Patents
Literature
343 results about "Complete sequence" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In mathematics, a sequence of natural numbers is called a complete sequence if every positive integer can be expressed as a sum of values in the sequence, using each value at most once. For example, the sequence of powers of two {1, 2, 4, 8, ...}, the basis of the binary numeral system, is a complete sequence; given any natural number, we can choose the values corresponding to the 1 bits in its binary representation and sum them to obtain that number (e.g. 37 = 100101₂ = 1 + 4 + 32). This sequence is minimal, since no value can be removed from it without making some natural numbers impossible to represent. Simple examples of sequences that are not complete include the even numbers, since adding even numbers produces only even numbers—no odd number can be formed.
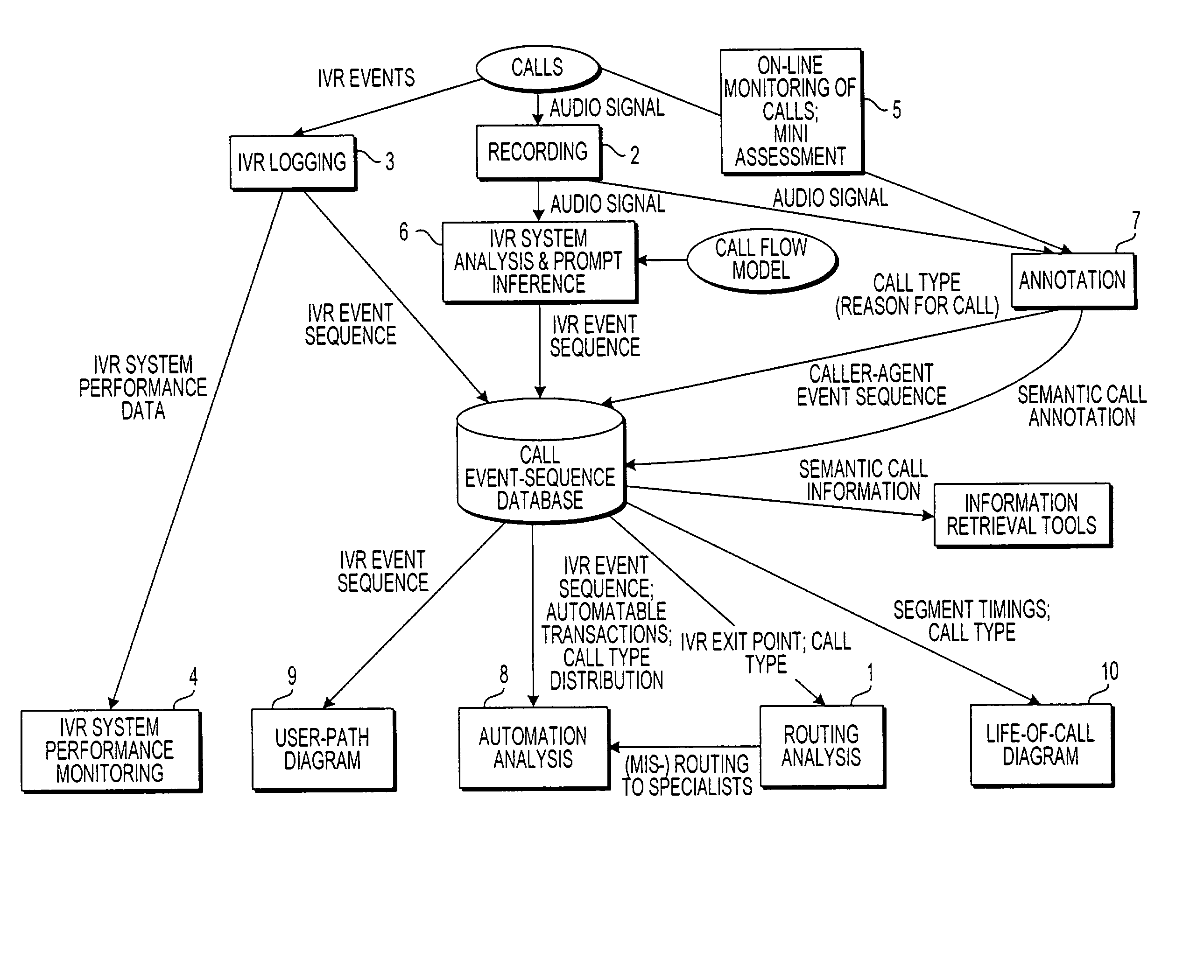
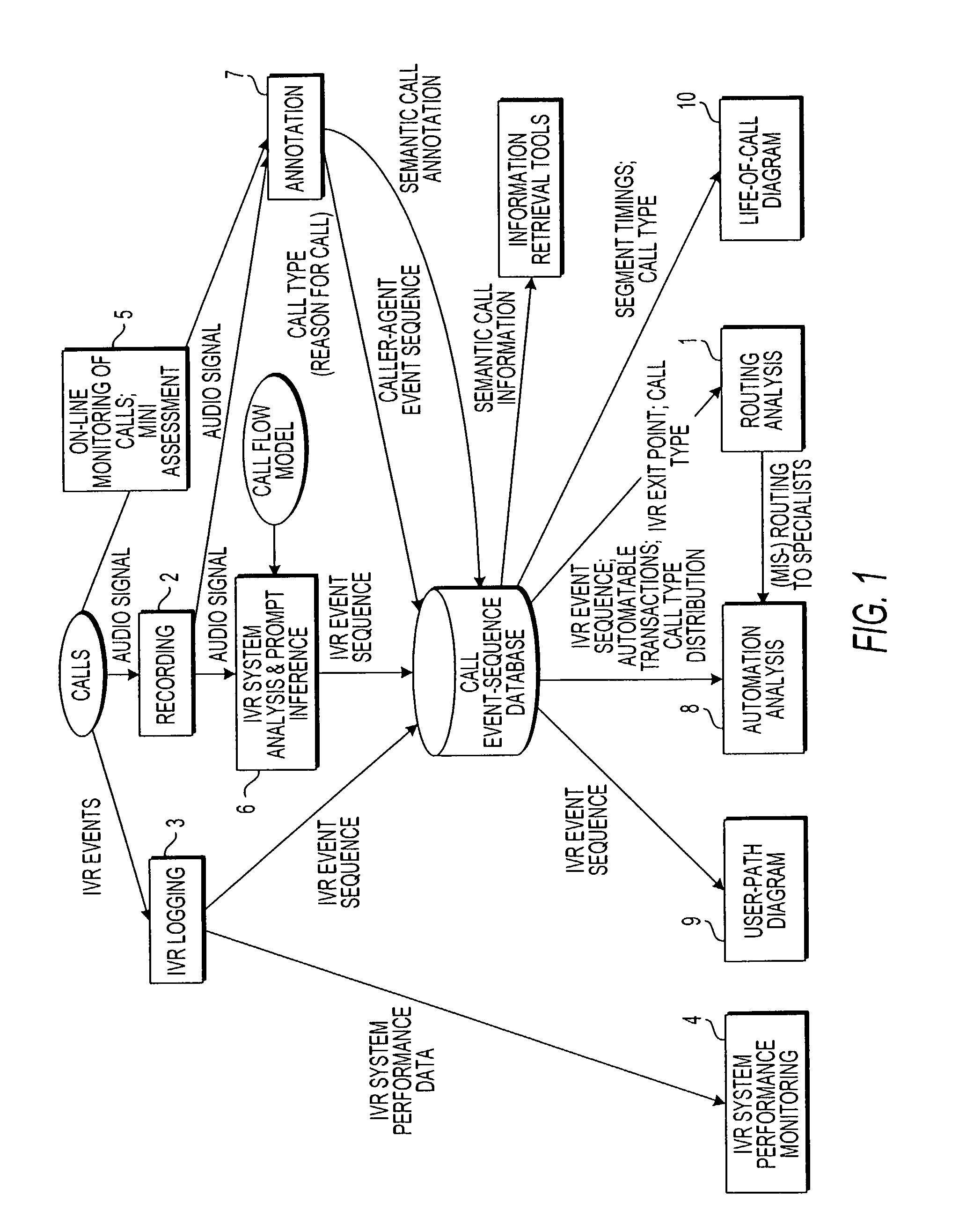
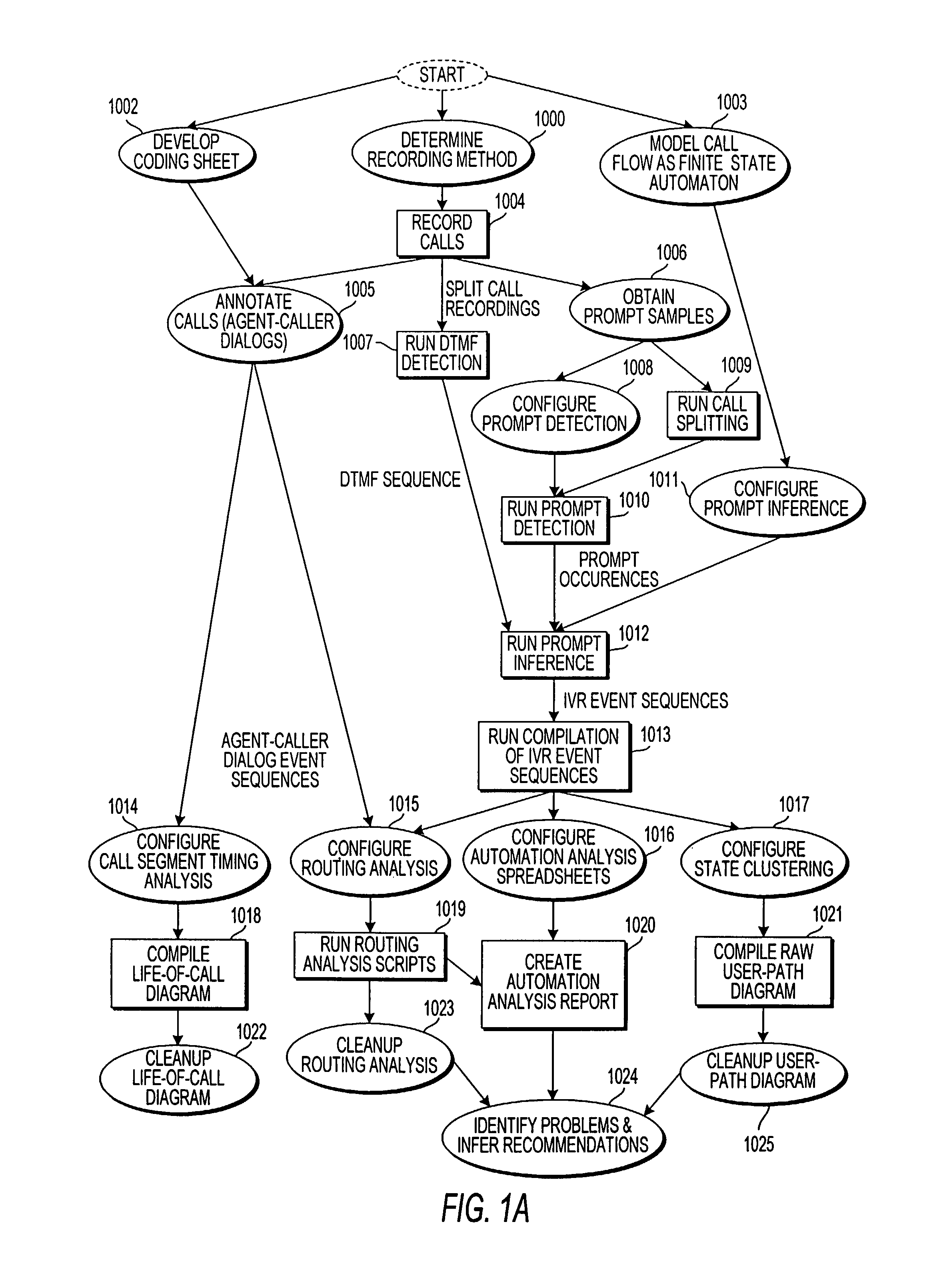
Apparatus and method for visually representing behavior of a user of an automated response system
InactiveUS7039166B1Overcome deficienciesSpeech analysisManual exchangesUser inputFinite-state machine
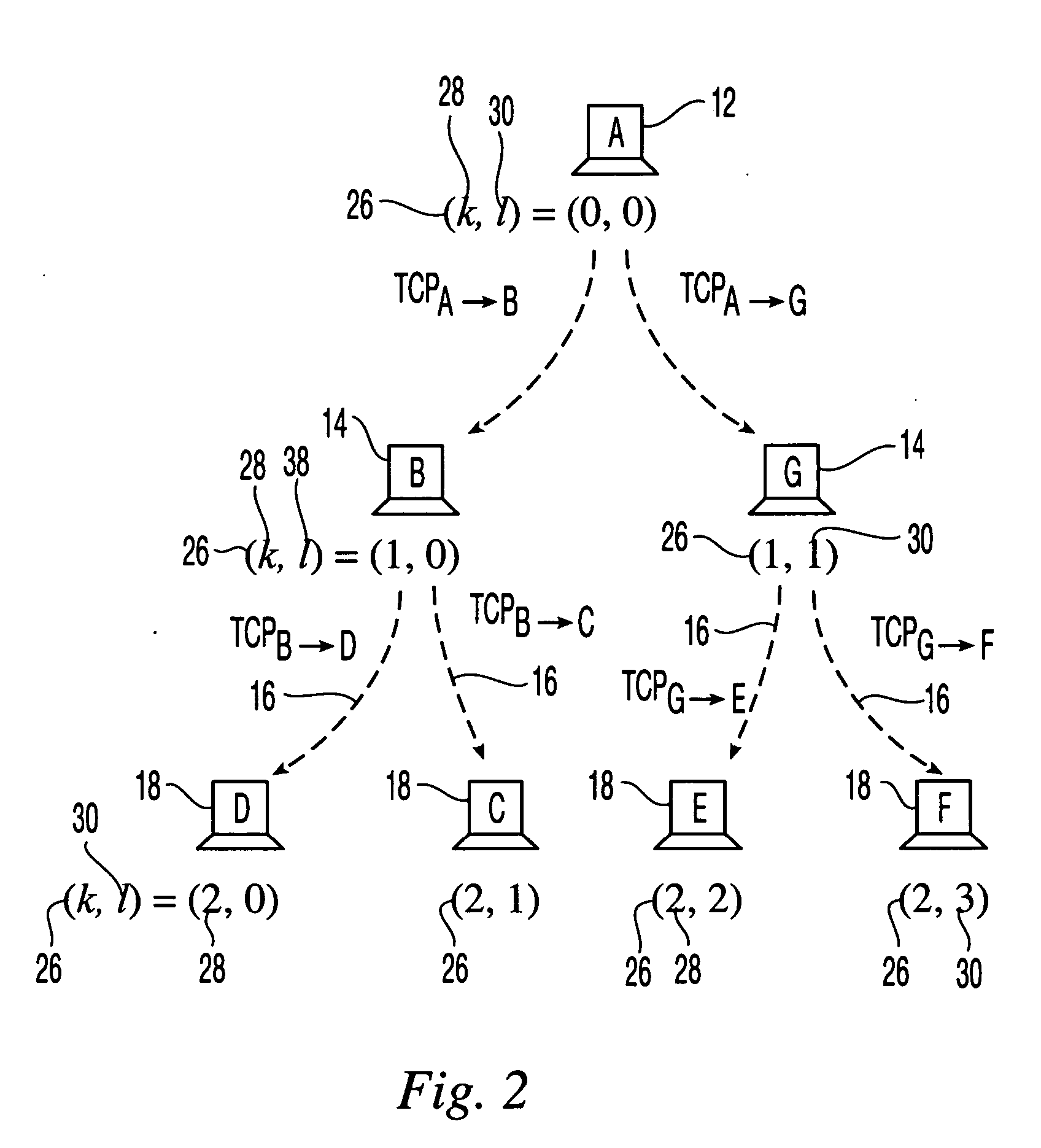
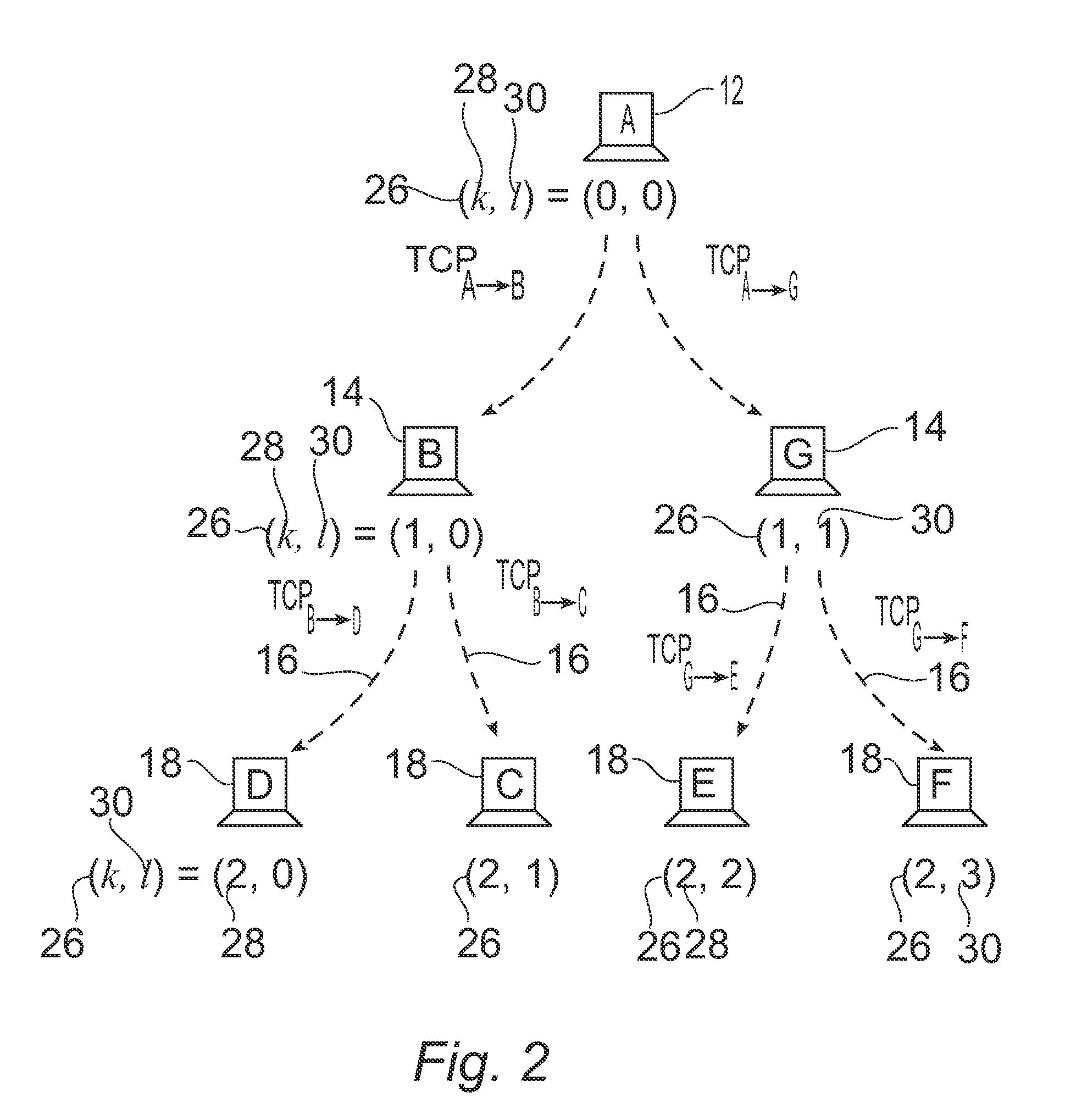
A system for visually representing user behavior within an interactive voice response (IVR) system of a call processing center generates a complete sequence of events within the IVR system for plural calls to the call processing center, the plurality of calls being recorded from end to end. A call flow of the IVR system is modeled as a non-deterministic finite-state machine, such that a start state of the finite-state machine represents a first prompt of the IVR system, other states of the finite-state machine represent subsequent prompts at which a branching occurs in the call flow of the IVR system, exit conditions are represented as end states, and transitions of the finite-state machine represent transitions between call flow states triggered by data inputted by a user or by internal processing of the IVR system. The complete sequences of events for the plural calls are provided to the finite-state machine to produce a two-way matrix of several counters. The data from the two-way matrix is represented as a state-transition diagram.
