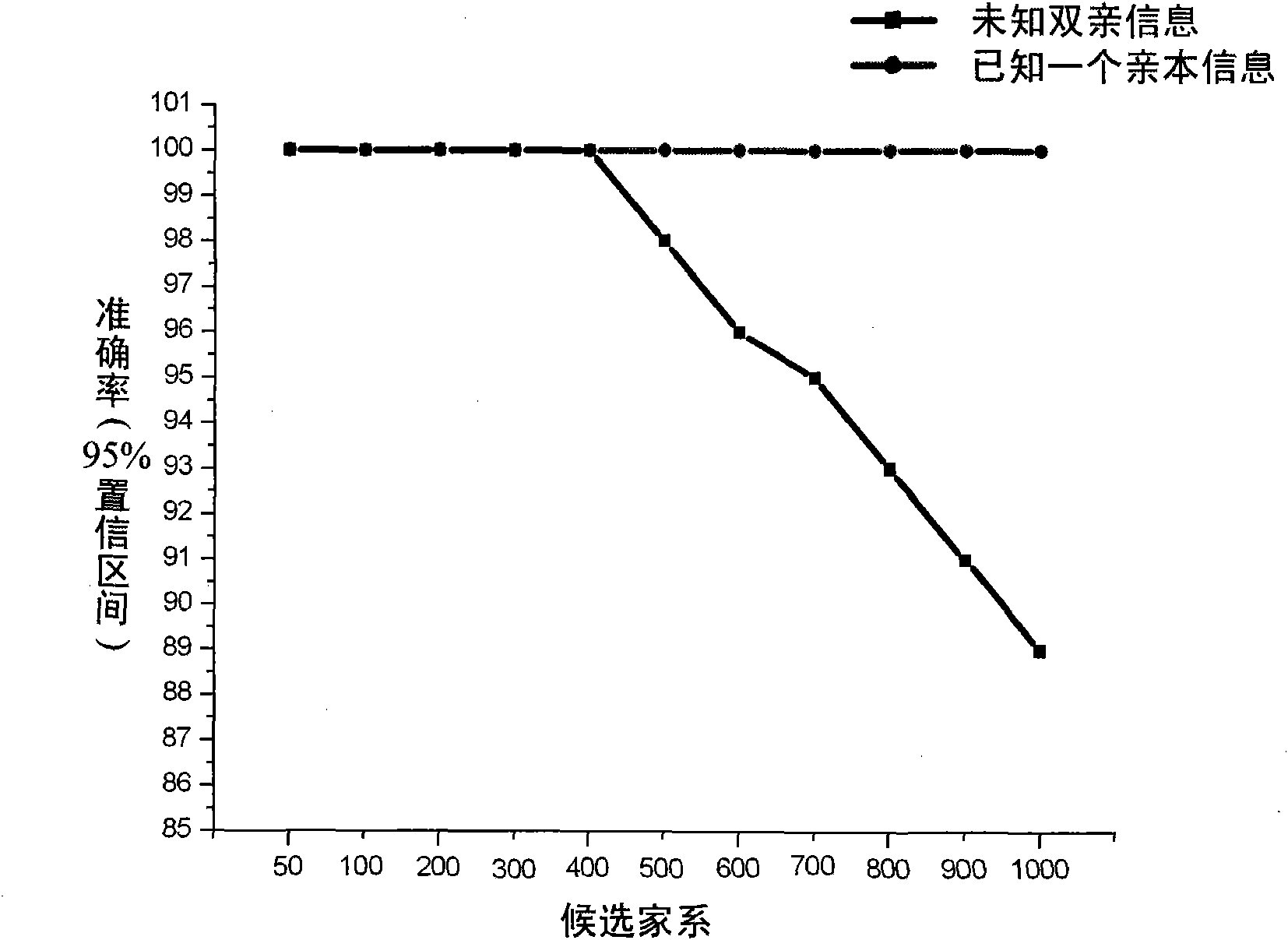
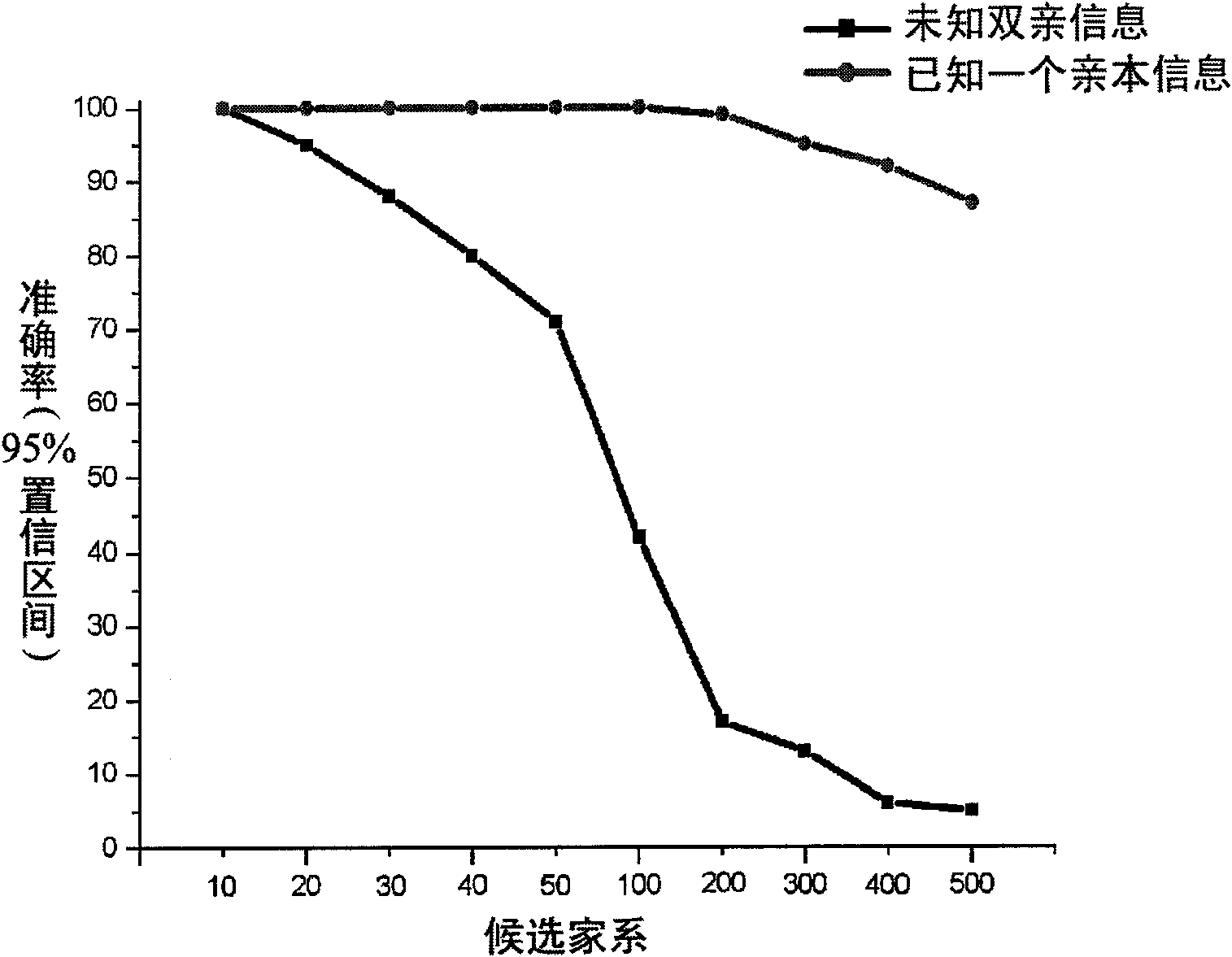
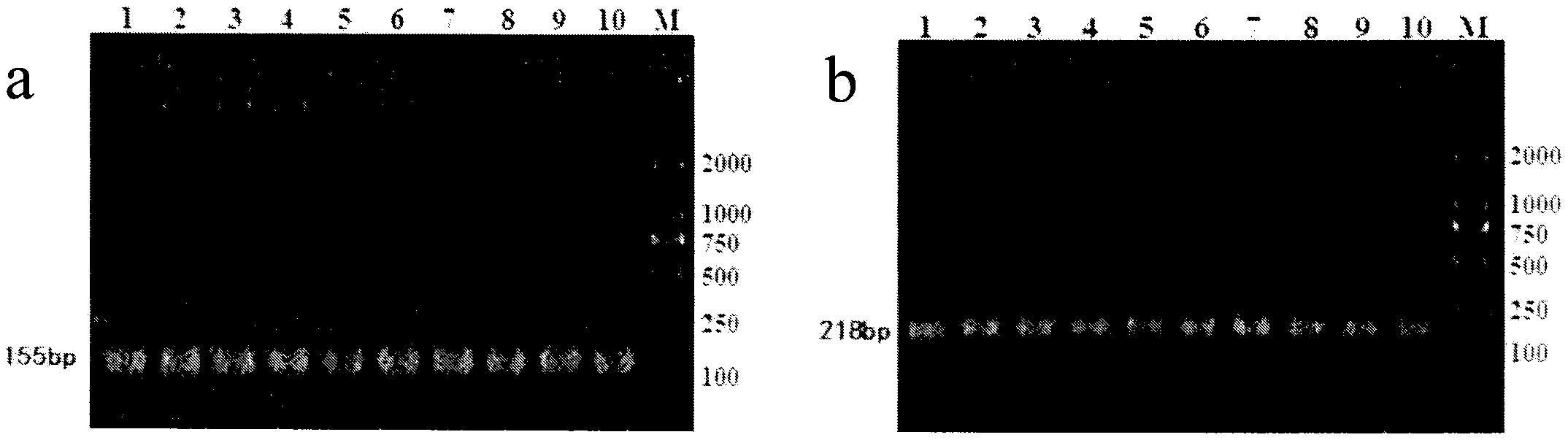
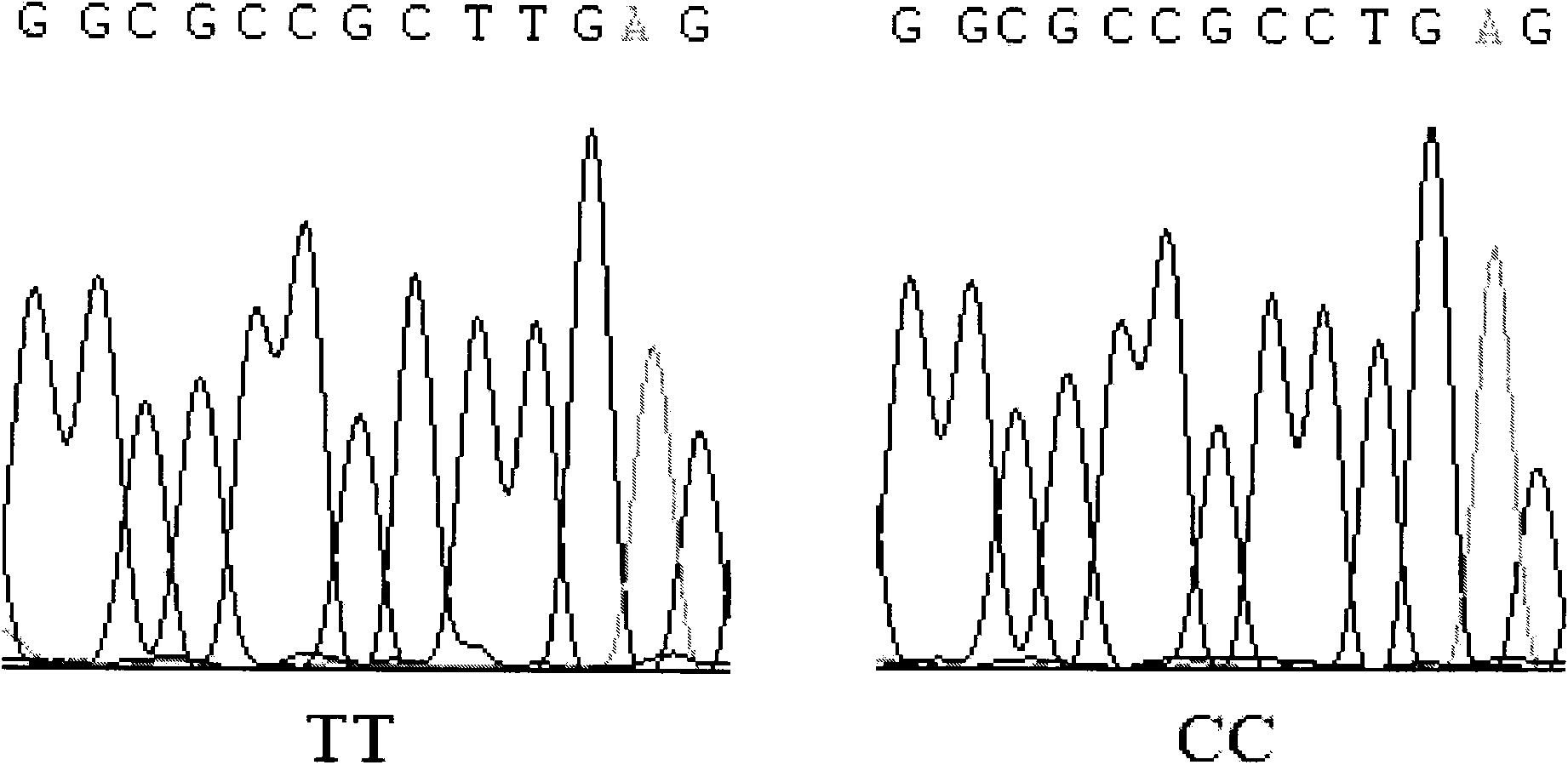
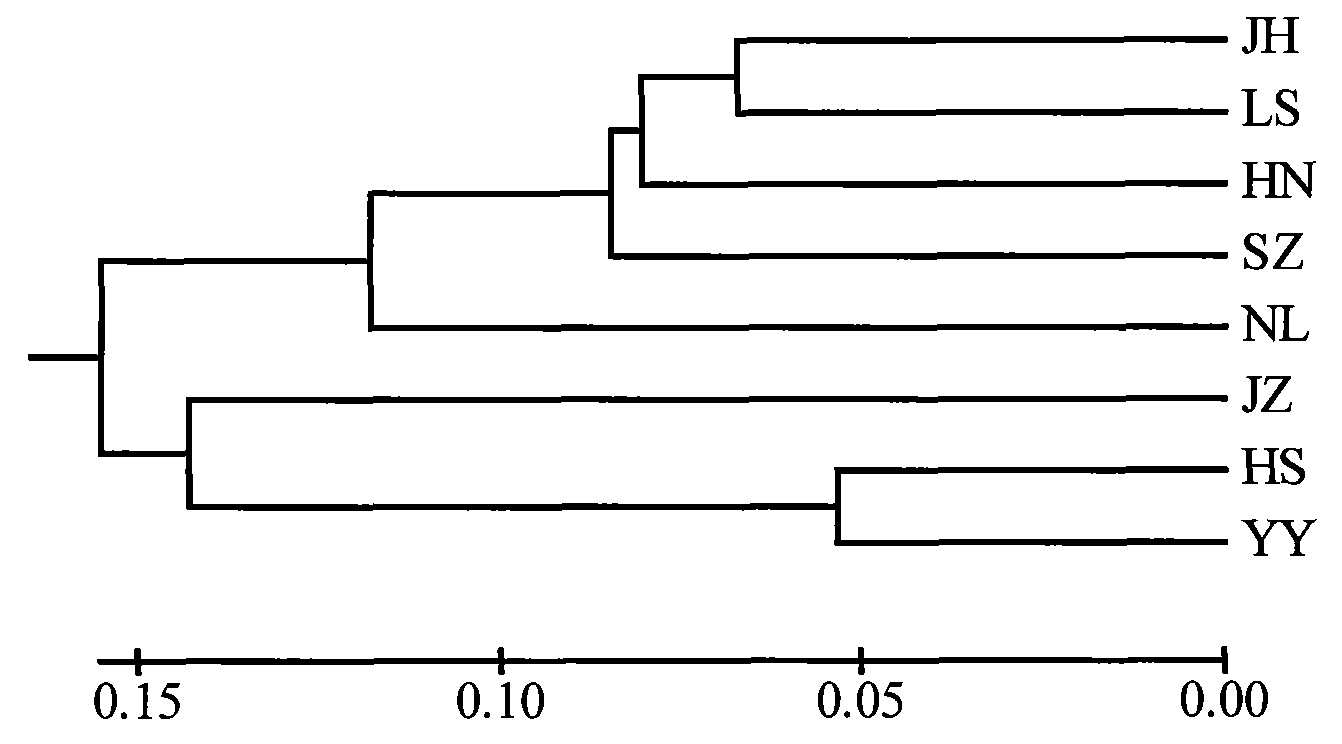
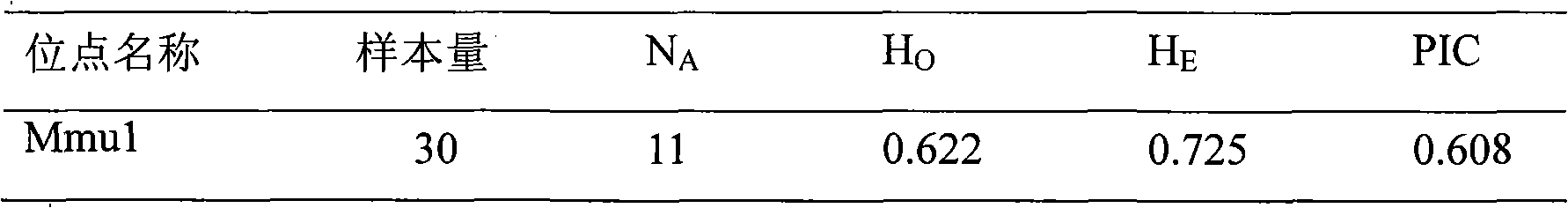
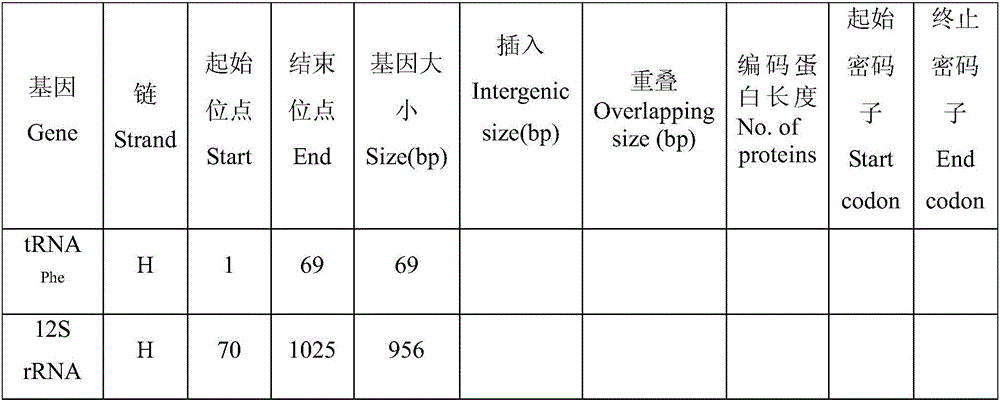
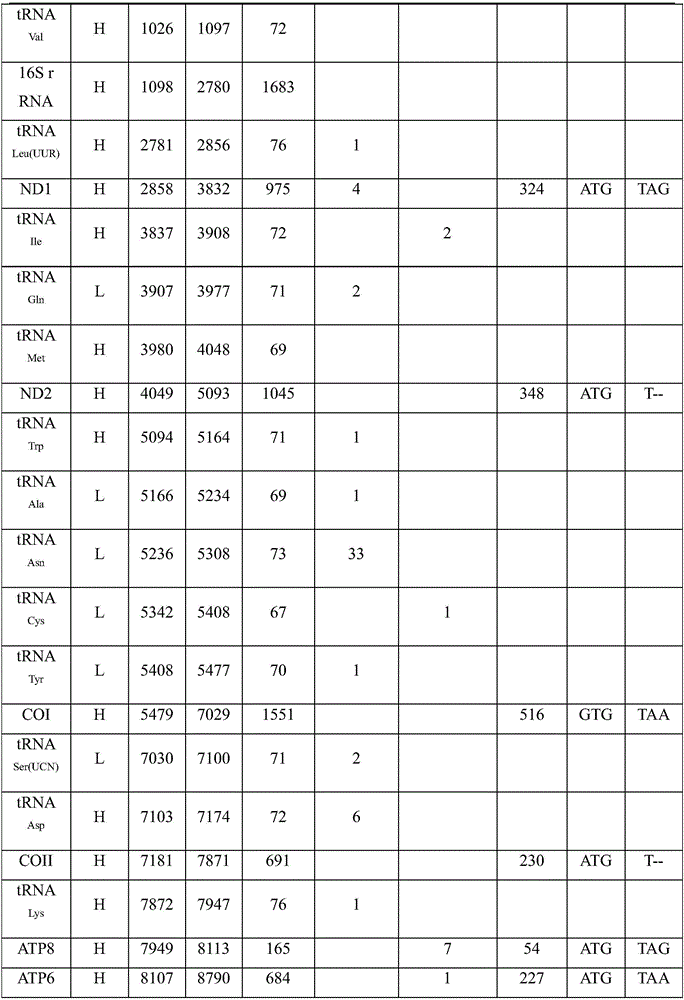
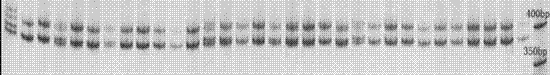
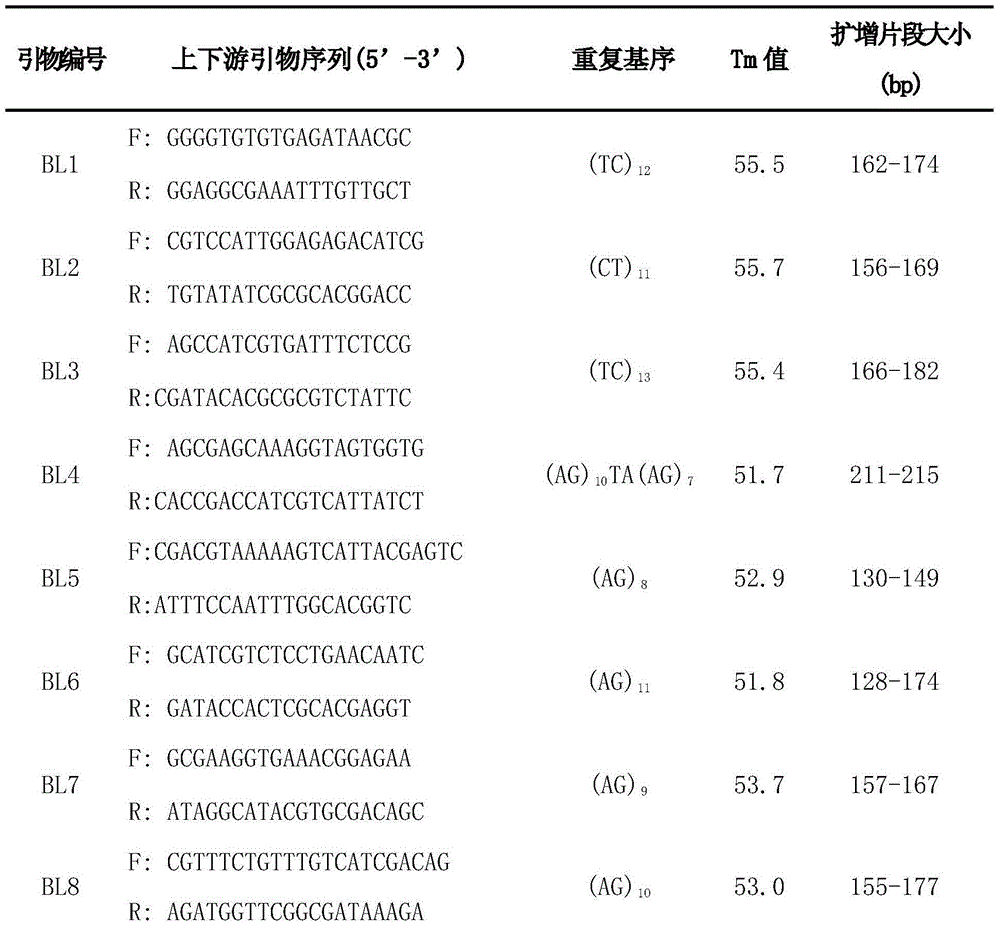
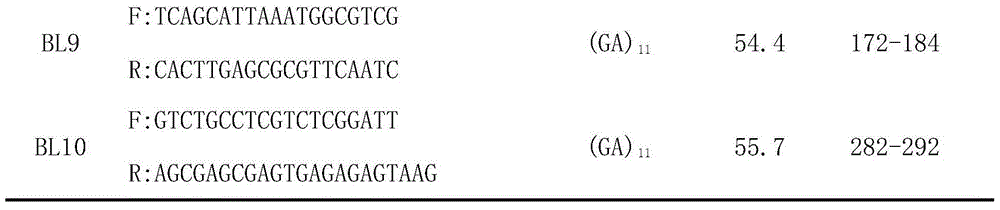
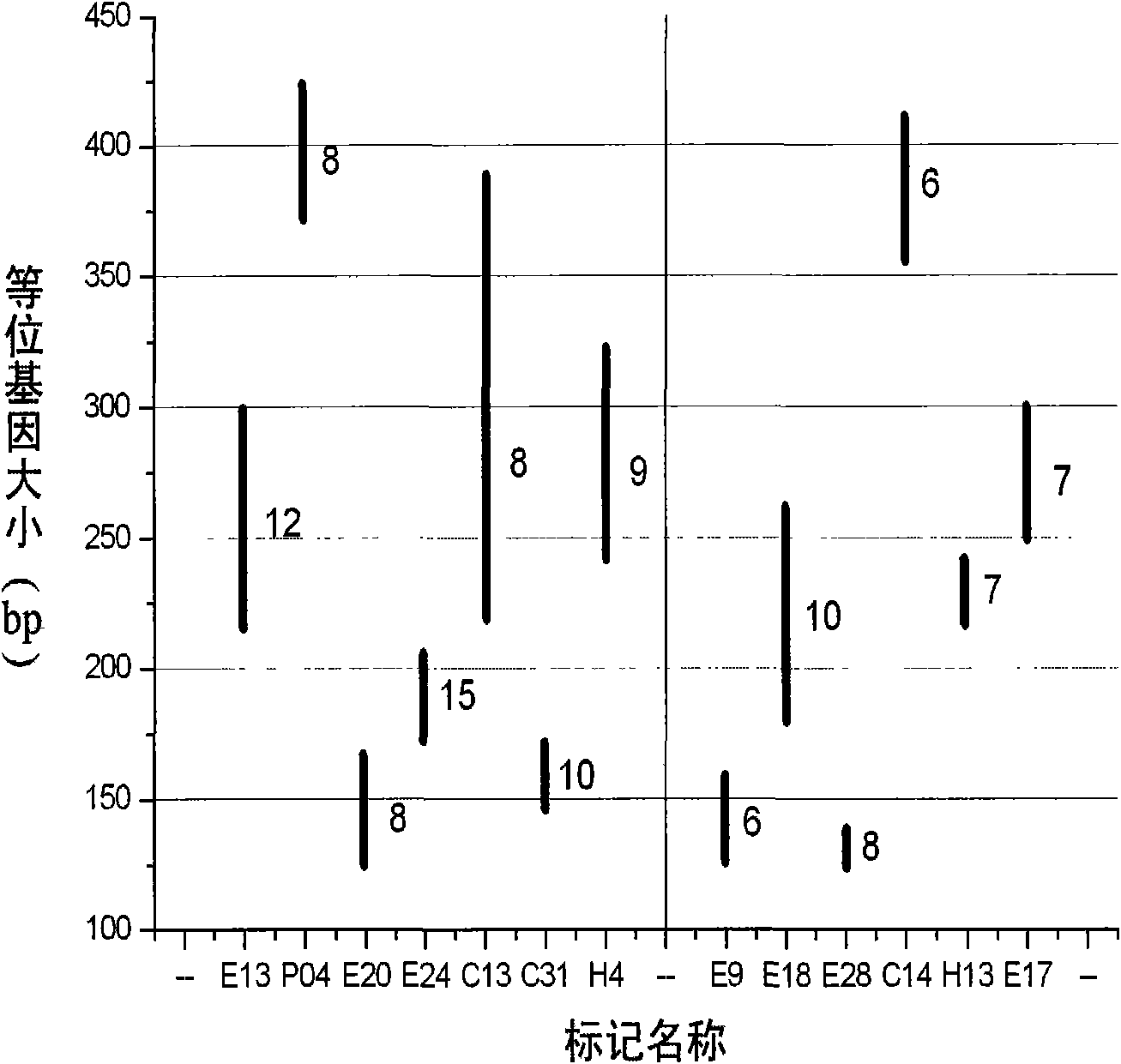
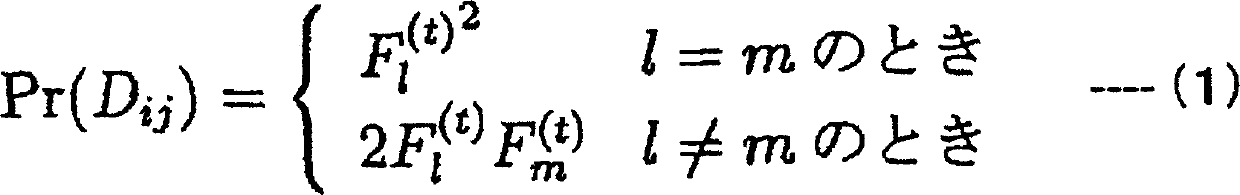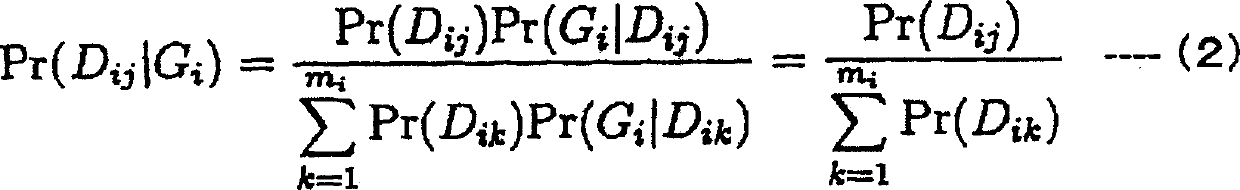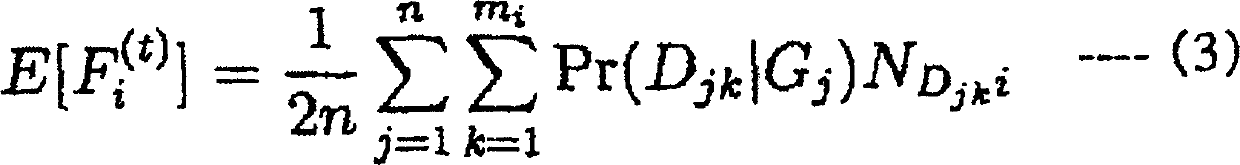Patents
Literature
