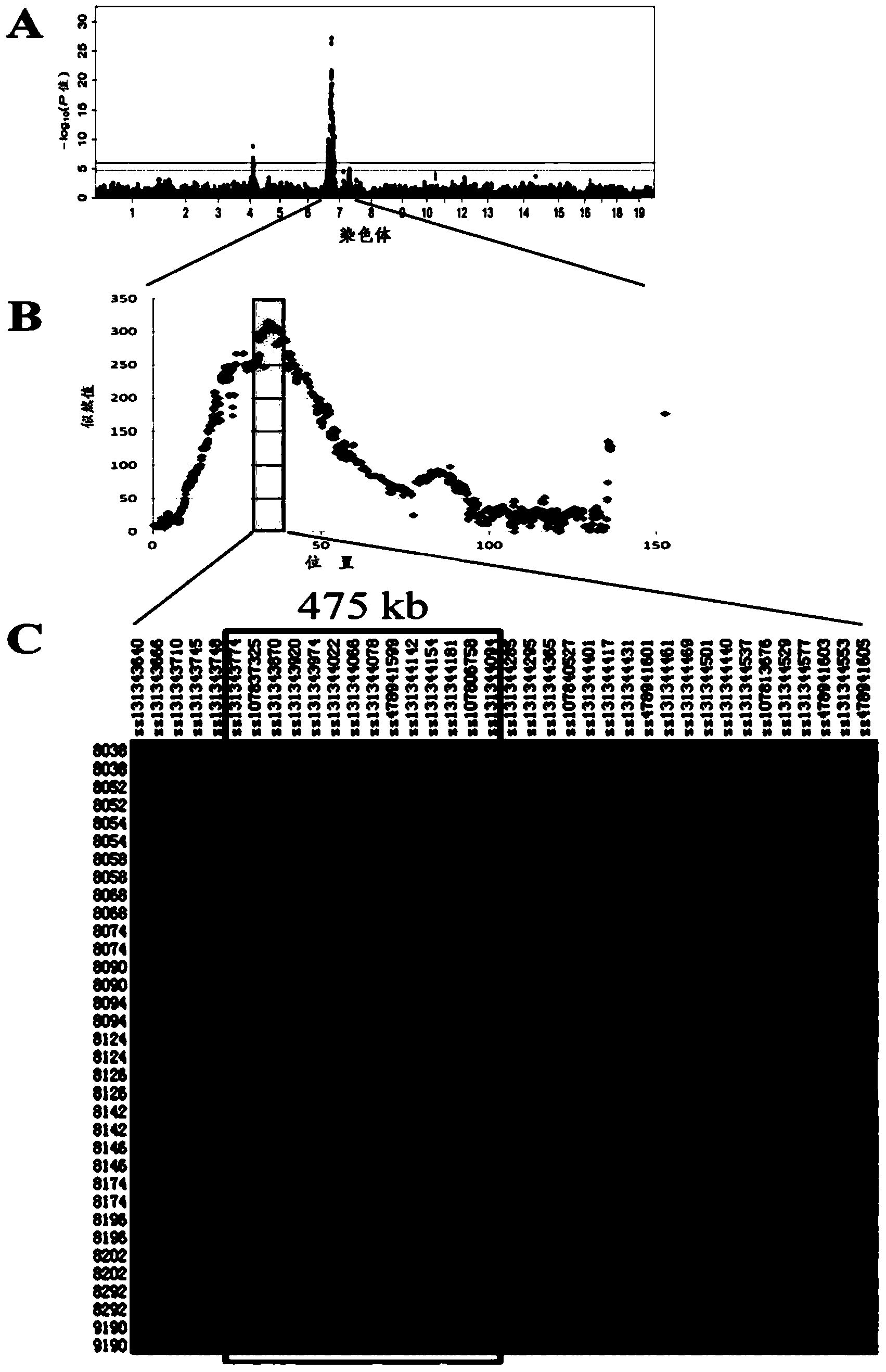
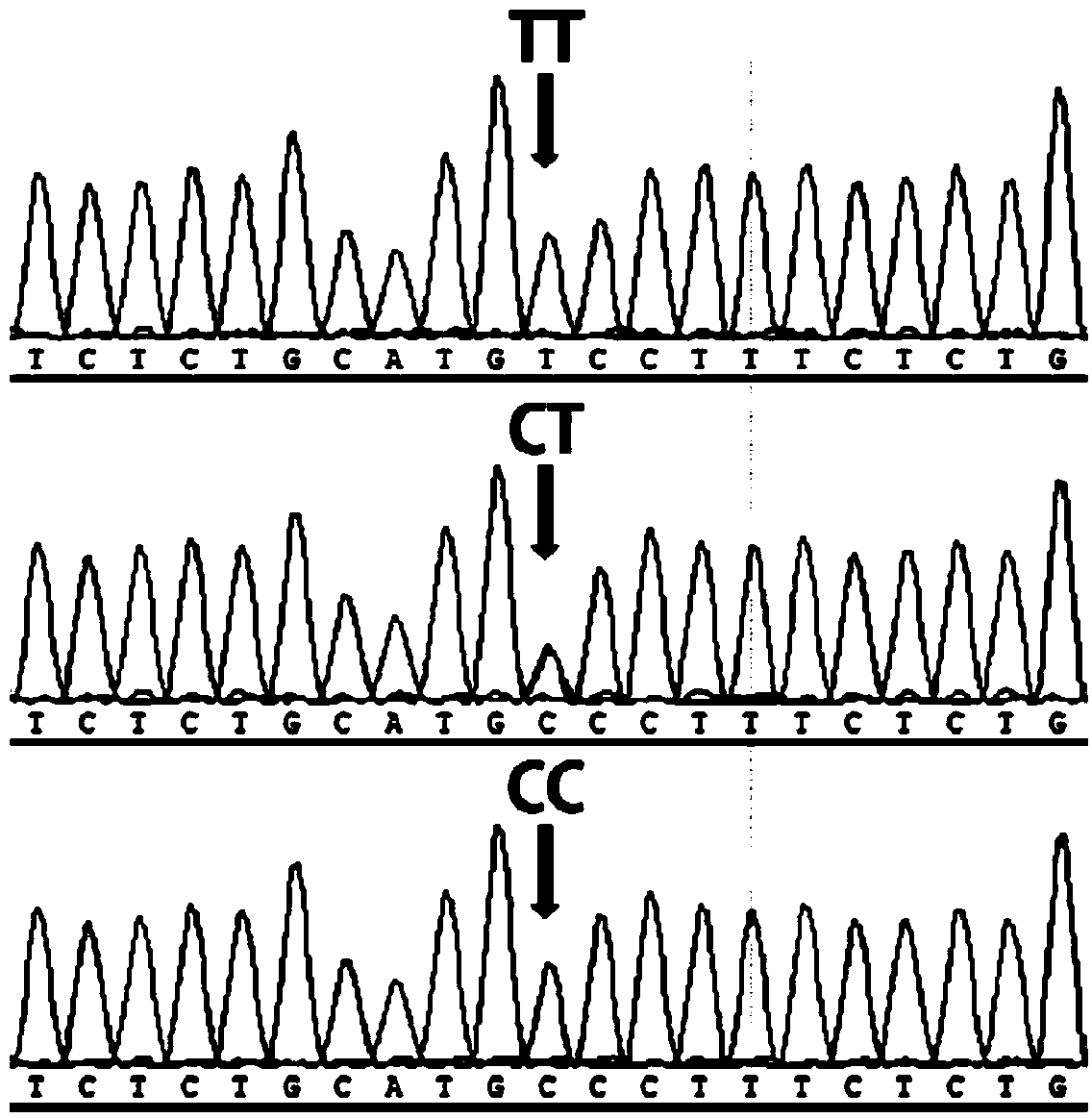
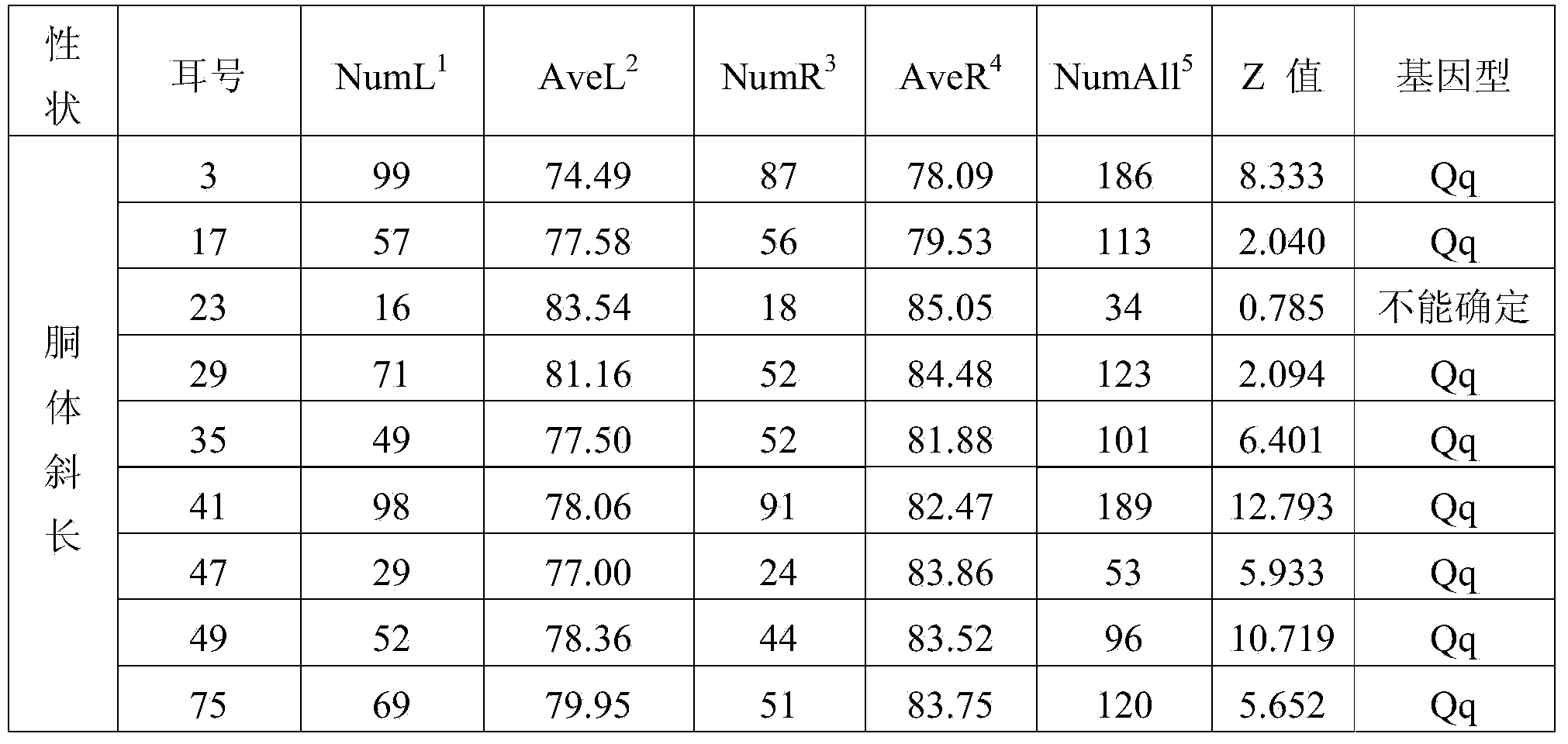
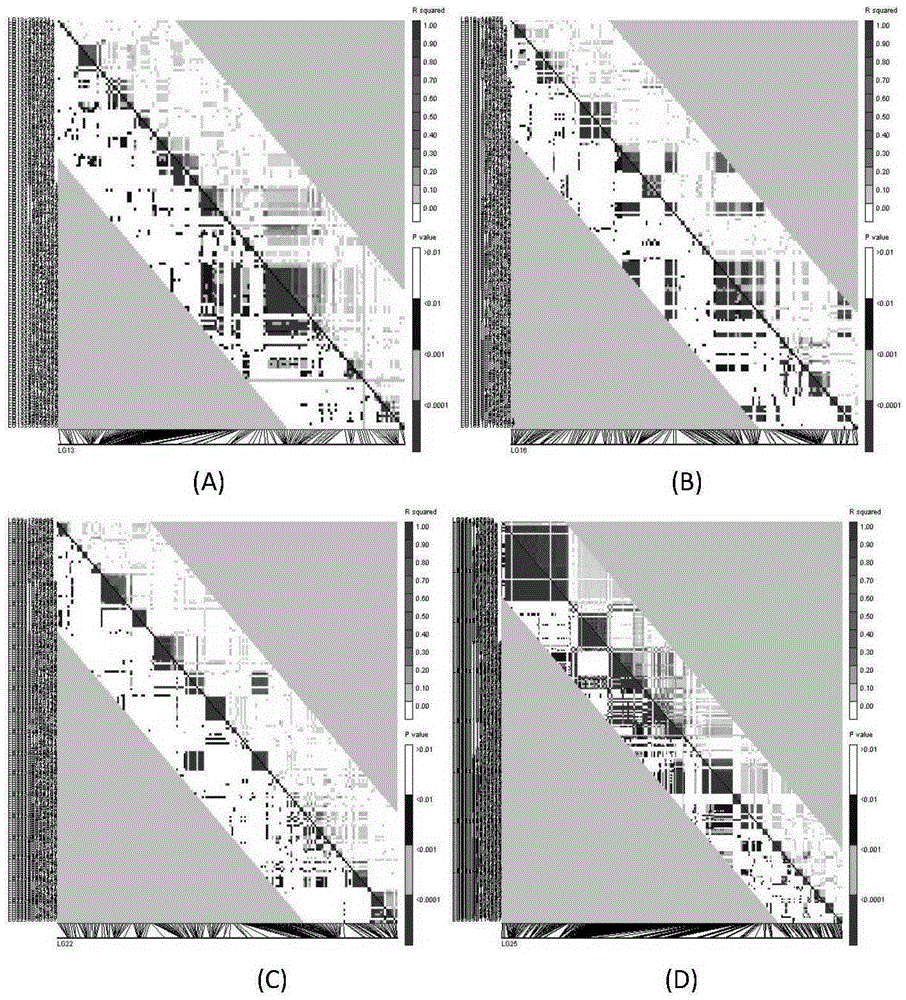
Patents
Literature
81 results about "Genetic linkage disequilibrium" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Linkage disequilibrium is influenced by many factors, including selection, the rate of genetic recombination, mutation rate, genetic drift, the system of mating, population structure, and genetic linkage. As a result, the pattern of linkage disequilibrium in a genome is a powerful signal of the population genetic processes that are structuring it.
Major SNP (single nucleotide polymorphism) marker influencing growth traits of pigs and application thereof in genetic improvement of productivity of breeding pigs
The invention provides a major SNP (single nucleotide polymorphism) marker influencing the growth traits of pigs. The SNP marker is located at a nucleotide sequence of an HMGA1 gene of a pig chromosome 7, a locus of the SNP marker is the nucleotide mutation of C857-G857 with an SEQ ID NO:1 sequence labeling position of 857, corresponding to a 34983991st nucleotide locus C> on chromosome 7 in a reference sequence in an international pig genome version 10.2, G mutation, or one of seven other loci completely linked with the locus. The invention also provides the application of the SNP marker in the genetic improvement of growth traits of breeding pigs, and provides the application of an SNP molecular marker with a linkage disequilibrium degree (r2) with the SNP marker of greater than 0.8 in the genetic improvement of growth traits of breeding pigs. The growth traits include one or more of the length and height of a living body of a pig, carcass length, carcass weight, daily gain and head weight. According to the invention, the breeding process of breeding pigs can be accelerated, the productivity of breeding pigs can be effectively improved, and remarkable economic benefits can be obtained.