Owner:CX360 INC +1
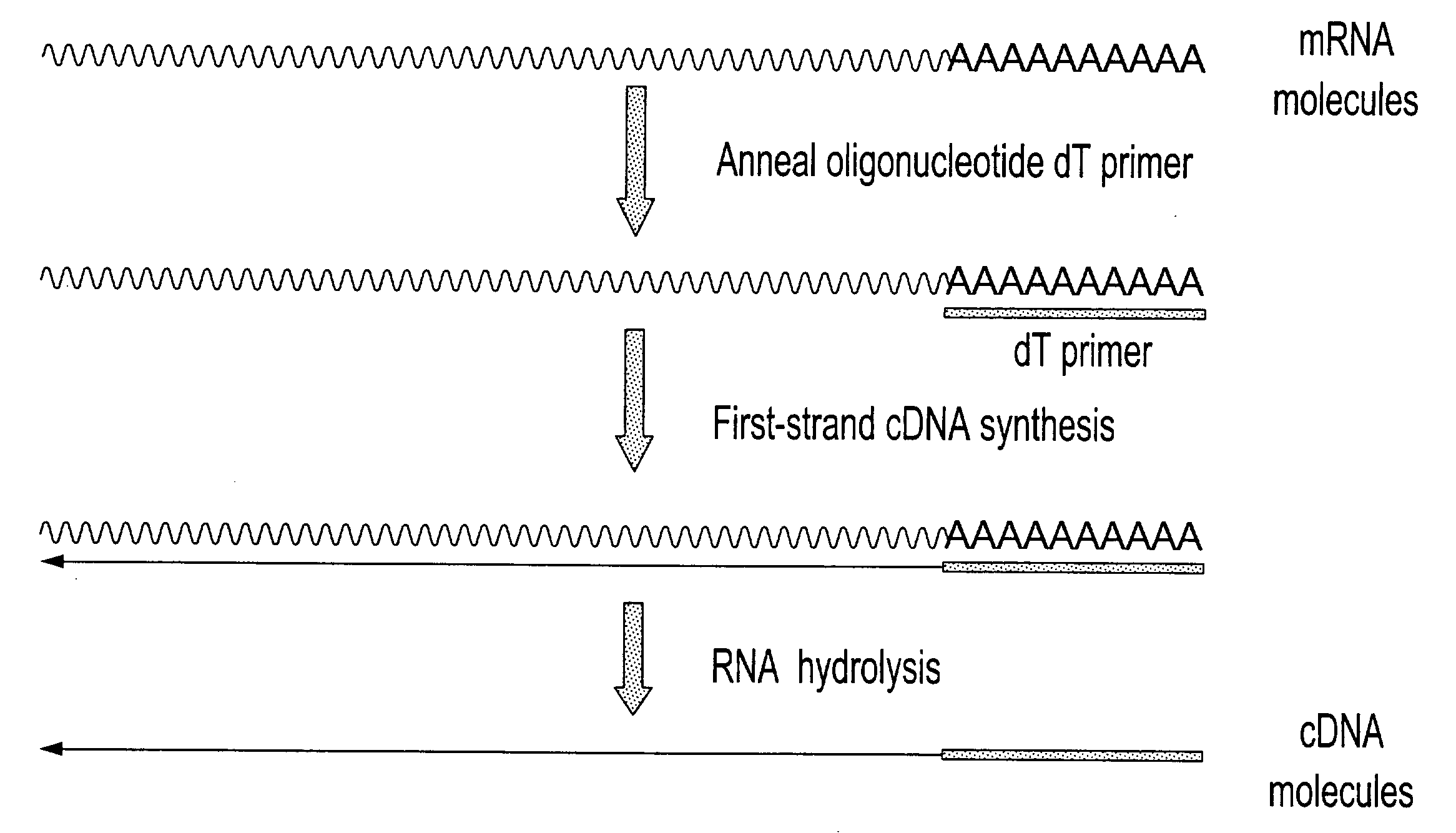
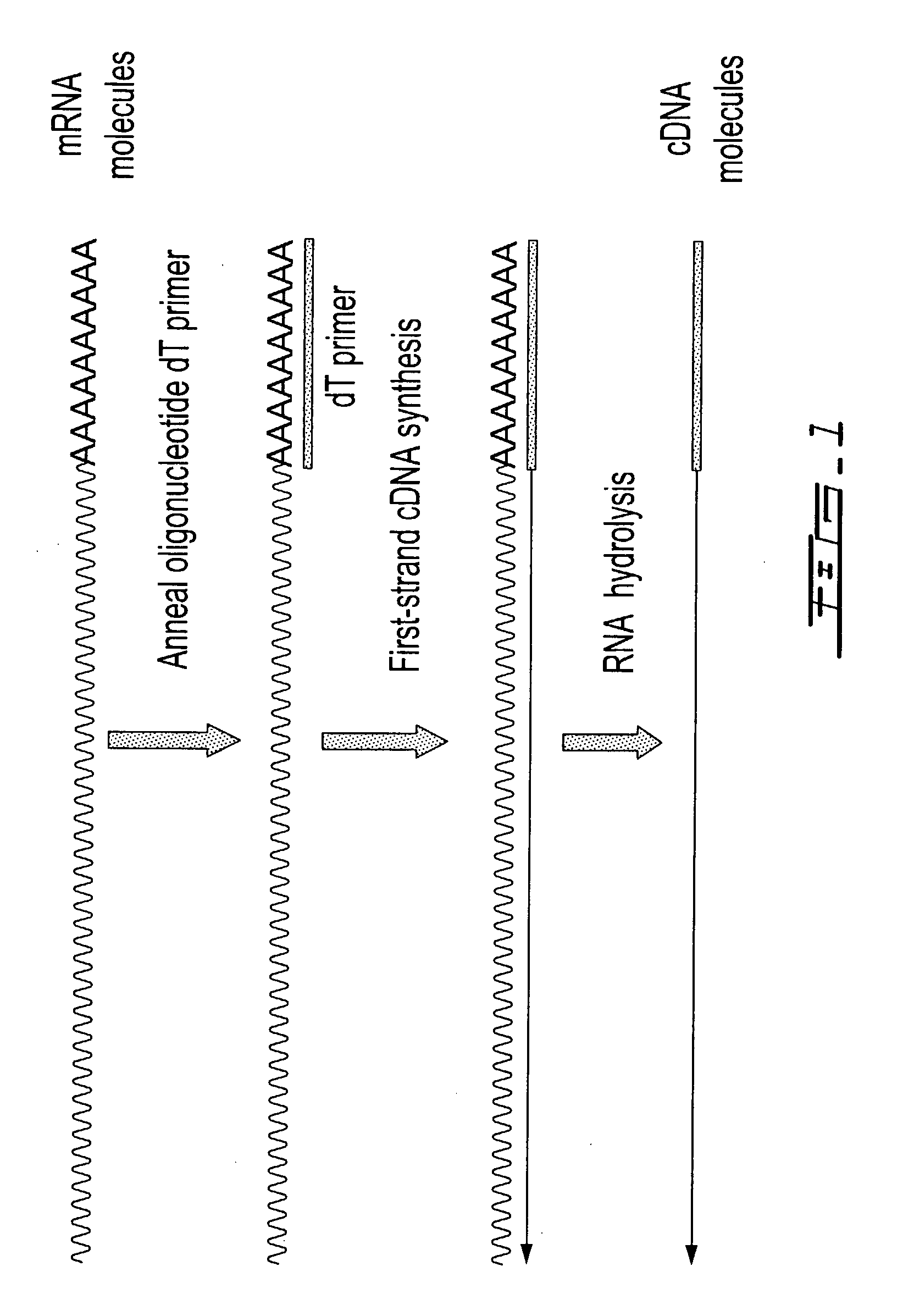
Method of sequencing genomes by hybridization of oligonucleotide probes
InactiveUS6018041ASugar derivativesMicrobiological testing/measurementMammalian genomeHomologous sequence
The conditions under which oligonucleotides hybridize only with entirely homologous sequences are recognized. The sequence of a given DNA fragment is read by the hybridization and assembly of positively hybridizing probes through overlapping portions. By simultaneous hybridization of DNA molecules applied as dots and bound onto a filter, representing single-stranded phage vector with the cloned insert, with about 50,000 to 100,000 groups of probes, the main type of which is (A,T,C,G)(A,T,C,G)N8(A,T,C,G), information for computer determination of a sequence of DNA having the complexity of a mammalian genome are obtained in one step. To obtain a maximally completed sequence, three libraries are cloned into the phage vector, M13, bacteriophage are used: with the 0.5 kb and 7 kbp insert consisting of two sequences, with the average distance in genomic DNA of 100 kbp. For a million bp of genomic DNA, 25,000 subclones of the 0.5 kbp are required as well as 700 subclones 7 kb long and 170 jumping subclones. Subclones of 0.5 kb are applied on a filter in groups of 20 each, so that the total number of samples is 2,120 per million bp. The process can be easily and entirely robotized for factory reading of complex genomic fragments or DNA molecules.
Owner:HYSEQ
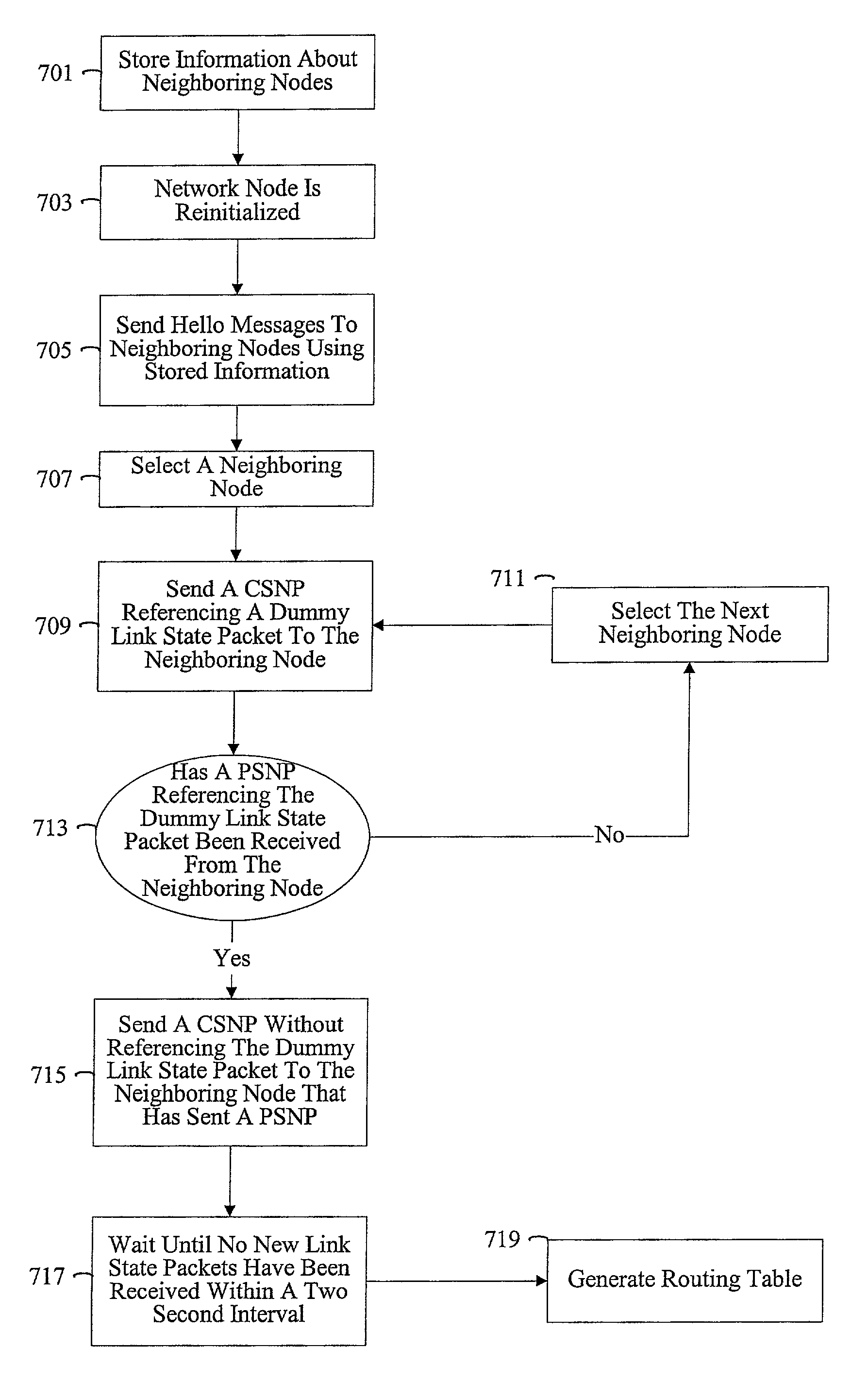
Methods and apparatus for requesting link state information
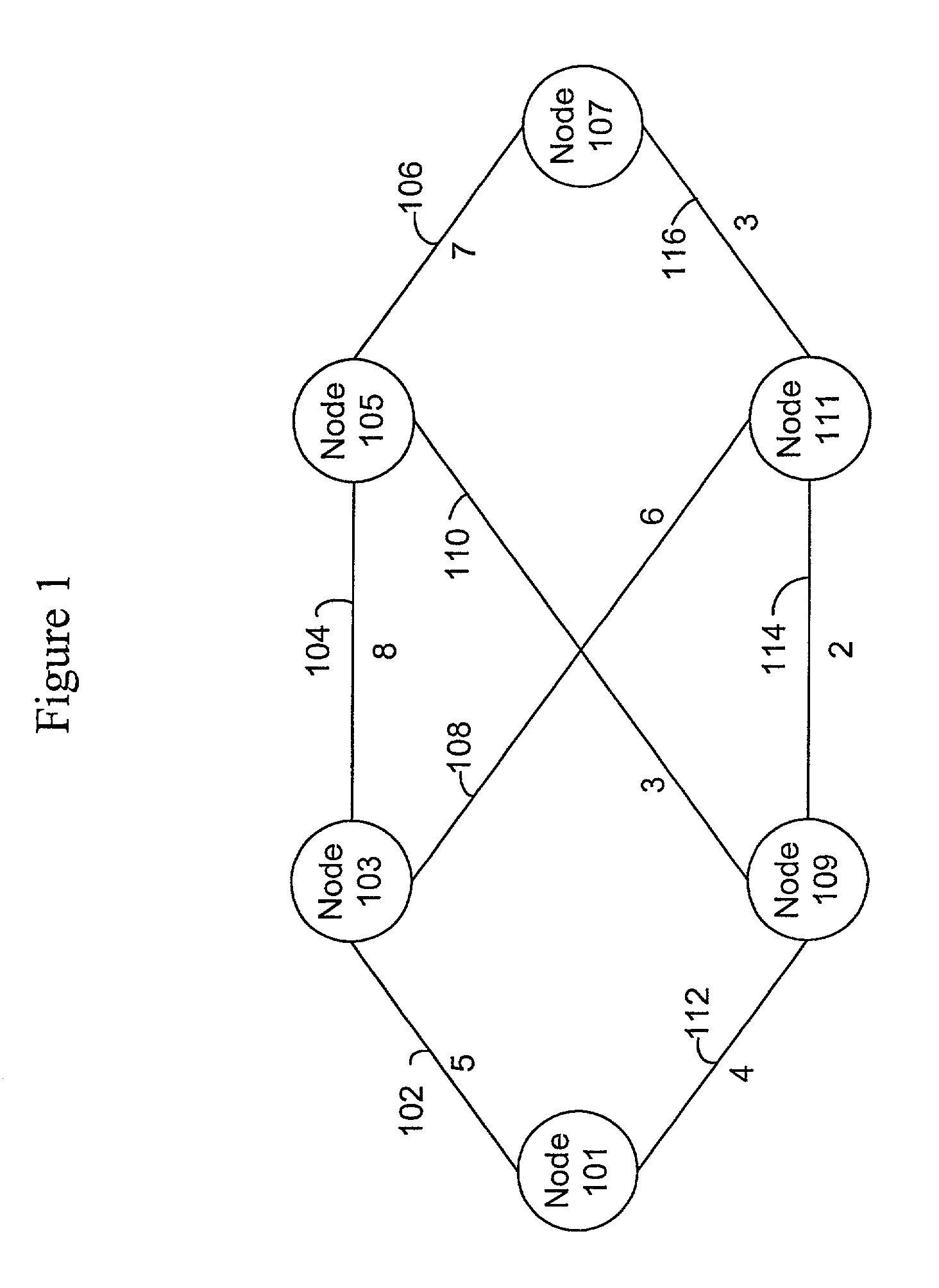
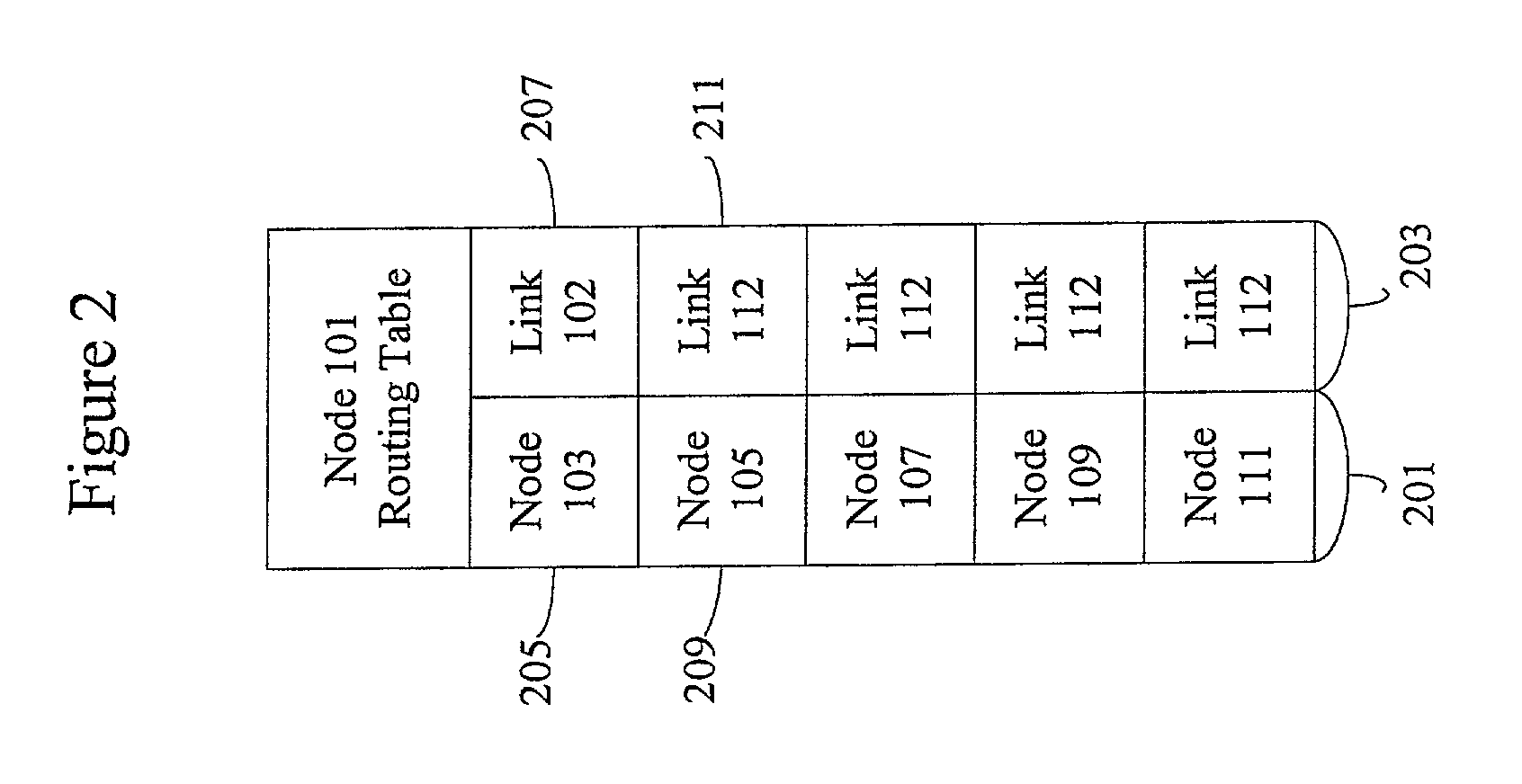
InactiveUS7174387B1Error detection/correctionMultiple digital computer combinationsComputer networkComplete sequence
Methods and apparatus are provided for optimizing the reintroduction of a network node into a network. Information about neighboring nodes is stored in persistent memory. The network node can then be reinitialized and reintroduced into the network. Upon reintroduction, the network node can transmit heartbeat messages such as Hello messages to its neighboring nodes using information stored in persistent memory. A link state packet request message such as a Complete Sequence Numbers Packet referencing dummy link state information is transmitted to a neighboring node. A partial packet request message such as a Partial Sequence Numbers Packet referencing the dummy link state packet from the neighboring node can acknowledge that the Complete Sequence Numbers Packet has been received.
Owner:CISCO TECH INC
Method and apparatus for group communication with end-to-end reliability
InactiveUS20050243722A1Avoid packet lossUndesirable characteristicSpecial service provision for substationError preventionComplete dataNetwork packet
The present invention addresses scalability and end-to-end reliability in overlay multicast networks. A simple end-system multicast architecture that is both scalable in throughput and reliable in an end-to-end way is used. In this architecture, the transfers between nodes use TCP with backpressure mechanisms to provide data packet transfers between intermediate nodes having finite-size forwarding buffers. There is also a finite-size backup buffer in each node to store copies of packets which are copied out from the receiver window to the forwarding buffers. These backup buffers are used when TCP connections are re-established to supply copies of data packets for the children nodes after their parent node fails, maintaining a complete sequence of data packets to all nodes within the multicast overlay network. The architecture provides end-to-end reliability, tolerates multiple simultaneous node failures and provides positive throughput for any group size and any buffer size.