105 results about "Genetic structure" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
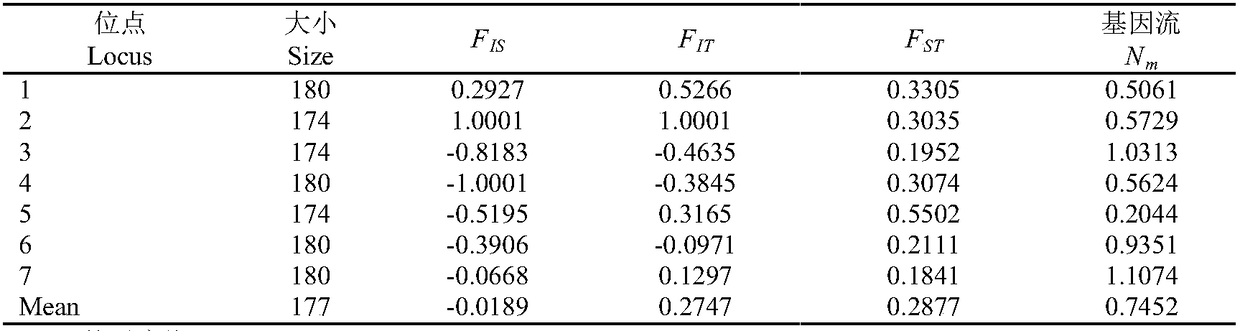
Genetic structure refers to any pattern in the genetic makeup of individuals within a population. Genetic structure allows for information about an individual to be inferred from other members of the same population. In trivial terms, all populations have genetic structure, because all populations can be characterised by their genotype or allele frequencies: if only 1% of a large sample of moths drawn from a single population have spotted wings, then it is safe to assume that any unknown individual is unlikely to have spotted wings.