Owner:JIANGXI AGRICULTURAL UNIVERSITY
Upland cotton SNP marker and application thereof
InactiveCN105349537AImprove detection efficiencyImprove work efficiencyNucleotide librariesMicrobiological testing/measurementAgricultural scienceHomologous sequence
The invention discloses an upland cotton SNP marker and application thereof, and belongs to the field of upland cotton SNP marker development. The upland cotton SNP marker is obtained by developing a single-copy SNP marker of an upland cotton genome via chip hybridization and homologous sequence comparison, and picking out SNP markers in a linkage disequilibrium recession distance via linkage disequilibrium analysis. The SNP marker provided by the invention can be applied to analysis of genetic diversity and group structure, or to germplasm identification of upland cotton, is single-copy, and has no remarkable linkage disequilibrium among markers, so that the efficiency of detection, adopting the SNP marker, during analysis of genetic diversity, group structure or fingerprint identification can be greatly improved, and the work efficiency is higher.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Chinese population linkage analysis single nucleotide polymorphism (SNP) marker sets and use method and application thereof
InactiveCN102121046AEnsure reliabilityEnsure comprehensivenessMicrobiological testing/measurementDNA/RNA fragmentationGenetic linkage disequilibriumInternational HapMap Project
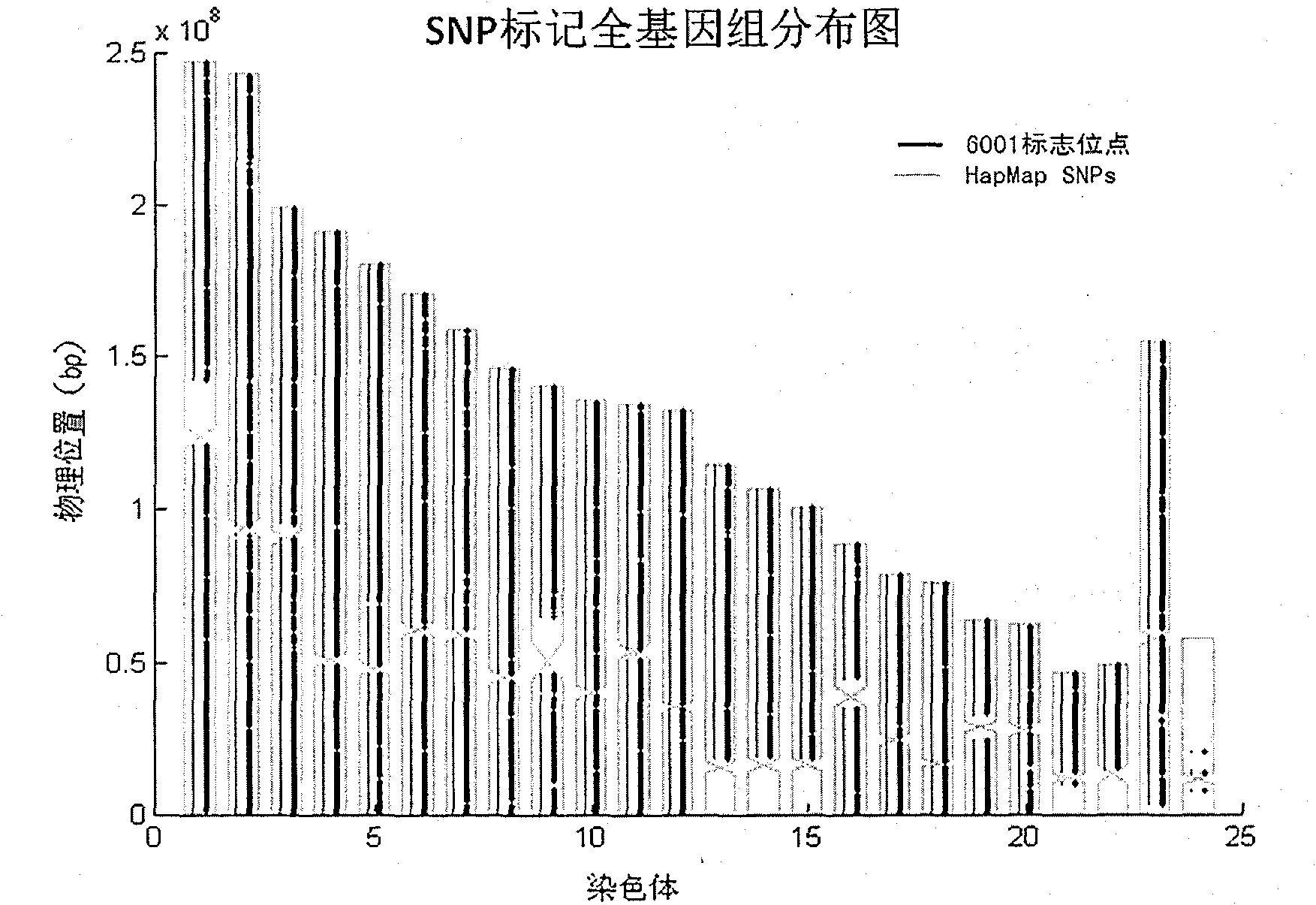
The invention relates to Chinese population linkage analysis single nucleotide polymorphism (SNP) marker sets and a use method and application thereof. On the basis of hundreds of millions of Chinese Han population data results in the mass data of the International HapMap Project, medium-density and high-density SNP marker sets for linkage analysis are constructed and optimized, according to the statistical comparisons of multiple parameters such as the linkage disequilibrium, the polymorphism level, the typing success rate, the distribution position and density of genomes and the functional characteristic, and the multi-level selections and experimental verifications. The two marker sets separately contain 3000 and 6001 loci, wherein the 6001 loci contain the 3000 loci. The SNP sets aim at the Han genetic background in design, have high polymorphism in Chinese and can realize the aim of efficiently marking the Chinese family sample genomes. The selection of polymorphic loci is based on the neutral evolution principle, and all the loci are in a non-gene function region, thus the influence of evolution on the gene function can be avoided. Meanwhile, the characteristics that the marking loci have high typing detectability and can uniformly cover the whole genomes can ensure that the whole genomes can be screened completely and new pathogenic genes can be located and found. The two sets of SNP markers are used to customize probes or chips and perform whole-genome genotyping to family samples; and the typing data are used for linkage analysis, and the haplotyping and fine locating of the linkage candidate region are also adopted, thus the use method has more accurate locating result than the traditional method while the cost is lower and the speed is higher. The distribution and coverage of the 6001 SNP marker set in human chromosomes are shown in the appended drawings.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION +1
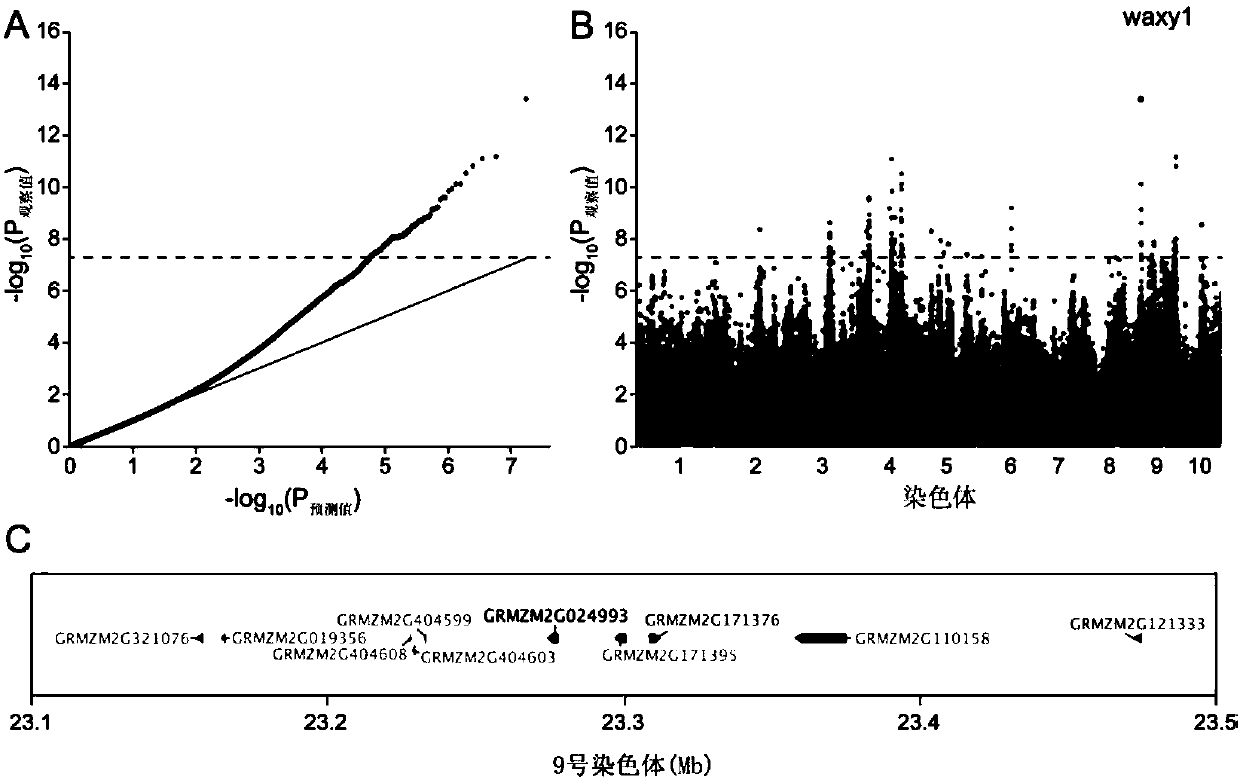
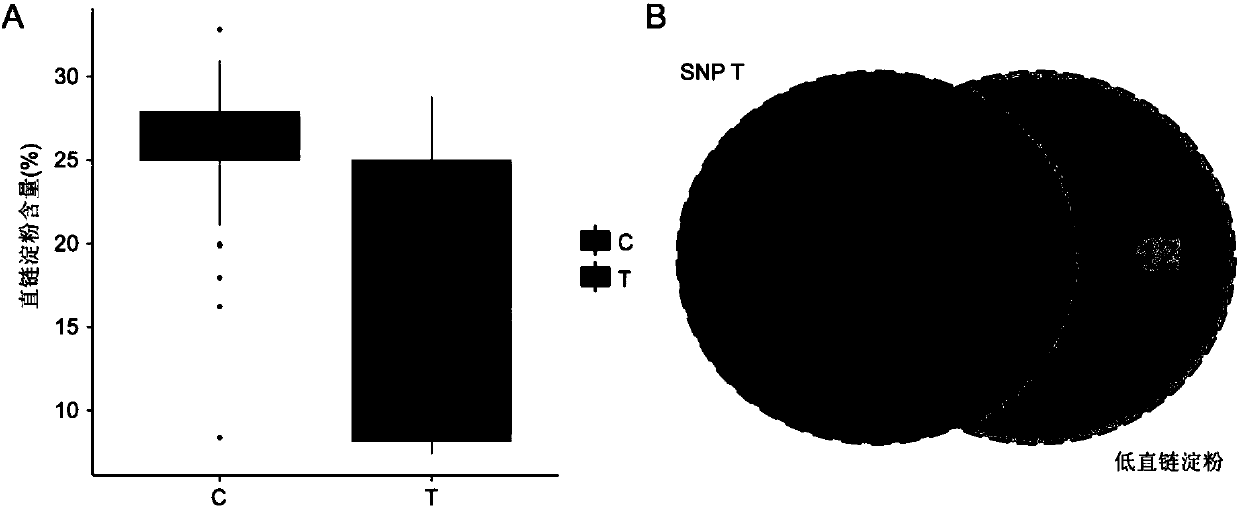
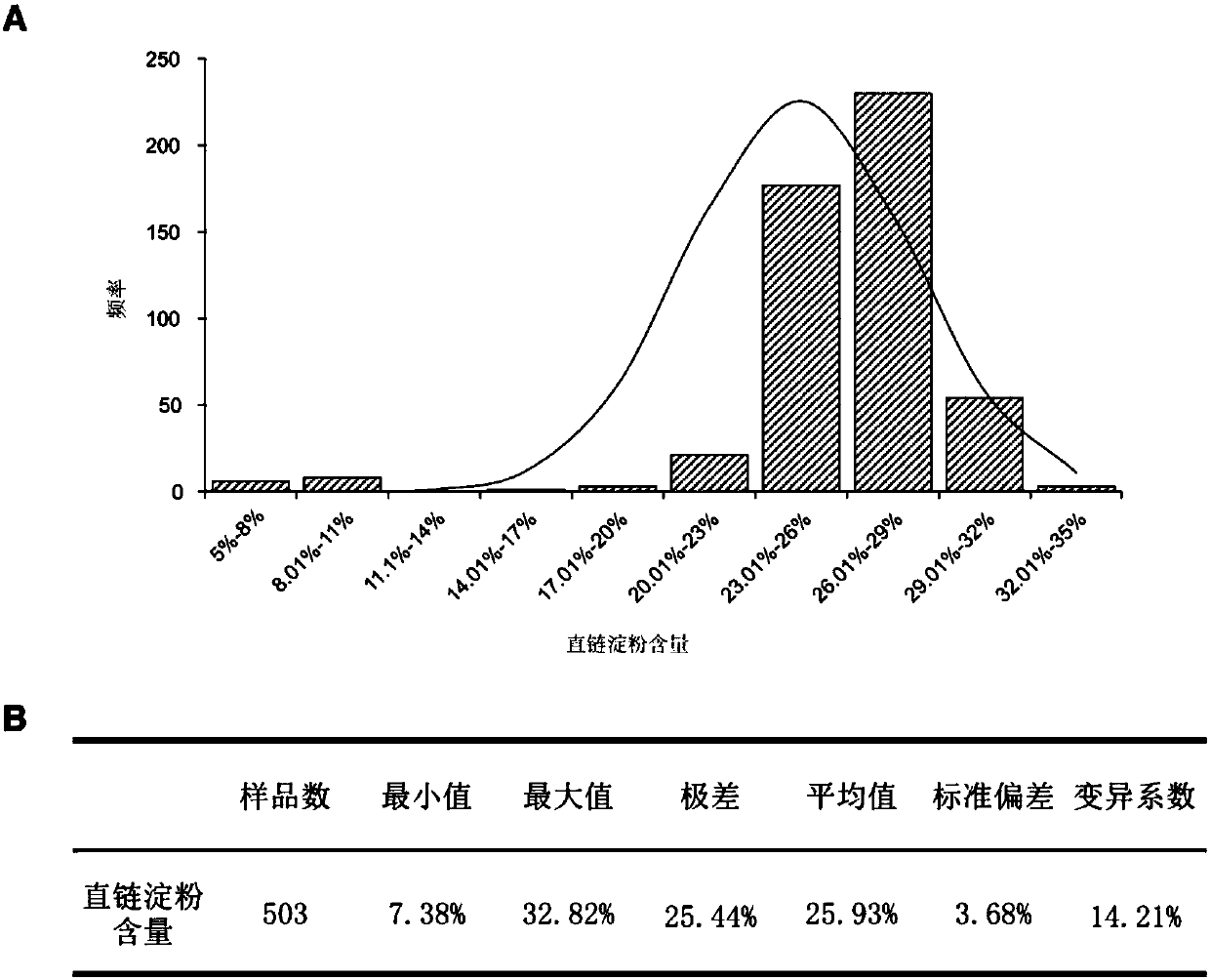
Method for developing waxy1-gene internal molecular markers on basis of association analysis and KASP
ActiveCN107619873AQuick digRapid phenotypeMicrobiological testing/measurementDNA/RNA fragmentationAmylaseCandidate Gene Association Study
The invention discloses a method for developing waxy1-gene internal molecule markers on the basis of association analysis and KASP. One indel and two snap markers located in waxy1 gene and remarkablyassociated with content of amylase are found out mainly through genome-wide association study and candidate gene association analysis techniques; the detected indel is marked as a major mutant type ofglutinous maize in China; glutinous maize materials can be identified through agarose gel by the markers developed on the basis of the indel, and convenience and rapidness is achieved as compared with that of original polyacrylamide gel identification; meanwhile, paired linkage disequilibrium test of 59 parts of waxy1-gene sequence regions of maize hybrid line materials is performed, close linkage of the indel marker and the two snp markers is tested, primers of the two snps are developed on the basis of the KASP technology, the glutinous maize materials can be identified rapidly, efficientlyand accurately, and molecular breeding of glutinous maize is facilitated.
Owner:SHANGHAI JIAO TONG UNIV
Markers and methods for assessing and treating severe or persistant asthma and TNF related disorders
InactiveUS20100331209A1Decreased asthma exacerbationPredict suitabilityNucleotide librariesMicrobiological testing/measurementMolecular Targeted TherapiesLinkage disequilibrium
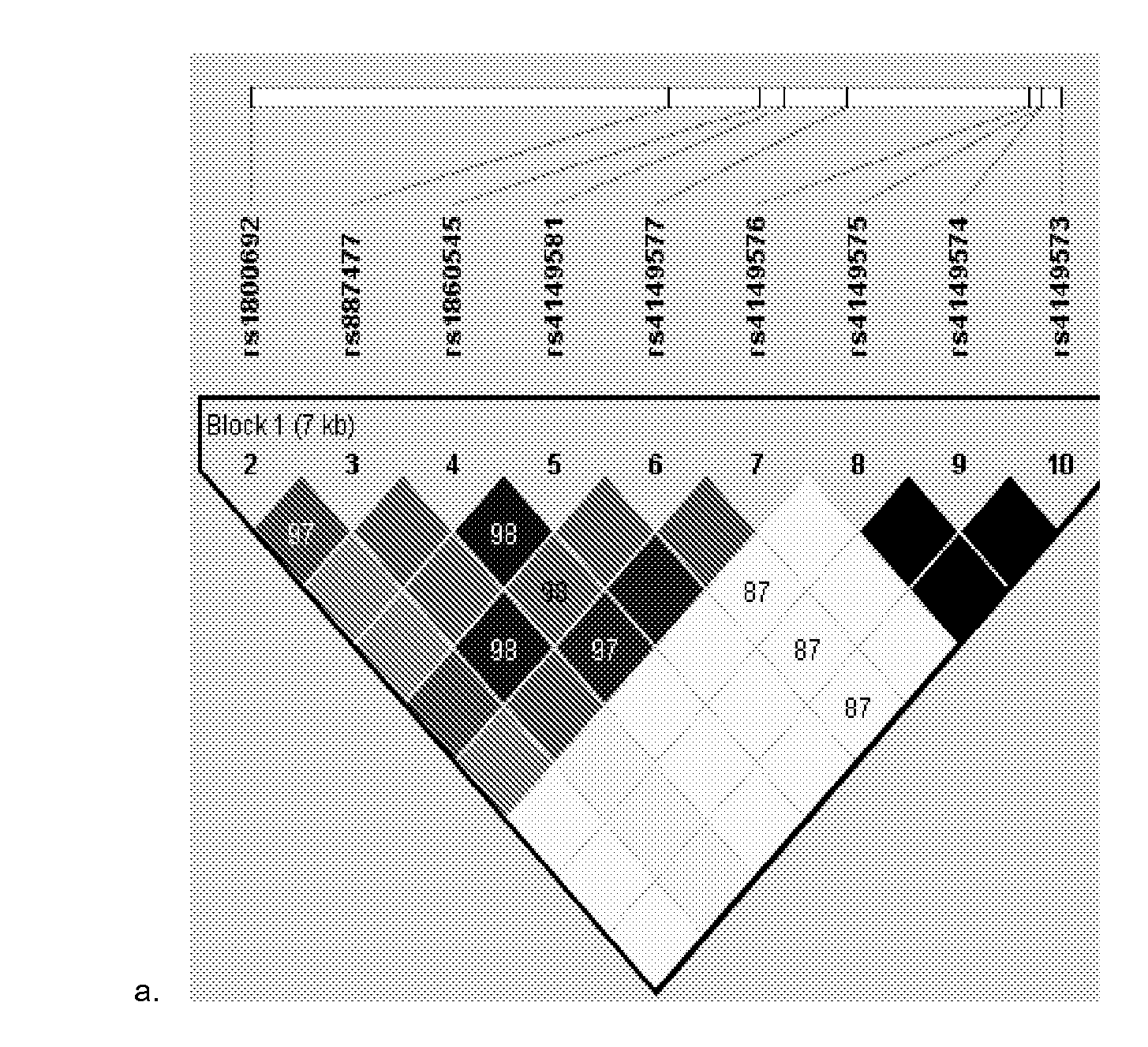
A method for assessment of the suitability of and / or effectiveness of a target therapy for a TNF-mediated-related disorder, such as severe or persistent asthma, in a subject evaluates the presence, absence, and / or magnitude of expression of one or more genes corresponding to contacting the sample with a panel of nucleic acid segments consisting of at least a portion of at least one member from the group consisting of the nucleotide sequences corresponding to at least one of TNFRSF1A SNP rs4149581 (SEQ ID NO:1), TNFRSF1 B SNP rs3766730 (SEQ ID NO:2) or TNFRSFI B SNP rs590977 (SEQ ID NO:3) SNPs which results in a determination that one or more of said SNPs in a sample are in linkage disequilibrium (LD). The method enables identification of the effectiveness of target therapies prior to or after starting a patient on such therapies.