Owner:IBM CORP
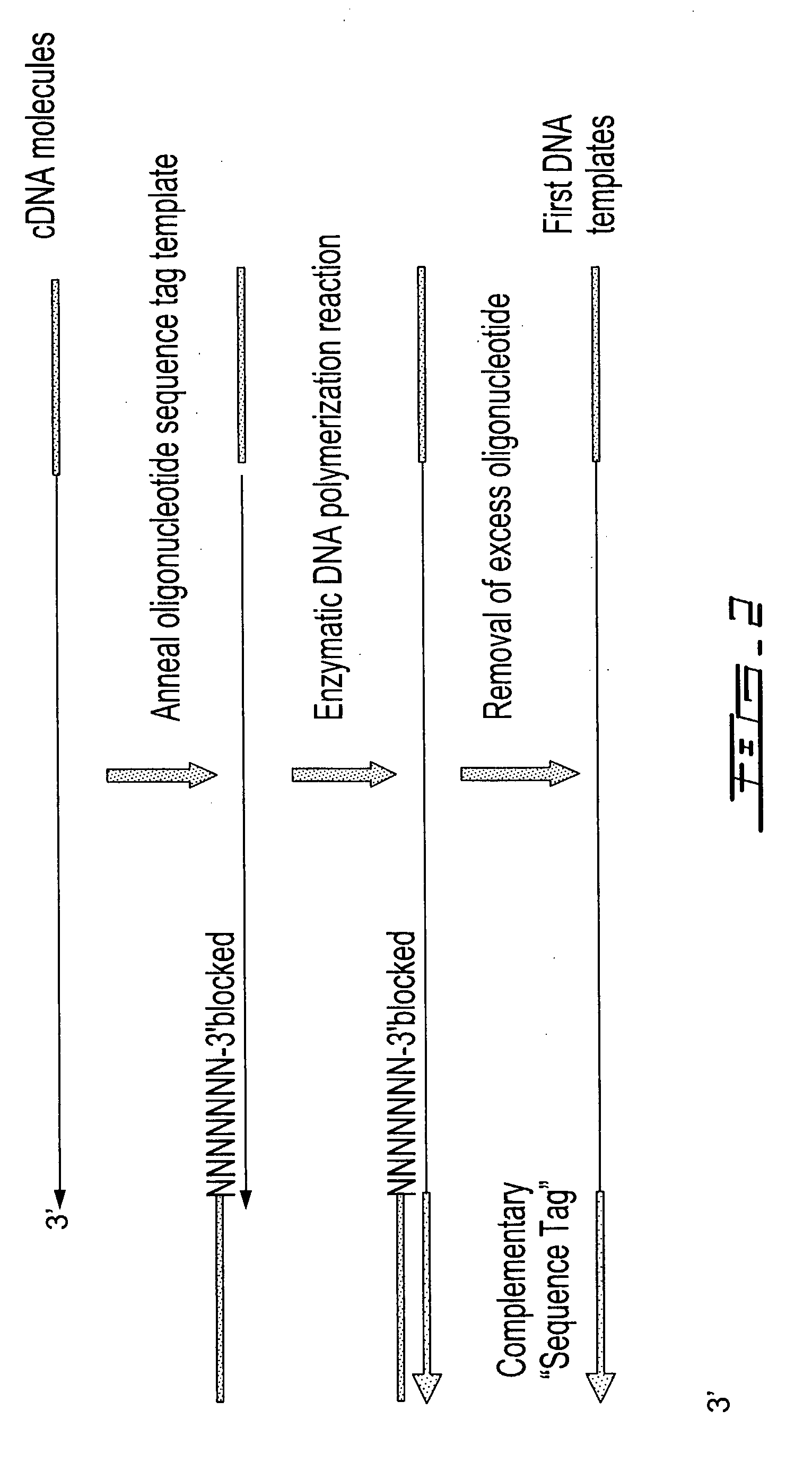
Selective terminal tagging of nucleic acids
ActiveUS8304183B2Sugar derivativesMicrobiological testing/measurementAmplification dnaComplete sequence
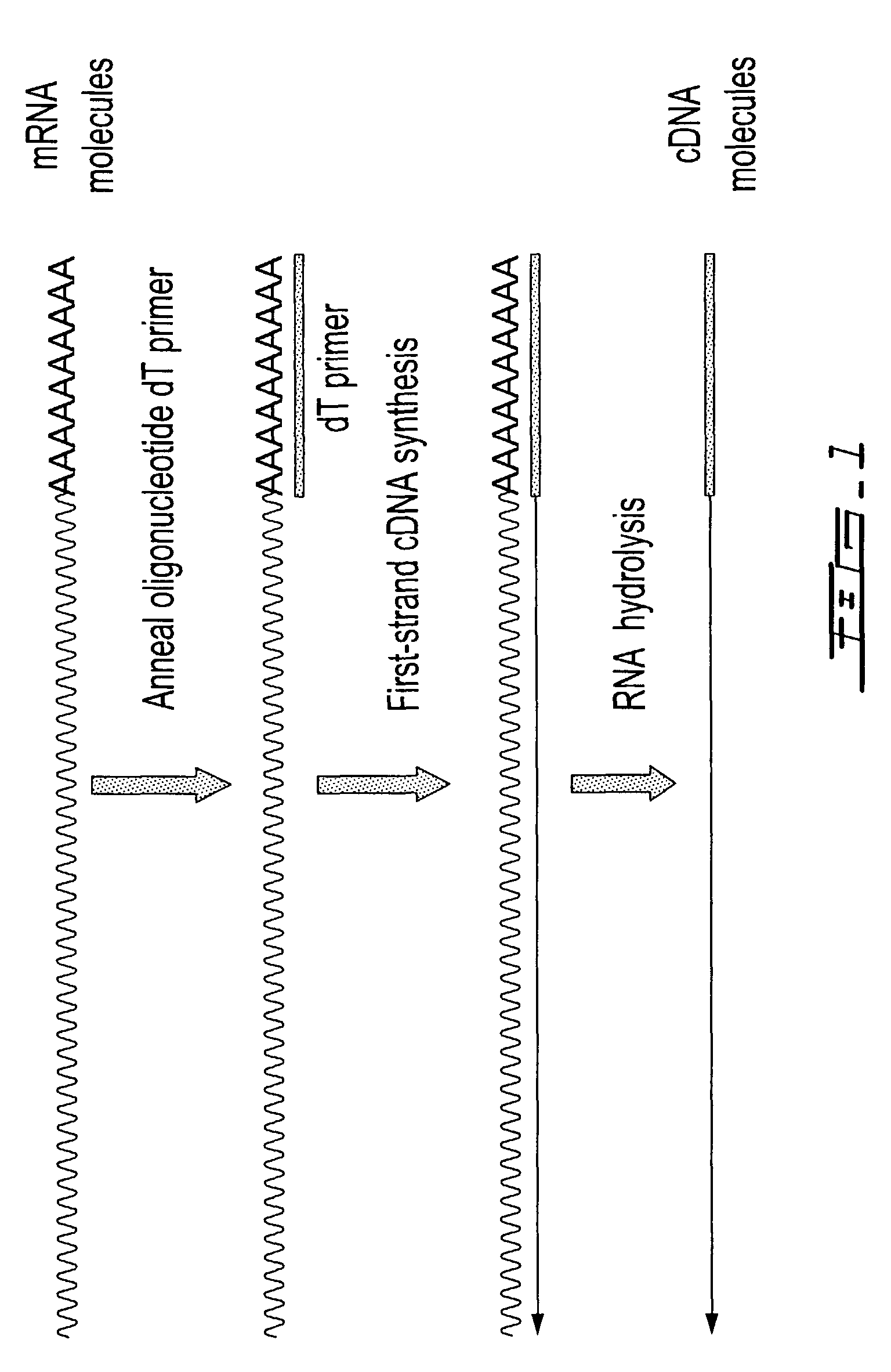
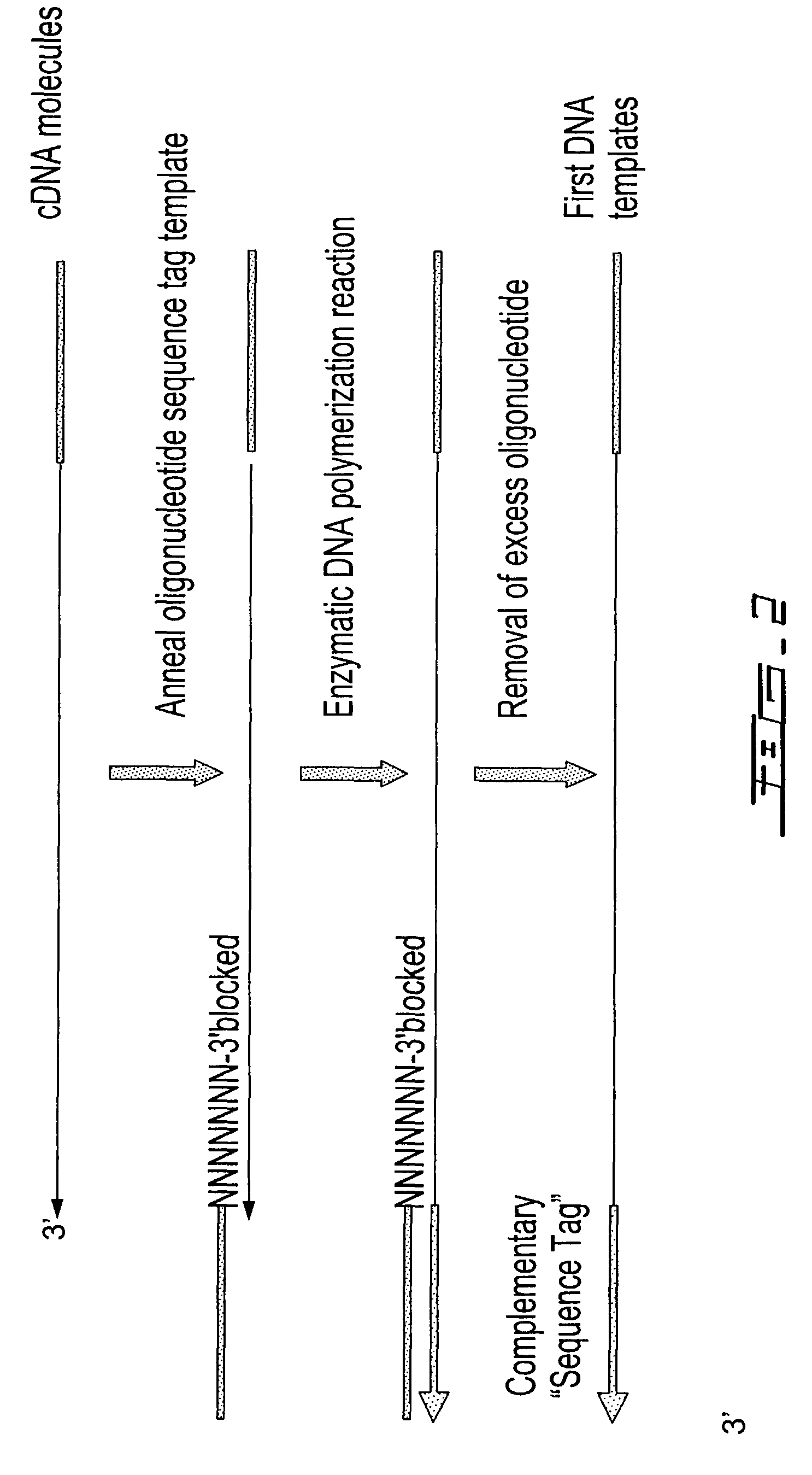
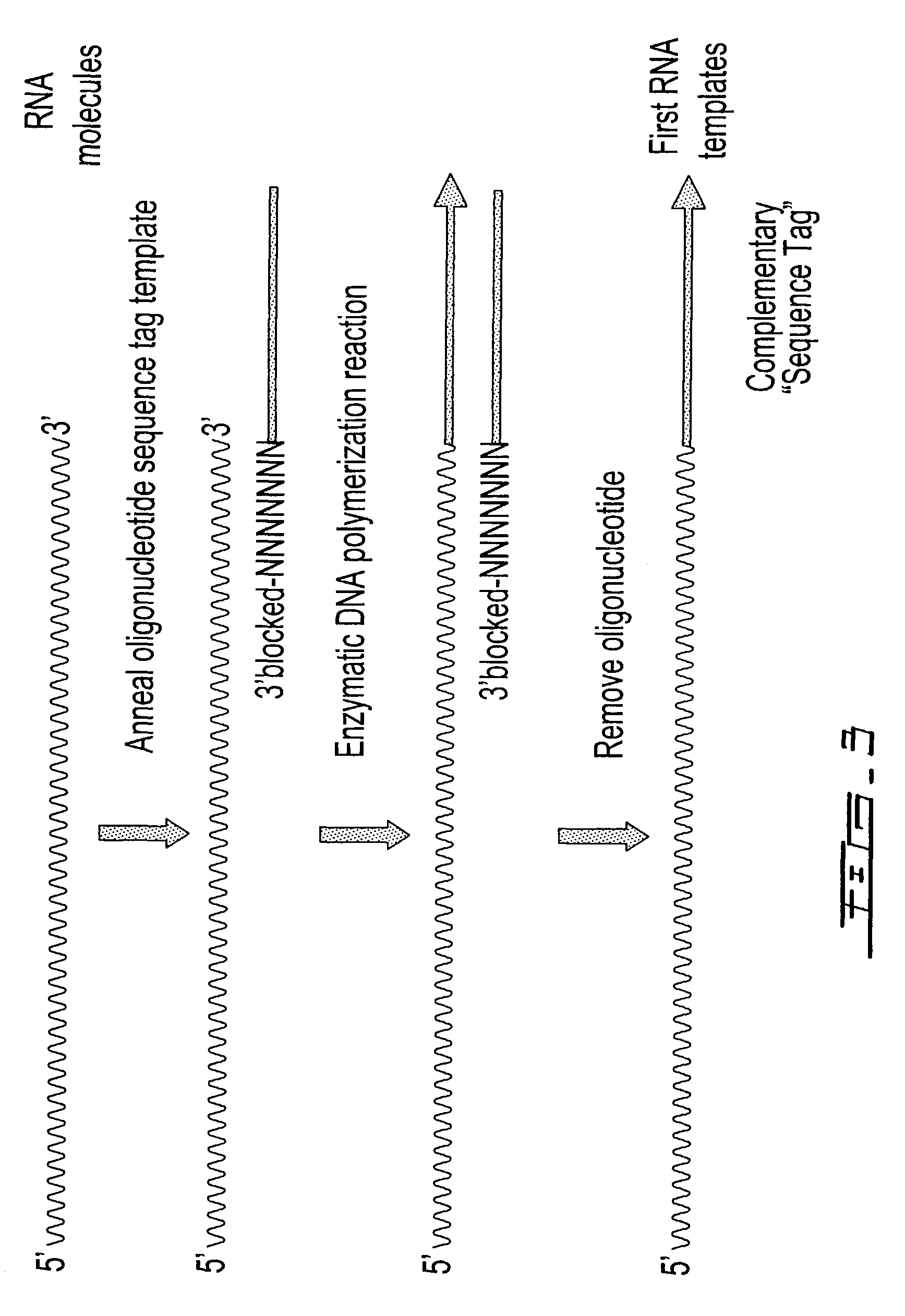
Methods are provided for adding a terminal sequence tag to nucleic acid molecules for use in RNA or DNA amplification. The tag introduced may be used as a primer binding site for subsequent amplification of the DNA molecule and / or sequencing of the DNA molecule and therefore provides means for identification and cloning of the 5′-end or the complete sequence of mRNAs.
Owner:CELLSCRIPT
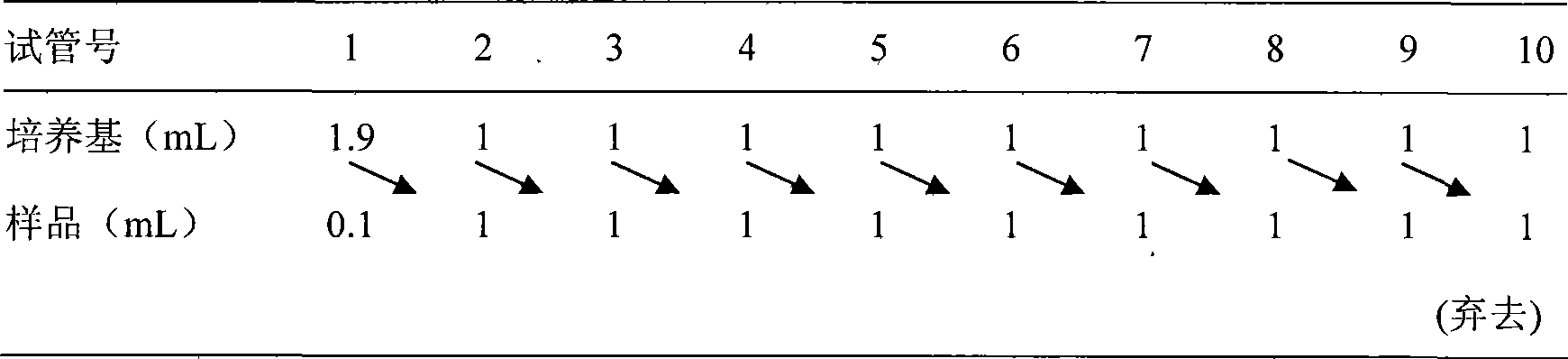
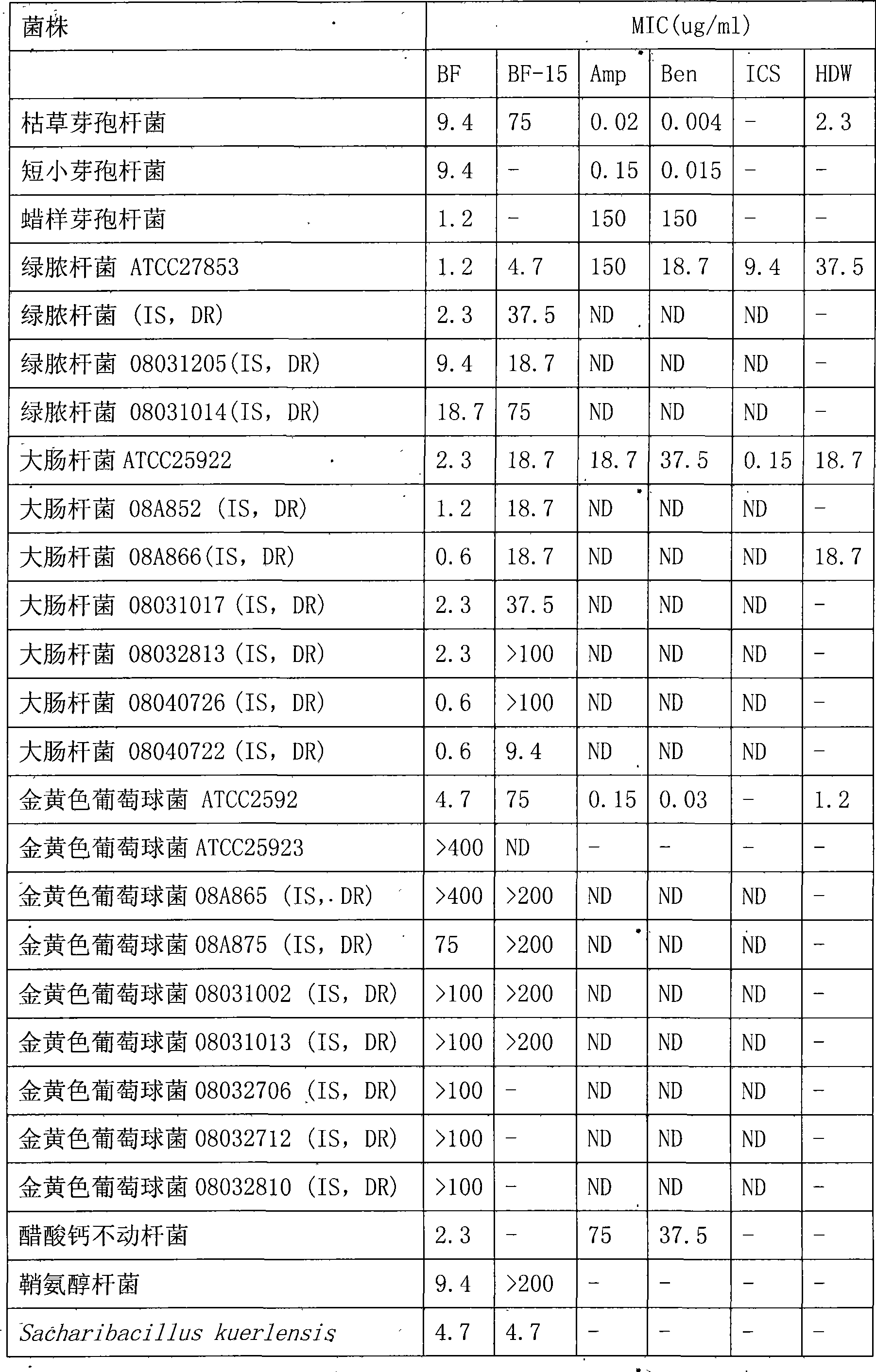
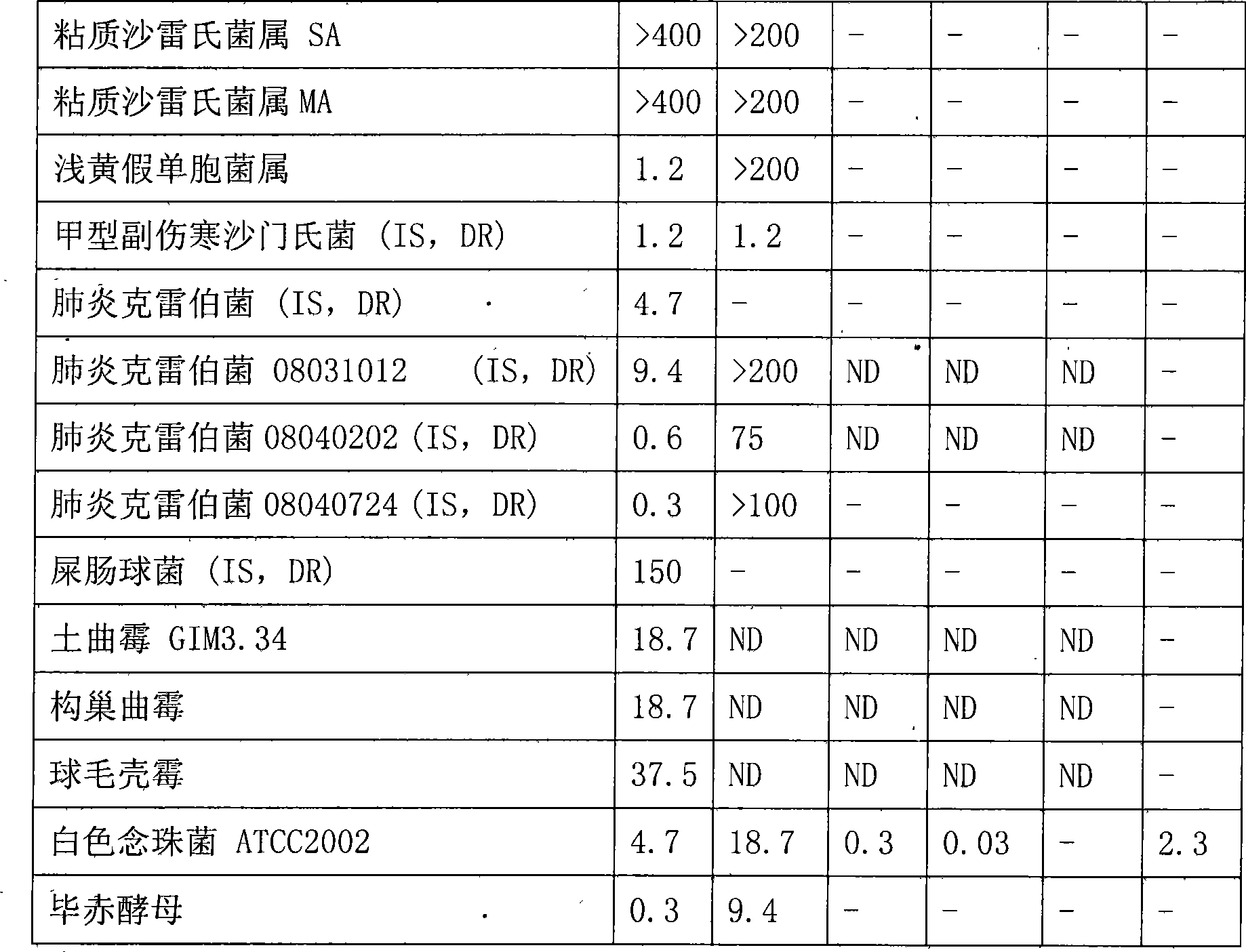
Bungarus fasciatus antibacterial peptide cathelicidin-BF, and genes and uses thereof
InactiveCN101412753ASmall molecular weightImprove the bactericidal effectFermentationAnimals/human peptidesArginineAntibiotic Y
The invention discloses cathelicidin-BF and a gene and application thereof, which belong to the field of biomedicine. The cathelicidin-BF is straight chain polypeptide and contains thirty amino acid residues, the molecular weight is 3,637.54Da, and the isoelectric point is 11.79. The complete sequence of the cathelicidin-BF is lysine-phenyl alanine-phenyl alanine-arginine-lysine-leucine- lysine-lysine-serine-valine-lysine-lysine-arginine-lactamine-lysine-glutamic acid-phenyl alanine- phenyl alanine-lysine-lysine-proline-arginine-valine-isoleucine-glycin-valine-serine-isoleucine- praline-phenyl alanine. The gene for encoding the cathelicidin-BF consists of 750 ribonucleotides, wherein 484th to 573rd ribonucleotides are used for encoding a mature peptide part. The cathelicidin-BF has small molecular weight, strong sterilization effect, and quick action time, and has quite strong killing function to a plurality of kinds of clinical drug-fast bacteria. In addition, the cathelicidin-BF also has the advantages of broad-spectrum antibiotics, salt independence and so on.