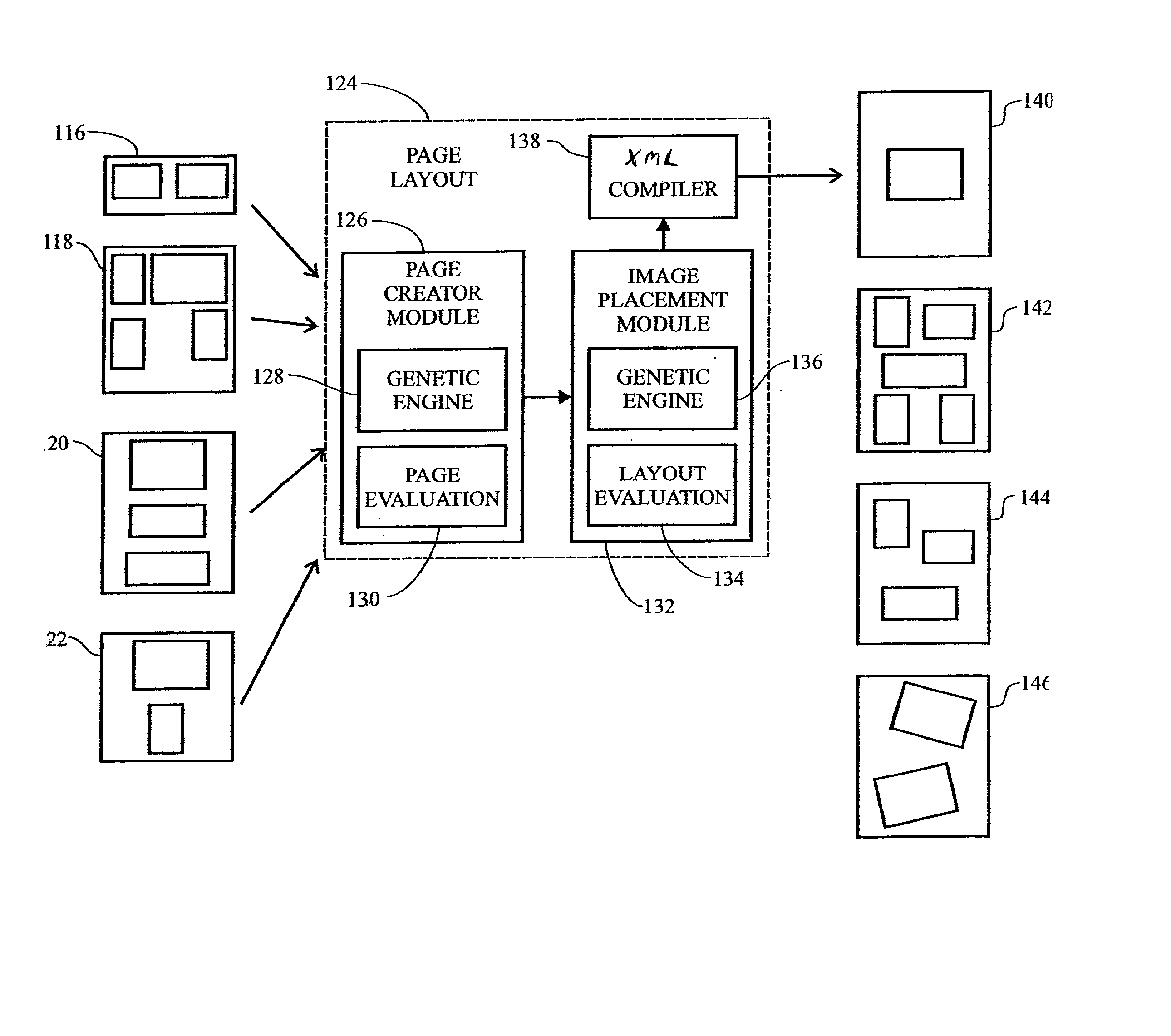
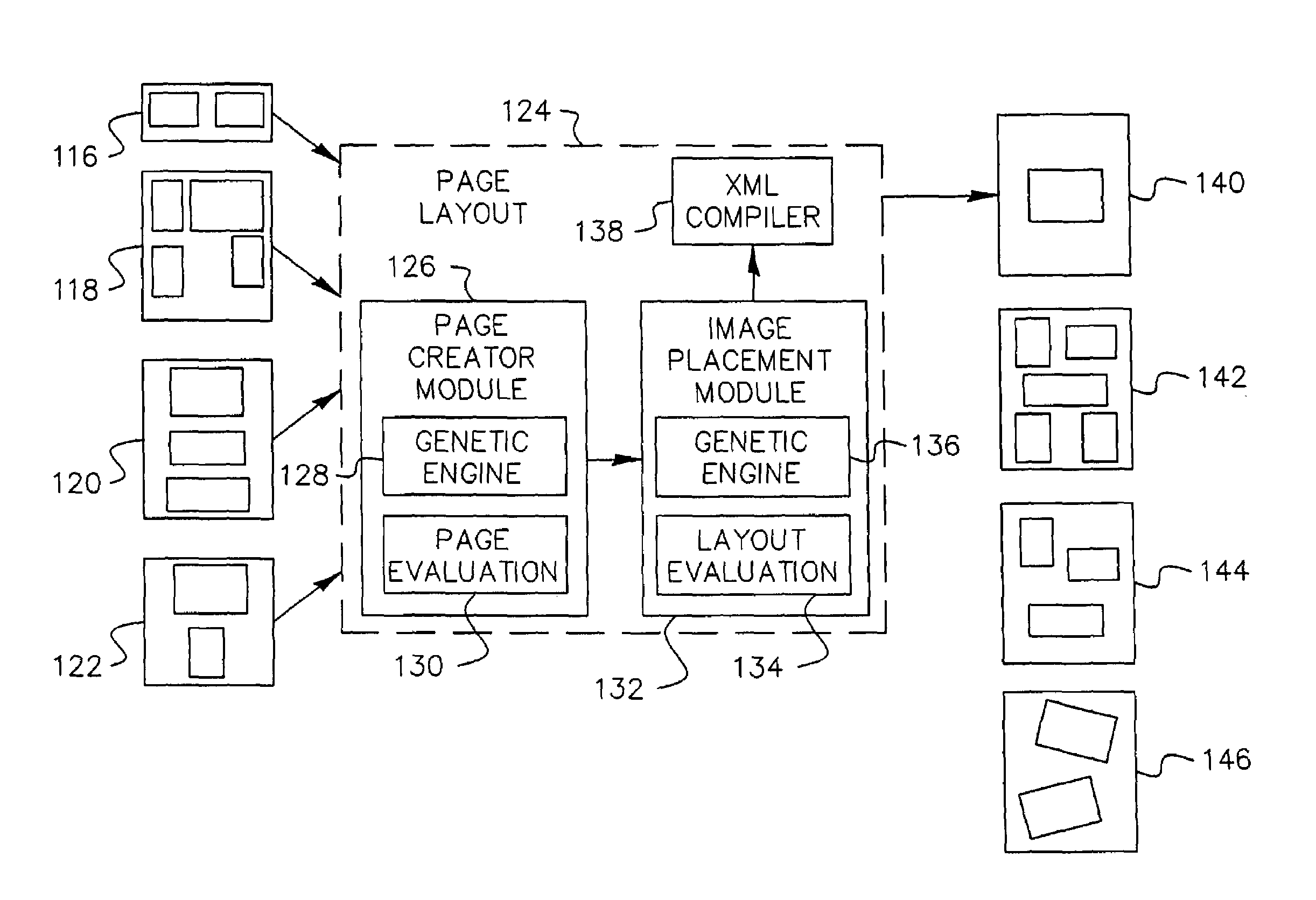
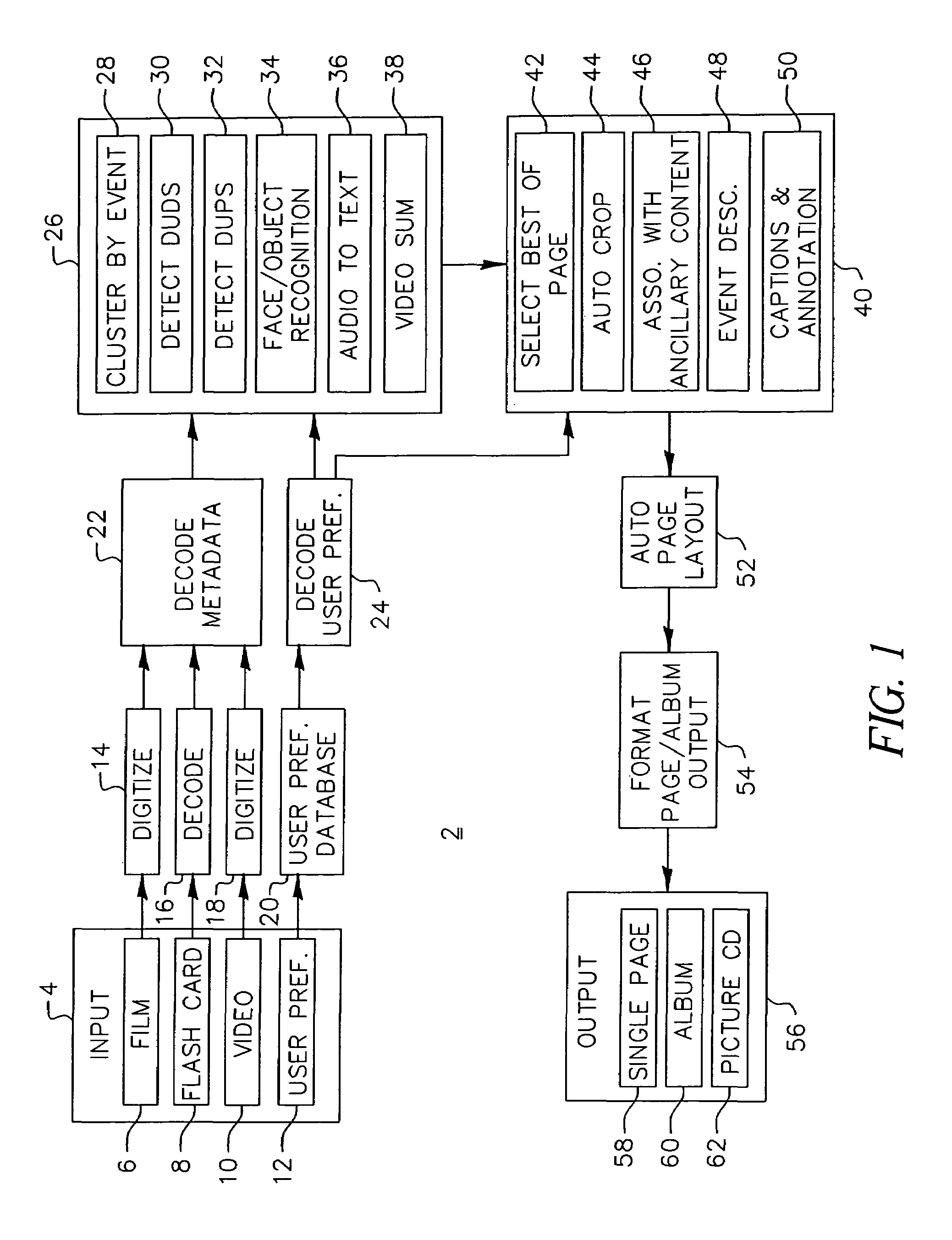
System and method for automatic layout of images in digital albums
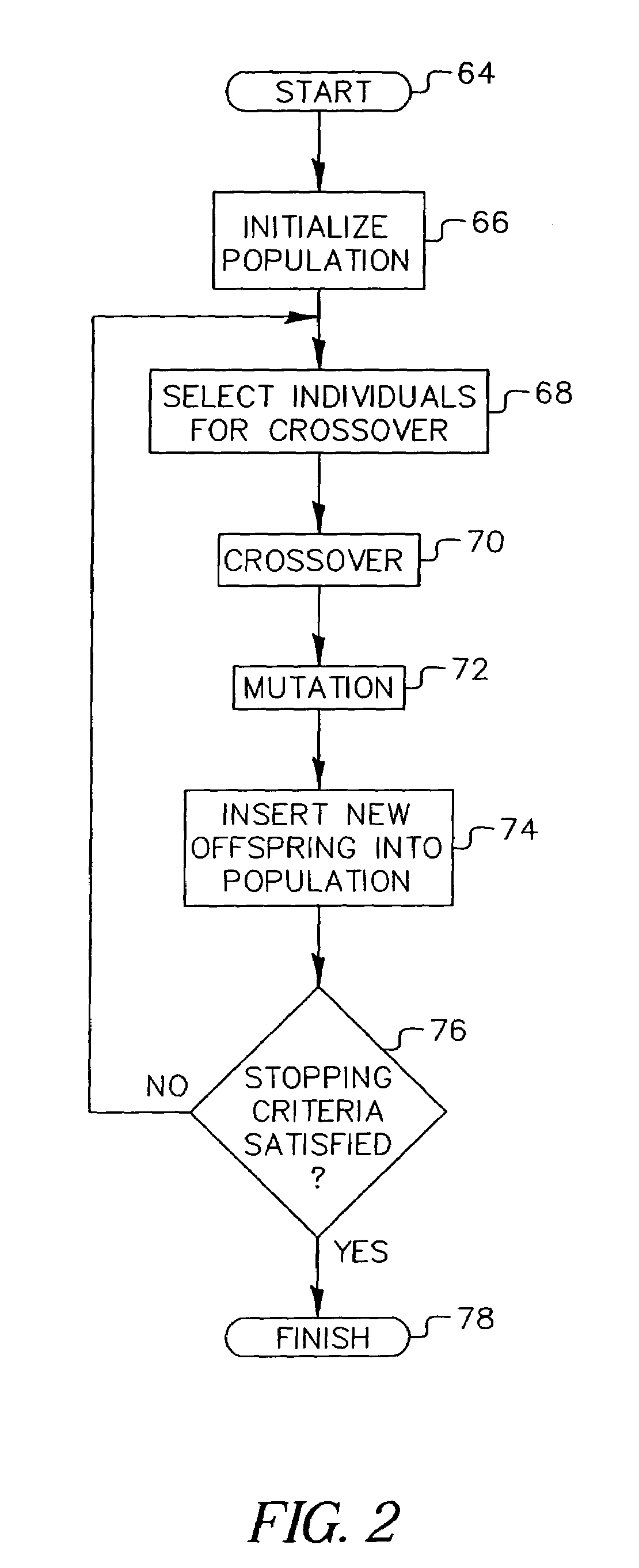
A system and method for automatic creation of digital image albums. A Page Creator Module utilizes a genetic engine and a layout evaluation module. The genetic engine evolves a group of images to a plurality of album pages, based on certain layout criteria. The evaluation module calculates layout criteria and compares them with user preferences. When an acceptable image / page layout has been generated, the image / page assignments are transferred to an Image Placement Module. The Image Placement Module utilizes a second genetic engine, which evolves various criteria to generate page layouts genetic structures. These structures define the location, scale, and rotation of images placed on a given page. A layout evaluation module calculates and compares these layouts with certain other preferences and page requirements. When a suitable layout has been generated, a final album output is generated, which may be displayed, printed, or otherwise transferred for subsequent utilization.
Owner:MONUMENT PEAK VENTURES LLC
System and method for automatic layout of images in digital albums
A system and method for automatic creation of digital image albums. A Page Creator Module utilizes a genetic engine and a layout evaluation module. The genetic engine evolves a group of images to a plurality of album pages, based on certain layout criteria. The evaluation module calculates layout criteria and compares them with user preferences. When an acceptable image / page layout has been generated, the image / page assignments are transferred to an Image Placement Module. The Image Placement Module utilizes a second genetic engine, which evolves various criteria to generate page layouts genetic structures. These structures define the location, scale, and rotation of images placed on a given page. A layout evaluation module calculates and compares these layouts with certain other preferences and page requirements. When a suitable layout has been generated, a final album output is generated, which may be displayed, printed, or otherwise transferred for subsequent utilization.
Owner:MONUMENT PEAK VENTURES LLC
Diagnostic decision support system and method of diagnostic decision support
InactiveUS20050216208A1Improve accuracyHigh precision analysisData processing applicationsHealth-index calculationClinical informationBiology
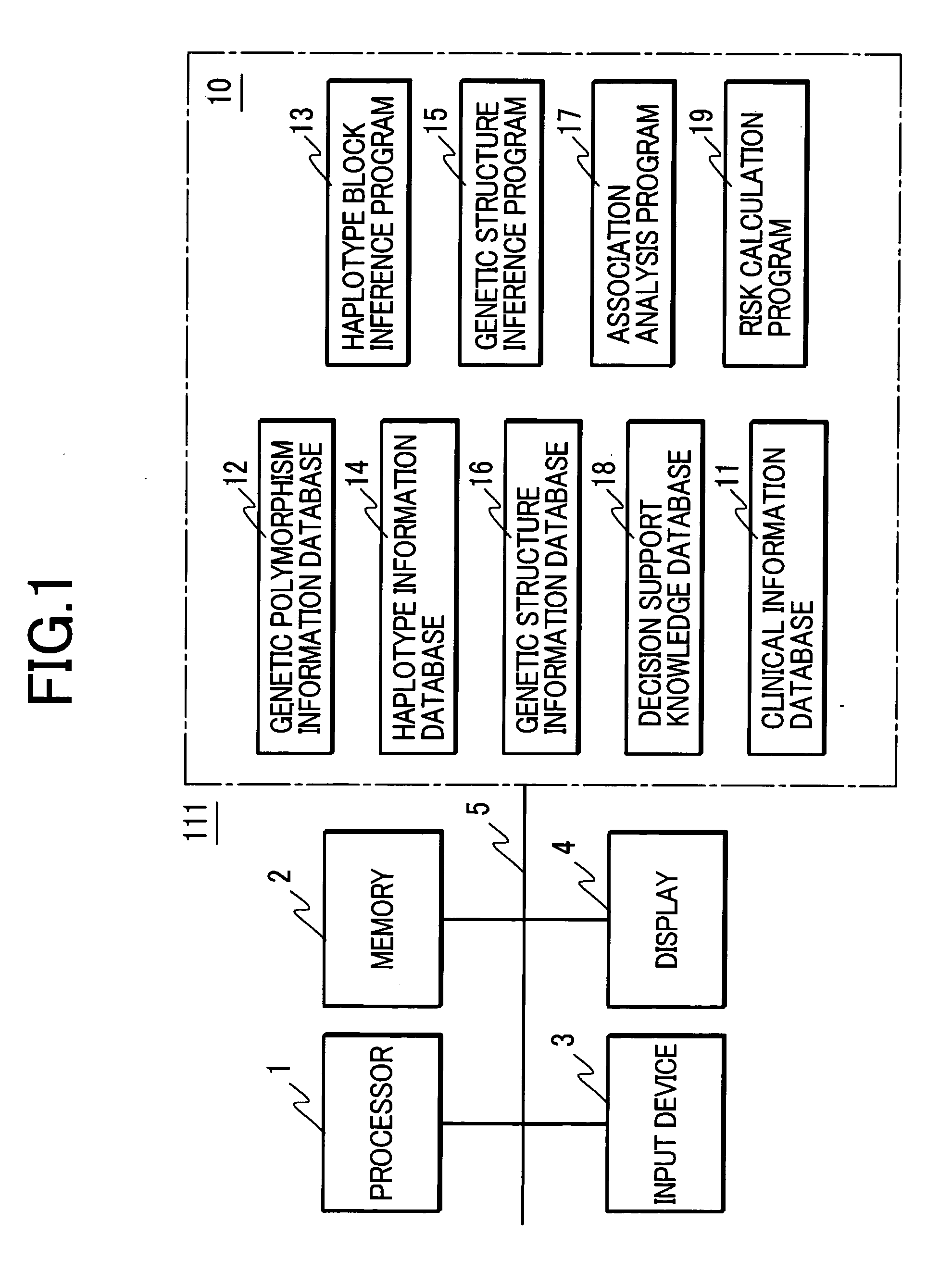
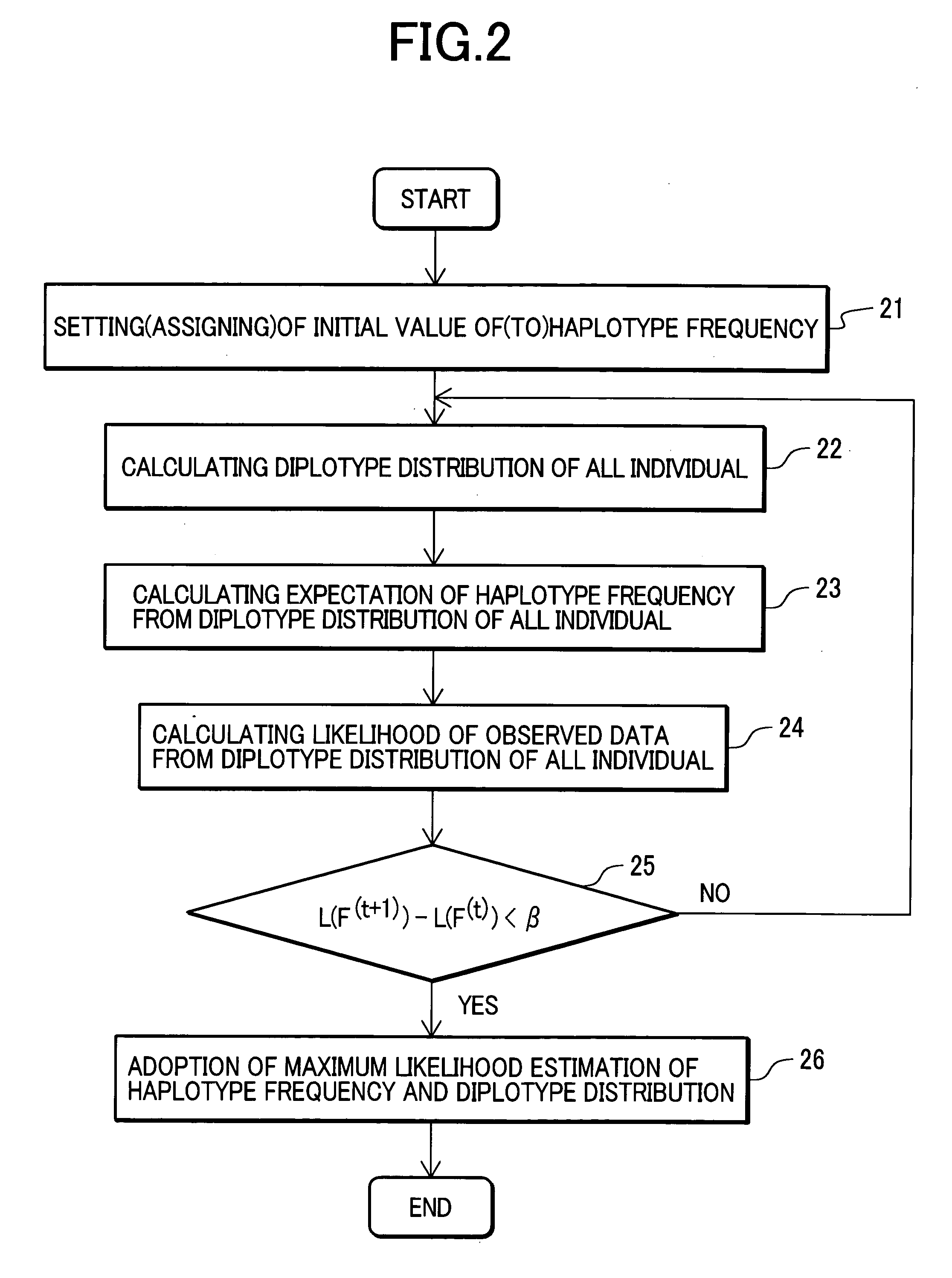
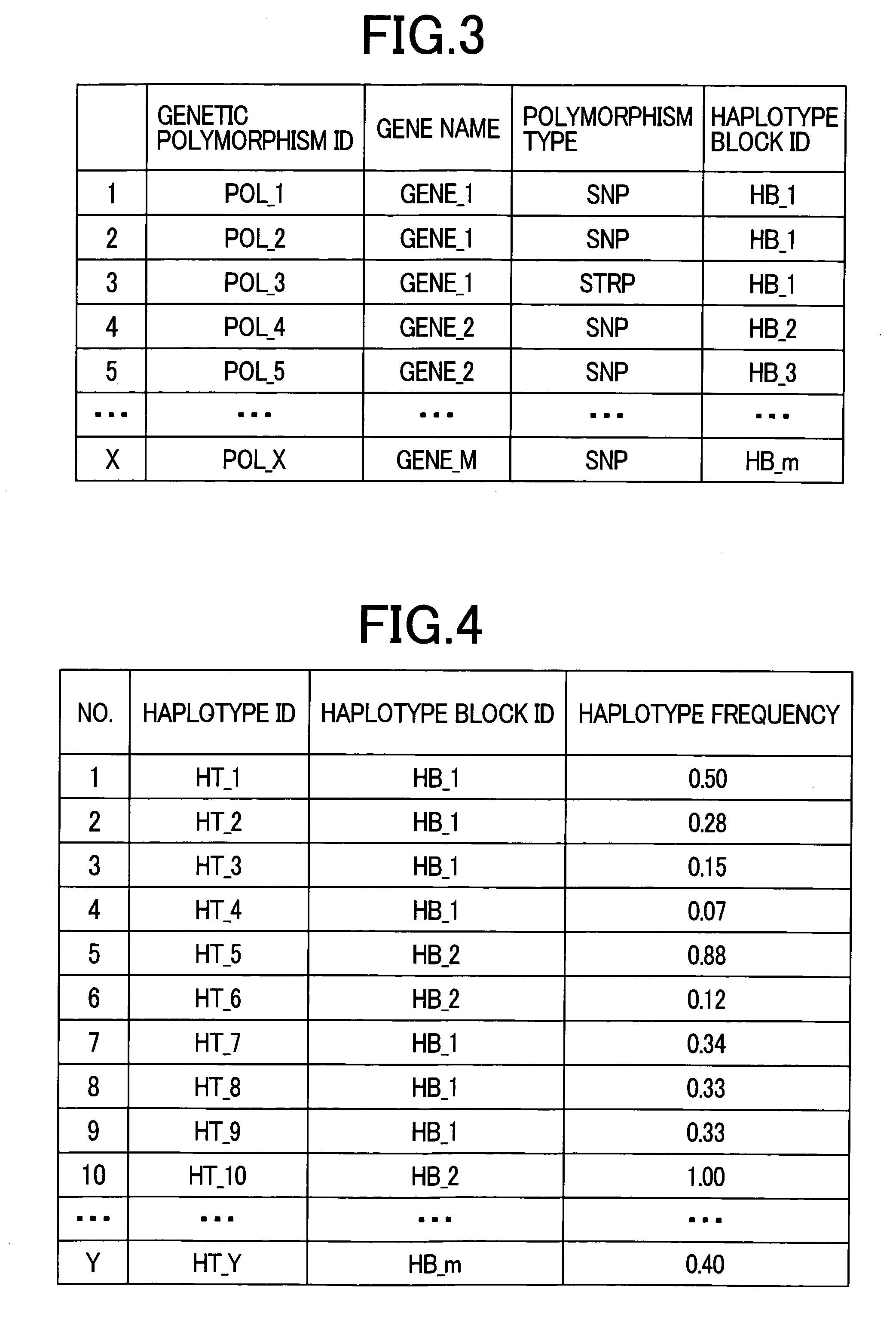
There is provided a system performing high-accuracy diagnostic decision support in consideration of the influence of a haplotype block and a genetic structure. Haplotype block inference means 13 infers the positions of haplotype blocks, and analyzes each of the haplotype blocks to infer a haplotype pattern of individuals with high accuracy. Genetic structure inference means 15 performs clustering the individuals on the basis of the haplotype pattern to divide a population into some subpopulations, and removes the influence of a genetic structure existing in the population. A genetic structure information database 16 and a clinical information database 11 are used to analyze association of clinical information with genetic information for providing high-accuracy diagnostic decision support knowledge. On the basis of the diagnostic decision support knowledge obtained by analyzing the association of clinical information with genetic information, risk calculation means 19 calculates a risk that a predetermined individual is affected by disease.