Owner:CENTOCOR ORTHO BIOTECH
Genetics of gender discrimination in date palm
InactiveUS20140208449A1Highly nutritious fruitMinimum careSugar derivativesMicrobiological testing/measurementGenetic linkage disequilibriumGermplasm
This invention relates to the genetics of gender discrimination in the dioecious date palm. Methods of the present invention involve analyzing DNA or RNA from a date palm plant, tissue, germplasm, or seed for the presence of (i) a nucleic acid sequence or genotype that identifies the sex of the plant, tissue, germplasm, or seed or (ii) a molecular marker in linkage disequilibrium with the nucleic acid sequence or genotype. Also disclosed are kits for selecting male and female date palm plants prior to flowering, methods of breeding a date palm plant, and a method of planting a date palm seed of a known sex.
Owner:CORNELL UNIVERSITY
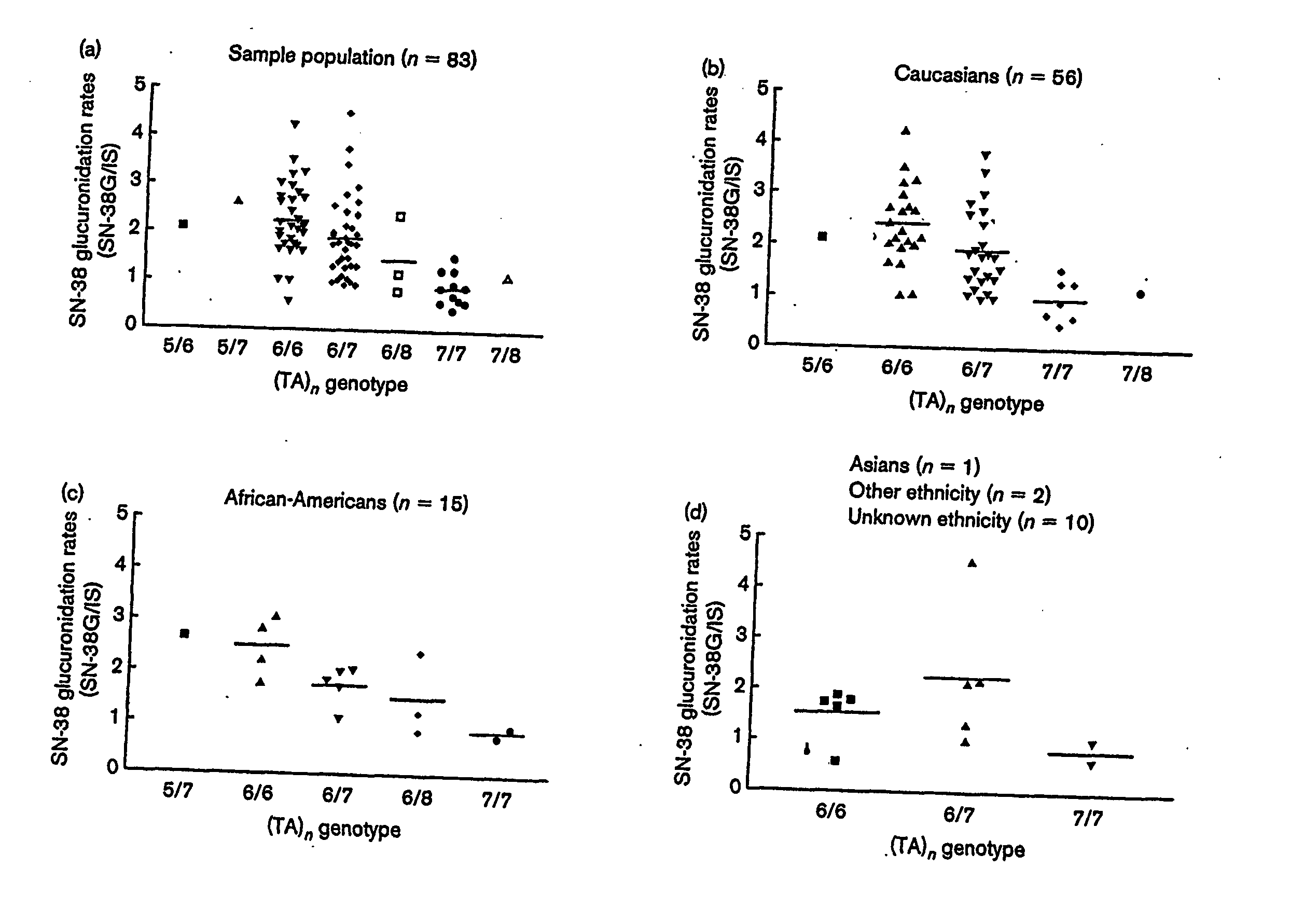
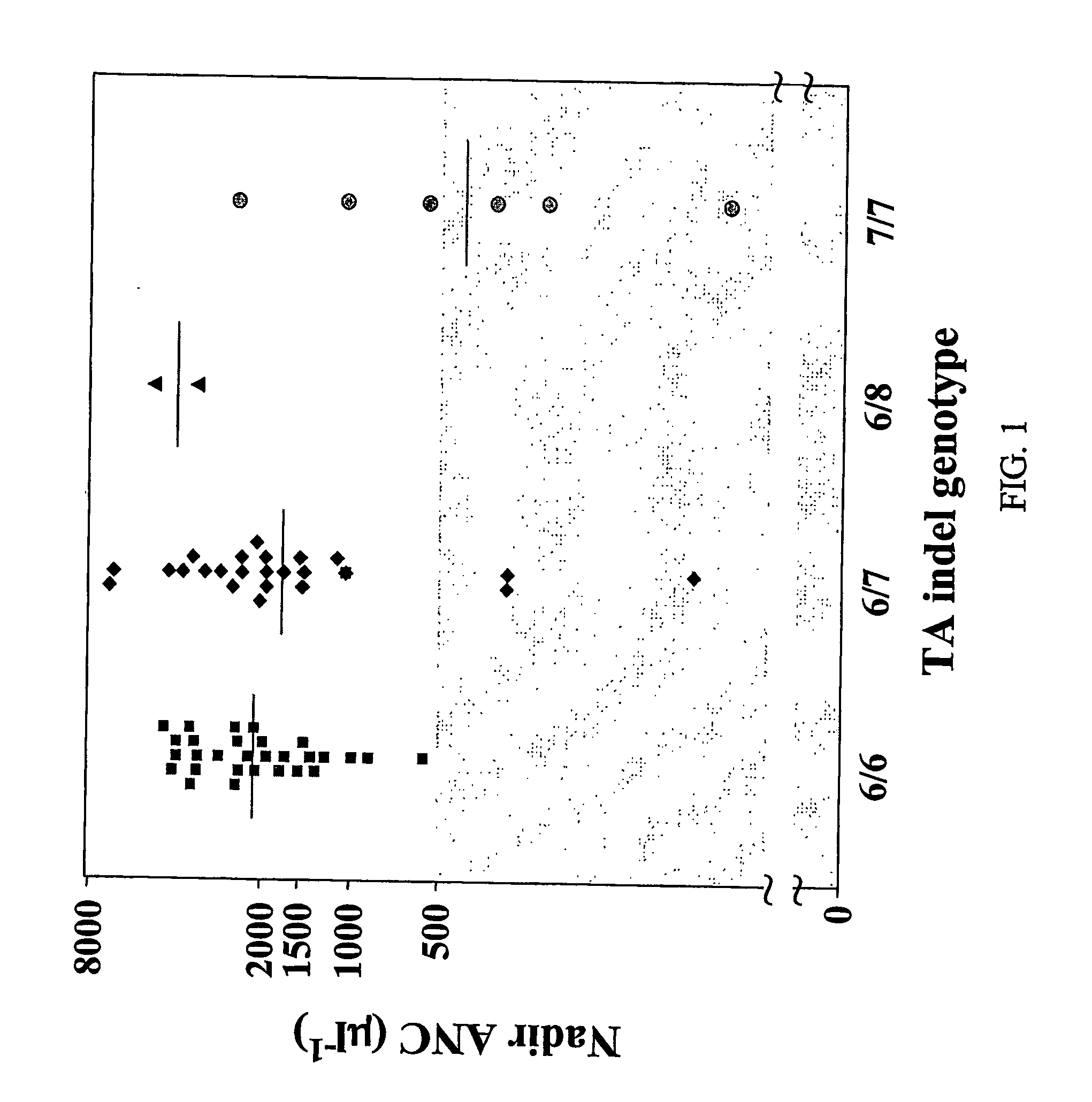
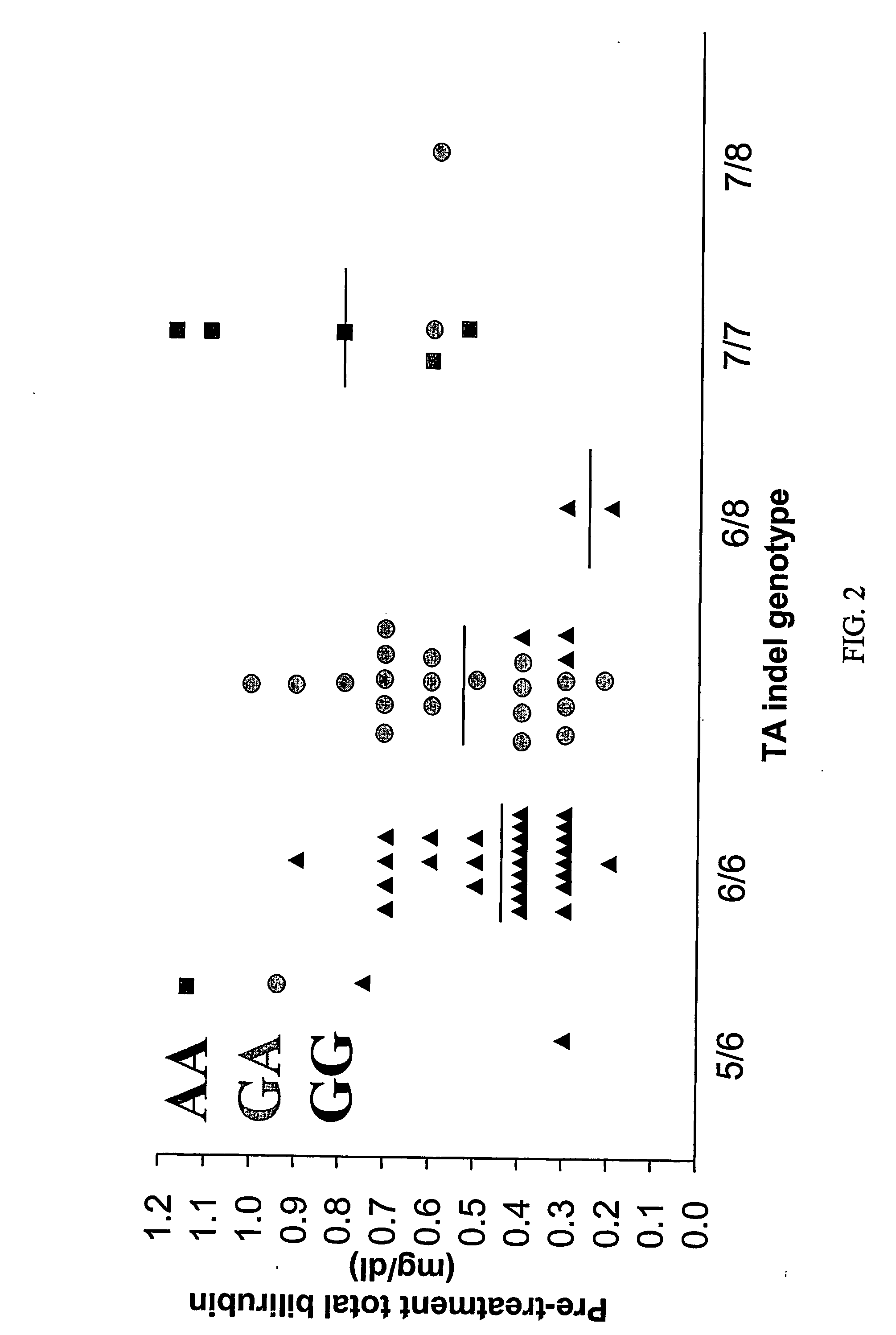
Methods and compositions for predicting irinotecan toxicity
The present invention concerns the methods and compositions for evaluating the risk of ironotecan toxicity in a cancer patient based on the genotype of the patient at position −3156 of the UGT1A1 gene or at any position in linkage disequilibrium with the −3156 variant.
Owner:UNIVERSITY OF CHICAGO
Cold-tolerance gene qCT-3-2HHZ for rice booting stage stable expression and molecular marking method thereof
ActiveCN105505922ARich diversityImprove cold resistanceMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyChilling injury
The invention provides a cold-tolerance gene qCT-3-2HHZ for rice booting stage stable expression and a molecular marking method thereof, and belongs to the field of rice high-yield stress-tolerance breeding and molecular genetics. According to the linkage and linkage disequilibrium phenomenon, the Huang zhanhua variety and eight associated breeding populations derived from donor parents rich in diversity are used, a whole-genome relational analysis and physical mapping method is adopted, the cold-tolerance gene qCT-3-2HHZ for the rice booting stage stability expression is precisely positioned and verified, and a practical economical marker ZCT-7 based on common PCR is further developed. The cold-tolerance gene qCT-3-2HHZ and the molecular marking method are applied to rice chilling-injury-tolerance assisted selection and pyramid breeding, defects of natural variation genes in the prior art can be effectively made up for, negative effects caused by assistant breeding conducted through primary positioning results can be remarkably reduced, genotype selection can be carried out on low-generation breeding populations in the seedling stage, cold-tolerance individuals are obtained, hybridization and transfer are facilitated in time, the process of identifying low-temperature resistance in the adult-plant stage is omitted, breeding efficiency is improved, and the breeding process is accelerated.