Owner:KUNMING INST OF ZOOLOGY CHINESE ACAD OF SCI
DNA sequence data compression system
InactiveCN102081707AGood repeatabilityEliminate redundancySequence analysisSpecial data processing applicationsComplete sequenceLossless compression
The invention discloses a DNA sequence data compression system which is a DNA sequence data lossless compression system based on an MA-ARV codebook. In the invention, a similar repeating segment of an MA-ARV code vector can be searched on a complete sequence, and a cultural meme heuristic optimization algorithm method (MA) is used for optimizing a construction process of the compression codebook so as to more comprehensively utilize the repeating characteristic of DNA sequence data and effectively eliminate redundancy.
Owner:SHENZHEN UNIV
Method for information storage with DNA (Deoxyribonucleic Acid)
InactiveCN106845158ASimple structureImprove continuityBioinformaticsSpecial data processing applicationsBinary informationDNA fragmentation
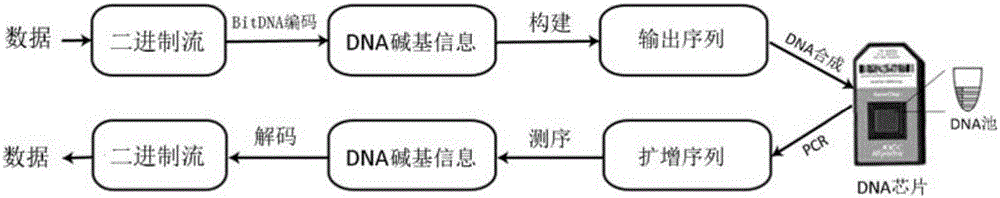
The invention relates to a method for information storage with DNA (Deoxyribonucleic Acid). The method comprises the following steps: (1) converting binary information of an original document of a computer into quaternary information, encoding, converting into a DNA complete sequence, wherein binary codes 00, 01, 10 and 11 are respectively and correspondingly converted into four deoxyribonucleotides of A, T, C and G; (2) separating the DNA complete sequence into a plurality of DNA fragments, and organizing and establishing an output DNA sequence which is 90-110nt in length and comprises an interpolation nucleotide encoding sequence consisting of DNA fragments, side primer sequences at two ends, and index encoding sequences on the inner sides of the primer sequences; (3) according to the output DNA sequence, synthesizing an artificial DNA sequence, and storing the artificial DNA sequence. The method provided by the invention has the remarkable advantages of being good in universality, being capable of simplifying calculation, improving continuity, storage efficiency and density of DNA information storage, reducing fault rates, lowering sequence synthesis and detection cost, and the like.
Owner:SUZHOU HONGXUN BIOTECH CO LTD
Method of sequencing of genomes by hybridization of oligonucleotide probes
InactiveUS6451996B1Sugar derivativesMicrobiological testing/measurementMammalian genomeHomologous sequence
The conditions under which oligonucleotides hybridize only with entirely homologous sequences are recognized. The sequence of a given DNA fragment is read by the hybridization and assembly of positively hybridizing probes through overlapping portions. By simultaneous hybridization of DNA molecules applied as dots and bound onto a filter, representing single-stranded phage vector with the cloned insert, with about 50,000 to 100,000 groups of probes, the main type of which is (A,T,C,G)(A,T,C,G)N8(A,T,C,G), information for computer determination of a sequence of DNA having the complexity of a mammalian genome are obtained in one step. To obtain a maximally completed sequence, three libraries are cloned into the phage vector, M13, bacteriophage are used: with the 0.5 kb and 7 kbp insert consisting of two sequences, with the average distance in genomic DNA of 100 kbp. For a million bp of genomic DNA, 25,000 subclones of the 0.5 kbp are required as well as 700 subclones 7 kb long and 170 jumping subclones. Subclones of 0.5 kb are applied on a filter in groups of 20 each, so that the total number of samples is 2,120 per million bp. The process can be easily and entirely robotized for factory reading of complex genomic fragments or DNA molecules.
Owner:COMPLETE GENOMICS INC
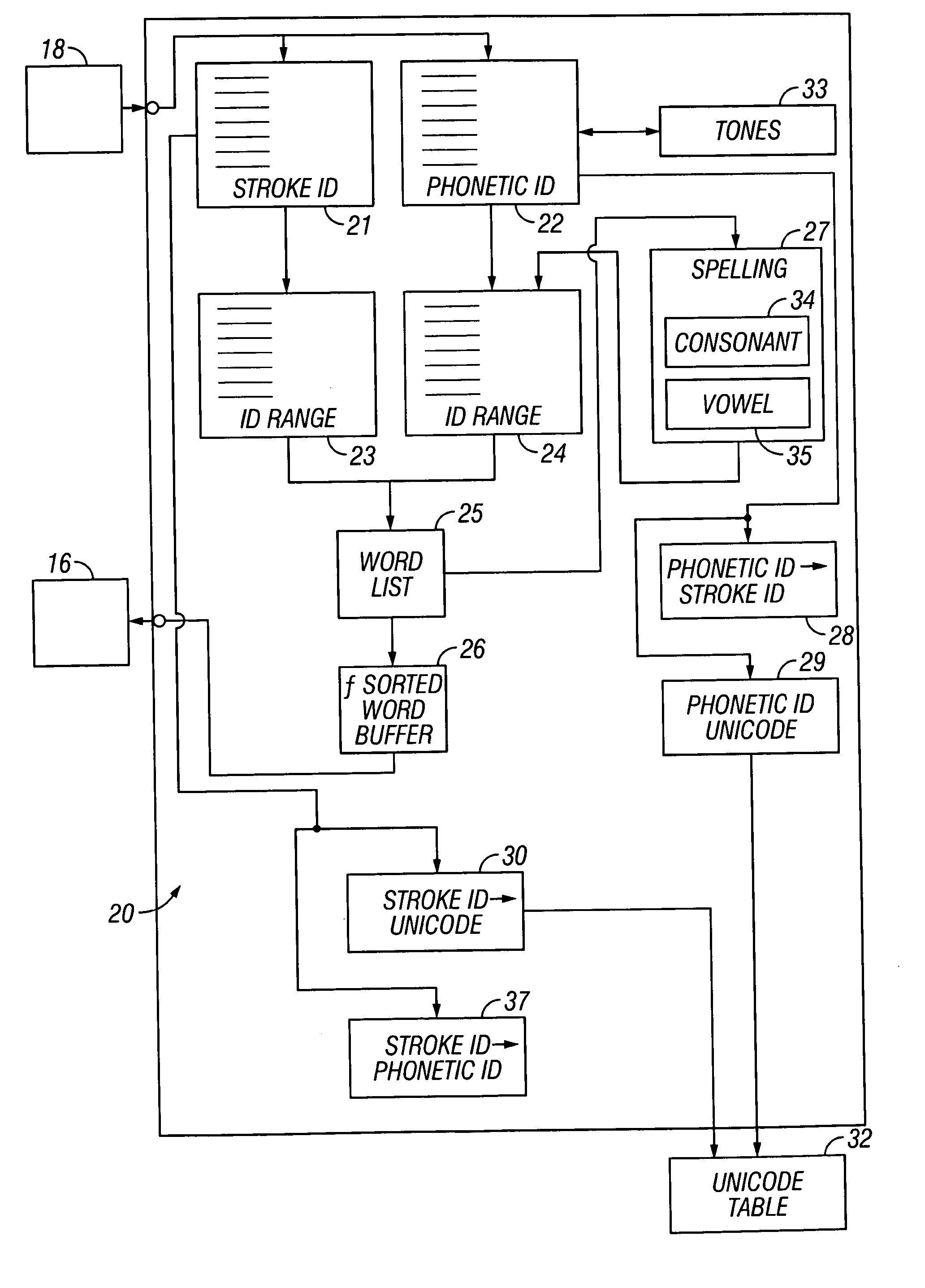
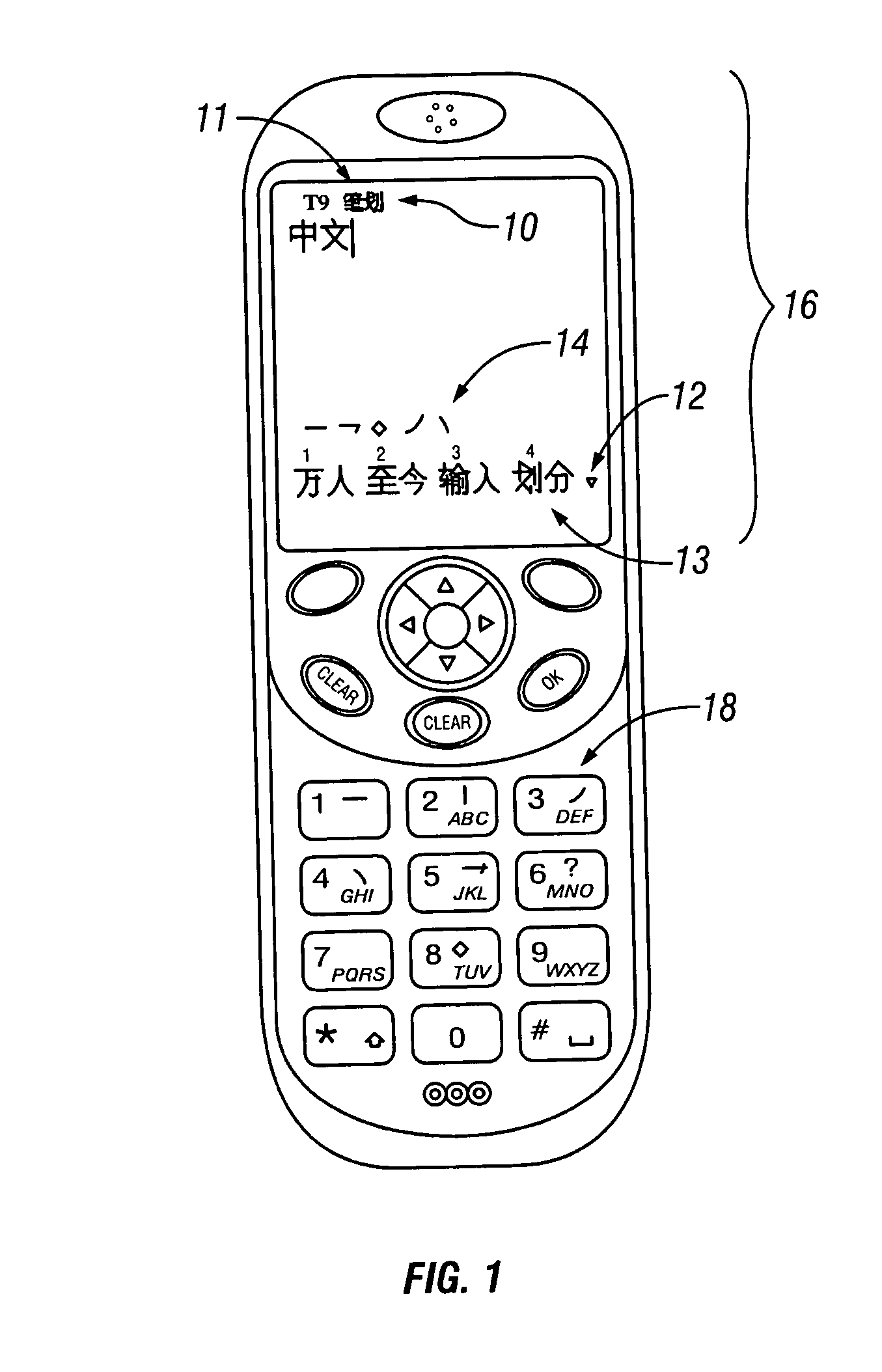
User interface and database structure for Chinese phrasal stroke and phonetic text input
InactiveUS20060018545A1Easy to learnText input faster and accurateCharacter and pattern recognitionNatural language data processingChinese character encodingHabit
The invention provides a stroke and phonetic text input entry system that has substantially the same definition of stroke match as that used in T9, where the input is a phrasal input rather than a character input. The invention solves the problem of Chinese phrasal stroke and phonetic text input by allowing users to enter an arbitrary number of strokes for each character in a phrase, where each character is separated by a delimiter. In this way, the invention provides a system that is easily learned and efficiently applied. Thus, the invention makes it possible for users to enter multiple characters while keeping their single character input habits. Each Chinese character has a standard stroke sequence in Guo Biao (GB), which is the standard for mainland China, or multiple sequences for BIG5 Chinese Character Encoding for Traditional (Complex) Characters, which is the de facto standard in Taiwan but not used in mainland China. With the invention, users do not have to enter the complete sequence for a single character, but instead can stop at any point and enter a delimiter which indicates the end of the previous character and the start of the next character. The whole stroke sequence entered by the user can then be split into a few groups that are separated by zero or more delimiters. Phrases can then be identified by user entry of groups of characters. The presently preferred phrase matching criteria are as follows: the first stroke group matches the leading stroke sequence of the first character of the phrase; the second stroke group matches the leading stroke sequence of the second character of the phrase, etc; the phrases that match the entered stroke sequence are presented to the user for selection. A user interface design for Chinese phrasal stroke text input is also provided.
Owner:TEGIC COMM
Method and apparatus for group communication with end-to-end reliability
InactiveUS7355975B2Avoid packet lossUndesirable characteristicSpecial service provision for substationError preventionComplete dataNetwork packet
Owner:INT BUSINESS MASCH CORP
Fast image splicing method of matching strategy fusion and low errors
InactiveCN107918927AQuality improvementHigh precisionImage enhancementImage analysisPattern recognitionReference image
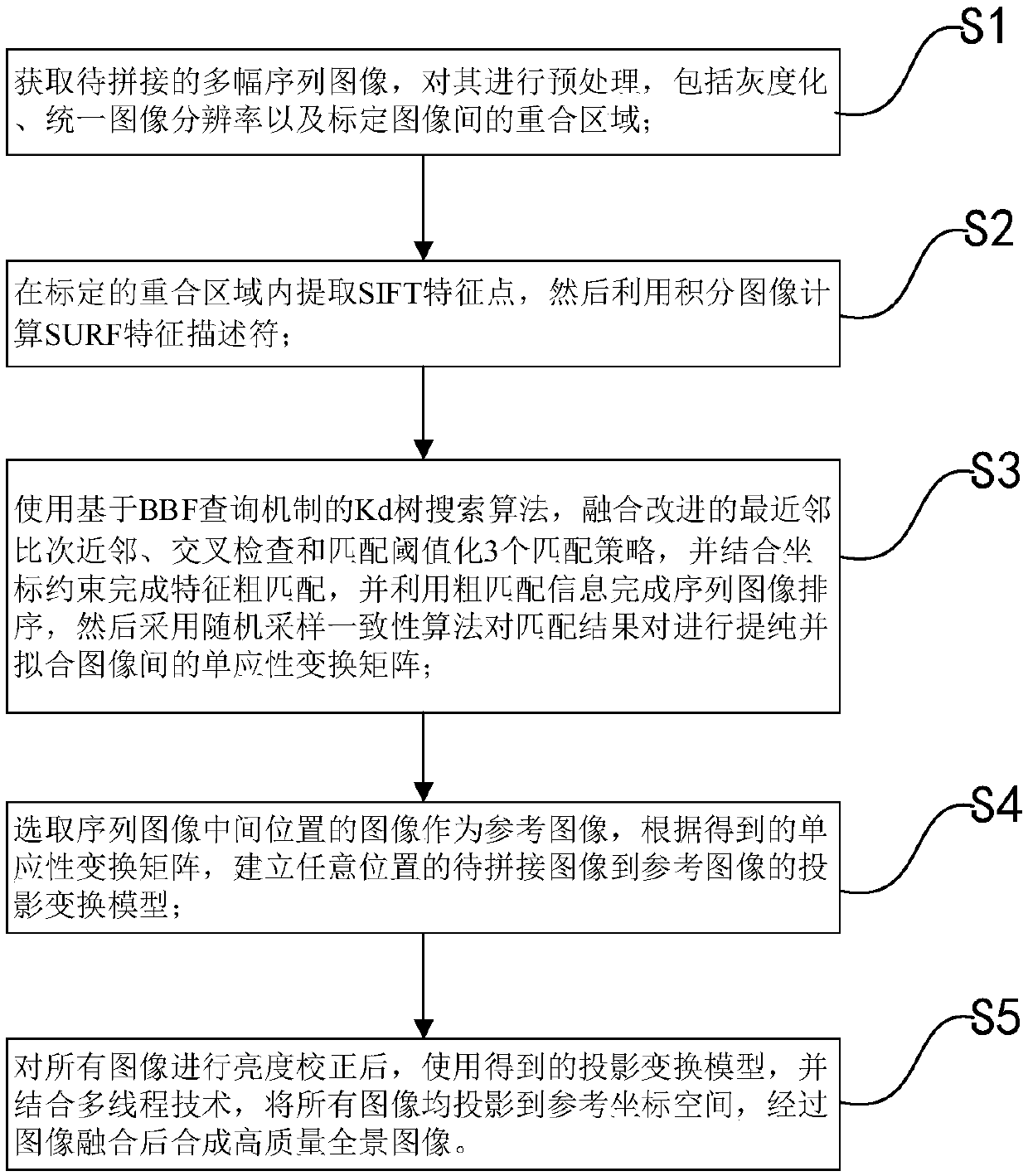
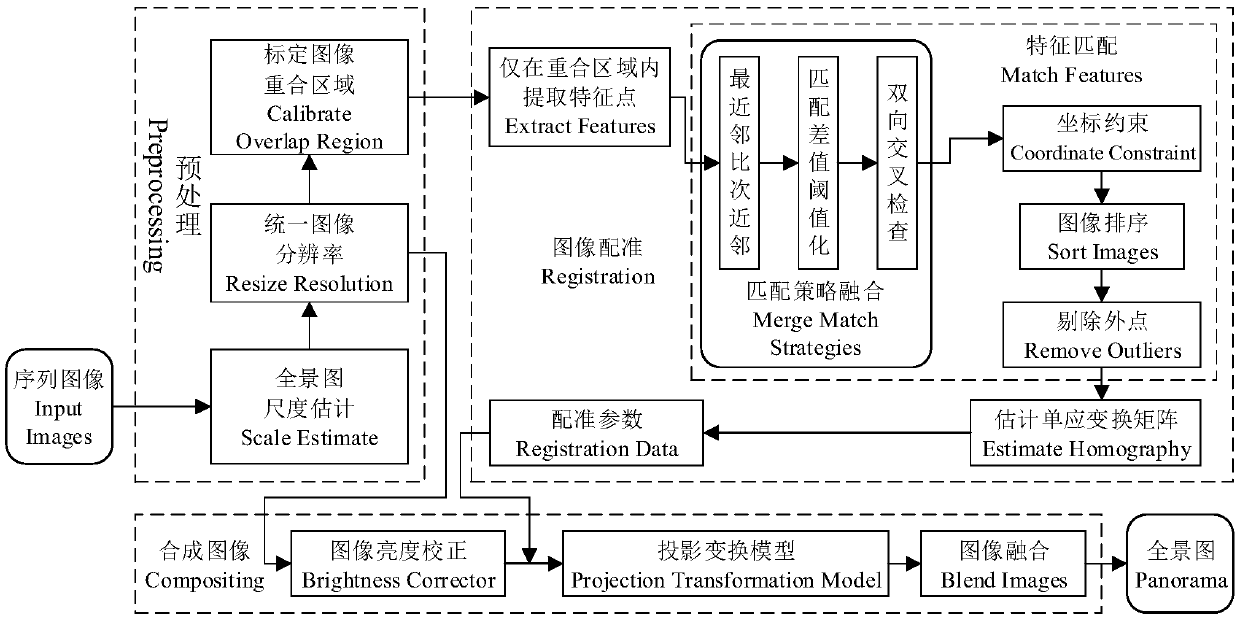
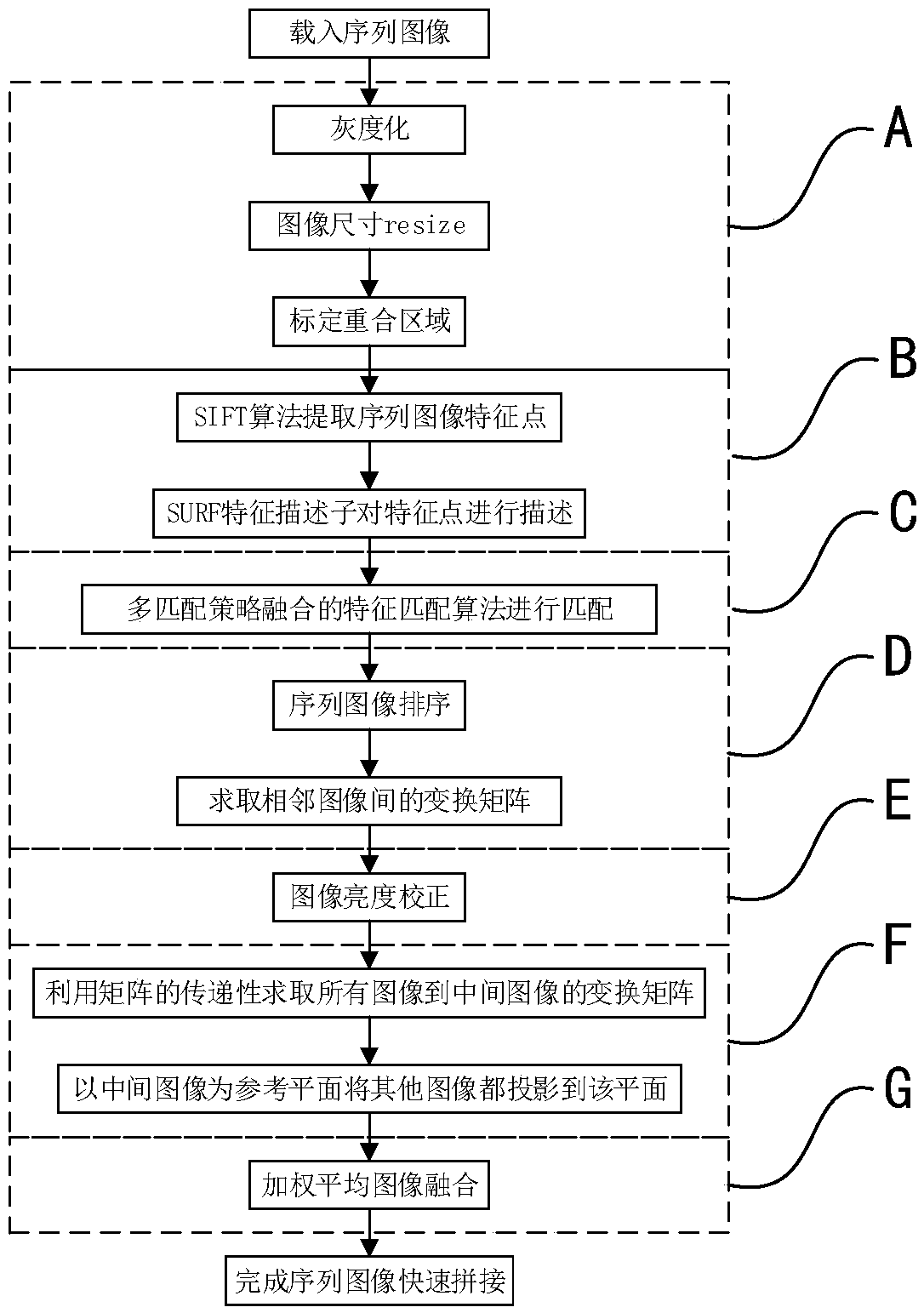
The invention discloses a fast image splicing method of matching strategy fusion and low errors. The method includes: S1, acquiring a plurality of to-be-spliced sequence images, and carrying out preprocessing and calibration of overlapped areas between the images thereon; S2, extracting SIFT feature points In the overlapped area, and calculating SURF feature descriptors; S3, completing feature rough-matching, utilizing rough-matching information to complete sequence image sorting, then adopting a random sample consensus algorithm to refine matching results, and fitting homography transformation matrices between the images; S4, selecting an image of a middle position of the sequence images to use the same as a reference image, and establishing a projection transformation model of any otherimage to the reference image; and S5, using the projection transformation model and combining multiple threads to project all the images to reference-image coordinate space after brightness correction, and synthesizing a high-quality panoramic-image after image fusion. The method greatly increases a correct-matching rate, can fast synthesize the high-quality panoramic-image of low errors, and hashigher practical values.