Owner:HITACHI LTD
Diagnostic decision support system and method of diagnostic decision support
InactiveCN1674028AHigh precisionEliminate the effects ofData processing applicationsHealth-index calculationDiseaseClinical information
There is provided a system performing high-accuracy diagnostic decision support in consideration of the influence of a haplotype block and a genetic structure. Haplotype block inference means 13 infers the positions of haplotype blocks, and analyzes each of the haplotype blocks to infer a haplotype pattern of individuals with high accuracy. Genetic structure inference means 15 performs clustering the individuals on the basis of the haplotype pattern to divide a population into some subpopulations, and removes the influence of a genetic structure existing in the population. A genetic structure information database 16 and a clinical information database 11 are used to analyze association of clinical information with genetic information for providing high-accuracy diagnostic decision support knowledge. On the basis of the diagnostic decision support knowledge obtained by analyzing the association of clinical information with genetic information, risk calculation means 19 calculates a risk that a predetermined individual is affected by disease.
Owner:HITACHI LTD
Enhancing the circulating half-life of antibody-based fusion proteins
InactiveUS20060194952A1Extended half-lifeReduced binding affinityHybrid immunoglobulinsSugar derivativesFc(alpha) receptorFc receptor
Disclosed are methods for the genetic construction and expression of antibody-based fusion proteins with enhanced circulating half-lives. The fusion proteins of the present invention lack the ability to bind to immunoglobulin Fc receptors, either as a consequence of the antibody isotype used for fusion protein construction, or through directed mutagenesis of antibody isotypes that normally bind Fc receptors. The fusion proteins of the present invention may also contain a functional domain capable of binding an immunoglobulin protection receptor.
Owner:MERCK PATENT GMBH
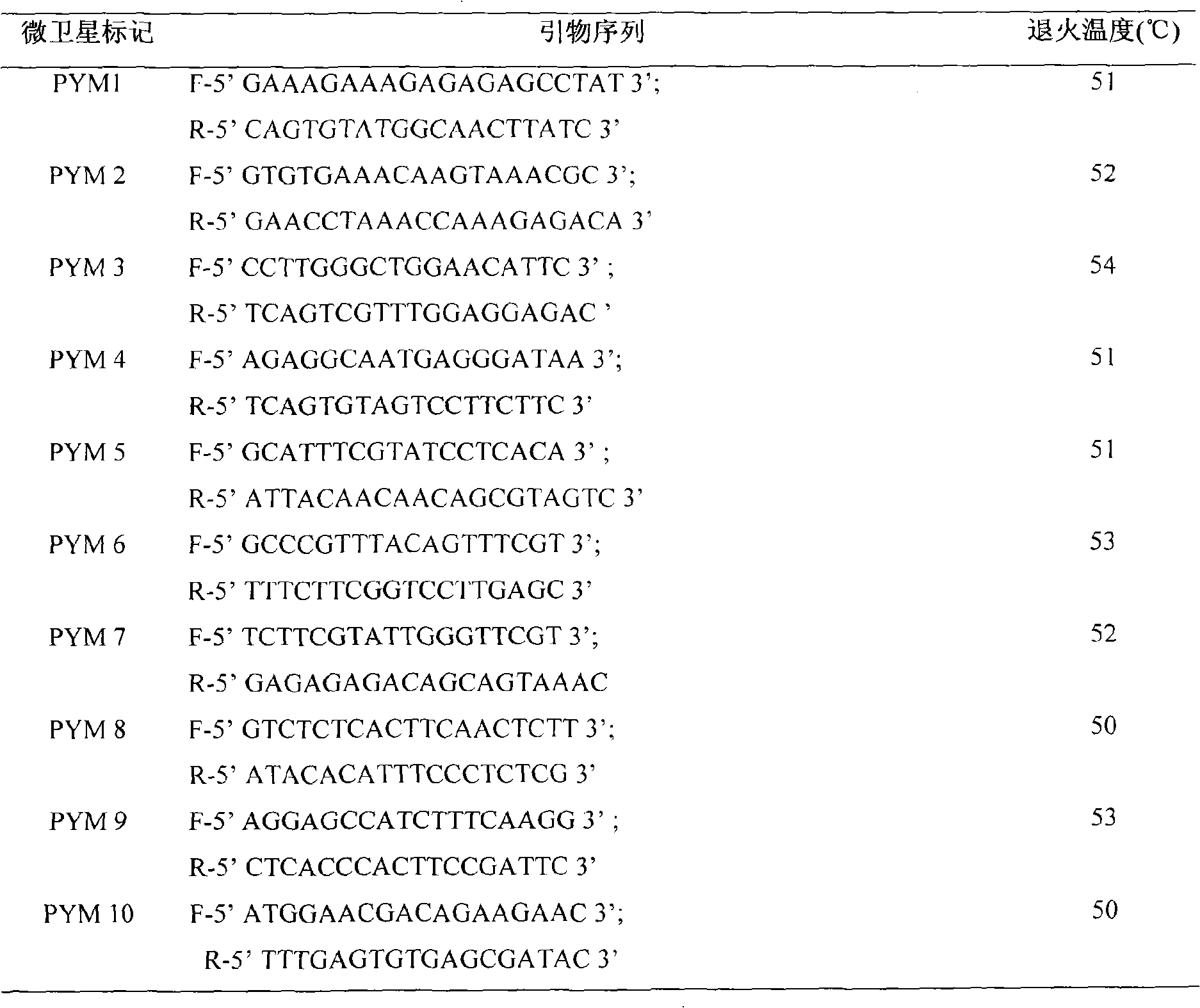
Porphyra yezoensis microsatellite marker screening method and use thereof
InactiveCN101550434AImprove reliabilityHigh yieldMicrobiological testing/measurementFermentationResource protectionGermplasm
The invention belongs to molecular biology DNA labeling technique and application field, concretely relates to a porphyra yezoensis microsatellite marker screening method, and a method for analyzing germplasm genetic diversity by the makers, thereby lay foundations for genetic structure analysis of porphyra yezoensis, germplasm resource protection and molecular marker auxiliary breeding.
Owner:OCEAN UNIV OF CHINA
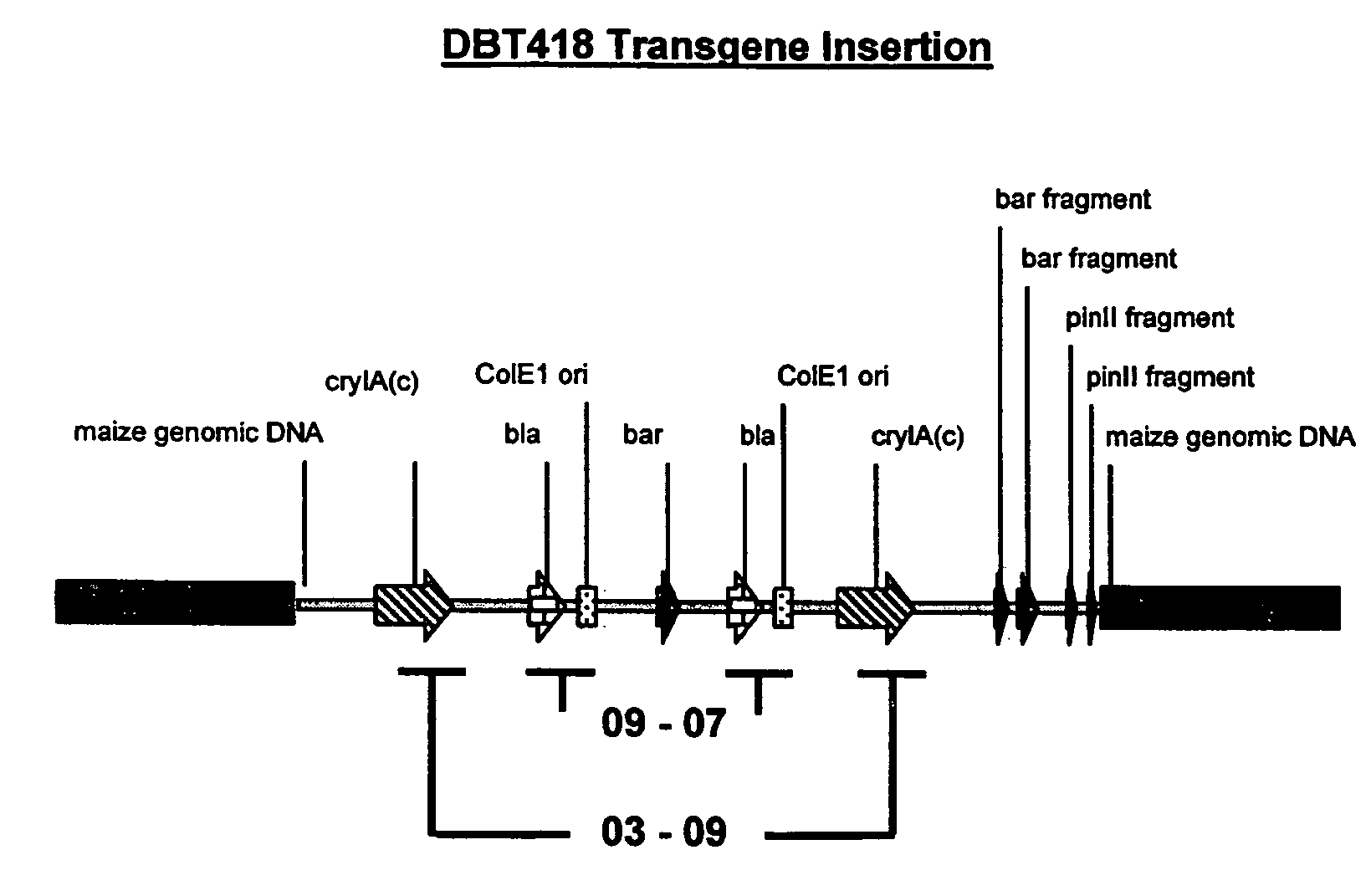
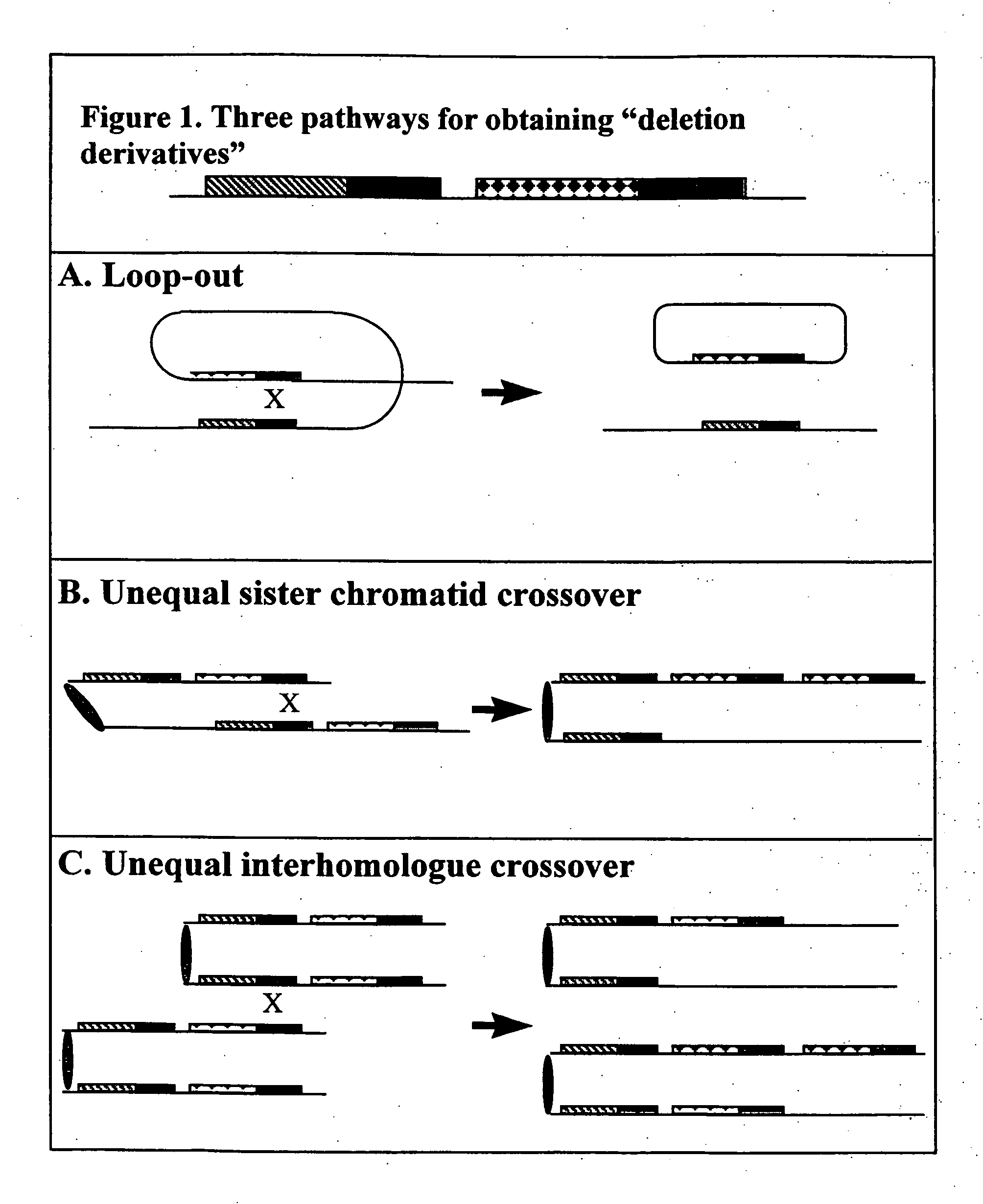
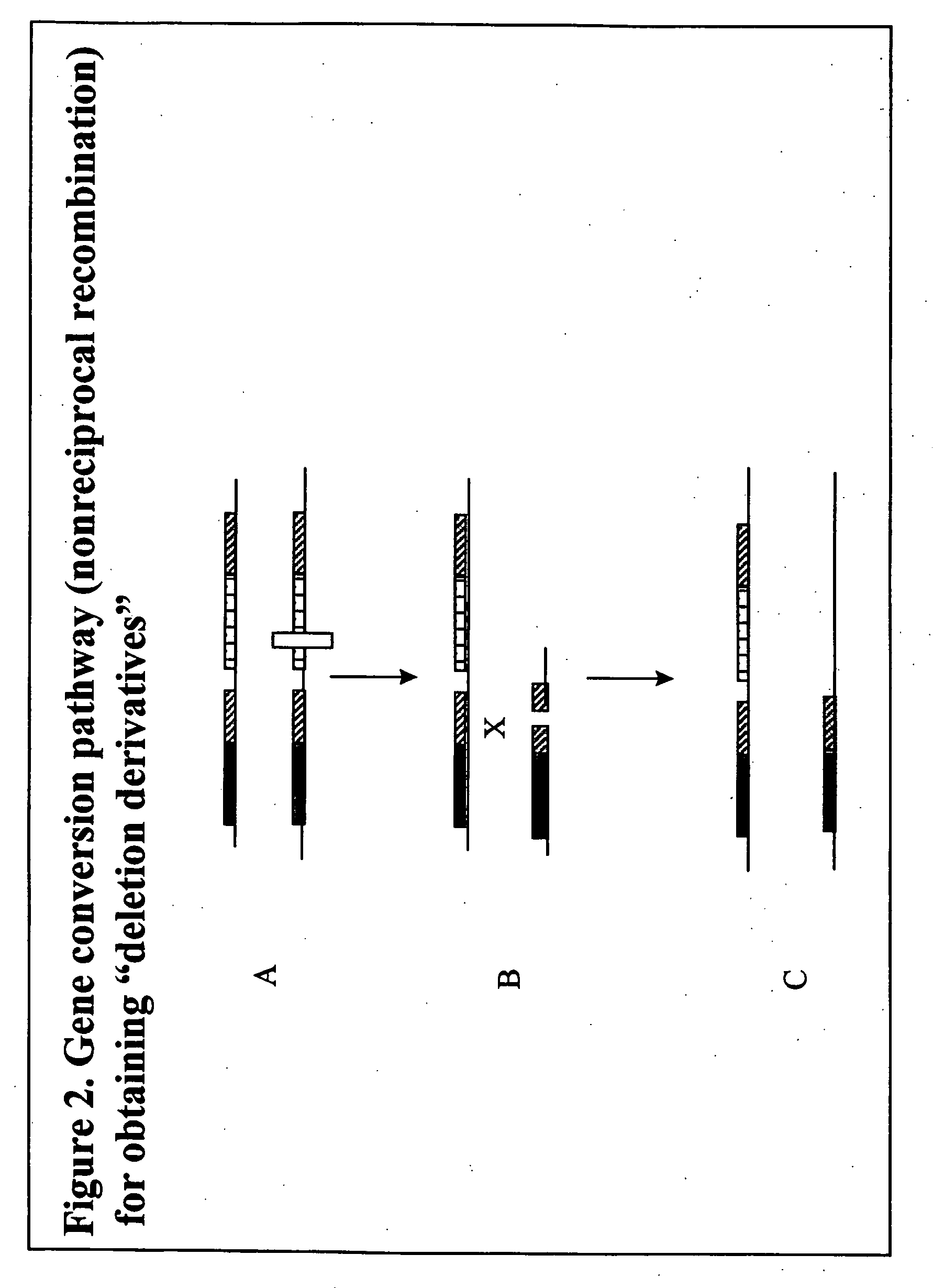
Homologous recombination-mediated transgene deletion in plant cells
InactiveUS20050060769A1Increasing transgene stabilityReduce generationOther foreign material introduction processesFermentationRecombinant TransgenesPlant cell
A process to prepare a recombined transgenic Zea mays plant or plant cell from a first transgenic Zea mays plant cell, wherein the transgene in the recombinant plant or plant cell has an altered genetic structure relative to the genetic structure of the transgene in the first transgenic plant cell, due to homologous recombination-mediated transgene deletion.
