Systems and methods for clonal replication and amplification of nucleic acid molecules for genomic and therapeutic applications
A technique for nucleic acid molecule and rolling circle amplification, applied in the field of replication and amplification of nucleic acid molecule
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
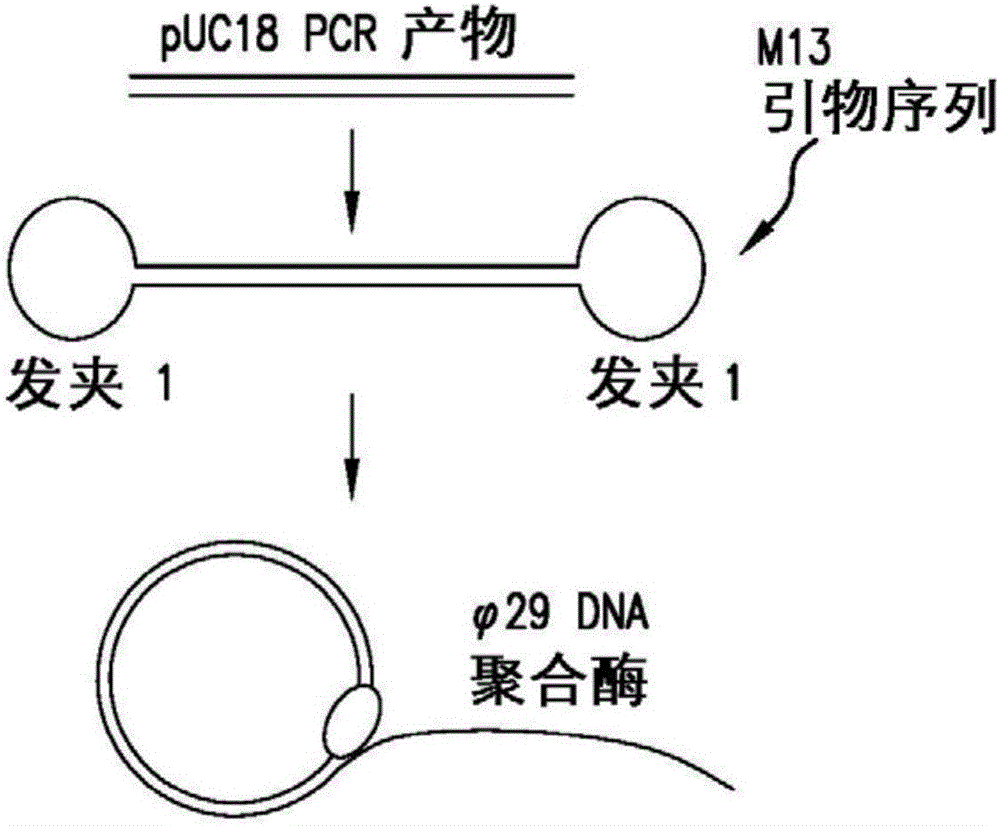
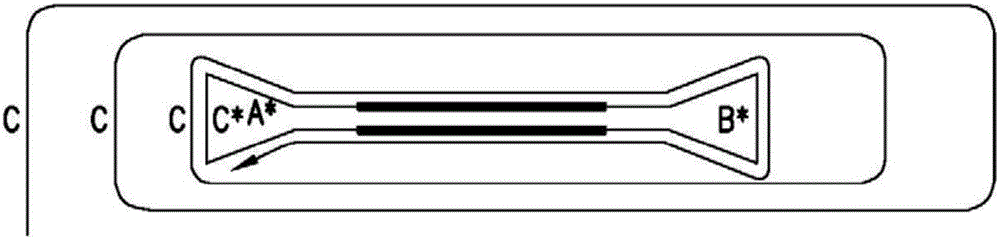
[0116] Demonstration of size independence for dumbbell-shaped templates containing two distinct hairpin structures. Use a set of primers (i.e. forward: 5'-GGA TCC GAA TTC GCT GAA GCC AGT TAC CTT CG and reverse: 5'-GGA TCC GAA TTCAGC CCT CCC GTA TCG TAG TT) to amplify the sample DNA, i.e. the pUC18 vector to A product with 425 base pairs was obtained. The 5'-end of each primer contains a BamHI restriction endonuclease site and an EcoRI restriction endonuclease site. The PCR product was then digested with EcoRI to provide a 5'-AATT overhang and purified with the QIAquick PCR purification kit. A 5'-AATT overhang is generated by heating to 50°C followed by cooling to allow the oligonucleotides to self-anneal at the underlined sequence to form a hairpin structure 1 (5'-AATT GCGAG TTG CGA GTT GTA AAA CGA CGG CCA GT CTCGC ). The loop structure contains the M13 universal primer sequence. The hairpin structure 1 and pUC18 PCR products were combined at a molar ratio of 10:1, res...