Owner:WUHAN UNIV OF TECH
Management in data structures
InactiveUS6298339B1Data processing applicationsDigital data information retrievalArray data structureTheoretical computer science
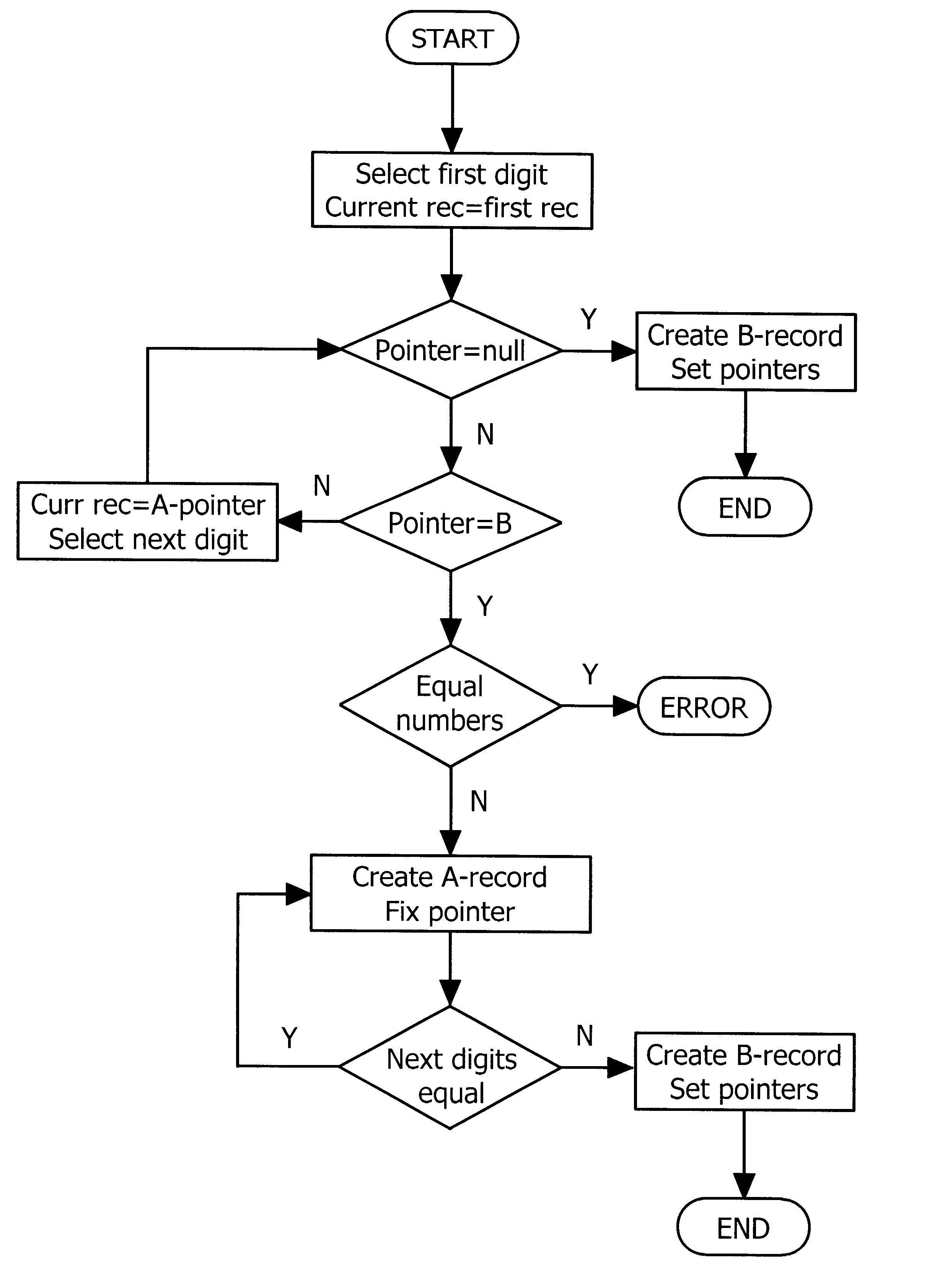
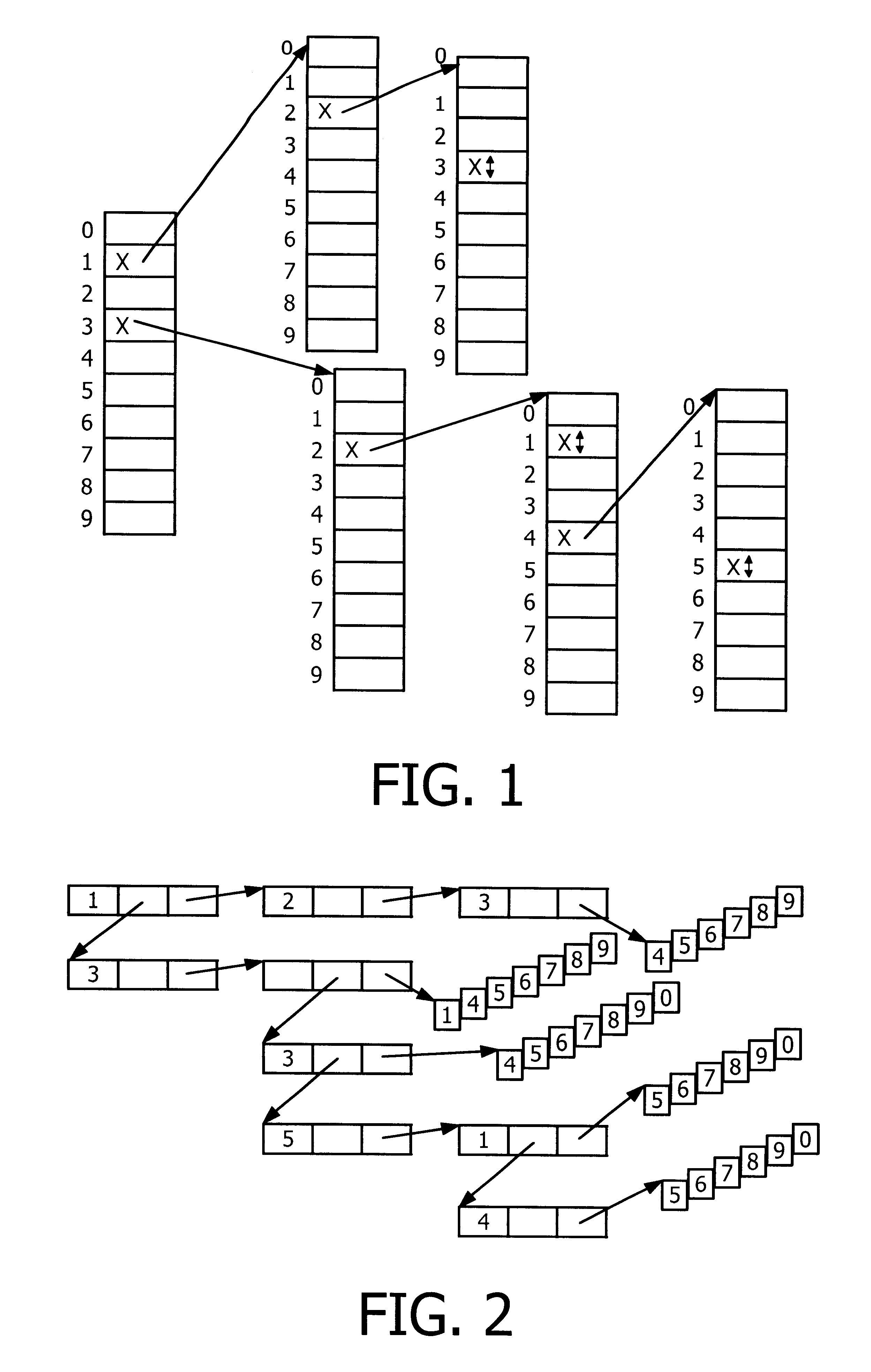
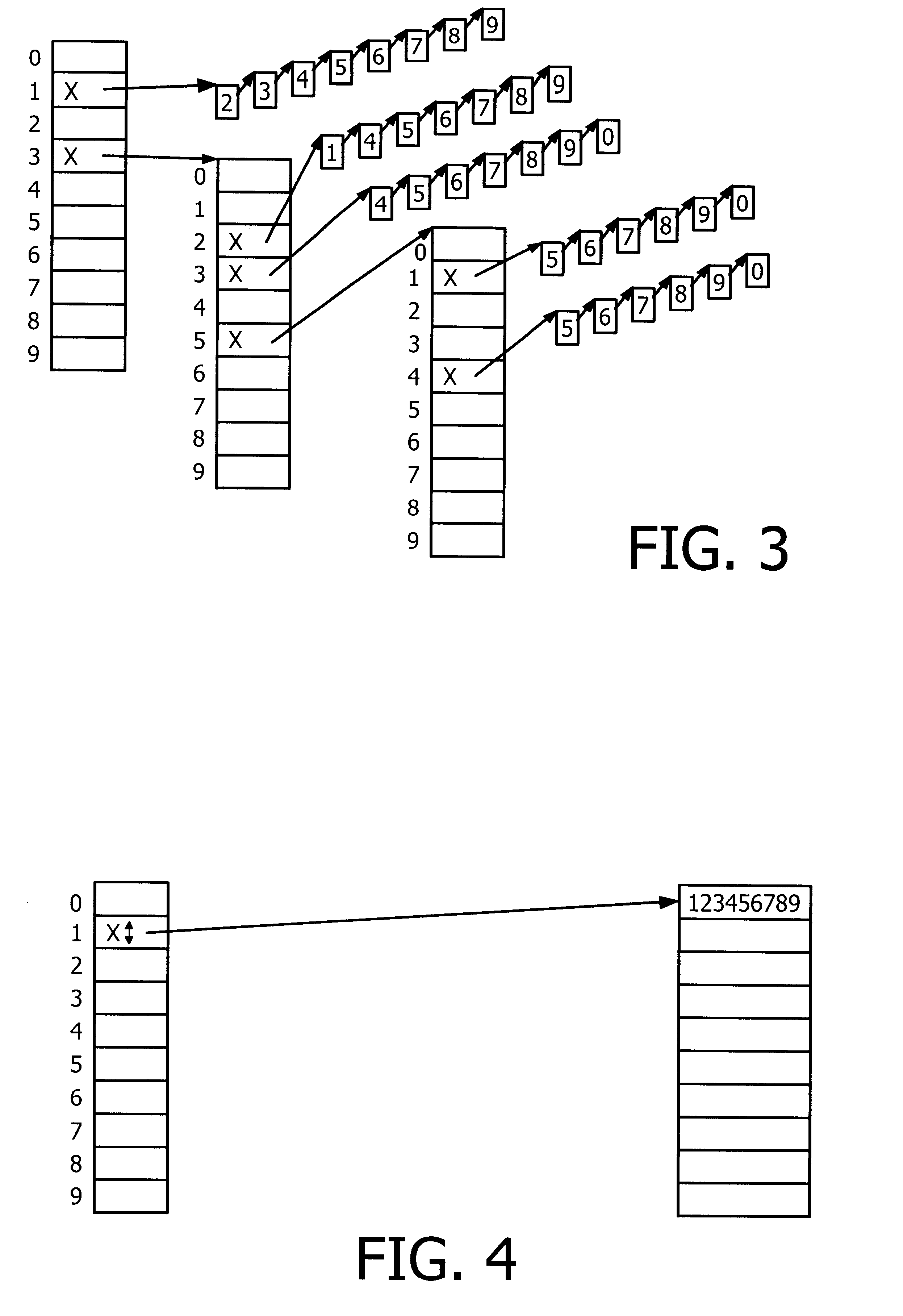
The present invention relates to a data structure and a method for, in a datastructure, managing sequences of symbols (e.g numbers or words). A solution with a double set of records is employed where the first set of records is defined by a linked list of records which are array records of pointers with one element for each symbol (e.g digit or letter). The second set of records is defined by strings where the complete sequences of symbols are stored. The managing of the data structures would suitably comprise storing, deleting and finding / searching the sequences of symbols.
Owner:TELEFON AB LM ERICSSON (PUBL)
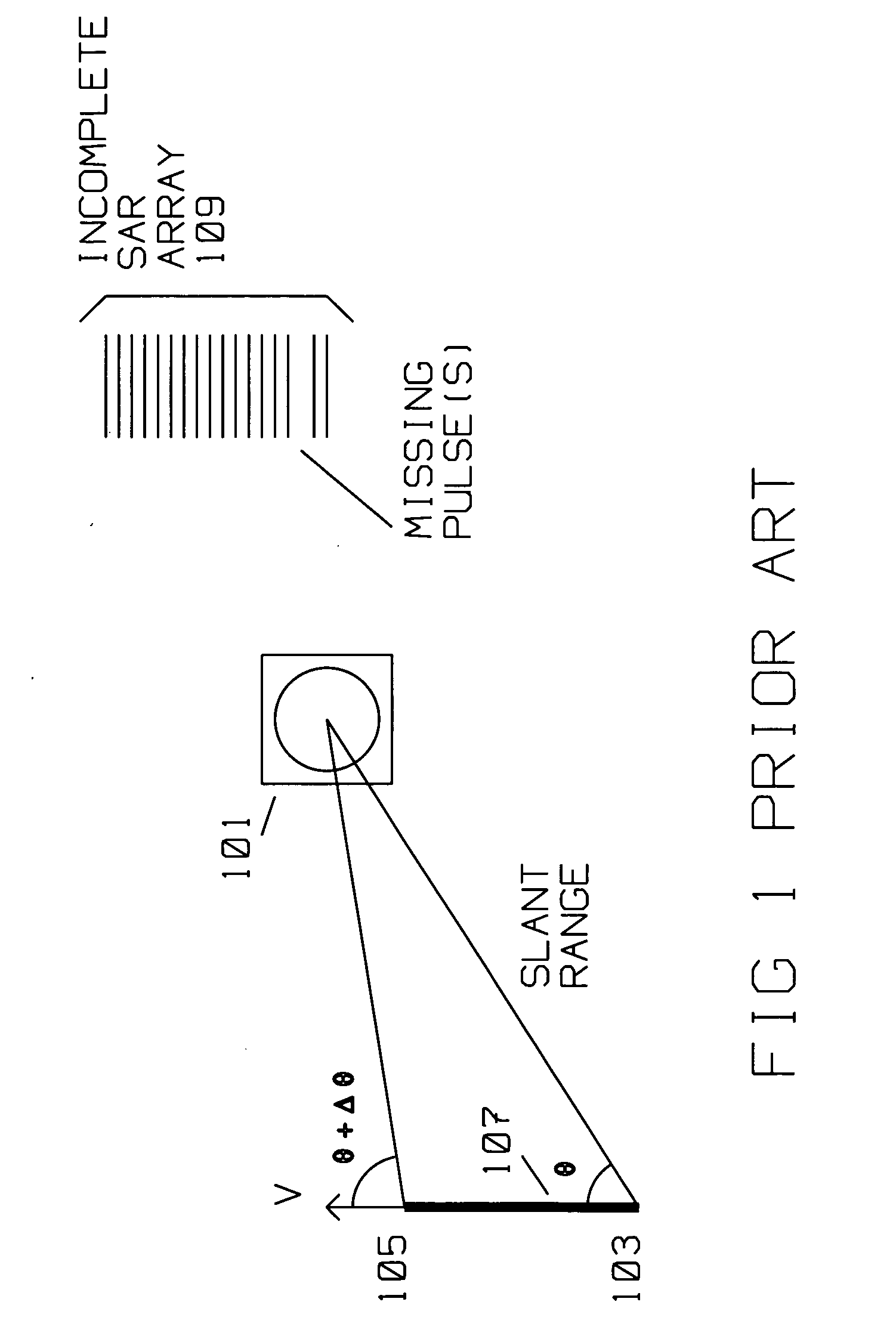
Interrupt SAR implementation for range migration (RMA) processing
ActiveUS20070159376A1Reduce restrictionsEnhance the imageRadio wave reradiation/reflectionRange migrationComplete sequence
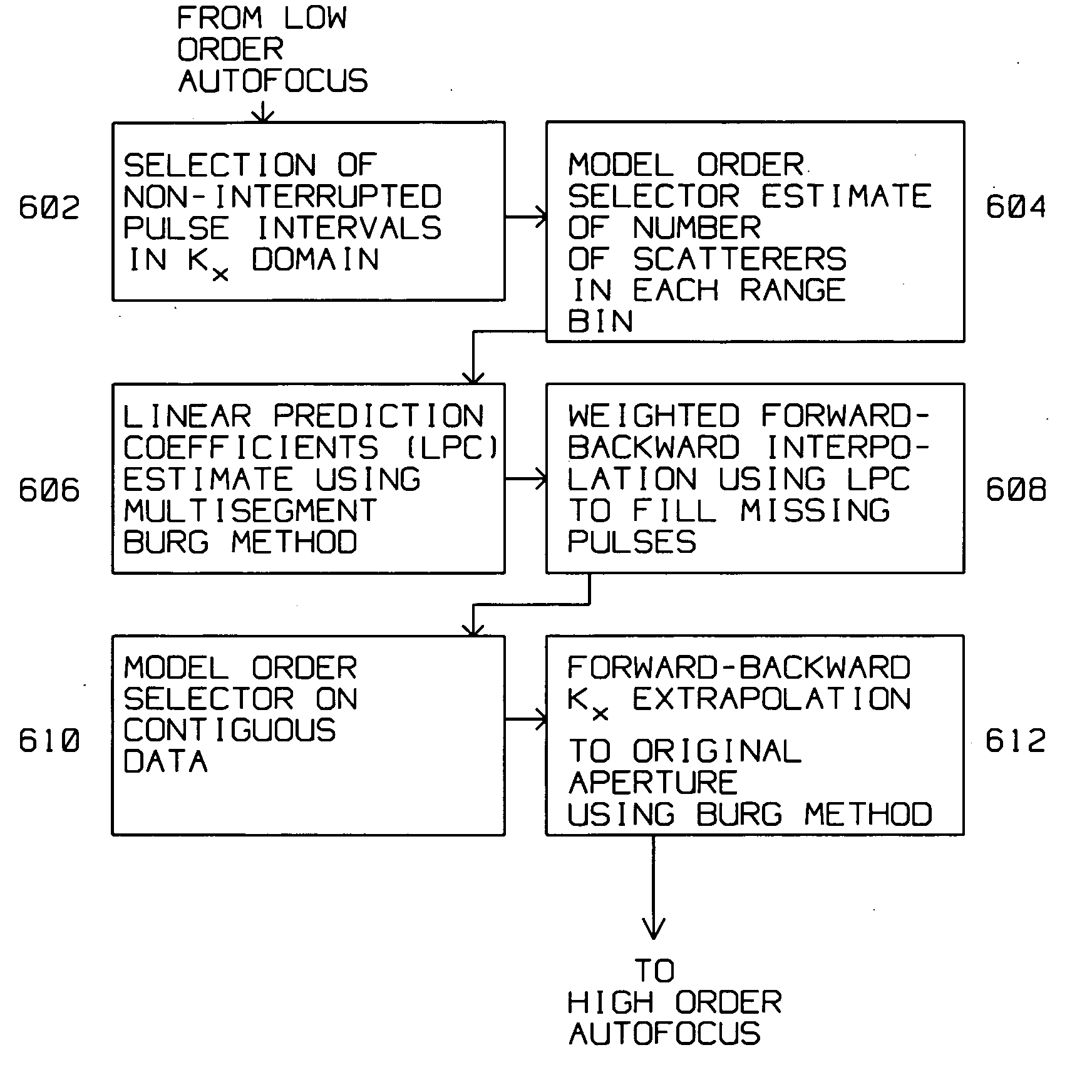
A moving radar (405) generates a synthetic aperture image from an incomplete sequence of periodic pulse returns. The incomplete sequence of periodic pulse returns has one or more missing pulses. The radar converts the incomplete sequence of pulse returns into a digital stream. A computer (403) processes the digital stream by computing an along track Fourier transform (402), a range compression (408), an azimuth deskew (410) and an image restoration and auto focus (412). The image restoration and autofocus (412) utilizes a low order autofocus (501), a gap interpolation using a Burg algorithm (503), and a high order autofocus (505) for generating an interpolated sequence. The interpolated sequence contains a complete sequence of periodic pulse returns with uniform spacing for generating the synthetic aperture image. The image restoration and autofocus (412) computes a linear prediction coefficients estimate using the Burg Algorithm (606). The linear prediction coefficients estimate (606) is used to compute a weighted forward-backward interpolation to generate the complete sequence of periodic pulse returns (608).
