcross progeny,4) Selecting, using molecular markers or a sequencing method, a backcross progeny having chromosomal regions derived from the background variety except for the module or locus to be improved, the molecular markers comprising genome molecular markers and module or locus molecular markers designed according to the selected module or locus,5) Self-crossing the selected backcross progeny to obtain an improved plant variety.The present disclosure also relates to a plant variety obtainable by said method.The method can select a plant in laboratory, obtaining a plant with a definite and improved trait and gene with high breeding efficiency, and achieving division of labour during the breeding process and accumulation of breeding advantages.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
Molecular marker linked to green retardation gene of pepper and application thereof
ActiveCN109207622ASolve the problem of long conventional breeding cycle and easy to be affected by the environmentSolve problems that are vulnerable to environmental influencesMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceF2 population
The present invention discloses a molecular marker linked to green retardation gene of hot pepper and its application. Two highly homozygous materials are used to construct the F2 population. The chromosomal regions closely linked to the brown (green-stagnant) gene of pepper fruits are obtained by BSR population mapping, and the candidate genes are identified by molecular markers in the candidateregion. A KASP molecular marker is designed according to the single base mutation of candidate gene. The genotype identification of 90 individual plants in F2 population by using this marker showed that the coincidence rate is 100%. The results not only contribute to the color identification and assisted breeding of pepper fruits, but also provide a basis for map-based cloning of green retardationgene and molecular mechanism of green retardation, which is worth popularizing widely.
Owner:HUNAN VEGETABLE RES INST
Detection of chromosomal region copy number changes to diagnose melanoma
Owner:RGT UNIV OF CALIFORNIA
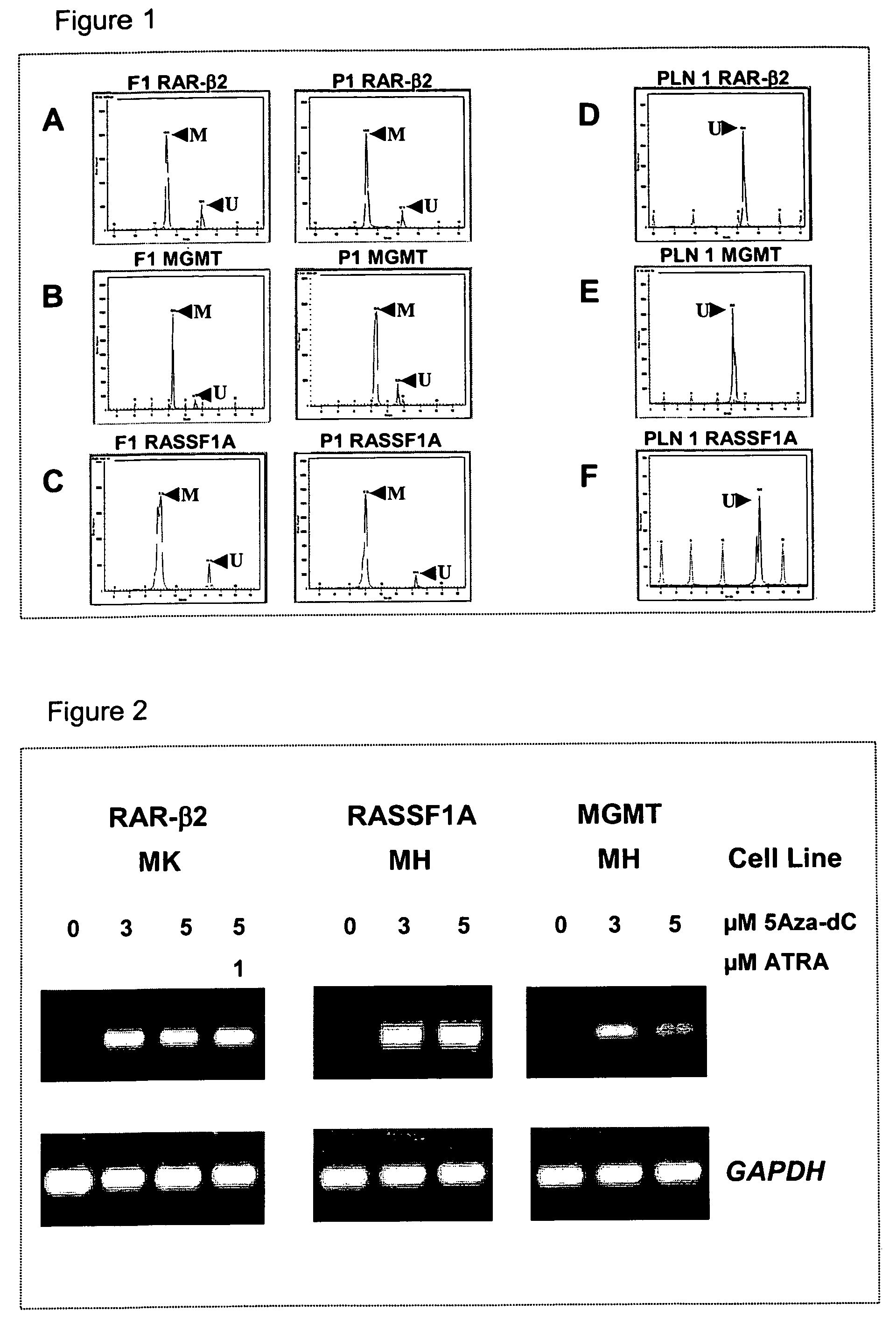
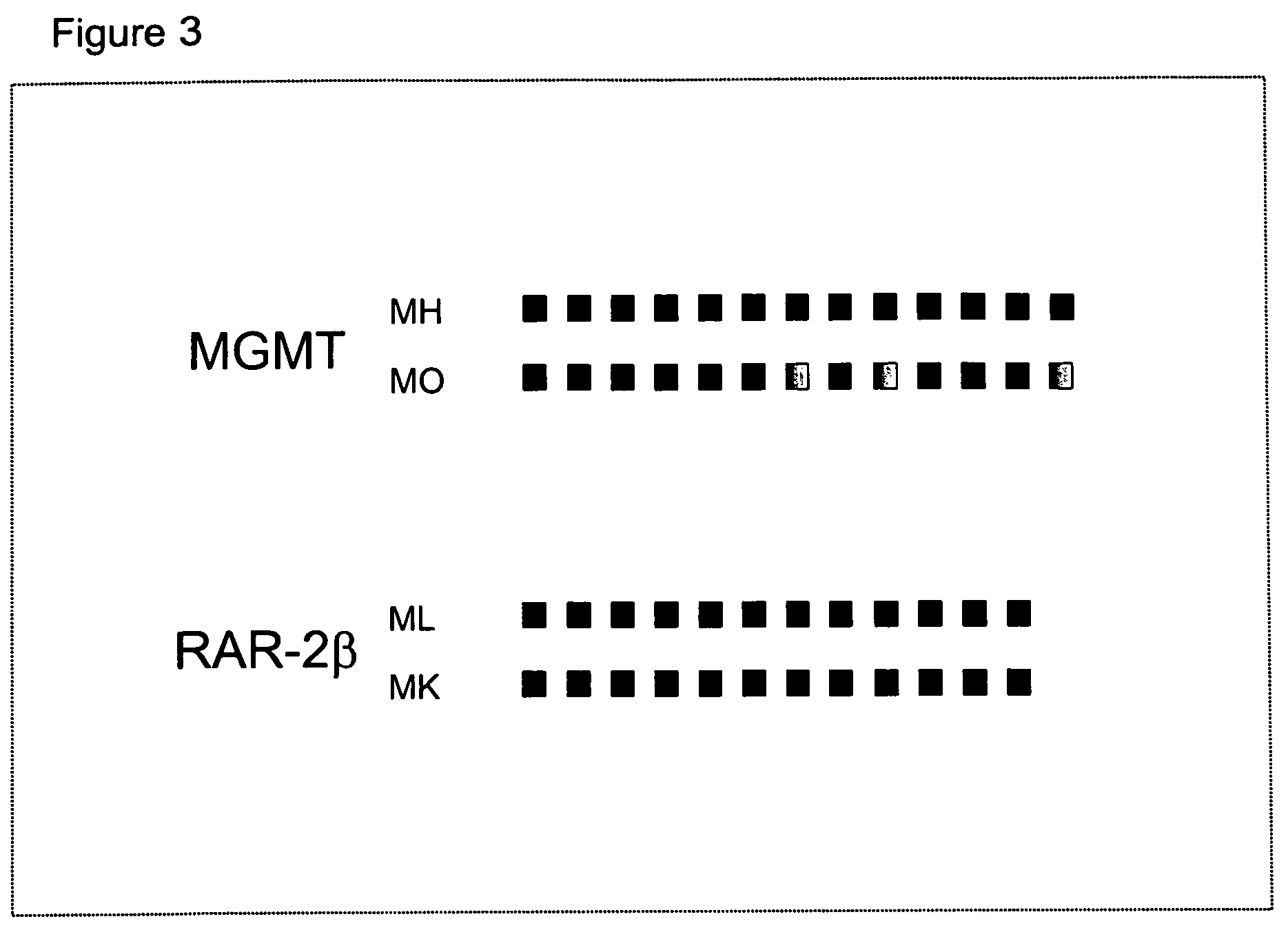
DNA markers for management of cancer
ActiveUS7718364B2Stage increaseMinimally invasiveSugar derivativesMicrobiological testing/measurementBlastomaSpecific chromosome
A method is provided for assessing allelic losses and hypermethylation of genes in CpG tumor promotor region on specific chromosomal regions in cancer patients, including melanoma, neuroblastoma breast, colorectal, and prostate cancer patients. The method relies on the evidence that free DNA and hypermethylation of genes in CpG tumor promotor region may be identified in the bone marrow, serum, plasma, and tumor tissue samples of cancer patients. Methods of melanoma, neuroblastoma, colorectal cancer, breast cancer and prostate cancer detection, staging, and prognosis are also provided.
