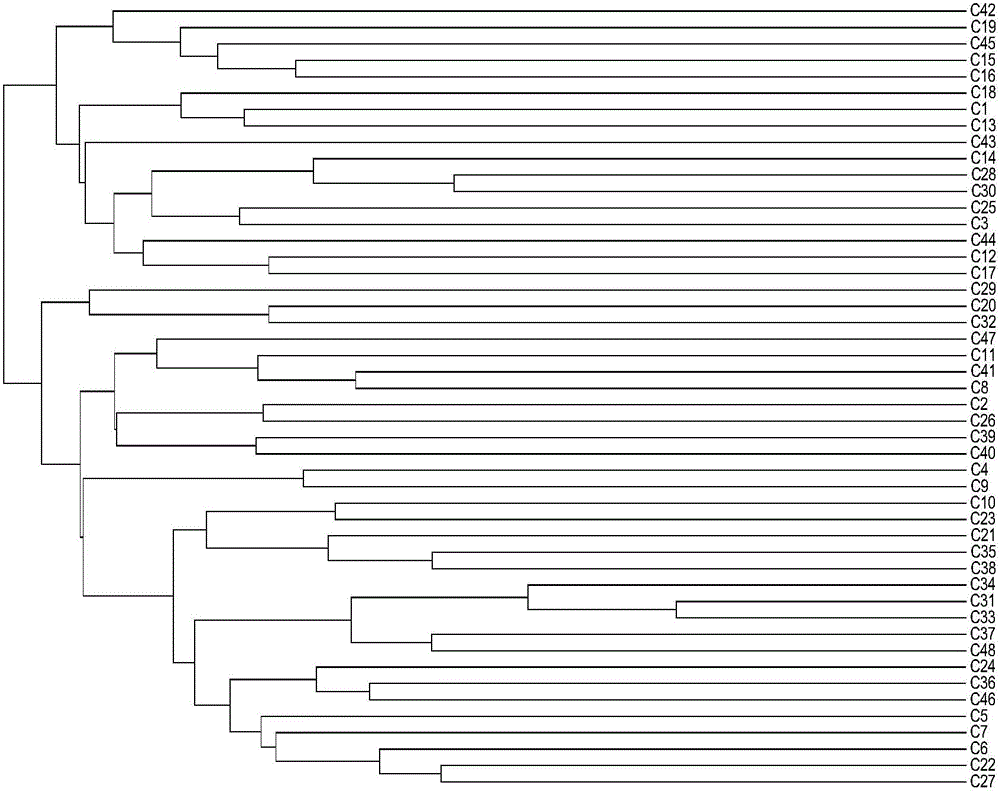
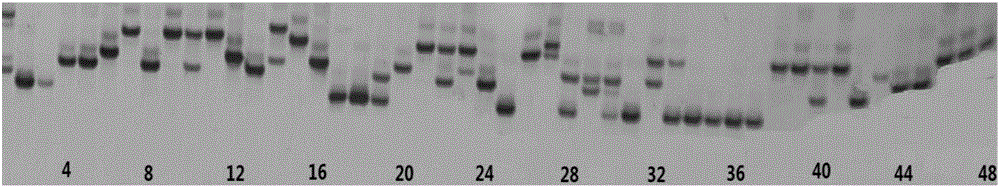
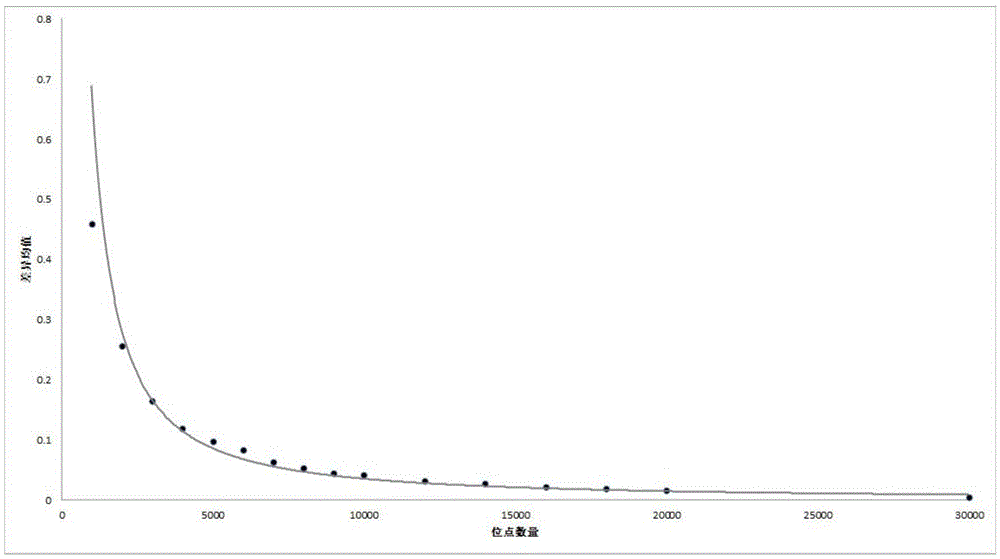
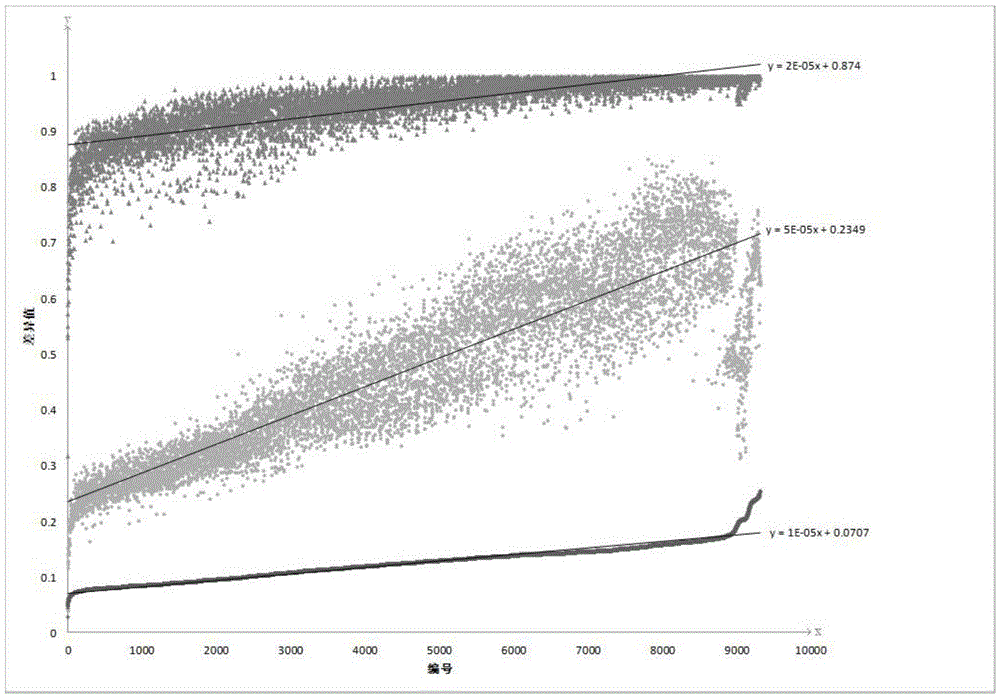
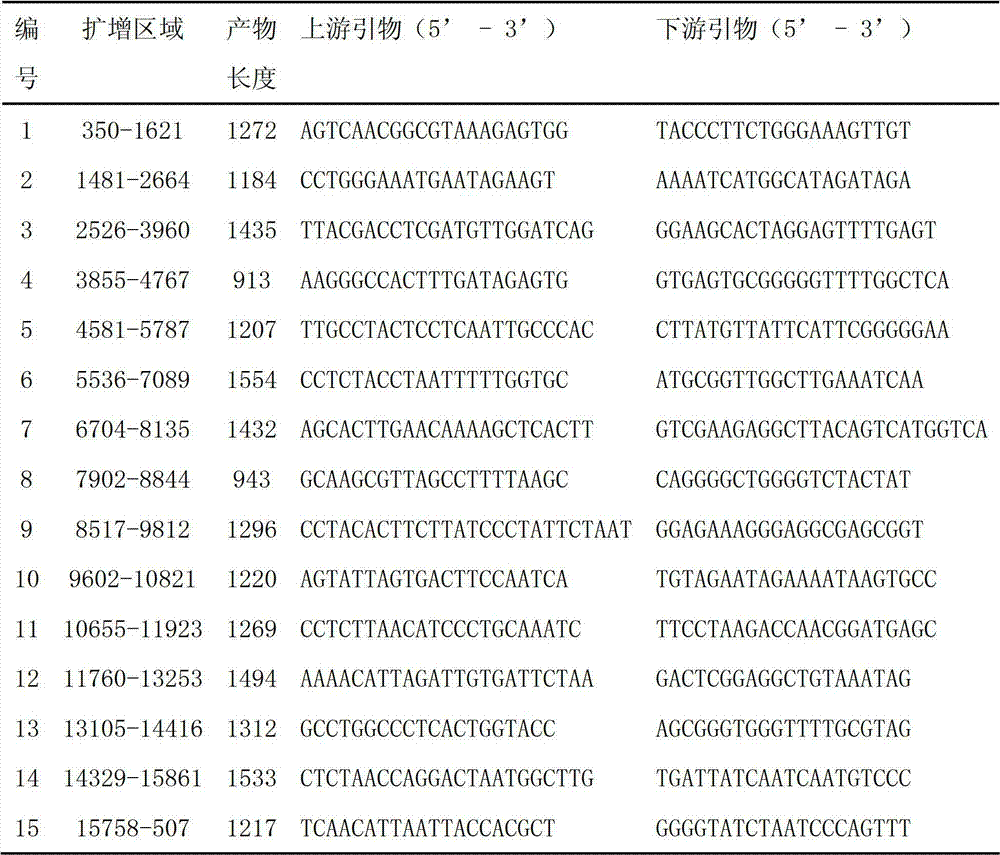
Patents
Literature
703 results about "Genetic diversity" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Genetic diversity is the total number of genetic characteristics in the genetic makeup of a species. It is distinguished from genetic variability, which describes the tendency of genetic characteristics to vary.
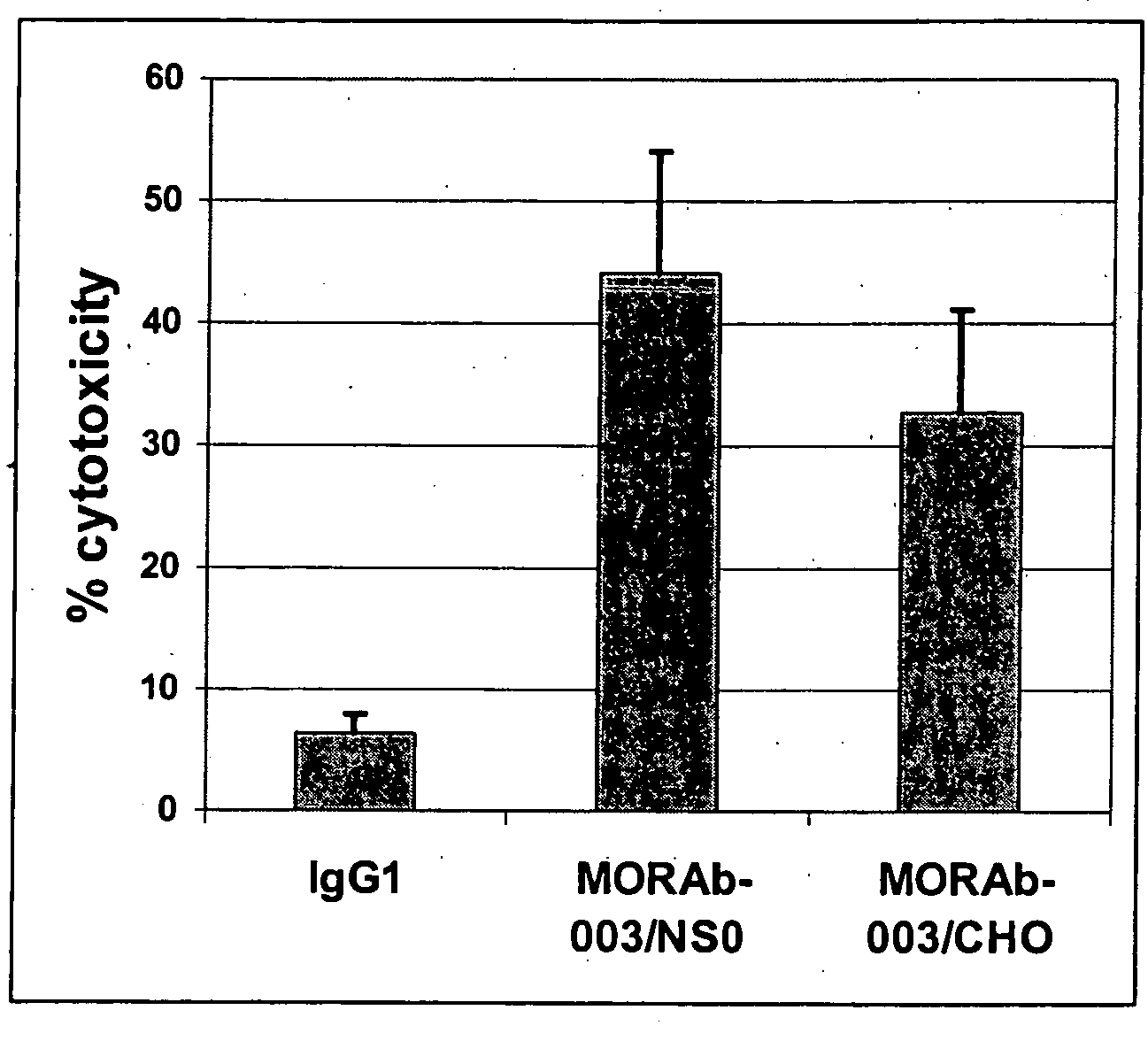
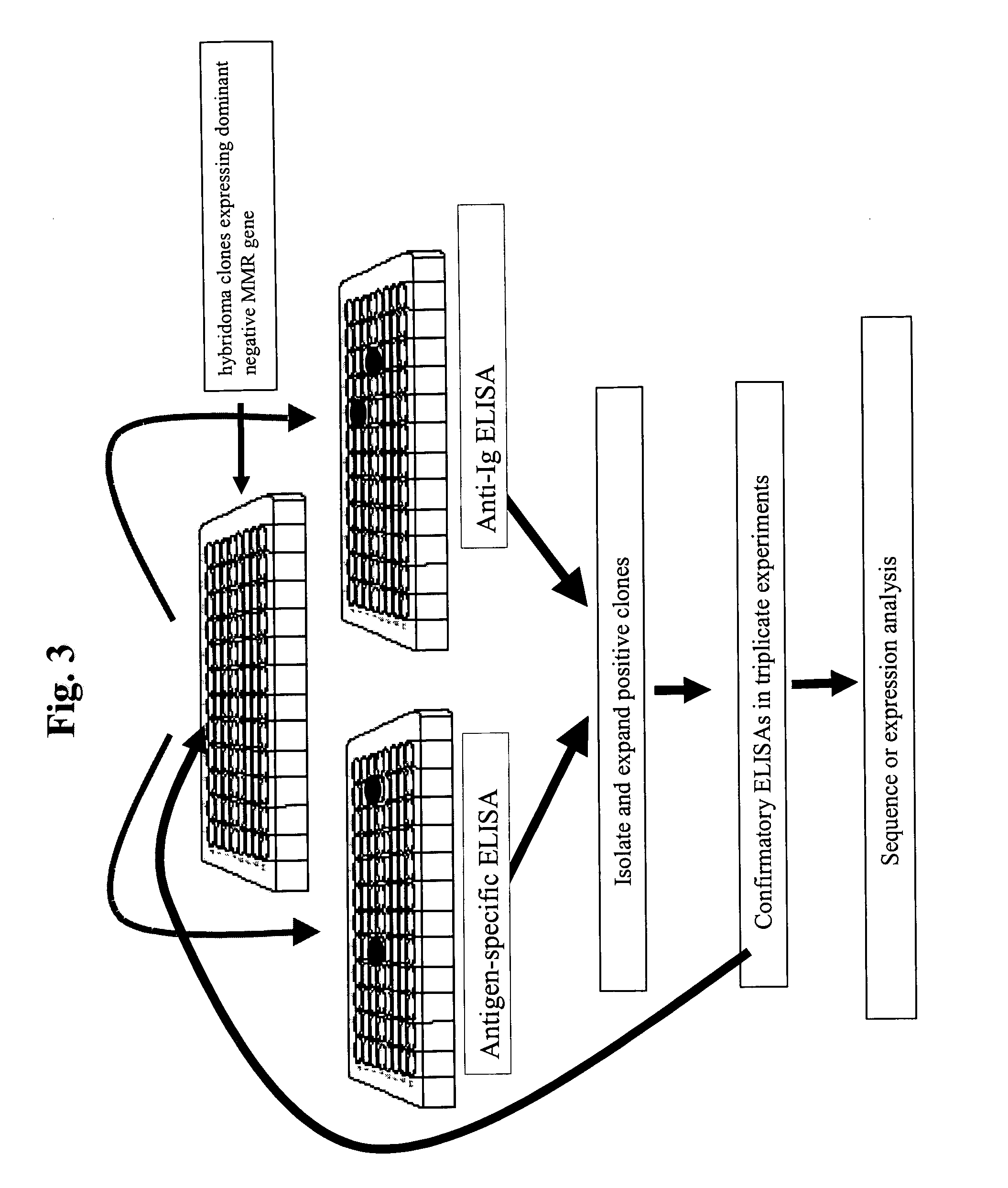
Antibodies and methods for generating genetically altered antibodies with enhanced effector function
InactiveUS20050054048A1Increase variabilityBetter pharmacokinetic profileAnimal cellsSugar derivativesGenetic diversityMonoclonal antibody
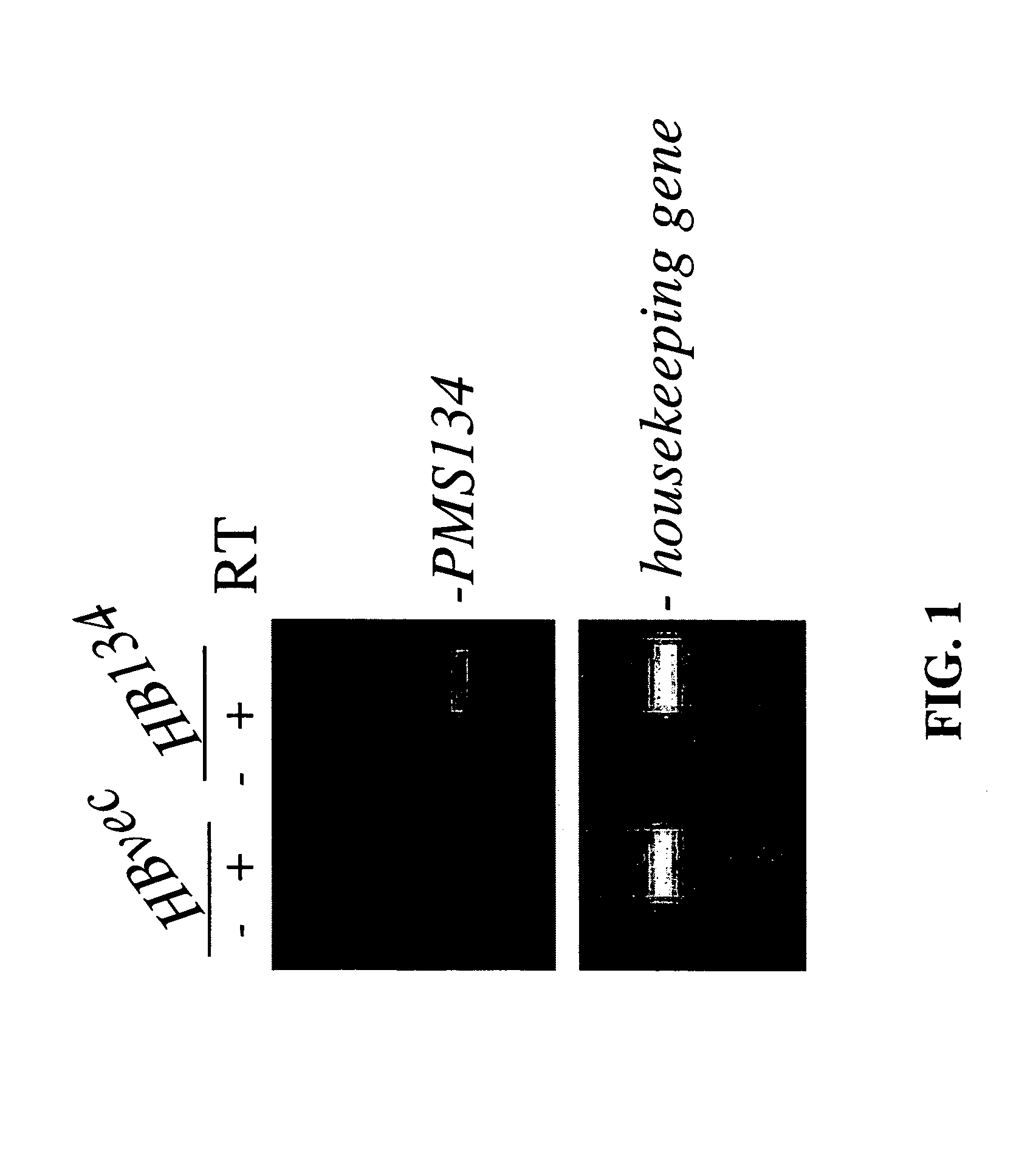