Owner:GILBERTSON LARRY A
Cognitive image filtering
InactiveUS7577631B2Accurate timingDiagnostic recording/measuringSensorsOperational systemImage formation
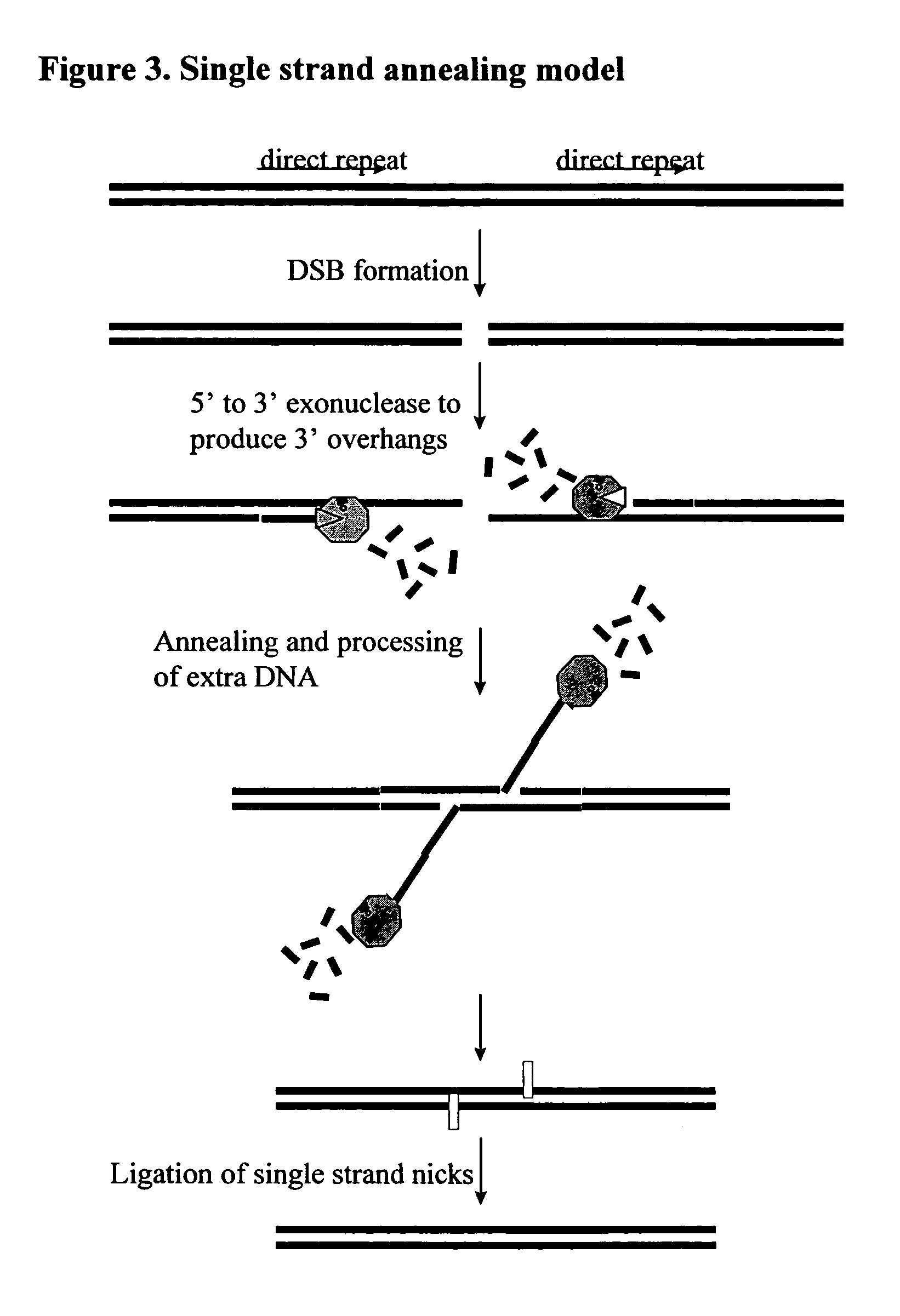
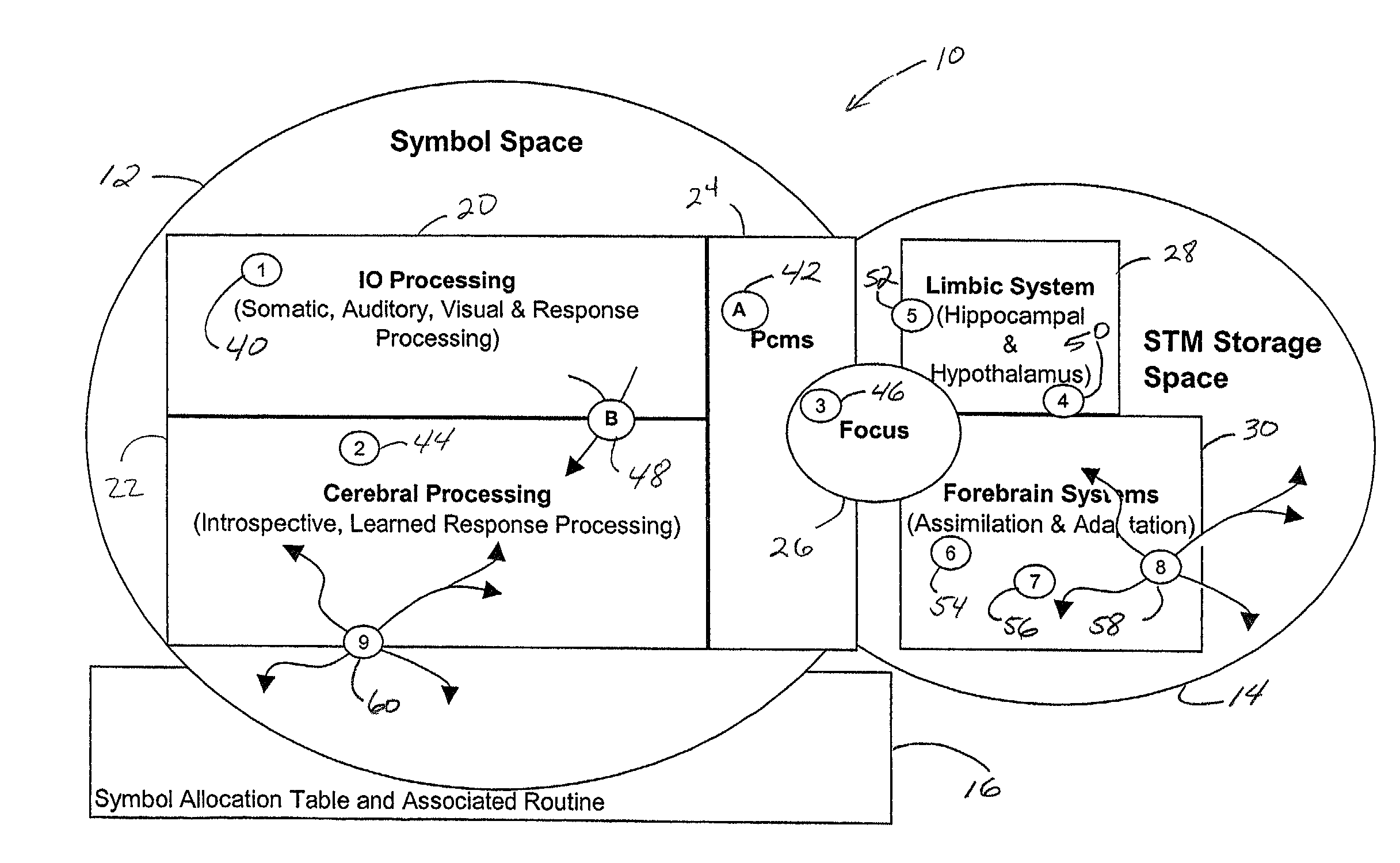
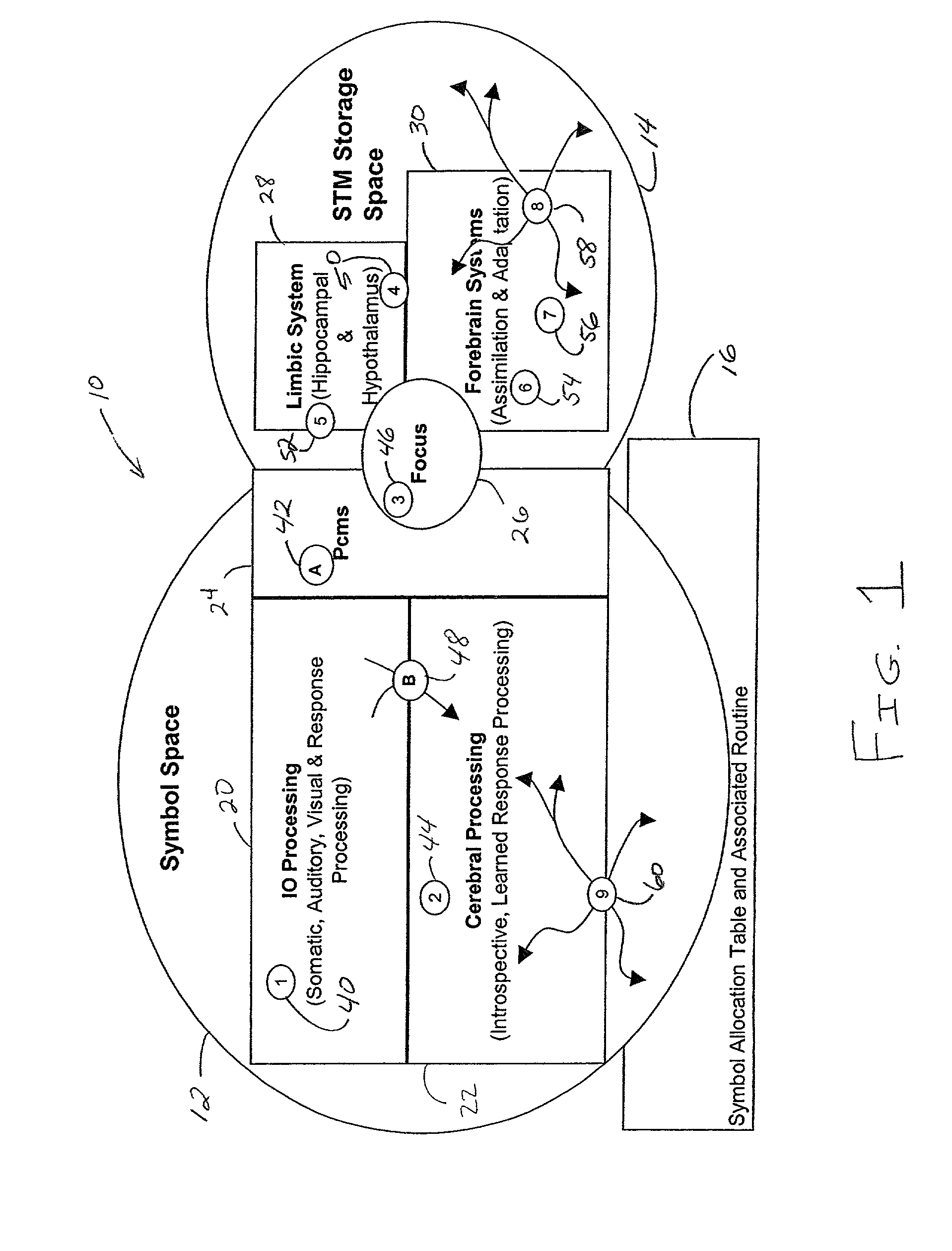
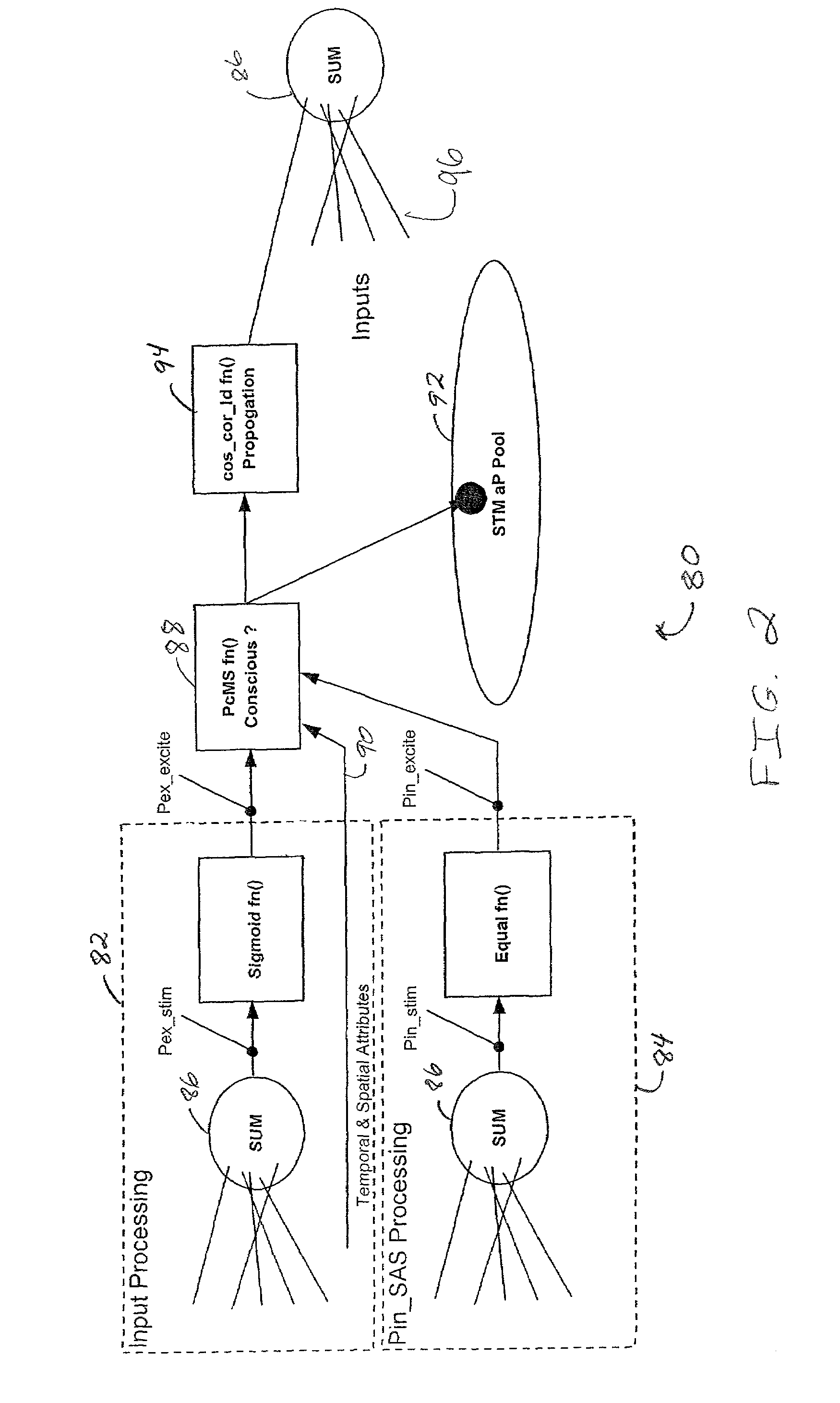
An operating system configured to support cognitive capable environments is described. The system comprises a memory structure and an I / O process configured to update inputs and outputs. The operating system further includes a process to determine changes to symbols within the symbol space due to the stimulus and create STM images of the inputs. Additional processes are configured to pass stimulations between connecting symbols, measure temporal and spatial properties of images, filter the STM images based upon the properties, and propagate stimuli through hereditary structures. Further processes analyze propagated symbol groups of STM images for novel distinctions, create new symbols, connect novel stimuli together for symbol groupings, update existing symbols to include connections to the new symbols, and assign weights to the connections. Additional processes form stimulus-response pairs from received images, provide time-based erosion of connection weights of symbols and adapt learned responses to long term memories.