Owner:JOHN WAYNE CANCER INST
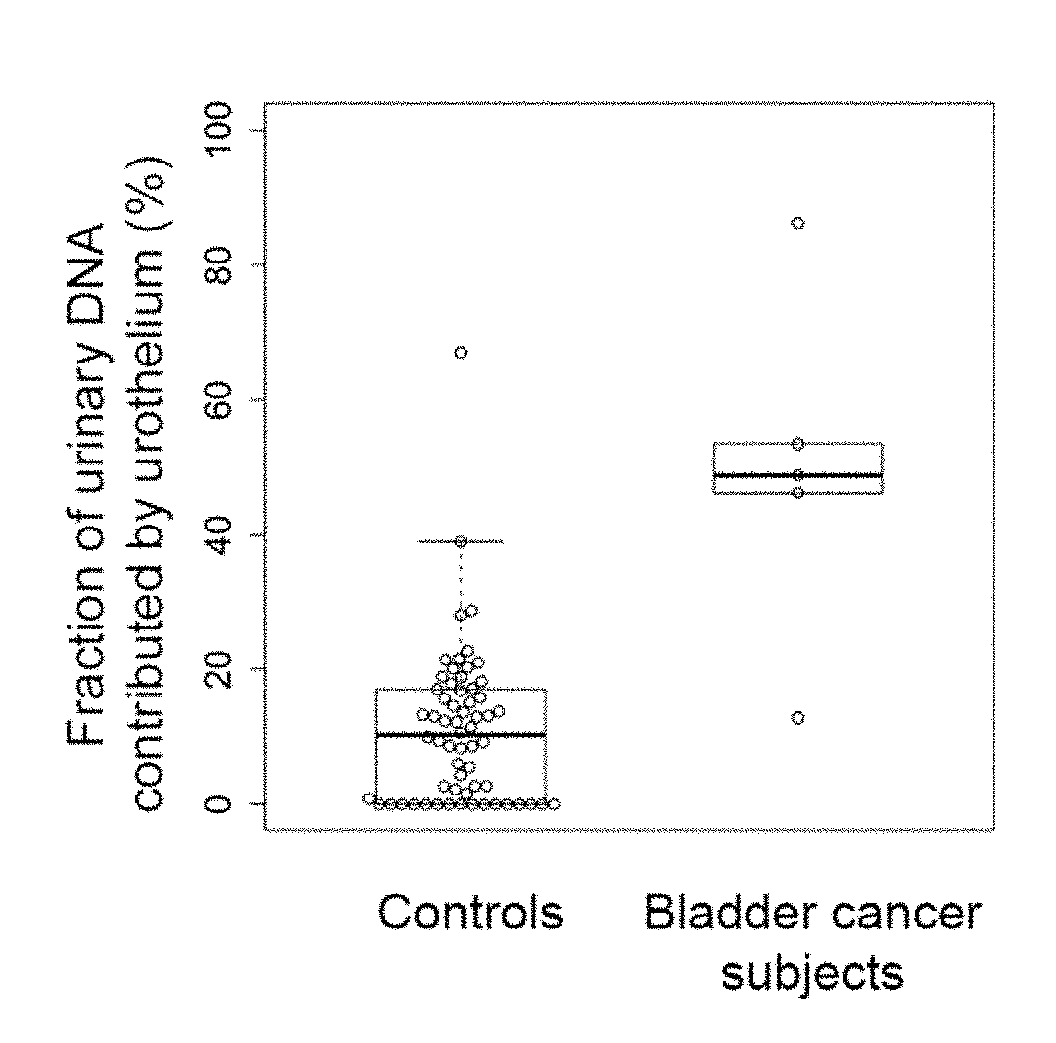
Analysis of cell-free DNA in urine and other samples
Diseases (e.g., cancer) of a particular organ can be detected by analyzing cell-free DNA. Some embodiments may use an organ-associated sample that is from a particular organ or passes through the particular organ, as may occur, for example, in urine, saliva, blood, and stool samples. In some embodiments, methylation levels of cell-free DNA can be measured in a sample. Tissue-specific methylation patterns can be used to determine fractional contributions from different tissue types. In other embodiments, sizes of organ-associated cell-free DNA can be measured. A statistical measure of the size profile may indicate that cell-free DNA fragments are collectively longer than expected for subjects with healthy tissue compared to non-healthy tissue. In other embodiments, two different samples can be analyzed to determine whether a particular organ has cancer. Cell-free DNA in a blood sample and organ-associated sample can both be analyzed to identify chromosomal regions exhibiting a copy number aberration.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
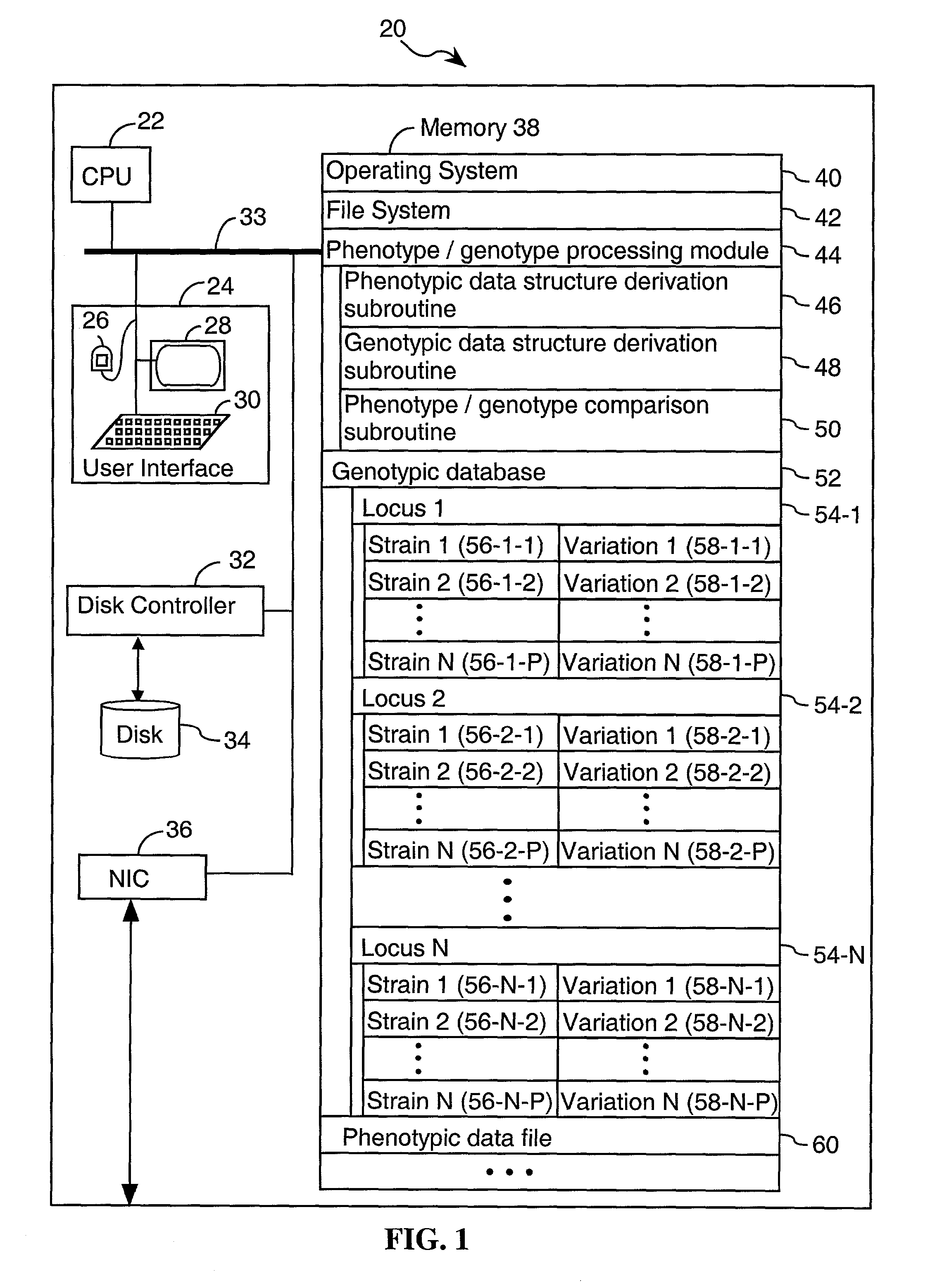