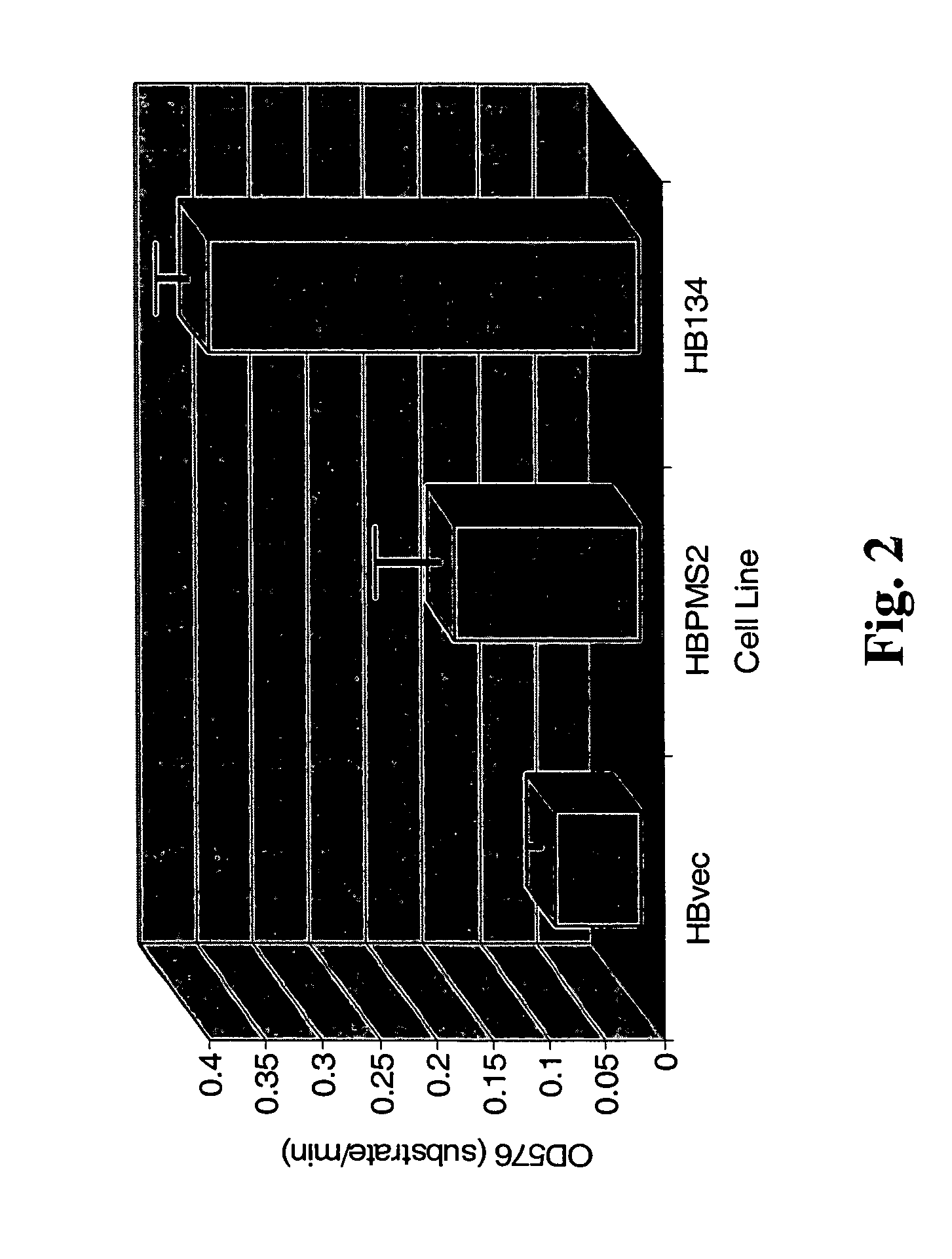
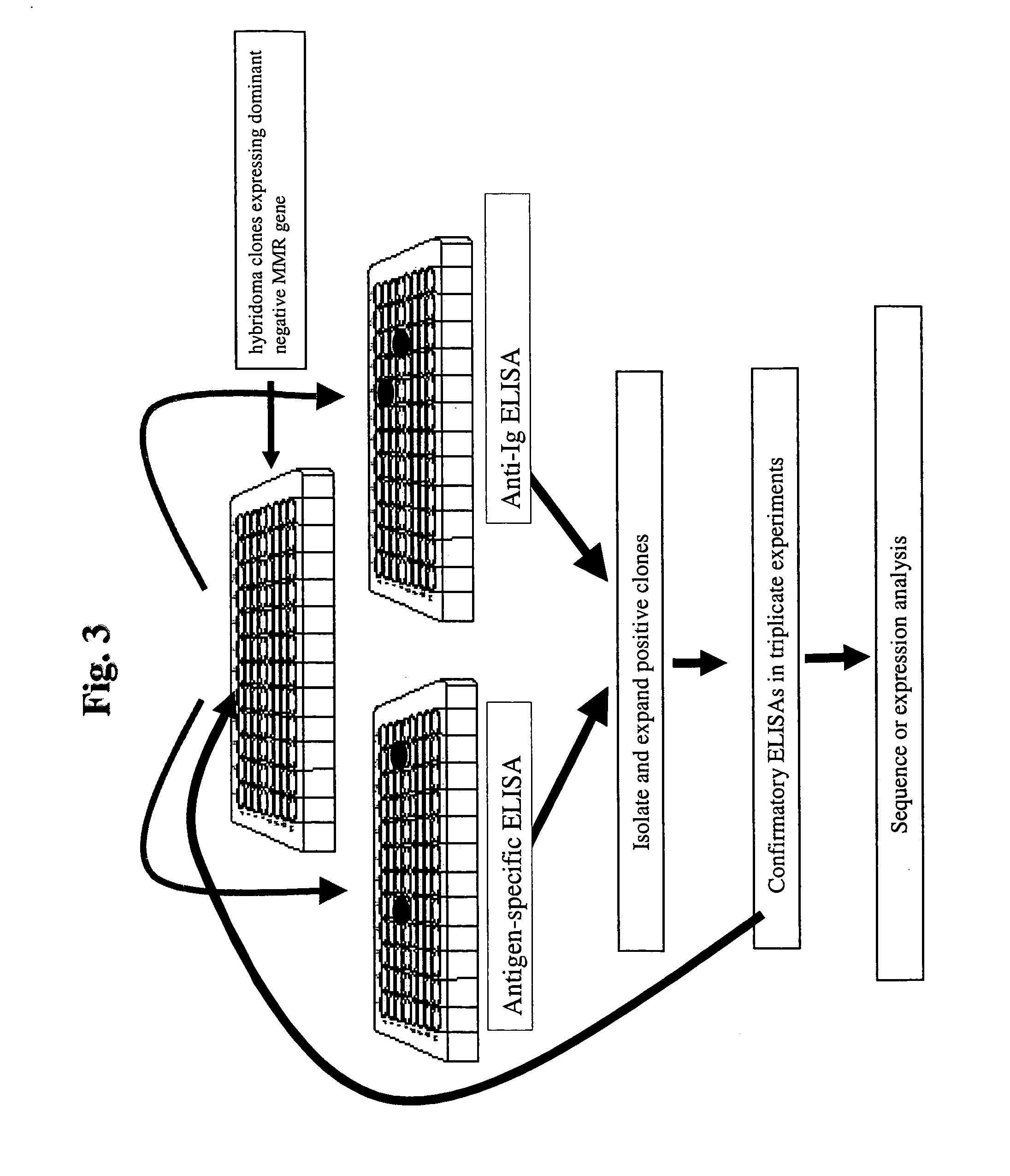
Dominant negative alleles of human mismatch repair genes can be used to generate hypermutable cells and organisms. By introducing these genes into cells and transgenic animals, new cell lines and animal varieties with novel and useful properties can be prepared more efficiently than by relying on the natural rate of mutation. These methods are useful for generating genetic diversity within immunoglobulin genes directed against an antigen of interest to produce altered antibodies with enhanced biochemical activity. Moreover, these methods are useful for generating antibody-producing cells with increased level of antibody production. The invention also provides methods for increasing the effector function of monoclonal antibodies and monoclonal antibodies with increased effector function.