Owner:FELDHAKE MICHAEL J
Litopenaeus vannamei osmotic regulation related functional gene EST-SSR markers and specific primers and detection method thereof
ActiveCN105969873AMicrobiological testing/measurementDNA/RNA fragmentationStructure analysisMicrosatellite
The invention discloses litopenaeus vannamei osmotic regulation related functional gene EST-SSR markers and specific primers and a detection method thereof. Litopenaeus vannamei genome DNA serves as a template, the specific primers of the litopenaeus vannamei osmotic regulation related functional gene EST-SSR markers are used for conducting PCR amplification on microsatellite locus sequences, and then typing is conducted on PCR amplification products through a sequenator. The specific primers and detection method of the litopenaeus vannamei osmotic regulation related functional gene EST-SSR markers can be used for population genetic structure analysis and molecular marker assisted breeding of litopenaeus vannamei and particularly have an important application value in breeding of good salt-tolerant varieties of the litopenaeus vannamei.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Multielement high flux genetic marking system and genetic analyzing method of Chinese mitten crabs
ActiveCN101613742AAvoid blindnessParsing Genetic DiversityMicrobiological testing/measurementDNA/RNA fragmentationPolycultureMicrosatellite
The invention relates to the field of genetic thremmatology of aquatic products, in particular to a multielement high flux genetic marking system and a genetic analyzing method of Chinese mitten crabs. The genetic marking system is a mitochondria sequence information system and a microsatellite molecule marking system, wherein the microsatellite molecule marking system comprises 7 EST-SSR systems marked 1and 6 EST-SSR marked systems 2. The method comprises the following steps: later generations produced by the same parent and different parents in a polyculture family are separated according to hereditary constitution and genetic diversity through information of sequences of three pairs of primer amplifications COI, Cytb and CR of mitochondria, and male parent discrimination is performed on later generations of the discriminated parents according to genetic distance and genetic diversity through the EST-SSR microsatellite molecule marking system, thereby completing the genealogy authentication during the genetic breeding of the Chinese mitten crabs and determining the inbreeding coefficients. The invention discriminates the genetic relationship among filial generations by using the multielement high flux genetic marking system on the basis of effectively utilizing species genetic information, and effectively avoids the inbreeding recession.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Method for preparing linen fiber morph-genetic structure C/Sn or C/Al compound material
The invention discloses a method for preparing a C / Sn or C / Al composite material of a morph-genetic fibrilia structure. Firstly, a nano SnO2 or Al2O3 precursor aqueous solution doped with Fe, La and Ce is prepared, and then fibrilia is taken and is pretreated; secondly, a fibrilia raw material or a crystalline structure C with the morph-genetic fibrilia structure is obtained and is put into the nano oxide precursor aqueous solution for dipping; and thirdly, the C / Sn or C / Al composite material of the morph-genetic fibrilia structure is obtained through high-temperature vacuum reaction hot-pressing sintering and annealing. The preparation method immerses a Sn or Al oxide precursor into or around a cavity of the fibrilia structure and utilizes a reactive hot pressing molding process to ensure that the nano precursor is dehydrated into an oxide which reacts with C and reduces the generated product to obtain metal Sn or Al; and the produced composite material has the characteristics of light mass, vibration elimination and good sound adsorption, antifrictional property and wear resistance, extends the application field of fibrilia composite materials, and has wider application prospect.
Owner:XI'AN POLYTECHNIC UNIVERSITY
Molecular marker method for breeding meat performance of Aletail sheep and application thereof
ActiveCN103525920AImprove meat performanceIncrease economic incomeMicrobiological testing/measurementSingle-strand conformation polymorphismCandidate Gene Association Study
The invention discloses a molecular marker method for breeding meat performance of Altay sheep and application thereof. The molecular marker method comprises the following steps: using a MyoD1 (Myogenic Determination) gene as a candidate gene which influences meat yield of the Altay sheep; detecting a genetic structure of the MyoD1 gene in the Altay sheep variety by utilizing a PCR-SSCP (Polymerase Chain Reaction-Single-strand Conformation Polymorphism) technology; analyzing and judging a correlation between the genetic structure of the MyoD1 gene and the meat yield of the sheet; by a DNA (Deoxyribose Nucleic Acid) sequence of the MyoD1 gene of the sheet, self-designing two pairs of specific primers; carrying out polymorphism detection on the MyoD1 gene of the Altay sheep in Sinkiang province by utilizing the PCR-SSCP technology; and carrying out correlation analysis with meat performance so as to find a molecular marker which is obviously related to the meat yield of the Altay sheep. The molecular marker method is used for improving meat performance of the Altay sheep to greatly improve the speed of improving and recovering excellent meat performance of the Altay sheep and has important significance and practical application value for improving economic income of farmers and herdsmen and quickening development of the mutton sheep industry in Sinkiang province.
Owner:XINJIANG AGRI UNIV
Macrolide efflux genetic assembly
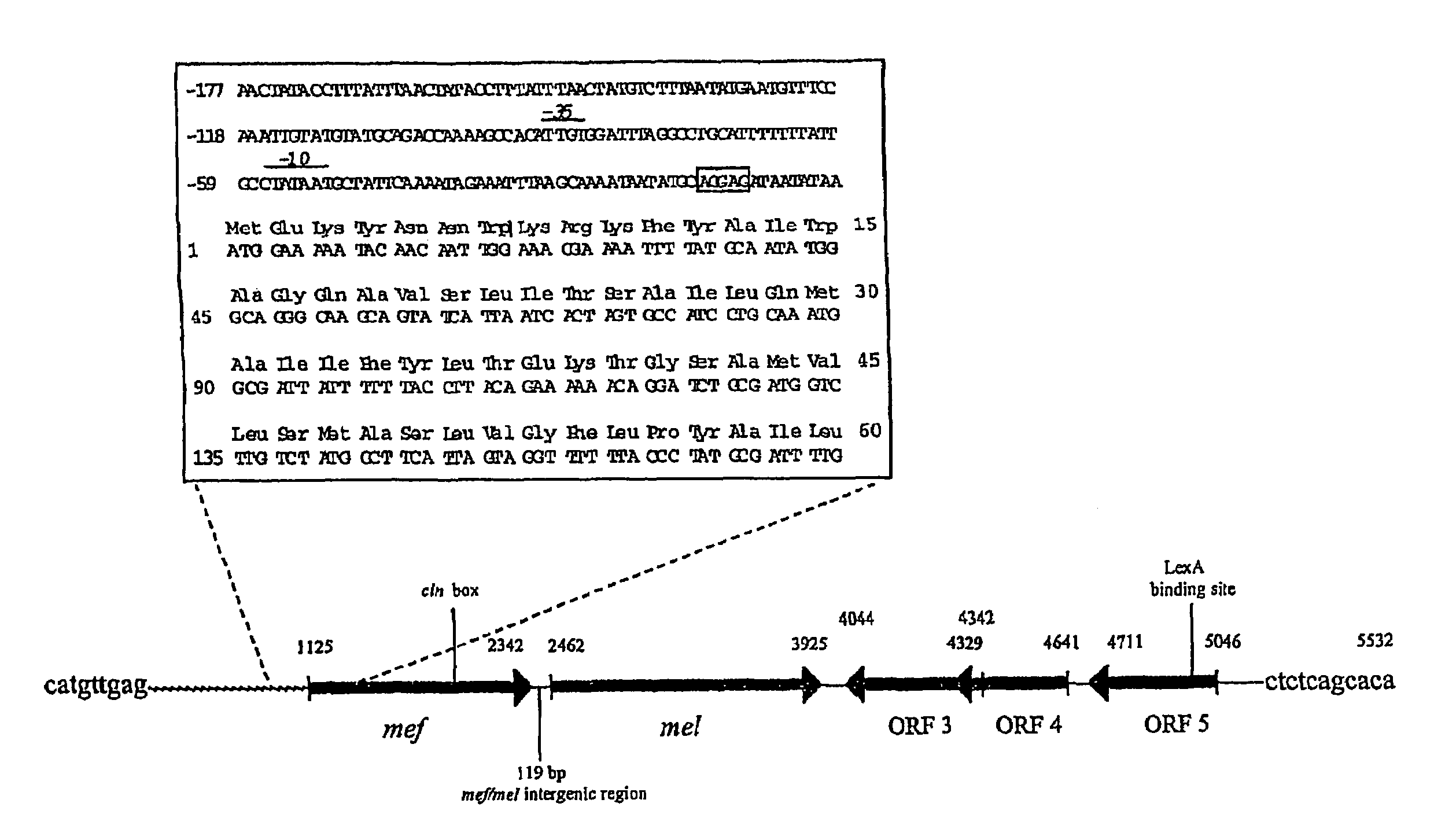
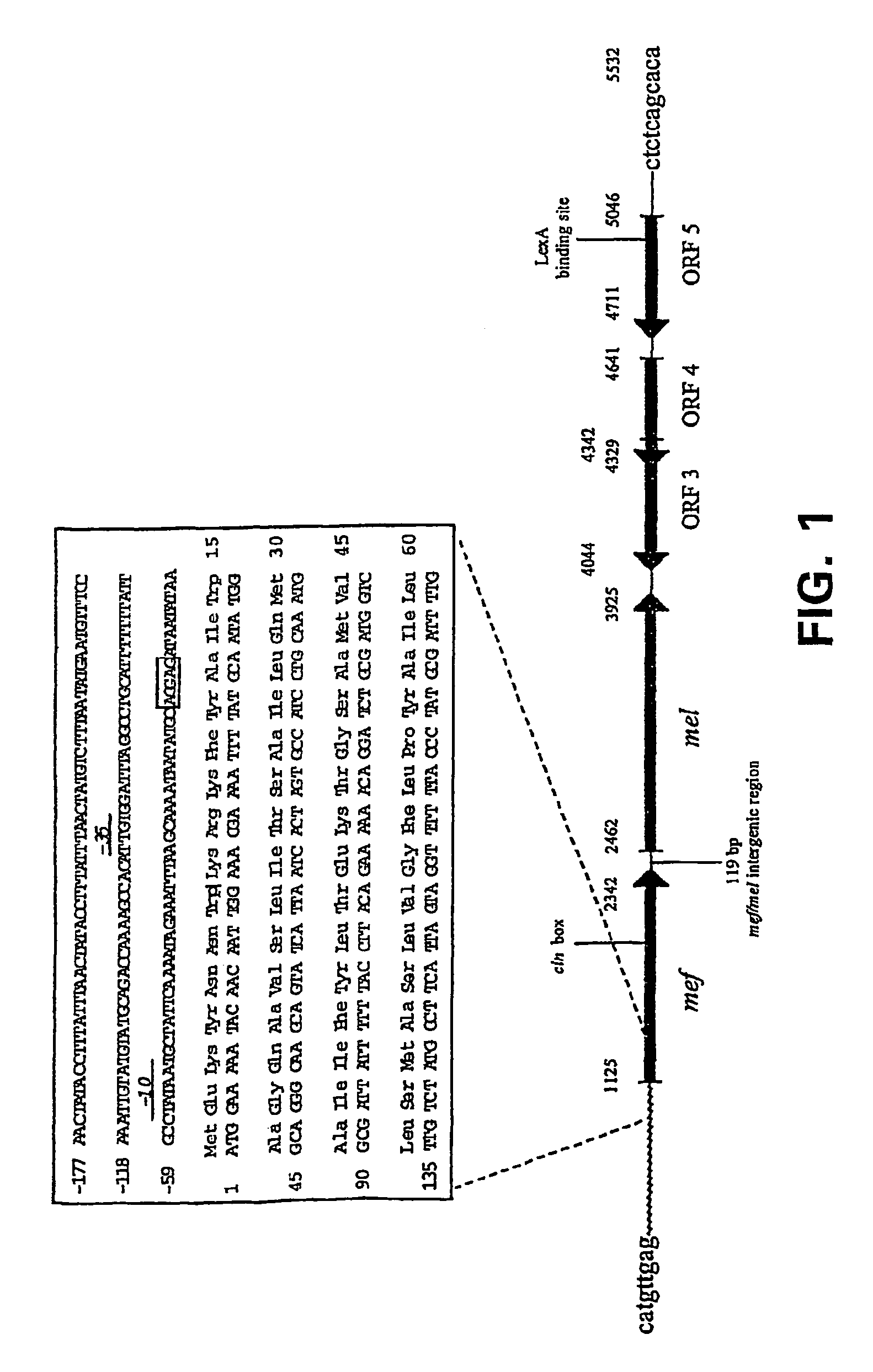
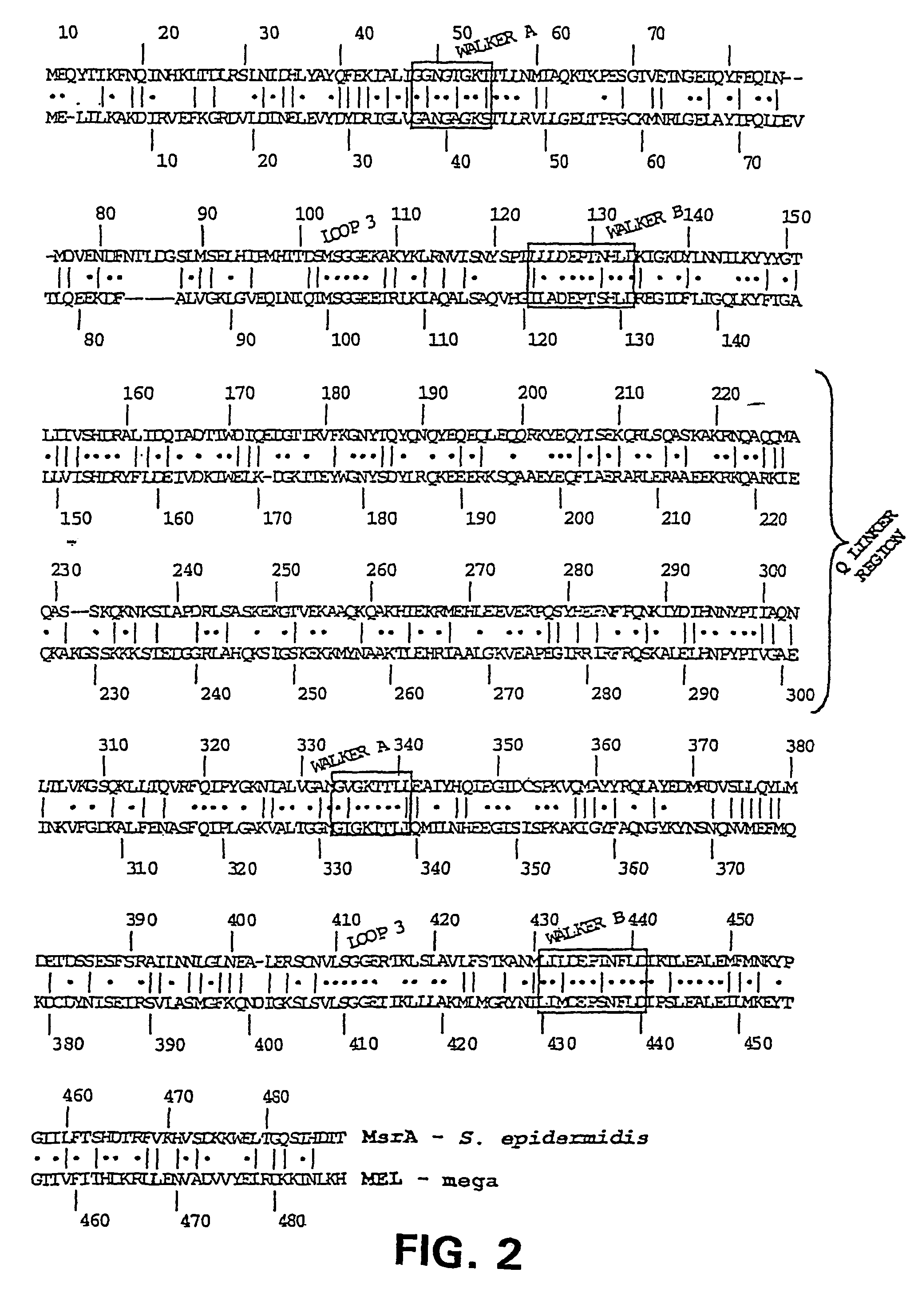
Macrolide resistance associated with macrolide efflux (mef) in Streptococcus pneumoniae has been defined with respect to the genetic structure and dissemination of a novel mefE-containing chromosomal insertion element. The mefE gene is found on the 5′-end of a 5.5 kb or 5.4 kb insertion designated mega (macrolide efflux genetic assembly) found in at least four distinct sites of the pneumococcal genome. The element is transformable and confers macrolide resistance to susceptible S. pneumoniae. The first two open reading frames (ORFs) of the element form an operon composed of mefE and a predicted ATP-binding cassette homologous to msrA. Convergent to this efflux operon are three ORFs with homology to stress response genes of Tn5252. Mega is related to mefA-containing element Tn1207.1. Macrolide resistance due to mega has been rapidly increased by clonal expansion of bacteria containing it and horizontally by transformation of previously sensitive bacteria.