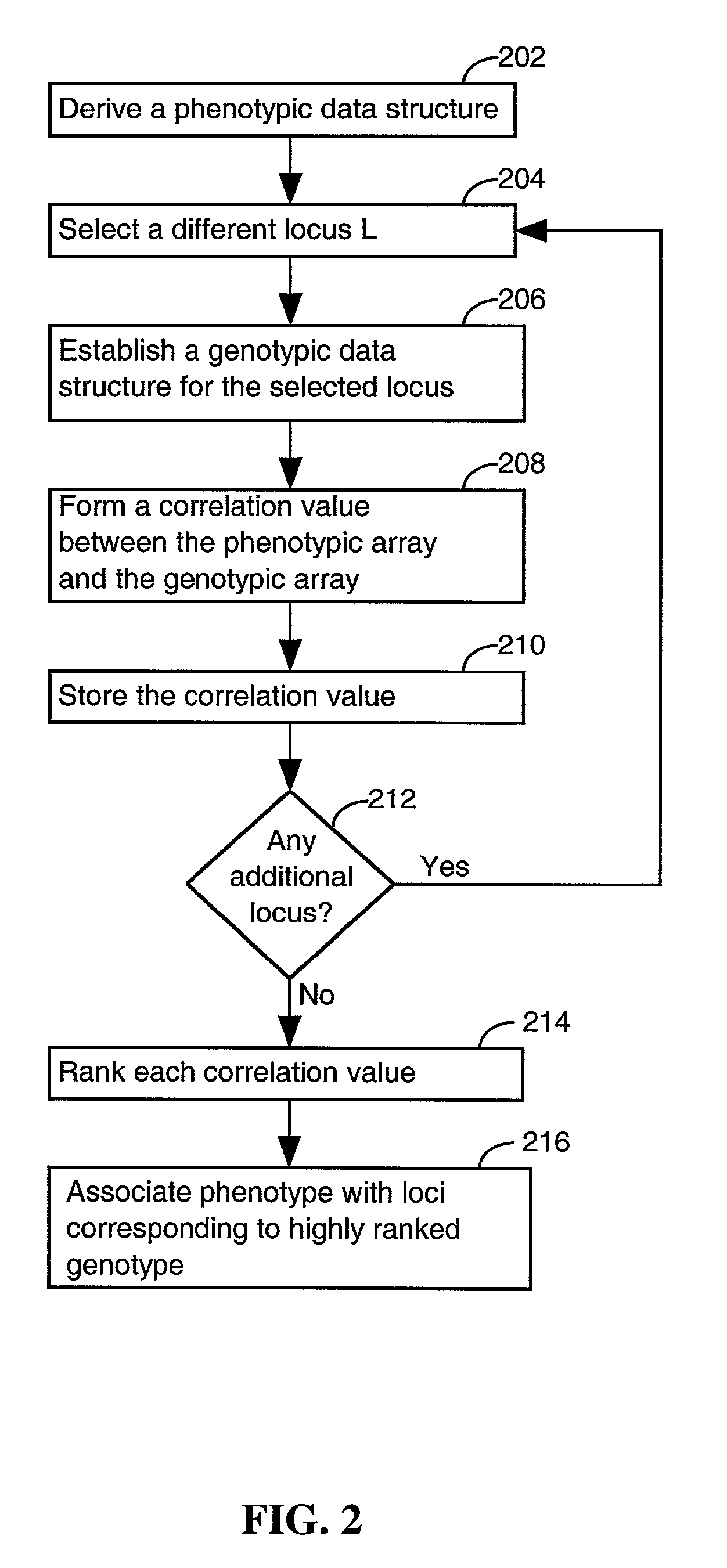
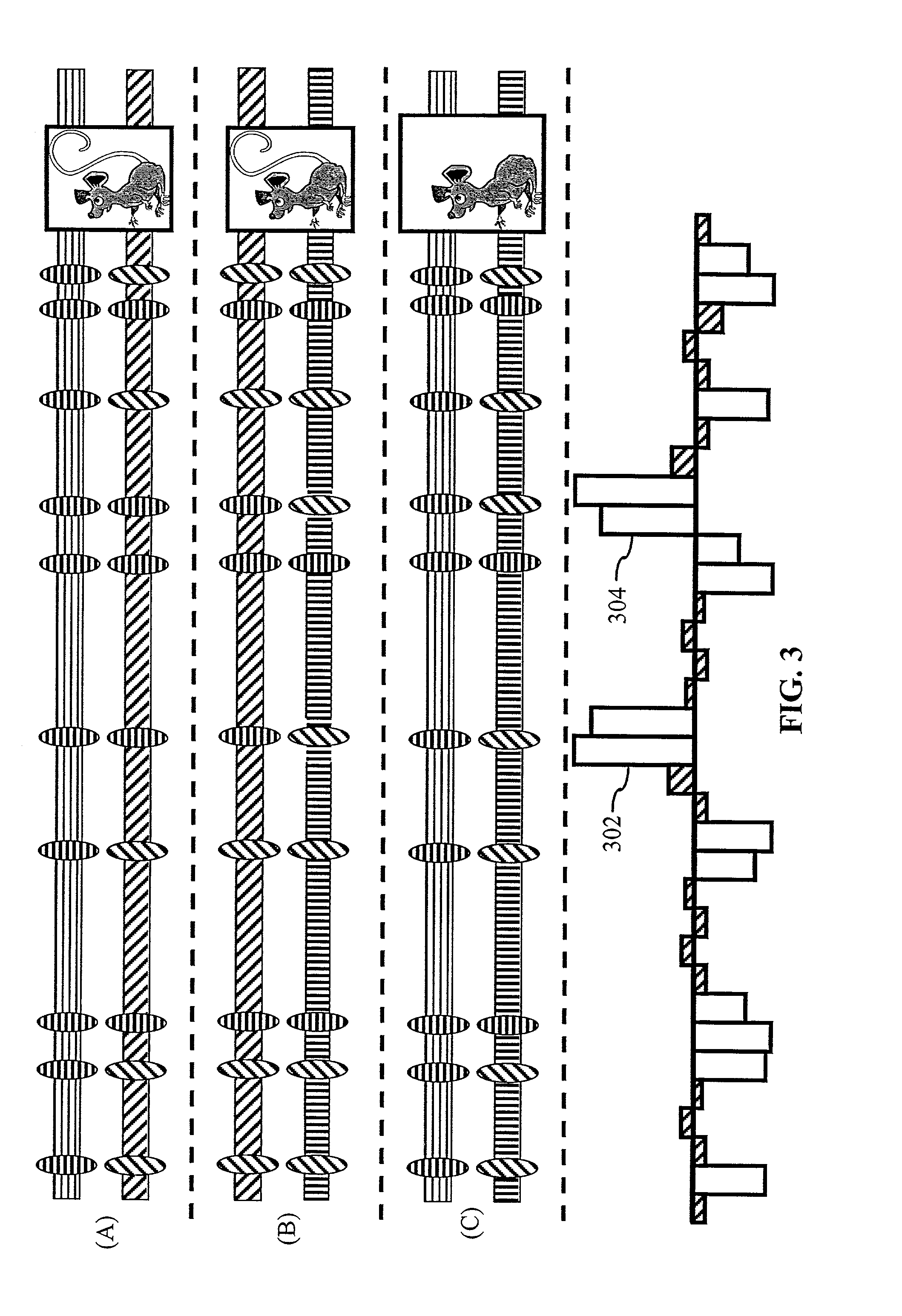
System and method for predicting chromosomal regions that control phenotypic traits
InactiveUS7698117B2Microbiological testing/measurementAnalogue computers for chemical processesBiological bodyPhenotypic trait
A method of associating a phenotype with one or more candidate chromosomal regions in a genome of an organism includes the step of deriving a phenotypic data structure that represents differences in phenotypes between different strains of the organism. Further, a genotypic data structure is established. The genotypic data structure corresponds to a locus selected from a plurality of loci in the genome of the organism. The genotypic data structure represents variations of at least one component of the locus between different strains of the organism. The phenotypic data structure is compared to the genotypic data structure to form a correlation value. The process of establishing a genotypic data structure and comparing it to the phenotypic data structure is repeated for each locus in the plurality of loci, thereby identifying one or more genotypic data structures that form a high correlation value relative to all other compared genotypic data structures. The loci that correspond to the one or more genotypic data structures having a high correlation value represent the one or more candidate chromosomal regions.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