Owner:EISAI INC
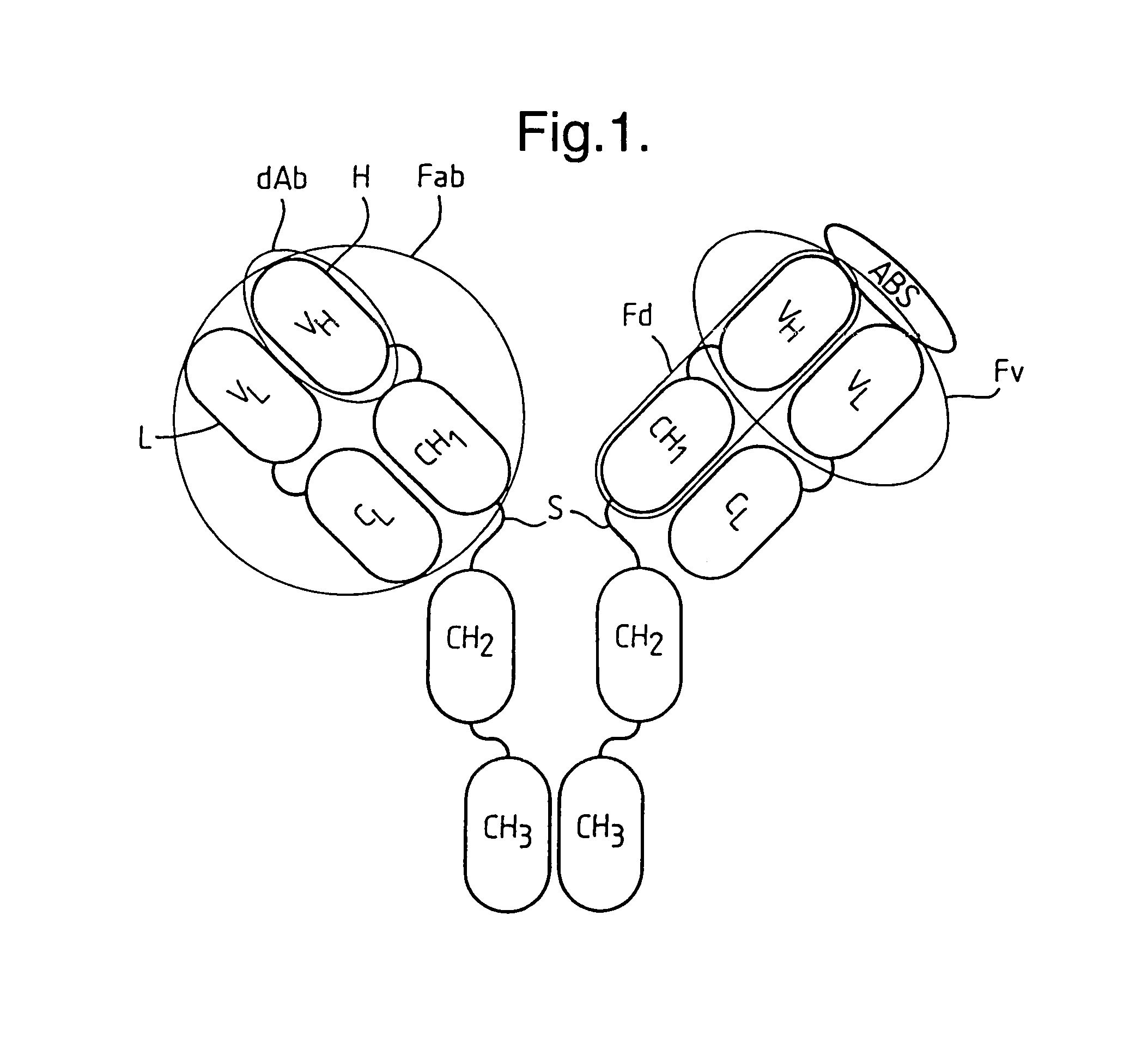
Methods for producing members of specific binding pairs
InactiveUS7063943B1Simple procedureSpeedSugar derivativesMicrobiological testing/measurementFree formGenetic diversity
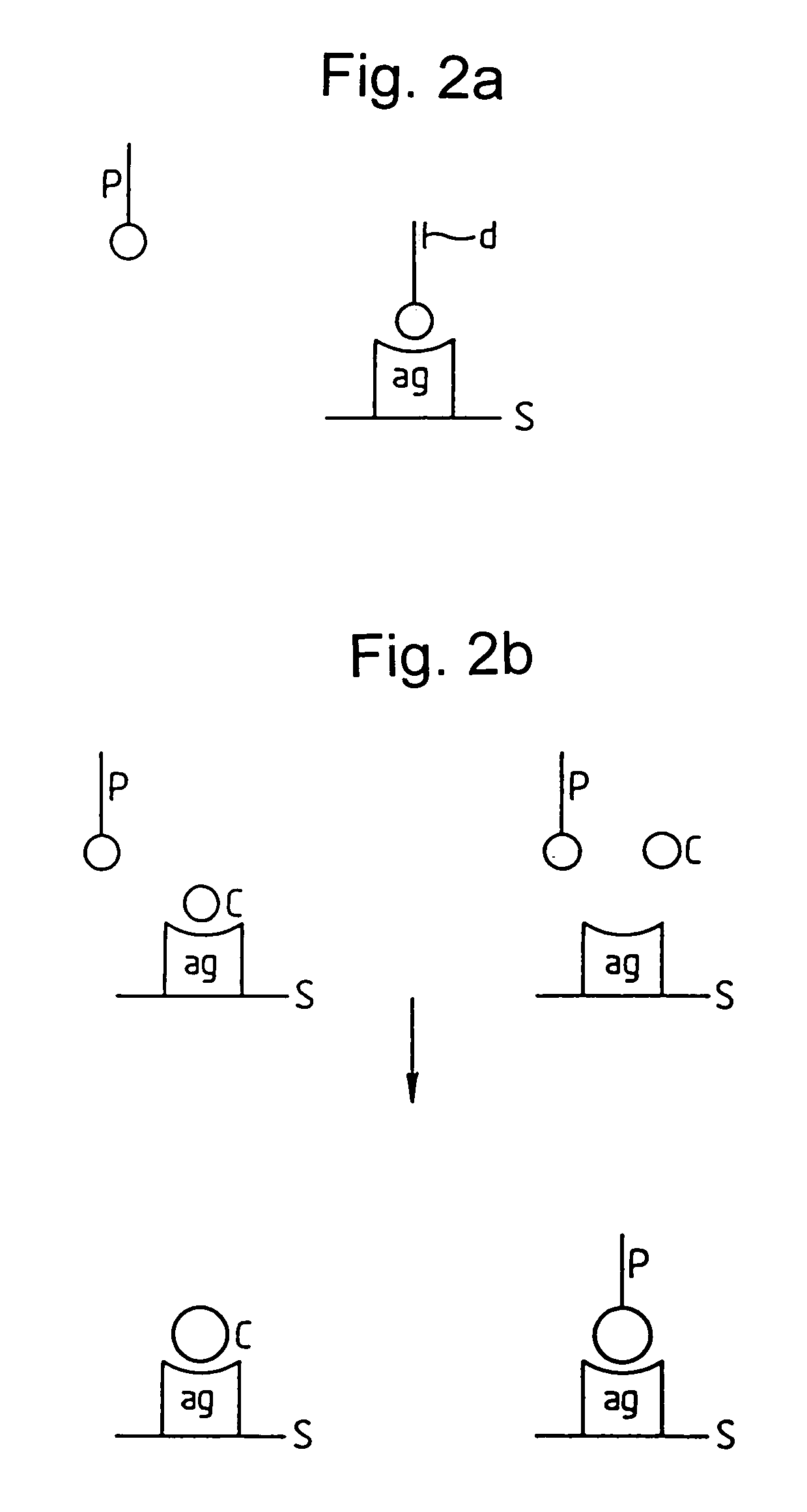
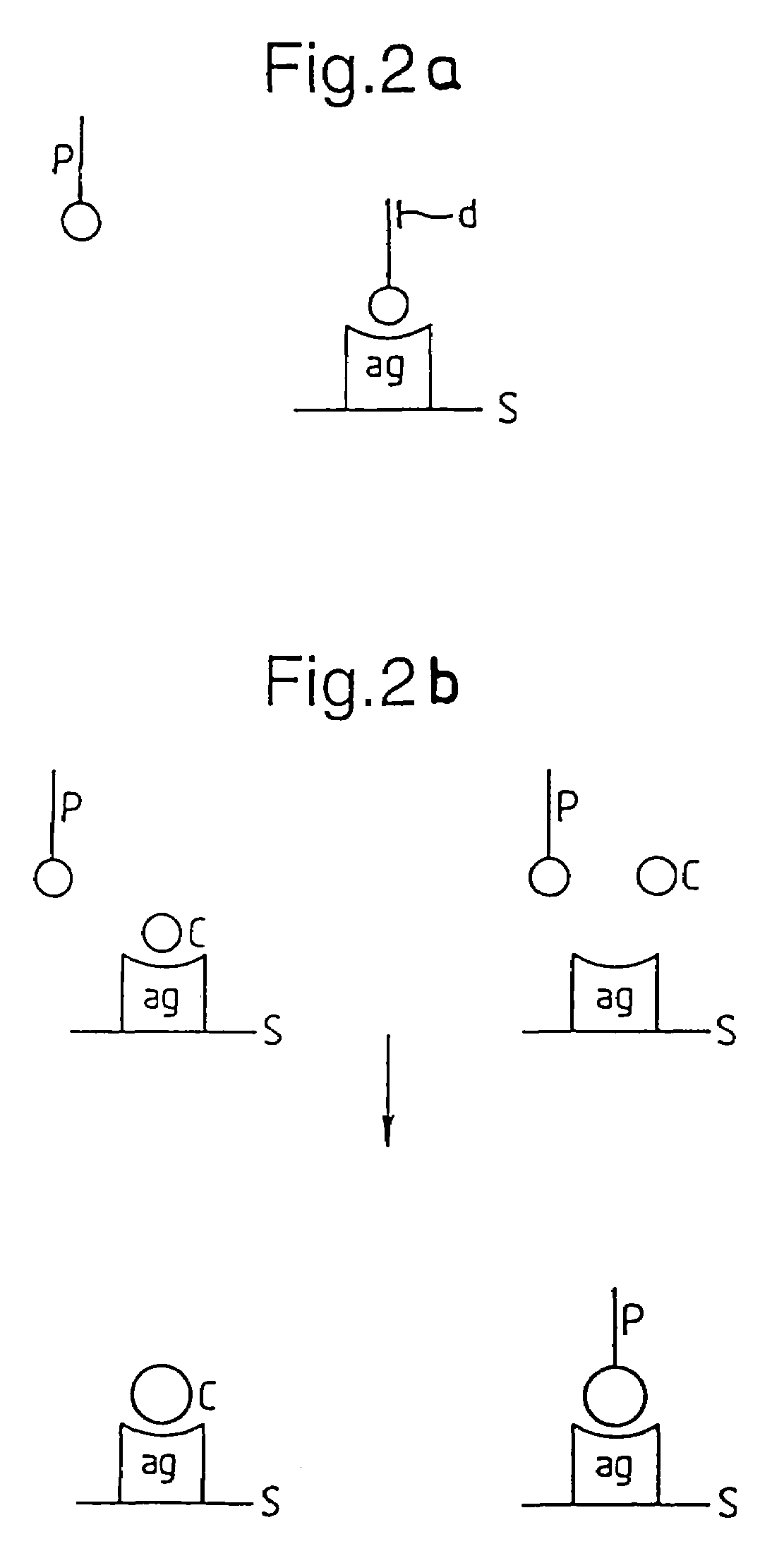
A member of a specific binding pair (sbp) is identified by expressing DNA encoding a genetically diverse population of such sbp members in recombinant host cells in which the sbp members are displayed in functional form at the surface of a secreted recombinant genetic display package (rgdp) containing DNA encoding the sbp member or a polypeptide component thereof, by virtue of the sbp member or a polypeptide component thereof being expressed as a fusion with a capsid component of the rgdp. The displayed sbps may be selected by affinity with a complementary sbp member, and the DNA recovered from selected rgdps for expression of the selected sbp members. Antibody sbp members may be thus obtained, with the different chains thereof expressed, one fused to the capsid component and the other in free form for association with the fusion partner polypeptide. A phagemid may be used as an expression vector, with said capsid fusion helping to package the phagemid DNA. Using this method libraries of DNA encoding respective chains of such multimeric sbp members may be combined, thereby obtaining a much greater genetic diversity in the sbp members than could easily be obtained by conventional methods.
Owner:MEDIMMUNE LTD
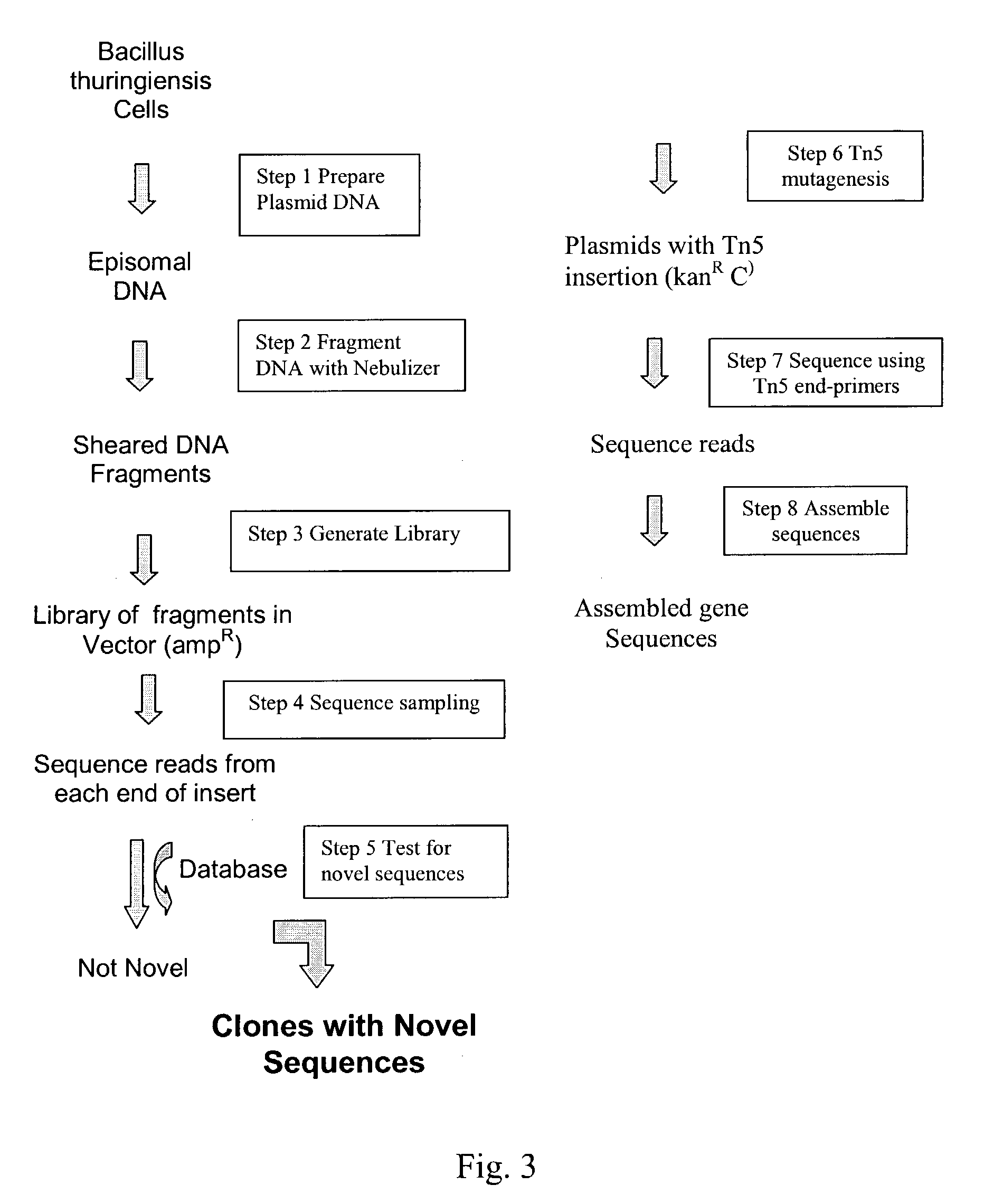
Integrated system for high throughput capture of genetic diversity
InactiveUS20040014091A1Rapid and highly efficient characterizationFast wayMicrobiological testing/measurementBiological testingGenetic diversitySequence database
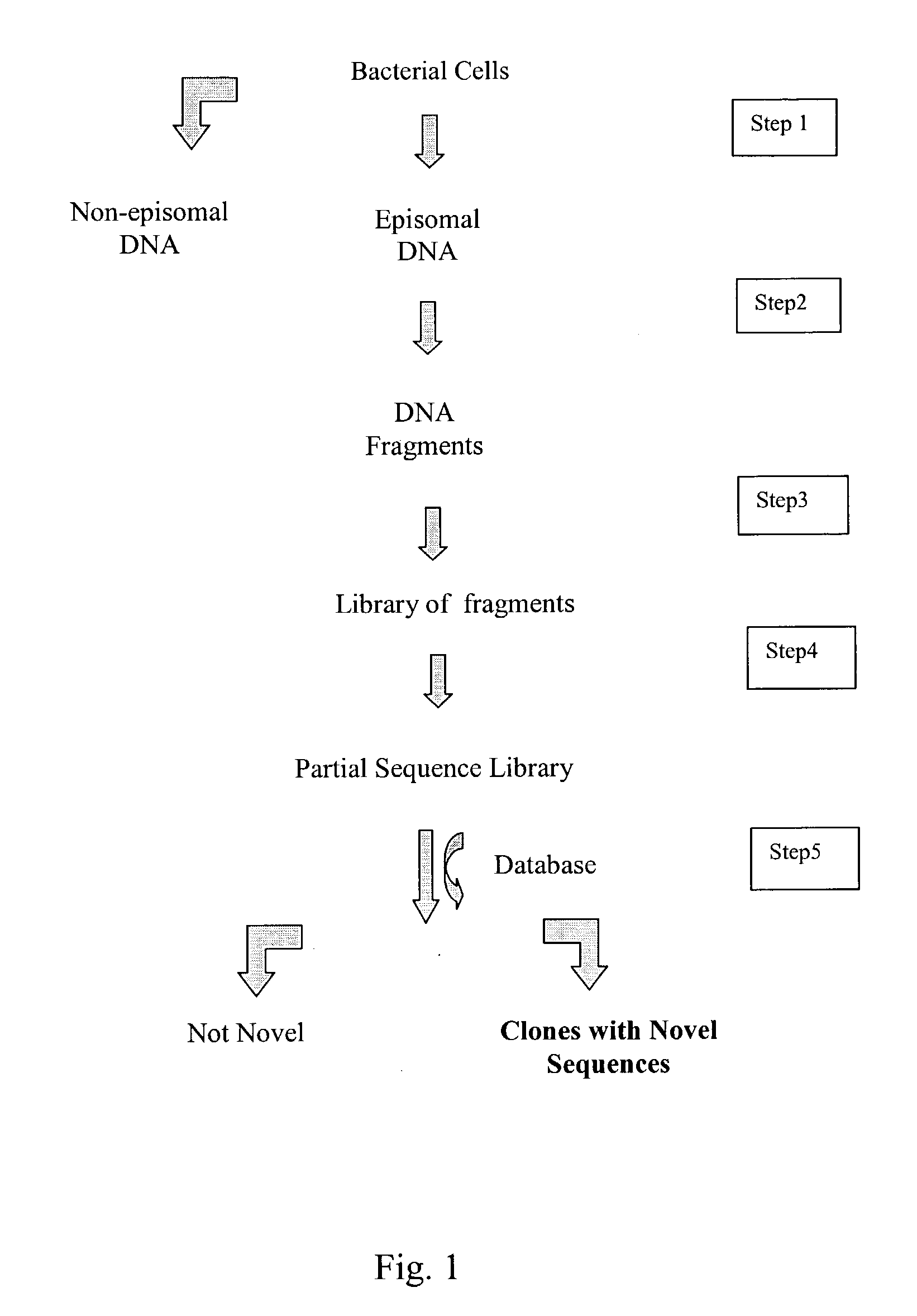
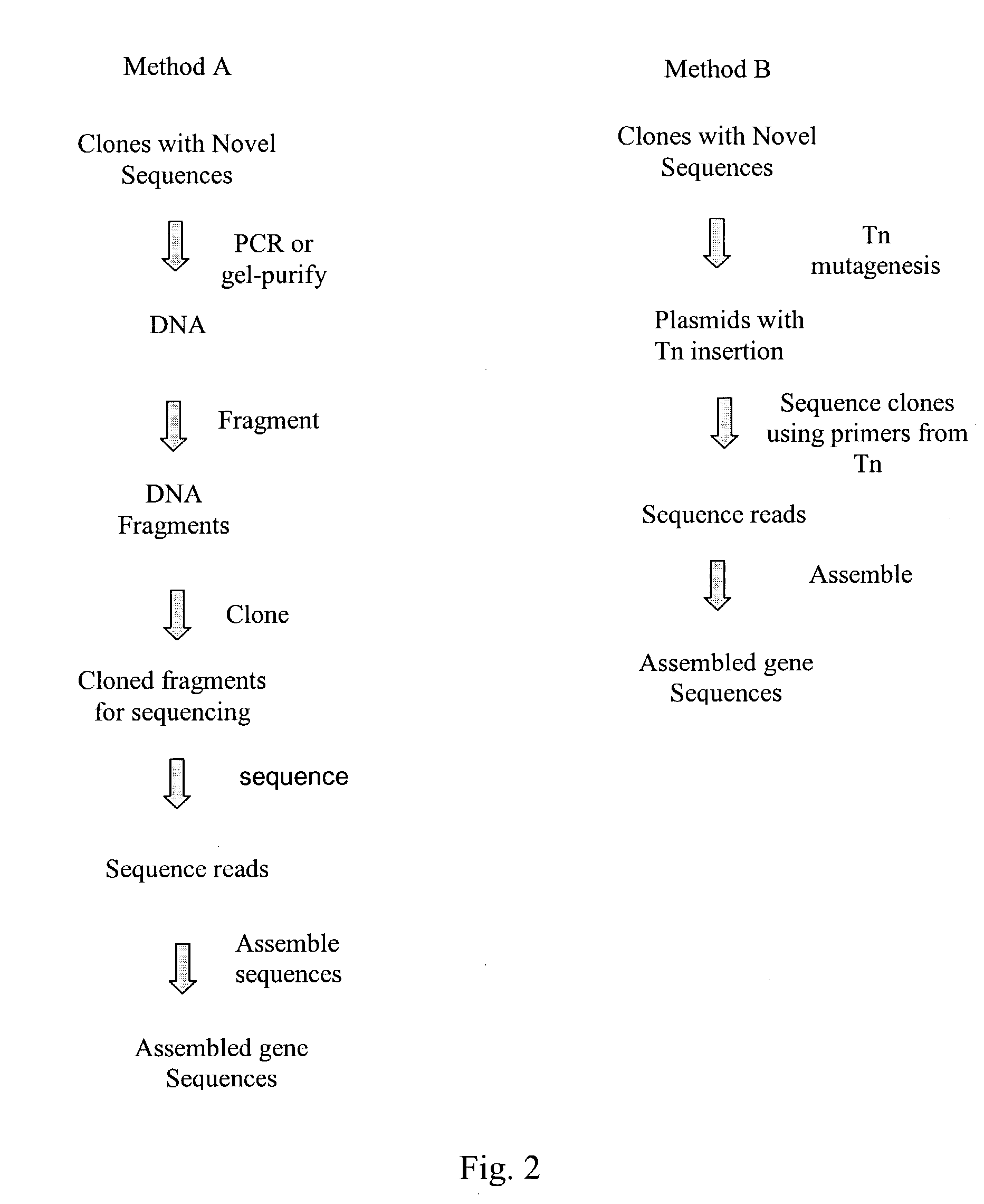
Compositions and methods for rapid and highly efficient characterization of genetic diversity in organisms are provided. The methods involve rapid sequencing and characterization of extrachromosomal DNA, particularly plasmids, to identify and isolate useful nucleotide sequences. The method targets plasmid DNA and avoids repeated cloning and sequencing of the host chromosome, thus allowing one to focus on the genetic, elements carrying maximum genetic diversity. The method involves generating a library of extrachromosomal DNA clones, sequencing a portion of the clones, comparing the sequences against a database of existing DNA sequences, using an algorithm to select said novel nucleotide sequence based on the presence or absence of said portion in a database, and identification of at least one novel nucleotide sequence. The DNA sequence can also be translated in all six frames and the resulting amino acid sequences can be compared against a database of protein sequences. The integrated approach provides a rapid and efficient method to identify and isolate useful genes. Organisms of particular interest include, but are not limited to bacteria, fungi, algae, and the like. Compositions comprise a mini-cosmid vector comprising a stuffer fragment and at least one cos site.
Owner:BASF AGRICULTURAL SOLUTIONS SEED LLC
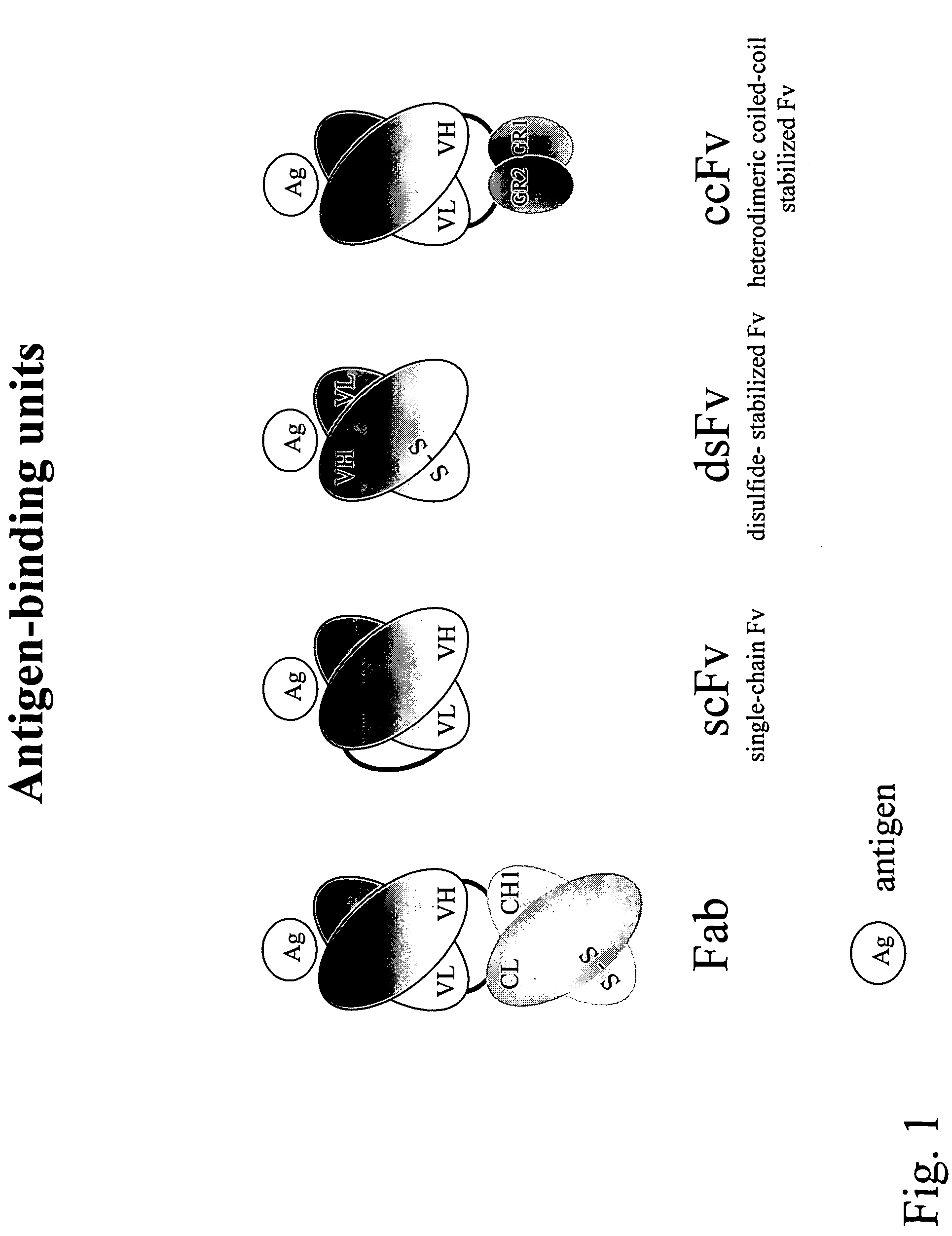
Compositions and methods for generating chimeric heteromultimers
InactiveUS7429652B2Facilitates high throughput productionAvoid assemblyFungiBacteriaGenetic diversityAntigen binding
Owner:ABMAXIS
Methods for producing members of specific binding pairs
InactiveUS7662557B2Simple procedureSpeedImmunoglobulins against blood group antigensAntibody mimetics/scaffoldsGenetic diversityDna encoding