Owner:AGRI GENOME INST OF SHENZHEN CHINESE ACADEMY OF AGRI SCI +1
Hair Shape Susceptibility Gene
InactiveUS20120329726A1Cosmetic preparationsPeptide/protein ingredientsAllele frequencyGenetic linkage disequilibrium
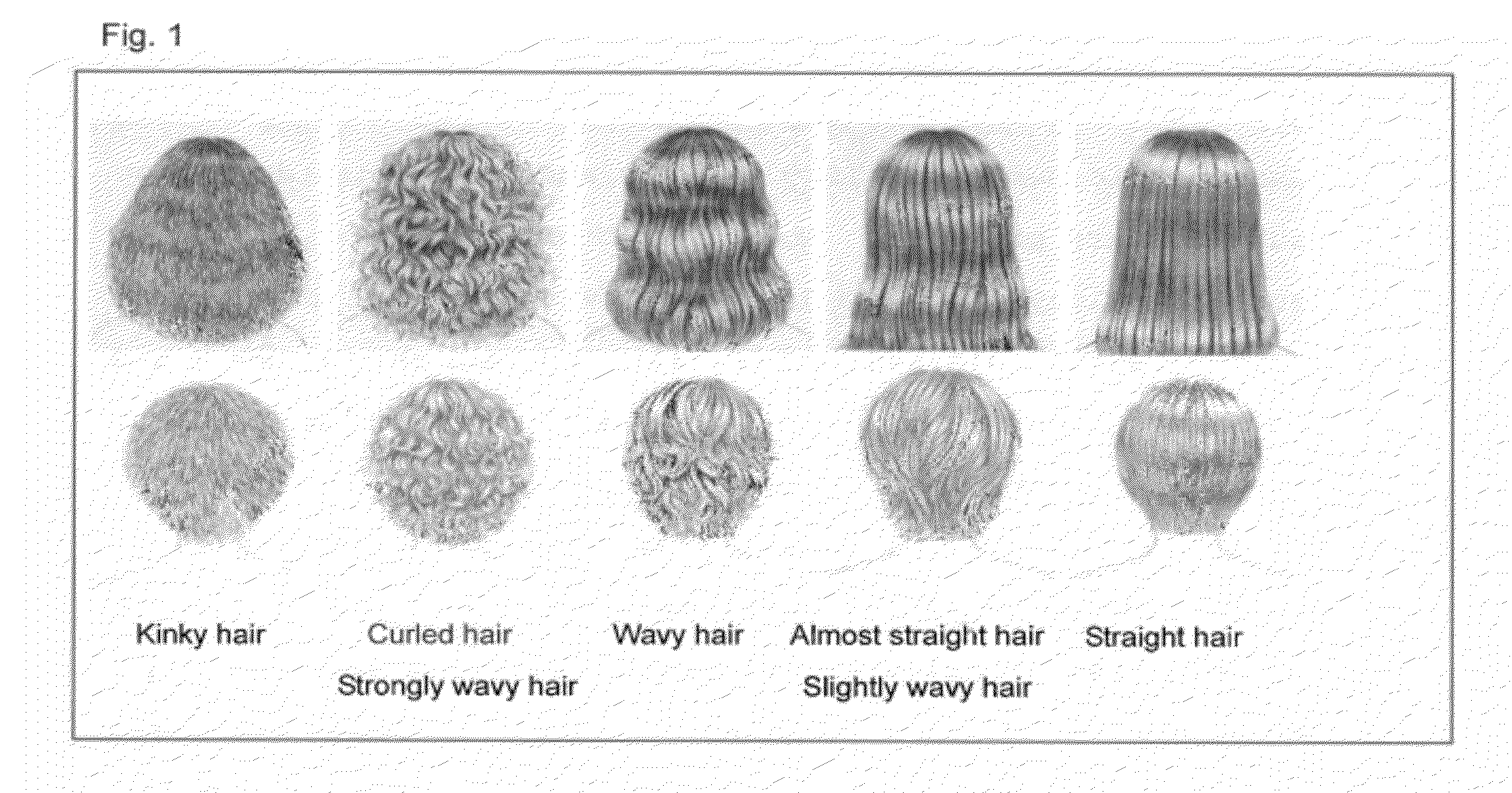
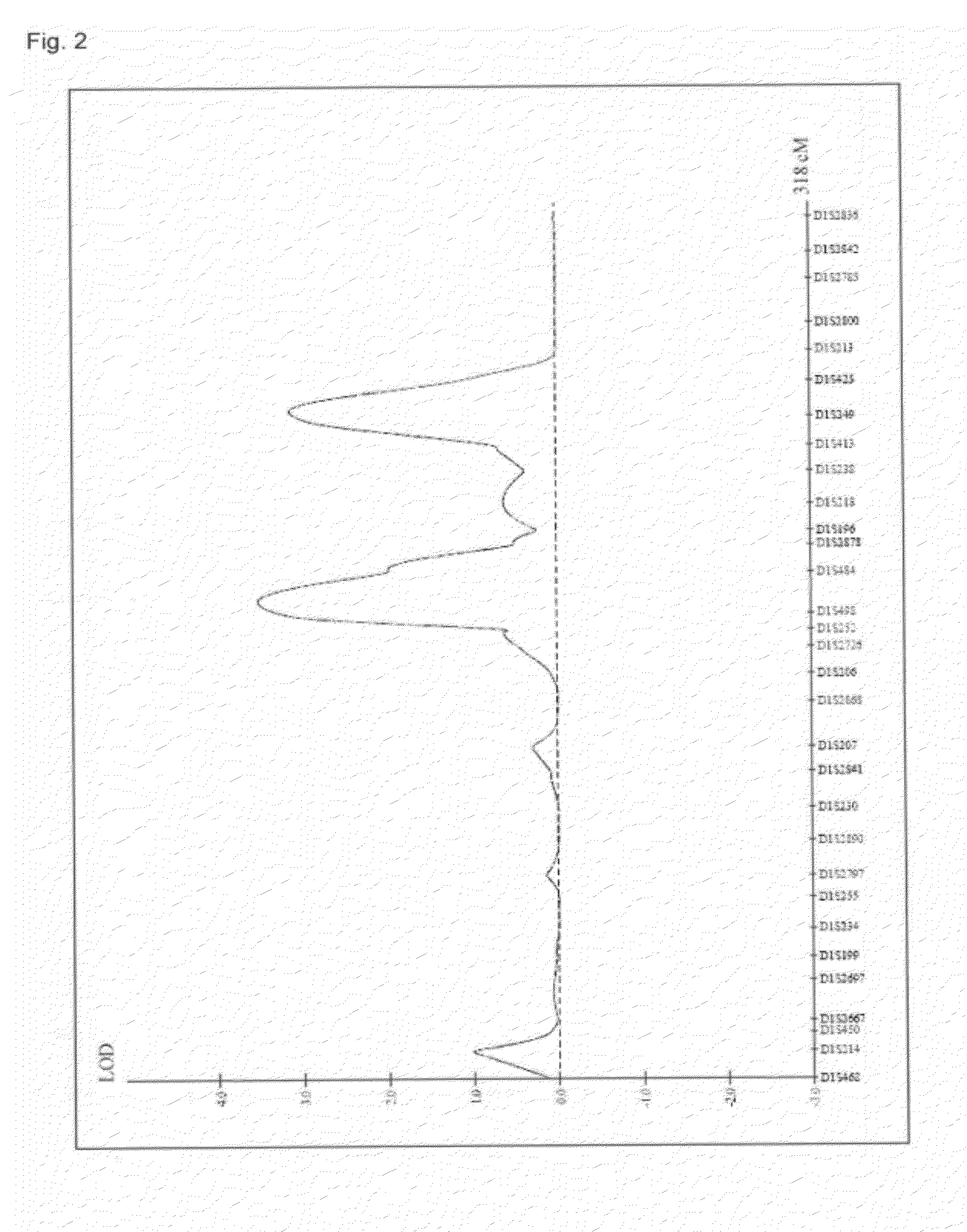
A genetic polymorphism and a hair shape susceptibility gene that are related to hair shape, and a method for determining the genetic susceptibility to hair shape in individual test subjects are provided. Disclosed is a hair shape susceptibility gene, which overlaps with a haplotype block in 1q21.3 region (D1S2696 to D1S2346) of human chromosome 1 and comprises a portion or the entirety of the base sequence of the haplotype block, wherein the haplotype block is determined by a linkage disequilibrium analysis conducted on a single nucleotide polymorphism (SNP) marker whose allele frequency differs statistically significantly between a group having a curly hair trait and a group having a non-curly hair trait, and consists of a base sequence set forth in any one of SEQ ID NO: 1 to NO: 5.
Owner:KAO CORP
Method for gene mapping from genotype and phenotype data
InactiveUS20050250098A1Model-free and computationally effectiveOvercomes drawbackMicrobiological testing/measurementBiostatisticsGene mappingGenetic linkage disequilibrium
A method for gene mapping from genotype and phenotype data utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. All marker patterns P that satisfy a certain pattern evaluation function e(P) are searched from the data, each marker mi of the data is scored by a marker score and the location of the gene is predicted as a function of the scores s(mi) of all the markers mi in the data.
Owner:LICENTIA OY
Phased Whole Genome Genetic Risk In A Family Quartet
An embodiment of the present invention is a method for resolving long-range haplotype phase based on family pedigree data, inheritance state determination, and population linkage disequilibrium data. A method according to an embodiment of the present invention provides for the evaluation of genome wide risk using phased haplotype data.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Methods and compositions of predicting activity of retinoid x receptor modulator
ActiveUS20150368720A1Chemical property predictionCompound screeningGenomic BiomarkerGenetic linkage disequilibrium
The present invention describes genomic biomarkers that have been discovered to correlate with varied individual responses (efficacy, adverse effect, and other end points) to therapeutic retinoid X receptor modulator, such as bexarotene, in treating diseases such as, non small cell lung cancer. The newly discovered biomarkers and others in linkage disequilibrium with them can be used in companion diagnostic tests which can help to predict drug responses and apply drugs only to those who will be benefited, or exclude those who might have adverse effects, by the treatment.
Owner:DENOVO BIOPHARMA HANGZHOU LTD
Methods and Workflows for Selecting Genetic Markers Utilizing Software Tool
InactiveUS20100153017A1Improve usabilityIncrease probabilityData visualisationProteomicsGenome mapGenetic linkage disequilibrium
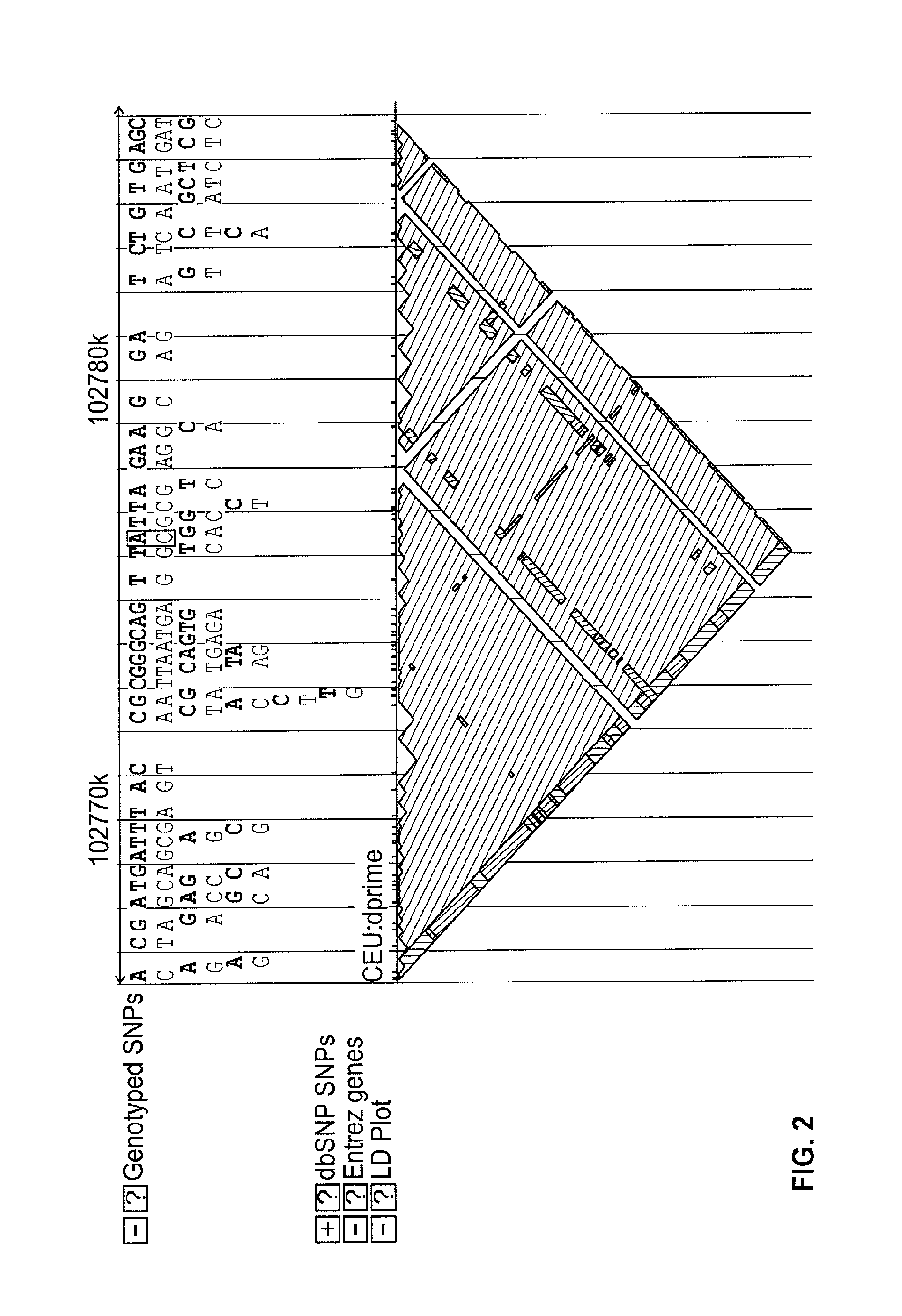
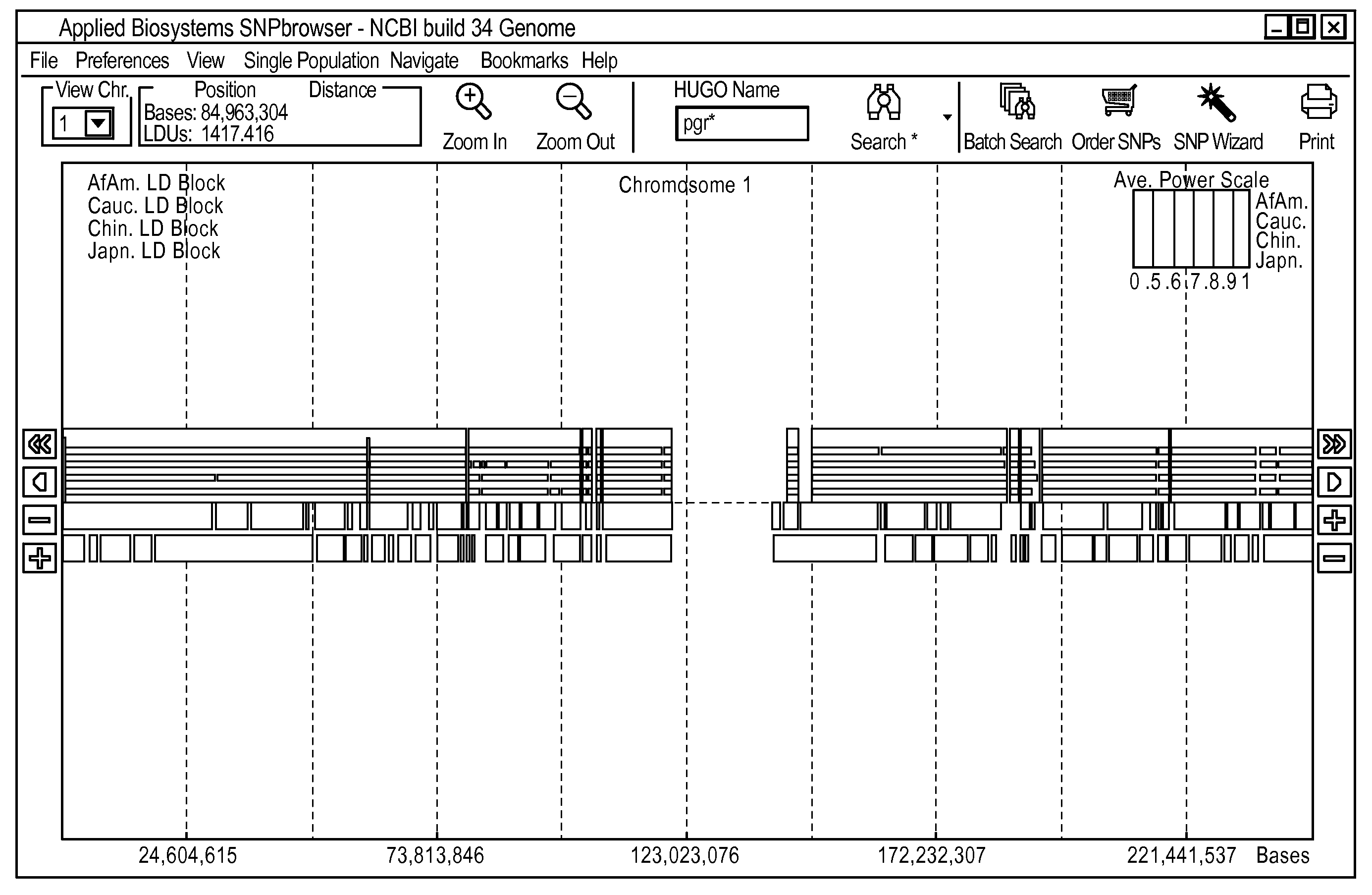
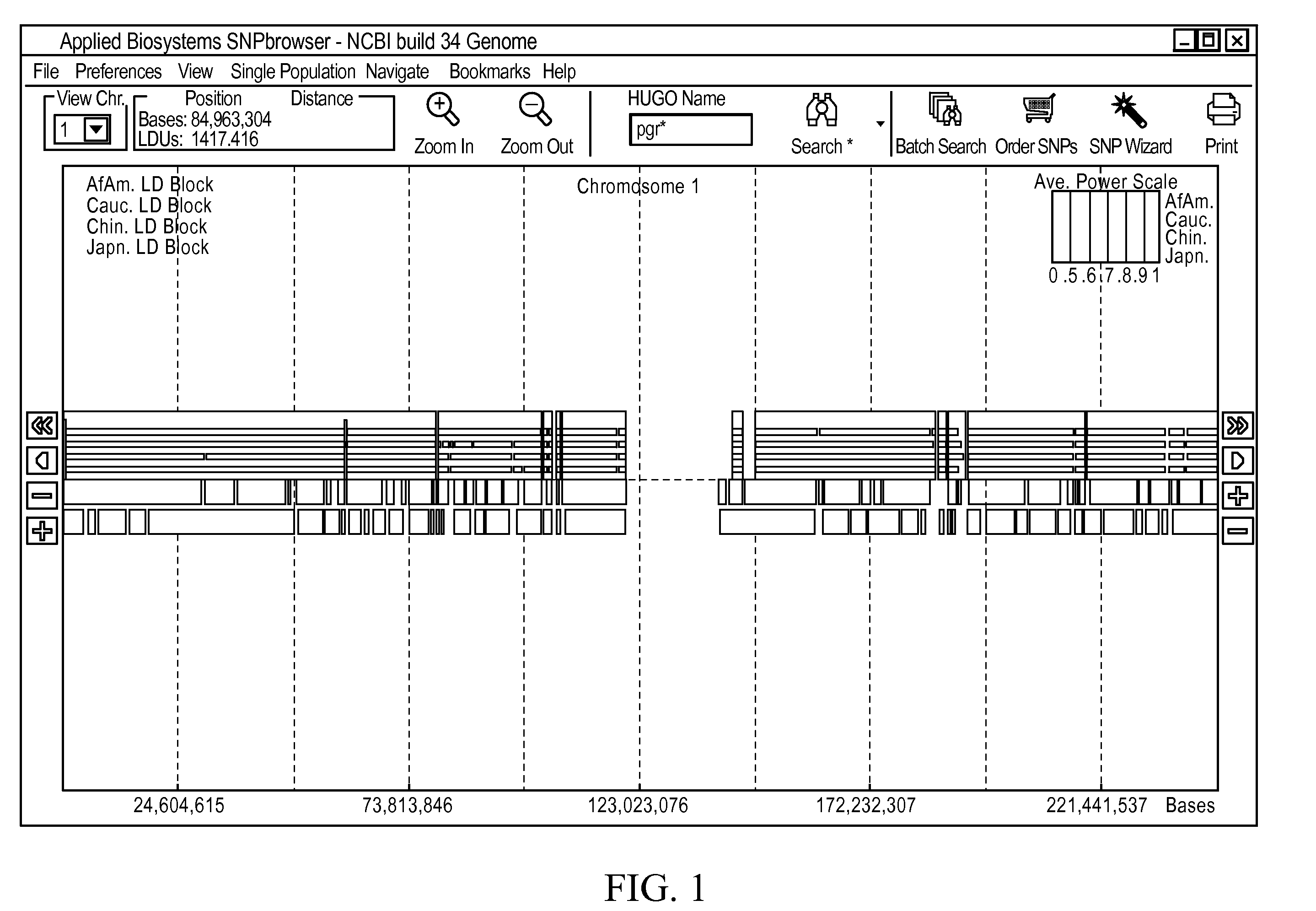
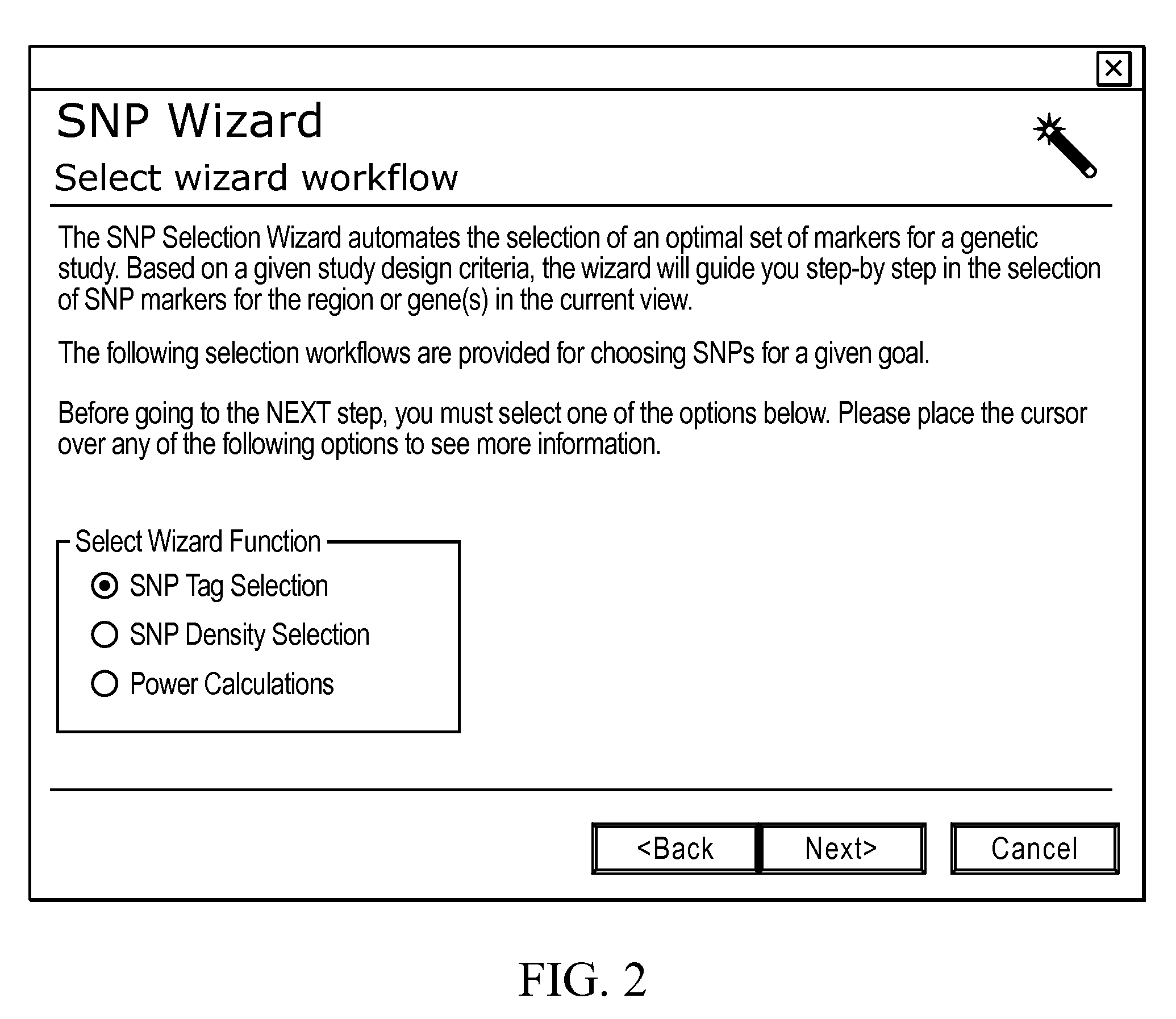
A visual tool facilitates selecting SNPs for genotyping experiments comprises a first memory containing a datastore of pre-calculated linkage disequilibrium map information; a second memory containing a datastore of haplotype block information; and a third memory containing at least one set of tagging SNPs. A graphical user interface provides visualization of SNPs, integrated with a physical genome map. A stepwise selection tool associated with the graphical user interface facilitates selection of tagging SNPs by selectively using the information in at least one of the first, second and third memories.
