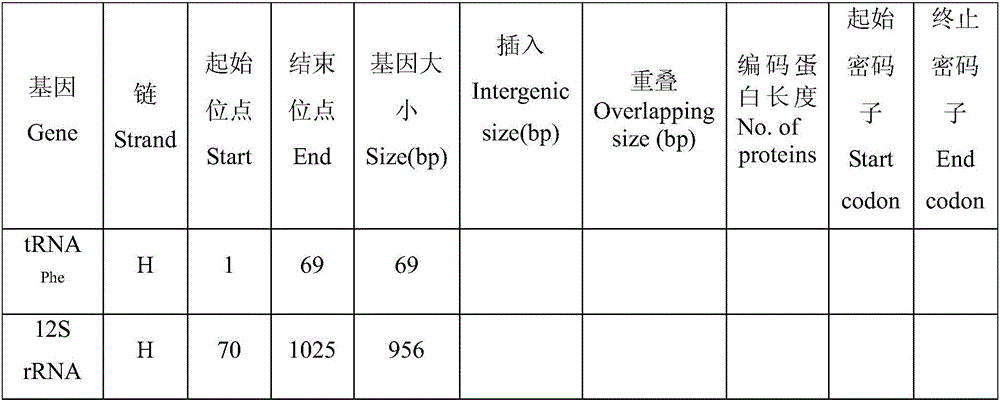
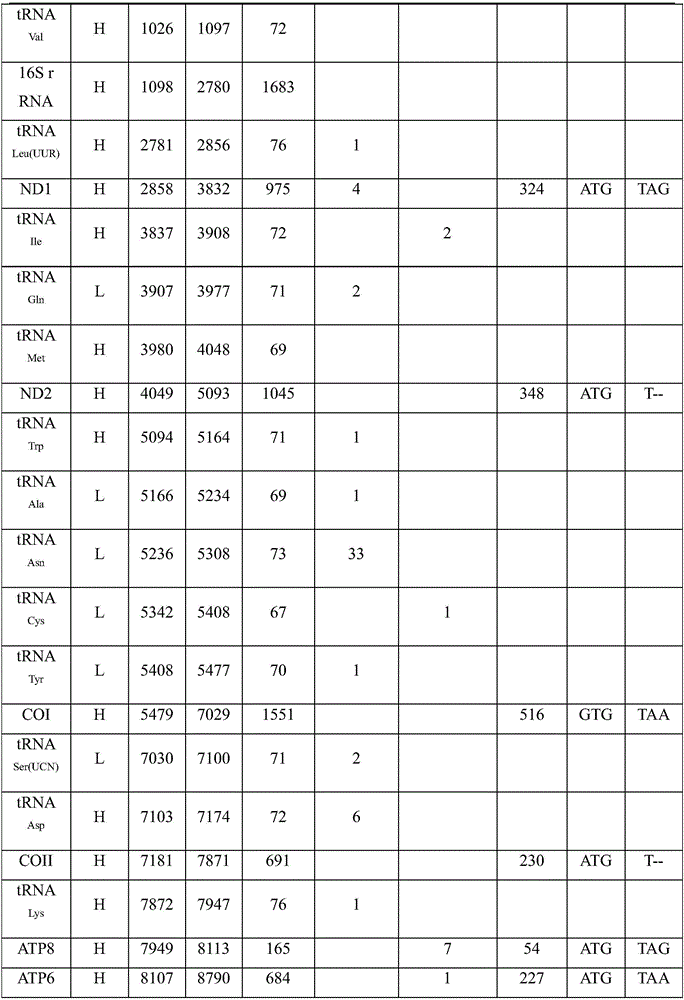
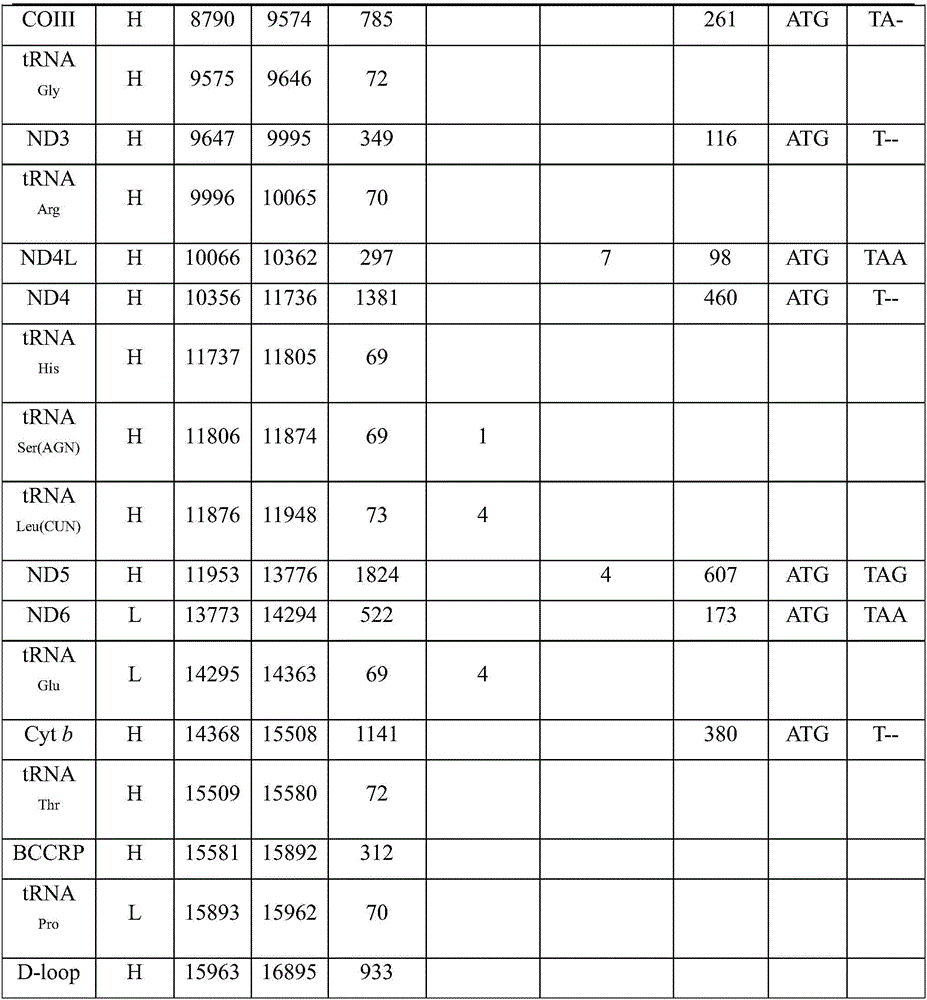
Complete sequence amplification method for mitochondrial genome of diptychus maculates steindachner
A technology of mitochondrial genome and double lip fish, applied in the field of molecular biology, can solve the problems of prone to errors, high fidelity, long amplified fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] The method for amplifying the complete mitochondrial genome sequence of P. chinensis provided in this example comprises the following steps:
[0066] (1) Amplification of four gene fragments of Cyt b, COI, ND4 and 16S rRNA in the mitochondria of P.
[0067] (2) LA-PCR amplification of the mitochondrial genome of P.
[0068] The method for amplifying the four gene fragments of Cyt b, COI, ND4 and 16S rRNA in the mitochondrial genome of the spotted fish comprises the following steps:
[0069] (1) For the screening of primers for amplifying the four gene fragments of Cyt b, COI, ND4 and 16S rRNA, the primers for amplifying the four genes of Cyt b, COI, ND4 and 16S rRNA were selected as follows:
[0070] Cyt b: The upstream primer is 5'-GACTTGAAAAACCACCGTTG-3', and the downstream primer is 5'-CTCCGATCTCCGGATTACAAGAC-3';
[0071] COI: The upstream primer is 5'-TCAACCAACCACAAAGACATTGGCAC-3' and the downstream primer is 5'-TAGACTTCTGGGTGGCCAAAGAATCA-3';
[0072] ND4: The up...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com