A member of a specific binding pair (sbp) is identified by expressing DNA encoding a genetically diverse population of such sbp members in recombinant host cells in which the sbp members are displayed in functional form at the surface of a secreted recombinant genetic display package (rgdp) containing DNA encoding the sbp member or a polypeptide component thereof, by virtue of the sbp member or a polypeptide component thereof being expressed as a fusion with a capsid component of the rgdp. The displayed sbps may be selected by affinity with a complementary sbp member, and the DNA recovered from selected rgdps for expression of the selected sbp members. A phagemid may be used as an expression vector, with said capsid fusion helping to package the phagemid DNA.
Owner:MEDIMMUNE LTD
Cell analysis in multi-through-hole testing plate
InactiveUS20050059074A1Simplify the test procedureEasy constructionSequential/parallel process reactionsLibrary tagsGenetic diversityTonicity
A spatially fixed library of mutated cells and a method for creating such a library. The library is created by creating a set of cells having a genetic diversity, causing the cells to multiply and express phenotypes, preparing a solution containing the cells diluted in a medium, populating a plurality of through-holes of a testing plate with the solution such that surface tension holds the solution in respective through-holes, and analyzing the phenotypes of the cells in the solution on a hole-by-hole basis. Cells expressing specified phenotypes may then be retrieved.
Owner:LIFE TECH CORP
DNA-based methods of geochemical prospecting
InactiveUS8071295B2Quick fixQuick snapSugar derivativesMicrobiological testing/measurementGenetic diversityForensic science
The present invention relates to methods for performing surveys of the genetic diversity of a population. The invention also relates to methods for performing genetic analyses of a population. The invention further relates to methods for the creation of databases comprising the survey information and the databases created by these methods. The invention also relates to methods for analyzing the information to correlate the presence of nucleic acid markers with desired parameters in a sample. These methods have application in the fields of geochemical exploration, agriculture, bioremediation, environmental analysis, clinical microbiology, forensic science and medicine.
Owner:TAXON BIOSCI
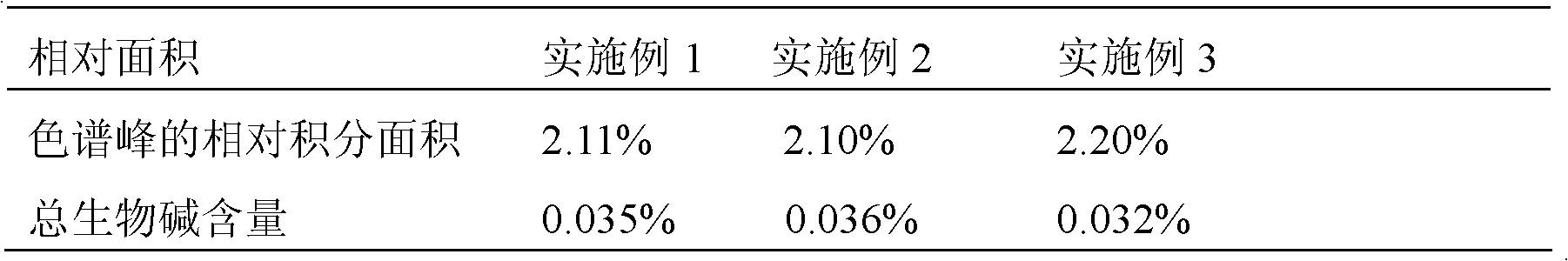
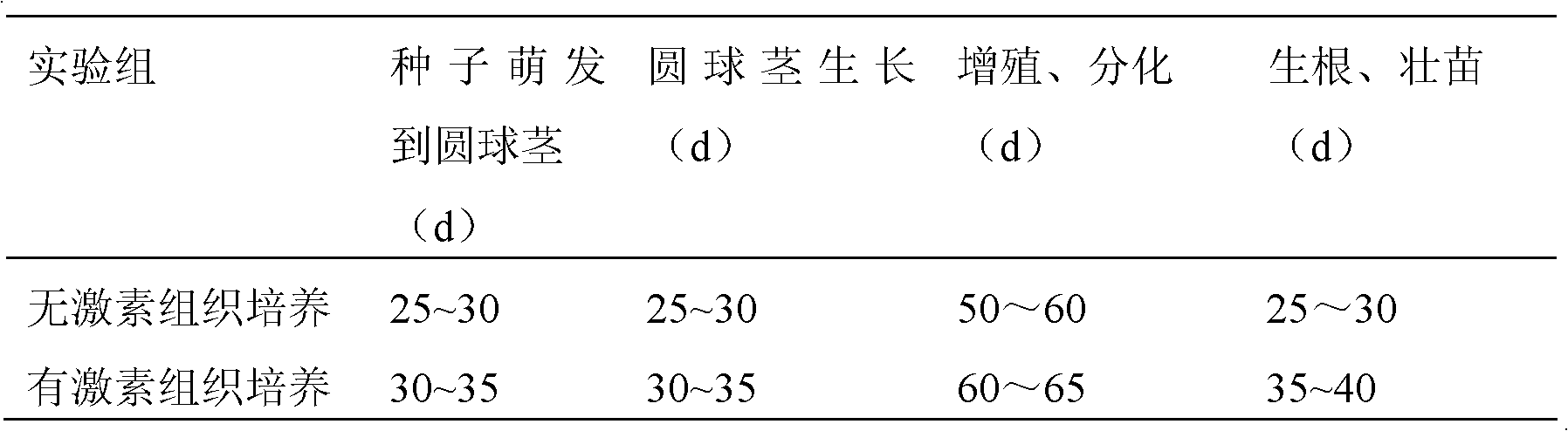
Hormone-free tissue culture method for dendrobium candidum
InactiveCN102613086ALow costReduce waste of resourcesPlant tissue cultureHorticulture methodsGenetic diversityPlant variety
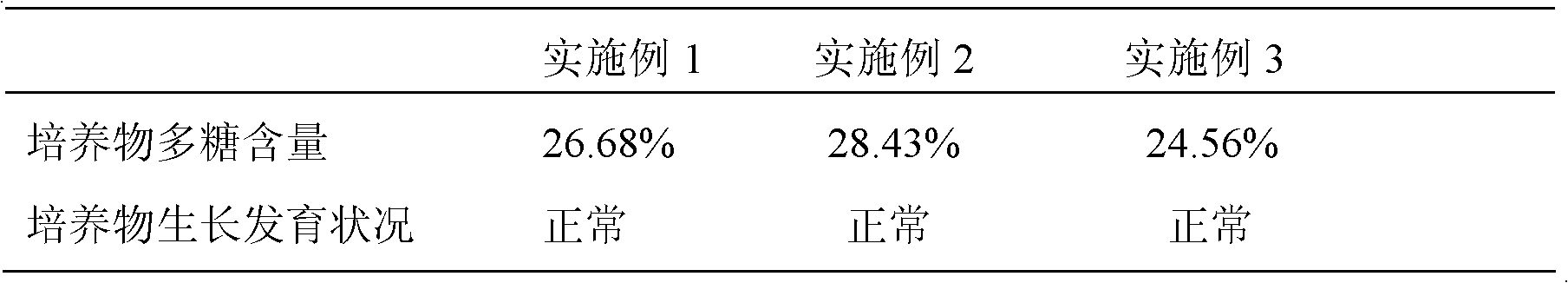
The invention discloses a hormone-free tissue culture method for dendrobium candidum, which comprises four tissue culture stages including germinating of seeds of the dendrobium candidum into spherical stems, multiplication and differentiation, and rootage and sound seedlings at stem sections; culture media used at the four stages include a hormone-free MS (Murashige and Skoog) culture medium, a hormone-free MS basal culture medium plus 15-20 g / l mashed potato culture medium, a hormone-free MS basal culture medium plus 15-20 g / l mashed banana plus 15-20 g / l mashed potato culture medium and a hormone-free 1 / 2 MS basal culture medium plus 15-20 g / l mashed banana plus 15-20 g / l mashed potato culture medium. The polysaccharide content and total alkaloid content in the culture of the dendrobium candidum obtained by hormone-free culture are increased compared with hormone-containing culture; the growth state of the dendrobium candidum is normal; the genetic diversity of the plant variety is protected; with the adoption of the method, the tissue culture of the dendrobium candidum is more convenient, green and environmentally friendly, and the resources are saved.