Owner:APPL BIOSYSTEMS INC
MUC13 gene determining susceptibility/resistance of piglet F4ac diarrhea and application thereof
InactiveCN101864423AReduce mortalityImprove resistance to diarrheal diseaseCell receptors/surface-antigens/surface-determinantsMicrobiological testing/measurementGenetic linkage disequilibriumDiarrhea
The invention discloses an MUC13 gene determining the susceptibility / resistance of piglet F4ac diarrhea and application thereof in boar genetic improvement, which adopts a large-scale white Dorec*Erhualian resource group and a wide cross group of 15 pig breeds at home and abroad, locks a target gene, i.e. the MUC13 gene coded with an ETEC F4ac receptor through the genome scanning chain positioning, high density SNP multi-point chain fine positioning in a destination field, breakpoint recombination analysis, wide cross group association analysis based on linkage disequilibrium, and separates out and clones the MUC13 gene. Five key mutational sites are identified in the MUC13 gene, and are coseparated with ETEC F4ac in vitro adherent phenotype in 144 purebred Duroc, Landrace and Yorkshire piglets from the core groups of five provinces, wherein the accuracy of judging the strongest relevant label of a resistance / susceptibility individual can be up to 97% (P=1.59*10-21). The invention provides a novel method for breeding and disease resisting of the piglet ETEC F4ac diarrhea.
Owner:JIANGXI AGRICULTURAL UNIVERSITY
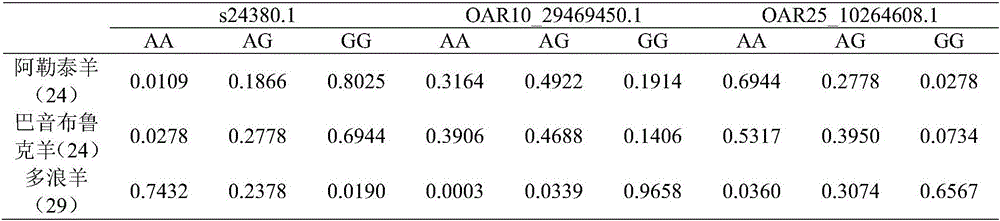
Duolang sheep SNP (single-nucleotide polymorphism) marker, and screening method and application thereof
ActiveCN106011259AImprove accuracyOvercoming the pitfalls of phenotypingMicrobiological testing/measurementDNA/RNA fragmentationGenetic linkage disequilibriumScreening method
The invention discloses a Duolang sheep SNP (single-nucleotide polymorphism) marker, and a screening method and application thereof, belonging to the field of animal molecular markers. A genetic differentiation coefficient method is utilized to perform screening on a commercialized sheep genome SNP chip and further remove high-linkage-disequilibrium SNP markers (r<2>>0.2) through linkage disequilibrium analysis, thereby obtaining the Duolang sheep SNP marker. The Duolang sheep SNP marker can be applied to identification of Duolang sheep species or animal products thereof on the premise of no obvious linkage disequilibrium among markers; and therefore, when the Duolang sheep SNP marker disclosed by the invention is used for identifying the Duolang sheep species or animal products thereof, no interference among sites exists, thereby greatly enhancing the Duolang sheep species identification accuracy and effectively lowering the identification cost.
Owner:新疆畜牧科学院生物技术研究所
Method for predicting the athletic performance potential of a subject
InactiveUS20110262915A1Reduce operating costsEnhance the imageMicrobiological testing/measurementTaming and training devicesGenetic linkage disequilibriumExercise performance
A method for predicting the athletic performance potential of a subject comprising the step of assaying a biological sample from a subject for a genetic variant in linkage disequilibrium with MSTN-66493737 (T / C) SNP. The invention also provides an assay for determining the athletic performance potential of a subject.
Owner:UNIV COLLEGE DUBLIN NAT UNIV OF IRELAND DUBLIN
Molecular marker method for two mutation sites of chicken MMP13 gene 5' control region and application of molecular marking method in chicken breeding
ActiveCN105420352AIncrease egg productionAchieve early selectionMicrobiological testing/measurementAnimal husbandryBreeding chickenGenetic linkage disequilibrium
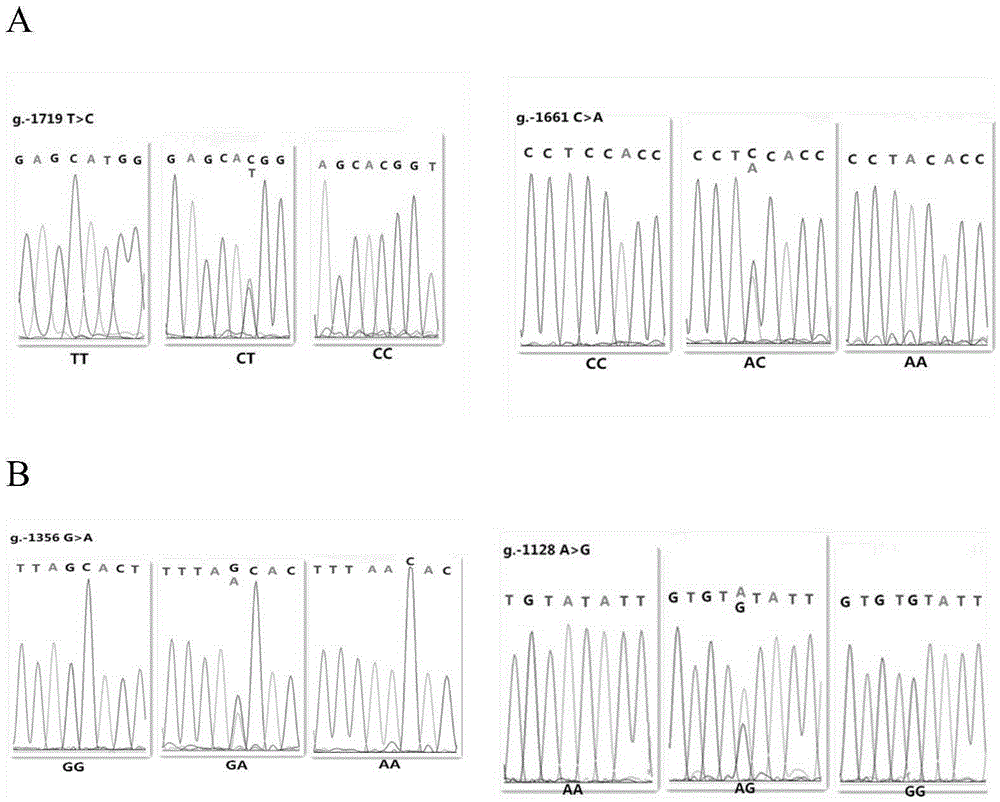
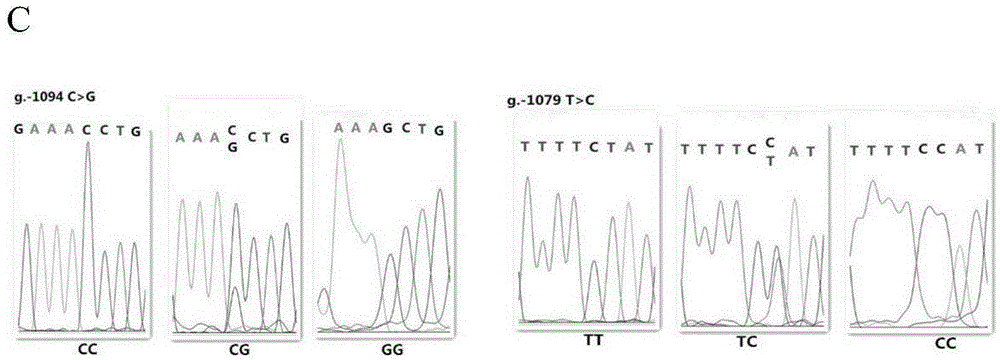
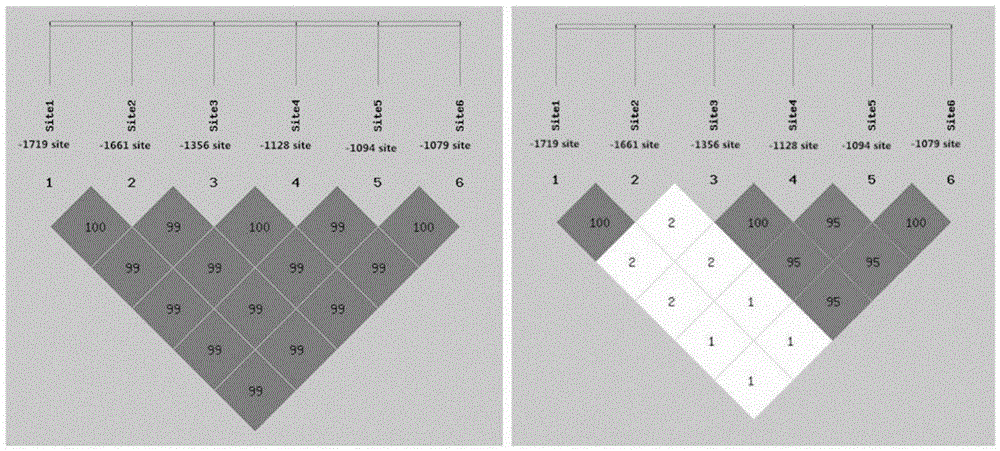
The invention relates to the field of molecular genetics, in particular to a molecular marker method for two mutation sites of a chicken MMP13 gene 5' control region and application of the molecular marker method in chicken breeding. It is found by the inventor that six mutation sites exist in the chicken MMP13 5' control region, namely, -1719 (T>C), -1661 (C>A), -1356 (G>A), -1128 (A>G), -1094 (C>A) and -1079 (T>C); linkage disequilibrium analysis is performed through the SHEsis online software, and it is found through the result that the sites -1719 and -1661, the sites -1356 and -1128 and the sites -1094 and -1079 are respectively in a completely-linked state in White Recessive Rock. The method is easy, convenient and fast to implement, beneficial for breeding chicken breeds laying eggs early at a high yield and capable of providing favorable help for the marker-assisted breeding work.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Method for effectively anchoring candidate gene region of peanut quantitative traits
InactiveCN109694924AAchieve positioningMicrobiological testing/measurementBiotechnologyGenetic linkage disequilibrium
The invention relates to the field of biotechnology, and in particular to a method for effectively anchoring a candidate gene region of a quantitative trait. The method is co-localized with a genome-wide association study and a quantitative trait localization method to achieve efficient anchoring of the quantitative trait candidate gene regions. The method achieves effective anchoring of the candidate gene region of the peanut quantitative trait by co-localization with the genome-wide association study and the quantitative trait localization. The method is exemplified by quantitative granule weight, firstly, through a simplified genome sequencing technology, the genome-wide SNP marker development of 165 cultivar peanut core collections in China is carried out; based on a principle of linkage disequilibrium, combined with peanut 100-fruit weight and 100-kernel weight breeding value, the related gene regions related to peanut weight traits are associated; in the previous reports, the QTLmapping results of the grain weight traits are converted to the corresponding genomic physical locations, the co-localization region of the two methods can be obtained, and the method can effectivelyand accurately realize the localization of the genome-wide particle weight-related trait genes.
Owner:SHANDONG PEANUT RES INST
Calcutta hemp InDel molecular marker as well as development method and application thereof
ActiveCN107447018AClear bandHigh polymorphismMicrobiological testing/measurementDNA/RNA fragmentationHemp plantGenetic linkage disequilibrium
The invention relates to the technical field of a molecular marker, in particular to a calcutta hemp InDel molecular marker as well as a development method and application thereof. The molecular markers can be used for identifying the allele polymorphism, identifying identical or relevant calcutta hemp plants, distinguishing the calcutta hemp plants and studying the genetic diversity in populations. The molecular marker can also be used for using inheritance and phenotype study of a statistical method, such as linkage analysis, QTL positioning, genealogy confirmation, population genetic polymorphism study, association mapping and linkage disequilibrium; the information can be used for calcutta hemp plant propagation and / or selection.