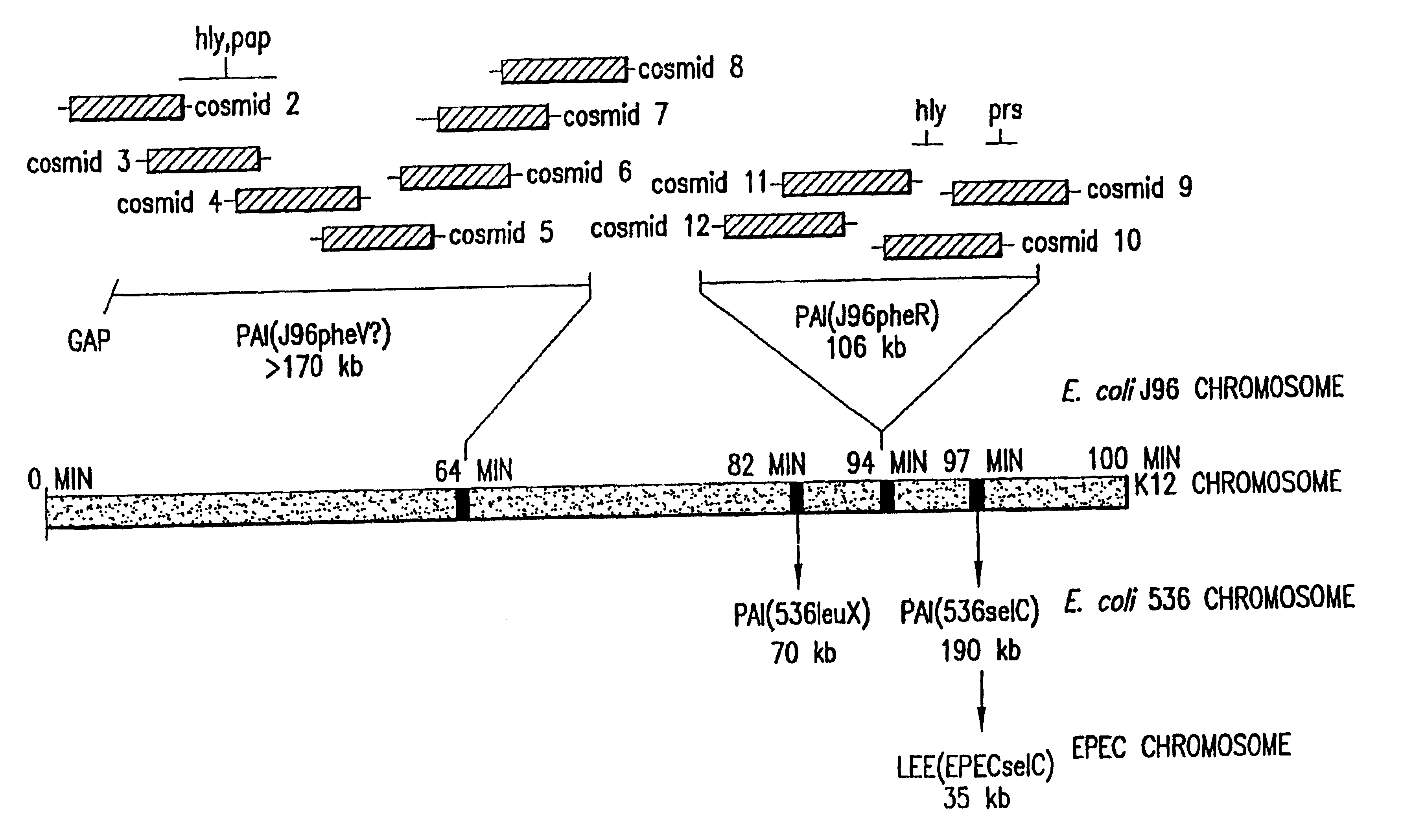
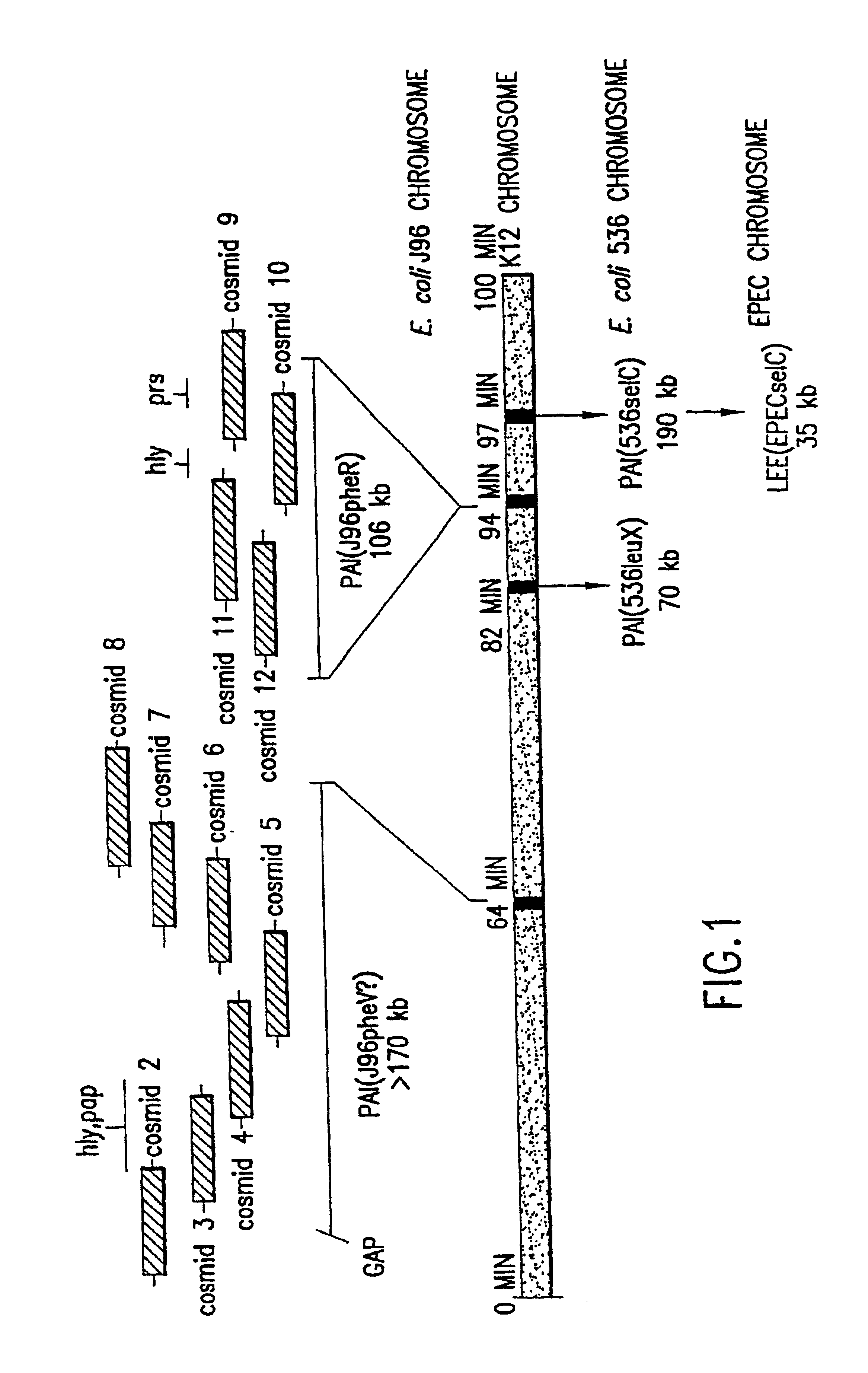
Nucleotide sequence of Escherichia coli pathogenicity islands
The present invention relates to novel genes located in two chromosomal regions within uropathogenic E. coli that are associated with virulence. These chromosomal regions are known as pathogenicity islands (PAIs). In particular, the present application discloses 142 sequenced fragments (contigs) of DNA from two pools of cosmids covering pathogenicity islands PAI IV and PAI V located on the chromosome of the uropathogenic Escherichia coli J96. Further disclosed are 351 predicted protein-coding open reading frames within the sequenced fragments.
Owner:HUMAN GENOME SCI INC +1
Somatic haploid human cell line
ActiveUS20170002380A1Speed up the conversion processEven mixtureMicrobiological testing/measurementStable introduction of DNAGenomic mutationChromosomal region
The invention provides for a somatic fully haploid, karyotypically stable human cell line, e.g. obtainable by targeted deletion of one or more disomic chromosomal regions of a somatic near-haploid human parental cell, a method of producing the same, as well as the use of the cell line for producing isogenic cell variants, comprising genomic mutations at different genomic target sites, and a library of such cell variants.
Owner:HORIZON DISCOVERY
Molecular markers linked to ppo inhibitor tolerance in soybeans
ActiveUS20100024080A1Increase probabilityHigh yieldBiocideMicrobiological testing/measurementGenotypingChromosomal region
This invention relates generally to the detection of genetic differences among soybeans. More particularly, the invention relates to soybean quantitative trait loci (QTL) for tolerance to protoporphyrinogen oxidase inhibitors, to soybean plants possessing these QTLs, which map to a novel chromosomal region, and to genetic markers that are indicative of phenotypes associated with protoporphyrinogen oxidase inhibitor tolerance. Methods and compositions for use of these markers in genotyping of soybean and selection are also disclosed.
Owner:PIONEER HI BRED INT INC
Method for gene mapping from genotype and phenotype data
InactiveUS20050250098A1Model-free and computationally effectiveOvercomes drawbackMicrobiological testing/measurementBiostatisticsGene mappingGenetic linkage disequilibrium
A method for gene mapping from genotype and phenotype data utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. All marker patterns P that satisfy a certain pattern evaluation function e(P) are searched from the data, each marker mi of the data is scored by a marker score and the location of the gene is predicted as a function of the scores s(mi) of all the markers mi in the data.