Owner:NANJING AGRICULTURAL UNIVERSITY
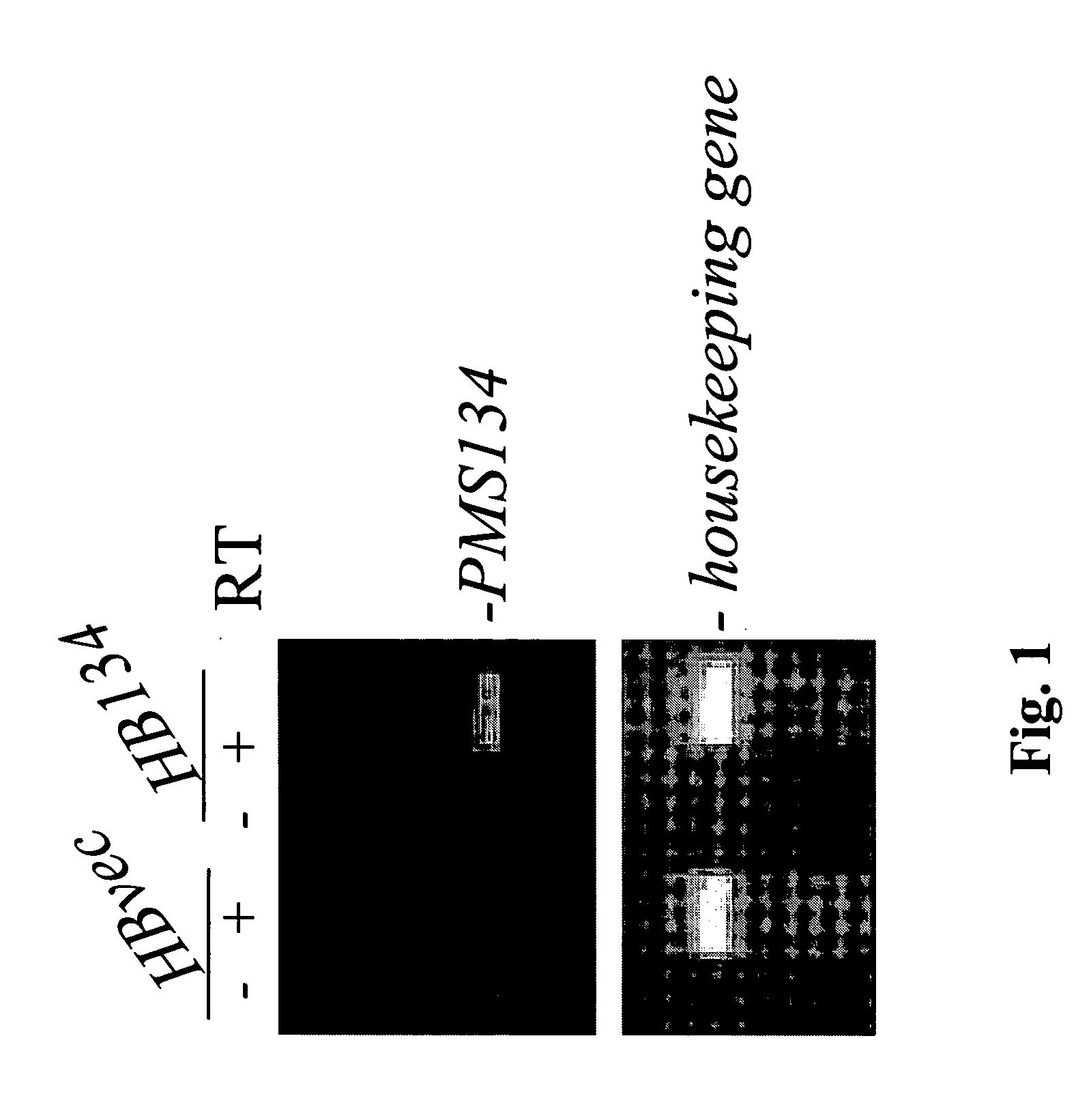
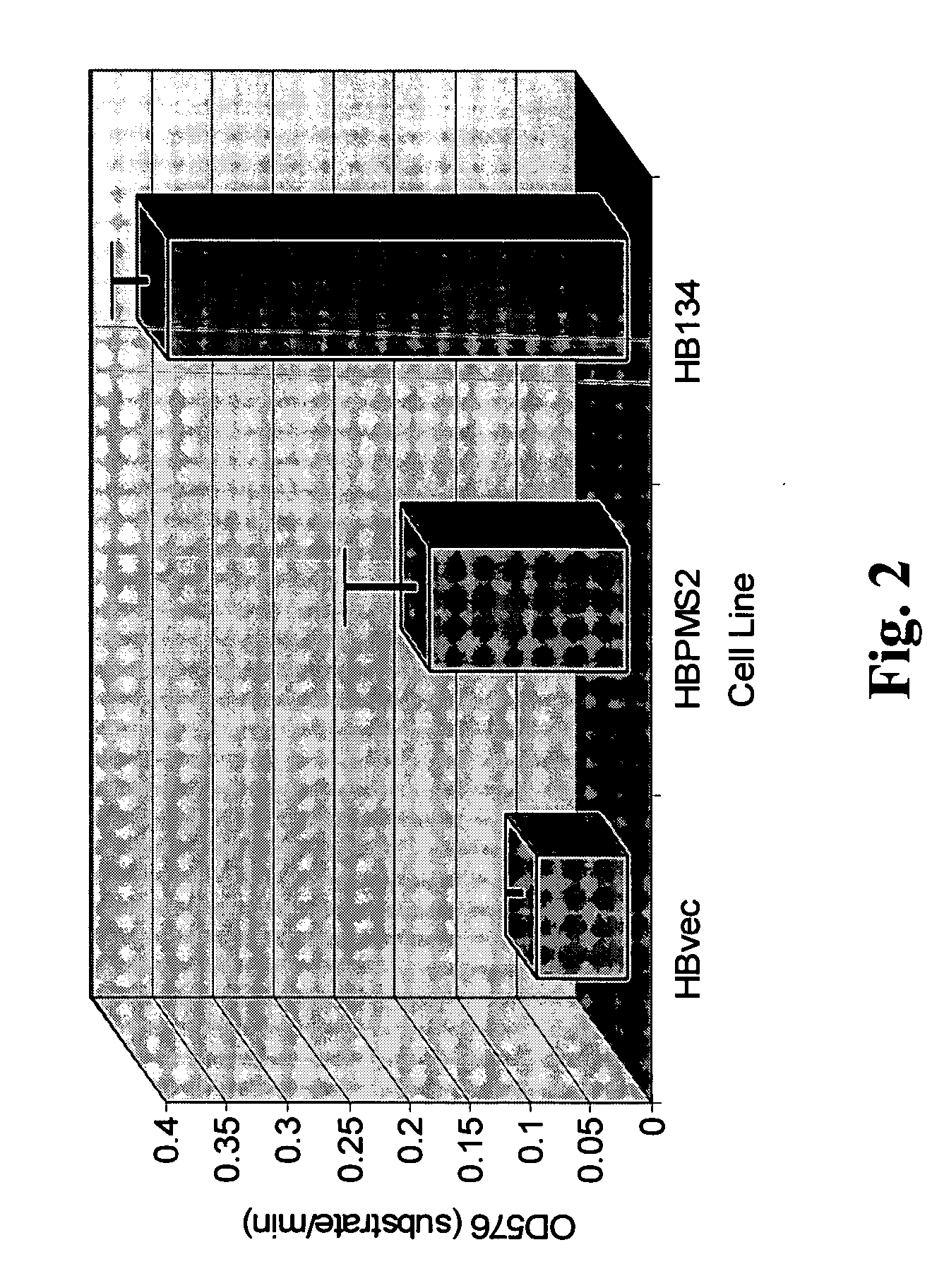
Genetically altered antibody-producing cell lines with improved antibody characteristics
InactiveUS20050048621A1Increase variabilityBetter pharmacokinetic profileAnimal cellsSugar derivativesBiological bodyGenetic diversity
Dominant negative alleles of human mismatch repair genes can be used to generate hypermutable cells and organisms. Cells may be selected for expression of activation-induced cytidine deaminase (AID), stimulated to produce AID, or manipulated to express AID for further enhancement of hypermutability. These methods are useful for generating genetic diversity within immunoglobulin genes directed against an antigen of interest to produce altered antibodies with enhanced biochemical activity. Moreover, these methods are useful for generating antibody-producing cells with increased level of antibody production.
Owner:EISAI INC
Upland cotton SNP marker and application thereof
InactiveCN105349537AImprove detection efficiencyImprove work efficiencyNucleotide librariesMicrobiological testing/measurementAgricultural scienceHomologous sequence
The invention discloses an upland cotton SNP marker and application thereof, and belongs to the field of upland cotton SNP marker development. The upland cotton SNP marker is obtained by developing a single-copy SNP marker of an upland cotton genome via chip hybridization and homologous sequence comparison, and picking out SNP markers in a linkage disequilibrium recession distance via linkage disequilibrium analysis. The SNP marker provided by the invention can be applied to analysis of genetic diversity and group structure, or to germplasm identification of upland cotton, is single-copy, and has no remarkable linkage disequilibrium among markers, so that the efficiency of detection, adopting the SNP marker, during analysis of genetic diversity, group structure or fingerprint identification can be greatly improved, and the work efficiency is higher.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
SSR (Simple Sequence Repeat) core primer group developed based on whole genome sequence of foxtail millet and application of SSR core primer group
ActiveCN103060318AImproved geneticsEvenly distributedMicrobiological testing/measurementDNA preparationAgricultural scienceGenetic diversity
The invention discloses an SSR (Simple Sequence Repeat) core primer group developed based on whole genome sequence of foxtail millet and an application of the SSR core primer group, and belongs to the technical field of molecular biology. The core primer group comprises 30 pairs of primers, wherein nucleotide sequences are represented by SEQ ID NO. 1-60. The primer has advantages of clear electrophoretic band and rich polymorphism, and is uniformly distributed and stable in amplification. The invention also provides the application of the SSR core primer group in identifying the genetic diversity and variety of the foxtail millet. The primer group can be used for precisely and quickly identifying the variety of the foxtail millet and precisely reflecting a genetic relationship among the varieties of the foxtail millet.