Owner:EMORY UNIVERSITY
Microsatellite molecular marker of mauremys mutica and application thereof
InactiveCN101880659AHighly polymorphicMicrobiological testing/measurementDNA/RNA fragmentationMicrosatelliteNucleotide sequencing
The invention discloses a microsatellite molecular marker of mauremys mutica. The nucleotide sequence of the microsatellite molecular marker is SEQ ID NO:1. The invention also discloses a primer of the microsatellite molecular marker of the mauremys mutica. The microsatellite molecular marker of the mauremys mutica can be used for carrying out genetic structure analysis, genetic map construction, gene location, auxiliary marking seed selection or auxiliary marking seed breeding on the mauremys mutica.
Owner:ZHEJIANG NORMAL UNIVERSITY
Complete sequence amplification method for mitochondrial genome of diptychus maculates steindachner
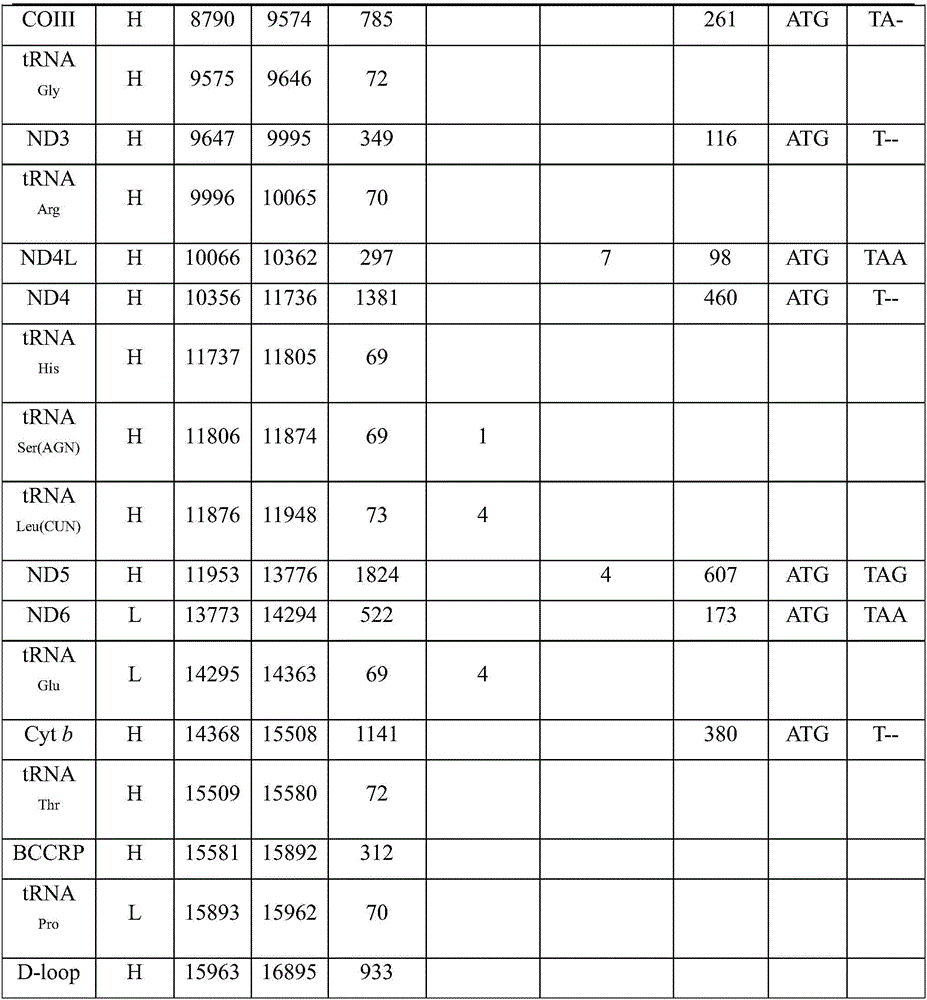
The invention discloses a complete sequence amplification method for a mitochondrial genome of diptychus maculates steindachner. The complete sequence amplification method specifically comprises the following steps: (1) amplifying four gene segments Cyt b, COI, ND4 and 16S rRNA of the mitochondrial genome of the diptychus maculates steindachner; and (2) carrying out LA-PCR amplification on the mitochondrial genome of the diptychus maculates steindachner. According to the complete sequence amplification method, the four gene segments Cyt b, COI, ND4 and 16S rRNA of the mitochondrial genome of the diptychus maculates steindachner are amplified, four pairs of LA-PCR primers are designed according to an obtained sequence and are used for carrying out large-segment sequence amplification, and 28 sequencing primers are utilized for carrying out PCR amplification on the other parts of the mitochondrial genome of the diptychus maculates steindachner by virtue of a primer PCR-Walking method so as to obtain a whole-genome sequence, with a length of 16895bp, of a mitochondrion of the diptychus maculates steindachner. The inheritance and evolution of the diptychus maculates steindachner are researched in a molecular level, and particularly mtDNA is used as a molecular marker, so that the basic data and technical support are provided for the researches of species identification, population genetic structures and system evolution of the diptychus maculates steindachner.
Owner:ZHEJIANG OCEAN UNIV
Linen fiber morph-genetic structure tin oxide or aluminium oxide compound material
InactiveCN101381098AIncrease dampingImprove sound absorptionTin oxidesAluminium oxides/hydroxidesFiberAluminium hydroxide
The invention discloses a method for preparing a tin oxide or alumina composite material with bastose morph-genetic structure. The method comprises the following steps: firstly, nanometer stannic hydroxide or aluminum hydroxide aqueous solution is prepared; secondly, bastose is taken out and is subjected to pretreatment to obtain a bastose material or a crystalline-state structure C with the bastose morph-genetic structure; and the bastose material or the crystalline-state structure C is placed in the hydroxid aqueous solution and is subjected to dipping, drying, intermediate temperate vacuum reaction, heat pressing, sintering and annealing to prepare a metal oxide / carbon-based composite material. With the preparation method, Sn or Al nanometer hydroxid is immersed into the inside or the periphery of a cavity of the bastose structure; a reaction, heat pressing and molding process is utilized to dewater the nanometer hydroxid to form metal oxide; and the metal oxide is compounded with carbon to prepare the metal oxide / carbon-based composite material with the bastose morph-genetic structure; and the composite material has the characteristics of light mass, vibration elimination, sound absorption and good friction reduction and abrasion resistance, broadens the application field of the bastose composite material and has wider application prospect.
Owner:XI'AN POLYTECHNIC UNIVERSITY
Method for developing SSR (Simple Sequence Repeat) molecular mark of nibea albiflora for population identification
InactiveCN105624322AImprove work efficiencyReduce screening costsMicrobiological testing/measurementDNA/RNA fragmentationGenomic DNAGrowing season
The invention discloses a method for developing an SSR (Simple Sequence Repeat) molecular mark of nibea albiflora for population identification. The method comprises the following steps: extracting RNA (Ribonucleic Acid) by using a nibea albiflora mixed tissue of a plurality of nibea albiflora, performing RNA sequencing, performing splicing to obtain a nibea albiflora expression gene sequence, and obtaining a potential SSR mark with a bioinformatics method; designing primers by utilizing an SSR flanking sequence, extracting genomic DNA (Deoxyribose Nucleic Acid) by using fins from a random sample, and screening the SSR primers; and performing PCR (Polymerase Chain Reaction) amplification on different nibea albiflora DNA templates by using each pair of the SSR primers, performing capillary electrophoresis detection on an amplification product by using a Qsep100 full-automatic nucleic acid analysis system, screening out primers with different amplification results, calculating allelic composition and allelic frequency by utilizing PopGene, and analyzing a nibea albiflora population relationship and a genetic structure. The SSR molecular mark method for identifying and verifying nibea albiflora groups, provided by the invention, is stable and reliable, is not influenced by ages, gender and growth seasons, and provides scientific bases for group analysis as well as species identification, breeding and evaluation of the nibea albiflora.
Owner:JIMEI UNIV
Frankliniella occidentalis arrhenotoky inbreeding population establishing method
ActiveCN104782574AGood for monitoring growth and developmentKeep dryAnimal husbandryMonosomal karyotypeClosest relatives
The invention discloses a frankliniella occidentalis arrhenotoky inbreeding population establishing method. According to the method, frankliniella occidentalis artificially bred is transferred into a centrifugal tube for single parthenogenetic breeding to establish an inbreeding population by controlling temperature, moisture and illumination manually. The method is characterized in that the frankliniella occidentalis arrhenotoky inbreeding population can be established, and obtained male offspring and multiple continuous generations of male and female offspring can be applied to karyotype analysis, microsatellite analysis and other experiments. The method has great significance for further learning the frankliniella occidentalis arrhenotoky features and establishing the inbreeding population and can further promote the analysis of genetic structure and sex determination mode.
Owner:QINGDAO AGRI UNIV
Haliotis discus hannai microsatellite molecular marker and preparation method thereof
ActiveCN103103182AGood repeatabilityMolecular markers are effective and reliableMicrobiological testing/measurementDNA/RNA fragmentationHaliotis discusPolymorphic microsatellites
The invention discloses a haliotis discus hannai microsatellite molecular marker which can analyze the genetic diversity of haliotis discus hannai wild population and identify a genetic relationship between cultured stocks and a preparation method thereof. According to the steps of establishing a haliotis discus hannai microsatellite (AC)n library, screening microsatellite sequence positive clone, designing a microsatellite primer, performing polymerase chain reaction (PCR) amplification and screening, ten highly polymorphic microsatellite molecular markers of Hd1, Hd2, Hd3, Hd4, Hd5, Hd6, Hd7, Hd8, Hd9 and Hd10 are obtained; and the polymorphic detection proves that the microsatellite molecular marker has the characteristic of high repeatability, is an effective and reliable molecular marker, can establish a haliotis discus hannai microsatellite DNA marker technology system and is used for evaluating the genetic diversity of haliotis discus hannai, analyzing the genetic structure and identifying the genetic relationship between the cultured stocks.
Owner:DALIAN HAIBAO FISHERY
Microsatellite fluorescence multi-PCR method for testing paternity of culter alburnus basilewsky
ActiveCN108611405AThe detection method is simpleConserve DNA samplesMicrobiological testing/measurementFluorescenceGenotype
Owner:ZHEJIANG INST OF FRESH WATER FISHERIES
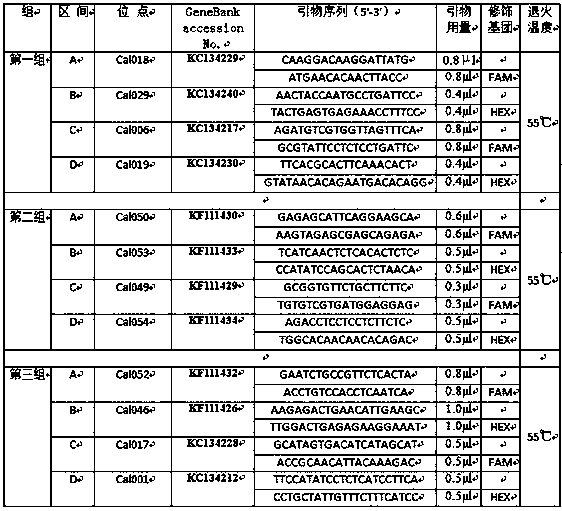
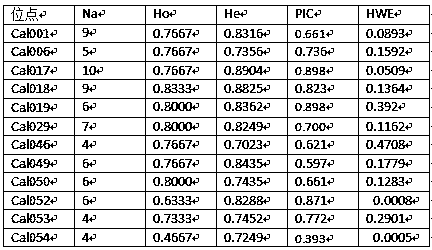
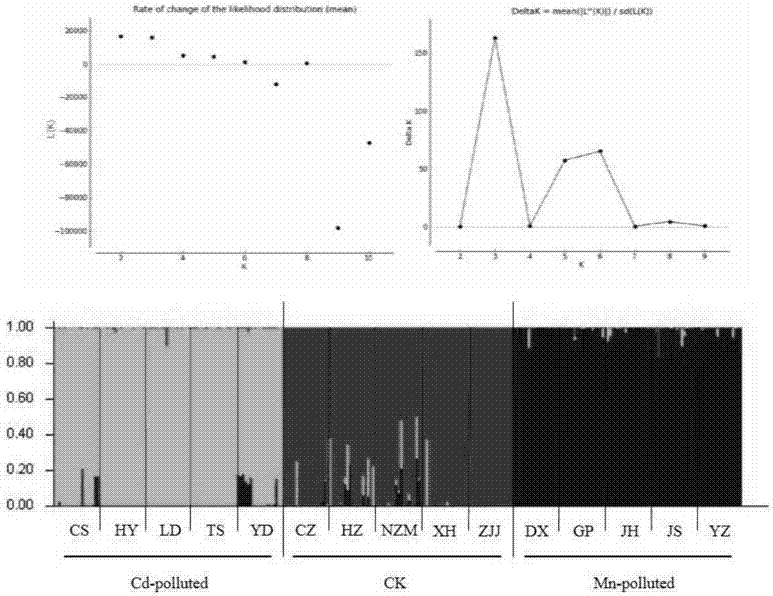
Method for screening manganese/cadmium absorption efficient ecological phytolacca americana
InactiveCN107475384AComparableEasy to operateMicrobiological testing/measurementSystems biologyManganeseScreening method
The invention relates to a plant restoration technology, and particularly discloses a method for screening manganese / cadmium absorption efficient ecological phytolacca americana. The method comprises the following steps: collecting in the wild leaves and seeds of phytolacca americana populations in a plurality of contaminated regions and uncontaminated regions in China and soil in the vicinity; detecting the manganese / cadmium content of the soil in the vicinity of the phytolacca americana populations in the regions, and selecting phytolacca americana populations in highly manganese contaminated, highly cadmium contaminated and uncontaminated regions respectively; performing AFLP (amplified fragment length polymorphism) molecular marker detection on the leaves of the selected phytolacca americana populations, and analyzing genetic structures thereof; selecting bred seedlings of the phytolacca americana populations to which different genetic differentiation occurs, culturing the seedlings for 50 days with manganese, cadmium and normal Hoagland solutions, and screening the manganese / cadmium absorption efficient ecological phytolacca americana by adopting the plant heights, fresh weights and leaf manganese / cadmium content of the phytolacca americana as indexes. According to the method, a manganese / cadmium absorption efficient ecological hyperaccumulator is used for treating manganese / cadmium-contaminated soil, and the heavy metal accumulation and restoration efficiency of the hyperaccumulator are improved.
Owner:NANJING AGRICULTURAL UNIVERSITY
Litopenaeus vannamei disease resistance related EST-SSR molecular marker and application thereof
ActiveCN108611429AMicrobiological testing/measurementClimate change adaptationDiseaseStructure analysis
The invention discloses a litopenaeus vannamei disease resistance related EST-SSR molecular marker and the application thereof. An SSR site is identified in a 5'-UTR area of litopenaeus vannamei disease resistance related genes LvIRF, specific primers are designed according to sequence characteristics of the area, and by virtue of PCR (Polymerase Chain Reaction) amplification and sequence-based typing, a set of analytical method for detecting polymorphism of the site is established. In addition, a number of repeats in a core repeat (CT) sequence in the site is directly associated with diseaseresistance of litopenaeus vannamei, so the site can serve as a rapid, efficient and convenient molecular marker so as to be applied to screening of the litopenaeus vannamei related to disease resistance. Moreover, the molecular marker can be combined with other molecular markers to be used for analyzing a population genetic structure of the litopenaeus vannamei and the like. The litopenaeus vannamei disease resistance related functional gene EST-SSR molecular marker and the application thereof provided by the invention can be directly used for selecting disease-resistant litopenaeus vannamei individuals, can also be used for population genetic structure analysis and molecular mark assisted breeding, and particularly have a wide application prospect in selective breeding of litopenaeus vannamei disease-resistant good varieties.
Owner:SUN YAT SEN UNIV
Triplex PCR system detecting method of panda microsatellite site
InactiveCN107190098AThe detection method is simpleConserve DNA samplesMicrobiological testing/measurementDNA paternity testingTriplex PCR
The invention belongs to the technical field of bioengineering, and particularly relates to a set of triplex PCR system detecting method of a panda microsatellite site. The set of triplex PCR system detecting method of the panda microsatellite site roughly includes the steps that 1, genome DNA is extracted; 2, the fragment sizes of amplification products of initial primers (a single pair of primers) are evaluated; 3, triplex PCR primer combinations are designed and optimized; 4, triplex PCR amplification is conducted, and conditions are optimized; 5, gene typing effects are detected, and a final combination mode is determined. The triplex PCR system detecting method of the panda microsatellite site is simple, compared with conventional panda microsatellite PCR systems, 66.7% of DNA samples and 66.7% of costs are saved, and the efficiency is improved by 200%. The triplex PCR system detecting method of the panda microsatellite site can be popularized in the aspects of evaluating panda paternity tests, genetic variation of populations and genetic structures.
Owner:四川省自然资源科学研究院 +1
Family selection breeding method of red-line strain siniperca chuatsi
InactiveCN108719144AFast growthRaise the ratioClimate change adaptationPisciculture and aquariaBody shapeBroodstock
The invention relates to a family selection breeding method of red-line strain siniperca chuatsi. The method comprises the following steps that 1, red-line siniperca chuatsi which is light yellow in color and has red body surface lines and obvious growth advantages is selected from a siniperca chuatsi population variant in body color to serve as standby parents; 2, after the selected female and male parents are subjected to adaptability breeding strengthening, artificial directional mating and induced spawning are carried out on each pair of male and female parents, and fertilized eggs produced by each pair of parent fishes are transferred into incubation barrel for independent breeding; 3, in the young fish stage and the adult fish stage of growth of siniperca chuatsi, a family high in red-line siniperca chuatsi proportion and high in weight gain rate is selected out to serve as a next-generation selection breeding basic family. Through the method, 4-6 generations of continuous selection breeding are carried out so that the red-line strain siniperca chuatsi bright in color, symmetric and attractive in body shape, strong in physique and obvious in variety feature can be bred. The method effectively solves the problems that the varieties of red-line siniperca chuatsi are mixed and degradation is serious; inbreeding is avoided, and the diversity of the genetic structure of the population is kept; selection breeding is carried out many times from the parents to fry, and the selection breeding efficiency is high.