Owner:LICENTIA OY
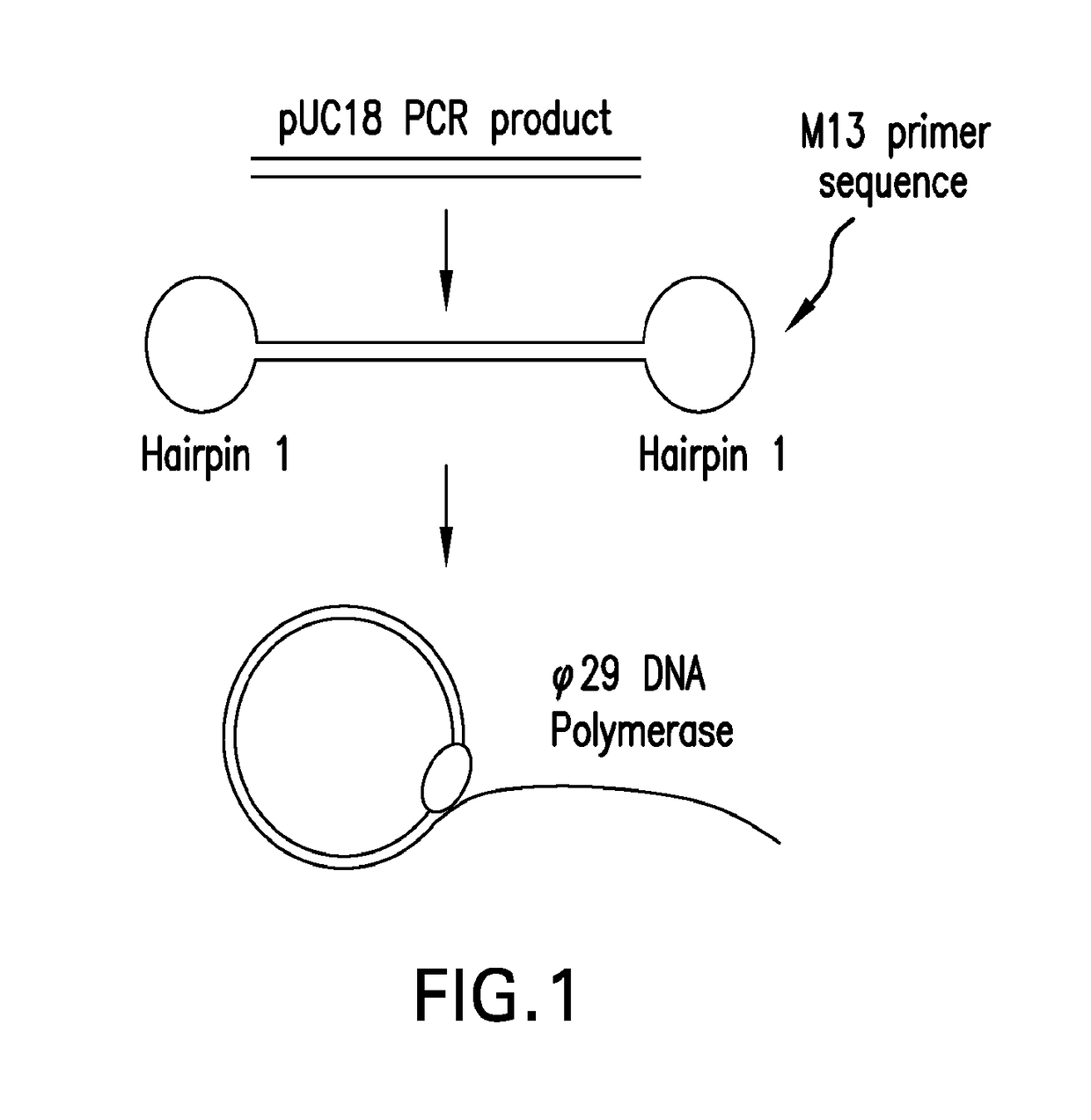
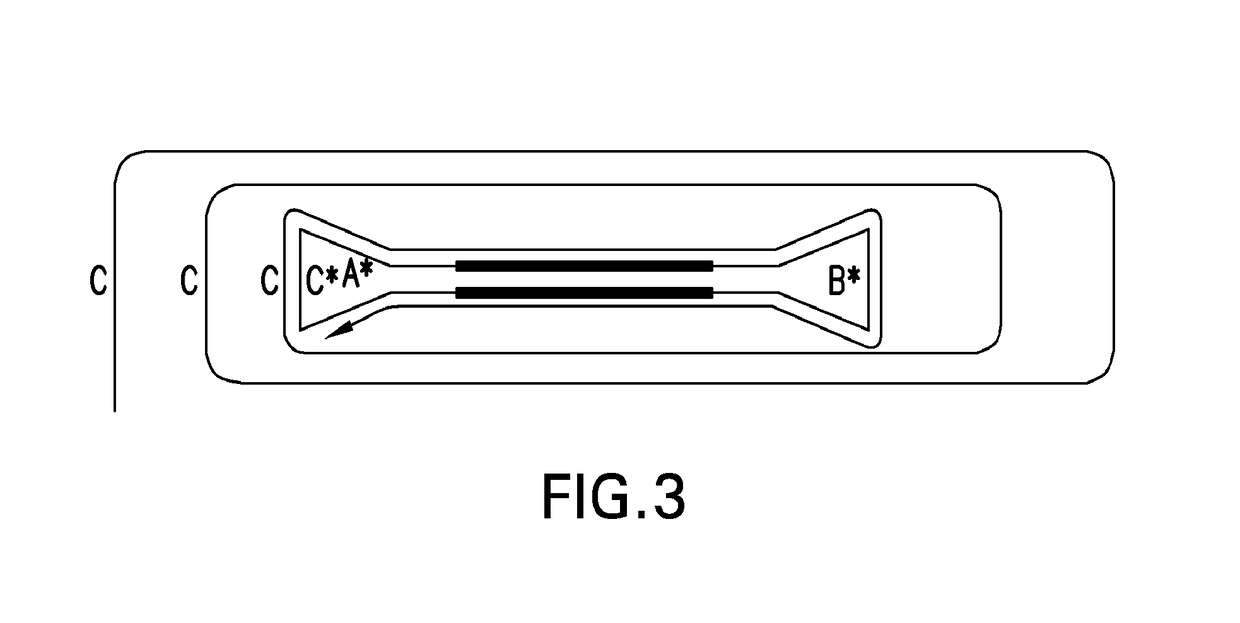
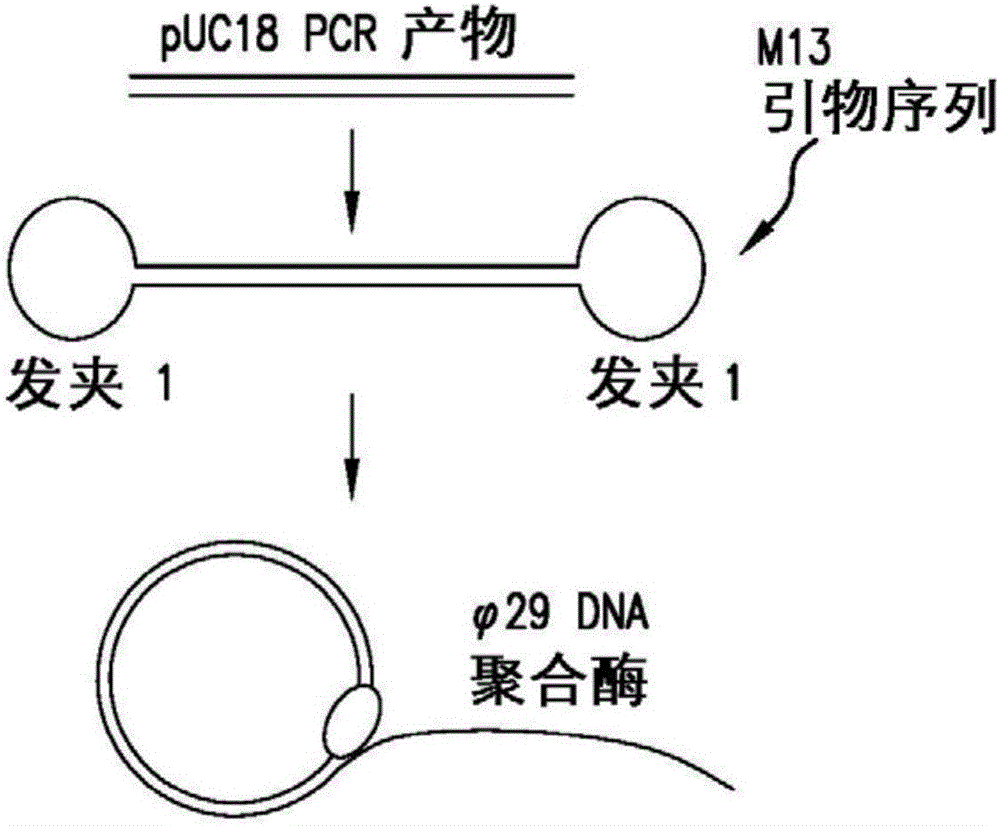
Systems and methods for clonal replication and amplification of nucleic acid molecules for genomic and therapeutic applications
PendingUS20170067097A1Efficient productionCreate efficientlyMicrobiological testing/measurementChromosomal regionHaplotype
The present invention provides for methods, reagents, apparatuses, and systems for the replication or amplification of nucleic acid molecules from biological samples. In one embodiment of the invention, the nucleic molecules are isolated from the sample, and subjected to fragmenting and joining using ligating agents of one or more hairpin structures to each end of the fragmented nucleic molecules to form one or more dumbbell templates. The one or more dumbbell templates are contacted with at least one substantially complementary primer attached to a substrate, and subjected to rolling circle replication or rolling circle amplification. The resulting replicated dumbbell templates or amplified dumbbell templates are used in numerous genomic applications, including whole genome de novo sequencing; sequence variant detection, structural variant detection, determining the phase of molecular haplotypes, molecular counting for aneuploidy detection; targeted sequencing of gene panels, whole exome, or chromosomal regions for sequence variant detection, structural variant detection, determining the phase of molecular haplotypes and / or molecular counting for aneuploidy detection; study of nucleic acid-nucleic acid binding interactions, nucleic acid-protein binding interactions, and nucleic acid molecule expression arrays; and testing of the effects of small molecule inhibitors or activators or nucleic acid therapeutics.