Owner:CROP RES INST SHANDONG ACAD OF AGRI SCI
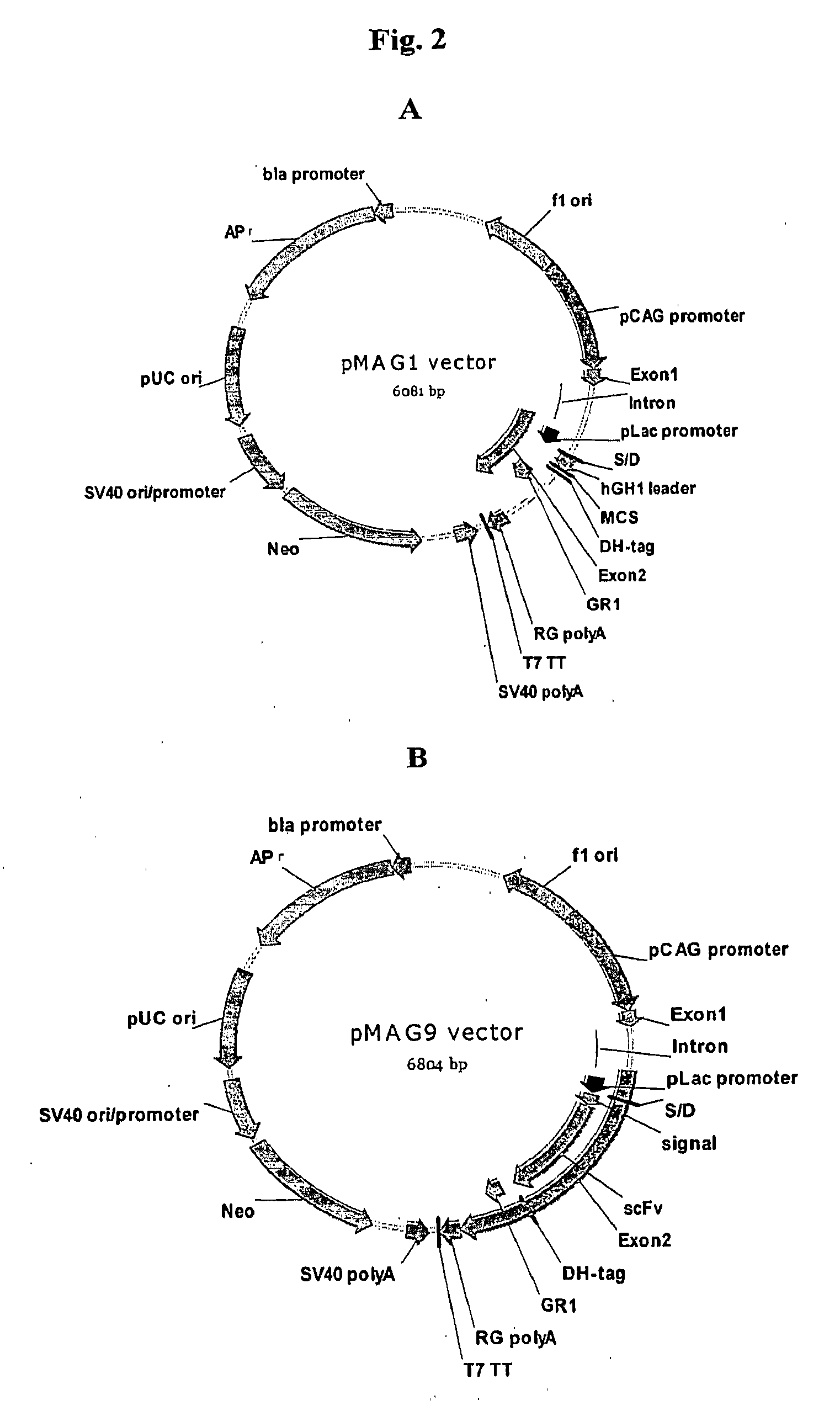
Eukaryotic cell display systems
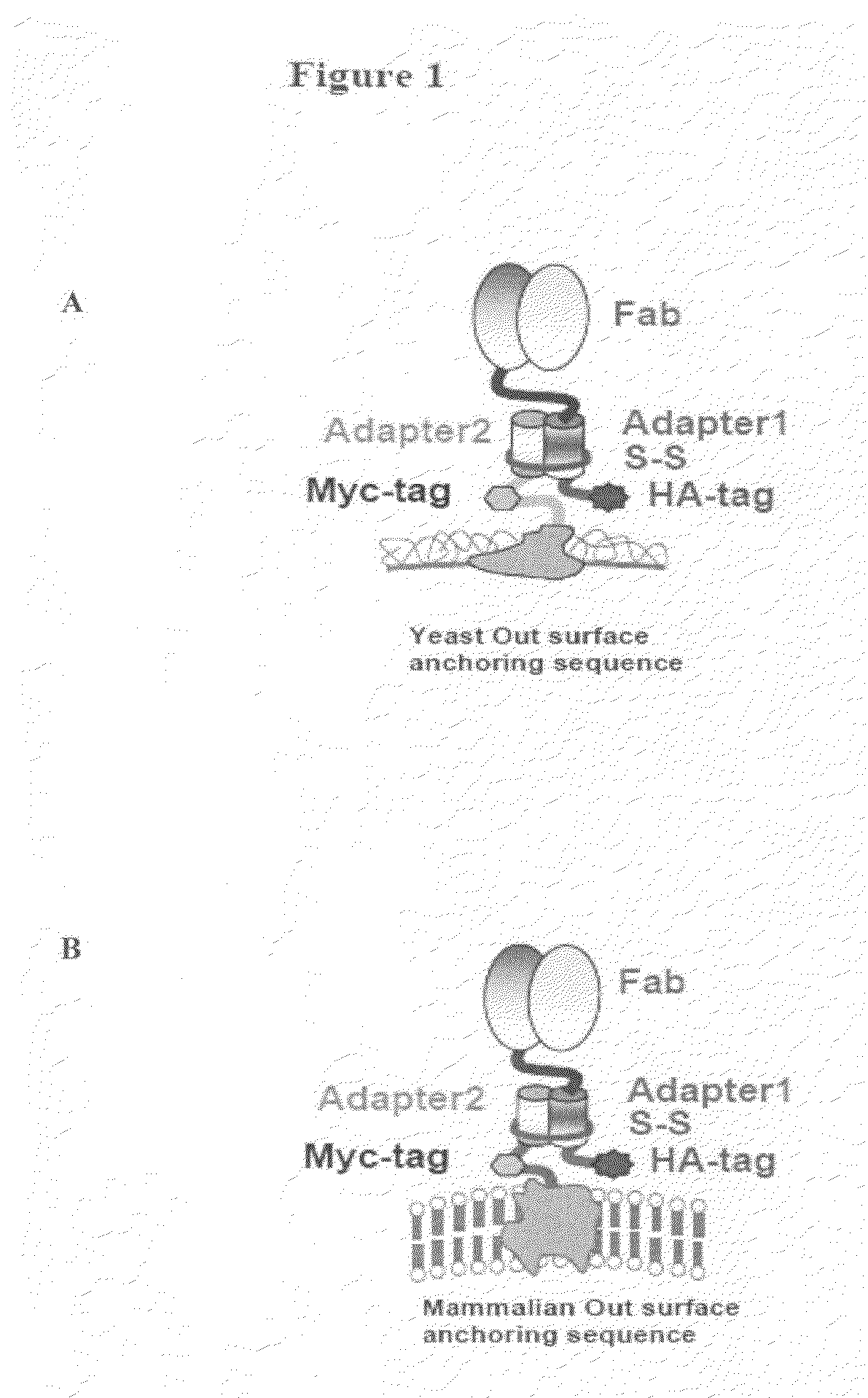
ActiveUS20090163379A1Facilitate direct expressionEfficient methodMicroorganism librariesLibrary creationYeastGenetic diversity
The present invention provides expression vectors and helper display vectors which can be used in various combinations as vector sets for display of polypeptides on the outer surface of eukaryotic host cells. The expression vector of the invention can be used alone for soluble expression without having to change or reengineer the display vectors. The display systems of the invention are particularly useful for displaying a genetically diverse repertoire or library of polypeptides on the surface of yeast cells, and mammalian cells.
Owner:MERCK SHARP & DOHME LLC
Porphyra yezoensis microsatellite marker screening method and use thereof
InactiveCN101550434AImprove reliabilityHigh yieldMicrobiological testing/measurementFermentationResource protectionGermplasm
The invention belongs to molecular biology DNA labeling technique and application field, concretely relates to a porphyra yezoensis microsatellite marker screening method, and a method for analyzing germplasm genetic diversity by the makers, thereby lay foundations for genetic structure analysis of porphyra yezoensis, germplasm resource protection and molecular marker auxiliary breeding.
Owner:OCEAN UNIV OF CHINA
SSR molecular marker primer based on transcriptome data of azalea as well as screening method and application of SSR molecular marker primer
InactiveCN105969767AAvoid it happening againEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGenetic diversity
The invention discloses an SSR molecular marker primer based on transcriptome data development and specific application of the SSR molecular marker primer, belonging to the technical field of biology. The primer is obtained based on development of a transcriptome sequence. By virtue of primer designs of carrying out batch screening on lots of sequencing data based on transcriptome sequencing to obtain an SSR sequence and carrying out SSR marking, the obstacles that the SSR molecular markers of the azalea are few, the development efficiency is low, and the like are overcome. The validity of the SSR primer is verified by virtue of different varieties of azaleas, and foundation is laid for the research of genetic diversity of the azaleas, the constitution of genetic linkage maps, the assistant breeding of the molecular marker, and the like.
Owner:HUANGGANG NORMAL UNIV
Watermelon complete genomic sequence information based analyzed and developed SSR core primer combinations and application thereof
ActiveCN102220315AHigh polymorphismGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide
Watermelon complete genomic sequence information based analyzed and developed SSR core primer combinations. The primer combinations comprise primers, of which nucleotide sequences as shown in the sequence table SEQ ID No.1-46. The obtained SSR core primer combinations in the invention can reflect the genetic diversity of watermelon germplasm resources to the maximum, has advantages of high polymorphism, good repeatability, reliable and stable marking, and are convenient for statistics. The set of primers provided by the invention are utilized to carry out a genetic diversity analysis, so as to accurately reflect the genetic relationship of tested materials to the maximum.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
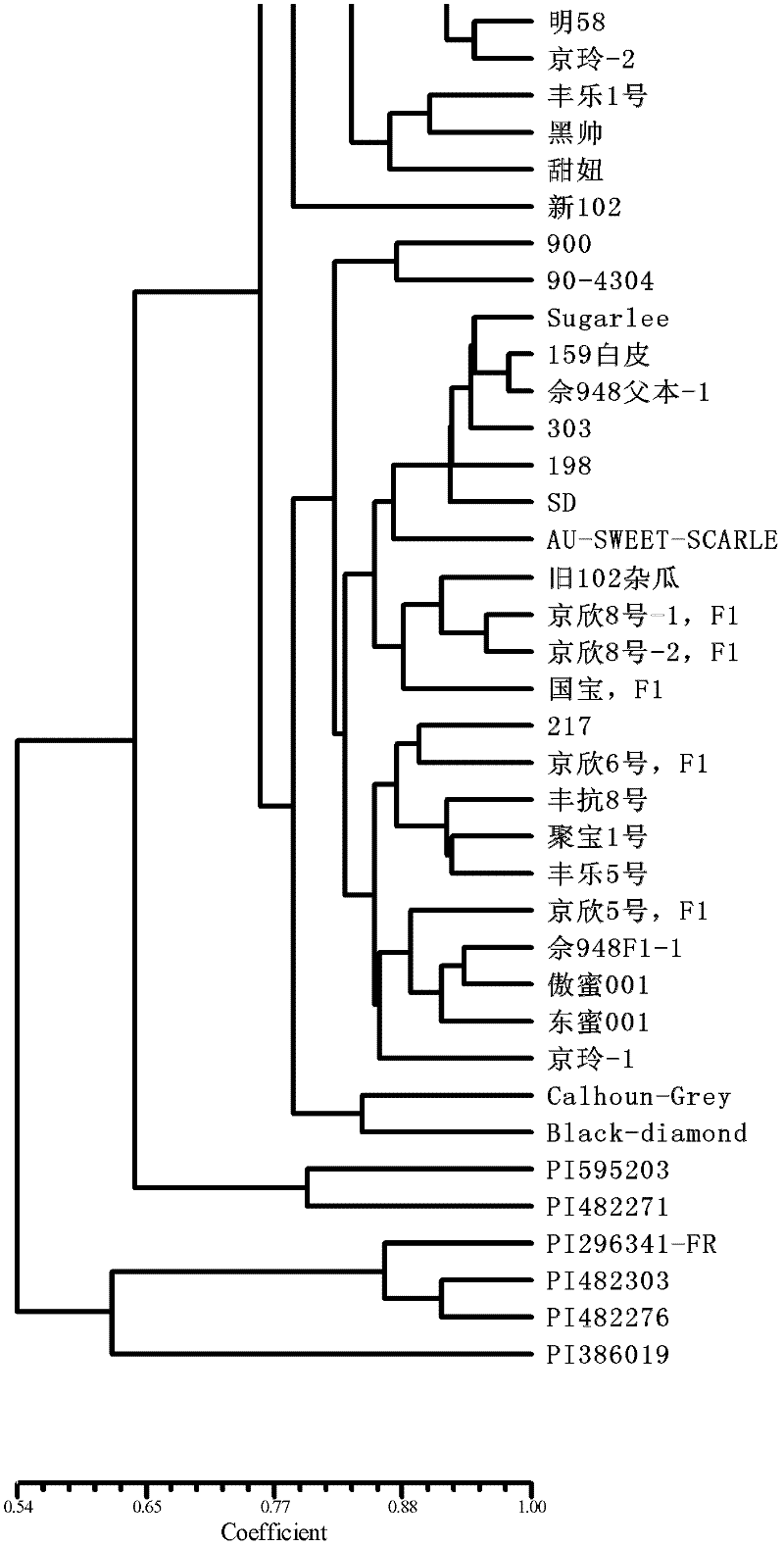
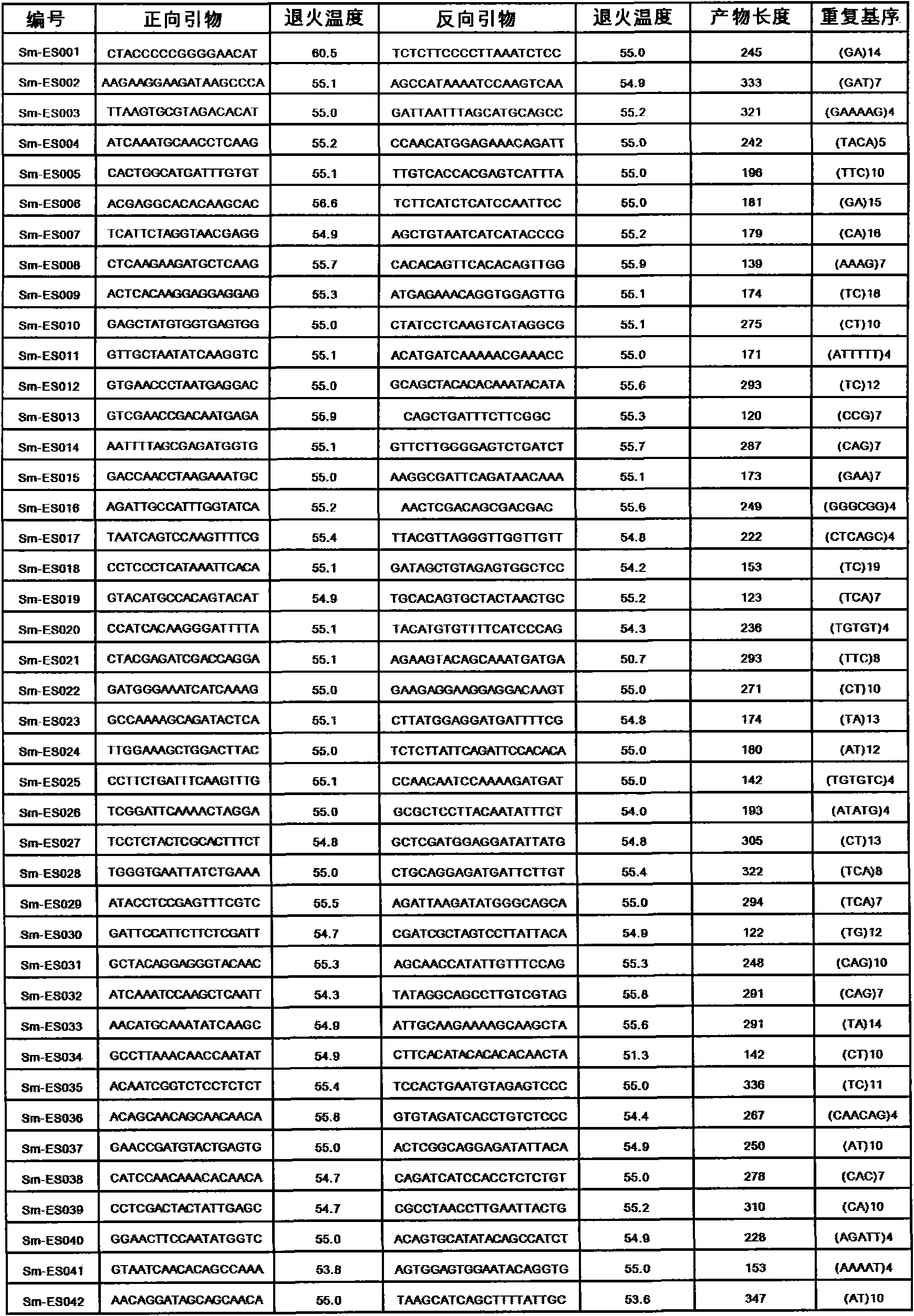
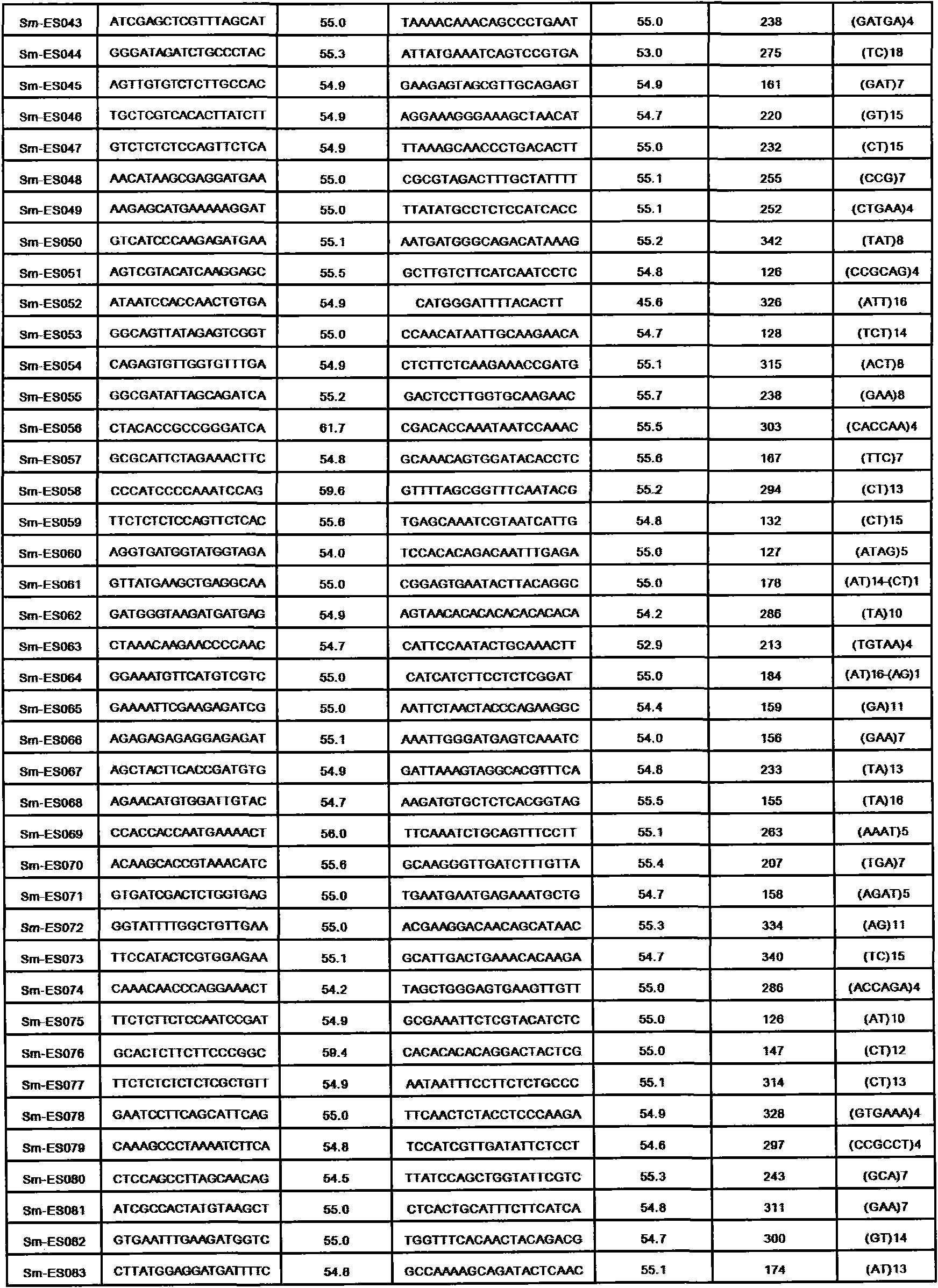
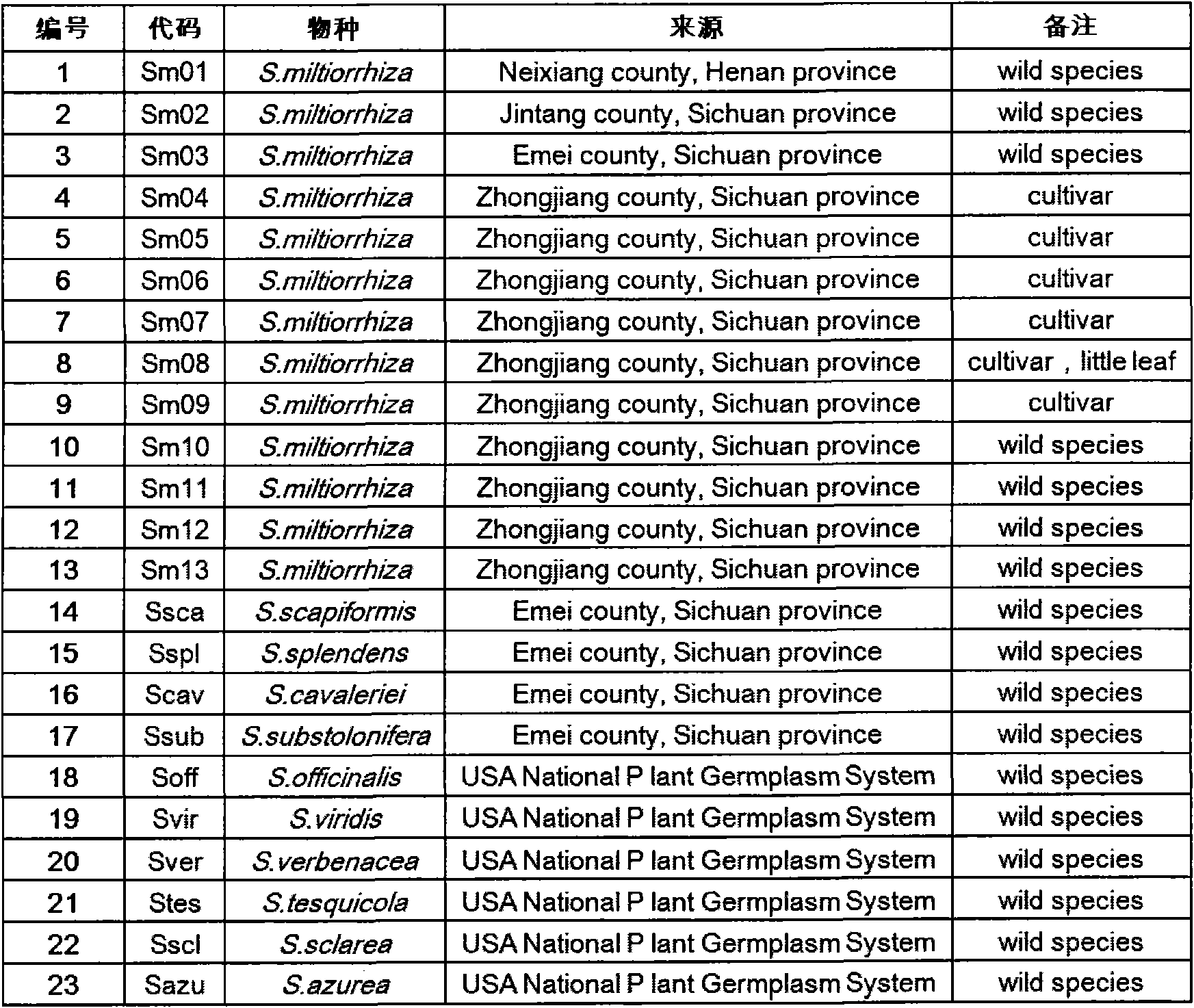
Method for preparing salvia miltiorrhiza EST-SSR molecular mark, specific primer and application thereof
The invention discloses a method for preparing a salvia miltiorrhiza EST-SSR molecular mark. The method mainly comprises the following steps of obtaining and pre-processing a salvia miltiorrhiza EST sequence, screening a SSR locus in the salvia miltiorrhiza EST sequence, designing and synthesizing the salvia miltiorrhiza EST-SSR primer, screening the salvia miltiorrhiza EST-SSR primer and the like. The method excavates the SSR locus aiming at the salvia miltiorrhiza EST and designs the PCR primer according to the flanking sequence of the SSR locus, and on the basis of this, the salvia specificEST-SSR mark system is established; and the method lays a solid foundation for genetic diversity evaluation, genetic relationship or idioplasmatic identification, genetic linkage description, molecular mark auxiliary selection, gene mapping cloning, functional genome analysis, related researches and the like of related species of the salvia miltiorrhiza and the salvia. The invention also discloses the EST-SSR locus specific primer obtained by the method for preparing the salvia miltiorrhiza EST-SSR molecular mark and the application thereof.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
Method of screening pacific oyster EST micro-satellite mark
The invention discloses a screening method of Pacific oyster EST micro satellite label, which comprises the following steps: finding micro satellite site with Pacific oyster EST sequence announced by GeneBank and micro satellite searching software SSRhunter; screening and separating 2-6 base repeat unit micro satellite fragment (repeat times >5); getting repeat ESTs sequence with micro satellite; designing primer at two sides flank sequence of micro satellite repeat sequence; detecting optimization primer as the micro satellite label further; proceeding overall merit for micro satellite repeatability, stability, polymorphism; getting micro satellite label. The invention can be used for Pacific oyster species resource and genetic heterogeneity, molecular population genetics, species resource determination, genetic atlas construct, functional gene research, Pacific oyster breeding and cultivation.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Primer set, kit and method for identifying different tobacco varieties
InactiveCN104726564AMicrobiological testing/measurementDNA/RNA fragmentationNicotiana tabacumAgricultural science
The invention discloses a primer set, an application of the primer set in identification of tobacco varieties and / or determination of genetic relationships of different tobacco varieties, a kit, an application of the kit in identification of the tobacco varieties and / or determination of the genetic relationships of the different tobacco varieties, a method for identifying the tobacco varieties and a method for determining the genetic relationships of the different tobacco varieties. The primer set comprises a primer 1, a primer 2, a primer 3 and a primer 4. By using the primer set to perform multiple PCR (polymerase chain reaction) and selectively perform single PCR, the different tobacco varieties can be quickly and efficiently identified and / or the genetic relationships and genetic diversities of the different tobacco varieties can be identified.
Owner:SOUTHWEST UNIVERSITY
Method for establishing SSR (Simple Sequence Repeat) fingerprint spectrum of broad beans
ActiveCN106755328ASignificant size differenceEffective monitoringMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCapillary electrophoresis