Owner:INST OF BAST FIBER CROPS CHINESE ACADEMY OF AGRI SCI
Reagents and methods for diagnosis of attention deficit hyperactivity disorder
InactiveUS20060204961A1Sugar derivativesMicrobiological testing/measurementGenetic linkage disequilibriumAllele
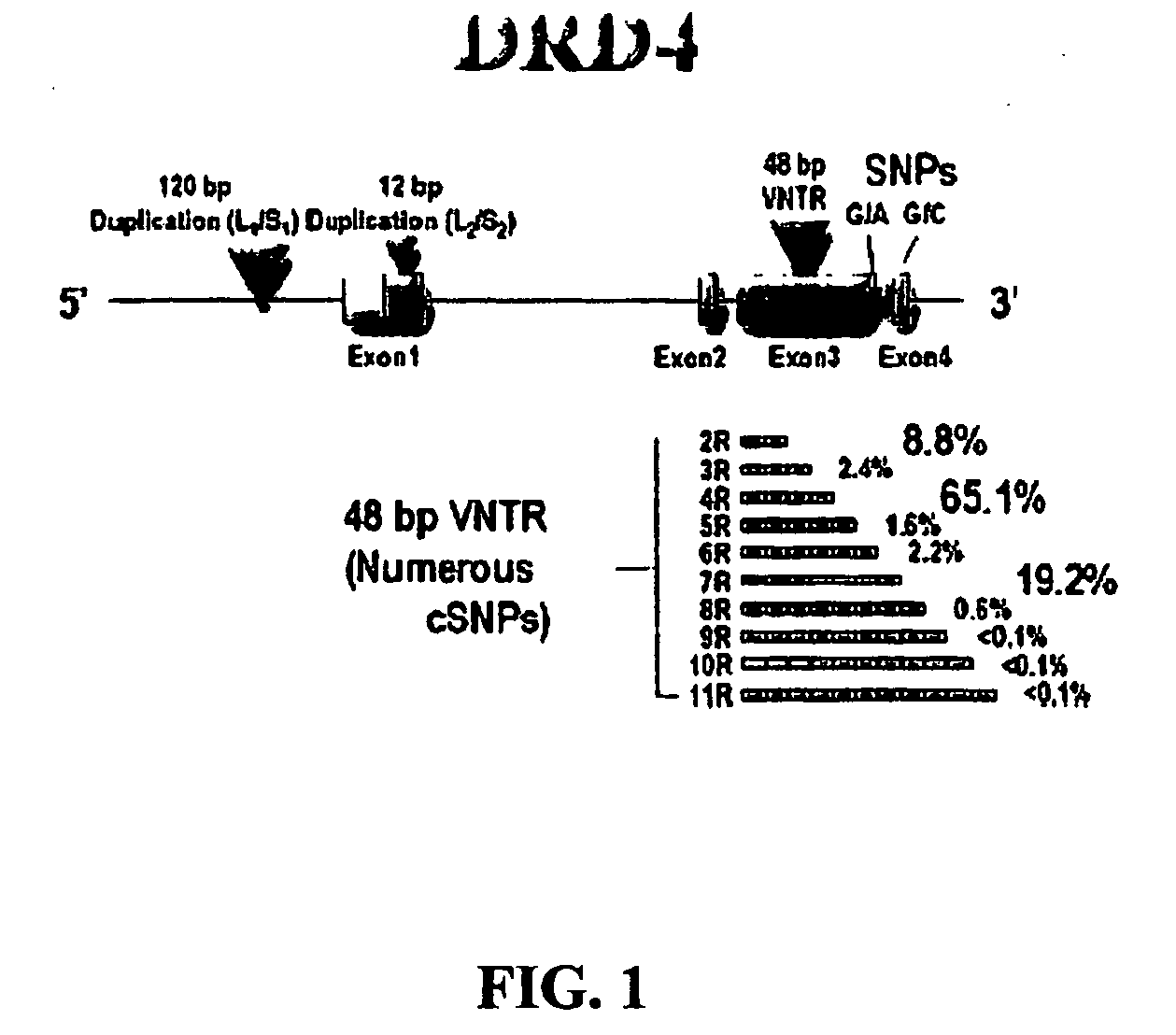
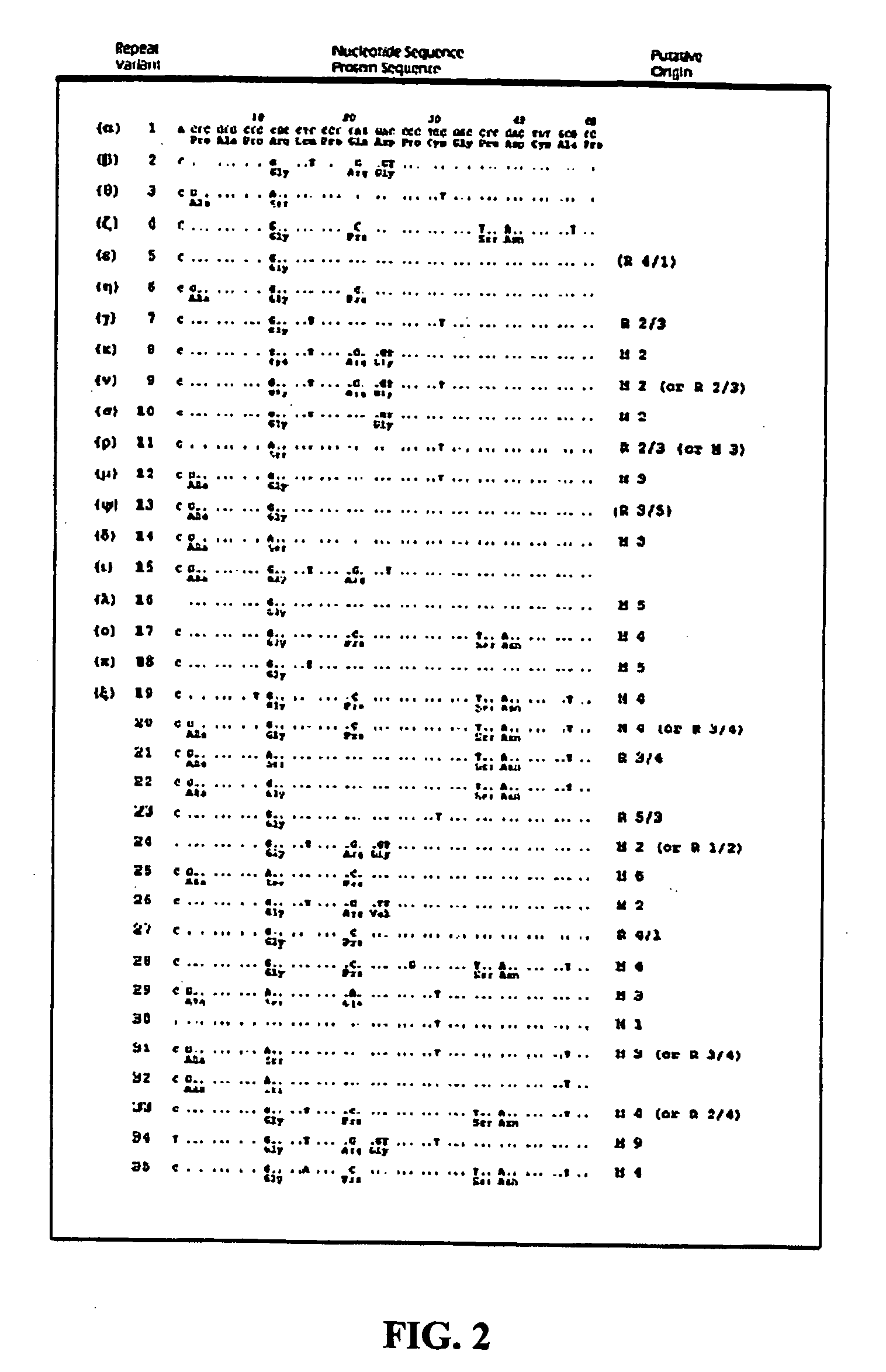
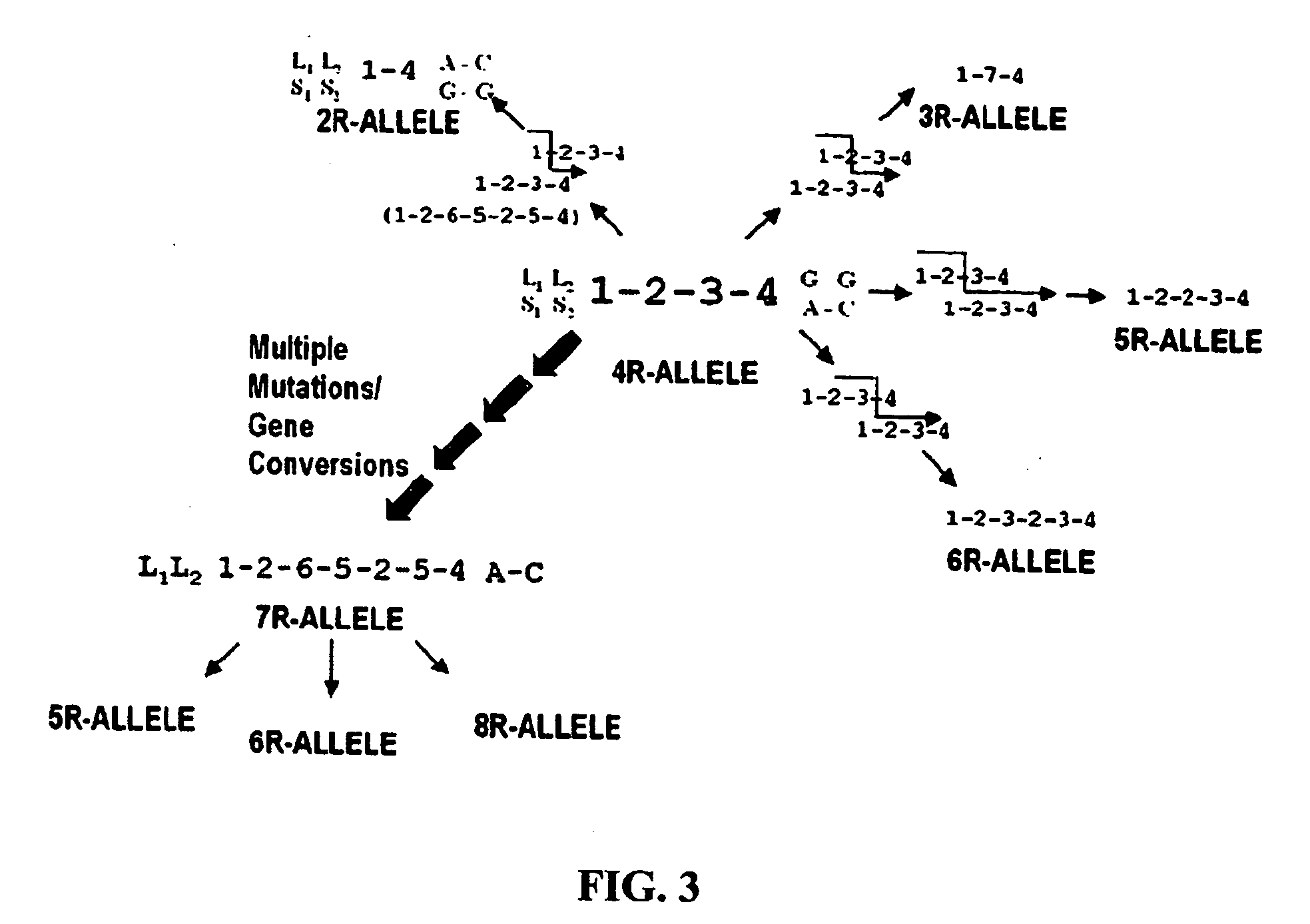
The subject invention provides reagents and methods for diagnosing Attention Deficit Hyperactivity Disorder (ADHD) in an individual based on the presence of specific alleles of the DRD4 gene or other markers found within a region of strong linkage disequilibrium to the DRD4 7R allele in the DNA of the individual.
Owner:RGT UNIV OF CALIFORNIA
Methods of diagnosis
InactiveUS20060234274A1Valuable informationMicrobiological testing/measurementGenetic linkage disequilibriumGenetics
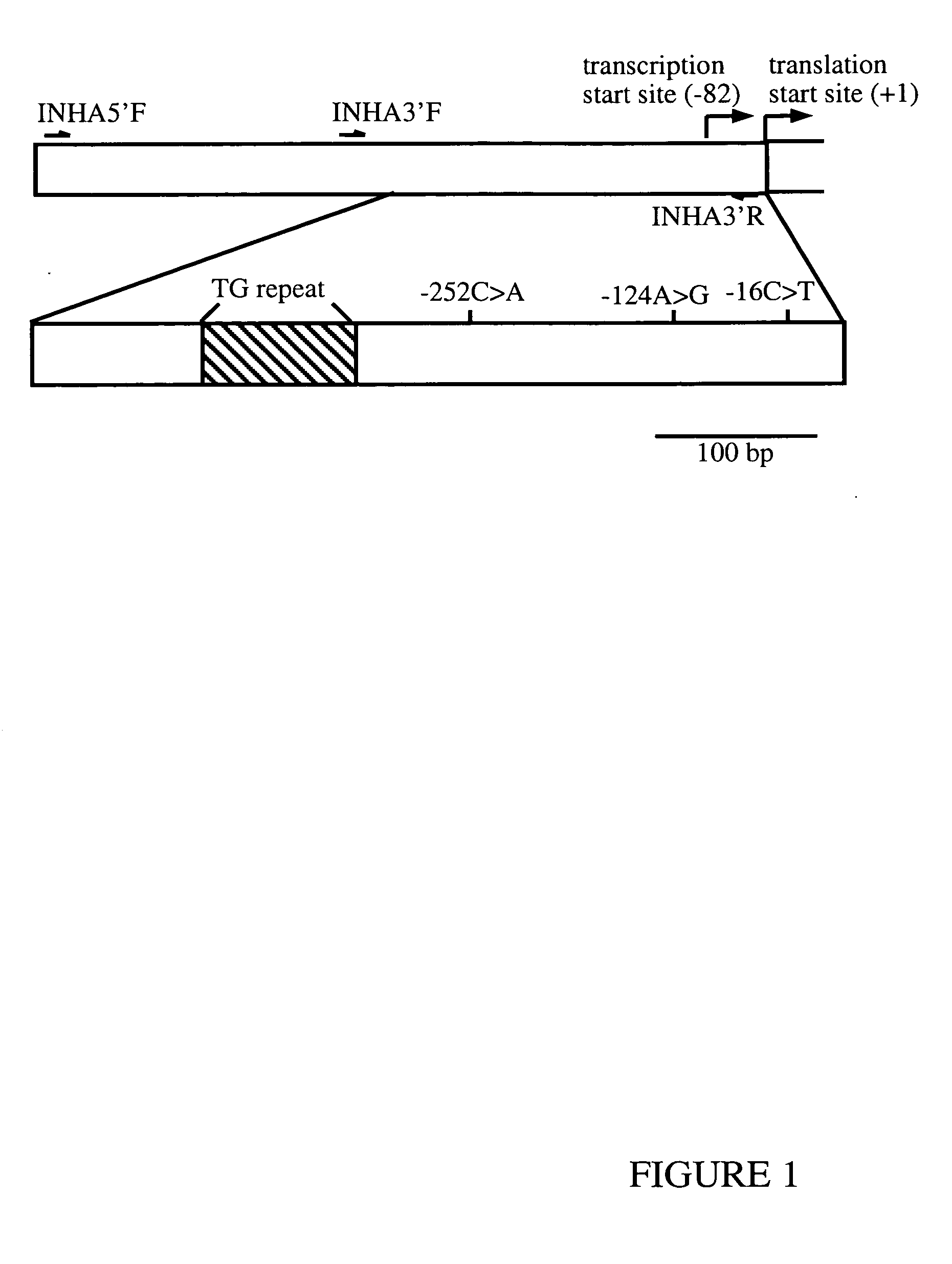
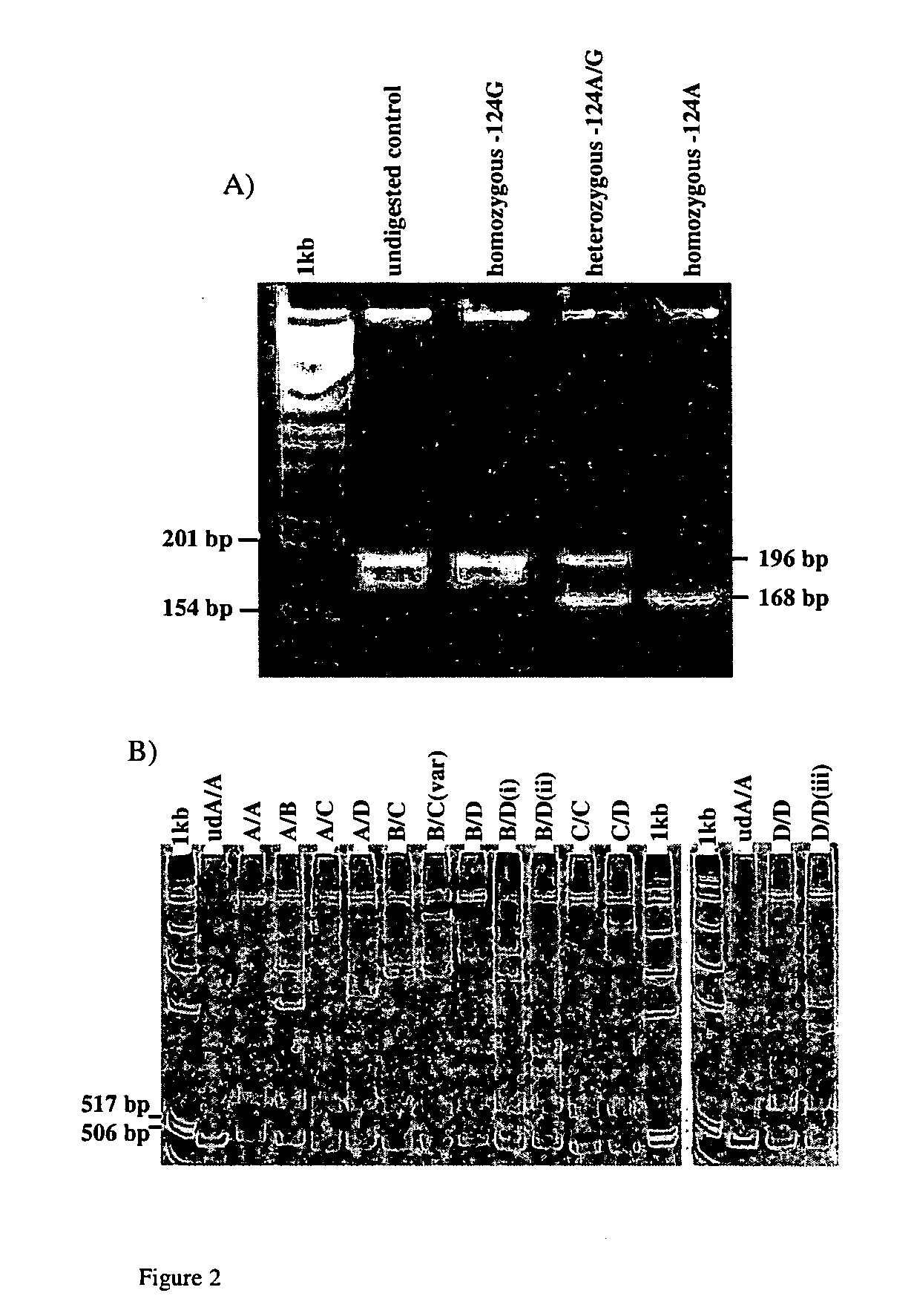
A method to detect whether a female subject is predisposed to POF, the method comprising analysing at least one or more polymorphism in the INHA gene chosen from the group consisting: −124A>G; −16C>T; TG repeat (as herein after described); and one or more polymorphism in the INHA gene which is in linkage disequilibrium with one or more of −124A>G, −16C>T or TG repeat (as herein after described).
Owner:AUCKLAND UNISERVICES LTD
Hair Shape Susceptibility Gene
A genetic polymorphism and a hair shape susceptibility gene that are related to hair shape, and a method for determining the genetic susceptibility to hair shape in individual test subjects are provided. Disclosed is a hair shape susceptibility gene, which overlaps with a haplotype block in the 1q32.1 to 1q32.2 region (D1S249 to D1S2891) of human chromosome 1 and comprises a portion or the entirety of the base sequence of the haplotype block, wherein the haplotype block is determined by a linkage disequilibrium analysis conducted on a single nucleotide polymorphism (SNP) marker whose allele frequency differs statistically significantly between a group having a curly hair trait and a group having a non-curly hair trait, and consists of a base sequence set forth in any one of SEQ ID NO:1 to NO:3.