Owner:RAYTHEON CO
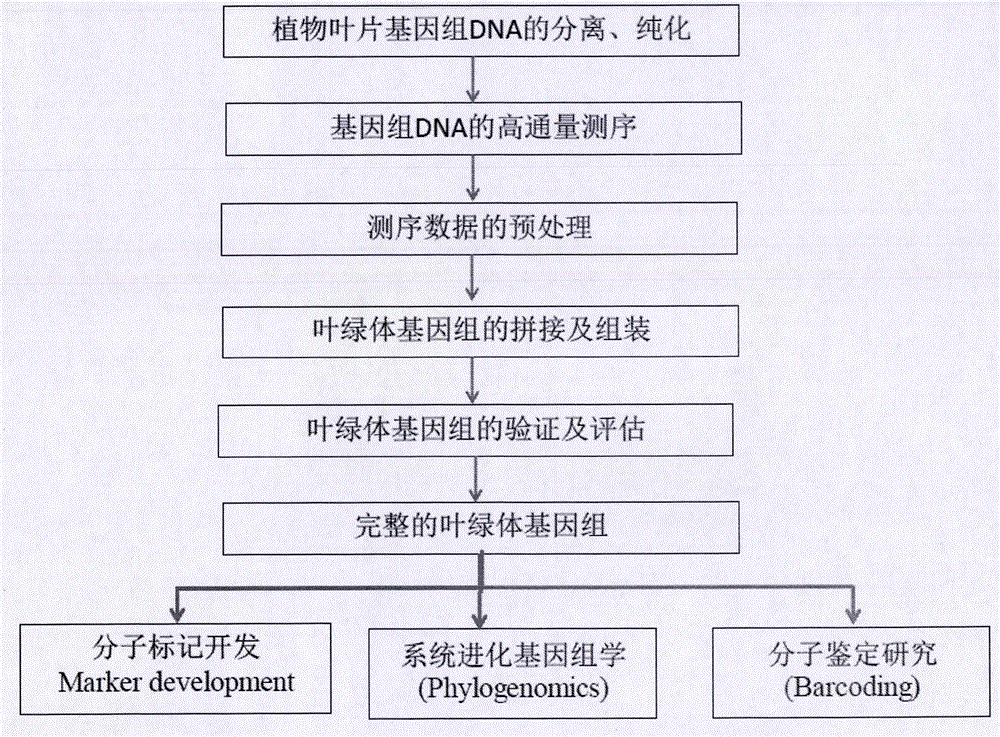
Simple, convenient, efficient and universal plant chloroplast genome sequencing method
PendingCN106834465AAccelerate research workImprove efficiencyMicrobiological testing/measurementSequence analysisHigh fluxInformatics
The invention discloses a simple, convenient, efficient and universal plant chloroplast genome sequencing method. The method comprises the following steps: directly performing high-flux sequencing by adopting genome DNA (Deoxyribonucleic Acid); capturing chloroplast reads by using a biology bioinformatics method; then, assembling and splicing to obtain a chloroplast genome complete sequence. Compared with the conventional method, the method has the advantages that a high-flux sequencing library is built directly by taking leaf genome DNA as a template, so that the efficiency and universality of the method are improved greatly; complicated PCR (Polymerase Chain Reaction) product sequencing and splicing and assembling of clone segments are not required, and splicing is directly performed from the start by using the reads obtained after high-coverage sequencing, so that test steps are reduced greatly, the test flow is simplified, and the method is simple, convenient, rapid and efficient.
Owner:NORTHWEST A & F UNIV
Amplification primer for grouper catostomidae mitochondrial genome complete sequence and design method thereof
InactiveCN101698842AEasy splicingNo mismatchMicrobiological testing/measurementDNA/RNA fragmentationGermplasmComplete sequence
The invention discloses an amplification primer for a grouper catostomidae mitochondrial genome complete sequence and a design method thereof, relates to groupers, and particularly relates to 16 pairs of amplification primers mainly used for specifically amplifying most grouper catostomidae mitochondrial genome complete sequences and a design method thereof. The invention provides an amplification primer for a grouper catostomidae mitochondrial genome complete sequence, a design method thereof and application thereof. The amplification primer for the grouper catostomidae mitochondrial genome complete sequence consists of 16 pairs of single-chain oligonucleotides, wherein each pair consists of one light-chain primer and one heavy-chain primer. The amplification primer for the grouper catostomidae mitochondrial genome complete sequence can be used in the aspects of variety authentication, geographical population authentication, germplasm resource evaluation and the like.
Owner:XIAMEN UNIV
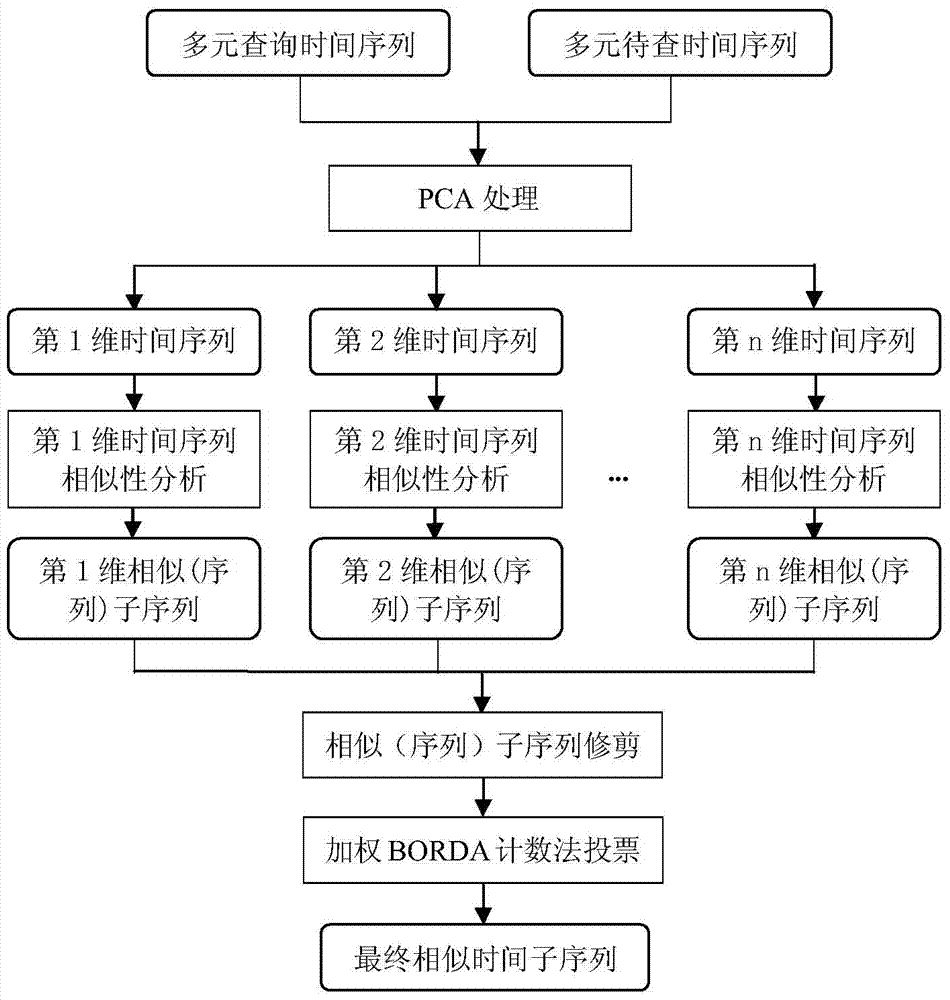
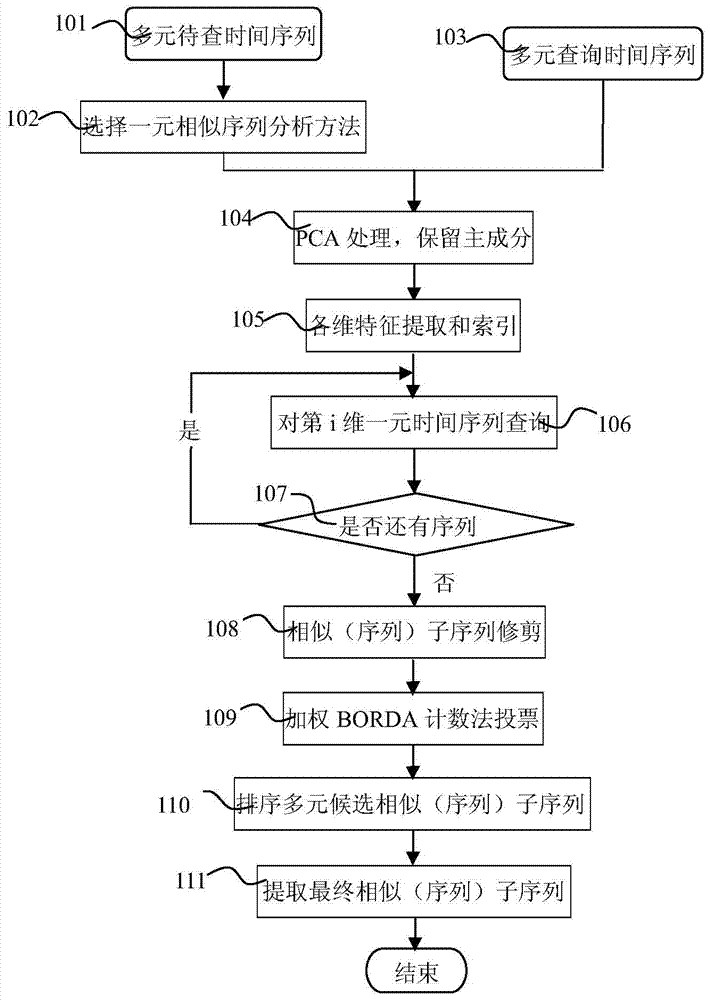
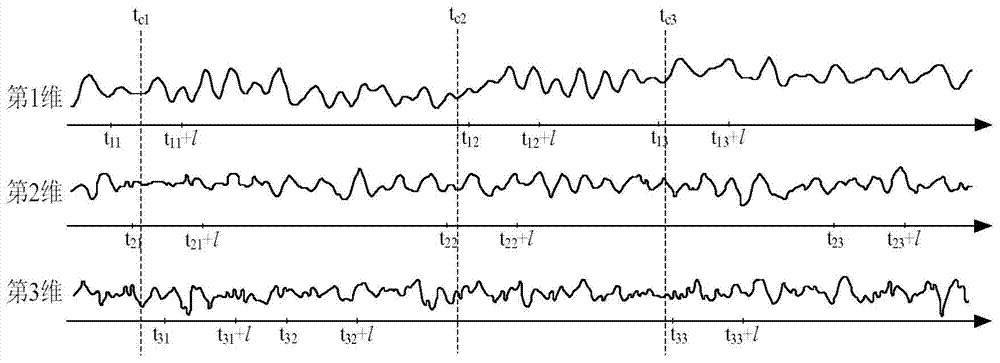
Polychronic time sequence similarity analysis method based on weighting BORDA counting method
The invention discloses a polychronic time sequence similarity analysis method based on a weighting BORDA counting method. The method comprises the steps of conducting PCA processing on polychronic sequences to be inquired and inquired sequences, reserving front p-dimensional principal component sequences with the characteristic value contribution rate reaching a certain threshold value (such as 80% and 95%), forming the p-dimensional principal component sequences, selecting a unitary time sequence similarity analysis method from existing time sequence similarity analysis methods according to the specific analysis requirements (such as sequence form similarity and distorted time shafts), utilizing the selected time sequence similarity analysis method for conducting unitary time sequence similar analysis on each-dimensional sequences of the p-dimensional principal component sequences, obtaining unitary similar (sequences) subsequences of the each-dimensional sequences, trimming the unitary similar (sequences) subsequences, generating polychromic candidate similar (sequences) subsequences, and utilizing the weighting BORDA counting method for conducting voting ranking on the candidate similar (sequences) subsequences to obtain the final similar (sequences) subsequences. The method is suitable for k-neighbor similarity inquiry of the complete sequences and the subsequences.
Owner:HOHAI UNIV
Selective terminal tagging of nucleic acids
ActiveUS20090227009A1Sugar derivativesMicrobiological testing/measurementNucleic acid detectionAmplification dna
Methods are provided for adding a terminal sequence tag to nucleic acid molecules for use in RNA or DNA amplification. The tag introduced may be used as a primer binding site for subsequent amplification of the DNA molecule and / or sequencing of the DNA molecule and therefore provides means for identification and cloning of the 5′-end or the complete sequence of mRNAs.