Owner:REDVAULT BIOSCI
Systems and methods for clonal replication and amplification of nucleic acid molecules for genomic and therapeutic applications
The present invention provides for methods, reagents, apparatuses, and systems for the replication or amplification of nucleic acid molecules from biological samples. In one embodiment of the invention, the nucleic molecules are isolated from the sample, and subjected to fragmenting and joining using ligating agents of one or more hairpin structures to each end of the fragmented nucleic molecules to form one or more dumbbell templates. The one or more dumbbell templates are contacted with at least one substantially complementary primer attached to a substrate, and subjected to rolling circle replication or rolling circle amplification. The resulting replicated dumbbell templates or amplified dumbbell templates are used in numerous genomic applications, including whole genome de novo sequencing; sequence variant detection, structural variant detection, determining the phase of molecular haplotypes, molecular counting for aneuploidy detection; targeted sequencing of gene panels, whole exome, or chromosomal regions for sequence variant detection, structural variant detection, determining the phase of molecular haplotypes and / or molecular counting for aneuploidy detection; study of nucleic acid - nucleic acid binding interactions, nucleic acid - protein binding interactions, and nucleic acid molecule expression arrays; and testing of the effects of small molecule inhibitors or activators or nucleic acid therapeutics.
Owner:REDVAULT BIOSCI
Predictive and therapeutic markers in ovarian cancer
ActiveUS8404829B2Improve the level ofReduced survivalSugar derivativesMicrobiological testing/measurementApoptosisIn vivo
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com