Owner:KAO CORP
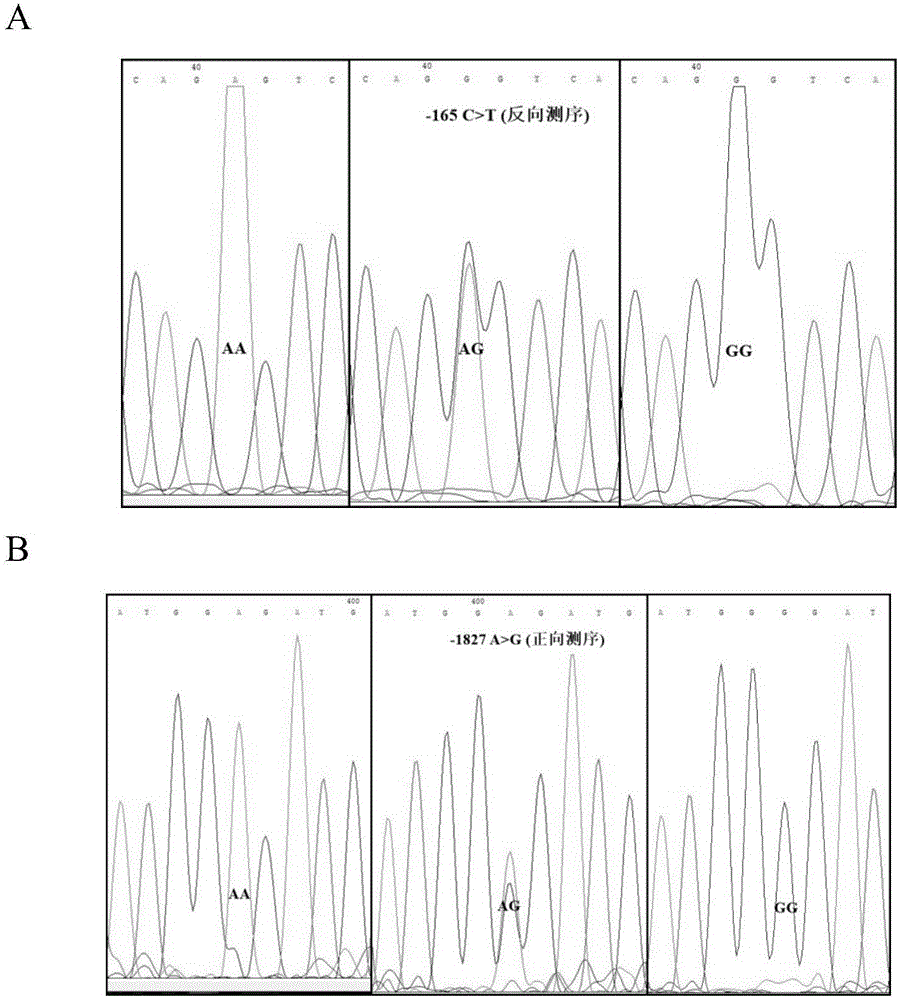
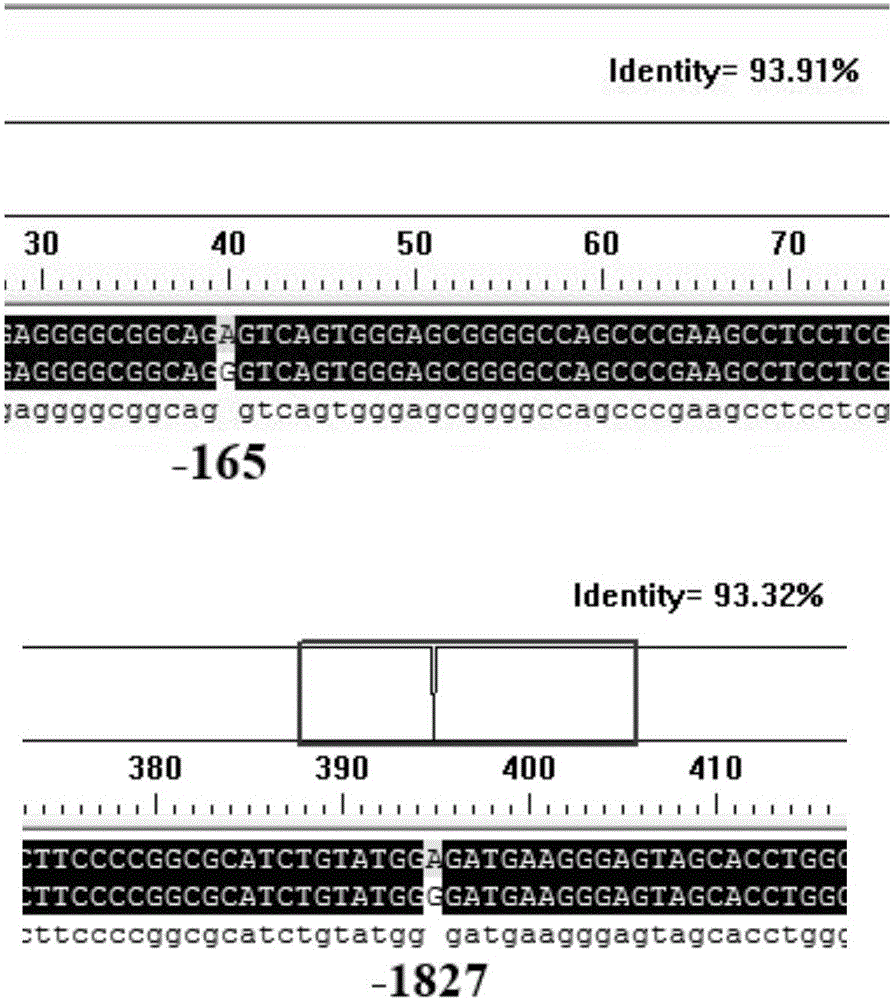
Molecular marking method for two mutation sites in chicken PTHLH gene 5' regulatory region and application thereof in chicken breeding
ActiveCN106048043AIncrease egg productionAchieve early selectionMicrobiological testing/measurementAnimal husbandryBreeding chickenGenetic linkage disequilibrium
The invention relates to a molecular marking method for two mutation sites in a chicken PTHLH gene 5' regulatory region and application thereof in chicken breeding. Two mutation sites (namely the -1827 (A>G) site and the -165(C>T) site) exist in the chicken PTHLH gene 5' regulatory region, SHEsis online software is used for linkage disequilibrium analysis, and the result shows that the two sites in recessive white plymouth rock are in a complete linkage state. The two sites are related with the first-egg age and egg laying quantity at the 32th week, the first-egg age of AC / GT gene type is remarkably earlier than that of the GT / GT type and the AC / AC type, and meanwhile the egg laying quantity at the 32th week is remarkably larger than that of the other two types. The method is simple, fast and beneficial for breeding chicken variety with the early first-egg age and the large egg laying quantity and provides favorable help for marker-assisted breeding work.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
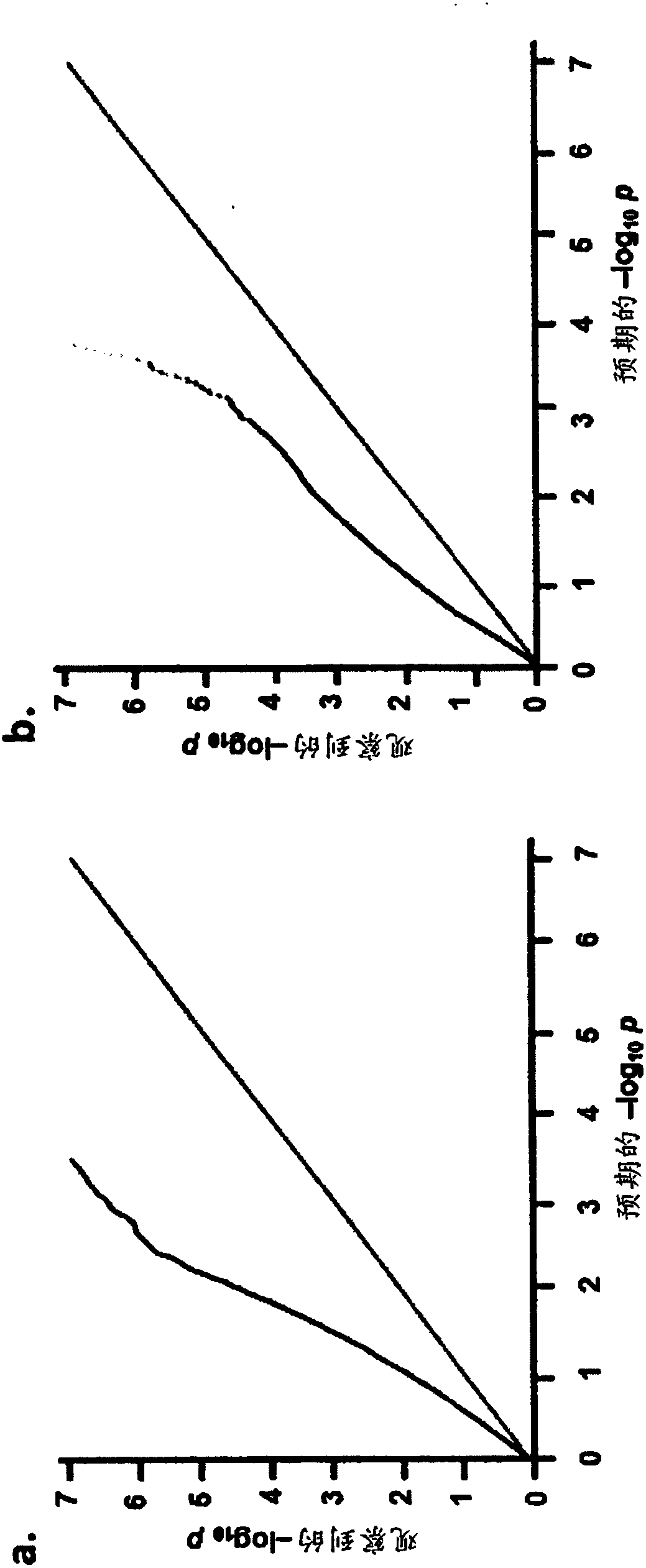
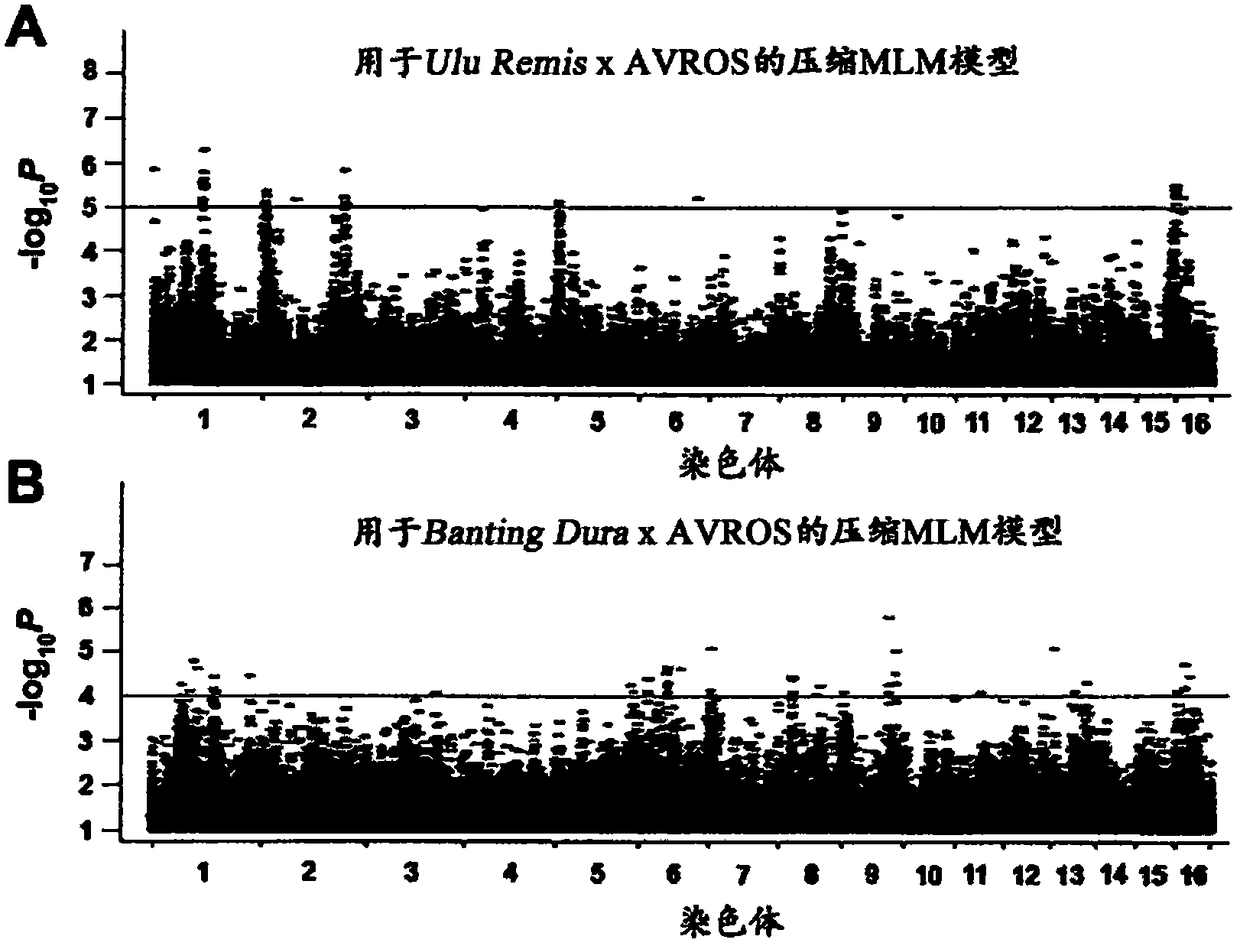
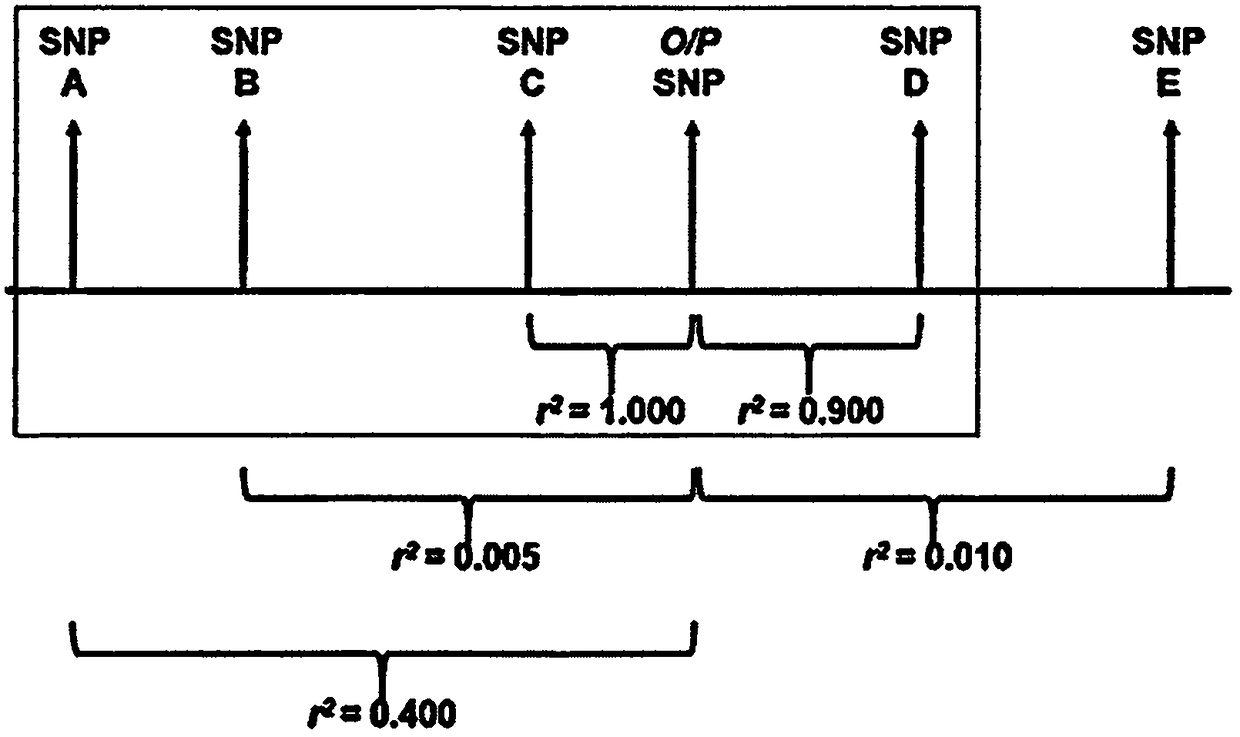
Methods and SNP detection kits for predicting palm oil yield of a test oil palm plant
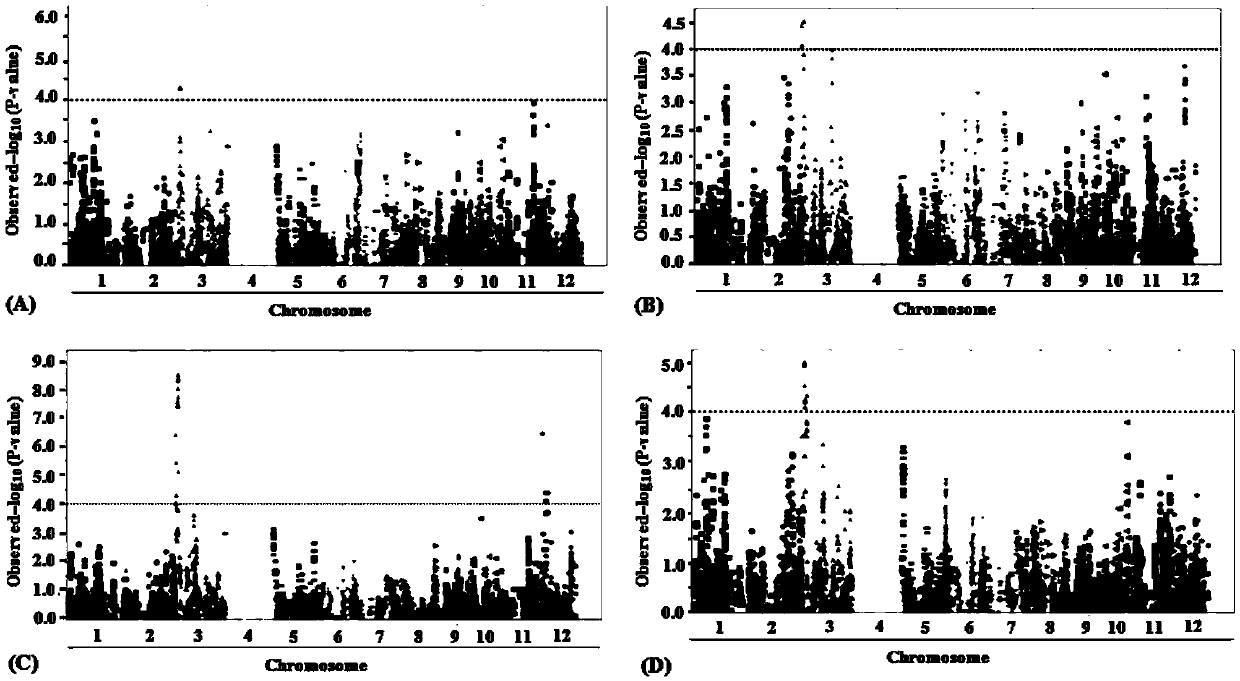
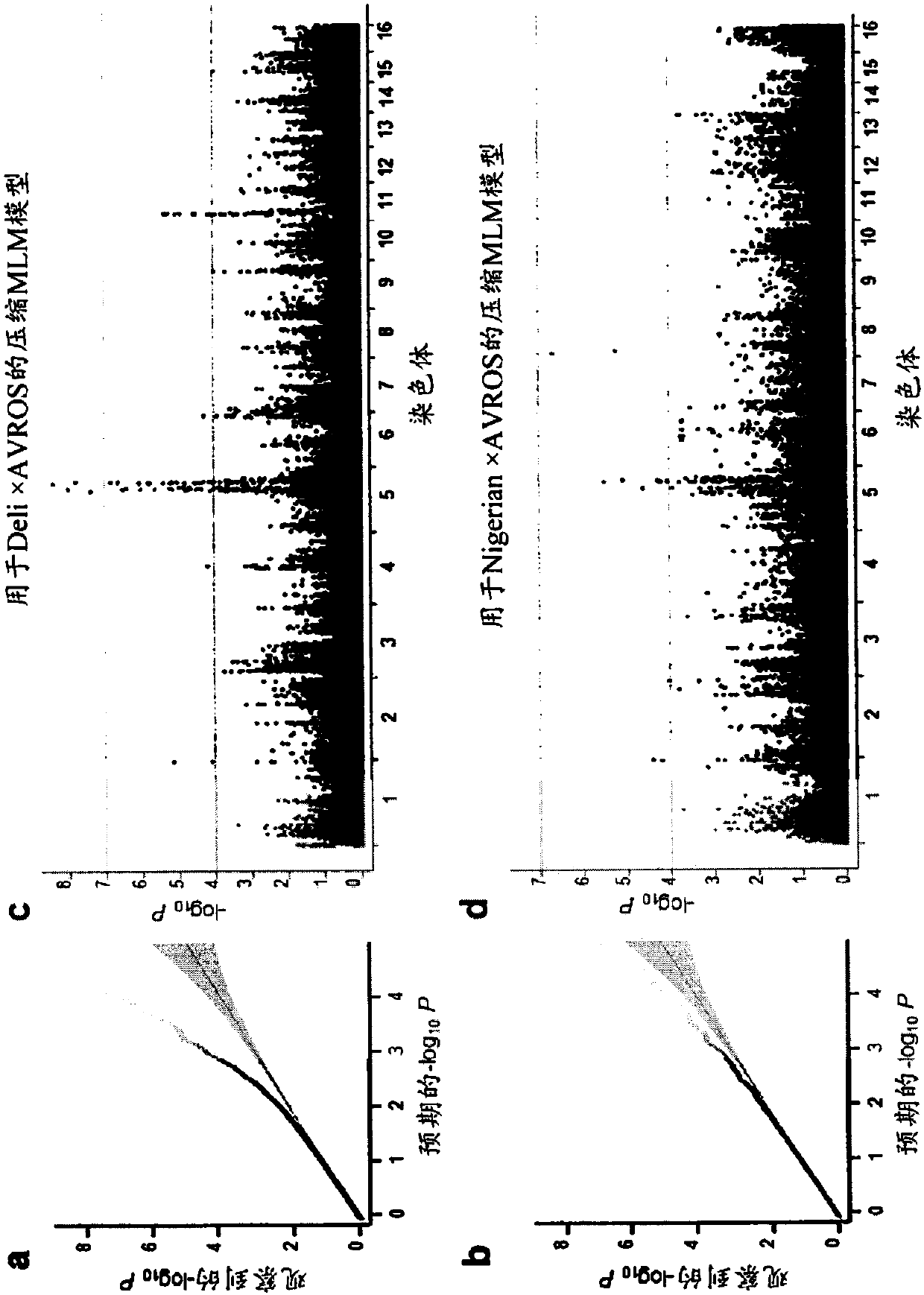
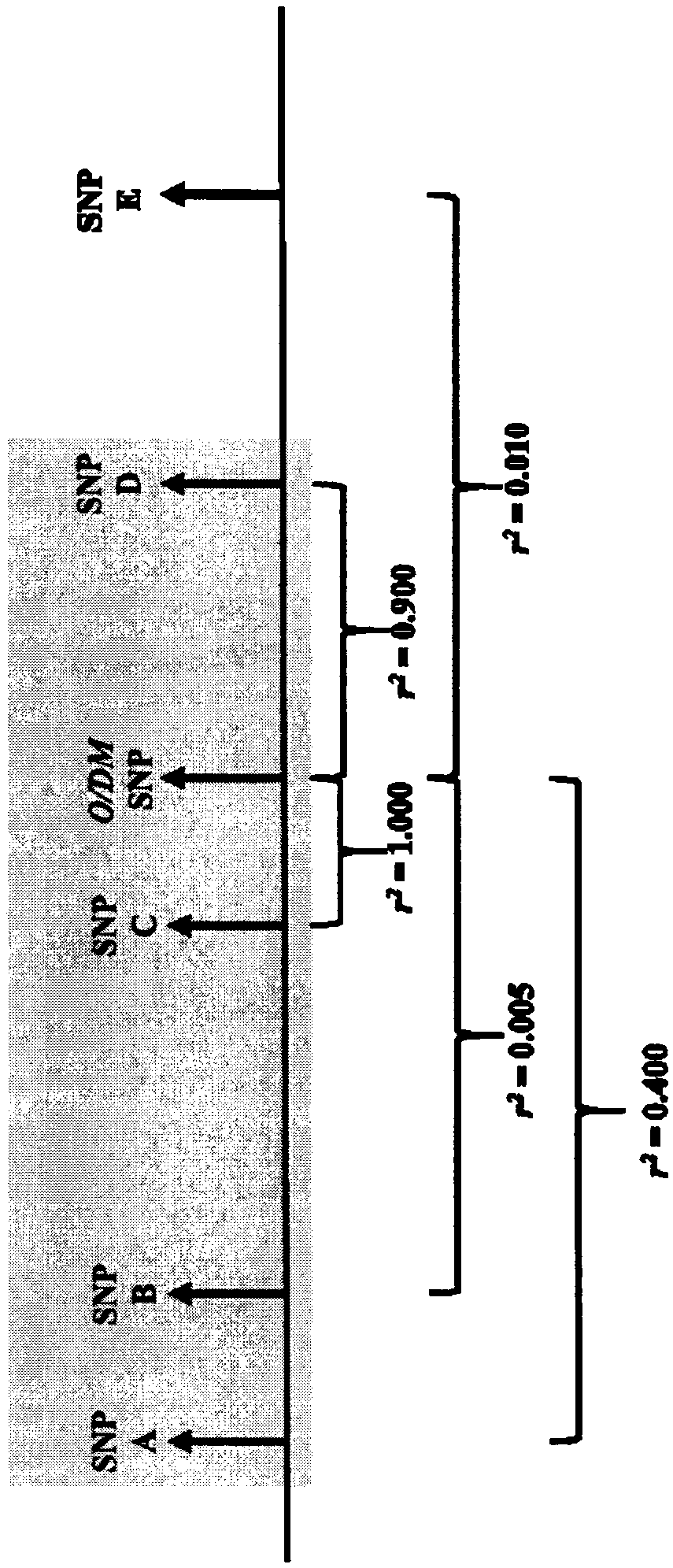
Methods for predicting palm oil yield of a test oil palm plant are disclosed. The methods comprise determining, from a sample of a test oil palm plant of a population, at least a first SNP genotype, corresponding to a first SNP marker, located in a first QTL for a high-oil- production trait and associated, after stratification and kinship correction, with the high-oil- production trait with a genome-wide -log10(p-value) of at least 4.0 in the population or having a linkage disequilibrium r2 value of at least 0.2 with respect to a first other SNP marker linked thereto and associated, after stratification and kinship correction, with the high-oil-production trait with a genome-wide -log10(p-value) of at least 4.0 in the population. The methods also comprise comparing the first SNP genotype to a corresponding first reference SNP genotype and predicting palm oil yield of the test plant based on extent of matching of the SNP genotypes.
Owner:SIME DARBY PLANTATION INTPROP SDN BHD
Method for cloning rice complex trait associated gene
InactiveCN102094001AEasy extractionQuick discoveryDNA preparationAgarose electrophoresisGenetic linkage disequilibrium
The invention provides a method for cloning a rice complex trait associated gene. A rice natural population of which the genome sequence is disclosed serves as a base; complex trait data among rice varieties is acquired; ECO-TILLING method analysis is performed on agarose electrophoresis by taking deoxyribonucleic acid (DNA) of the material as a template; generated polymorphism strips are converted into genetic markers; and linkage disequilibrium analysis is performed by association analysis software to determine a gene associated with traits and clone the gene. By the method for cloning the rice complex trait associated gene, the gene associated with the complex traits can be found with high efficiency, low cost and high speed. The method has great significance for quickening clone of the plant complex trait associated gene.
Owner:SHANGHAI AGROBIOLOGICAL GENE CENT
Method for gene mapping from chromosome and phenotype data
InactiveUS20050064408A1Microbiological testing/measurementProteomicsGene mappingGenetic linkage disequilibrium