Owner:CELLSCRIPT
SNP (single nucleotide polymorphism) marker for evaluating growth performance of ctenopharyngodonidella, primer and evaluation method
ActiveCN106755527AEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationGenetic exchangeIntein
The invention provides an SNP (single nucleotide polymorphism) marker (comprising SNP1 and SNP4) for evaluating growth performance of ctenopharyngodonidella and a primer pair for amplifying a gene segment where an SNP locus is positioned by virtue of methods of molecular genetics and molecular biology. SNP1 is positioned at the 940th locus in a promoter of a complete sequence of a MyoD (myogenic determining factor) gene of the ctenopharyngodonidella, and a base T is inserted or deficient at the position; SNP4 is positioned in a 107th locus in a first intron of the complete sequence of the MyoD gene of the ctenopharyngodonidella, and a base at the position is A or T. The growth performance of the ctenopharyngodonidella is determined by detecting haplotypes of the two SNP loci. The adopted haplotypes are mutated according to bases generated in the MyoD gene, so that genetic exchange and further phenotype verification are avoided. By virtue of the haplotypes of the SNP marker, rapidly growing ctenopharyngodonidella can be simply and rapidly identified, and rapidly growing ctenopharyngodonidella of a new line can also be guided to be bred.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
Tobacco PDS gene fragment-containing cucumber mosaic virus RNA2 attenuated mutant type plasmid vector and application thereof
ActiveCN108486148AMeet basic requirementsSsRNA viruses positive-senseVirus peptidesNicotiana tabacumPlasmid Vector
The invention discloses a tobacco PDS gene fragment-containing cucumber mosaic virus RNA2 attenuated mutant type plasmid vector and application thereof. The plasmid vector contains a CMVFny strain RNA2 complete sequence, and a tobacco PDS gene 199bp fragment is inserted behind a 2a protein termination codon. The plasmid vector and a wild CMVFny RNA1 and wild CMVFny RNA3-containing plasmid are inoculated to nicotiana benthamiana through an agrobacterium infiltration method, so that weakly pathogenic symptoms can be displayed, and the capacity of inducing gene silencing can be judged with nakedeyes without depending on an instrument; great convenience is provided for deep researches on attenuated vaccines.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
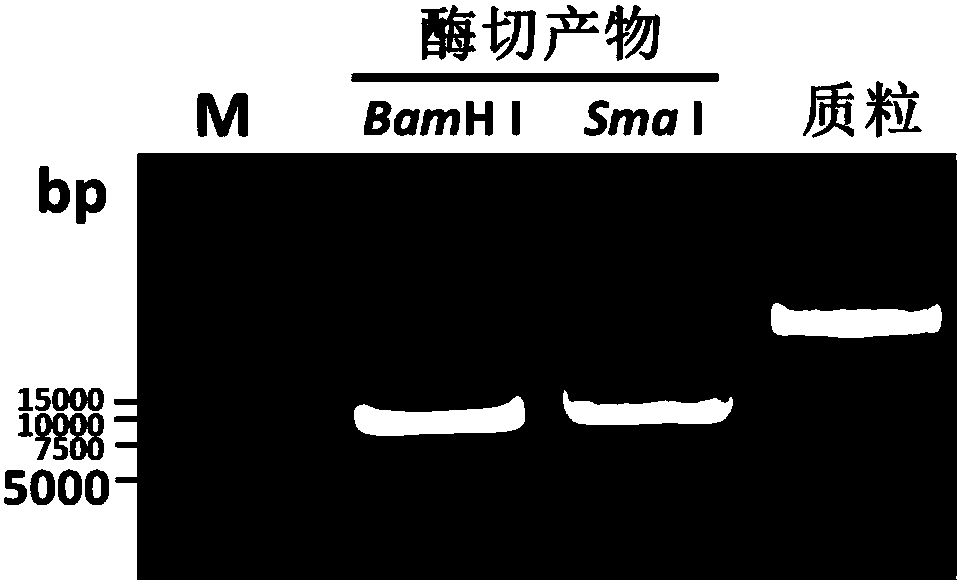
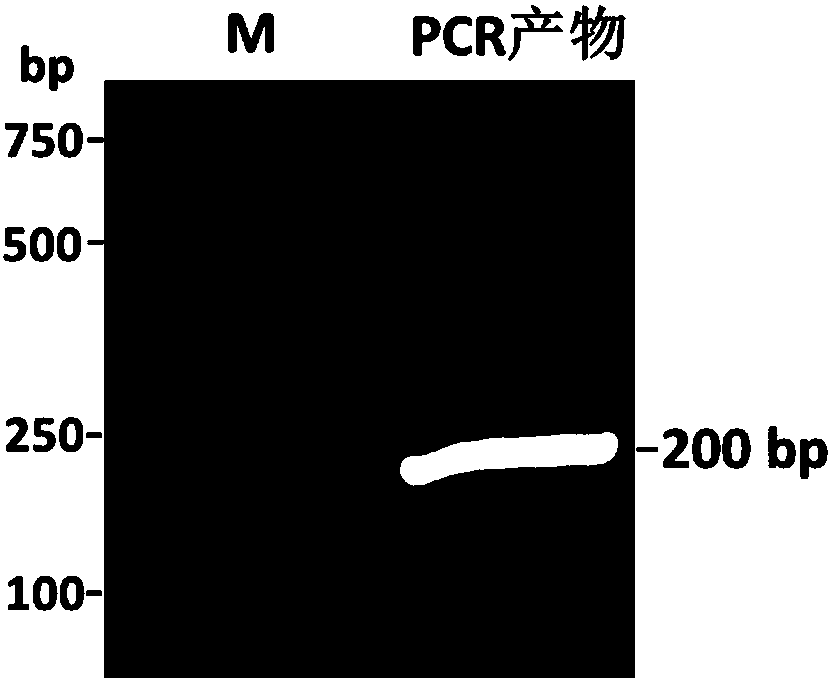
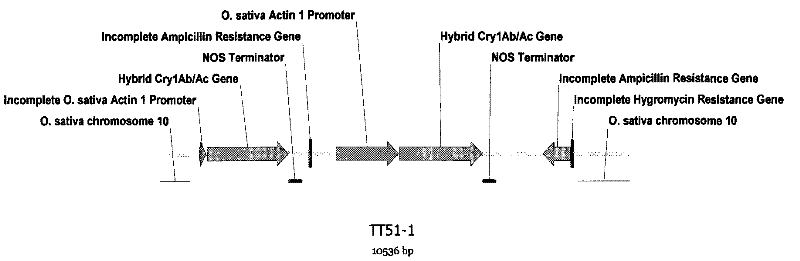
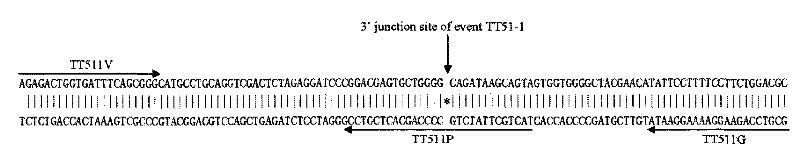
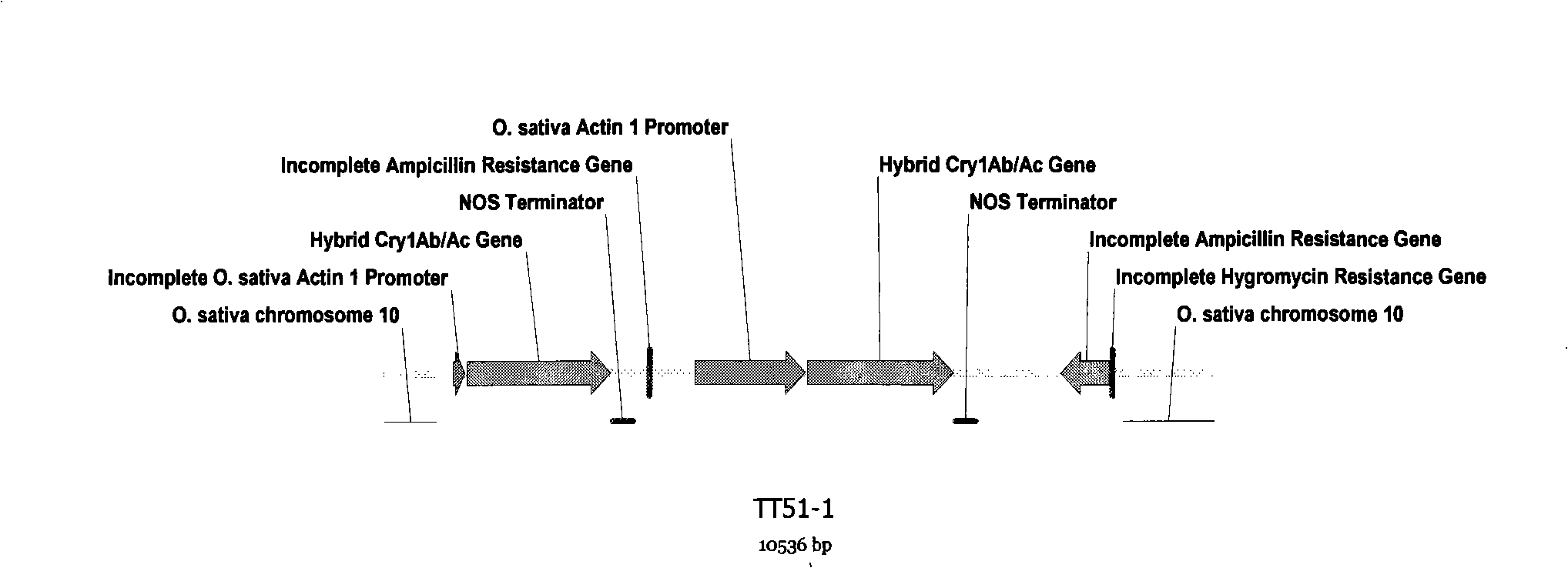
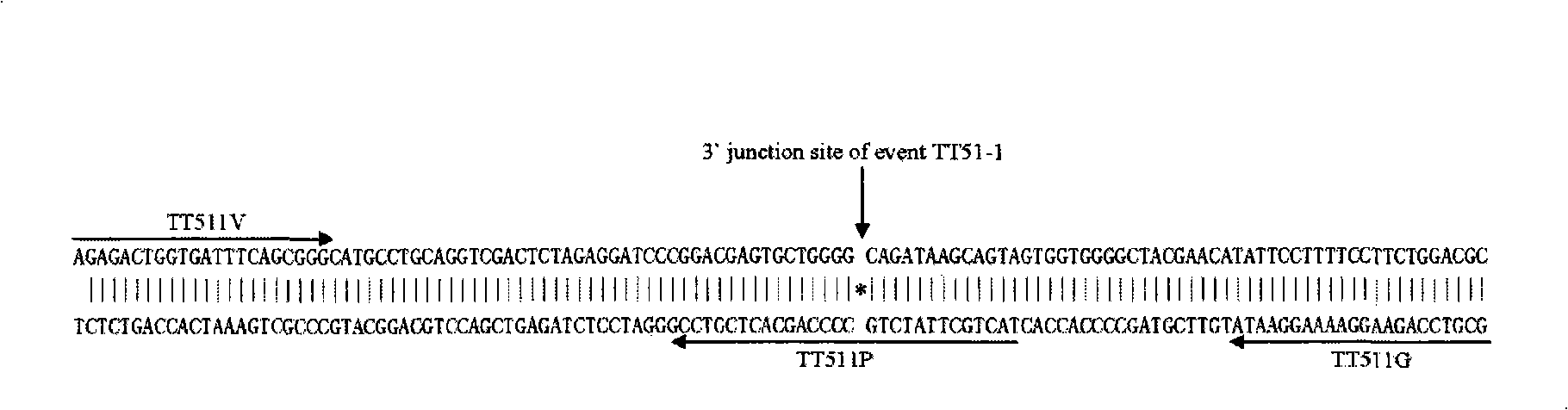
Transgenic rice TT51-1 transformation event foreign vector integration site complete sequence and use thereof
InactiveCN101302520BMicrobiological testing/measurementFermentationGenetically modified riceAgricultural science
The invention discloses an event whole sequence with an integration site of a foreign vector transferred by transgenic paddy rice TT51-1 and application of the event whole sequence, relating to safety evaluation and detection of transgenic paddy rice in the biological engineering technical field. The invention uses transgenic insect-resistant rice variety TT51-1 as a material, extracts a gene group DNA as a template, and divides into four sections to enlarge TT51-1 event whole sequence containing a side paddy rice gene group sequence which is shown in SEQ NO.1, and the whole sequence is formed commonly by a basic group of between first and 673rd from the paddy rice gene group, a basic group of between 674th and 9364th from a vector sequence and a basic group of between 9365th and 10536th from the paddy rice gene group; the whole sequence is applicable to carrying out detection, monitoring and safety management of the transgenic paddy rice TT51-1(including parent strain, hybridism F1 and later generation) and products (including plant, tissue, paddy, rice and products thereof).
Owner:INST OF OIL CROPS RES CHINESE ACAD OF AGRI SCI
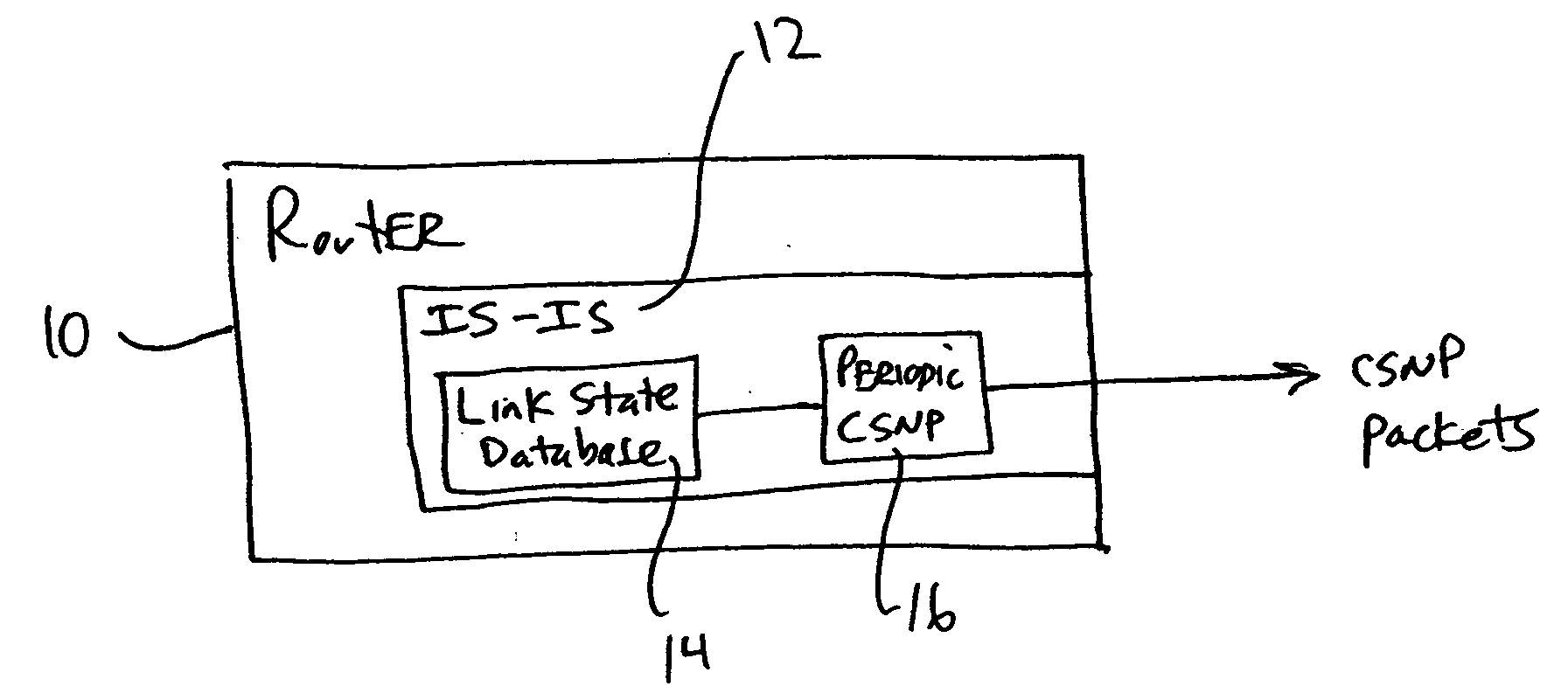
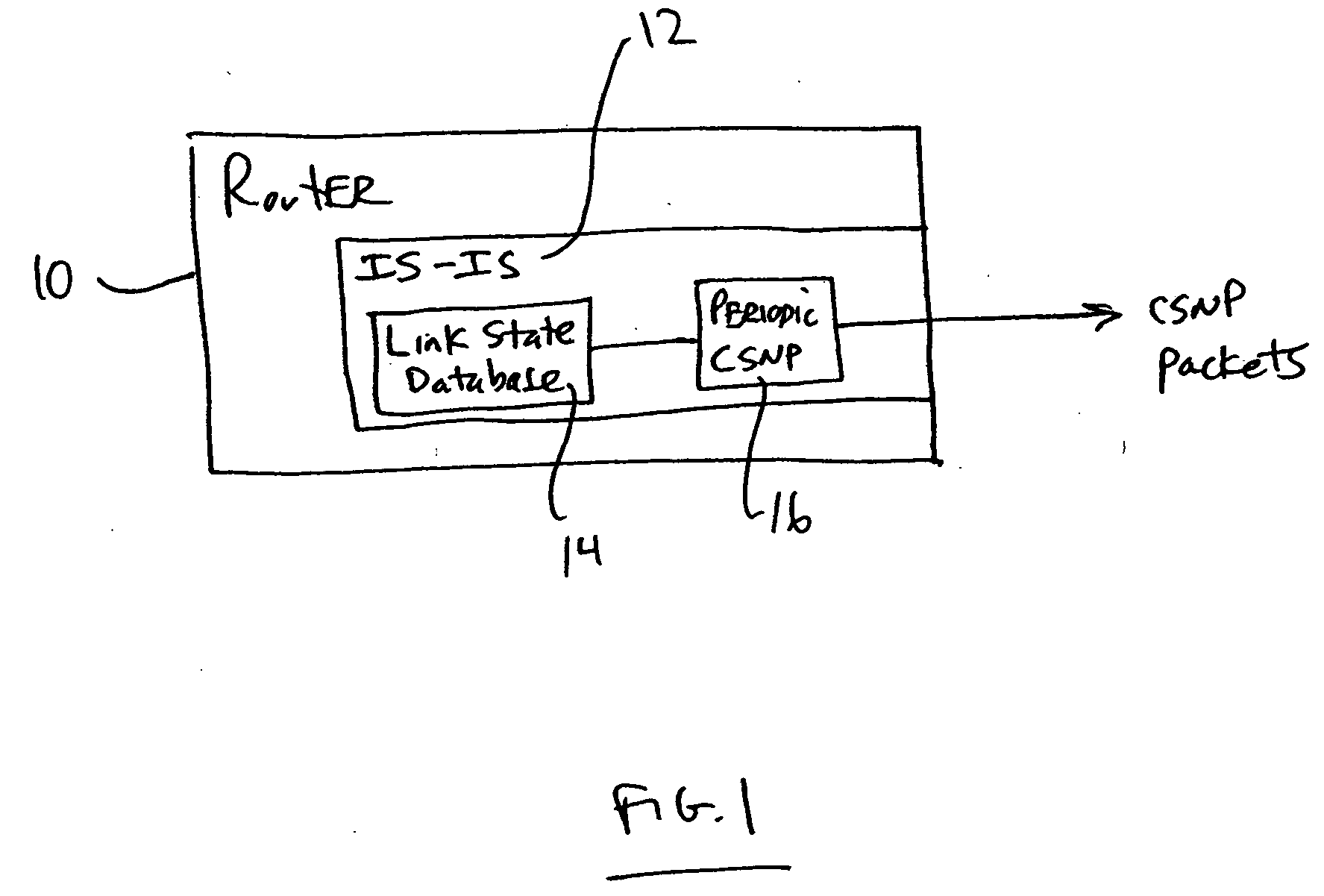
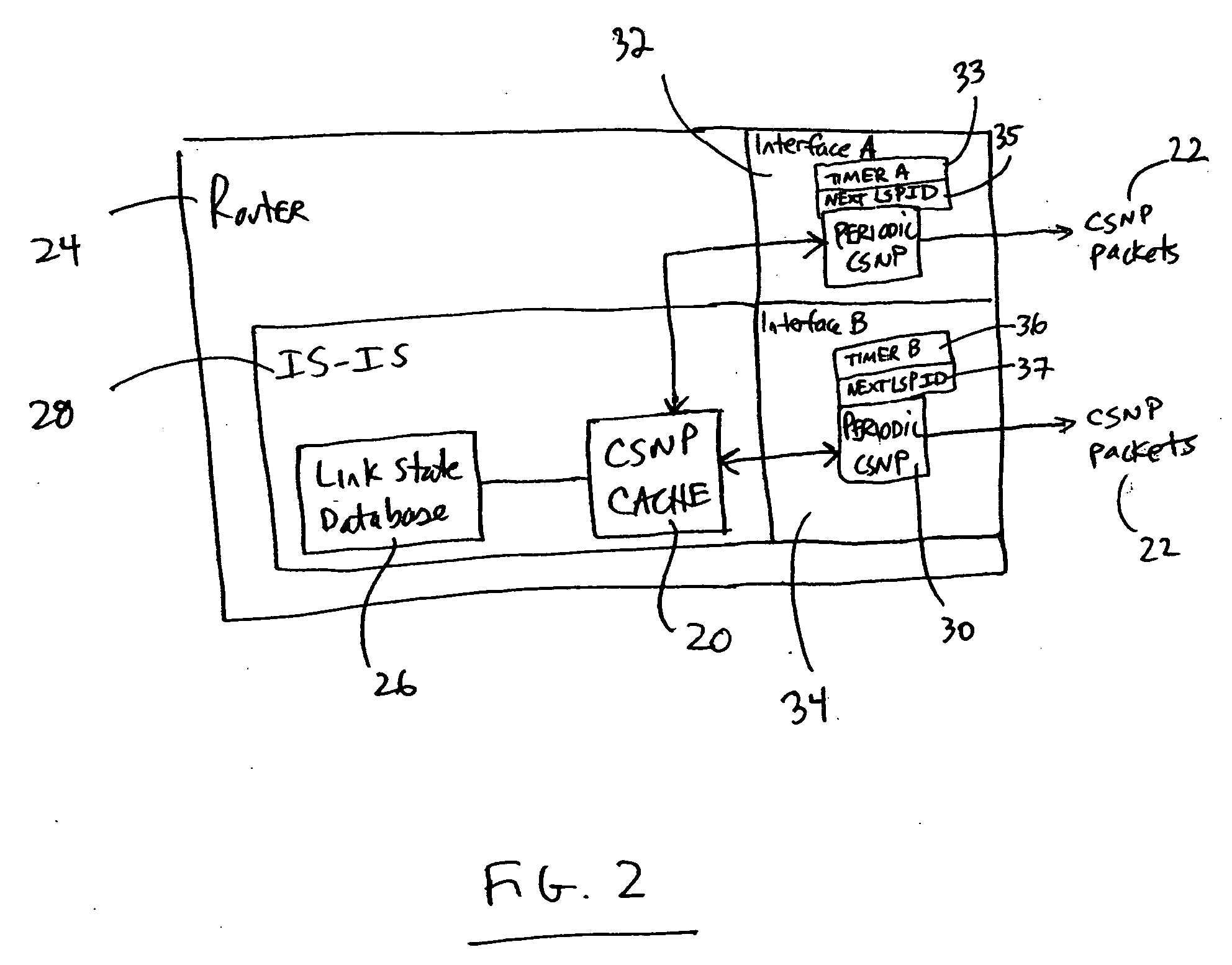
CSNP cache for efficient periodic CSNP in a router
A networking device such as a router may include, in one embodiment, a database storing a plurality of link state entries, and a cache operatively coupled with the database, the cache storing entries relating to the link state entries of the database. The networking device may also include a module for sending, over a network, packets including link state data, the module operatively coupled with the cache. In one example, the module accesses the cache to create one or more packets including link state data. Embodiments of the invention may be used for forming CSNP packets (complete sequence number packets) without the need for having to repeatedly walking a link state database in order to form the CNSP packets.
Owner:CISCO TECH INC
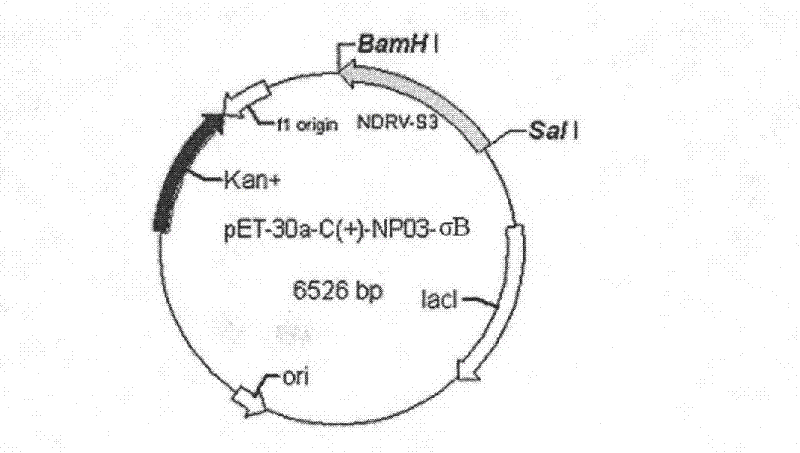
Novel duck reovirus recombinant sigma B protein antigen, preparation method and application
InactiveCN102675469ANo cross reactionHigh detection sensitivityPeptide preparation methodsFermentationInclusion bodiesNucleotide
The invention discloses preparation of novel duck reovirus recombinant sigma B protein antigen, and application of the novel duck reovirus recombinant sigma B protein antigen in detection of a novel duck reovirus antibody. With RNA (ribonucleic acid) of the novel duck reovirus (NDRV) as a template, a nucleotide complete sequence of an NDRV S3 gene coding region can be amplified through a reverse-translation polymerase-chain reaction, is directionally subcloned to a fusion expression vector of the prokaryotic expression vector pET-30a-C(+) of escherichia coli, and transformed into the host cell of the Escherichia coli BL21 (DE3), and is subjected to inducible expression with IPTG (isopropyl-beta-d-thiogalactoside), purification of Ni-NTA Agarose, inclusion body refolding to obtain the expressed NDRV alpha B recombinant protein antigen. The expression mode of the protein is fusion protein (His-alpha B) with molecular weight being about 44kU; and immunoblotting assays prove that the protein has immunoreactivity similar to natural protein..
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
Transgenic rice TT51-1 transformation event foreign vector integration site complete sequence and use thereof
InactiveCN101302520AMicrobiological testing/measurementGenetic engineeringGenetically modified riceAgricultural science
The invention discloses an event whole sequence with an integration site of a foreign vector transferred by transgenic paddy rice TT51-1 and application of the event whole sequence, relating to safety evaluation and detection of transgenic paddy rice in the biological engineering technical field. The invention uses transgenic insect-resistant rice variety TT51-1 as a material, extracts a gene group DNA as a template, and divides into four sections to enlarge TT51-1 event whole sequence containing a side paddy rice gene group sequence which is shown in SEQ NO.1, and the whole sequence is formed commonly by a basic group of between first and 673rd from the paddy rice gene group, a basic group of between 674th and 9364th from a vector sequence and a basic group of between 9365th and 10536th from the paddy rice gene group; the whole sequence is applicable to carrying out detection, monitoring and safety management of the transgenic paddy rice TT51-1(including parent strain, hybridism F1 and later generation) and products (including plant, tissue, paddy, rice and products thereof).