The invention provides a method for establishing an SSR (Simple Sequence Repeat) fingerprint spectrum of broad beans, and belongs to the technical field of plant variety identification and breeding. The method provided by the invention provides a marker combination consisting of 21 pairs of broad bean genome SSR primers, and the nucleotide sequences of which are shown in SEQ ID NO. 1 to 42 respectively; the broad bean varieties are identified by PCR (Polymerase Chain Reaction) amplification technology, non-denaturing polyacrylamide gel electrophoresis or capillary electrophoresis detection technology, and the identification of broad bean seed varieties and genetic diversity assessment work can be completed in a short time. The method has the advantages of time saving, high efficiency, fastness, accuracy, and convenience in operation, and the identification results are not easily affected by the environment. The method provided by the invention has the advantages of effectively monitoring the authenticity of the broad bean seeds, revealing the genetic variation and genetic relationship of the varieties from the DNA level, protecting the crop varieties, preventing the fake and inferior varieties from entering the market, and also providing a technical support for the reasonable utilization of excellent germplasm during the breeding of broad bean varieties, and has good application prospects.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Method for identifying rice DNA identities and application thereof
InactiveCN105550537AReduce volatilityEasy to useSequence analysisSpecial data processing applicationsGenetic diversityGenotype
The invention provides a method for identifying rice DNA identities, comprising following steps: obtaining standard gene fingerprint data of rice through detecting the genotypes of the genetic diversity molecular markers of the whole genome of the rice, identifying the DNA identities of the rice, or further, identifying the DNA identities of the rice through feature gene fingerprint data. According to the method provided by the invention, the DNA identities of the rice can be identified rapidly and accurately; and the method is applicable for new rice product identification, seed supervision and management or variety right protection.
Owner:CHINA NAT SEED GRP
Seedless litchi graft stock selecting technology
InactiveCN1887048AGrafting affinity is goodImprove survival rateHorticultureRootstockGenetic diversity
The present invention discloses the seedless litchi graft stock selecting technology. The technological process of selecting seedless litchi graft stock includes the following steps: 1. separating the simple sequence repeating (SSR) sequence through a combined selective amplifying microsatellite (SAM) and database searching method; 2. designing SSR primer with the flanking sequence of SSR sequence; 3. screening out the useful primer to obtain litchi specificity SSR marker; 4. applying the litchi specificity SSR marker in the genetic diversity analysis of partial litchi germplasm; 5. data analysis; and 6. selecting the litchi germplasm with near affinity as stock for seedless litchi grafting propagation based on the analysis result. The said process results in high survival rate and can obtain high quality seedless litchi variety.
Owner:INST OF TROPICAL BIOSCI & BIOTECH CHINESE ACADEMY OF TROPICAL AGRI SCI +1
Method for breeding blue crab families
ActiveCN103250666AIncrease specificationImprove survival rateClimate change adaptationPisciculture and aquariaMatingGenetic diversity
A method for breeding blue crab families comprises the following steps: S1, collecting blue crabs bred in different populations of China to construct base groups, and conducting genetic diversity comprehensive evaluation, S2, respectively selecting parent crabs from the different populations, establishing the families according to the complete diallel cross mode to grow larvae and breed in a standardizing mode, and calculating hybrid heterosis rates, S3, utilizing the crabs with outstanding hybrid heterosis to mate and combine, constructing the families through indoor artificial directional mating to grow the larvae and breed in the standardizing mode, and twice screening stocking families, S3, over-wintering mated female crabs of parent groups bred indoors of the stocking families, S4, selecting the parent crabs with high egg-laying rates from each family in following spring to grow the larvae and breed in the standardizing mode, and utilizing the breeding method to continue to screen the families. By means of the method for breeding the blue crab families, the growth rate and the survival rate of the blue crabs are used as indexes, blue crab families breeding is conducted, family economic characters are comprehensively evaluated, according to breeding results, blue crab norms are improved in the harvesting process, the breeding survival rate is improved, so that breeding benefits are greatly increased, and deviation caused by single character breeding is avoided.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Quality assurance and identification method for high-quality strawberries
ActiveCN105203672AScientific and reliable quality evaluation systemEffective reflection of total antioxidant activityComponent separationMicrobiological testing/measurementEnzyme GeneGenetic diversity
The invention relates to a quality assurance and identification method for high-quality strawberries, belonging to the technical field of biology. The quality assurance and identification method comprises the following steps: determining phenols and flavor substances of different varieties of strawberries by virtue of HPLC-MS and GC-MS, and establishing a chemical fingerprint chromatography database; determining the total antioxidant activity of the various strawberries by virtue of an ORAC method; analyzing blastogenesis diversity of the strawberries by utilizing RFLP, RAPD and SSR methods and combining key enzyme genes in synthetic routes of anthocyanin and polyphenol; establishing a spectrum-effect relationship between chemical fingerprint chromatography and biological activity information of the different varieties of strawberries, and analyzing the relation between hereditary basis and functional quality character of the strawberries. The high-quality variety of the strawberries is selected by combining chemical fingerprint chromatography, the effectiveness evaluation, genetic diversity analysis and the spectrum-effect relationship and is subjected to authenticity identification, so that a scientific, reliable, uniform and standard strawberry quality evaluation system is established, and technical support and scientific basis are provided for the quality control of the strawberries.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Acipenser dabryanus microsatellite marker as well as screening method and application of acipenser dabryanus microsatellite molecular marker
InactiveCN106434949APromote rapid developmentCutting costsMicrobiological testing/measurementDNA/RNA fragmentationNucleotideGenetic diversity
The invention belongs to the field of microsatellite molecular markers and particularly relates to an acipenser dabryanus microsatellite marker as well as a screening method and application of the microsatellite molecular marker, wherein the acipenser dabryanus microsatellite marker has the nucleotide sequence of any one or more of SEQ ID No.1-8. In the invention, the development of microsatellite molecular markers using the high-throughput sequencing technology is faster and costs less than a traditional method; and moreover, the functional traits of corresponding EST sequence can be found while the microsatellite sites are obtained, and the gene relevance is relatively strong. In the invention, through the developed microsatellite sites, basic data is provided for the genetic diversity of acipenser dabryanus, and the study fields such as genetic diversity analysis, genetic map establishment, gene location, variety identification, germplasm conservation, quantitative character gene analysis and parent-child relationship identification can be carried out. According to actual needs, any one or more markers disclosed by the invention can be adopted, or any one or more pairs of specific primers corresponding to related markers can be analyzed and studied.