The present invention relates to a method for gene mapping from chromosome and phenotype data, which utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. The method according to the invention is based on discovering and assessing tree-like patterns in genetic marker data. It extracts, essentially in the form of substrings and prefix trees, information about the historical recombinations in the population. This infor-mation is used to locate fragments potentially inherited from a common diseased founder, and to map the disease gene into the most likely such fragment. The method measures for each chromosomal location the disequilibrium of the prefix tree of marker strings starting from the location, to assess the distribution of disease-associated chromosomes.
Owner:LICENTIA OY
SNP molecular markers, primer pairs and kit of FABP4 gene related to beef quality in Yanbian yellow cattle and application of SNP molecular marker, primer pair and kit
ActiveCN110438243AMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide
The invention provides SNP molecular markers, primer pairs and a kit of FABP4 gene related to beef quality in Yanbian yellow cattle, and application of the SNP molecular markers, the primer pairs andthe kit, and belongs to the technical field of beef quality screening in yellow cattle. The SNP molecular markers of the FABP4 gene contain a nucleotide sequence of a polymorphism at the 3496 bp as anA / C site, a polymorphism at the 3533 bp as an A / T site, a polymorphism at the 3711 bp as a G / C site, a polymorphism at the 3745 bp as a T / C site and a polymorphism at the 3767 bp as a T / C site of theFABP4 gene. The above five polymorphic sites are located in the third exon, and are in a strong linkage disequilibrium state with similar genetic effects. When the SNP molecular markers are applied to screen breeding fat cattle, fat content and marble pattern are selected as the detection indicators for screening breeding fat cattle while the genotype is heterozygous.
Owner:YANBIAN UNIV
SSR markers for plants and uses thereof
Simple sequence repeat (SSR) markers identified in Jatropha curcas and useful for the molecular genotyping of plants. are described. These markers may be used for identifying allele polymorphisms, identifying identical or related plants, differentiating plants and studying genetic diversity in a population. The markers may also be used in genetic and phenotype studies using statistical methods, for example, linkage analysis, association mapping, linkage disequilibrium and the like. The information may be used for breeding and / or selection of plants.
Owner:ACGT INTELLECTUAL
Methods for predicting palm oil yield of a test oil palm plant
Owner:SIME DARBY PLANTATION INTPROP SDN BHD
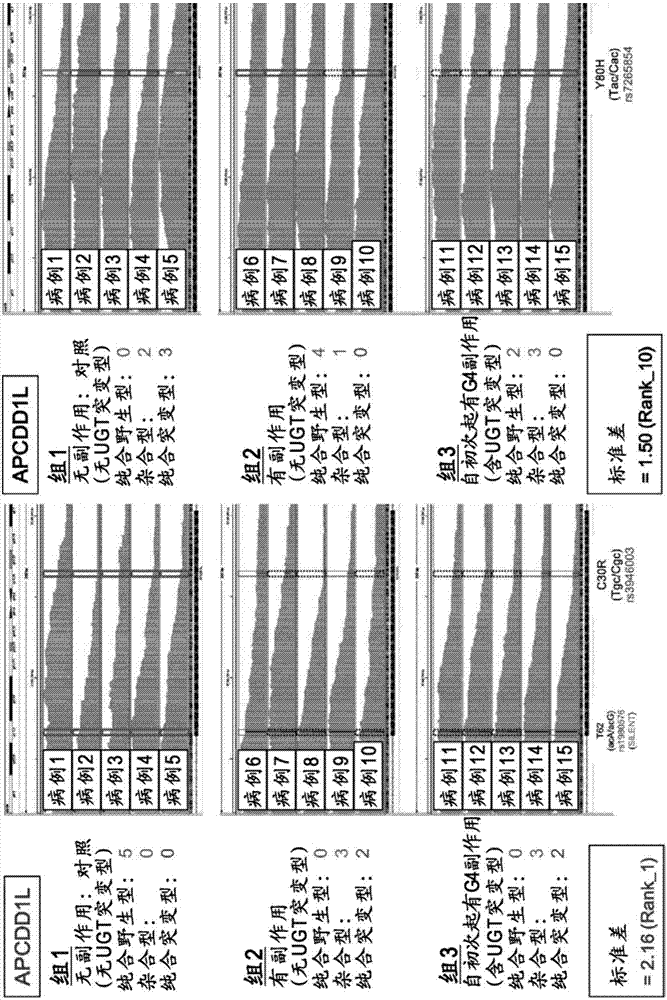
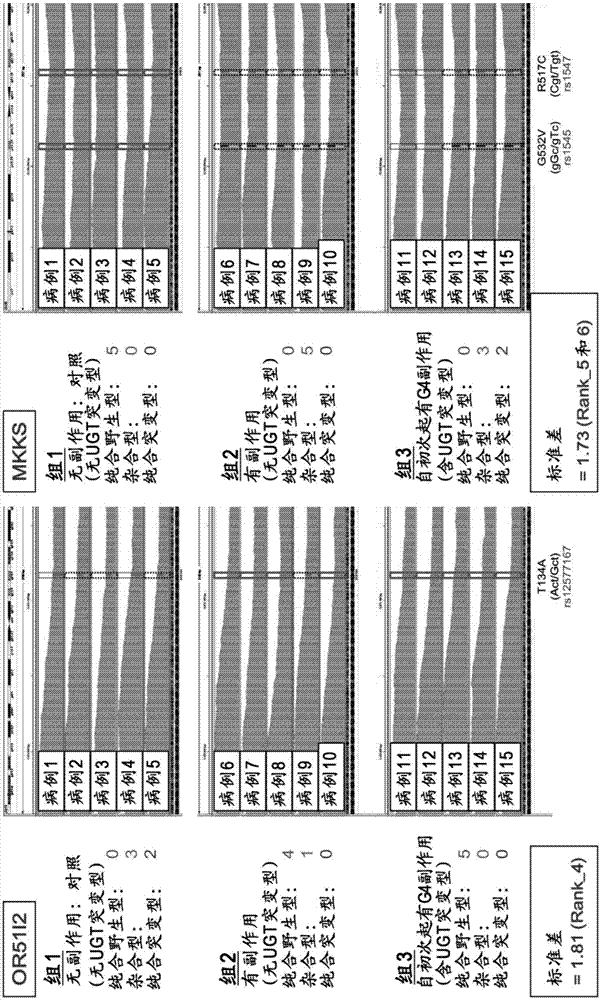
Method for assisting prediction of risk of occurrence of side effect of irinotecan
ActiveCN107208163AMicrobiological testing/measurementRecombinant DNA-technologySide effectGenetic linkage disequilibrium
The present invention addresses the problem of providing a simple and efficient means for predicting the risk of occurrence of a side effect of irinotecan by analyzing a single nucleotide polymorphism in a region encoding a specific gene. The prediction of the risk of occurrence of a side effect of irinotecan is assisted by analyzing: a single nucleotide polymorphism in a region encoding the APCDD1L gene, the R3HCC1 gene, the OR51I2 gene, the MKKS gene, the EDEM3 gene, or the ACOX1 gene which are present on genomic DNA in a biological sample collected from a subject; or a single nucleotide polymorphism which is in linkage disequilibrium with or genetically linked to the single nucleotide polymorphism, thereby determining whether the single nucleotide polymorphism is homozygous or heterozygous for a variant-type, or homozygous for a wild-type and assisting prediction of the risk of occurrence of a side effect of irinotecan.
Owner:YAMAGUCHI UNIV +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com