Owner:INST OF OIL CROPS RES CHINESE ACAD OF AGRI SCI
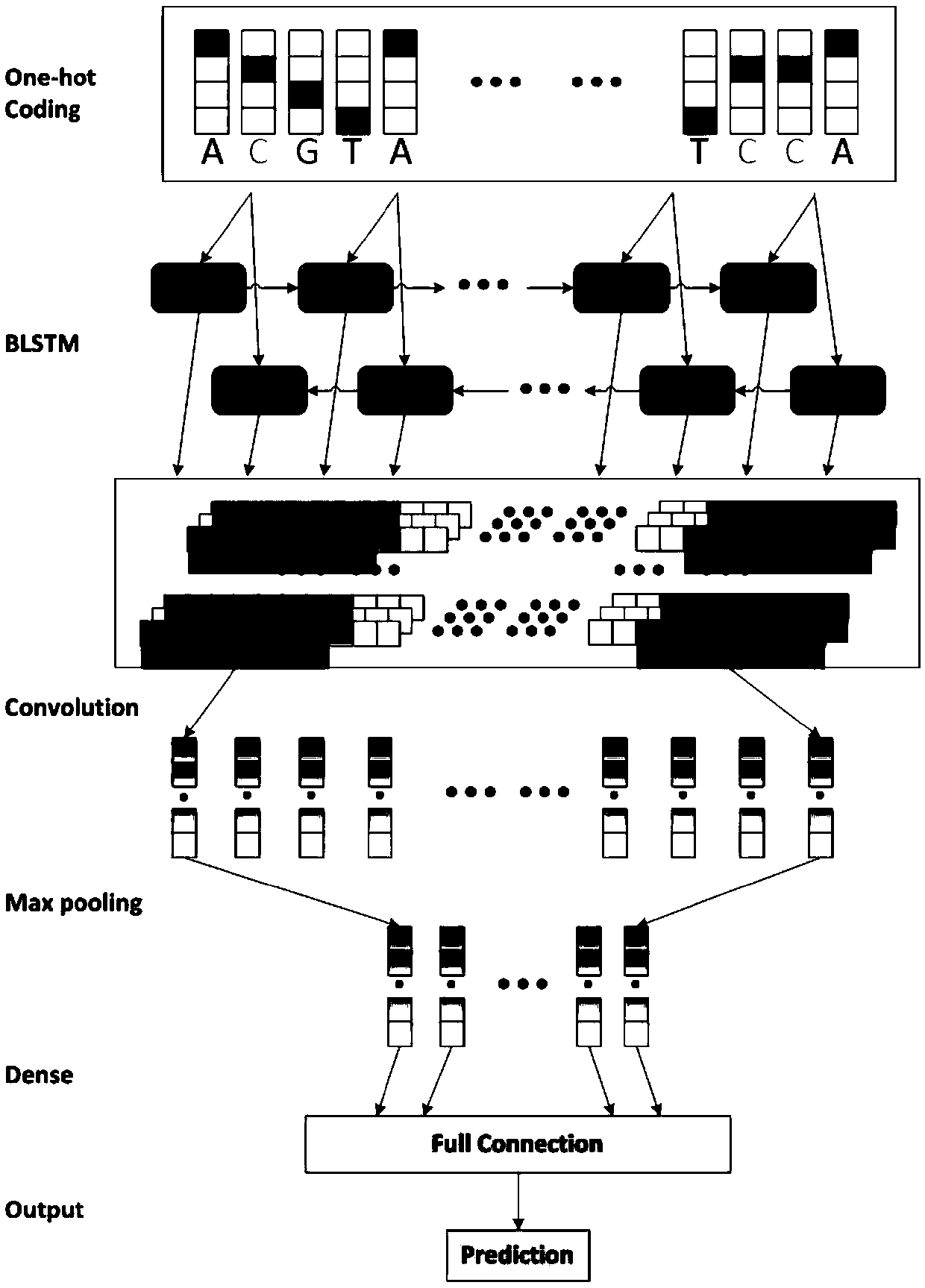
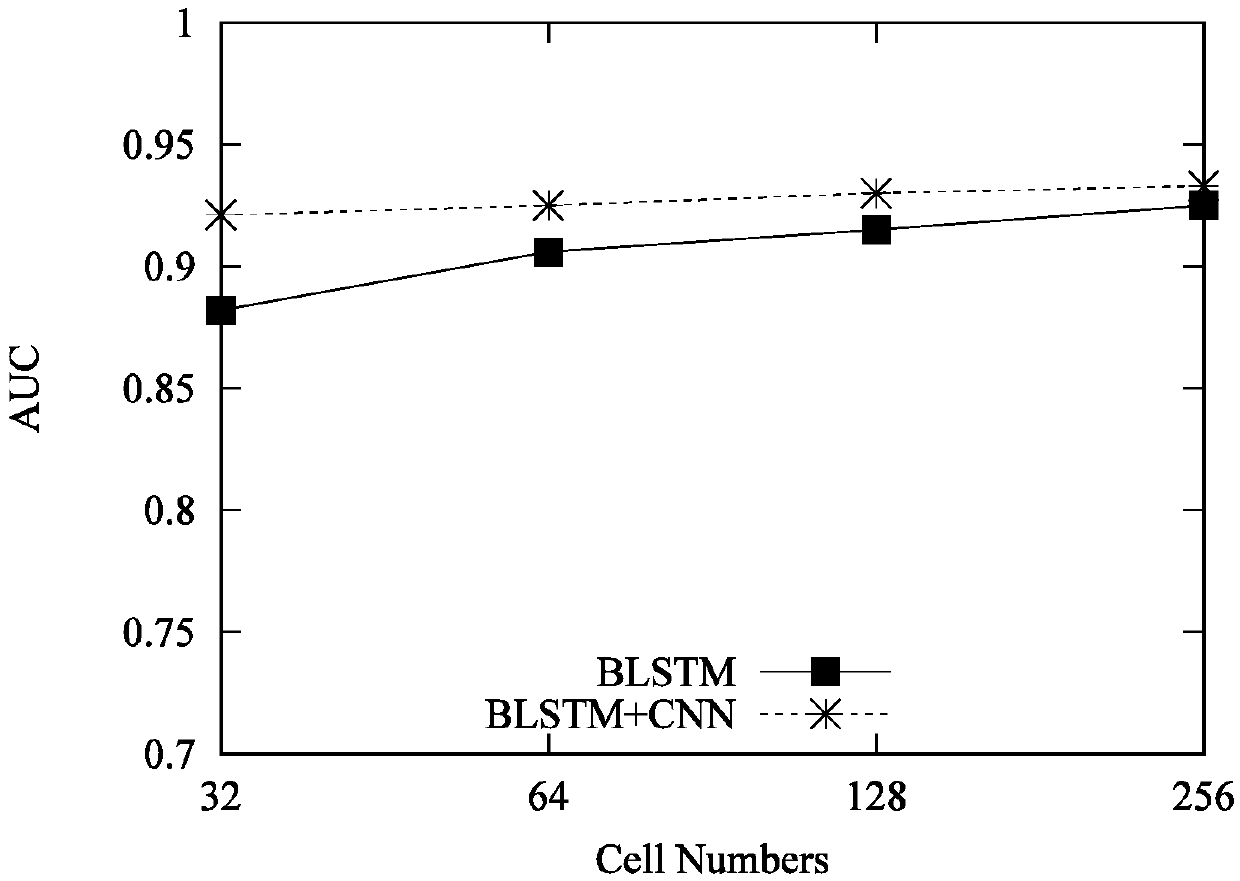
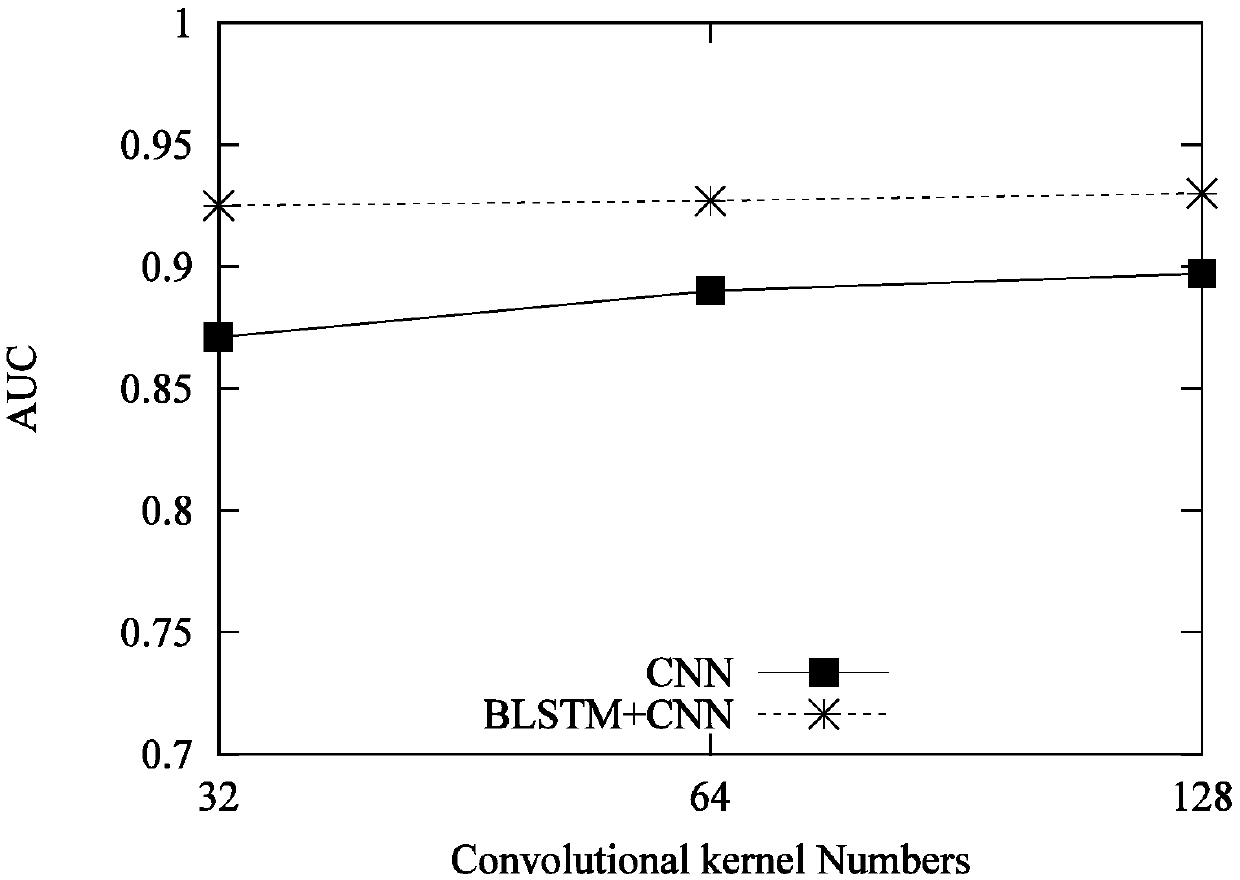
Bidirectional LSTM and CNN model for predicting DNA-protein binding
InactiveCN109559781AEasy to learnImprove predictive performanceSequence analysisNeural architecturesAlgorithmComplete sequence
The invention provides a bidirectional LSTM and CNN model for predicting DNA-protein binding. The model includes an input layer, a BLSTM layer, a convolutional layer, a maximum pooling layer, a full connection layer, and an output layer. Each input sequence is expressed as a four-line binary matrix by the input layer through single thermal coding. In the BLSTM layer, each LSTM model in a previouslayer will receive information of interest on DNA from an input sequence, and encode and interpret contributions from past historical information to a hidden state; then, the BLSTM module is propagated to the next BLSTM module, wherein a matrix scanned and input by each convolution kernel in the convolution layer is used for motif discovery, and information with different intensities is associatedwith potential sequence patterns; the maximum pooling layer is used for maximizing an output signal of each convolution kernel to form a complete sequence; the output layer performs non-linear conversion to determine DNA-protein binding feature information.
Owner:CHENGDU UNIV OF INFORMATION TECH
A digital control code encoder and method for establishing digital control system software based on the same
The present invention discloses a numeric control code compiler, and also discloses a method for creating numeric control system software by using said numeric control code compiler. It is characterized by that it includes the following several steps: (1), constructing PIDP_NC code compiler; (2), in upper-position machine adopting PIDP_NC code compiler to compile NC program into object code file with specific data format and complete sequence execution signification; (3), upper-position machine can feed the PIDP object codes into lower-position machine circulation buffer zone in batches according to sequence; and (4), lower-position machine can read out PIDP object codes from circulation buffer zone according to sequence, identify the data in PIDP and judge the significance correspondent to NC command, at the same time obtain correspondent data and call correspondent motion control function according to significance of command so as to implement processing control.
Owner:SHENZHEN INSTITUTE OF INFORMATION TECHNOLOGY
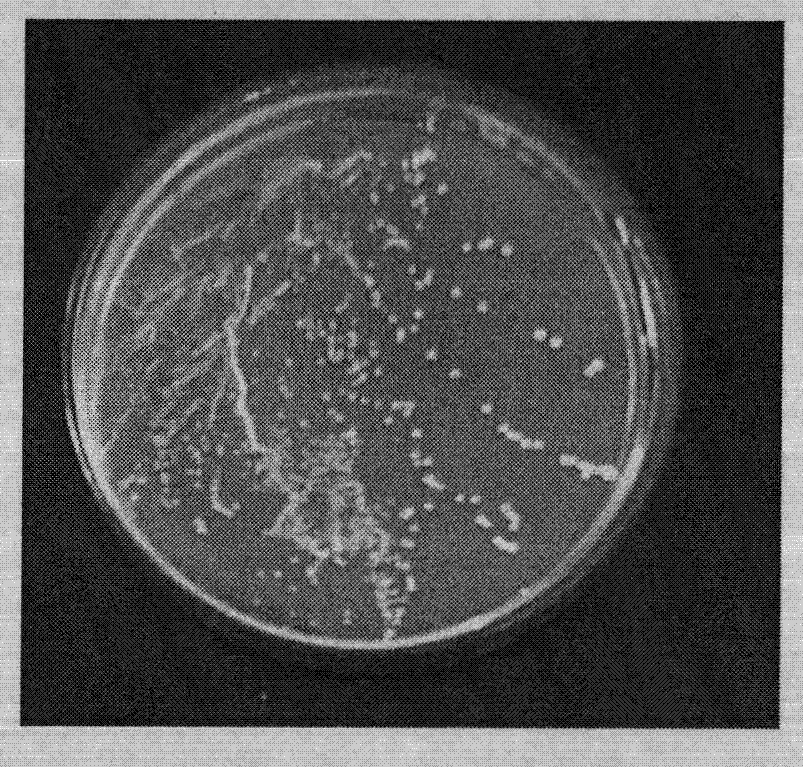
Lactobacillus rhamnosus, cultivation of lactobacillus rhamnosus and microcapsule method
InactiveCN103031263AShorten the growth cycleReproduce fastBacteriaMicroorganism based processesLactobacillus rhamnosusBacteroides dorei
The invention discloses lactobacillus rhamnosus, cultivation of the lactobacillus rhamnosus and a microcapsule method. The bacterial strain is preserved in China General Microbiological Culture Collection Center (CGMCC); a registering number is CGMCC NO.3994; a Latin name of the bacterial strain is Lactobacillus rhamnosus LC-STH-13; morphological characteristics are gram-positive bacteria, short-rod bacterial thallus, no motion, no gemma, circular bacterial colony, milk white, tidy edges, smooth surfaces and protuberant cores. The invention further discloses a 16S ribosome DNA (Deoxyribose Nucleic Acid) (16S rDNA) complete sequence of the lactobacillus rhamnosus. The lactobacillus rhamnosus has the advantages that a culture is short in growth cycle, high in breeding speed and- produced lactic acid concentration. The lactobacillus rhamnosus can -colonize and survive in an intestinal tract, improves a mucosal barrier function of a gastrointestinal tract, has a function of improving the gastrointestinal tract, and restrains the colonization of harmful bacteria; and after being embedded by a microcapsule, the lactobacillus rhamnosus has stronger ability to resist the poor environment.
Owner:哈尔滨美华生物技术股份有限公司
Applications of odorrana grahami polypeptide AH90 and CW49 for promoting skin regeneration
ActiveCN102657845APromote repairReduced thickness of hyperplasiaAmphibian material medical ingredientsPeptide/protein ingredientsCuticleComplete sequence
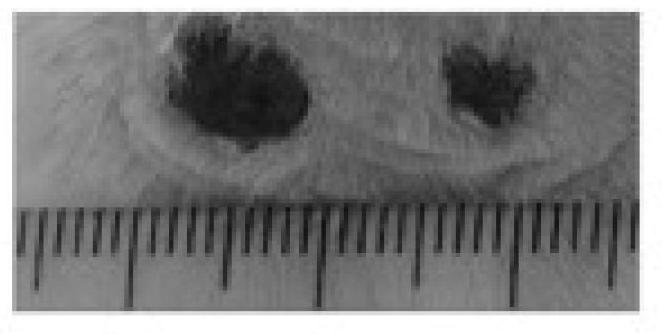
The invention belongs to the technical field of polypeptide drugs in biochemistry and particularly relates to applications of odorrana grahami polypeptide AH90 and CW49 for promoting skin regeneration. The polypeptide AH90 which has the molecular weight of 2657.18 Da, the isoelectric point of 10.26 and the complete sequence primary structure of ATAWDFGPHGLLPIRPIRIRPLCG is used as a drug to be applied to treatments of body-surface traumas, burns and skin ulcer, reduction of scar formation and acceleration of scar repair. The polypeptide CW49 which has the molecular weight of 1210.43 Da, the isoelectric point of 8.29 and the complete sequence primary structure of APFRMGICTTN is used as a drug to be applied to treatments of body-surface traumas, burns and skin ulcer, reduction of scar formation and acceleration of scar repair. The odorrana grahami polypeptide AH90 and CW49 for promoting skin regeneration can promote skin regeneration and reduce epidermis proliferation thickness during the skin regeneration and can be used for preparing drugs for treating body-surface traumas, burns and skin ulcer, reducing scar formation and accelerating scar repair.
Owner:KUNMING INST OF ZOOLOGY CHINESE ACAD OF SCI
Rana pleuraden antioxidant peptide and genes and application thereof
ActiveCN101638432ASimple structureEasy to synthesizePeptide/protein ingredientsAntinoxious agentsRana pleuradenComplete sequence
The invention relates to rana pleuraden antioxidant peptide and genes and an application thereof, belonging to the technical field of biomedical science. The rana pleuraden antioxidant peptide is thesingle chain polypeptide through genetic encoding of Chinese amphibians rana pleuraden, wherein, the molecular weight is 1653.88 daltons, the isoelectric point is 8.22, and the complete sequence of the rana pleuraden antioxidant peptide is NH2-GIRPTYNRQCEIGF-COOH; the gene for encoding is composed of 316 nucleotides, wherein, the encoding mature part is the 130-171th nucleotide. The synthetic ranapleuraden antioxidant peptide features strong antioxidant activity, can be applicable to the preparation of medicines of skin antioxidant protection and free radicals in vivo elimination, and has theadvantages of simple sequence and convenient synthesis.
Owner:KUNMING INST OF ZOOLOGY CHINESE ACAD OF SCI
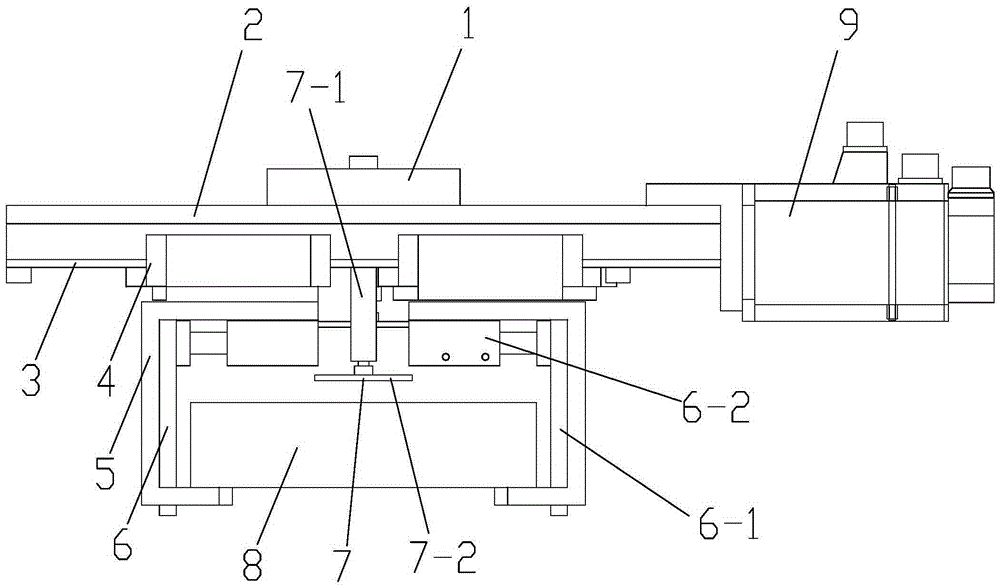
Complete-sequence iron core lamination robot gripper
The invention belongs to the technical field of iron core production equipment and relates to a complete-sequence iron core lamination robot gripper which comprises a top plate, wherein a connector for being connected with an arm of a robot is arranged at the top part of the top plate; a slide rail is arranged on the lower surface of the top plate; a screw rod is arranged at the middle part of the slide rail; two sliding blocks are arranged on the slide rail and fixedly connected with two clamping teeth respectively; a motor is installed at the end part of the top plate; the output end of the motor is connected with the screw rod; the screw rod drives the two sliding blocks to move towards opposite directions; and a lifting device is arranged between the two clamping teeth. The complete-sequence iron core lamination robot gripper has compact, reasonable and efficient layout, can efficiently complete iron core lamination work with high quality, is high in automation degree, improves the production efficiency, solves the problem about low efficiency, poor performance and rusted iron cores caused by artificial lamination, improves the production efficiency and the quality of a product and reduces used persons.
Owner:WUXI PUTIAN IRON CORE CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com