Owner:FISHERIES INST SICHUAN ACADEMY OF AGRI SCI
Amplimer of large yellow croaker mitochondrion complete genome sequence and application thereof
ActiveCN102776183AEasy splicingIncrease success rateMicrobiological testing/measurementDNA preparationGerm plasmGene orders
The invention discloses an amplimer of a large yellow croaker mitochondrion complete genome sequence and an application thereof. The 15 pairs of amplimer are applied, a gene order is respectively shown as SEQ ID No: 1-30, the amplimer is further used for obtaining a Tai Qu family large yellow croaker mitochondrion complete genome sequence and a Min-Yue East family large yellow croaker mitochondrion complete genome sequence which are respectively shown as SEQ ID No: 31 and SEQ ID No: 32, and an amplimer of large yellow croakers in a high variation region is obtained. The amplimer and the application have the advantages that the Tai Qu family large yellow croaker and Min-Yue East family large yellow croaker mitochondrion complete genome sequences can be quickly and efficiently obtained respectively through the 15 pairs of amplimers; and the amplimer of the large yellow croakers in the high variation region provides support for the research of species identification, geographical population identification, germ plasm resources and genetic diversity of the large yellow croakers, and further provide the basis for developing an identification technology of Tai Qu family large yellow croakers and Min-Yue East family large yellow croakers.
Owner:NINGBO UNIV
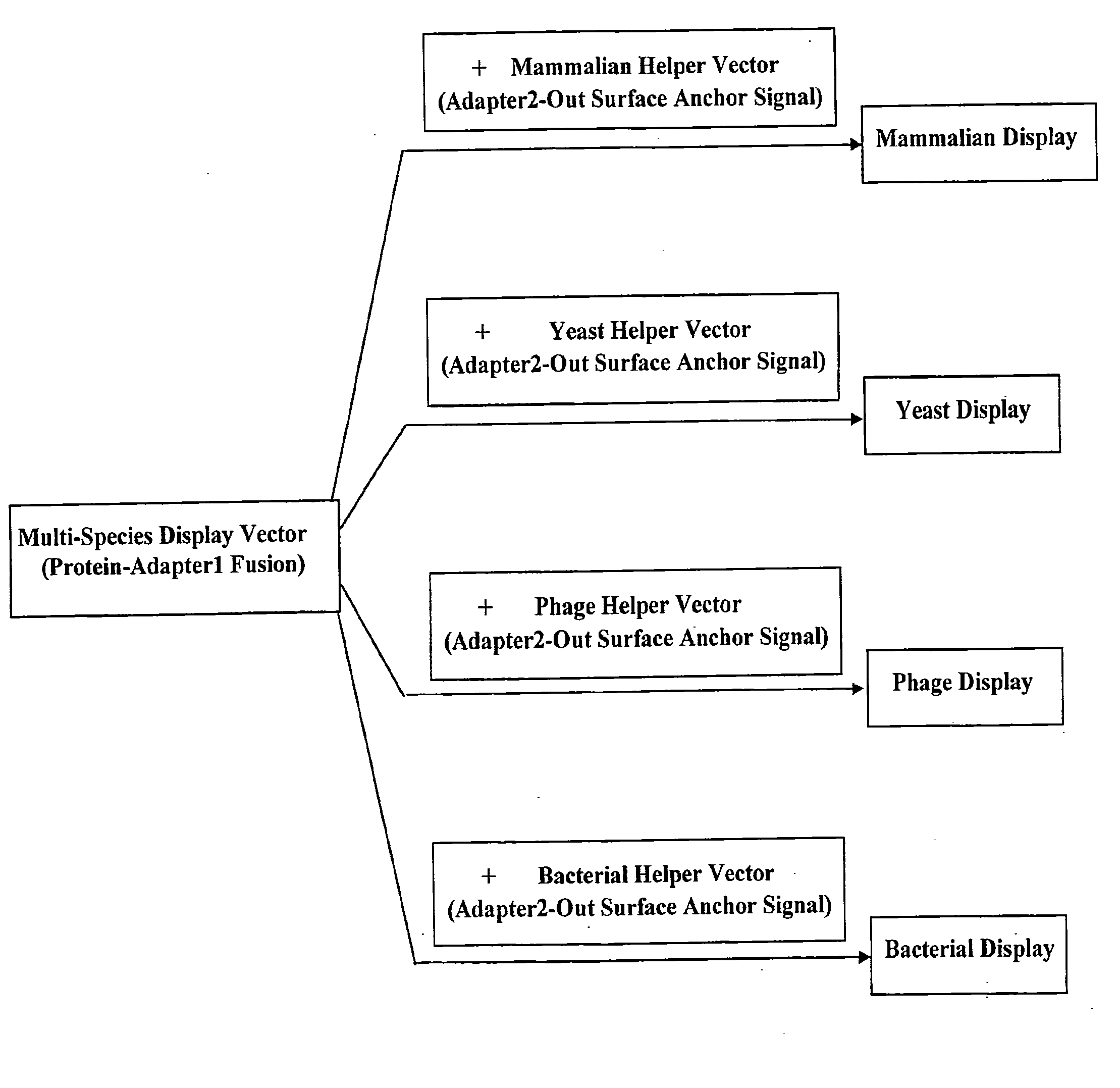
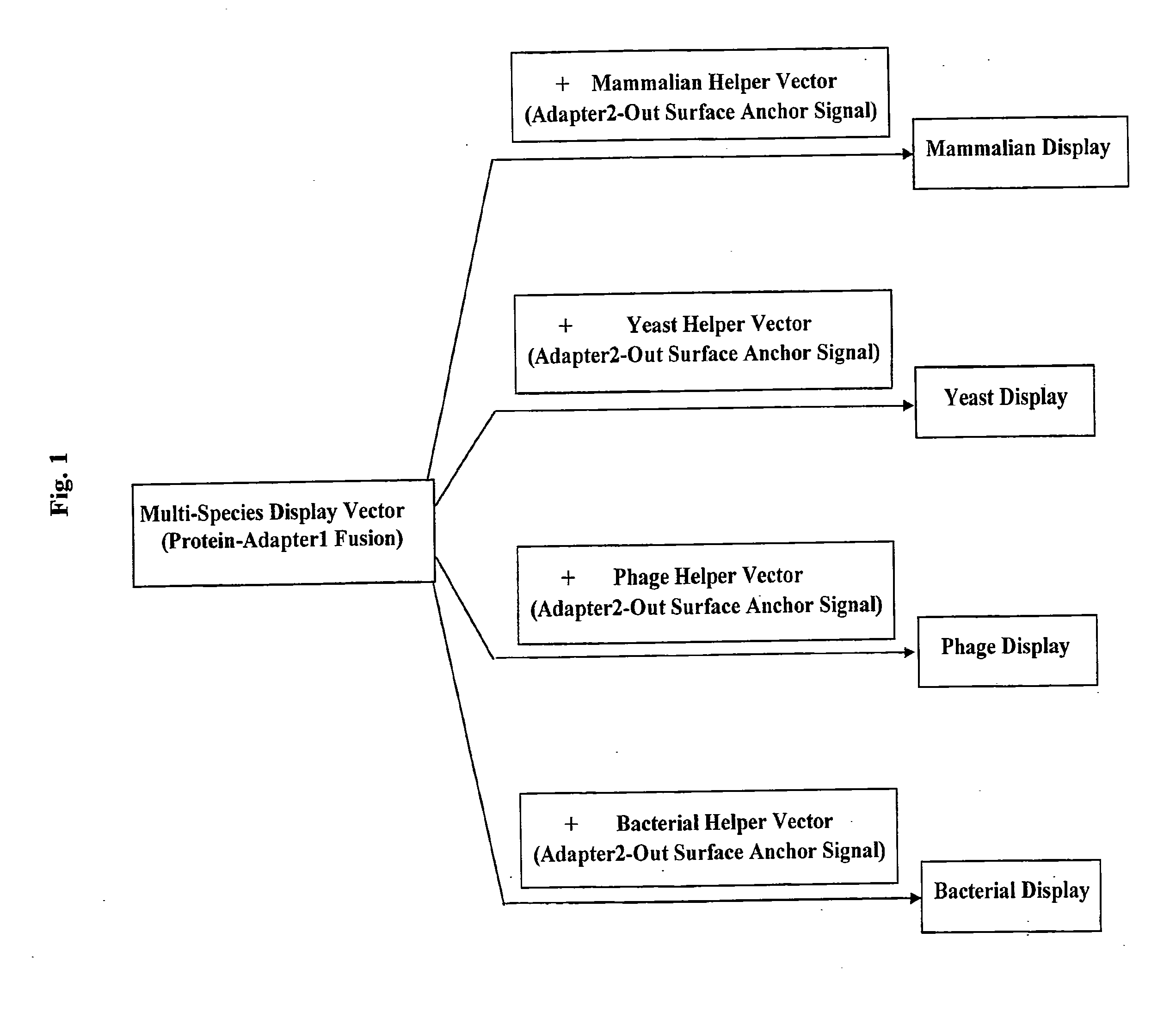
Cross-Species and Multi-Species Display Systems
The present invention provides expression vectors and helper display vectors which can be used in various combinations as vector sets for multi-species and cross-species display of polypeptides on the outer surface of prokaryotic genetic packages and / or eukaryotic host cells. The multi-species and cross-species display systems can be practiced using the vector sets of the invention without having to change or reengineer the display vectors. The display systems of the invention are particularly useful for displaying a genetically diverse repertoire or library of polypeptides on the surface of phage, bacterial host cells, yeast cells, and mammalian cells.
Owner:MERCK & CO INC
Detecting method for Litopenaeus vannamei Boone LvE165 microsatellite DNA marker
InactiveCN102146460AEasy identificationEasy to detectMicrobiological testing/measurementGenetic diversityMicrosatellite
The invention belongs to the field of molecular biology DNA marking technology and application, in particular to a detecting method for a Litopenaeus vannamei Boone LvE165 microsatellite DNA marker. The invention provides a Litopenaeus vannamei Boone LvE165 microsatellite specific DNA primer, a PCR (Polymerase Chain Reaction) system utilizing the primer and a detecting method for the Litopenaeus vannamei Boone LvE165 microsatellite DNA marker. The method comprises the following steps of: extracting a Litopenaeus vannamei Boone genome and diluting for later use; designing specific primers at both ends of a sequence by utilizing a Litopenaeus vannamei Boone LvE165 microsatellite DNA core sequence; and then carrying out PCR amplification on genome DNA of different groups of Litopenaeus vannamei Boone or individuals in the group by using the primer, analyzing a product and confirming the genotype of each individual so as to obtain a polymorphism genetic variation map. The invention is mainly applied to Litopenaeus vannamei Boone germplasm resource and genetic diversity analysis, molecular population genetics, construction of the genetic map, and the like.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
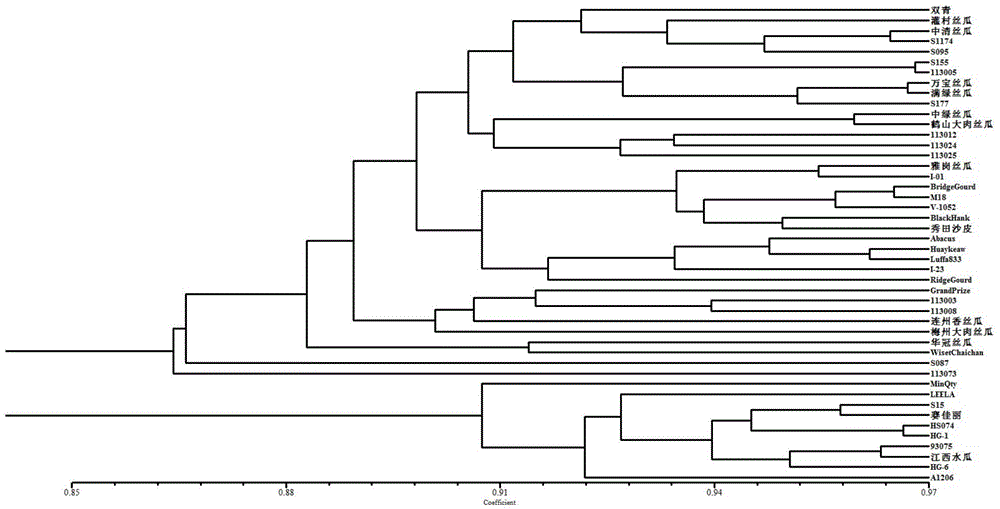
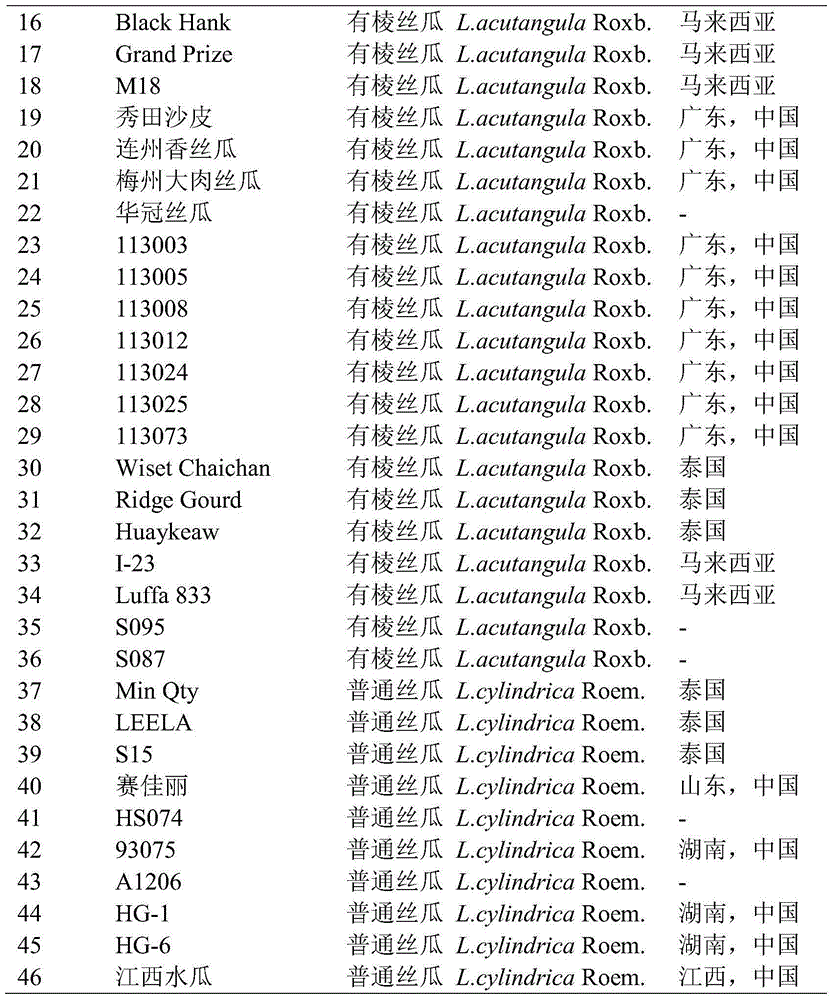
EST-SSR core primer group developed on basis of transcriptome sequence of towel gourd and application thereof
ActiveCN104004833APolymorphism richStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGourd
The invention discloses an EST-SSR core primer group developed on the basis of the transcriptome sequence of towel gourd. The EST-SSR core primer group comprises 50 pairs of primers with nucleotide sequences as shown in SEQ ID No. 1-100 in a sequence table. The 50 pairs of EST-SSR core primers have the advantages of abundant polymorphism, stable amplification, good repeatability, convenience in statistics, etc.; meanwhile, since the primers are originated from the expressed gene of towel gourd and can reflect variation of functional sequences in a genome, so germplasm genetic diversity of towel gourd can be better distinguished. The core primer group can be used in fields like identification of the variety of towel gourd, genetic genealogical analysis of varieties and analysis of germplasm genetic diversity, is beneficial for protection of legal interests of breeders, producers and consumers and promotes sound development of genetic breeding and production of towel gourd.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
Antibodies and methods for generating genetically altered antibodies with high affinity
InactiveUS7235643B2Increase variabilityBetter pharmacokinetic profileAnimal cellsSugar derivativesGenetic diversityMonoclonal antibody
Dominant negative alleles of human mismatch repair genes can be used to generate hypermutable cells and organisms. By introducing these genes into cells and transgenic animals, new cell lines and animal varieties with novel and useful properties can be prepared more efficiently than by relying on the natural rate of mutation. These methods are useful for generating genetic diversity within immunoglobulin genes directed against an antigen of interest to produce altered antibodies with enhanced biochemical activity. Moreover, these methods are useful for generating antibody-producing cells with increased level of antibody production. The invention also provides methods for increasing the affinity of monoclonal antibodies and monoclonal antibodies with increased affinity.
Owner:EISAI INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com