Owner:FISHERIES RES INST ANHUI ACAD OF AGRI SCI
Method for development and application of endangered rhododendron molle polymorphism SSR molecular marker
ActiveCN105624282AMicrobiological testing/measurementDNA/RNA fragmentationGenomic sequencingGenetics
The invention discloses a method for development and application of endangered rhododendron molle polymorphism SSR molecular markers. The method comprises the following steps: developing by using a simplified genome sequencing technique so as to obtain 11687 pairs of rhododendron molle SSR molecular markers, and identifying so as to obtain 16 pairs of polymorphism SSR molecular markers, wherein the polymorphism SSR molecular markers can be used for detecting SSR locus information of rhododendron molle genomes; the effective number of alleles of the rhododendron molle genomes is 3-15; the heterozygosity (He) of the rhododendron molle genomes is 0.489-0.908. The invention provides a series of SSR primers which can be applied to study of population genetics, phylogenetics, conservation biologics and the like, the polymorphism SSR molecular markers can be used for evaluating and analyzing genetic diversity and genetic structures of endangered rhododendron molle, and scientific basis and reference can be provided for protection of the specie.
Owner:JIANGXI NORMAL UNIV
Genetic diversity analysis method for inbreeding male giant freshwater prawn
InactiveCN108642187AQuality improvementMicrobiological testing/measurementDNA/RNA fragmentationFresh water organismAllele frequency
The invention discloses a genetic diversity analysis method for a inbreeding male giant freshwater prawn, which comprises the following steps of: step 1, acquiring inbreeding giant freshwater prawn samples with different traits, and extracting the DNA; step 2, selecting a microsatellite marker combination and designing a specific PCR primer set for amplification; step 3, evaluating the number of PCR products and observing the genotype and size of the alleles; step 4, recording the allele number A, the allele duplication number G, the observed heterozygosity Ho, the desired heterozygosity He, and the accuracy deviation value P based on the Hardy-Weinberg equilibrium law of each prawn, and analyzing the difference of the microsatellite sites; calculating the gene flow Nm according to the frequency parameters of the population allele; gene spacing is determined by using the gene Nei coefficient method. According to the invention, based on the microsatellite analysis method, the method provides a genetic diversity analysis method for a inbreeding male giant freshwater prawn, which is very suitable for researching the genetic structure of different near-parent male prawn groups, and provides a high-quality male parent for the giant freshwater prawns.
Owner:GUANGXI ACADEMY OF FISHERY SCI
Homologous recombination mediated transgene deletion in plant cells
InactiveUS20080178348A1Reduce generationImprove stabilityTissue cultureOther foreign material introduction processesRecombinant TransgenesPlant cell
A process to prepare a recombined transgenic Zea mays plant or plant cell from a first transgenic Zea mays plant cell, wherein the transgene in the recombinant plant or plant cell has an altered genetic structure relative to the genetic structure of the transgene in the first transgenic plant cell, due to homologous recombination-mediated transgene deletion.
Owner:MONSANTO TECH LLC
Sebastes schlegeli microsatellite DNA (Deoxyribonucleic Acid) molecular marker
InactiveCN103484454AMicrobiological testing/measurementDNA/RNA fragmentationStructure analysisMicrosatellite
The invention discloses a sebastes schlegeli microsatellite DNA (Deoxyribonucleic Acid) molecular marker. The molecular marker comprises construction of sebastes schlegeli microsatellite (CA) 16 and (GA) 16 enriched libraries and screening and sequencing of a microsatellite sequence-containing positive clone and determines 10 microsatellite markers SS101, SS102, SS103, SS104, SS105, SS106, SS107, SS108, SS109 and SS110 with rich polymorphism. The invention provides 10 sebastes schlegeli new microsatellite sites and a primer sequence and an amplification method for amplifying the 10 microsatellite sites; the molecular marker can be applied to population genetic structure analysis, paternity identification, molecular marker-assisted selection and the like of the sebastes schlegeli, and is a reliable and effective molecular marker.
Owner:AQUATIC PROD RES INST SHANDONG PROV
Microsatellite primer used for analyzing diversity of pachycrepoideus vindemmiae, purpose and analysis method
ActiveCN103981257AGood polymorphismMicrobiological testing/measurementDNA/RNA fragmentationMicrosatelliteDNA extraction
The invention relates to a microsatellite primer used for analyzing diversity of pachycrepoideus vindemmiae, a purpose and an analysis method. The analysis method comprises the following steps: 1)extracting genome DNA; 2)detecting a TP-M13-SSR molecular marker; and 3)analyzing a genetic structure. DNA is extracted from a sample, the site of a microsatellite is screened, so that the primer is designed. The primer has the advantages of stable PCR amplification product and good polymorphism, and can be used for analysis of diversity and a population genetic structure.
Owner:ANHUI NORMAL UNIV
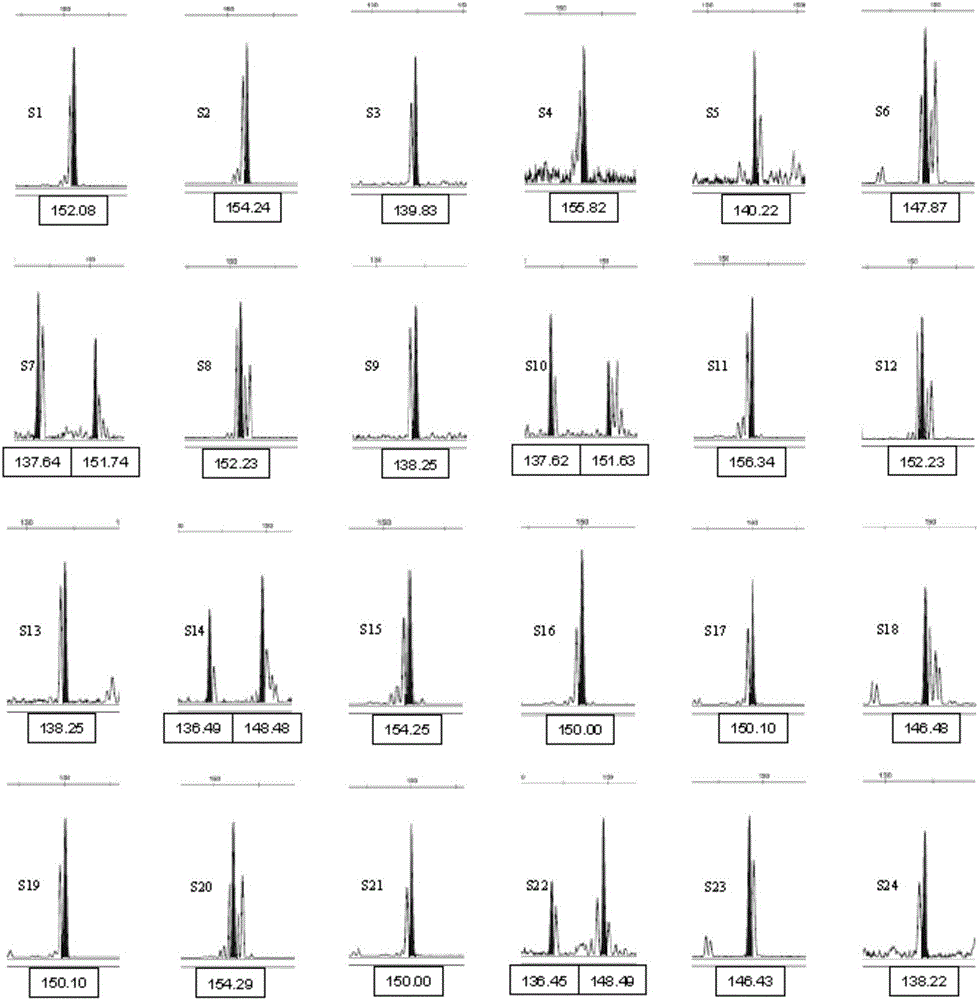
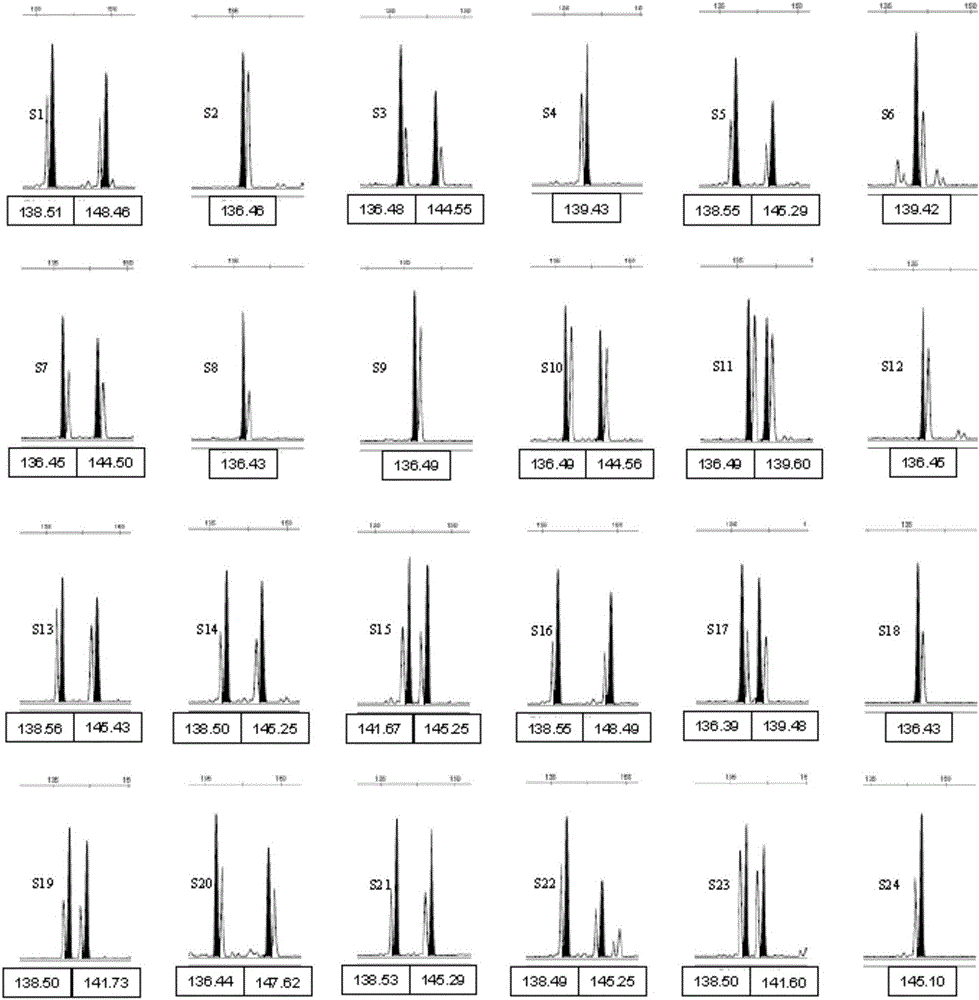
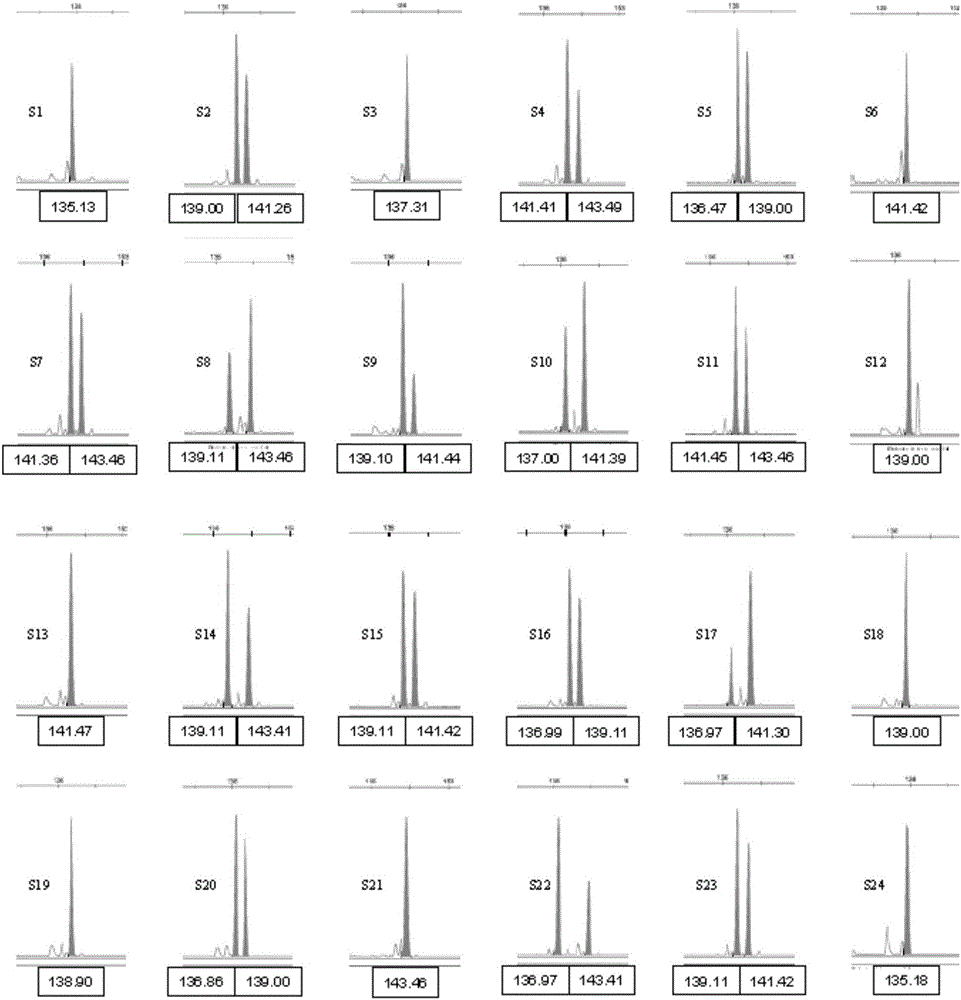
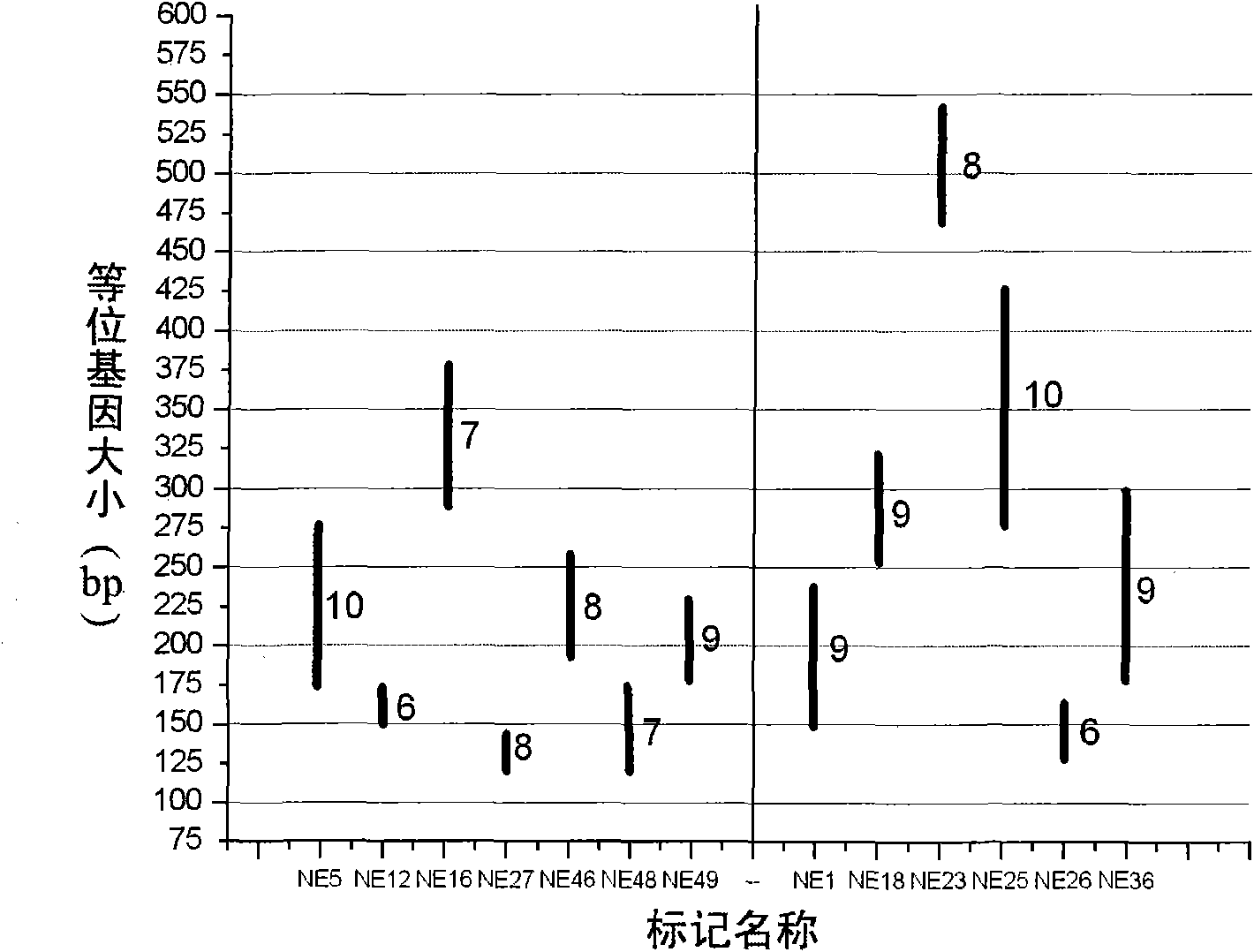
Polynary high-throughput genetic marking system and genetic analysis method for blue crabs
ActiveCN101565744AAvoid blindnessClear genetic relationshipMicrobiological testing/measurementDNA/RNA fragmentationMitophagyPortunus trituberculatus
The invention relates to the field of aquaculture genetics and breeding, and in particular to a polynary high-throughput genetic marking system and a genetic analysis method for blue crabs. The polynary high-throughput genetic marking system comprises a mitochondrial sequence information system and a simple sequence repeat molecular marking system, wherein the simple sequence repeat molecular marking system consists of seven SSR marking systems 1 and six SSR marking systems 2. The method comprises the following steps: using information on sequences of three-pair primer amplified COI, 16S and CR of a mitochondrion as a molecular marker; separating the offspring of the same female parent and the offspring of different female parents in a mixed breed pedigree according to genetic structures and genetic diversity; and performing male parent separation of the offspring subjected to female parent separation according to the genetic direction and the genetic diversity by using simple sequence repeat molecular markers to complete the pedigree verification in the genetic and breeding process of the blue crabs and the confirmation of an inbreeding coefficient. On the basis of the effective use of genetic information of species, the method uses the polynary high-throughput genetic marker system to differentiate the genetic relationship between offspring and effectively avoids inbreeding depression at the same time.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com