Embodiment 2
[0120] Genomic DNA can also be used as a starting sample. By way of example and without limitation, purified genomic DNA from HapMap sample NA18507 can be obtained from Coriell Cell Repositories and sheared using standard next-generation sequencing methods (i.e., using a Covaris E210R device), then Fragments were size selected in size increments of 0.5 kb, 1.0 kb, 2.5 kb, 5.0 kb, 7.5 kb, and 10.0 kb. Similar to above, DNA samples can be fragmented to generate DNA fragments of different sizes, the starting number of fragments quantified, hpA / hpB ligated using the same conditions, and enriched for hpA-fragment-hpB dumbbell templates . Enrichment factors will be determined using double-label fluorescence microscopy to count co-localized fluorescent signals and comparing this number to the total number of fluorescent signals. The Nikon Eclipse Microscopy Analysis Tool can perform many analyses, including intensity measurements, colocalization of multiple fluorescent signals, and...
Embodiment 3
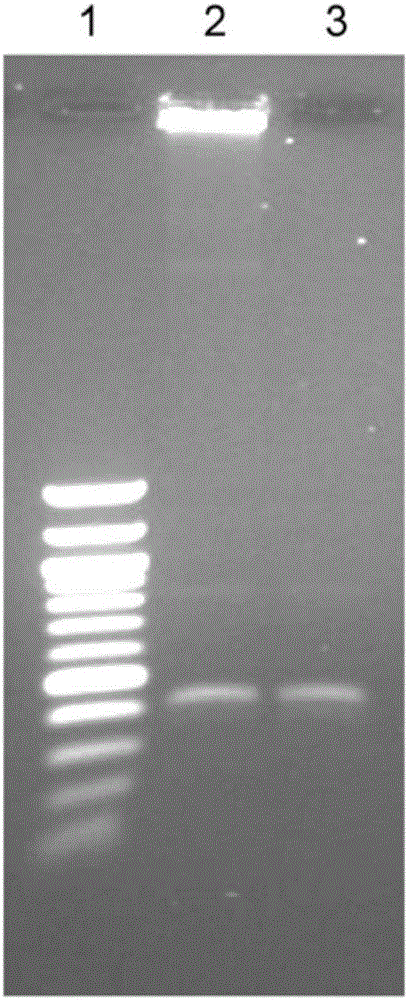
[0124] Dumbbell templates for replication are also formed from large fragmented dA-tailed genomic DNA. Here, hairpins were ligated by TA cloning and blunt end ligation. TA cloning methods are well integrated into most current NGS platforms. We have engineered hairpin 2 (HP2) (5'- / Phos- CTTTTTTCTTTCTTTTCTGGGTTGCGTCTGTTCGTCT AGAAAAGAAAGAAAAAG T). Human genomic DNA (500ng) was fragmented using Covaris G-tubes to achieve a tightly defined population of fragment lengths, such as Figure 5 Lanes 1 and 2 are shown. This genomic DNA was then end-repaired and dA-tailed using the End Preparation Module of the NEBNext Ultra DNA Library Prep Kit. HP2 was self-annealed similarly to HP1 and ligated to repaired genomic DNA using Blunt / TA Ligase Mater Mix (5:1 molar ratio). Excess HP2 and unligated genomic DNA were removed using exonuclease III and VII.
[0125] The resulting dumbbell-shaped template was purified using Qiaex ii beads and an RCR reaction using a unique primer (5'-AAAA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com